Abstract
Background and Aims.
Colorectal cancer (CRC) is a heterogeneous disease that can develop via several pathways. Different CRC subtypes, identified based on tumor markers, have been proposed to reflect these pathways. We evaluated the significance of these previously proposed classifications to survival.
Methods.
Participants in the population-based Seattle Colon Cancer Family Registry were diagnosed with invasive CRC from 1998 through 2007 in western Washington State (n=2706), and followed for survival through 2012. Tumor samples were collected from 2050 participants and classified into 5 subtypes based on combinations of tumor markers: type 1 (microsatellite instability [MSI] high, CpG island methylator phenotype [CIMP] positive, positive for BRAF mutation, negative for KRAS mutation); type 2 (microsatellite stable [MSS] or MSI-low, CIMP-positive, positive for BRAF mutation, negative for KRAS mutation); type 3 (MSS or MSI-low, non-CIMP, negative for BRAF mutation, positive for KRAS mutation); type 4 (MSS or MSI-low, non-CIMP, negative for mutations in BRAF and KRAS); and type 5 (MSI-high, non-CIMP, negative for mutations in BRAF and KRAS). Multiple imputation was used to impute tumor markers for those missing data on 1-3 markers. We used Cox regression to estimate hazard ratios (HR) and 95% confidence intervals (CI) for associations of subtypes with disease-specific and overall mortality, adjusting for age, sex, body mass, diagnosis year, and smoking history.
Results.
Compared to participants with type 4 tumors (the most predominant), participants with type 2 tumors had the highest disease-specific mortality (HR=2.20, 95% CI: 1.47-3.31); subjects with type 3 tumors also had higher disease-specific mortality (HR=1.32, 95% CI: 1.07-1.63). Subjects with type 5 tumors had the lowest disease-specific mortality (HR=0.30, 95% CI: 0.14-0.66). Associations with overall mortality were similar to those with disease-specific mortality.
Conclusions.
Based on a large, population-based study, CRC subtypes, defined by proposed etiologic pathways, are associated with marked differences in survival. These findings indicate the clinical importance of studies into the molecular heterogeneity of CRC.
Keywords: oncogene, methylation, serrated colorectal cancer, prognostic factor
INTRODUCTION
Increasing evidence indicates that colorectal cancer (CRC) is a biologically heterogeneous disease that can develop via a number of distinct pathways involving different combinations of genetic and epigenetic changes.1,2 Proposed subtype classifications for CRC, based on the presence of microsatellite instability (MSI), the CpG island methylator phenotype (CIMP), and somatic mutations in BRAF and KRAS, are thought to approximate these distinct pathways.1,2 In particular, CRC reflective of the “traditional” adenoma-carcinoma pathway has been described as typically demonstrating absent (microsatellite stable, MSS) to low-level MSI (MSI-low) without CIMP and without somatic BRAF or KRAS mutations; CRC resulting from a “serrated” pathway has been described as frequently BRAF-mutated and CIMP-positive; and an additional pathway has been suggested for KRAS-mutated CRC that is MSS/MSI-low and CIMP-low.2,3
The biologic distinctions between CRC subtypes resulting from different etiologic pathways may plausibly translate to differences in survival. As tumor markers that may reflect such different pathways, MSI, CIMP, BRAF-mutation, and KRAS-mutation status have each been studied extensively, with evidence of differences in the distribution of tumor site, sex, age and stage at diagnosis, and survival.4-22 However, the significance of subtype classifications based on combinations of these four tumor markers with respect to survival has been minimally described.3,23 In the only prior study to evaluate differences in survival across CRC subtypes defined by these four tumor markers in combination, Samadder et al. suggested that CRC with a BRAF-mutated/CIMP-high phenotype, suggestive of the serrated pathway, was associated with modestly worse survival than CRC with a MSS/CIMP-negative/BRAF-mutation negative/KRAS-mutation negative phenotype, suggestive of the traditional pathway.3
Using data from the population-based Seattle Colon Cancer Family Registry (SCCFR) and the Postmenopausal Hormones Supplemental Study to the SCCFR (PMH-SCCFR),24,25 we further explored the relationship between CRC molecular subtypes, defined by common tumor marker combinations, and survival.
METHODS
Study population
A description of the study populations has been published elsewhere.24,25 Briefly, SCCFR study participants included persons diagnosed with incident invasive CRC between January 1998 and June 2002 who, at the time of diagnosis, were aged 20-74 years and resided in King, Pierce, or Snohomish counties of Washington State (Supplementary Table 1). Over this same period, women aged 50-74 at CRC diagnosis and residing in 10 surrounding counties were also recruited for participation in the PMH-SCCFR. During a second SCCFR recruitment phase (diagnosis dates April 2002 to July 2007), eligible participants were identified as individuals diagnosed at ages 18-49 with invasive CRC within the combined 13-county region. All cases were identified through the population-based Surveillance, Epidemiology, and End Results (SEER) cancer registry serving western Washington State. Eligibility was limited to English speakers with publicly-available telephone numbers. Of 3,525 eligible individuals contacted, 302 (9%) were deceased, 401 (11%) refused participation, 92 (3%) were lost to follow-up prior to interview, and 24 (1%) completed only a partial interview. Among participants who completed the interview (N=2,706), adequate tumor specimens were available for 77% (N=2,080). Participants for whom tumor specimens were not obtained were excluded from this analysis.
This study was approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center in accordance with assurances filed with and approved by the U.S. Department of Health and Human Services.
Tumor characteristics
DNA extracted from paraffin-embedded formalin-fixed diagnostic tumor tissue specimens was used in tumor marker testing. Testing for MSI was based on a 10-gene panel in DNA from tumor and normal surrounding tissue (BAT25, BAT26, BAT40, MYCL, D5S346, D17S250, ACTC, D18S55, D10S197, BAT34C4) for the majority of cases (N=1,430):24,26 tumors were classified as MSI-high if instability was observed for ≥30% of markers, and MSS/MSI-low if instability was observed in <30% of markers. For other cases (N=534), MSI status was based on immunohistochemistry testing of four markers (MLH1, MSH2, MSH6, PMS2): cases whose tissue exhibited positive staining for all markers were considered MSS/MSI-low, whereas cases negative for the expression of at least one marker were considered MSI-high.27,28 Tumor DNA was tested for the p.V600E BRAF mutation (N=1,948) using a fluorescent allele-specific PCR assay as described previously;29 this mutation accounts for ~90% of BRAF mutations in CRC.30 Mutations in KRAS codons 12 and 13 were identified through forward and reverse sequencing of amplified tumor DNA (N=1,894);8,31 mutations in this hotspot region account for ~80% of KRAS mutations in CRC.32,33 CIMP testing was completed for a large subset of cases (N=1,508) based on a validated quantitative DNA methylation assay using a five-gene panel (CACNA1G, IGF2, NEUROG1, RUNX3, SOCS1).34-36 As described elsewhere,34 tumors were classified as CIMP-positive if the percentage of methylated reference (PMR) ratio was ≥10 for at least three of five markers and as non-CIMP if the PMR ratio was ≥10 for fewer than three markers; PMR is calculated as the amount of methylated tumor DNA at a specific locus (normalized to input bisulfite DNA amount measured at ALU repetitive elements) divided by the ALU-normalized amount in a methylated reference sample, multiplied by 100. Tumor site and stage information was available from SEER.
Subtype classifications
Tumor subtypes were defined as follows, consistent with previously-suggested classifications:1,2 1) “type 1” (i.e., MSI-high, CIMP-positive, BRAF-mutated, KRAS-mutation negative); 2) “type 2” (i.e., MSS/MSI-low, CIMP-positive, BRAF-mutated, KRAS-mutation negative); 3) “type 3” (i.e., MSS/MSI-low, non-CIMP, BRAF-mutation negative, KRAS-mutated); 4) “type 4” (i.e., MSS/MSI-low, non-CIMP, BRAF-mutation negative, KRAS-mutation negative); and 5) “type 5” (i.e., MSI-high, non-CIMP, BRAF mutation-negative, KRAS-mutation negative). Other marker combinations were grouped together as an “other” category for tabulations. In sensitivity analyses, we explored changes to the type 3 subtype classification for comparison to previous reports,3 removing cases for whom all methylation markers had a PMR ratio <10 from this subgroup.
Of the N=2080 cases for whom tumor tissue was available, N=30 were excluded due to insufficient tissue or uninformative assays. Multiple imputation was used to approximate tumor marker status for cases with one (N=564), two (N=104), or three missing markers (N=38):37,38 the imputation model included variables for MSI, BRAF- and KRAS-mutation status, methylation status for the five genes used in classifying CIMP, stage, histology, sex, age at diagnosis, diagnosis year, body mass index (BMI), height, smoking history, use of non-steroidal anti-inflammatory drugs at diagnosis, history of endoscopic screening prior to diagnosis, education, race, first line of therapy, time from diagnosis to interview, censoring indicators, and analysis time. Iterative rounds of imputation (N=25) were performed using the mi command in STATA SE version 13.1 (College Station, Texas). Tumor subtype classifications were thus determined on the basis of assayed and, as necessary, imputed tumor markers. In addition to analyses utilizing these imputed data, we conducted sensitivity analyses using a complete-case approach, wherein only cases with complete tumor marker data were included.
Outcome information
Vital status, death date, and cause of death were determined through linkage to SEER and the National Death Index. CRC-specific deaths included those with an underlying cause attributed to ICD-10 codes C18.0-C20.0 or C26.0.39 Vital status linkage was performed periodically, with the most recent linkage capturing deaths occurring through December, 2012.
Statistical analysis
We used Cox proportional hazards regression to evaluate relative differences in survival after diagnosis by tumor subtype, using the type 4 subtype as the referent category. The time axis was defined as days since diagnosis, with left truncation to account for time between diagnosis and enrollment (mean=8.6 months). We conducted separate analyses for CRC-specific and overall survival. Participants alive at their last vital status assessment were censored at that date; in analyses of CRC-specific survival, persons who died due to causes other than CRC were censored at the time of death. Proportional hazards assumptions were assessed by testing for a non-zero slope of the scaled Schoenfeld residuals on ranked failure times.40
Regression models included adjustment terms for age (continuous and ten-year categories), sex, BMI (continuous), diagnosis year, and cigarette smoking history (never, former, current smoker). In secondary analyses, we further adjusted for stage via stratification of the baseline hazards. We also assessed potential confounding by several additional characteristics: tumor site, family history of CRC, race, education, history of endoscopy screening prior to diagnosis, non-steroidal anti-inflammatory drug use at the time of diagnosis, and receipt of chemotherapy as first course of treatment. However, these latter factors were not retained in our analytic models as adjustment for each variable had minimal impact on point estimates (i.e., <5% change). In sensitivity analyses, we also evaluated associations separately by sex and study phase [first (1998-2002), second (2002-2007)]. To account for multiple comparisons we used Hochberg’s step-up method to control for the family-wise error rate of 0.05 across each family of pairwise comparisons across subtypes (i.e., 5 tests per family).41
RESULTS
Among the N=2,080 cases with available tumor tissue, 99% (N=2,050) had information on at least one tumor marker and were included in the analysis; 65% (N=1,344) had complete data on all tumor markers. Approximately 16% of cases had tumors that were MSI-high, 13% had tumors that were BRAF-mutated, 31% had KRAS-mutated tumors, and 18% had CIMP-positive tumors. Among those with complete tumor marker data, 7% (N=100) were classified as having the type 1 subtype, 4% (N=55) had type 2 CRC, 26% (N=353) had type 3 CRC, 47% (N=631) had type 4 CRC, and 4% (N=50) were classified as having type 5 CRC; approximately 12% exhibited other tumor marker combinations (Supplementary Figure 1). Cases with types 1 or 2 CRC, particularly those with type 2 CRC, had the highest mean age at diagnosis and were most likely to be female (Table 1). Type 1, 2, and 5 tumors were rarely located outside the proximal colon (≤20%). Cases with type 2 CRC were least likely to have been diagnosed with stage I disease and had the lowest 5-year survival (46%). Cases with missing data on one to three tumor markers were younger at diagnosis and more likely to have stage IV CRC relative to other case groups.
Table 1.
Demographic and clinical characteristics by colorectal cancer (CRC) case group*
|
Type 1
MSI-high, BRAF- mutated, KRAS- mutation negative, CIMP-positive (N=100, 7%) |
Type 2
MSS/MSI-low, BRAF- mutated, KRAS- mutation negative, CIMP-positive (N=55, 4%) |
Type 3
MSS/MSI-low, BRAF-mutation negative, KRAS- mutated, non-CIMP (N=353, 26%) |
Type 4
MSS/MSI-low, BRAF- & KRAS- mutation negative, non-CIMP (N=631,47%) |
Type 5
MSI-high, BRAF- & KRAS-mutation negative, non- CIMP (N=50, 4%) |
Other
(N=155, 12%) |
Unknown†
(N=706) |
X2
p- value |
|
|---|---|---|---|---|---|---|---|---|
| Age at diagnosis | ||||||||
| Mean (SD) | 67.3 (5.3) | 63.6 (8.4) | 61.4 (9.0) | 60.1 (9.8) | 56.0 (12.2) | 60.7 (10.9) | 52.4 (12.1) | |
| <40 | 0 (0) | 1 (2) | 5 (1) | 22 (3) | 5 (10) | 8 (5) | 87 (12) | <0.01 |
| 40-49 | 1 (1) | 2 (4) | 37 (10) | 59 (9) | 10 (20) | 13 (8) | 292 (41) | |
| 50-59 | 7 (7) | 11 (20) | 93 (26) | 201 (32) | 15 (30) | 39 (25) | 102 (14) | |
| 60-69 | 51 (51) | 27 (49) | 140 (40) | 222 (35) | 10 (20) | 58 (37) | 137 (19) | |
| ≥70 | 41 (41) | 14 (25) | 78 (22) | 127 (20) | 10 (20) | 37 (24) | 88 (12) | |
| Sex | ||||||||
| Male | 17 (17) | 16 (29) | 149 (42) | 333 (53) | 21 (42) | 71 (46) | 318 (45) | <0.01 |
| Female | 83 (83) | 39 (71) | 204 (58) | 298 (47) | 29 (58) | 84 (54) | 388 (55) | |
| Race | ||||||||
| White | 95 (95) | 52 (95) | 318 (90) | 575 (91) | 43 (86) | 145 (94) | 471 (67) | 0.05 |
| African-American | 2 (2) | 3 (5) | 9 (3) | 27 (4) | 3 (6) | 1 (1) | 19 (3) | |
| Asian | 2 (2) | 0 (0) | 16 (5) | 11 (2) | 2 (4) | 3 (2) | 18 (3) | |
| >1 race | 1 (1) | 0 (0) | 6 (2) | 2 (0.3) | 0 (0) | 2 (1) | 9 (1) | |
| Other / Unknown | 0 (0) | 0 (0) | 4 (1) | 16 (3) | 2 (4) | 4 (3) | 189 (27) | |
| CRC family history | ||||||||
| No | 85 (85) | 47 (85) | 295 (84) | 538 (85) | 33 (66) | 128 (83) | 596 (84) | 0.02 |
| Yes | 15 (15) | 8 (15) | 58 (16) | 93 (15) | 17 (34) | 27 (17) | 110 (16) | |
| Stage at diagnosis | ||||||||
| I | 47 (47) | 11 (20) | 132 (38) | 281 (45) | 25 (50) | 60 (39) | 257 (37) | <0.01 |
| II-III | 52 (52) | 37 (67) | 174 (49) | 282 (45) | 24 (48) | 85 (55) | 338 (48) | |
| IV | 1 (1) | 7 (13) | 46 (13) | 66 (10) | 1 (2) | 10 (6) | 107 (15) | |
| Unknown | 0 | 0 | 1 | 2 | 0 | 0 | 4 | |
| 1st treatment course | ||||||||
| Received chemo | 44 (46) | 37 (71) | 203 (59) | 344 (67) | 24 (50) | 88 (59) | 435 (63) | 0.05 |
| No chemo | 52 (54) | 15 (29) | 142 (41) | 271 (33) | 24 (50) | 62 (41) | 256 (37) | |
| Unknown | 4 | 3 | 8 | 16 | 2 | 5 | 15 | |
| Tumor site | ||||||||
| Right colon: | 93 (93) | 43 (80) | 136 (39) | 132 (22) | 42 (84) | 111 (73) | 250 (36) | <0.01 |
| Cecum | 36 | 17 | 72 | 54 | 20 | 38 | 81 | |
| Ascending colon | 33 | 17 | 33 | 30 | 11 | 37 | 82 | |
| Hepatic flexure | 11 | 2 | 9 | 10 | 3 | 11 | 25 | |
| Transverse colon | 11 | 6 | 19 | 31 | 8 | 19 | 41 | |
| Splenic flexure | 2 | 1 | 3 | 13 | 0 | 6 | 21 | |
| Left colon: | 6 (6) | 9 (17) | 102 (29) | 218 (35) | 4 (8) | 18 (12) | 187 (27) | |
| Descending colon | 2 | 0 | 19 | 22 | 0 | 7 | 26 | |
| Sigmoid colon | 4 | 9 | 83 | 196 | 4 | 11 | 161 | |
| Rectal: | 1 (1) | 2 (4) | 112 (32) | 268 (43) | 4 (8) | 24 (16) | 258 (37) | |
| Rectosigmoid Junction | 1 | 1 | 28 | 67 | 2 | 8 | 61 | |
| Rectum | 0 | 1 | 84 | 201 | 2 | 16 | 197 | |
| Unknown | 0 | 0 | 3 | 7 | 0 | 2 | 2 | |
| MSI status | ||||||||
| MSS/MSI-L | 0 (0) | 55 (100) | 353 (100) | 631 (100) | 0 (0) | 84 (54) | 535 (86) | -- |
| MSI-H | 100 (100) | 0 (0) | 0 (0) | 0 (0) | 50 (100) | 71 (46) | 85 (14) | |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 | 86 | |
| BRAF-mutation status | ||||||||
| Wildtype | 0 (0) | 0 (0) | 353 (100) | 631 (100) | 50 (100) | 118 (76) | 533 (92) | -- |
| Mutated | 100 (100) | 55 (100) | 0 (0) | 0 (0) | 0 (0) | 37 (24) | 51 (8) | |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 | 102 | |
| KRAS-mutation status | ||||||||
| Wildtype | 100 (100) | 55 (100) | 0 (0) | 631 (100) | 50 (100) | 75 (48) | 396 (72) | -- |
| Mutated | 0 (0) | 0 (0) | 353 (100) | 0 (0) | 0 (0) | 80 (52) | 154 (28) | |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 | 156 | |
| CIMP status | ||||||||
| Non-CIMP | 0 (0) | 0 (0) | 353 (100) | 631 (100) | 50 (100) | 71 (46) | 127 (77) | -- |
| CIMP-positive | 100 (100) | 55 (100) | 0 (0) | 0 (0) | 0 (0) | 84 (54) | 37 (23) | |
| Missing | 0 | 0 | 0 | 0 | 0 | 0 | 542 | |
| 5-yr survival (%) | ||||||||
| Overall | 80.5 | 46.2 | 67.8 | 78.0 | 84.1 | 71.8 | 75.3 | |
| Disease-specific | 89.5 | 49.2 | 72.4 | 82.5 | 93.1 | 79.7 | 78.7 |
Cases missing data on all 4 markers used in subtype classification are excluded from all analyses (N=616).
Cases of “unknown” subtype have missing data on 1 to 3 markers used in subtype classification and are re-allocated to subtype groups through multiple imputation in analyses.
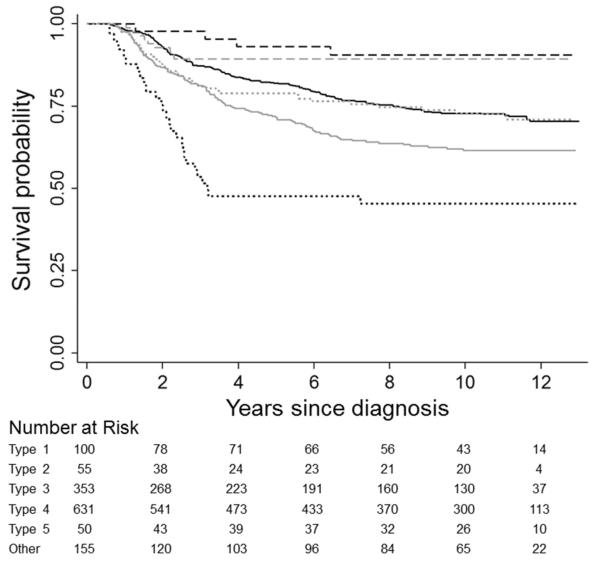
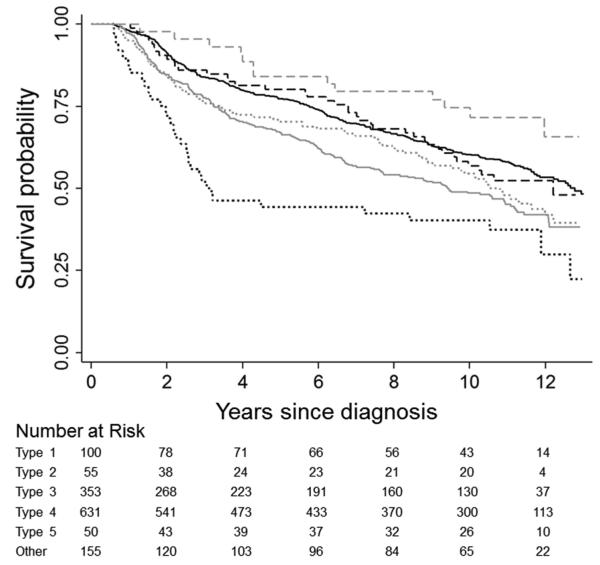
Kaplan-Meier curves illustrate unadjusted differences in CRC-specific (Figure 1) and overall survival (Figure 2) across subtypes. Observed patterns of survival differences were maintained in multivariable-adjusted analyses (Table 2). With respect to both outcomes, mortality rates were highest for type 2 CRC (HR=2.20, 95% CI: 1.47-3.31, and HR=1.55, 95% CI: 1.08-2.22 for CRC-specific and overall mortality, respectively) and lowest for type 5 CRC (HR=0.30, 95% CI: 0.14-0.66, and HR=0.61, 95% CI: 0.39-0.96); however, after accounting for multiple comparisons, associations with overall mortality were not statistically significant for these subgroups. CRC-specific survival was similarly favorable for both MSI-high subtypes (i.e., types 1 and 5). CRC-specific mortality was statistically significantly higher in the type 3 versus type 4 subgroup (HR=1.32, 95% CI: 1.07-1.63); a similar association was noted with respect to overall mortality.
Figure 1.
Kaplan-Meier survival curves comparing disease-specific survival in colorectal cancer patients by tumor subtype: type 1 (dashed black), type 2 (dotted black), type 3 (solid gray), type 4 (solid black), type 5 (dashed gray), some other tumor marker combination (dotted gray). Subtypes are defined as follows: type 1 = MSI-high, BRAF-mutated, KRAS-mutation negative, CIMP+; type 2 = MSS/MSI-low, BRAF-mutated, KRAS-mutation negative, CIMP+; type 3 = MSS/MSI-low, BRAF-mutation negative, KRAS-mutated, non-CIMP; type 4 = MSS/MSI-low, BRAF-mutation negative, KRAS-mutation negative, non-CIMP; type 5 = MSI-high, BRAF-mutation negative, KRAS-mutation negative, non-CIMP
Figure 2.
Kaplan-Meier survival curves comparing overall survival in colorectal cancer patients by tumor subtype: type 1 (dashed black), type 2 (dotted black), type 3 (solid gray), type 4 (solid black), type 5 (dashed gray), some other tumor marker combination (dotted gray). Subtypes are defined as follows: type 1 = MSI-high, BRAF-mutated, KRAS-mutation negative, CIMP+; type 2 = MSS/MSI-low, BRAF-mutated, KRAS-mutation negative, CIMP+; type 3 = MSS/MSI-low, BRAF-mutation negative, KRAS-mutated, non-CIMP; type 4 = MSS/MSI-low, BRAF-mutation negative, KRAS-mutation negative, non-CIMP; type 5 = MSI-high, BRAF-mutation negative, KRAS-mutation negative, non-CIMP
Table 2.
Colorectal cancer (CRC)-specific and overall mortality by tumor subtype
| Subtype* | Raw N Case Participants (col%)† |
Raw N CRC-Specific Deaths (col%)† |
CRC-Specific Mortality |
Raw N Total Deaths (col%)† |
Overall Mortality |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR‡ | (95% CI) | HR§ | (95% CI) | HR‡ | (95% CI) | HR§ | (95% CI) | |||||||
| Primary Analysis¶ | ||||||||||||||
| Type 1 | 100 | (5) | 9 | (2) | 0.41 | (0.22-0.75) | 0.54 | (0.29-0.99) | 42 | (5) | 0.88 | (0.65-1.20) | 1.05 | (0.78-1.44) |
| Type 2 | 55 | (3) | 26 | (4) | 2.20 | (1.47-3.31) | 1.84 | (1.21-2.78) | 32 | (3) | 1.55 | (1.08-2.22) | 1.40 | (0.98-2.01) |
| Type 3 | 353 | (26) | 112 | (30) | 1.32 | (1.07-1.63) | 1.25 | (1.01-1.54) | 173 | (28) | 1.26 | (1.07-1.49) | 1.23 | (1.04-1.46) |
| Type 4 | 631 | (49) | 154 | (52) | 1.0 | (ref) | 1.0 | (ref) | 263 | (49) | 1.0 | (ref) | 1.0 | (ref) |
| Type 5 | 50 | (5) | 4 | (1) | 0.30 | (0.14-0.66) | 0.42 | (0.19-0.93) | 14 | (3) | 0.61 | (0.39-0.96) | 0.74 | (0.47-1.17) |
| Other | 155 | (12) | 36 | (11) | 1.05 | (0.76-1.44) | 1.18 | (0.87-1.62) | 74 | (13) | 1.14 | (0.90-1.43) | 1.25 | (0.99-1.57) |
| Complete-Case Analyses** | ||||||||||||||
| Type 1 | 100 | (5) | 9 | (2) | 0.43 | (0.22-0.85) | 0.56 | (0.28-1.11) | 42 | (5) | 0.94 | (0.67-1.32) | 1.12 | (0.80-1.58) |
| Type 2 | 55 | (3) | 26 | (4) | 2.72 | (1.78-4.17) | 2.40 | (1.56-3.70) | 32 | (3) | 1.79 | (1.23-2.59) | 1.65 | (1.14-2.41) |
| Type 3 | 353 | (26) | 112 | (30) | 1.54 | (1.20-1.97) | 1.44 | (1.12-1.83) | 173 | (28) | 1.40 | (1.15-1.68) | 1.36 | (1.12-1.65) |
| Type 4 | 631 | (49) | 154 | (52) | 1.0 | (ref) | 1.0 | (ref) | 263 | (49) | 1.0 | (ref) | 1.0 | (ref) |
| Type 5 | 50 | (5) | 4 | (1) | 0.31 | (0.12-0.85) | 0.46 | (0.17-1.26) | 14 | (3) | 0.70 | (0.41-1.20) | 0.84 | (0.49-1.45) |
| Other | 155 | (12) | 36 | (11) | 1.02 | (0.71-1.47) | 1.27 | (0.87-1.84) | 74 | (13) | 1.21 | (0.94-1.57) | 1.38 | (1.06-1.79) |
Subtype classifications abbreviated as follows: Type 1 = MSI-high, BRAF-mutated, KRAS-mutation negative, CIMP-positive; Type 2 = MSS/MSI-low, BRAF-mutated, KRAS-mutation negative, CIMP-positive; Type 3 = MSS/MSI-low, BRAF-mutation negative, KRAS-mutated, non-CIMP; Type 4 = MSS/MSI-low, BRAF-mutation negative, KRAS-mutation negative, non-CIMP; Type 5 = MSI-high, BRAF-mutation negative, KRAS-mutation negative, non-CIMP
Case counts and numbers of deaths by subtype are based on observed, non-imputed data. Column percents reflect imputed distributions.
Adjusted for age at diagnosis, sex, BMI, diagnosis year, and smoking history.
Adjusted for stage at diagnosis is addition to age at diagnosis, sex, BMI, diagnosis year, and smoking history.
Multiple imputation-based analysis in which missing tumor marker data was inferred based on known variables and then used to derive tumor subtype.
Complete-case analysis. Cases missing data on any tumor markers are excluded.
Adjustment for stage at diagnosis had a modest impact on observed associations. Most point estimates were slightly attenuated with stage-adjustment, but patterns of survival differences across subtypes persisted (Table 2). Sensitivity analyses restricted to cases with complete tumor marker data showed more pronounced survival differences across subtypes (Table 2). In sensitivity analyses excluding cases with a PMR ratio <10 on all CIMP markers from the type 3 subgroup, the poor survival profile of this group persisted (CRC-specific survival HR=1.44, 95% CI: 1.04-1.98, overall survival HR=1.35, 95% CI: 1.04-1.74, not shown). In other sensitivity analyses, patterns of survival differences by subtype were similar across strata defined by sex and study phase; in particular, in all strata, the type 2 case group was associated with the poorest survival (not shown).
DISCUSSION
In this large population-based cohort of individuals with incident invasive CRC, we found important differences in survival across CRC subtypes defined on the basis of pre-specified combinations of MSI, CIMP, BRAF-mutation, and KRAS-mutation status. Patients with MSI-high subtypes of disease (i.e., types 1 and 5) had the most favorable survival, whereas those with type 2 CRC (MSS/MSI-low, CIMP-positive, BRAF-mutated, KRAS mutation negative) had the highest mortality. Observed survival differences were consistent with differences in the distribution of stage across subtypes and stage-adjustment did diminish the strength and statistical significance of most findings; however, patterns of differences in survival were maintained after stage-adjustment. These findings contribute to a small but growing literature supporting the significance of CRC-subtype classifications defined by combinations of these tumor markers.
The subtypes evaluated in the present analysis are based on classifications first proposed by Jass in 2007.1 Jass’ types 1 and 2 correspond to the type 1 and 2 subtypes evaluated here, respectively, and were originally proposed as reflecting a serrated morphology, with origins in serrated polyps. Jass’ type 3, similar to our type 3 subtype but restricted to CIMP-low tumors, was proposed as reflecting an alternate serrated pathway, with origins in KRAS-mutated adenomas, whereas Jass’ type 4 subtype, consistent with our type 4 subtype, was proposed to reflect CRC arising from the traditional adenoma-carcinoma sequence. Jass’ type 5 subtype, also consistent with our type 5 subtype classification, was suggested to be indicative of possible Lynch Syndrome as is reflected in the high prevalence of CRC family history in our type 5 case group.
To our knowledge, only one prior study has evaluated survival differences across CRC subtypes derived from the classifications proposed by Jass.3 Samadder et al. noted differences in age at diagnosis, tumor site, and grade across three CRC subtypes defined by combinations of MSI, CIMP, BRAF, and KRAS status in the Iowa Women’s Health Study (IWHS); however, no significant differences in subtype-specific survival were observed.3 Noted limitations of the IWHS include restricted demographics and sample size. Also, the tumor subtypes of greatest significance in the present analysis were not distinguished by Samadder et al.: the authors combined type 1 and 2 case groups into a single serrated subtype classified without regard to MSI, and did not evaluate the type 5 subtype as a distinct case group.3 Although we found type 1 and 2 CRC subtypes to be similar with the respect to their later age at diagnosis and proximal site distribution, we identified very different survival trajectories for these subtypes. This suggests that MSI status is a clinically-relevant marker of distinction in individuals with CRC suggestive of the serrated pathway. The observed favorable survival profile of the type 5 subtype further supports the need to distinguish MSI-high cases in CRC-subtype classification.
Most prior studies assessing the prognostic significance of MSI, CIMP, and BRAF- and KRAS-mutations in CRC have evaluated these markers individually.4-15 MSI status is most consistently associated with survival:15,42 in a recent meta-analysis, MSI-high CRC was associated with 40% better overall survival than MSS CRC (95% CI: 31-47%).15 The BRAF V600E mutation has also consistently been associated with poor survival relative to CRC that is not BRAF mutated.9-14,17-19 In contrast, studies of CRC survival in relation to CIMP18-20 and KRAS-mutation status4-8,10,11 have been inconsistent. Studies assessing associations between pairwise combinations of markers and CRC survival further support our findings of a complex interplay among these markers. In particular, previous studies have suggested that the prognostic significance of BRAF-mutation status is more pronounced in, if not restricted to, patients with MSS/MSI-low CRC.9,11,12,19,20,43 Other studies have reported higher mortality in MSS/CIMP-positive CRC relative to CRC with other MSI/CIMP combinations.44,45
The biologic basis for the observed differences in subtype-specific CRC survival remains an important topic for future research. Although the type 2 and 3 subtypes were diagnosed at an advanced stage, our finding that the higher mortality in these subtypes persisted after controlling for stage suggests that these are more inherently aggressive tumors and not simply tumors that were diagnosed late. Differences in response to available cancer therapies may also contribute to subtype-specific survival differences. Over the time period during which study participants were diagnosed with CRC, testing for the tumor markers in the present analysis was not clinically indicated for treatment decision-making. However, differential response to 5-fluorouracil-based chemotherapy by MSI46,47 and CIMP status48 has been reported, and differential response to newer anti-EGFR therapies (e.g., cetuximab) on the basis of KRAS and BRAF are well-documented.10,49,50 Thus, the relationship of these subtype classifications to CRC treatment response merits further investigation.
The results of the present investigation should be interpreted in the context of study limitations. Information on the clinical management of CRC patients included in the analysis was limited; however, as described above, treatments were unlikely to differ across the evaluated CRC subtypes over the study period beyond any differences due to stage at diagnosis, diagnosis year, and tumor site. Tumor marker data were missing for a substantial proportion of cases. Participants for whom no tumor marker data was available were excluded from the analysis and were, on average, younger at diagnosis (mean age=53 versus 58 in included cases), more likely to be non-white (39% versus 17%), had lower five-year overall survival (63% versus 74%), and later stage at diagnosis (21% versus 11% distant stage). The prevalence of stage IV disease was lower in our study population than is reflected in SEER estimates for the study area,51 further suggesting an exclusion of late-stage disease. Thus, it is plausible that the distribution of tumor subtypes among excluded cases differed from that among included cases. We used multiple imputation to account for missing tumor marker data in cases with information on one to three tumor markers (N=706). Simulation studies comparing multiple imputation to complete-case analyses suggest that excluding observations with missing data can lead to considerable bias in regression coefficients and that such bias can be reduced via multiple imputation.37,38 The fact that there were only modest differences in point estimates from multiple imputation versus complete-case analyses reflects the robustness of our conclusions to various analytic approaches. When tumor marker data were available, those data were based on single assays for each marker and, thus, do not capture information on intra-tumoral heterogeneity. Lastly, the tumor markers evaluated in the present analysis represent only a subset of those that might be relevant to CRC survival and subtype classification. It is likely that some etiologic and clinical heterogeneity remains within each of the evaluated CRC subtypes. Characterization of additional somatic mutations (e.g., in KRAS codon 61), gene amplifications (e.g., in EGFR), methylation sites (e.g., in CDKN2A), and other molecular alterations was beyond the scope of the present analysis, but could facilitate more refined and detailed classification of homogeneous CRC subtypes.
Important strengths of the present study include a long follow-up and large study population, which allowed for the evaluation of survival outcomes in less common CRC subtypes. The two smallest subtypes evaluated in the present analysis (i.e., types 2 and 5) demonstrated the most pronounced differences in survival. Further evaluation of these important CRC subtypes will require larger sample size.
Here we extend previous reports regarding the relevance of CRC subtypes defined jointly by MSI, CIMP, and BRAF- and KRAS-mutation status. Our findings suggest that the biologic distinctions between these subtypes translate to important differences in survival and highlight a poorer survival for CRC demonstrating the type 2, serrated-like phenotype. These results support the value of considering these four markers in combination, in addition to their individual value as predictive and prognostic markers for CRC.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by the National Cancer Institute at the National Institutes of Health (K07CA172298 to A.I.P., U01CA74794, R01CA076366, R01CA118699, R01CA107333, K05CA142885 to P.A.N.), the National Center for Advancing Translational Science, National Institutes of Health (KL2TR000421 to A.N.B.H.), and through cooperative agreements with members of the Colon Cancer Family Registry and Principal Investigators. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The sponsors had no role in the design and conduct of the study; no role in the collection, management, analysis, and interpretation of data; no role in preparation, review, or approval of the manuscript; and no role in the decision to submit the manuscript for publication.
Abbreviations
- CRC
colorectal cancer
- MSI
microsatellite instability
- MSS
microsatellite stable
- CIMP
CpG island methylator phenotype
- HR
hazard ratio
- CI
confidence interval
- SCCFR
Seattle Colon Cancer Family Registry
- SEER
Surveillance, Epidemiology, and End Results
- PMR
percentage of methylated reference
- BMI
body mass index
- IWHS
Iowa Women’s Health Study
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Outside the present work, Dr. P. Limburg reported grant support received or pending from Olympus America, BENEO-Orafti Group, Bayer Healthcare, Fujinon, Inc., Boston Scientific, Astra Zeneca, and Ironwood Pharmaceuticals; he also reported having received payment for lectures from the American School of Oncology and Imedex, and royalties and stock from Exact Sciences. Dr. D. Weisenberger reported serving as a consultant to Zymo Research and receiving payment for patents and royalties from Epigenomics. Dr. P. Laird also reported receiving royalties from Epigenomics, as well as receiving payment for lectures from the American Society for Clinical Oncology and for travel from the American Association for Cancer Research.
REFERENCES
*Author names in bold designate shared co-first authorship.
- 1.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–30. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 2.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–100. doi: 10.1053/j.gastro.2009.12.066. [DOI] [PubMed] [Google Scholar]
- 3.Samadder NJ, Vierkant RA, Tillmans LS, et al. Associations Between Colorectal Cancer Molecular Markers and Pathways With Clinicopathologic Features in Older Women. Gastroenterology. 2013;145:348–56. doi: 10.1053/j.gastro.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samowitz WS, Curtin K, Schaffer D, et al. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: A population-based study. Cancer Epidemiology Biomarkers & Prevention. 2000;9:1193–1197. [PubMed] [Google Scholar]
- 5.Andreyev HJ, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. Br J Cancer. 2001;85:692–6. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogino S, Meyerhardt JA, Irahara N, et al. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res. 2009;15:7322–9. doi: 10.1158/1078-0432.CCR-09-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M, Qian ZR, Nishihara R, Meyerhardt JA, Haigis KM, Fuchs CM, Ogino S. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF-wild-type colorectal cancers. Clin Cancer Res. 2012;18:4753–63. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phipps AI, Buchanan DD, Makar KW, et al. KRAS-mutation status in relation to colorectal cancer survival: the joint impact of correlated tumour markers. Br J Cancer. 2013;108:1757–64. doi: 10.1038/bjc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phipps AI, Buchanan DD, Makar KW, et al. BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2012;21:1792–8. doi: 10.1158/1055-9965.EPI-12-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 11.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–74. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 12.Farina-Sarasqueta A, van Lijnschoten G, Moerland E, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 13.Kalady MF, Dejulius KL, Sanchez JA, et al. BRAF mutations in colorectal cancer are associated with distinct clinical characteristics and worse prognosis. Dis Colon Rectum. 2012;55:128–33. doi: 10.1097/DCR.0b013e31823c08b3. [DOI] [PubMed] [Google Scholar]
- 14.Ogino S, Shima K, Meyerhardt JA, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18:890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guastadisegni C, Colafranceschi M, Ottini L, et al. Microsatellite instability as a marker of prognosis and response to therapy: A meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46:2788–98. doi: 10.1016/j.ejca.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Wang C, van Rijnsoever M, Grieu F, et al. Prognostic significance of microsatellite instability and Ki-ras mutation type in stage II colorectal cancer. Oncology. 2003;64:259–65. doi: 10.1159/000069311. [DOI] [PubMed] [Google Scholar]
- 17.French AJ, Sargent DJ, Burgart LJ, et al. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008;14:3408–15. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–9. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 19.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–6. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S, Cho NY, Choi M, et al. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol Int. 2008;58:104–13. doi: 10.1111/j.1440-1827.2007.02197.x. [DOI] [PubMed] [Google Scholar]
- 21.Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–70. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 22.Nash GM, Gimbel M, Cohen AM, et al. KRAS mutation and microsatellite instability: two genetic markers of early tumor development that influence the prognosis of colorectal cancer. Ann Surg Oncol. 2010;17:416–24. doi: 10.1245/s10434-009-0713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zlobec I, Bihl MP, Foerster A, et al. Stratification and prognostic relevance of Jass's molecular classification of colorectal cancer. Front Oncol. 2012;2:7. doi: 10.3389/fonc.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–43. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 25.Newcomb PA, Zheng Y, Chia VM, et al. Estrogen plus progestin use, microsatellite instability, and the risk of colorectal cancer in women. Cancer Res. 2007;67:7534–9. doi: 10.1158/0008-5472.CAN-06-4275. [DOI] [PubMed] [Google Scholar]
- 26.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 27.Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 28.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchanan DD, Sweet K, Drini M, et al. Risk factors for colorectal cancer in patients with multiple serrated polyps: a cross-sectional case series from genetics clinics. PLoS One. 2010;5:e11636. doi: 10.1371/journal.pone.0011636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 31.Alsop K, Mead L, Smith LD, et al. Low somatic K-ras mutation frequency in colorectal cancer diagnosed under the age of 45 years. Eur J Cancer. 2006;42:1357–61. doi: 10.1016/j.ejca.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 32.The Cancer Genome Atlas cBioPortal for Cancer Genomics. 2013 http://www.cbioportal.org/public-portal/index.do. Computational Biology Center, Memorial Sloan-Kettering Cancer Center.
- 33.Imamura Y, Lochhead P, Yamauchi M, Kuchiba A, Qian ZR, Liao X, Nishihara R, Jung S, Wu K, Nosho K, Wang YE, Peng S, Bass AJ, Haigis KM, Meyerhardt J, Chan AT, Fuchs CS, Ogino S. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 35.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–6. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–82. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Heijden GJMG, Donders ART, Stijnen T, et al. Imputation of missing values is superior to complete case analysis and the missing-indicator method in multivariable diagnostic research: A clinical example. J Clin Epidemiol. 2006;59:1102–9. doi: 10.1016/j.jclinepi.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 38.Azur MJ, Stuart EA, Frangakis C, et al. Multiple imputation by chained equations: what is it and how does it work? Int J Methods Psychiatr Res. 2011;20:40–9. doi: 10.1002/mpr.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.World Health Organization . International Classification of Diseases. WHO; Geneva: 2007. [Google Scholar]
- 40.Therneau TM, Grambsch PM. Modeling survival data: extending the Cox model. Springer; New York: 2000. [Google Scholar]
- 41.Farcomeni A. A review of modern multiple hypothesis testing, with particular attention to the false discovery proportion. Stat Methods Med Res. 2008;17:347–88. doi: 10.1177/0962280206079046. [DOI] [PubMed] [Google Scholar]
- 42.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–18. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 43.Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite Instability and BRAF Mutation Testing in Colorectal Cancer Prognostication. J Natl Cancer Inst. 2013;105:1151–1156. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward RL, Cheong K, Ku SL, et al. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003;21:3729–36. doi: 10.1200/JCO.2003.03.123. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez JA, Krumroy L, Plummer S, et al. Genetic and epigenetic classifications define clinical phenotypes and determine patient outcomes in colorectal cancer. Br J Surg. 2009;96:1196–204. doi: 10.1002/bjs.6683. [DOI] [PubMed] [Google Scholar]
- 46.Bertagnolli MM, Niedzwiecki D, Compton CC, et al. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J Clin Oncol. 2009;27:1814–21. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–26. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jover R, Nguyen TP, Perez-Carbonell L, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140:1174–81. doi: 10.1053/j.gastro.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin AY, Buckley NS, Lu AT, et al. Effect of KRAS mutational status in advanced colorectal cancer on the outcomes of anti-epidermal growth factor receptor monoclonal antibody therapy: a systematic review and meta-analysis. Clin Colorectal Cancer. 2011;10:63–9. doi: 10.3816/CCC.2011.n.009. [DOI] [PubMed] [Google Scholar]
- 50.Bokemeyer C, van Cutsem E, Rougier P, et al. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: Pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer. 2012;48:1466–75. doi: 10.1016/j.ejca.2012.02.057. [DOI] [PubMed] [Google Scholar]
- 51. Surveillance Epidemiology and End Results (SEER) Program ( www.seer.cancer.gov) SEER*Stat Database. Incidence - SEER 13 Regs Research Data, Nov 2011 Sub, Vintage 2009 Pops (1992-2009) <Katrina/Rita Population Adjustment> - Linked To County Attributes - Total U.S., 1969-2010 Counties. In: National Cancer Institute D, Surveillance Research Program, Cancer Statistics Branch, ed, released April 2012, based on the November 2011 submission.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.