Abstract
Koolpinyah virus (KOOLV) isolated from healthy Australian cattle and Yata virus (YATV) isolated from a pool of Mansonia uniformis mosquitoes in the Central African Republic have been tentatively identified as rhabdoviruses. KOOLV was shown previously to be related antigenically to kotonkon virus, an ephemerovirus that has caused an ephemeral fever-like illness in cattle in Nigeria, but YATV failed to react antigenically with any other virus tested. Here we report the complete genome sequences of KOOLV (16133 nt) and YATV (14479 nt). Each has a complex genome organisation, with multiple genes, including a second non-structural glycoprotein (GNS) gene and a viroporin (α1) gene, between the G and L genes as is characteristic of ephemeroviruses. Based on an analysis of genome organisation, sequence identity and cross-neutralisation, we demonstrate that both KOOLV and YATV should be classified as two new species in the genus Ephemerovirus.
Keywords: Koolpinyah virus, Yata virus, rhabdovirus, ephemerovirus, genome sequence, phylogeny
Ephemeroviruses are arthropod-borne rhabdoviruses that infect ruminants (Walker et al., 2012). Bovine ephemeral fever virus (BEFV), which causes an acute disabling illness in cattle and water buffalo, has significant economic impacts in many tropical, sub-tropical and temperate regions of Africa, the Middle-East, Asia and Australia (Walker, 2005). Other known ephemeroviruses include Berrimah virus (BRMV), Kimberley virus (KIMV), Adelaide River virus (ARV), Obodhiang virus (OBOV) and kotonkan virus (KOTV) which have been isolated from healthy cattle or haematophagous insects (mosquitoes or biting midges) in Africa or Australia (Blasdell et al., 2012a; Blasdell et al., 2012b; Cybinski & Zakrzewski, 1983; Gard et al., 1983; Gard et al., 1984; Kemp et al., 1973). Of these, only KOTV has to date been associated with clinical disease in cattle (Tomori et al., 1974). Koolpinyah virus (KOOLV) was isolated on two occasions, in 1985 and 1986, from the blood of sentinel cattle at Berrimah (12° 26′ S, 130° 55′ E), near Darwin, in the Northern Territory of Australia (Gard et al., 1992). At the time of the isolations, there was evidence of seroconversion to KOOLV antibody in other cattle at the same site and sheep infected experimentally with the virus also seroconverted. KOOLV was shown to have typical rhabdovirus morphology and to cross-react in indirect immunofluorescence (IFA) and virus neutralisation tests with KOTV, which had been isolated from a mixed pool of biting midges (Culicoides spp.) collected at Ibadan, Nigeria, in 1967 (Bauer and Murphy, 1975; Kemp et al., 1973). A serological survey conducted in Nigeria indicated a high prevalence of KOTV antibody in cattle, sheep and some other mammals (Kemp et al., 1973), and seroconversion to KOTV neutralising antibody in cattle was reported to be associated with an acute bovine ephemeral fever-like illness with low levels of mortality in heifers that had been imported to Nigeria from Europe (Kemp et al., 1973). KOTV was also shown to cause disease in a calf following experimental infection (Tomori et al., 1974). Recently, analysis of the complete genome sequence has indicated that KOTV is most closely related to BEFV and other ephemeroviruses (Blasdell et al., 2012a) and it has now been classified as a species in the genus Ephemerovirus.
Yata virus (YATV) was isolated in 1969 from a pool of mosquitoes (Mansonia uniformis) collected in the village of Birao (10° 17′ N, 22° 47′ E) in the Central African Republic (Karabatsos, 1985). It was found to cause mortality following intracerebral or intraperitoneal inoculation of suckling mice but no information is currently available on its natural host range, pathogenicity or geographic distribution. Although reported to be a possible rhabdovirus (Brown et al., 1979), YATV failed to react in complement-fixation (CF) or IFA tests with a wide range of rhabdoviruses (Calisher et al., 1989; Tesh et al., 1983), including KOTV and other ephemeroviruses, and its correct classification and natural ecology have remained unclear. This study aimed to resolve the relationships of KOOLV and YATV to ephemeroviruses and other members of the Rhabdoviridae.
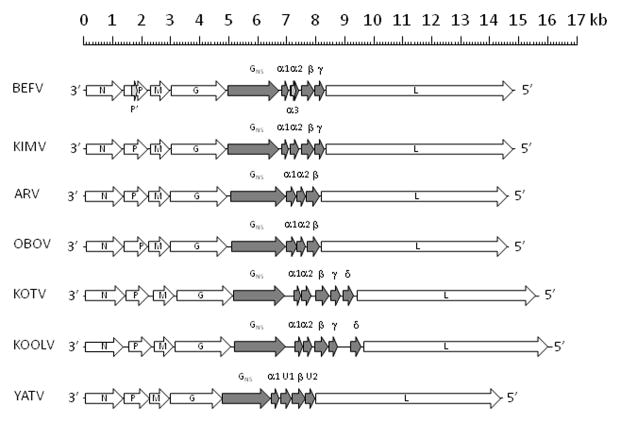
KOOLV (isolate DPP819) and YATV (isolate DakArB2181) were prepared for sequencing as described previously (Vasilakis et al., 2014). The complete sequences of the KOOLV and YATV genomes were determined using the Illumina MiSeq (2×75 paired-ends) and Illumina HiSeq (50 paired-ends) platforms, respectively. Genome assembly and open reading frame (ORF) identification was performed as described previously (Vasilakis et al., 2014). The average read coverage was approximately 7,400 for KOOLV and approximately 870 for YATV, but with poor coverage at the extreme termini. Terminal sequences for YATV were determined by 5′ and 3′ RACE and Sanger sequencing of the amplicon as described previously (Gubala et al., 2010; Li et al., 2005) (primers available on request). The organisation and features of the genomes are shown in Figure 1.
Figure 1.
Schematic representation of the genome organisations of BEFV, KIMV, ARV, OBOV, KOTV, KOOLV and YATV. Block arrows indicate the location of ORFs, including alternative ORFs (light shading) and ephemerovirus accessory genes (dark shading).
The 16,133 nt KOOLV genome is the largest yet reported of any rhabdovirus. The genome organization (3′-l-N-P-M-G-GNS-α1-α2-β-γ-δ-L-t-5′ in negative polarity) is most similar to that of KOTV; the N, P, M, G and L ORFs encode the canonical rhabdovirus structural proteins, ORFs GNS, α1 α2, β, and γ encode accessory proteins common to BEFV and other ephemeroviruses, and ORF δ is an accessory gene that occurs only in KOOLV and KOTV. Each ORF is flanked by conserved transcription initiation (UUGUC) and transcription termination/polyadenylation (GUACUUUUUUU or UAUCUUUUUUU) sequence motifs, except for the α1 and α2 ORFs which, as in other ephemeroviruses, occur consecutively in the same transcriptional unit. Like KOTV, the KOOLV genome is unusual amongst ephemeroviruses in that each transcriptional unit features very long 3′ non-coding regions (>40 nt) prior to the polyadenylation motif. In KOOLV, the longest of these occurs following the GNS ORF (227 nt) and following the γ ORF (438 nt). In KOTV, the corresponding regions following these ORFs are in total somewhat shorter (316 nt and 131 nt), accounting for much of the 263 nt difference in the overall length of the KOOLV and KOTV genomes.
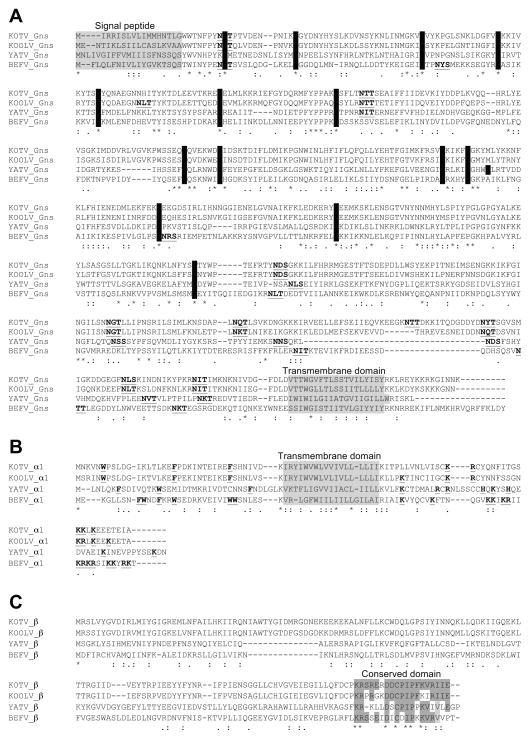
The 14,479 nt YATV genome is organised similarly to those of other ephemeroviruses, comprising ORFs encoding the five canonical rhabdovirus structural proteins (N, P, M, G and L) and five ORFs between the G and L coding for non-structural proteins (3′-l-N-P-M-G-GNS-α1-U1-β-U2-L-t-5′ in negative polarity). The GNS ORF follows the G ORF and encodes a class 1 transmembrane glycoprotein that shares significant sequence identity with the GNS proteins of ephemeroviruses, including a set of 14 cysteine residues that are likely to form seven conserved disulphide bridges (Figure 2A). However, YATV is unique in that there is a single unpaired cysteine residue (C247) in the GNS ectodomain. The α1 ORF encodes a predicted 11.9 kDa transmembrane protein which, like the BEFV α1 protein (Joubert et al., 2014), has the structural characteristics of class I viroporins (Figure 2B). The β ORF encodes a 16.5 kDa protein of unknown function that shares a conserved C-terminal domain with the ephemerovirus β proteins (Figure 2C). The U1 ORF encodes a 14.4 kDa protein that has no remarkable structural features and although it is located in the same position in the genome as the ephemerovirus α2 ORFs, U1 shares no evident sequence identity or predicted structural homology with these proteins. Furthermore, whereas the ephemerovirus α1 and α2 ORFs occur as consecutive ORFs within a single transcriptional unit, the YATV U1 ORF occurs in an independent transcriptional unit flanked by conserved transcription initiation (UUGUC) and transcription termination/polyadenylation (UACUUUUUUU) sequence motifs. Similarly, the U2 ORF encodes a 13.8 kDa protein with no remarkable structural characteristics that, although similar in size and also encoded in an independent transcriptional unit downstream of the β gene, shares no identifiable homology with the ephemeroviruses γ or δ proteins. Thus, although the YATV genome shares many features with those of other ephemeroviruses, it has some unique distinguishing characteristics.
Figure 2.
Clustal X alignments of the deduced amino acid sequences of KOTV, KOOLV, YATV and BEFV accessory proteins. A. GNS proteins showing predicted signal peptide (SignalP) and transmembrane (TMHMM) domains (shaded in grey), cysteine residues in the ectodomain (shaded in black) and potential N-glycosylation sites (bold and underlined). B. α1 proteins showing characteristics of Class I viroporins including predicted transmembrane domains (shaded in grey), large aromatic residues in the N-terminal domain and basic residues in the C-terminal domain (bold and underlined). C. β proteins showing the conserved domain near the C-terminus (shaded).
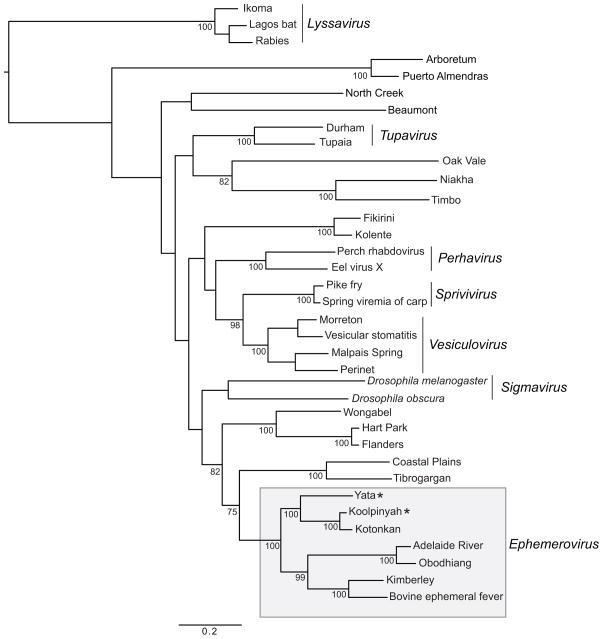
To infer the phylogenetic relationships among KOOLV, YATV and other ephemeroviruses, we prepared a data set that contained complete L gene amino acid sequences from all known and proposed ephemeroviruses, as well as a minimum of two representative sequences (where available) from the majority of the remaining animal rhabdovirus genera. A protein sequence alignment was constructed using the MUSCLE program with default setting (Edgar, 2004), with ambiguous regions of the alignment then removed using GB locks (Talavera and Castresana, 2007). The final sequence alignment for phylogenetic analysis comprised 36 taxa and 947 amino acids. A phylogenetic tree of these data was inferred using the maximum likelihood method available in PhyML version 3.0 (Guindon et al., 2010), incorporating the WAG+Γ model of amino acid substitution and 1,000 bootstrap replicates. The results of this analysis indicated that KOOLV, KOTV and YATV formed a well supported clade within genus Ephemerovirus, with KOOLV and KOTV falling as sister taxa and YATV as more divergent (Figure 3). MUSCLE alignments of the full genome sequences, individual genes and predicted proteins of all described ephemeroviruses were performed in Geneious 7.1.3. Pairwise alignments of full genome sequences revealed nucleotide sequence identities of 71.5% for KOOLV and KOTV, 52.0% for KOOLV and YATV and 52.5% for KOTV and YATV. A multiple alignment of all known and proposed ephemerovirus genomes gave sequence identities of >41% between KOOLV, YATV and all other ephemeroviruses. Nucleotide and amino acid sequence identities for individual genes were consistently high between KOOLV and KOTV and were the highest for any pair of viruses for the N, P, G, GNS, α1 and α2 genes and also for nucleotide sequence identity in the L gene. In contrast, sequence identities were relatively low between YATV and all other ephemeroviruses (Table S1).
Figure 3.
Maximum likelihood phylogeny of the conserved regions of the L protein from all described ephemeroviruses (boxed), and select representatives of other rhabdovirus genera. The positions of YATV and KOOLV are indicated with a *. Bootstrap values > 70% are shown, and the scale bar represents the number of amino acid substitutions per site.
Neutralisation tests were conducted in Vero cells as described previously using KOOLV, KOTV and YATV, and the corresponding immune mouse ascites fluids (IMAFs) (Blasdell et al., 2012a). No cross-reactions were observed between YATV and either of the other viruses. KOOLV and KOTV were also clearly distinguished using KOTV IMAF but a low-level cross reaction was observed (Table 1). Gard et al. (1992) have shown previously that KOOLV and KOTV can be clearly distinguished in cross-neutralisation tests using KOOLV IMAF. We also tested this antiserum but it failed to react against homologous virus, likely due to long-term storage.
Table 1.
Virus neutralisation tests of KOOLV, KOTV and YATV.
| Virus | Antiserum (IMAF)* | |
|---|---|---|
| KOTV | YATV | |
| KOOLV | 1/20 | <1/20 |
| KOTV | 1/640 | <1/20 |
| YATV | <1/20 | 1/320 |
The only available KOOLV IMAF failed to neutralize the homologous virus.
Currently, the genus Ephemerovirus is comprised of the species Bovine ephemeral fever virus, Adelaide River virus, Berrimah virus, Kotonkan virus and Obodhiang virus (Adams and Carstens, 2012; Adams et al., 2013; Dietzgen et al., 2012), and it has recently been proposed that Kimberley virus (including KIMV from Australia and Malakal virus from Sudan) should be assigned as a new species (Blasdell et al., 2012b). Based on solid two-way cross-reactions with several ephemeroviruses in IFA tests (Calisher et al., 1989), Puchong virus (PUCV) also appears to be a member of the genus. So far, all ephemeroviruses have been isolated either from cattle and/or arthropod vectors (Walker, 2005). Like BEFV, BRMV, KIMV and ARV, KOOLV has been isolated from cattle (Gard et al., 1992); and like MALV, OBOV and PUCV, YATV has been isolated from the mosquito Mansonia uniformis (Schmidt et al., 1965). Both BEFV and KIMV have also been isolated from mosquitoes (Culex annulirostris, Anopheles bancroftii) and, although BEFV, KIMV and KOTV have also been isolated from biting midges (Culicoides spp.), there is evidence suggesting that mosquitoes rather than midges are likely to be the primary vectors (St George, 2009). Thus, the natural ecology of KOOLV and YATV appears to be consistent with that of other ephemeroviruses. The genome organisations of KOOLV and YATV are also consistent with their assignment as ephemeroviruses, featuring a complex region between the G and L ORFs in which the GNS ORF is followed by the α1 ORF and several other small ORFs (Walker et al., 2011). Phylogenetic analysis also clearly demonstrates that both viruses fall within the ephemerovirus clade. Based on these characteristics, we propose that both KOOLV and YATV should be assigned as new members of the genus Ephemerovirus.
Current species demarcation criteria for ephemeroviruses are based on low or no cross-neutralisation, up to 91% sequence identity in the N protein and variations in genome organisation and transcription control sequences (Dietzgen et al., 2012). YATV clearly fulfils these criteria as genetic analysis indicates that it is highly divergent (<52.5% full genome sequence identity; <51.2% N protein sequence identity) from other known ephemeroviruses and contains ORFs that encode proteins that do not have homologues in other ephemeroviruses. Previous serological analysis also found no serological cross reaction between YATV and other known rhabdoviruses based on IFA and CF tests (Calisher et al., 1989; Tesh et al., 1983) and we have now shown that it does not cross-react in neutralisation tests with either KOOLV or KOTV, to which it is most closely related. The case for species assignment of KOOLV is more complex. It does not appear to meet the current species demarcation criteria, having the same transcription control sequences, a very similar genome organisation and an N protein with 92.7% sequence identity to KOTV. As little is known about the distribution of these viruses and KOOLV has only been isolated in Australia and KOTV in Nigeria, it could be argued that these closely related viruses actually represent geographic variants of the same species. However, the level of full genome nucleotide sequence identity between these two viruses is considerably lower (71.5%) than that found between KIMV from Australia and MALV from Sudan (90.6%), which are considered isolates of the same species (Blasdell et al., 2012b). Indeed, the level sequence identity is more similar than that found between two other pairs of closely related ephemerovirus species: ARV and OBOV (70.2% identity), which were isolated in Australia and Sudan, respectively, (Blasdell et al., 2012a) and BEFV and BRMV (72.0%, unpublished data) which are sympatric in at least part of their respective ranges (Walker, 2005). In addition, our serological analysis found a low-level, one-way cross-neutralisation and >16-fold difference in neutralisation titres between KOOLV and KOTV. Furthermore, unlike KOTV, which is a known pathogen of cattle (Tomori et al., 1974), KOOLV has only been isolated from healthy cattle (Gard et al., 1992). This is similar to BEFV and BRMV; BEFV is a common bovine pathogen whilst BRMV is closely related both genetically and serologically but has never been linked to disease (Cybinski et al., 1992; Cybinski et al., 1990; Gard et al., 1983). Therefore, we suggest that ephemerovirus species demarcation criteria should be reviewed to allow consideration of KOOLV as a distinct ephemerovirus species.
Little is known of the vertebrate host range, principal vectors or geographic distribution of most ephemeroviruses. There is evidence that multiple sequential infections of cattle with ephemerovirus can occur (Cybinski, 1987) and reports suggest that bovine ephemeral fever does occur in the absence of seroconversion to BEFV (St. George et al., 1977; Uren et al., 1987). There is also evidence that cross-reactive antibody to KIMV and other ephemeroviruses may drive the evolution of BEFV (Trinidad et al., 2014). Further studies should be conducted to better define the ecology and pathogenicity of these viruses and their contribution to the disease burden of livestock.
Supplementary Material
Highlights.
Koolpinyah and Yata viruses are rhabdoviruses isolated from cattle and mosquitoes
We determine the complete genome sequences of these poorly characterised viruses
We show that they have similar genome structure and sequence to bovine ephemeral fever virus
We show they cluster phylogenetically as ephemeroviruses but are distinct serologically
Each should be classified as new ephemerovirus species of unknown pathogenic potential
Acknowledgments
This work was supported in part by a grant from the Institute for Human Infections and Immunity, University of Texas Medical Branch (NV), and NIH contract HHSN272201000040I/HHSN27200004/D04 (NV, RBT). ECH is supported by an NHMRC Australia Fellowship. We thank Brandy Russell, Centres for Disease Control, Fort Collins, Colorado, for the supply of KOOLV IMAF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MJ, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses, 2012. Archiv Virol. 2012;157:1411–1422. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MJ, King AM, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses, 2013. Archiv Virol. 2013;158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- Bauer SP, Murphy FA. Relationship of two arthropod-borne rhabdoviruses (Kotonkan and Obodhiang) to the rabies serogroup. Infect Immun. 1975;12:1157–1172. doi: 10.1128/iai.12.5.1157-1172.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasdell KR, Voysey R, Bulach D, Joubert DA, Tesh RB, Boyle DB, Walker PJ. Kotonkan and Obodhiang viruses: African ephemeroviruses with large and complex genomes. Virology. 2012a;425:143–153. doi: 10.1016/j.virol.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Blasdell KR, Voysey R, Bulach DM, Trinidad L, Tesh RB, Boyle DB, Walker PJ. Malakal virus from Africa and Kimberley virus from Australia are geographic variants of a widely distributed ephemerovirus. Virology. 2012b;433:236–244. doi: 10.1016/j.virol.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Brown F, Bishop DHL, Crick J, Francki RIB, Holland JJ, Hull R, Johnson KA, Martelli GP, Murphy FA, Obijeski JF, Peters D, Pringle CR, Reichmann ME, Schneider LG, Shope RE, Simpson DIH, Summers DF, Wagner RR. Rhabdoviridae. Intervirology. 1979;12:1–7. doi: 10.1159/000149062. [DOI] [PubMed] [Google Scholar]
- Calisher CH, Karabatsos N, Zeller H, Digoutte JP, Tesh RB, Shope RE, Travassos da Rosa AP, St George TD. Antigenic relationships among rhabdoviruses from vertebrates and hematophagous arthropods. Intervirology. 1989;30:241–257. doi: 10.1159/000150100. [DOI] [PubMed] [Google Scholar]
- Cybinski DH. Homologous and heterologous antibody reactions in sera from cattle naturally infected with bovine ephemeral fever group viruses. Vet Microbiol. 1987;13:1–9. doi: 10.1016/0378-1135(87)90092-7. [DOI] [PubMed] [Google Scholar]
- Cybinski DH, Davis SS, Zakrzewski H. Antigenic variation of the bovine ephemeral fever virus glycoprotein. Archiv Virol. 1992;124:211–224. doi: 10.1007/BF01309803. [DOI] [PubMed] [Google Scholar]
- Cybinski DH, Walker PJ, Byrne KA, Zakrzewski H. Mapping of antigenic sites on the bovine ephemeral fever virus glycoprotein using monoclonal antibodies. J Gen Virol. 1990;71:2065–2072. doi: 10.1099/0022-1317-71-9-2065. [DOI] [PubMed] [Google Scholar]
- Cybinski DH, Zakrzewski H. The isolation and preliminary characterization of a rhabdovirus in Australia related to bovine ephemeral fever virus. Vet Microbiol. 1983;8:221–235. doi: 10.1016/0378-1135(83)90075-5. [DOI] [PubMed] [Google Scholar]
- Dietzgen RG, Calisher CH, Kurath G, Kuzman IV, Rodriguez LL, Stone DM, Tesh RB, Tordo N, Walker PJ, Wetzel T, Whitfield AE. Rhabdoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego: 2012. pp. 654–681. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard GP, Cybinski DH, St George TD. The isolation in Australia of a new virus related to bovine ephemeral fever virus. Aust Vet J. 1983;60:89–90. doi: 10.1111/j.1751-0813.1983.tb05882.x. [DOI] [PubMed] [Google Scholar]
- Gard GP, Cybinski DH, Zakrzewski H. The isolation of a fourth bovine ephemeral fever group virus. Aust Vet J. 1984;61:332. doi: 10.1111/j.1751-0813.1984.tb07147.x. [DOI] [PubMed] [Google Scholar]
- Gard GP, Melville LF, Calisher CH, Karabatsos N. Koolpinyah: a virus related to kotonkan from cattle in northern Australia. Intervirology. 1992;34:142–145. doi: 10.1159/000150274. [DOI] [PubMed] [Google Scholar]
- Gubala A, Davis S, Weir R, Melville L, Cowled C, Walker P, Boyle D. Ngaingan virus, a macropod-associated rhabdovirus, contains a second glycoprotein gene and seven novel open reading frames. Virology. 2010;399:98–108. doi: 10.1016/j.virol.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Joubert DA, Blasdell KR, Audsley MD, Trinidad L, Monaghan P, Dave KA, Lieu KG, Amos-Ritchie R, Jans DA, Moseley GW, Gorman JJ, Walker PJ. Bovine ephemeral fever rhabdovirus α1 protein has viroporin-like properties and binds importin β1 and importin 7. J Virol. 2014;88:1591–1603. doi: 10.1128/JVI.01812-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabatsos N, editor. International Catalogue of Arboviruses Including Certain other Viruses of Vertebrates. American Society for Tropical Medicine and Hygiene; San Antonio: 1985. [DOI] [PubMed] [Google Scholar]
- Kemp GE, Lee VH, Moore DL, Shope RE, Causey OR, Murphy FA. Kotonkan, a new rhabdovirus related to Mokola virus of the rabies serogroup. Amer J Epidemiol. 1973;98:43–49. doi: 10.1093/oxfordjournals.aje.a121531. [DOI] [PubMed] [Google Scholar]
- Li Z, Yu M, Zhang H, Wang HY, Wang LF. Improved rapid amplification of cDNA ends (RACE) for mapping both the 5′ and 3′ terminal sequences of paramyxovirus genomes. J Virol Meth. 2005;130:154–156. doi: 10.1016/j.jviromet.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Schmidt JR, Williams MC, Lulu M, Mivule A, Mujombe E. East African Virus Research Institute Report. 1965. Viruses isolated from mosquitoes collected in the Southern Sudan and Western Ethiopia; p. 24. [Google Scholar]
- St George TD. In: Ryan P, Aaskov J, Russell R, editors. Evidence that mosquitoes are the vectors of bovine epmemeral fever virus; 10th Symposium on Arbovirus Research in Australia; Coffs Harbour: QIMR/QUT; 2009. pp. 161–164. [Google Scholar]
- St George TD, Standfast HA, Christie DG, Knott SG, Morgan IR. The epizootiology of bovine ephemeral fever in Australia and Papua - New Guinea. Aust Vet J. 1977;53:17–28. doi: 10.1111/j.1751-0813.1977.tb15812.x. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. System Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tesh RB, Travassos da Rosa AP, Travassos da Rosa JS. Antigenic relationship among rhabdoviruses infecting terrestrial vertebrates. J Gen Virol. 1983;64:169–176. doi: 10.1099/0022-1317-64-1-169. [DOI] [PubMed] [Google Scholar]
- Tomori O, Fagbami A, Kemp G. Kotonkan virus: experimental infection of white Fulani calves. Bull Epizoot Dis Afr. 1974;22:195–200. [PubMed] [Google Scholar]
- Trinidad L, Blasdell KR, Joubert DA, Davis SS, Melville L, Kirkland PD, Coulibaly F, Holmes EC, Walker PJ. Evolution of bovine ephemeral fever virus in the Australian episystem. J Virol. 2014;88:1525–1535. doi: 10.1128/JVI.02797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren MF, St George TD, Kirkland PD, Stranger RS, Murray MD. Epidemiology of bovine ephemeral fever in Australia 1981–1985. Aust J Biol Sci. 1987;40:125–136. doi: 10.1071/bi9870125. [DOI] [PubMed] [Google Scholar]
- Vasilakis N, Castro-Llanos F, Widen SG, Aguilar PV, Guzman H, Guevara C, Fernandez R, Auguste AJ, Wood TG, Popov V, Mundal K, Ghedin E, Kochel TJ, Holmes EC, Walker PJ, Tesh RB. Arboretum and Puerto Almendras viruses: two novel rhabdoviruses isolated from mosquitoes in Peru. J Gen Virol. 2014;95:787–792. doi: 10.1099/vir.0.058685-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker PJ. Bovine ephemeral fever in Australia and the world. Curr Top Microbiol Immunol. 2005;292:57–80. doi: 10.1007/3-540-27485-5_4. [DOI] [PubMed] [Google Scholar]
- Walker PJ, Blasdell KR, Joubert DA. Ephemeroviruses: arthropod-borne rhabdoviruses of ruminants, with large and complex genomes. In: Dietzgen RG, Kuzman IV, editors. Rhabdoviruses: Molecular Taxonomy, Evolution, Genomics, Ecology, Cytopathology and Control. Horizon Scientific Press; Norwich: 2012. pp. 60–88. [Google Scholar]
- Walker PJ, Dietzgen RG, Joubert DA, Blasdell KR. Rhabdovirus accessory genes. Virus Res. 2011;162:110–125. doi: 10.1016/j.virusres.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.