Abstract
For more than a decade, mitochondria-targeted nucleases have been used to promote double-strand breaks in the mitochondrial genome. This was done in mitochondrial DNA (mtDNA) homoplasmic systems, where all mtDNA molecules can be affected, to create models of mitochondrial deficiencies. Alternatively, they were also used in a heteroplasmic model, where only a subset of the mtDNA molecules were substrates for cleavage. The latter approach showed that mitochondrial-targeted nucleases can reduce mtDNA haplotype loads in affected tissues, with clear implications for the treatment of patients with mitochondrial diseases. In the last few years, designer nucleases, such as ZFN and TALEN, have been adapted to cleave mtDNA, greatly expanding the potential therapeutic use. This chapter describes the techniques and approaches used to test these designer enzymes.
1. MITOCHONDRIAL DNA
Mitochondria are dynamic membranous organelles that satisfy almost all of the energy requirements of a cell through the generation of ATP. This takes place through the actions of the five enzyme complexes of the oxidative phosphorylation system (OXPHOS complexes I–V) located in the mitochondrial inner membrane. Mitochondria carry their own multicopy genome and human mitochondrial DNA (mtDNA) is a circular, double-stranded, supercoiled molecule present in hundreds to several thousands of copies per cell. It is 16,569 bp in length and encodes for 37 genes (22 tRNAs, 2 rRNAs, and 13 proteins). All the protein-coding genes encode subunits of the OXPHOS enzyme complexes. Complex II is the only OXPHOS complex contain entirely of nuclear encoded subunits; complexes I and III–V comprise both nuclear- and mtDNA-encoded subunits.
Mutations in mtDNA can inhibit OXPHOS biogenesis through disruption of structural subunits or impairment of tRNAs or RNAs required for mitochondrial translation. Single nucleotide variants (SNVs) and rearrangements like deletions and duplications have been associated with disease. As mtDNA is a multicopy genome, mutant alleles are normally present in conjunction with wild-type alleles, a phenomenon-termed heteroplasmy. In general, mutant alleles have to be present on 70–95% of mtDNA molecules before a pathogenic phenotype is apparent (Gardner, Craven, Turnbull, & Taylor, 2007). Mutant alleles that are present in 100% of mtDNA molecules are termed homoplasmic but these are associated with fewer mitochondrial disorders than heteroplasmic mutations.
Reductions in the availability of cellular ATP can influence many metabolic and signaling pathways and trigger apoptosis, thus, the maintenance of OXPHOS function is critical for cellular physiology and development. Manipulation of the mitochondrial genome provides a mechanism for enhancing or reducing OXPHOS biogenesis. Mitochondria-targeted restriction endonucleases (REs) have been adopted as a useful tool for mitochondrial genome manipulation. In their simplest form, these are recombinant REs with mitochondrial localization signals (MLSs) that direct import the enzymes to the mitochondrial matrix where they can access mtDNA and create site-specific double-strand breaks (Srivastava & Moraes, 2001). Cleavage of mtDNA in this manner leads primarily to the degradation of the target mtDNA species and if present, expansion of heteroplasmic species lacking the sequence (Bayona-Bafaluy, Blits, Battersby, Shoubridge, & Moraes, 2005). Mutations in mtDNA such as deletions and SNVs can also be introduced at low frequency using restriction enzymes (Fukui & Moraes, 2009). This ability to manipulate mtDNA has meant that mitochondria-targeted restriction enzymes have become valuable tools to both disrupt OXPHOS biogenesis in disease models and enhance OXPHOS biogenesis through the clearance of heteroplasmic mtDNA mutations in therapeutic models.
2. MITOCHONDRIA-TARGETED RESTRICTION NUCLEASES TO CLEAVE mtDNA AND MODEL OXPHOS DISEASES
Restriction endonucleases (REs) have been used to create models of mtDNA-derived OXPHOS dysfunction in many model systems including models of mitochondrial disease, neurodegenerative disorders (Parkinson’s, Alzheimer’s, Huntington’s diseases, and ALS), and the aging process (Pinto & Moraes, 2013).
In Drosophila melanogaster, targeting of wild-type mtDNA using XhoI has been used to produce models of disease by selecting for rare mtDNA molecules lacking the single restriction site (Xu, DeLuca, & O’Farrell, 2008). In an inducible transgenic model, activating XhoI in germline cells severely compromised fertility but about 1% of females produced escaper progeny at low frequency. These flies all carried homoplasmic single-base mutations that ablated the XhoI restriction site. Screening identified three protein sequence variants: p.A302T that appeared wild type; p.R301L with male sterility; and p.R301S with developmental dysfunction and male sterility.
One of the first mouse models that took advantage of restriction enzymes was a transgenic mouse expressing mitochondria-targeted PstI in muscle (Srivastava & Moraes, 2005). Murine mtDNA contains two PstI restriction sites at 8,424 nt and 12,242 nt. It was anticipated that homologous recombination between two double-strand breaks at these sites might recapitulate the human common deletion, m.8483_13459del4977, that accumulates in disease and aging. A synthetic mammalianized gene coding for the PstI RE (NCBI P00640) was constructed with a human COX8A N-terminal mitochondrial localization signal (MLS) to direct the enzyme to mitochondria (mito-PstI). This construct was cloned into a pSMA plasmid downstream of the human skeletal muscle actin (SMA) promoter. The final construct pSMA-PstI was used to generate transgenic mice with the expression of mito-PstI in muscle. The resulting offspring showed early signs of myopathy, with mitochondrial abnormalities and defects in the OXPHOS complex I+III and complex IV activities due to mtDNA depletion and the presence of low levels of mtDNA with a large-scale deletion between the PstI sites.
The creation of inducible mouse models has allowed the careful control of mito-PstI expression using the Tet-on/Tet-off system. The expression of the tTA activator protein (or rtTA in case of the tet-on system) can be driven by different promoters, restricting the expression to discrete cells, for example, CamKIIα for neurons, dopamine transporter (DAT) for dopaminergic cells, and Rosa26 for ubiquitous expression. In addition, temporal control can be achieved through control of the administration of doxycycline (Dox) which is generally added to the food (Fukui & Moraes, 2009).
To study the molecular mechanisms underlying the generation and accumulation of mtDNA deletions in neurons in vivo, mice expressing neuron-specific mito-PstI were created by crossing the TRE-mito-PstI mouse with a model carrying the tTA gene under the control of the Cam-KIIa promoter (Fukui & Moraes, 2009). In the absence of Dox, CamKIIα-tTA drives the expression of mito-PstI in forebrain neurons. The constant expression of mito-PstI starting at 21 days after birth, led to the generation of a family of partially deleted mtDNA closely resembling the mtDNA deletions that occur in normal human aging. Interestingly, in these mice deletion breakpoints with features of homologous recombination and non-homologous end joining were observed, probably resulting from the action of the mtDNA repair machinery.
To study the effect of chronic and low-level mtDNA damage, a second TRE-mito-PstI line expressing lower levels of mito-PstI was bred with CamKIIα-tTA mice. This approach allowed comparisons of the susceptibility of different brain regions to mtDNA damage (Pickrell, Fukui, Wang, Pinto, & Moraes, 2011). This system was also used to analyze the effect of mtDNA damage in mouse models of neurodegenerative diseases like Alzheimer’s and Parkinson’s diseases. The CamKIIα-tTA/TRE-PstI mice bred to a mouse model of Alzheimer’s disease (AD-mice) carrying mutant APP and mutant presenilin 1 (Pinto, Pickrell, Fukui, & Moraes, 2013). In this case, the induction regimen was different from the previous case in that mito-PstI was suppressed in utero until 6 months of age. After this suppression period, mito-PstI was expressed for 2 months, and the mice were sacrificed and analyzed at 8 months of age. Surprisingly, we identified a decrease in amyloid plaques and oligomers, which was linked to a decrease in ROS. Although respiratory chain defects are commonly described as ROS inducers, a decrease in the levels of OXPHOS complexes would reduce the possibility of electrons reacting with oxygen (Pinto et al., 2013).
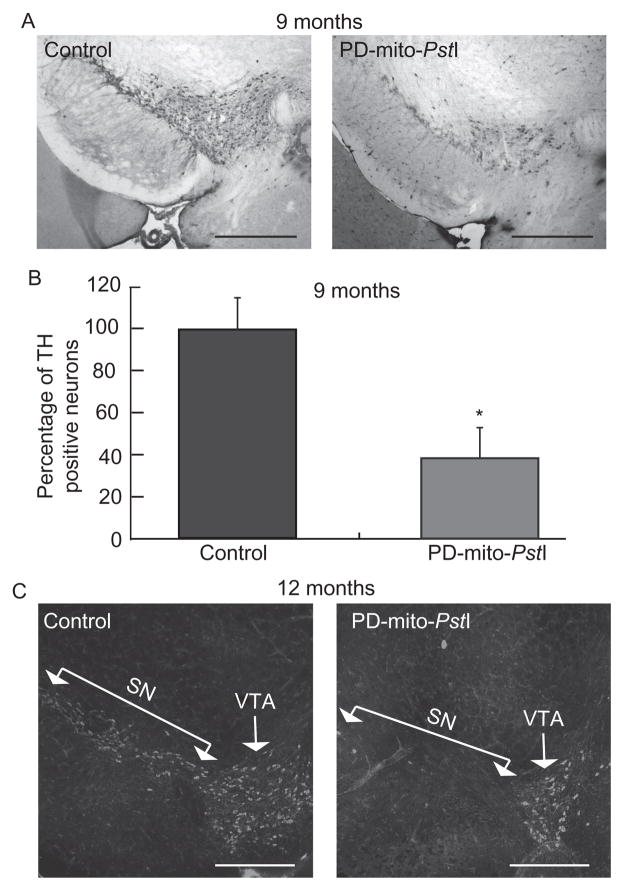
The TRE-PstI line with high expression of mito-PstI was also bred with a mouse in which tTA was expressed under the control of the DAT promoter, creating a model where mito-PstI cut mtDNA in dopaminergic cells (Pickrell, Pinto, Hida, & Moraes, 2011). Chronic OXPHOS dysfunction in dopaminergic cells was sufficient to induce neurodegeneration, striatal dopamine depletion, and consequent motor impairment, all typical of Parkinson’s disease (PD) pathology. Interestingly, not all dopaminergic cell populations were equally affected by the mtDNA damage (Fig. 18.1).
Figure 18.1.
Progressive loss of A9 tyrosine hydroxylase-positive neurons in mice expressing mito-PstI in dopaminergic neurons. (A) Representative immunohistochemical identification of TH+ neuron sections used for quantification at 9 months of age. (B) Quantification of A9 TH+ neuronal populations in 9-month-old animals; PD-mito-PstI mice showed an ~40% reduction in the amount of TH+ neurons compared to controls. (C) Representative immunohistochemical staining of TH in the midbrain of 12-month-old animals; PD-mito-PstI showed the absence of SN region and a milder degeneration of dopaminergic neurons in VTA. Values are mean±SEM (control n =4–5, PD-mito-PstI=4–5, *p <0.05). Scale bars, 5 μm. Adapted from Pickrell, Pinto, et al. (2011).
The TRE-mito-PstI mouse was also used in a Tet-on system through mating with a mouse line expressing a reverse tetracycline transactivator (rtTA) protein under the control of the ubiquitous Rosa26 promoter (Wang et al., 2013). The experimental animals were fed with a Dox diet for only 5 days at 3 months of age to induce transient systemic expression of mito-PstI. They were analyzed after an additional 3 months of suppression of mito-PstI expression. This regimen resulted in a myopathy with onset 3 months after the mtDNA lesion occurred. Further experiments suggested that the phenotype was due to a reduction in the muscle satellite cells’ pool (Wang et al., 2013). Because we observed similar phenotypes in other tissues, this ubiquitous induction will be also a valuable tool to study the behavior of different cells and tissues after mtDNA DSB in particular to study the link between mtDNA damage and aging, which has always been a controversial topic in the field.
3. mtDNA HETEROPLASMY AND APPROACHES TO ALTER THE BALANCE BETWEEN WILD-TYPE AND MUTANT mtDNA
Pathogenic mutations in mtDNA can affect mitochondrial tRNA, rRNA, and protein-encoding genes. As mentioned earlier, for heteroplasmic mutations which are the most prevalent type in diseases caused by mtDNA mutations, high loads of a mutant allele are necessary to cause pathology. This is due to threshold effects where the copies of the wild-type allele can compensate for the biochemical defect caused by the copies of the mutant allele. In general, mtDNA deletions cause disease at lower tissue mutation loads than point mutations, ~60% versus ≥80% (Russell & Turnbull, 2014). These thresholds can be observed among members of the same family, where small differences in mutation load can be the difference between normal life and a devastating disease (Hao, Bonilla, Manfredi, DiMauro, & Moraes, 1995). Even within the tissue of the same individual, higher mutation loads are associated with strong biochemical phenotypes (Sciacco, Bonilla, Schon, DiMauro, & Moraes, 1994).
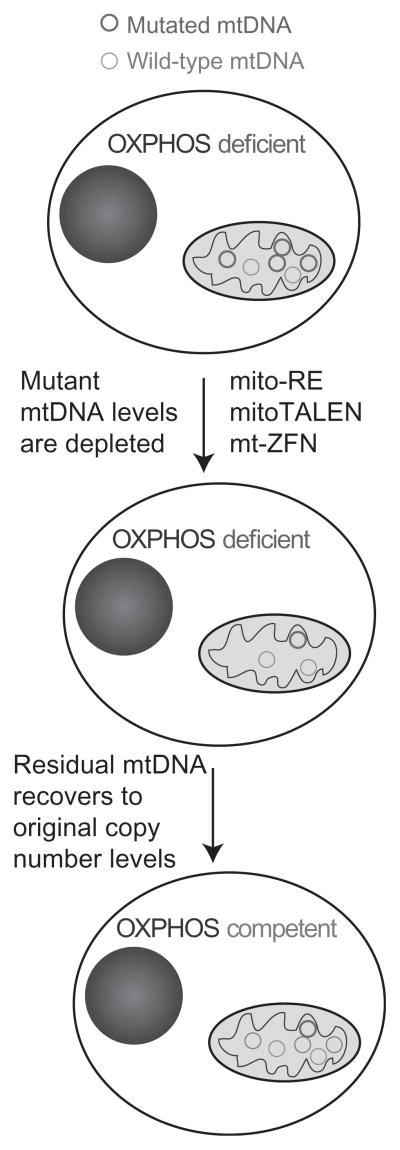
One of the consequences of heteroplasmy and threshold effects is that a reduction in pathology can be achieved without completely removing mutant alleles, simply reducing the levels of the mutant allele below threshold levels can ameliorate OXPHOS dysfunction (Bacman, Williams, Pinto, Peralta, & Moraes, 2013; Hayashi et al., 1991; Smith & Lightowlers, 2011; Wenz, Williams, Bacman, & Moraes, 2010). This can be achieved using mitochondria-targeted endonucleases that have recognition sequences that encompass one of a pair of heteroplasmic alleles. Recognition and cleavage by the enzyme lead to a reduction in the relative level of the target allele through cleavage-stimulated mtDNA degradation (Bayona-Bafaluy, Blits, et al., 2005). As illustrated in Fig. 18.2, mitochondrial-targeted endonucleases can have different architectures. mtDNA copy number is robustly controlled and mtDNA depletion stimulates mtDNA replication through mechanisms that remain poorly defined (Moraes, 2001). Only the uncleaved mtDNA can replicate and repletion to normal mtDNA levels subsequently results in an increased relative abundance of the non-target mtDNA without the endonuclease recognition site (Bayona-Bafaluy, Blits, et al., 2005). The following section will discuss the use of mitochondria-targeted endo-nucleases for the modulation of heteroplasmy through heteroplasmy shift and the application of this approach to alter the balance between target and non-target mtDNA.
Figure 18.2.
Changing mtDNA heteroplasmy with mitochondrial-targeted nucleases. Irrespective of the type of nuclease, the approach requires a highly specific nuclease that can recognize one haplotype but not the other(s). Upon double-strand breaks, linearized mtDNA is quickly degraded (Bayona-Bafaluy, Blits, et al., 2005), leading to an mtDNA depletion, which can be severe if the susceptible mtDNA species was abundant. Nonetheless, the residual mtDNA will replicate to make up for the depletion. Because of the specificity of the digestion, most residual mtDNA will not be susceptible to the endonuclease. (See the color plate.)
4. mtDNA HETEROPLASMY SHIFT USING RESTRICTION ENDONUCLEASES
Heteroplasmy shift is an attractive strategy for the genetic therapy of mtDNA disorders and our group and others have investigated its potential in many model systems. A mouse–rat cybrid cell line containing both mouse and rat mtDNA was the first cell line used to demonstrate the efficacy of the approach (Srivastava & Moraes, 2001). Mouse mtDNA contains two PstI sites while rat mtDNA contains none. After the expression of PstI targeted to mitochondria (mito-PstI), a significant shift in the relative abundance of rat versus mouse mtDNA was achieved, providing the first proof of principle for using mitochondria-targeted endonucleases for mtDNA heteroplasmy shift. Soon after a human cybrid cell line carrying the heteroplasmic mutation, m.8993T>C mutation in MT-ATP6 was investigated (Tanaka et al., 2002). At mutation loads >90%, this mutation is associated with a devastating pediatric encephalomyelopathy called Leigh syndrome; at mutation loads >75%, the mutation causes NARP syndrome, characterized by neuropathy, ataxia, and retinitis pigmentosa. The mutant allele m.8993C creates a SmaI restriction site and a mitochondria-targeted form of the enzyme was created by the addition of the yeast CoxIV N-terminal MLS. Complete elimination of m8993C and restoration of ATP levels were achieved after five cycles of transfection and selection of cybrids with an initial mutation load of 98%. The isoschizomer of SmaI, XmaI, has also been used to target m.8993T>C in human cybrids (Alexeyev et al., 2008). In this study, an adenovirus vector was used. Interestingly, it was found that virus production was improved by the inclusion of a gene for M.SmaI; the SmaI/XmaI methylase with the authors speculating this may temper aberrant endonuclease activity in packaging cells. At a viral multiplicity of infection of 50, m.8993T>C was effectively eliminated from cells with an initial mutation load of 78%, restoring OXPHOS function.
Mitochondria-targeted REs have also been used effectively in mouse models. Our group has focused on the heteroplasmic BALB/NZB asymptomatic mouse that carries both the BALB and NZB wild-type mtDNA haplotypes (Jenuth, Peterson, Fu, & Shoubridge, 1996). These two mtDNAs differ in 93 positions (90 SNVs and 3 indels). The BALB haplotype contains an ApaLI restriction site that is absent in the NZB haplotype. A mitochondria-targeted form of the enzyme was created by adding a human COX8A N-terminal MLS to a mammalianized gene for ApaLI with a C-terminal HA tag for immuno-detection (Bayona-Bafaluy, Blits, et al., 2005). Inducible expression of mito-ApaLI in cells derived from these mice enabled examination of the transient depletion and kinetics of heteroplasmy shift. The extent of mtDNA depletion was found to correlate with initial levels of the target BALB mtDNA and repletion was complete within 24 h or approximately one cell cycle. Injection of the construct in brain and muscle using adenovirus type 5 vector (Ad5) demonstrated for the first time that the approach could be translated to animal models (Bayona-Bafaluy, Blits, et al., 2005).
The large number of differences between the two mtDNA haplotypes in the BALB/NZB mouse allowed us to extend the approach to a multiple “cleavage site model,” using a restriction enzyme that recognizes different numbers of sites on each haplotype (Bacman, Williams, Hernandez, & Moraes, 2007). BALB mtDNA contains three restriction sites for ScaI, whereas NZB mtDNA has five. The same construct design as mito-ApaLI was used but with a mammalianized ScaI enzyme. Again an Ad5 vector was used, but this time systemic injection in the jugular vein was used to target liver and local injection was used for muscle. In both tissues, we were able to induce heteroplasmy shift toward BALB mtDNA that has fewer restriction sites, although the magnitude of the shift was lower than for the single differential cleavage site model of ApaLI.
The ability to induce heteroplasmy shift in specific tissues using systemic injection with different vector systems has also been demonstrated using jugular injection of cardiotropic AAV6 and hepatotropic Ad5 to deliver mito-ApaLI (Bacman, Williams, Garcia, & Moraes, 2010). For the former, anesthetized adult female mice (6–8 weeks old) were injected in the right external jugular vein. Injections of 1.0 or 5.0 × 1011 vg (viral genomes) of rAAV6-mito-ApaLI-HA or rAAV6-AP (alkaline phosphatase) control diluted in 100 μl of phosphate-buffered saline (PBS) were performed.
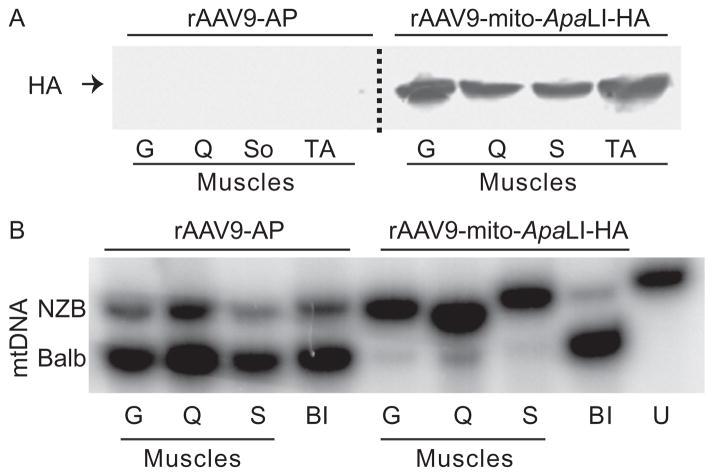
We also used an AAV9 vector under the control of the CMV promoter to deliver the transgene to all skeletal muscles (Bostick, Ghosh, Yue, Long, & Duan, 2007; Ghosh, Yue, Long, Bostick, & Duan, 2007; Inagaki et al., 2006). Systemic delivery was achieved in newborn mice at P2–P3. Mice were subjected to i.p. (intraperitoneal) or i.v. (temporal vein) injection. Each mouse was injected with 5 × 1011 viral genome (vg) in a 50-μl final volume in PBS. Injections were carried out using a short insulin syringe with a 31-G needle (Becton Dickenson). Tail tissue was obtained before performing the injections to determine day 0 heteroplasmy levels and at 6, 12, and 24 weeks postinjection. In neonatal mice, both i.p. and i.v. injections of rAAV9 vectors carrying mito-ApaLI showed heteroplasmy changes in all striated muscle (Bacman, Williams, Duan, & Moraes, 2012). Figure 18.3 illustrates the expression of the recombinant AAV9-mito-ApaLI in different muscle types and the dramatic change in heteroplasmy toward the uncleaved NZB haplotype.
Figure 18.3.
Recombinant rAAV9-mito-ApaLI-HA induces a shift in mtDNA heteroplasmy in muscle. Neonatal mice heteroplasmic for the mtDNA haplotypes NZB and BALB were injected i.p. with rAAV9 expressing either a control alkaline phosphatase (AP) or a mito-ApaLI-HA. (A) The expression of ApaLI-HA was analyzed by anti-HA Western blotting 6 weeks after i.p. injections in P2–P3 mice. Homogenates from different skeletal muscles showed expression. (B) NZB/BALB mtDNA heteroplasmy quantified using last-cycle hot PCR/RFLP with DNA samples from mice injected with rAAV9-mito-ApaLI-HA at 6 weeks i.p. postinjections. Increases in the percentage of NZB mtDNA were observed in all skeletal muscle when compared to the samples obtained from the tail before injection. Gastrocnemius (G), quadriceps (Q), soleus (S), tibialis anterior (TA), before injection (BI), and uncut DNA (U). Adapted from Bacman et al. (2012). (See the color plate.)
5. DESIGNER ENDONUCLEASES FOR THE MODULATION OF mtDNA HETEROPLASMY
Although REs-targeted mitochondria have been efficient to change mtDNA heteroplasmy both ex vivo and in vivo, their application is limited due to the fact that very few clinically relevant heteroplasmic mtDNA mutations create restriction sites for naturally occurring RE. To solve this problem, our group and others have investigated the use of endonucleases with modular DNA recognition domains, which can be designed to target almost any sequence. To date, the most promising systems available are zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). Both systems share a common basic structure utilizing sequencing-independent endonuclease domain from FokI coupled to a sequence-specific modular DNA-binding domain. As FokI creates double-strand breaks as a dimer, both enzyme systems require the design of pairs of monomers that bind the region of interest tail–tail in close proximity enabling the dimerization of FokI domains and double-strand cleavage between the monomer-binding sites. The principal differences between the systems are in the modularity of DNA sequence recognition. Both systems employ tandem repeats of modular DNA-binding domains to create sequence-specific DNA-binding domains. In ZFNs, each individual zinc-finger domain recognizes 3 bp of DNA, and for TALENs, each TALE domain recognizes 1 bp.
While the various genome editing tools available, ZFN, TALENs, and CRISPR have all been successfully utilized in nuclear genome editing; their application to mtDNA heteroplasmy modulation has various limitations. The first is that many successful applications in the nuclear genome result from the targeting of specific genes where an optimal recognition site can be selected from within a very large region of DNA-avoiding sites with similarity to other regions of the genome. In contrast, many of the heter-oplasmy shift models require discrimination between only one base from identical regions on non-target mtDNA. In addition, import of endonucleases into mitochondria can also prove problematic, especially for ZFN which have an inherent tropism for the nucleus.
6. mtDNA HETEROPLASMY SHIFT USING ZINC-FINGER NUCLEASES
Cys2–His2 zinc-finger domains as used in ZFNs are small 34-amino acid protein motifs containing an antiparallel β-sheet and an α-helix stabilized by a zinc ion. Each domain binds a triplet DNA sequence and combinations of zinc-finger domains can be designed to bind many specific sequences. The addition of C-terminal FokI nuclease domain and the design of pairs of proteins that bind DNA in close proximity tail–tail enable the cleavage of DNA at specific sequences. ZFNs have an innate tropism for the nucleus, most likely driven by association with the nuclear import machinery. To overcome this and enable efficient trafficking to mitochondria, it is necessary to incorporate nuclear export sequences into ZFN monomers (Minczuk, Papworth, Kolasinska, Murphy, & Klug, 2006). The murine minute virus NS2 sequence is commonly used, inserted internally between an MLS and the DNA-binding domain (Eichwald, Daeffler, Klein, Rommelaere, & Salome, 2002). Import efficiencies vary for different ZFNs in an, as yet, uncharacterized way. We have found that for some nucleases several MLS sequences need to be tested and that occasionally the addition of tandem MLS can also help. ZFNs’ technology has been used to target pathogenic mitochondrial genomes in cybrid cells, such as cells harboring the m.8993T>G NARP/Leigh’s syndrome mutation or the “common deletion” (m.8483_13459del4977; Gammage, Rorbach, Vincent, Rebar, & Minczuk, 2014; Minczuk et al., 2006; Minczuk, Papworth, Miller, Murphy, & Klug, 2008). To reduce off-site cleavage, almost all recombinant nucleases utilize obligate hetero-dimeric FokI domains (Doyon et al., 2011; Miller et al., 2007). These domains have modified protein sequences that create a different dimerization face on each monomer and prevent dimerization of identical monomers. This modification has been found to reduce self–self dimerization by approximately 100-fold.
An interesting advance in the development of ZFNs was the development of nucleases containing quasi-dimeric FokI domains joined by a linker (Minczuk et al., 2008). This approach enables the use of a single DNA-binding domain with two tandem FokI domains joined by a 35-amino acid flexible linker. This single chain ZFN was able to induce heteroplasmy shift in cybrids carrying the m.8993T>G mtDNA mutation.
7. HETEROPLASMY SHIFT USING TAL EFFECTOR NUCLEASES
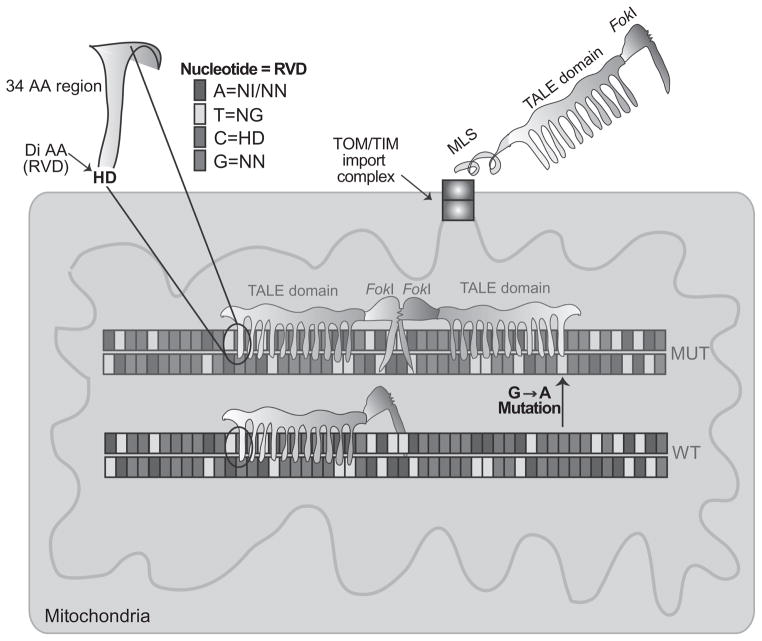
TAL effector nucleases (TALENs) are made of DNA-binding modules fused to a nuclease domain from a FokI restriction enzyme. Each protein module contains 34 amino acids, and the 12th and 13th residues are the key site to identify the targeted DNA, known as repeated variable di-residues (RVDs, Fig. 18.4). TALENs require two monomers to bind to the DNA, as previously mentioned, FokI works as a dimer (Boch et al., 2009; Hockemeyer et al., 2011; Moscou & Bogdanove, 2009; Pan et al., 2013; Sung et al., 2013).
Figure 18.4.
Development of mito-TALENs to alter mtDNA heteroplasmy in vivo. TALENs are enzymes used by the Xanthomonas bacteria to bind and modify specific DNA regions. A nonspecific FokI cleavage domain can be fused to the TALE DNA-binding domain to create a nuclease. However, FokI requires dimerization, and therefore, two TALEN monomers are required for effective cleavage. This requirement provides increased specificity for the DSB. The sequence that binds DNA corresponds to an approximate 34-amino acid sequence: LTPEQVVAIASHDGGKQALETVQRLLPVLC QAHG. The residues at the 12th and the 13th positions are known as repeat variable di-residue or RVD. The figure illustrates how different RVDs are specific to one of the four bases. To direct TALENs to mitochondria, we fused a mitochondrial localization signal (MLS) at the N-terminus of the fusion protein. The TALENs are imported through the TOM/TIM import complexes unfolded, but gain their final folding in the mitochondrial matrix, with the help of molecular chaperones. The MLS is cleaved by a mitochondrial peptidase and the mature protein is ready to bind DNA. When dimers of FokI are formed, the mito-TALEN cleaves the mtDNA haplotype, which is mostly degraded. To target mutant mtDNAs that differ by a single-base pair (in this example, a G>A transition), one of the TALEN monomers must be able to bind only to the mutated sequence and not to the wild type.
We have recently developed mitochondria-targeted TALENs or mito-TALENs (Bacman et al., 2013) directed to the mtDNA “common deletion” (m.8483_13459del4977) and Leber’s hereditary optic neuropathy plus dystonia mtDNA mutation m.14459G>A in MT-ND6 (Bacman et al., 2013; Jun, Trounce, Brown, Shoffner, & Wallace, 1996). Figure 18.4 illustrates the general architecture of mito-TALENs, how they are designed to recognize specific mtDNA mutations and how dimerization is necessary for the nuclease domain (FokI) to be active.
The recognition sequences for our TALENs against the common deletion (Δ5-mito-TALEN) and the m.14459G>A mutation (14459A-mito-TALEN) are given in Table 18.1.
Table 18.1.
DNA recognition sequences used to develop specific mito-TALENs to two pathogenic human mtDNA mutations.
TALEN pairs were designed so that cleavage would only occur in the mutated mtDNA molecules.
| Target | mtDNA recognition sequence |
|---|---|
| Common deletion (Δ5-mito-TALEN) |
|
| m.14459G>A (14459A-mito-TALEN) |
|
TALEN monomer-binding sites are underlined. In the upper panel, the residual 13-bp repeat unit that flanks deletion breakpoints in full-length mtDNA is in bold. In the lower panel, 14459A is in bold. Numbering is given relative to the rCRS.
The TALE DNA-binding domains were designed by Cellectis Bioresearch to target the specified mtDNA regions. The following principles were applied when designing the TALE domains: (1) basic TAL-binding codes were used (Cermak et al., 2011); (2) the DNA-binding domain was kept relatively short (10–16 repeats); and (3) whenever possible the critical discriminating base pairs at position 0 or 1 were used (Bacman et al., 2013). The plasmid constructs for mito-TALEN monomers were assembled using the In-Fusion HD cloning kit from Clontech Laboratories by subcloning into pVAX. Generic commercial TALEN constructs typically contain nuclear localization signals and these need to be removed when subcloning for use in mitochondrial studies. We have found that import of TALEN monomers into mitochondria is variable and that some TALE domains lead to poor import. Strikingly, we have found that this to be the case even when the only differences between monomers are the number of repeats and the RVDs. MLSs from human ATP5B, COX8A, and SOD2 can all direct TALEN monomers to mitochondria. However, for reason that we do not understand, an MLS that works for one TALEN monomer may not work for another, even though the mito-TALEN overall sequences are almost identical. Altering the MLS is currently the best solution when poor import is encountered.
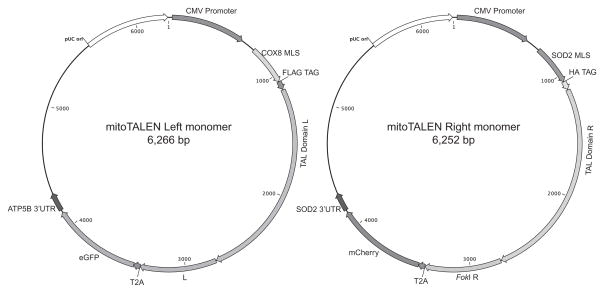
We also included a unique immuno-tag in the N-terminus of each TAL domain (HA or FLAG), a 3′ UTR from ATP5B or SOD2 mRNA known to localize to ribosomes contacting mitochondria and an independent fluorescent marker to select for expression and sorting (eGFP in one monomer and mCherry in the other). We also added a picornaviral 2A-like sequence (T2A′) between the mito-TALEN and the fluorescent marker (Szymczak et al., 2004). Obligate heterodimeric FokI domains (Doyon et al., 2011; Miller et al., 2007) were utilized in all mito-TALENs (Bacman et al., 2013). The general structure of the plasmids is depicted in Fig. 18.5.
Figure 18.5.
Plasmids expressing mito-TALENS were designed to allow mitochondrial localization and fluorescence selection. The figure illustrates the structure of the two plasmids coding for the two monomers required for mito-TALEN function. The two FokI moieties form obligatory heterodimers. The T2A sequence allows fluorescent markers to be expressed from the same transcripts. (See the color plate.)
8. SINGLE-STRAND ANNEALING ASSAY TO ANALYZE THE EFFICACY AND SPECIFICITY OF DESIGNER NUCLEASE
This assay provides important information about the quality of the TALENs that we used to create the final versions of the mito-TALENs. Single-strand annealing in Saccharomyces cerevisiae was performed as previously described (Liddell, Manthey, Pannunzio, & Bailis, 2011). Two haploid strains are mated to carry out the assay. The first expresses the plasmid encoding the TALEN and the second a plasmid carrying a nonfunctional/split LacZ gene (β-galactosidase). The LacZ gene is interrupted by a spacer flanked by duplications of 70–220 bp. The TALEN recognition site is cloned between these two repeats. The efficiency of cleavage by the TALEN in the yeast diploid nucleus is measured by assaying β-galactosidase enzyme activity resulting from single-strand annealing and homologous recombination between the two duplications. Typically, this is done with blue/white colony screening, but spectrophotometric assays are also used.
9. THE USE OF CYBRID CELLS TO TEST APPROACHES TO CHANGE MITOCHONDRIAL DNA HETEROPLASMY
To test a reagent that can alter mtDNA heteroplasmy, it is important to use appropriate cell lines that optimally harbor high levels of mutated mtDNA. Transmitochondrial cybrids are eukaryotic cell lines produced by the fusion of a nuclear donor and a cytoplast containing mitochondria from a different donor (Bacman & Moraes, 2007). Although any cell harboring heteroplasmic mtDNA mutations can be used to test mitochondrial-targeted nucleases, it may be useful to have lines with different levels of heteroplasmy. In addition, fibroblasts often have lower mtDNA mutation loads and are biochemically normal. Furthermore, cells should be amenable to good transfection efficiencies. We found that transmitochondrial cybrids based on the human osteosarcoma 143B(TK−) fulfill the criteria and can be produced relatively easy. By producing and isolating different clones, there are higher chances of obtaining lines with the desired mtDNA mutation load. Moreover, further manipulation, such as transient depletion of mtDNA by ethidium bromide treatment, can help in the isolation of clones with different mtDNA mutation loads.
There are several techniques that have been used to produce trans-mitochondrial cybrids, among them cell fusion is one of the most popular ones (Clayton, Teplitz, Nabholz, Dovey, & Bodmer, 1971; Hayashi et al., 1991). A large number of pathogenic mtDNA mutations, have been produced using this procedure, including rearrangements (Bacman et al., 2013; Hayashi et al., 1991; Porteous et al., 1998), point mutations in tRNA (Bacman, Atencio, & Moraes, 2003; Bornstein et al., 2005; Chomyn, 1996; Hao & Moraes, 1997; King, Koga, Davidson, & Schon, 1992; Toompuu, Tiranti, Zeviani, & Jacobs, 1999), rRNA (Inoue et al., 1996), and protein-coding genes (Bacman et al., 2013; Bruno et al., 1999; Jun et al., 1996). Most of these approaches used Rho-zero cells (ρ′; cells devoid of mtDNA) as nuclear donors that were repopulated with exogenous mtDNA, resulting in transmitochondrial cybrids (King & Attardi, 1989).
10. IMMUNODETECTION AND MITOCHONDRIAL LOCALIZATION IN CELLS AND TISSUES
Before attempting to change mtDNA heteroplasmy, it is important to demonstrate that the recombinant mito-TALENs localize to mitochondria. Besides performing Western blots to assure that the full-length (tagged) protein is expressed in transfected cells, immunocytochemistry assays are performed to test for mitochondrial localization of TALEN monomers from all constructs. Although different cell types were used, Cos7 and HeLa cells were the chosen ones as they are easy to transfect cells and have distinct mitochondrial distribution making it easy to colocalize with the transfected plasmid. Cells were plated on glass coverslips and in the following day they were transfected with the respective plasmids. After transfection, cells are incubated for 30 min at 37 °C with 200 nM MitoTracker Red CMXRos (Invitrogen) and fixed with 4% paraformaldehyde in PBS for 20 min. After a brief treatment with methanol (5 min), primary antibodies to HA (Roche; 1:200) or FLAG (Sigma-Aldrich; 1:200) in 5% BSA in PBS were incubated overnight at 4 °C in a humid chamber. The next day, coverslips were incubated for 2 h at room temperature with Alexa Fluor 488-conjugated goat antibody anti-rat IgG (Molecular Probes; 1:200) or Alexa Fluor 488-conjugated goat antibody anti-mouse IgG (Molecular Probes; 1:200). Images were recorded using confocal microscopy.
11. CHANGING mtDNA HETEROPLASMY IN CULTURED CELLS WITH mito-TALENs
Cells with heteroplasmic mtDNA mutations were transfected with GenJet DNA In Vitro Transfection Reagent version II (SignaGen Laboratories) following the protocol suggested by the manufacturers (3:1 ratio of DNA/GenJet and 1–2 μg DNA for a 6-well dish transfection). We scaled up and transfected a 70–80% confluent T-75 flask with 30 μg total DNA. When the two mito-TALEN monomers were cotransfected, 15 μg of each plasmid was used for the transfection (Bacman et al., 2013). The expectation was that cells subjected to mito-TALEN would change mtDNA heteroplasmy in a predictable manner.
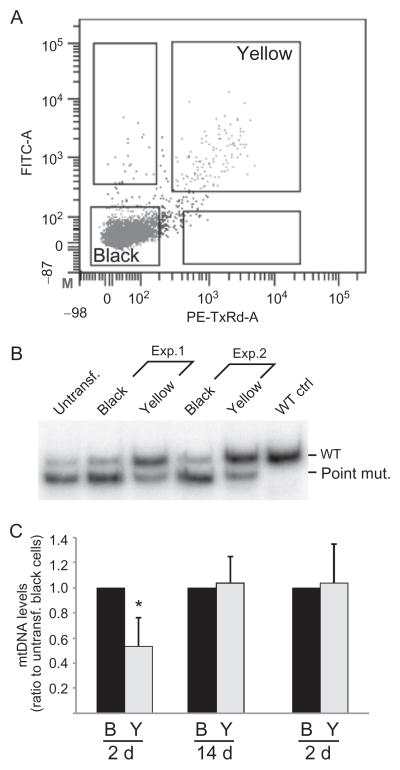
Fluorescent-activated cell sorting (FACs) is an excellent tool to isolate different cell populations after transfection with the mito-TALENs. This was possible due to the fact that each plasmid coding for the TALEN monomer posses a fluorescence protein, eGFP or mCherry, that can be used to sort the cells after transient transfection. We were able to cell sort the population that did not get the TALEN transfection (untransfected), the populations that incorporated only one TALEN monomer individually (green-eGFP or red-mCherry) or the population that incorporated both TALENs in the same cell. Approximately 48 h after transfection, cells were collected in DMEM with no addition of serum or antibiotics, sorted and collected in PBS, using a FACSAria IIU by gating on single-cell fluorescence using a 561-nm laser and 600LP, 610/20 filter set for mCherry and a 488-nm laser and 505LP, 530/30 filter set for eGFP (Bacman et al., 2013). Cells were separated based on having only one fluorescent marker, or having both, which were referred to as “yellow” cells (Fig. 18.6A). Sorted cells not expressing fluorescent markers were likely untransfected and were referred to as “black” cells (Fig. 18.6A). FAC-sorted cells were allowed to grow in complete medium or were directly subjected to DNA extraction.
Figure 18.6.
Analyzing mtDNA heteroplasmy changes by mito-TALENs. We transfected cells harboring high levels of a pathogenic mutation (m.14459G>A). Because the two plasmids had fluorescent markers, we sorted the cells for double transfectants 48 h after transfection (panel A). Cells gated for both fluorophores were termed “yellow, Y.” Control cells “black, B” showed no fluorescence. These cells had their DNA isolated and analyzed by RFLP after “last-cycle hot” PCR (panel B). Total mtDNA levels were also determined by qPCR (panel C). Adapted from Bacman et al. (2013). (See the color plate.)
DNA from sorted cell populations was subjected to “last-cycle hot PCR” (Moraes et al., 1992), which visualizes only nascent amplicons and avoid interference from heteroduplexes formed during the final PCR cycles. Following by a digestion with appropriate diagnostic restriction endonuclease, samples were electrophoresed in 8% acrylamide/polyacrylamide (BioRad) in Tris Boric EDTA. The radioactive signal was quantified using a Cyclone Phosphorimager System (Perkin Elmer; Bacman et al., 2007; Bayona-Bafaluy, Muller, & Moraes, 2005).
As an example, to determine the levels of them.14459G>A mutation by the “last-cycle hot” PCR, total DNA extracted from FAC-sorted cells was used as template, and PCR was performed with the following mtDNA primers: F-BclI-mut-F, 5′-CCCCCATGCCTCAGGATACTCCTCAATAGTGATC-3′ and 14579B, 5′-TGATTGTTAGCGGTGTGGTCGGGTGTGT-3′. The “F” primer creates a BclI restriction site in combination with the amplified mutated mtDNA. The use of mismatch primers is useful when a specific mutation does not create or destroys a site that can be recognized by commercially available enzymes. PCR products were digested with BclI and resolved in a 12% acrylamide/polyacrylamide gel. Radioactive signal was quantified using a Cyclone Phosphorimaging System (Perkin Elmer; Bacman et al., 2013). As illustrated in Fig. 18.6B–C, the comparison of yellow and black cells showed a marked reduction in mtDNA mutation load in “yellow” cells.
12. EVALUATION OF THE mtDNA CONTENT
Because these approaches actively eliminate mtDNA, it is crucial to evaluate mtDNA levels after transfection/infection. This can be done by Southern blot or by qPCR. This analysis is particularly important when the levels of the target mtDNA population is very high (e.g. 90%). A precipitous decline in mtDNA levels can have deleterious consequences for the cell.
Southern blot can be used to estimate the mtDNA/nuclear DNA ratio. In the case of mouse mtDNA, 5 μg of total DNA from cells (Bayona-Bafaluy, Blits, et al., 2005) or tissues (Bacman et al., 2007, 2010) was digested with SacI (NEB), or any other enzyme that linearizes mtDNA. In this case, SacI digests mtDNA at position 9047 and produces a 16.5 kb fragment. After digestion, samples were electrophoresed on a 0.7–0.8% agarose gel and transferred to a zeta-probe membrane (BioRad). mtDNA was detected by hybridization with random prime-labeled probes with dCTP P32 (Perkin Elmer), corresponding to positions 5556–6268 of the mouse mtDNA (Bacman, Williams, & Moraes, 2009). To quantify the nuclear DNA, a 1-kb PCR product corresponding to nts 502–1515 of the murine rDNA repeat unit of the 18S rDNA (GeneBank BK000964) was hybridized to the same membranes. Blots were stripped and hybridized with the 18S rDNA gene (GeneBank BK000964). The radioactive signal was quantified using a Cyclone Phosphorimager System (Perkin Elmer; Bacman et al., 2009; Bayona-Bafaluy, Blits et al., 2005).
For human mtDNA, PvuII provides a good digestion to vizualize both the mtDNA (16.5 kb) and the 18S rDNA (12 kb). Species-specific probes are recommended. MtDNA regions close to the ND1 region are usually used as probes as this region is rarely deleted.
Real-time PCR can also be used to determine relative mtDNA levels. We performed quantitative PCRs using SYBR/ROX chemistry (172–5264; BioRad, SsoAdvanced SYBR Green; Bacman et al., 2012) on a BioRad CFX96/C1000 qPCR machine using the manufacturer’s software to calculate ΔΔCT values. In the case of cells harboring a heteroplasmic mtDNA deletion, we determined the levels of different mtDNA species between sorted cell populations or tissues by quantifying the levels of deletion breakpoint/actin, wild-type/actin, and total mtDNA/actin. Total DNA can be extracted from cells or snap-frozen tissues and treated with RNAase A (Qiagen) at 37 °C for 30 min (1 ml/10 mg DNA) followed by sodium acetate and ethanol precipitation. In the case of point mutants, we determined the levels of total mtDNA, soon after the mito-TALEN treatment and at later points (Fig. 18.6C).
Total mtDNA content in human cells were evaluated using the following primers:
ND1 3281F: 5′-CAGCCTGACCCATAGCCATA-3′,
ND1 3364B: 5′-ATTCTCCTTCTGTCAGGTCGAA-3′.
Loading control primers used were:
β-Actin Exon 6F: 5′-GCGCAAGTACTCTGTGTGGA-3′,
β-Actin Exon 6B: 5′-CATCGTACTCCTGCTTGCTG-3′,
GAPDH Exon 5F: 5′-GCAGTGGCAAAGTGGAGATT-3′,
GAPDH Exon 5B: 5′-GAATTTGCCGTGAGTGGAGT-3′.
13. FUTURE PERSPECTIVES
Mitochondrial-targeted nucleases have proven useful to create models of mitochondrial DNA diseases. However, their ability to change mtDNA heteroplasmy levels and reduce mutation loads efficiently has opened a new therapeutic window. It is now important to demonstrate that the approach can be used safely in vivo. The ability to pack these large genes, which code for a dimeric protein, remains a challenge. The development of additional architectures for specific DNA editing may accelerate this field. However, certain requirements, such as RNA mitochondrial import when using CRISPR editing, may limit this approach.
Acknowledgments
This work is supported by the US National Institutes of Health Grants 5R01EY010804, 1R01AG036871, and 1R01NS079965, the Muscular Dystrophy Association, the United Mitochondrial Disease Foundation, the JM Foundation, and the Florida Biomedical Research Foundation.
Footnotes
The authors declare no competing financial interests.
References
- Alexeyev MF, Venediktova N, Pastukh V, Shokolenko I, Bonilla G, Wilson GL. Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Therapy. 2008;15(7):516–523. doi: 10.1038/gt.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Atencio DP, Moraes CT. Decreased mitochondrial tRNALys steady-state levels and aminoacylation are associated with the pathogenic G8313A mitochondrial DNA mutation. The Biochemical Journal. 2003;374(Pt. 1):131–136. doi: 10.1042/BJ20030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Moraes CT. Transmitochondrial technology in animal cells. Methods in Cell Biology. 2007;80:503–524. doi: 10.1016/S0091-679X(06)80025-7. [DOI] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Duan D, Moraes CT. Manipulation of mtDNA heteroplasmy in all striated muscles of newborn mice by AAV9-mediated delivery of a mitochondria-targeted restriction endonuclease. Gene Therapy. 2012;19(11):1101–1106. doi: 10.1038/gt.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Garcia S, Moraes CT. Organ-specific shifts in mtDNA heteroplasmy following systemic delivery of a mitochondria-targeted restriction endonuclease. Gene Therapy. 2010;17(6):713–720. doi: 10.1038/gt.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Hernandez D, Moraes CT. Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a ‘differential multiple cleavage-site’ model. Gene Therapy. 2007;14(18):1309–1318. doi: 10.1038/sj.gt.3302981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Moraes CT. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Research. 2009;37(13):4218–4226. doi: 10.1093/nar/gkp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nature Medicine. 2013;19(9):1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby BJ, Shoubridge EA. Selection of a mtDNA sequence variant in hepa-tocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Human Molecular Genetics. 2001;10(22):2469–2479. doi: 10.1093/hmg/10.22.2469. [DOI] [PubMed] [Google Scholar]
- Bayona-Bafaluy MP, Blits B, Battersby BJ, Shoubridge EA, Moraes CT. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(40):14392–14397. doi: 10.1073/pnas.0502896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Bafaluy MP, Muller S, Moraes CT. Fast adaptive coevolution of nuclear and mitochondrial subunits of ATP synthetase in orangutan. Molecular Biology and Evolution. 2005;22(3):716–724. doi: 10.1093/molbev/msi059. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bornstein B, Mas JA, Patrono C, Fernandez-Moreno MA, Gonzalez-Vioque E, Campos Y, et al. Comparative analysis of the pathogenic mechanisms associated with the G8363A and A8296G mutations in the mitochondrial tRNA(Lys) gene. The Biochemical Journal. 2005;387(Pt. 3):773–778. doi: 10.1042/BJ20040949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick B, Ghosh A, Yue Y, Long C, Duan D. Systemic AAV-9 transduction in mice is influenced by animal age but not by the route of administration. Gene Therapy. 2007;14(22):1605–1609. doi: 10.1038/sj.gt.3303029. [DOI] [PubMed] [Google Scholar]
- Bruno C, Martinuzzi A, Tang Y, Andreu AL, Pallotti F, Bonilla E, et al. A stop-codon mutation in the human mtDNA cytochrome c oxidase I gene disrupts the functional structure of complex IV. American Journal of Human Genetics. 1999;65(3):611–620. doi: 10.1086/302546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn A. Platelet-mediated transformation of human mitochondrial DNA-less cells. Methods in Enzymology. 1996;264:334–339. doi: 10.1016/s0076-6879(96)64031-2. [DOI] [PubMed] [Google Scholar]
- Clayton DA, Teplitz RL, Nabholz M, Dovey H, Bodmer W. Mitochondrial DNA of human–mouse cell hybrids. Nature. 1971;234(5331):560–562. doi: 10.1038/234560a0. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nature Methods. 2011;8(1):74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- Eichwald V, Daeffler L, Klein M, Rommelaere J, Salome N. The NS2 proteins of parvovirus minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. Journal of Virology. 2002;76(20):10307–10319. doi: 10.1128/JVI.76.20.10307-10319.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Analytical Biochemistry. 1983;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Human Molecular Genetics. 2009;18(6):1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammage PA, Rorbach J, Vincent AI, Rebar EJ, Minczuk M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Molecular Medicine. 2014;6(4):458–466. doi: 10.1002/emmm.201303672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JL, Craven L, Turnbull DM, Taylor RW. Experimental strategies towards treating mitochondrial DNA disorders. Bioscience Reports. 2007;27(1–3):139–150. doi: 10.1007/s10540-007-9042-3. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Yue Y, Long C, Bostick B, Duan D. Efficient whole-body transduction with trans-splicing adeno-associated viral vectors. Molecular Therapy. 2007;15(4):750–755. doi: 10.1038/sj.mt.6300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Bonilla E, Manfredi G, DiMauro S, Moraes CT. Segregation patterns of a novel mutation in the mitochondrial tRNA glutamic acid gene associated with myopathy and diabetes mellitus. American Journal of Human Genetics. 1995;56(5):1017–1025. [PMC free article] [PubMed] [Google Scholar]
- Hao H, Moraes CT. A disease-associated G5703A mutation in human mitochondrial DNA causes a conformational change and a marked decrease in steady-state levels of mitochondrial tRNA(Asn) Molecular and Cellular Biology. 1997;17(12):6831–6837. doi: 10.1128/mcb.17.12.6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(23):10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nature Biotechnology. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki K, Fuess S, Storm TA, Gibson GA, McTiernan CF, Kay MA, et al. Robust systemic transduction with AAV9 vectors in mice: Efficient global cardiac gene transfer superior to that of AAV8. Molecular Therapy. 2006;14(1):45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Takai D, Soejima A, Isobe K, Yamasoba T, Oka Y, et al. Mutant mtDNA at 1555 A to G in 12S rRNA gene and hypersusceptibility of mitochondrial translation to streptomycin can be co-transferred to rho 0 HeLa cells. Biochemical and Biophysical Research Communications. 1996;223(3):496–501. doi: 10.1006/bbrc.1996.0923. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nature Genetics. 1996;14(2):146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- Jun AS, Trounce IA, Brown MD, Shoffner JM, Wallace DC. Use of transmitochondrial cybrids to assign a complex I defect to the mitochondrial DNA-encoded NADH dehydrogenase subunit 6 gene mutation at nucleotide pair 14459 that causes Leber hereditary optic neuropathy and dystonia. Molecular and Cellular Biology. 1996;16(3):771–777. doi: 10.1128/mcb.16.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science. 1989;246(4929):500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- King MP, Koga Y, Davidson M, Schon EA. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Molecular and Cellular Biology. 1992;12(2):480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddell L, Manthey G, Pannunzio N, Bailis A. Quantitation and analysis of the formation of HO-endonuclease stimulated chromosomal translocations by single-strand annealing in Saccharomyces cerevisiae. Journal of Visualized Experiments. 2011;(55) doi: 10.3791/3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nature Biotechnology. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Minczuk M, Papworth MA, Kolasinska P, Murphy MP, Klug A. Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(52):19689–19694. doi: 10.1073/pnas.0609502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M, Papworth MA, Miller JC, Murphy MP, Klug A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Research. 2008;36(12):3926–3938. doi: 10.1093/nar/gkn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes CT. What regulates mitochondrial DNA copy number in animal cells? Trends in Genetics. 2001;17(4):199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- Moraes CT, Ricci E, Petruzzella V, Shanske S, DiMauro S, Schon EA, et al. Molecular analysis of the muscle pathology associated with mitochondrial DNA deletions. Nature Genetics. 1992;1(5):359–367. doi: 10.1038/ng0892-359. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Pan Y, Xiao L, Li AS, Zhang X, Sirois P, Zhang J, et al. Biological and biomedical applications of engineered nucleases. Molecular Biotechnology. 2013;55(1):54–62. doi: 10.1007/s12033-012-9613-9. [DOI] [PubMed] [Google Scholar]
- Pickrell AM, Fukui H, Wang X, Pinto M, Moraes CT. The striatum is highly susceptible to mitochondrial oxidative phosphorylation dysfunctions. The Journal of Neuroscience. 2011;31(27):9895–9904. doi: 10.1523/JNEUROSCI.6223-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell AM, Pinto M, Hida A, Moraes CT. Striatal dysfunctions associated with mitochondrial DNA damage in dopaminergic neurons in a mouse model of Parkinson’s disease. The Journal of Neuroscience. 2011;31(48):17649–17658. doi: 10.1523/JNEUROSCI.4871-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto M, Moraes CT. Mitochondrial genome changes and neurodegenerative diseases. Biochimica et Biophysica Acta. 2013;1842:1198–1207. doi: 10.1016/j.bbadis.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto M, Pickrell AM, Fukui H, Moraes CT. Mitochondrial DNA damage in a mouse model of Alzheimer’s disease decreases amyloid beta plaque formation. Neurobiology of Aging. 2013;34(10):2399–2407. doi: 10.1016/j.neurobiolaging.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteous WK, James AM, Sheard PW, Porteous CM, Packer MA, Hyslop SJ, et al. Bioenergetic consequences of accumulating the common 4977-bp mitochondrial DNA deletion. European Journal of Biochemistry. 1998;257(1):192–201. doi: 10.1046/j.1432-1327.1998.2570192.x. [DOI] [PubMed] [Google Scholar]
- Russell O, Turnbull D. Mitochondrial DNA disease-molecular insights and potential routes to a cure. Experimental Cell Research. 2014;325(1):38–43. doi: 10.1016/j.yexcr.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacco M, Bonilla E, Schon EA, DiMauro S, Moraes CT. Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Human Molecular Genetics. 1994;3(1):13–19. doi: 10.1093/hmg/3.1.13. [DOI] [PubMed] [Google Scholar]
- Shoubridge EA, Johns T, Karpati G. Complete restoration of a wild-type mtDNA genotype in regenerating muscle fibres in a patient with a tRNA point mutation and mitochondrial encephalomyopathy. Human Molecular Genetics. 1997;6(13):2239–2242. doi: 10.1093/hmg/6.13.2239. [DOI] [PubMed] [Google Scholar]
- Smith PM, Lightowlers RN. Altering the balance between healthy and mutated mitochondrial DNA. Journal of Inherited Metabolic Disease. 2011;34(2):309–313. doi: 10.1007/s10545-010-9122-6. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Moraes CT. Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Human Molecular Genetics. 2001;10(26):3093–3099. doi: 10.1093/hmg/10.26.3093. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Human Molecular Genetics. 2005;14(7):893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, et al. Knockout mice created by TALEN-mediated gene targeting. Nature Biotechnology. 2013;31(1):23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nature Biotechnology. 2004;22(5):589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Borgeld HJ, Zhang J, Muramatsu S, Gong JS, Yoneda M, et al. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. Journal of Biomedical Science. 2002;9(6 Pt 1):534–541. doi: 10.1159/000064726. [DOI] [PubMed] [Google Scholar]
- Toompuu M, Tiranti V, Zeviani M, Jacobs HT. Molecular phenotype of the np 7472 deafness-associated mitochondrial mutation in osteosarcoma cell cybrids. Human Molecular Genetics. 1999;8(12):2275–2283. doi: 10.1093/hmg/8.12.2275. [DOI] [PubMed] [Google Scholar]
- Wang X, Pickrell AM, Rossi SG, Pinto M, Dillon LM, Hida A, et al. Transient systemic mtDNA damage leads to muscle wasting by reducing the satellite cell pool. Human Molecular Genetics. 2013;22(19):3976–3986. doi: 10.1093/hmg/ddt251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Williams SL, Bacman SR, Moraes CT. Emerging therapeutic approaches to mitochondrial diseases. Developmental Disabilities Research Reviews. 2010;16(2):219–229. doi: 10.1002/ddrr.109. [DOI] [PubMed] [Google Scholar]
- Xu H, DeLuca SZ, O’Farrell PH. Manipulating the metazoan mitochondrial genome with targeted restriction enzymes. Science. 2008;321(5888):575–577. doi: 10.1126/science.1160226. [DOI] [PMC free article] [PubMed] [Google Scholar]