Chromatin compaction mediated by CHD3.1 must be counteracted by ACF1–SNF2H and RNF20 in order to allow DNA double-strand break repair in heterochromatin of postreplicative cells.
Abstract
Heterochromatin is a barrier to DNA repair that correlates strongly with elevated somatic mutation in cancer. CHD class II nucleosome remodeling activity (specifically CHD3.1) retained by KAP-1 increases heterochromatin compaction and impedes DNA double-strand break (DSB) repair requiring Artemis. This obstruction is alleviated by chromatin relaxation via ATM-dependent KAP-1S824 phosphorylation (pKAP-1) and CHD3.1 dispersal from heterochromatic DSBs; however, how heterochromatin compaction is actually adjusted after CHD3.1 dispersal is unknown. In this paper, we demonstrate that Artemis-dependent DSB repair in heterochromatin requires ISWI (imitation switch)-class ACF1–SNF2H nucleosome remodeling. Compacted chromatin generated by CHD3.1 after DNA replication necessitates ACF1–SNF2H–mediated relaxation for DSB repair. ACF1–SNF2H requires RNF20 to bind heterochromatic DSBs, underlies RNF20-mediated chromatin relaxation, and functions downstream of pKAP-1–mediated CHD3.1 dispersal to enable DSB repair. CHD3.1 and ACF1–SNF2H display counteractive activities but similar histone affinities (via the plant homeodomains of CHD3.1 and ACF1), which we suggest necessitates a two-step dispersal and recruitment system regulating these opposing chromatin remodeling activities during DSB repair.
Introduction
Heterochromatin, the transcriptionally silent or inert component of the eukaryotic genome, represents a challenging environment for DNA double-strand break (DSB) response pathways (Goodarzi and Jeggo, 2012a, 2013; Soria et al., 2012; Price and D’Andrea, 2013) and correlates with increased somatic mutation in cancer (Schuster-Böckler and Lehner, 2012). During heterochromatic DSB repair, alterations, including nucleosome respacing, are necessary before DNA religation can take place by either nonhomologous end joining (NHEJ) or homologous recombination (HR; Goodarzi and Jeggo, 2012a). Essential is the ataxia telangiectasia mutated (ATM) protein kinase, which phosphorylates KAP-1, a key component of the heterochromatic superstructure (Ziv et al., 2006; Goodarzi et al., 2008, 2009). KAP-1 binds to sequence-specific KRAB (Krüppel-associated box)-containing repressors and recruits heterochromatin-promoting activities including ATP-dependent nucleosome remodeling enzymes, such as CHD3, a class II CHD (chromodomain helicase, DNA-binding protein) family enzyme that is part of the nucleosome remodeling and deacetylase complex (Stanley et al., 2013). KAP-1 requires SUMOylation to interact with CHD3 isoform 1 (CHD3.1), which possesses a small ubiquitin-like modifier (SUMO)–interacting motif (Schultz et al., 2001, 2002). DSB-induced KAP-1 S824 phosphorylation (pKAP-1) by ATM perturbs interactions between SUMOylated KAP-1 and the CHD3.1 SUMO-interacting motif, triggering CHD3.1 dispersal away from DSB sites, localized chromatin relaxation, and DSB repair (Goodarzi et al., 2011). This additionally requires H2AXS139p (γ-H2AX), MDC1, RNF8, RNF168, and 53BP1 to form ionizing radiation (IR)–induced foci (IRIF), which concentrate sufficient ATM activity to maintain densely localized pKAP-1 at heterochromatic DSB sites to counter constitutive pKAP-1 dephosphorylation by protein phosphatase 4 (Noon et al., 2010; Lee et al., 2012). Heterochromatic DSB repair also requires the Artemis nuclease, which has an unknown DSB-processing function downstream of chromatin relaxation (Riballo et al., 2004; Woodbine et al., 2011).
Mechanistically, how nucleosome compaction is altered after CHD3.1 dispersal is not known. CHD-class enzymes adjust linker DNA length between nucleosomes, increasing per capita histone occupancy and disfavoring DNA sequence-positioned nucleosome deposition (Moshkin et al., 2012; Stanley et al., 2013). One explanation for CHD3.1 dispersal before DSB-induced chromatin relaxation is that CHD-class activity counters another process attempting to adjust nucleosome spacing. Of the major chromatin remodeling classes, only ISWI (imitation switch)-class activity counters CHD-class enzymes directly to reduce nucleosome occupancy and favor sequence-directed nucleosome positioning (Moshkin et al., 2012; Stanley et al., 2013). A study of purified Drosophila melanogaster ISWI and CHD homologues demonstrates that although both activities “mobilize” nucleosomes, they do so in an opposing manner with each reversing the “products” of the other (Brehm et al., 2000). This suggested to us that ISWI activity might underlie heterochromatin relaxation after CHD dispersal.
Several ISWI-class complexes are implicated in DSB signaling or repair, most sharing SNF2H (also called SMARCA5) as a catalytic subunit (Xiao et al., 2009; Lan et al., 2010). In complex with Williams syndrome transcription factor (WSTF), SNF2H promotes H2AXY142p, a modification assisting γ-H2AX maintenance but not induction (Xiao et al., 2009). In complex with ACF1, SNF2H is recruited transiently to microirradiation-induced DSB tracks and improves NHEJ and HR in reporter-based assays (Lan et al., 2010; Smeenk et al., 2013). SNF2H is implicated in DSB repair by regulating BRCA1 and/or RAD51 retention, via PARP1-dependent recruitment (Smeenk et al., 2013), SIRT6-dependent processes (Toiber et al., 2013), and SUPT16H-dependent processes (Oliveira et al., 2014). Studies have suggested that the role of ACF1–SNF2H is regulated by the RNF20-RNF40 ubiquitin ligase, although their functional relationship is unclear (Moyal et al., 2011; Nakamura et al., 2011; Oliveira et al., 2014). Notably, ACF1–SNF2H activity alleviates barriers posed by heterochromatin to enable DNA replication (Collins et al., 2002). This, together with its dynamic response to DSBs, made SNF2H a strong candidate for the ISWI-class activity that we postulated counters CHD3.1 during DSB-induced heterochromatic relaxation. Here, we unveil that, selectively in nondividing cells, ACF1–SNF2H (ISWI class) chromatin remodeling respaces heterochromatic nucleosomes after CHD3.1 dispersal to enable Artemis-dependent repair of IR-induced DSBs. Our data suggest a concept wherein nucleosome arrangements incompatible with DSB repair are temporarily “reversed” via not a one but a two-step process of dispersing and recruiting chromatin remodelers with counteractive activity.
Results
SNF2H is required for ATM- and Artemis-dependent heterochromatic DSB repair
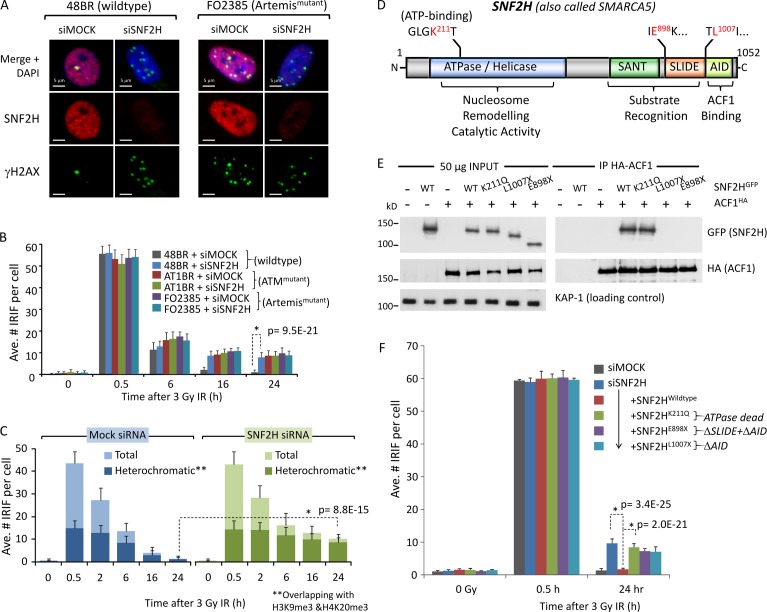
To address whether SNF2H plays a role in heterochromatic DSB repair via NHEJ, we first determined whether SNF2H depletion impacted DSB repair requiring ATM, RNF168, or Artemis, as loss of any of these factors results in heterochromatic DSB repair defects (Riballo et al., 2004; Goodarzi et al., 2008; Noon et al., 2010; Woodbine et al., 2011). We used IRIF enumeration in primary human fibroblasts to monitor repair (the advantages of which are for review in Goodarzi and Jeggo [2012b] and Löbrich et al. [2010]). Using G0-arrested cells also enables the interrogation of SNF2H specifically within the context of NHEJ. 1BR3 (wild type), 48BR (wild type), AT1BR (ATM mutated), RIDDLE (RNF168 mutated), CJ179 (Artemis mutated), and F02385 (Artemis mutated) patient-derived primary fibroblasts were transfected with scrambled (mock) or SNF2H siRNA (pool of two distinct sequences), irradiated, harvested, and stained for γ-H2AX. After validation of SNF2H knockdown by immunoblotting and assessing by immunofluorescence (IF), IRIF were enumerated only in cells with confirmed loss of >75% SNF2H expression relative to scrambled siRNA controls (Fig. 1, A and B; and Fig. S1, A–E); results were reproducible with individual siRNAs (Fig. S1 F). ATM, RNF168, or Artemis mutated cells displayed (expected) defects, such that ∼15% of lesions persisted at 24 h after IR (Fig. 1, A and B; and Fig. S1, D and E). SNF2H depletion produced a comparable defect and was not additive with defects in ATM, RNF168, or Artemis mutated cells, indicative of SNF2H functioning epistatically with these proteins in heterochromatic DSB repair (Riballo et al., 2004; Goodarzi et al., 2008; Noon et al., 2010).
Figure 1.
SNF2H chromatin remodeling activity is required for ATM- and Artemis-dependent DSB repair in heterochromatin. (A) SNF2H was depleted by siRNA (mock = scrambled siRNA) in wild-type (48BR) or Artemis mutant (FO2385) quiescent primary human fibroblasts. Cells were irradiated with 3 Gy IR and immunostained 24 h later for SNF2H (red), γ-H2AX (green), and DAPI (blue). Bars, 5 µm. (B) The mean number of γ-H2AX per nucleus from cells prepared as in A and harvested at 0.5, 6, 16, and 24 h after 3 Gy IR was enumerated for three independent experiments. (C) Quiescent 48BR cells were treated with scrambled (mock) or SNF2H siRNA, irradiated, and harvested as indicated. Cells were then stained with γ-H2AX (green) and H3K9me3 + H4K20me3 (red) and imaged by confocal microscopy. Regions of green- and red-positive signal were identified by software, isolating overlap of γ-H2AX foci in an H3K9me3 + H4K20me3–positive (heterochromatic) region. The heterochromatic foci were enumerated relative to total number. (D) A schematic diagram of human SNF2H. N, N terminus; C, C terminus. (E) The indicated GFP-tagged SNF2H or HA-tagged ACF1 constructs were transfected into 1BRhTERT cells; 24 h later, whole cell extracts were prepared and incubated with anti–HA-agarose for 4 h at 4°C. Washed immunoprecipitates and 50 µg of input lysate were then immunoblotted for the indicated proteins. (F) Confluent 1BRhTERT cells were transfected with SNF2H-B siRNA and, 48 h later, then split to 75% confluency and transfected with the indicated SNF2HGFP constructs. 16 h later, cells were irradiated as indicated, harvested, and immunostained for γ-H2AX and either SNF2H or FLAG. The mean number of γ-H2AX per nucleus was scored in cells with confirmed knockdown or construct expression for three independent experiments. P-values (standard two-tailed Student’s t test) are indicated for significance of relevant data points. Error bars show SD. Ave., average.
To substantiate whether SNF2H depletion results in a specifically heterochromatic DSB repair defect, we repeated the aforementioned experiment and coimmunostained for γ-H2AX with both H3K9me3 and H4K20me3, markers of heterochromatin used previously in NIH3T3 cells to monitor DSB repair (Figs. 1 C and S1 G; Goodarzi et al., 2008; Noon et al., 2010; Brunton et al., 2011). Using confocal microscopy and computer-assisted analysis, we refined this method for primary human fibroblasts (Fig. S1 G). Total IRIF and those overlapping with H3K9me3- and H4K20me3-positive regions (considered heterochromatic DSBs) were enumerated in irradiated cells ± mock/SNF2H siRNA. The majority of persisting IRIF in SNF2H-depleted cells overlapped with H3K9me3- and H4K20me3-positive regions, consistent with a role for SNF2H in heterochromatic DSB repair (Fig. 1 C).
Artemis-dependent DSB repair requires SNF2H catalytic activity, ACF1, and RNF20
SNF2H has an ATPase/helicase domain required for nucleosome remodeling, a substrate recognition module comprised of SANT (Swi3, Ada2, N-Cor, and TFIIIB) and SLIDE (SANT-like ISWI) domains and a C-terminal ACF1-interacting domain (AID; Fig. 1 D). We generated siRNA-resistant human SNF2H cDNA constructs (Fig. S2, B, E, and F) and analyzed DSB repair in SNF2H-depleted cells reexpressing SNF2H. Immortalized cells were used because plasmid expression in slowly dividing primary cells was prohibitive. Cells were treated with SNF2H siRNA, transfected with siRNA-resistant, GFP-tagged SNF2H cDNAs, irradiated, harvested, and immunostained for IRIF enumeration (Fig. S2 F). Whereas wild-type SNF2H restored normal DSB repair kinetics in SNF2H-depleted cells (confirming siRNA specificity), catalytically inactive SNF2H (ATP binding site mutant K211Q) was unable to do so (Fig. 1, E and F; and Fig. S2 F), indicating that catalytic function is important for DSB repair. Truncation mutants lacking the AID domain (L1007X) or SLIDE and AID domains (E898X) also failed to restore normal DSB repair, suggesting the importance of interactions with ACF1 (Fig. 1, E and F).
To explore a role for ACF1 in Artemis-dependent DSB repair, both ACF1 isoforms were depleted by siRNA from wild-type or Artemis mutated primary fibroblasts. ACF1 depletion resulted in a comparable and nonadditive DSB repair defect to that of Artemis mutation and/or SNF2H depletion, suggesting that ACF1 functions within the same repair process (Figs. 2 A and S1, C and F). ACF1 isoform 1 (ACF1.1) is comprised of a DNA-binding WAC (WSTF, ACF1, and CBP146) domain, central DTT-BAZ (bromodomain adjacent to zinc finger) module (required to bind SNF2H), and a tandem plant homeodomain (PHD)–bromodomain (required for chromatin binding), whereas ACF1 isoform 2 (ACF1.2) lacks exon 12 but is otherwise identical (Fig. 2 B). Notably, ACF1.2 has never been characterized functionally in DNA repair. GFP-tagged, siRNA-resistant ACF1.1 and ACF1.2 immunoprecipitated equally well with SNF2H and restored normal DSB repair after endogenous ACF1 depletion (Fig. 2, C and D; and Fig. S2, C, E, and F). In contrast, ACF1.1ΔBAZ mutants were unable to interact with SNF2H by immunoprecipitation (IP) and failed to restore normal DSB repair, further highlighting the importance of the ACF1–SNF2H interaction in DSB repair (Fig. 2, C and D; and Fig. 1, E and F).
Figure 2.
ACF1 and RNF20 enable SNF2H and Artemis-dependent DSB repair. (A) ACF1 was depleted by siRNA (mock = scrambled siRNA) in wild-type (48BR) or Artemis mutant (FO2385) quiescent primary human fibroblasts before being treated, immunostained, and analyzed as in Fig. 1 A. (B) Schematic diagram of human ACF1. (C) The indicated FLAG-tagged SNF2H or GFP-tagged ACF1 constructs were transfected into 1BRhTERT cells; 24 h later, whole cell extracts were prepared and incubated with anti–FLAG-Sepharose for 3 h at 4°C. Washed GFP immunoprecipitates and 50 µg of input lysate were then immunoblotted for the indicated proteins. (D) HeLa cells were transfected with ACF1-B siRNA and, 48 h later, then split to 75% confluency and transfected with the indicated ACF1GFP constructs. 16 h later, cells were irradiated as indicated, harvested, and immunostained for γ-H2AX; the mean number of γ-H2AX per nucleus was scored in cells with confirmed construct expression for three independent experiments. (E) RNF20 was depleted by siRNA (mock = scrambled siRNA) in wild-type (48BR) or Artemis mutant (FO2385) quiescent primary human fibroblasts before being treated, immunostained, and analyzed as in Fig. 1 A. (F) Schematic diagram of human RNF20. BromoD, bromodomain; CC, coiled coil. (G) The indicated HA-tagged RNF20 constructs were transfected into 1BRhTERT cells alongside wild-type Myc-tagged RNF40; 24 h later, whole cell extracts were prepared and immunoblotted for the indicated proteins. (H) HeLa cells were transfected with RNF20-A siRNA and, 48 h later, then split to 75% confluency and transfected with the indicated RNF20HA constructs alongside wild-type RNF40Myc. 16 h later, cells were irradiated as indicated, harvested, and immunostained for γ-H2AX and either RNF20 or HA; the mean number of γ-H2AX per nucleus was scored in cells with confirmed knockdown or construct expression for three independent experiments. P-values (standard two-tailed Student’s t test) are indicated. Error bars show SD. Ave., average.
The role of ACF1 in DSB repair requires the H2BK120 ubiquitin-ligase activity of the RNF20-RNF40 heterodimer (Moyal et al., 2011; Nakamura et al., 2011). We next tested whether RNF20-RNF40 impacted Artemis-dependent heterochromatic DSB repair. RNF20 depletion by siRNA induced a comparable and nonadditive repair defect to Artemis mutation or SNF2H–ACF1 depletion, suggesting that RNF20 also functions within the same repair process (Figs. 2 E and S1, A, B, and F). Expression of siRNA-resistant, HA-tagged RNF20 restored normal DSB repair kinetics after endogenous RNF20 depletion (Fig. 2, F–H; and Fig. S2, D and E). RNF20S172A+S553A phosphomutant failed to complement the RNF20 siRNA-induced DSB repair defect, suggesting that ATM-mediated RNF20 phosphorylation (described previously in Moyal et al., 2011) is part of the mechanism of action.
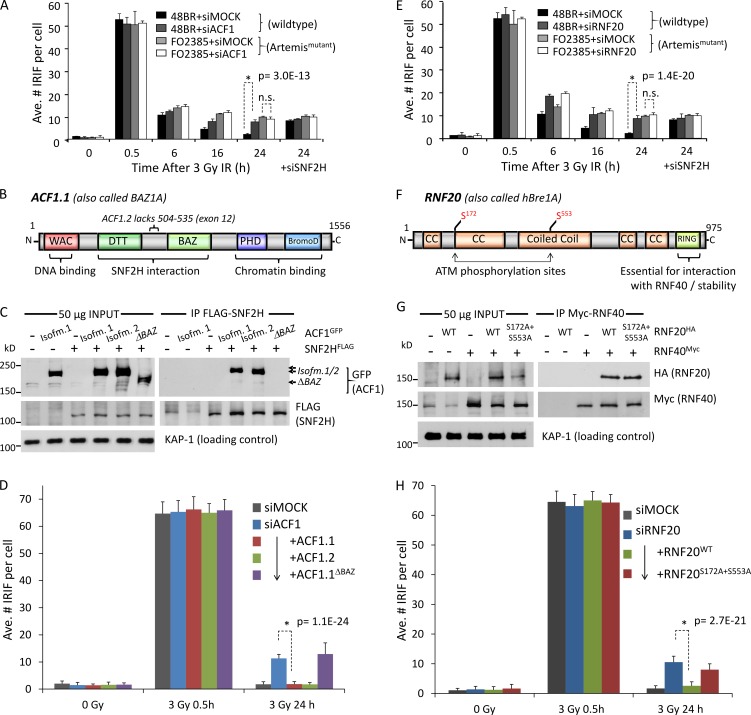
ACF1–SNF2H functions independently of pKAP-1 and CHD3.1 dispersal
Previous work defined the key events in heterochromatic DSB repair as ATM activation, γ-H2AX/MDC1/RNF8/RNF168/53BP1 foci formation, pKAP-1–mediated CHD3.1 dispersal, chromatin relaxation, and Artemis-dependent NHEJ (or HR in G2 phase; Ziv et al., 2006; Goodarzi et al., 2008, 2011; Beucher et al., 2009; Noon et al., 2010; Woodbine et al., 2011). No substantial impact of SNF2H, ACF1, or RNF20 depletion on ATMS1981p, γ-H2AX/53BP1 formation, pKAP-1 induction, or pKAP-1 foci formation over a range of IR doses or times was observed (Fig. 3, A and B; and Fig. S1 H). In the case of SNF2H only, a reduction in γ-H2AX signal magnitude but not IRIF number was seen in >90% SNF2H depletion, at a level long past that needed to elicit a DSB repair defect (>60% SNF2H depletion; Fig. S1 H). No impact on 53BP1 foci formation was observed after SNF2H loss, even at late times (Fig. S2, A and F), which fits with what has been documented previously (Smeenk et al., 2013). The phenotype of SNF2H, ACF1, or RNF20 depletion contrasted loss of ATM (in which no pKAP-1 forms; Goodarzi et al., 2008) or RNF8 depletion (in which pKAP-1 occurs but fails to form focally; Noon et al., 2010) and mirrored the effects of Artemis mutation (Fig. 3, A and B; Woodbine et al., 2011). Using a detergent extraction method described previously (Goodarzi et al., 2011), ATM-dependent CHD3.1 dispersal after IR was also found not to be impacted by SNF2H, ACF1, or RNF20 depletion (Fig. 3 C). These data suggest that RNF20, ACF1, and SNF2H function downstream of pKAP-1–mediated CHD3.1 dispersal.
Figure 3.
SNF2H-ACF1 and RNF20 enable DSB-induced chromatin relaxation downstream of KAP-1 phosphorylation and CHD3.1 dispersal. (A) SNF2H was depleted by siRNA (mock = scrambled siRNA) in wild-type 48BR cells and irradiated with 0, 1, 2, 4, and 8 Gy IR before being harvested 0.5 h later and immunoblotted for the indicated proteins. (B) Wild-type (48BR), ATM mutant (AT1BR), or Artemis mutant (FO2385) quiescent primary human fibroblasts were treated with SNF2H, ACF1, RNF20, or RNF8 siRNA (as indicated) before being irradiated and harvested as indicated. Cells were immunostained for pKAP-1 (red) and γ-H2AX (green). Bars, 5 µm. (C) 48BR cells were treated with siRNA as in B and were irradiated with 0, 1, 4, and 16 Gy IR ± DMSO or 10 µM Ku55933 ATM inhibitor (ATMi). Cells were incubated with 0.1% (vol/vol) Triton X-100 in PBS for 30 s before being fixed and immunostained for CHD3.1 (red) or γ-H2AX (green). Nuclear CHD3.1 signal was quantified as in Goodarzi et al. (2011); data from three independent experiments were plotted together. Solid bars indicate means with SD. A.U., arbitrary unit. (D) HeLa cells were treated with siRNA as indicated. 48 h later, cells were treated with 200 ng/ml neocarzinostatin (NCS) ± ATM inhibitor (as indicated). 0.5 after NCS treatment, nuclei were purified and treated with MNase, and DNA were isolated as described in Goodarzi et al. (2011). 2.5 µg DNA was resolved by 1.2% agarose gel and visualized by ethidium bromide staining. (E) The quantification of signal in each lane of the gel in D. Data are expressed as the percentages of the total signal (for a given lane) across the distance from the well to end of the gel. Calibrated kilobase pair sizes are indicated. The experimental dataset shown in this figure is representative of four independent repeat experiments, all showing the same result/trends.
Chromatin relaxation triggered by DSBs produces an increased DNA susceptibility to micrococcal nuclease (MNase) digestion, translating on agarose gels as elevated lower molecular weight DNA fragments; this effect is both ATM- and pKAP-1–dependent (Fig. 3 D; Ziv et al., 2006; Goodarzi et al., 2011). These assays generally use the radiomimetic drug neocarzinostatin (NCS), as large doses (analogous to levels needed for pulsed-field gel electrophoresis) of DSBs must be delivered over a very short period of time, which is not always feasible for IR sources. Notably, depletion of SNF2H, ACF1, or RNF20 blocked DSB-induced increased MNase susceptibility, indicating that they are required for chromatin relaxation (Fig. 3, D and E). Depletion of CHD3 produced a constitutive relaxation phenotype (as seen in Goodarzi et al., 2011) with no impact observed by SNF2H codepletion, suggesting possibly that SNF2H acts to relax a similar chromatin component that CHD3 compacts.
ACF1–SNF2H and RNF20 function cooperatively in heterochromatin relaxation
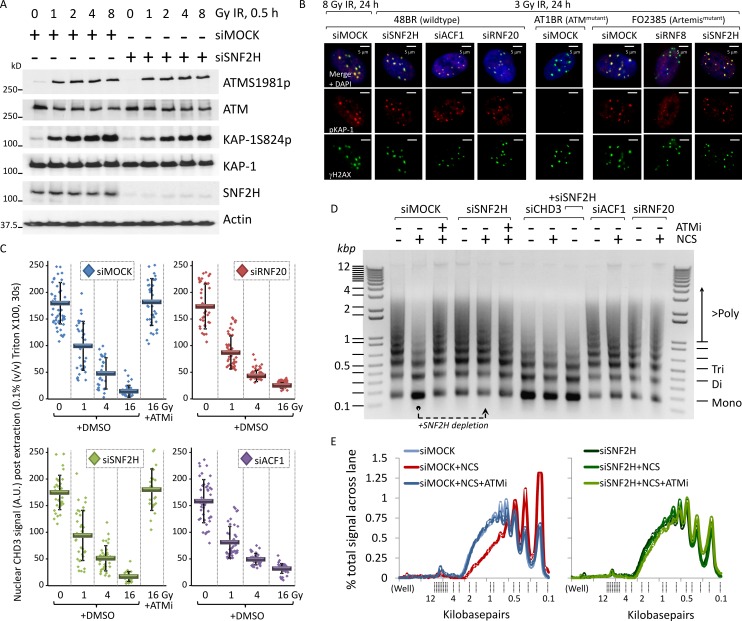
To consolidate our results using alternative systems, we took advantage of the differential properties of heterochromatin and euchromatin during NIH3T3 cell fractionation. Heterochromatin is enriched for H3K9me3 and contains sparse protranscriptional H3K4me3 or H4K8ac (Fig. 4 A). Because these marks are not altered by acute DSB induction (Tjeertes et al., 2009; Goodarzi et al., 2011), they can be used to distinguish heterochromatic and euchromatic nucleosomes. Adapting a method used in Goodarzi et al. (2008), we fractionated confluence-arrested NIH3T3 cells such that the most nuclease-resistant fraction (C5) was enriched for H3K9me3 and was largely free of H3K4me3 or H4K8ac, indicative of it containing mostly heterochromatic nucleosomes (also see Materials and methods for full explanation of segregation; Fig. 4 B). IR mobilized H3K9me3-enriched/H4K8ac-sparse nucleosomes from fraction C5 into more nuclease-susceptible fractions C1–C4, which were positive for γ-H2AX (Fig. 4 B, red arrows), fitting with the widely held notion that DSB-associated chromatin is more accessible (Soria et al., 2012; Stanley et al., 2013). In contrast, γ-H2AX–modified, H3K9me3-enriched nucleosomes could be observed in fraction C5 from RNF20- or SNF2H-depleted cells (Fig. 4 C, red arrows), suggesting that steps leading to full chromatin relaxation (i.e., solubility) were attenuated in the absence of RNF20 or SNF2H.
Figure 4.
SNF2H and RNF20 enable specifically heterochromatic relaxation after DSB induction. (A) Confluence-arrested NIH3T3 cells immunostained for H3K9me2/3, H3K9me3, H4K8ac, and/or H3K4me3, as indicated. Bars, 5 µm. (B) Confluence-arrested NIH3T3 cells were exposed to 0 or 40 Gy IR and, 1 h later, fractionated into extracts (for a full explanation of each fraction, see Materials and methods) and immunoblotted for the indicated proteins. (C) Confluence-arrested NIH3T3 cells were treated with scrambled (Mock), SNF2H, or RNF20 siRNA and, 48 h later, were irradiated, fractionated and immunoblotted as in B. Red arrows highlight the H3K9me3 signal in the C5 fraction undergoing a dynamic change in response to stimuli.
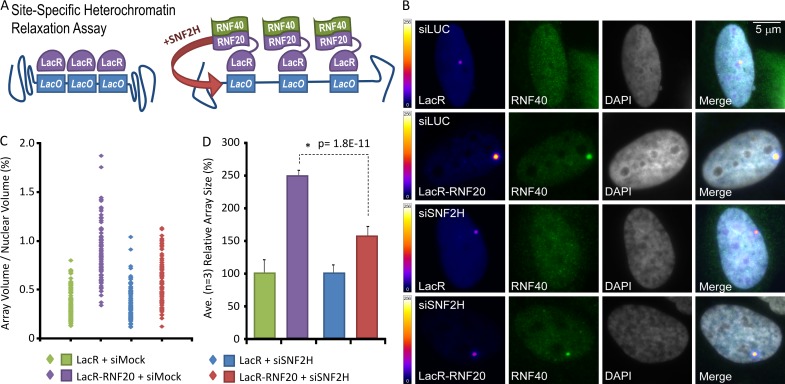
We next aimed for direct proof that cooperative RNF20 and SNF2H activity underlies heterochromatin relaxation. Human RNF20 fused to LacR was coexpressed with RNF40 in cells with a LacO array integrated within a heterochromatic region, a system first described in Janicki et al. (2004; Fig. 5 A). Both LacR and LacR-RNF20 localized to the array, visible as a single focus, whereas RNF40 accumulated at the array only in LacR-RNF20 fusion-expressing cells (Figs. 5 B and S3 A). Array volumes (as a function of total nuclear volume, and a readout for chromatin relaxation) expanded ∼2.5-fold when LacR-RNF20 was localized to it, compared with LacR alone (Fig. 5, C and D; and Fig. S3 A). RNF20-dependent array expansion was muted significantly by SNF2H depletion. These data provide direct evidence that RNF20 elicits SNF2H-dependent nucleosome relaxation within heterochromatin.
Figure 5.
RNF20 promotes site-specific heterochromatin relaxation in an SNF2H-dependent manner. (A) Schematic for site-specific chromatin relaxation assay. (B) Cells containing an integrated LacO array within a region of heterochromatin were treated ± siRNA toward luciferase (siLuc) or SNF2H (siSNF2H) and transfected with RNF40Myc and either LacR or LacR-RNF20, as indicated. Color scales indicate false-colored lookup table of pixel intensities (8 bit, pixel range = 1–256). (C and D) The nuclear volume of the LacR-signal (array) as a percentage of the overall nuclear volume was plotted for each condition, either as individual data points (C) or a mean of three independent experiments (D). P-values (standard two-tailed Student’s t test) are indicated. Error bars show SD.
CHD3 dispersal is insufficient for chromatin relaxation in nonreplicating cells
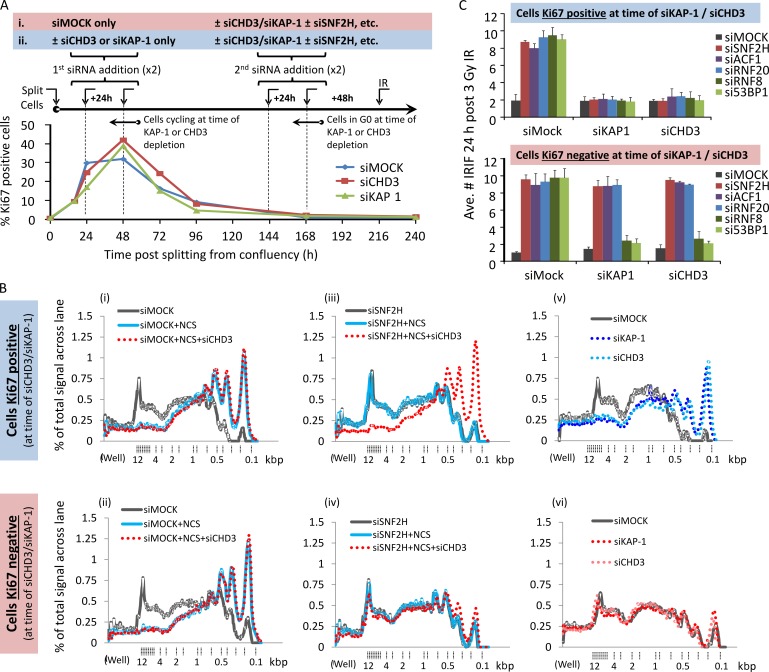
Our data suggested that RNF20 and ACF1–SNF2H function cooperatively but downstream of CHD3.1 dispersal. If correct, CHD3.1 depletion alone should not trigger chromatin relaxation in the absence of SNF2H; however, our data in Fig. 3 D argued against this. The inconsistency was explained and proved very informative once DNA replication was taken into account. For instance, in the case of Fig. 3 D, HeLa cells in a state of proliferation were used at the time of siRNA transfection. During DNA replication, newly synthesized chromatin must be recompacted and, without key heterochromatin building factors (e.g., KAP-1 or CHD3.1), remains in a more open state (Rowbotham et al., 2011). Thus, in dividing KAP-1/CHD3-depleted cells, the heterochromatic barrier to DSB repair is never “rebuilt,” and so, further relaxation is superfluous for repair (Goodarzi et al., 2008, 2011). To address this, we reoptimized the MNase assay for primary fibroblasts and compared CHD3/KAP-1 depletion either with or without additional SNF2H depletion between confluence-arrested and Ki67-negative (i.e., G0 phase) cells at the time of CHD3/KAP-1 siRNA-mediated knockdown, versus those logarithmically dividing and Ki67 positive (Figs. 6 A and S3 B). Both Ki67-positive and -negative cells showed an SNF2H-dependent increase in smaller sized DNA fragments after NCS treatment (Fig. 6 B, i–iv; and Fig. S3 C). However, cells that were Ki67 negative during CHD3 depletion showed a clear SNF2H dependence for chromatin relaxation, whereas cells Ki67 positive during CHD3 depletion could relax their chromatin after NCS treatment regardless of SNF2H status. This suggests that SNF2H is essential for DSB-induced chromatin relaxation so long as heterochromatin is unperturbed and/or assembled “normally” before CHD3 loss. In support of this, general MNase susceptibility was altered substantially by KAP-1/CHD3 depletion in Ki67-positive cells, whereas no impact was observed in Ki67-negative cells (Fig. 6 B, v and vi). This fits with the idea that heterochromatic compaction is largely stable in nondividing cells, even after de novo building factors are lost (Maison et al., 2011).
Figure 6.
SNF2H–ACF1 and RNF20 remain essential for heterochromatic relaxation and DSB repair in the absence of CHD3 in nonproliferating cells. (A) Quiescent, confluence-arrested 48BR cells were split to ∼50% confluency and monitored over the course of 10 d (240 h) for proliferation as measured by Ki67 signal by IF. Two different schemes of siRNA treatment during this time were used: in the first (i) scenario, cells were subject only to mock siRNA while in a proliferative state (24–72 h after splitting); in the second (ii), either KAP-1 or CHD3 siRNA were added to cells while they were proliferating. In both cases, after 7 d (144 h), cells achieved confluency, and they were subjected to further CHD3/KAP-1 siRNA in combination with siRNA targeting SNF2H, ACF1, RNF20, RNF8, or 53BP1. At 9 d (216 h) after splitting, once complete knockdown of all targets was achieved, cells were irradiated with 3 Gy IR harvested a day later. The percentage of cells positive for Ki67 was plotted over time (>250 cells monitored per condition, per experiment). (B) Cells obtained from either treatment schemes i or ii as outlined in A were treated with NCS and processed for the chromatin relaxation assay as in Fig. 3 (D and E). (C) Cells obtained from either treatment schemes i or ii as outlined in A were immunostained for γ-H2AX (green) and the relevant target of siRNA (e.g., in SNF2H-depleted cells, cells were counterstained with SNF2H in the red channel to confirm knockdown). The mean number of IRIF per nucleus was scored in cells with confirmed knockdown for three independent experiments. Error bars show SD. Ave., average.
To substantiate this, we monitored IRIF persistence in cells either Ki67 positive or negative at the time of KAP-1/CHD3 depletion and treated subsequently (when all cells were Ki67 negative) with SNF2H, ACF1, or RNF20 siRNA. Depletion of RNF8 or 53BP1 was used as a positive control for activities effectively bypassed by KAP-1 or CHD3 loss (Noon et al., 2010). SNF2H, ACF1, RNF20, RNF8, or 53BP1 depletion all caused late-repairing IRIF to persist, fitting with roles in heterochromatic DSB repair (Fig. 6 C; data here and in Noon et al., 2010). If KAP-1/CHD3 was depleted from proliferating cells, the need for SNF2H, ACF1, RNF20, RNF8, or 53BP1 in DSB repair was bypassed. In contrast, when KAP-1/CHD3 were depleted from nonproliferating cells, only RNF8 and 53BP1 were rendered dispensable, whereas SNF2H, ACF1, and RNF20 remained important for DSB resolution. This fits with the observed patterns of heterochromatin compaction in Fig. 6 B (graph v) and supports a model whereby the recruitment of ISWI-class activity is a key part of the ATM-dependent chromatin relaxation pathway.
ACF1–SNF2H activity directly counters CHD3.1 activity in heterochromatin
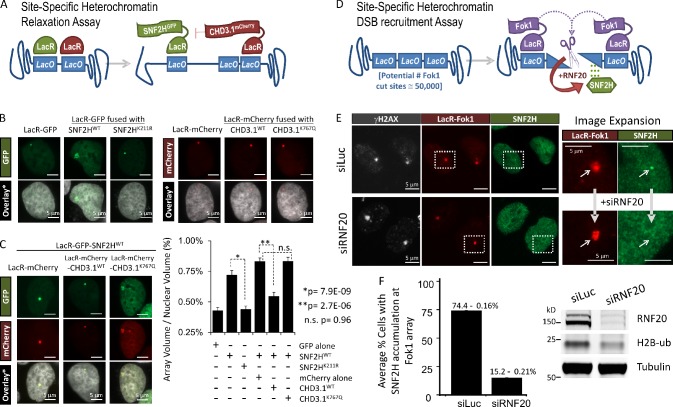
For additional proof that the activities of CHD3.1 and ACF1–SNF2H counteract one another within heterochromatin, we expressed LacR-fused CHD3.1mCherry and/or LacR-fused SNF2HGFP at the heterochromatic LacO array described in Fig. 5 (Fig. 7 A). Tethering CHD3.1 to the array had no effect on its volume, most likely as the array was already in a fully compacted state (Fig. 7 B). In contrast, tethering wild-type but not catalytically inactive SNF2H (K211R), or GFP alone, resulted in a marked expansion of the array, indicative of chromatin relaxation; this relaxation effect was attenuated strongly in the presence of tethered wild-type but not catalytically inactive (K767Q) CHD3.1 (Fig. 7 B). These data support the notion that ISWI- and CHD-class chromatin remodeling activities oppose one another in heterochromatin.
Figure 7.
Active CHD3.1 opposes SNF2H chromatin remodeling activity in heterochromatin, which requires RNF20 to localize to heterochromatic DSBs. (A) Schematic of the site-specific heterochromatin relaxation assay. (B) Representative images of cells containing a LacO array integrated within a region of heterochromatin and transfected with LacR-GFP, LacR-GFP-SNF2H (wild type or K211R), LacR-mCherry, or LacR-mCherry-CHD3.1 (wild type or K767Q), as indicated. Bars, 5 µm. (C) Cells from B were transfected with LacR-GFP-SNF2H wild type and either LacR-mCherry or LacR-mCherry-CHD3.1 (wild type or K767Q), as indicated. The nuclear volume of the LacR-GFP signal, as a percentage of the overall nuclear volume was plotted for each condition (from B and C), either as a mean of three independent experiments (∼75 cells for each condition). P-values (standard two-tailed Student’s t test) are indicated. Bars, 5 µm. Error bars show SD. (D) Schematic of the site-specific heterochromatin DSB recruitment assay. (E) U2OS 2-6-3 cells treated with either RNF20 (siRNF20) or luciferase (siLuc) siRNA and stably expressing ER-Fok1-mCherry-LacR-DD were induced with 300 nM 4-OHT and 1 µM Shield-I for 5 h. Subsequently, cells were preextracted using 0.25% Triton X-100 in CSK buffer for 10 min, fixed with formaldehyde, and immunostained with SNF2H (green) and γ-H2AX (greyscale). Boxes are enlarged in the right images. Arrows point to the site of DSB induction at the array. Bars, 5 µm. (F) The mean percentages of cells with SNF2H foci present at Fok1/γ-H2AX foci were quantified (130 cells for each condition from two independent experiments). siRNA efficiency was assessed by immunoblotting.
Localization of SNF2H to heterochromatic DSBs requires RNF20
Another important question is how ACF1–SNF2H activity is actually targeted to heterochromatic DSBs. To address this, we exploited a site-specific DSB induction system whereby LacR-fused Fok1 nuclease was targeted to the same LacO array described in the previous paragraph, inducing highly localized DSBs within heterochromatin (Fig. 7, D and E). After extraction, discrete SNF2H foci were observed to overlap with γ-H2AX at the LacR-Fok1–bound array. RNF20 depletion ablated SNF2H accumulation at γ-H2AX foci, indicating that RNF20 enables physical relocalization of ISWI activity to DSBs in heterochromatin.
The PHD fingers of CHD3.1 and ACF1 have similar histone preferences
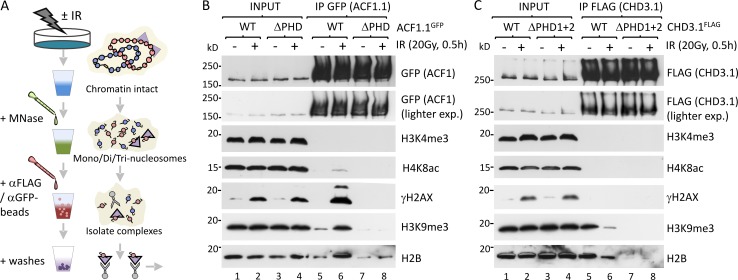
Our final objective was to dissect the molecular mechanism by which the ISWI and CHD branches of DSB-induced chromatin relaxation interface with one another. Both ACF1 and CHD3.1 possess PHD fingers (two, in the case of CHD3.1) that rely upon Zn2+ coordination to mediate histone interactions often in a methylation-specific manner (Taverna et al., 2007). H3K4me3 is a well-known binding target for PHDs and is a known, indirect consequence of RNF20 activity (Kim et al., 2009; Faucher and Wellinger, 2010). We speculated whether RNF20 promoted de novo H3K4me3 at heterochromatic DSBs to provide a binding platform for the ACF1 PHD finger; however, an IP approach to analyze H4K8ac (euchromatic) and H3K9me3 (heterochromatic) nucleosomes separately showed no change in H3K4me3 after IR (Fig. S3 D). Sequence analysis revealed that, compared with known H3K4me3-binding proteins (bromodomain PHD finger transcription factor [BPTF]), the ACF1 and CHD3.1 PHD fingers lack key aromatic cage residues required for H3K4me3 binding (Ruthenburg et al., 2011) and are most similar to the CHD4 PHD fingers that binds H3K4me0/H3K9me3 preferentially (Fig. S3 E; Mansfield et al., 2011). Thus, we hypothesized that ACF1 and CHD3.1 may both prefer unmodified H3K4, as would be found in heterochromatin, and that similar substrate binding preferences may drive ACF1 to bind sites vacated by CHD3.1. To explore these possibilities, key Zn2+ coordination residues were mutated (ΔPHD) within the PHDs of ACF1.1 and CHD3.1, which were then immunoprecipitated from nucleosome-solubilized extracts of cells ±IR and immunoblotted for histone marks (Fig. 8, A–C; and Fig. S2 E). Both wild-type ACF1.1 and CHD3.1 showed a preference for H3K9me3-enriched nucleosomes essentially devoid of H3K4me3 or H4K8ac. Notably, CHD3.1’s interaction with H3K9me3-modified nucleosomes was decreased after IR (Fig. 8 C, lanes 5 and 6), whereas ACF1.1’s interaction was increased (Fig. 8 B, lanes 5 and 6). ACF1.1ΔPHD displayed reduced affinity for overall nucleosomes (monitored by H2B) and negligible affinity for H3K9me3-modified (i.e., heterochromatic) nucleosomes. A slight increase in H2B but not H3K9me3 signal was observed after IR (Fig. 8 B, lanes 7 and 8), suggesting that although ACF1 interactions with heterochromatin rely upon its PHD finger, interactions with other chromatin compartments may not. CHD3.1ΔPHD1+2 displayed no detectable affinity for nucleosomes at all, irrespective of IR (Fig. 8 C, lanes 7 and 8), suggesting that CHD3.1’s PHD fingers are critical for direct chromatin interactions.
Figure 8.
The PHD fingers of ACF1 and CHD3.1 confer similar binding specificity for nucleosomes. (A) A schematic for experiments in B and C. (B) HeLa cells were transfected with wild-type ACF1.1 or ACF1.1 with C/H→A mutations in both Zn-3 and Zn-4 of the PHD finger (ACF1.1ΔPHD). 24 h later, cells were irradiated as indicated, and nucleosome-solubilized (MNase digested) whole cell extracts were prepared, immunoprecipitated with GFP-agarose, and immunoblotted for the indicated proteins. exp., exposure. (C) HeLa cells were transfected with wild-type CHD3.1 or CHD3.1 with C/H→A mutations in both Zn-1 and Zn-2 of the PHD finger 1 and both Zn-3 and Zn-4 of the PHD finger 2 (CHD3.1ΔPHD1+2). 24 h later, cells were irradiated as indicated, and nucleosome-solubilized (MNase digested) whole cell extracts were prepared, immunoprecipitated with GFP-agarose, and immunoblotted for indicated proteins.
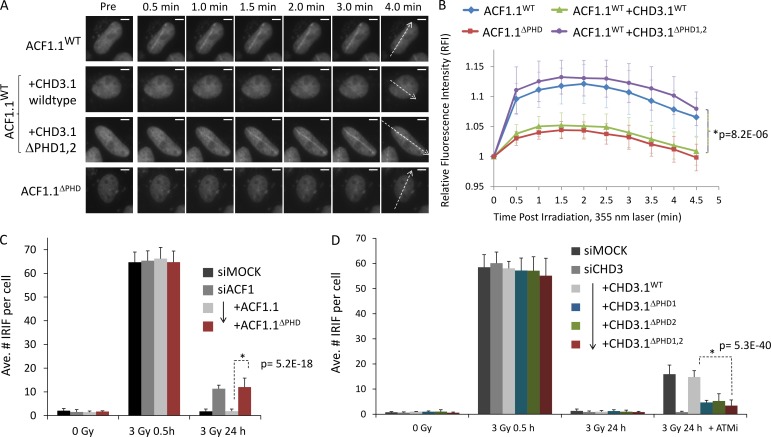
To explore the significance of the ACF1.1 and CHD3.1 PHD fingers further, we examined the recruitment of wild-type or PHD mutated ACF1.1GFP to laser microirradiation-induced DSB tracks (Fig. 9, A and B). We used a 355-nm laser (in BrdU-sensitized cells) at a localized damage level equivalent to 8–10 Gy within the damage track (dose estimation methodology described in Bekker-Jensen et al., 2006). Wild-type ACF1.1GFP was recruited clearly to DSB tracks within 30 s of microirradiation (Fig. 9, A [top row] and B [blue line]). By comparison, ACF1.1ΔPHD accumulation was threefold lower relative to the wild type under the same conditions (Fig. 9, A [bottom row] and B [red line]). Comparable trends were obtained using a much more densely damaging 405-nm laser (in Hoechst-sensitized cells), with the recruitment of ACF1.1ΔPHD being reduced and delayed compared with ACF1.1 and supporting findings with the 355-nm laser system (Fig. S3, F and G). To test the idea of competitive binding, wild-type ACF1.1GFP recruitment to 355-nm laser-induced microirradiation DSB tracks was monitored in cells ± overexpression wild-type or PHD mutated CHD3.1FLAG. Wild-type but not PHD mutant CHD3.1FLAG attenuated the recruitment of ACF1.1GFP to tracks, suggesting that an abundance of CHD3.1 with a functional PHD blocked binding sites necessary for ACF1.1 retention (Fig. 9, A and B).
Figure 9.
The PHD fingers of ACF1 and CHD3.1 confer similar binding specificity for nucleosomes. (A) HeLa cells were transfected as in Fig. 8 (B and C) with the indicated constructs and incubated with BrdU for 16–24 h before irradiation with a directed 355-nm laser. GFP signal was imaged live over time. The dotted arrows indicate path of laser microirradiation through nuclei. Bars, 5 µm. (B) The relative fluorescence intensity increase for GFP signal obtained in A was quantified. P-values (standard two-tailed Student’s t test) are indicated. (C) HeLa cells were transfected with ACF1-B siRNA and, 48 h later, then split to 75% confluency and transfected with the indicated ACF1.1GFP constructs. 16 h later, cells were irradiated as indicated, harvested, and immunostained for γ-H2AX; the mean number of γ-H2AX per GFP-positive nucleus was scored for three independent experiments. (D) HeLa cells were transfected with CHD3 siRNA and, 48 h later, then split to 75% confluency and transfected with the indicated CHD3.1FLAG constructs. 16 h later, cells were incubated ± ATM inhibitor (ATMi), irradiated as indicated, harvested, and immunostained for 53BP1 and FLAG; the mean number of 53BP1 per FLAG-positive nucleus was scored for three independent experiments. Ave., average. Error bars show SD. P-values (standard two-tailed Student’s t test) are indicated.
Finally, we theorized that if PHD fingers were important for the localization and/or activity of each protein, ACF1.1ΔPHD would be unable to support heterochromatic DSB repair, whereas CHD3.1ΔPHD1+2 might permit repair in the absence of ATM signaling. Indeed, ACF1-depleted cells expressing the ACF1.1ΔPHD showed a comparable DSB repair defect to ACF1 depletion alone, whereas CHD3.1ΔPHD1+2-expressing cells showed normal DSB repair even in the absence of ATM activity (Fig. 9, C and D). Cells expressing CHD3 mutated for single PHD fingers displayed intermediate effects, suggesting a small degree of functional redundancy between CHD3’s PHD fingers in this context. These data indicate that ACF1.1 and CHD3.1 both require functional PHD fingers for their roles in DSB repair.
Discussion
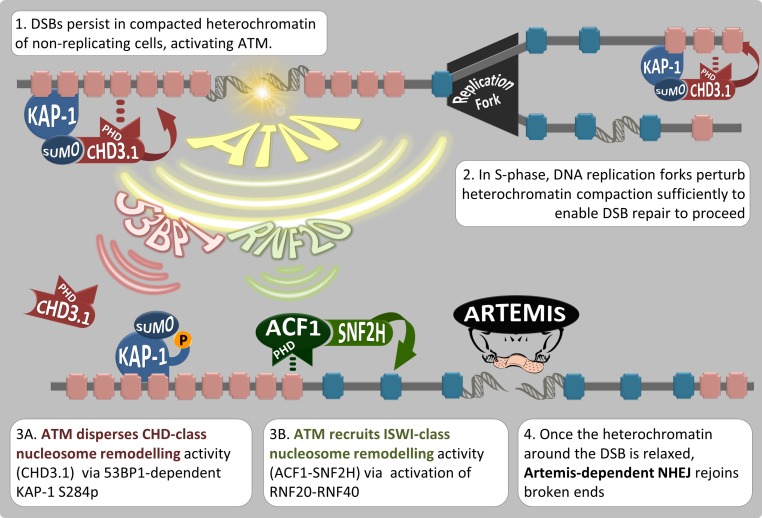
Our data suggest that (a) RNF20-dependent ACF1–SNF2H (ISWI class) chromatin remodeling is required for Artemis-dependent DSB repair in heterochromatin; (b) RNF20 enables the physical retention of SNF2H at heterochromatic DSBs, but otherwise does not impact pKAP-1 induction or CHD3.1 dispersal directly; (c) ACF1–SNF2H is the ISWI-class activity that actually decompacts heterochromatic nucleosomes after CHD3.1 dispersal; (d) this is essential for DSB repair in nonreplicating cells where chromatin is unperturbed by DNA replication processes (Löbrich et al., 2010; Goodarzi and Jeggo, 2013); and (e) the PHD fingers of both CHD3.1 and ACF1 have similar histone-binding preferences (enriched for H3K9me3, depleted for H3K4me3 and H4K8ac) and are required for the roles of these proteins in heterochromatic DSB repair. The accrual of ISWI and dispersal of CHD class II chromatin remodeling activity are both ATM dependent. Beyond that, however, these pathways appear to separate in their mechanism of activation, with the ISWI-accrual branch being RNF20 dependent and the CHD-dispersal branch involving γ-H2AX, MDC1, RNF8, RNF168, 53BP1, and pKAP-1. Once initiated, these mechanisms converge to generate a chromatin environment favorable for the repair of DSBs via the Artemis nuclease (Fig. 10).
Figure 10.
Model for heterochromatin nucleosome relaxation and Artemis-dependent DSB repair. (1) Artemis-dependent DSB repair stalls within the KAP-1– and CHD3.1-rich heterochromatin of nondividing cells. (2) During DNA replication, the heterochromatic superstructure is perturbed, and newly synthesized strands remain in an open configuration, bypassing the need for additional chromatin relaxation during repair of DSBs incurred during S phase. (3) ATM protein kinase activity triggers two signaling axes. The first (3A) enables the dispersal of class II CHD chromatin remodeling activity (CHD3.1) via the IRIF mediator proteins (such as 53BP1) and densely localized pKAP-1. The other (3B) promotes the gain of ISWI-class chromatin remodeling activity via RNF20-RNF40 activation and the recruitment/activation of ACF1–SNF2H, which we propose occupies PHD finger binding sites vacated by CHD3.1. (4) These mechanisms converge to enable heterochromatin relaxation, which is favorable for the repair of DSBs via Artemis. P, phosphorylation.
Recent studies have found SNF2H to impact HR pathways via BRCA1 foci and/or RAD51 filament formation, alongside ACF1, RNF20, RNF168, the SIRT6 histone deacetylase, and the SUPT16H component of the FACT histone chaperone complex (Lan et al., 2010; Nakamura et al., 2011; Smeenk et al., 2013; Toiber et al., 2013; Oliveira et al., 2014; Pessina and Lowndes, 2014). It will be important to determine which of these HR processes may also be relevant to the role of SNF2H in Artemis-dependent DSB repair. Artemis is also required for HR-mediated DSB repair in G2 phase (Beucher et al., 2009). Therefore, as a corollary to these findings, it is possible that the events enabled by ACF1–SNF2H activity in G0/G1 phase are required equally in G2.
We interpret the negative effect of SNF2H depletion on γ-H2AX foci intensity (Fig. S1 H) to be explained by SNF2H role in the WICH complex, which promotes H2AXY142p and γ-H2AX signal maintenance but not induction (Xiao et al., 2009). This effect on γ-H2AX signal only appears after the vast majority of SNF2H is depleted from the cell, suggesting perhaps the WSTF–SNF2H (WICH) complex forms “preferably” over the ACF1–SNF2H (ACF [ATP-utilizing chromatin assembly and remodeling factor]) complex when SNF2H levels are limiting; thus, ACF-associated defects are observable after only partial (60–90%) SNF2H depletion compared with putative WICH-associated defects that require near total loss of SNF2H (>90%). Whatever the case, SNF2H depletion conditions that generated a clear ATM/Artemis-dependent heterochromatic DSB repair defect (Fig. 1) had no impact on γ-H2AX→53BP1→pKAP-1 foci formation and CHD3.1 dispersal, arguing against a role for SNF2H in those processes in this context. Furthermore, ACF1 depletion or AID domain deletion of SNF2H, which would not impact any role of SNF2H with WSTF in the WICH complex, recapitulates the same DSB repair defects that are epistatic with SNF2H depletion to perturb Artemis-dependent DSB repair.
ACF1 and/or SNF2H accumulation is observable only at very densely (laser) induced DSBs (this work; Lan et al., 2010; Smeenk et al., 2013; Oliveira et al., 2014) and suggests that ISWI activity is recruited to DSBs only transiently or at a very small stoichiometric amount, perhaps as a result of extreme potency; this has been proposed previously (Lan et al., 2010). We suggest that this stoichiometric imbalance is perhaps another mechanistic reason for CHD3.1 dispersal, which is abundant at KAP-1–rich heterochromatic DSBs and would otherwise overwhelm any other activity.
PHD fingers are important histone modification reader domains, conferring methylation-specific chromatin binding affinity to many proteins (Taverna et al., 2007), including proteins recruited to DSBs, such as SPOC1 (Mund et al., 2012). In the case of SPOC1, its PHD finger confers H3K4me2/3-specific binding required for its role DSB repair, which is to negatively regulate NHEJ while enhancing HR (Mund et al., 2012). SPOC1 promotes heterochromatization at DSBs insofar as it interacts with KAP-1 (preventing its phosphorylation at S824 by ATM), HP1, and H3K9 methyltransferases. Thus, the SPOC1 pathway is seemingly the inverse of the pathway we describe here, wherein RNF20 and ACF1–SNF2H serve to transiently alleviate the heterochromatic barrier to DSB repair and promote Artemis-dependent NHEJ, whereas SPOC1 promote a heterochromatin configuration and impairs NHEJ. Given the differential specificity of the PHD fingers of SPOC1 compared with ACF1 and CHD3.1 for H3K4me3, we consider it unlikely that these two pathways overlap broadly within chromatin, the expectation being that SPOC1 would preferentially bind euchromatin typical regions. Although often viewed in association with H3K4me3 binding (Ruthenburg et al., 2011), PHD fingers may also bind H3K4me0 selectively in conjunction with the heterochromatin-typical mark H3K9me3 (Mansfield et al., 2011). We now demonstrate two cases (ACF1.1 and CHD3.1) of PHD-mediated H3K4me0/H3K9me3-preferred binding during the DSB response.
Our data suggest that the need to disperse CHD factors before ISWI factors can occupy and manipulate the same chromatin environment is driven by (a) a similar histone modification binding preference between the two enzymes, coupled with (b) an “adversarial” relationship between each enzyme’s activity on chromatin. This mechanism is supported by previous work indicating that purified Drosophila ISWI and CHD homologues display opposing in vitro activities (Brehm et al., 2000; Moshkin et al., 2012), and is logical in that it minimizes energy consumption by avoiding counterproductive remodeling events occurring within damaged heterochromatin.
Overall, this study reveals that nucleosome arrangements incompatible with DSB repair processes are temporarily reversed via a two-step system of dispersing and recruiting opposing chromatin remodelers, all under the singular control of ATM-dependent DSB response signaling. This concept, demonstrated here for heterochromatin, may apply quite widely to other chromatin environments subject to alteration during a DSB response. Although not in the context of DNA damage-induced modifications, the idea that certain chromatin regions require the activity of more than one nucleosome-remodeling enzyme to regulate accessibility was demonstrated recently by chromatin IP in murine model systems, lending credence to the concept we propose for DSB repair (Morris et al., 2014).
Materials and methods
Reagents and tissue culture
ATM inhibitor KU-55933 (EMD Millipore) was used at 10 µM. NCS was obtained from Sigma-Aldrich. 1BR3 (WT), 48BR (WT), AT1BR (containing an ATM genomic deletion [specific genetic lesion not known], no transcript, and null expression), RIDDLE (RNF168 biallelic mutation producing two frame-shifted, prematurely stopped protein products: A133fsX and Q442fsX), CJ179 (Artemis genomic deletion [specific genetic lesion not known], no transcript, and null expression), and F02385 (Artemis genomic deletion [specific genetic lesion not known], no transcript, and null expression) primary human fibroblasts were as in Riballo et al. (2004), Noon et al. (2010), and Woodbine et al. (2011). NIH3T3 and human U2OS-2-6-3 cells containing 200 copies of a LacO (256×)/TetO (96×)-containing cassette of ∼4 Mbp (obtained from S. Janicki, Wistar Institute, Philadelphia, PA and as in Janicki et al., 2004) were cultured in DMEM with 10% (vol/vol) FCS, GlutaMAX (Gibco), penicillin, and streptomycin. HeLa cells were cultured in MEM with 10% (vol/vol) FCS, l-glutamine, penicillin, and streptomycin. IR was performed with γ rays (137Cs) delivered by Gammacell 1000 Elite (MDS Nordion).
Transient knockdown of protein expression and siRNA-resistant construct expression
All siRNA-mediated knockdown was achieved using Metafectene Pro (Biontex)–mediated transfection (according to the manufacturer’s instructions) using 25–100 pmol of siRNA duplexes per 2 × 105 cells. With the exception of single siRNAs necessary for use in combination with siRNA-resistant plasmids, all siRNAs were used as a 1:1 pool of A + B sequences, as outlined below. Stealth siRNA oligonucleotides were obtained from Invitrogen. siRNA (pools) to RNF8 and 53BP1 were validated in Noon et al. (2010) and to CHD3 was as in Goodarzi et al. (2011). siRNA sequences were as follows: SNF2H A, 5′-CCGGGCAAAUAGAUUCGAGUAUUUA-3′; SNF2H B, 5′-CAGGGAAGCUCUUCGUGUUAGUGAA-3′; SNF2H mouse, 5′-GGGAGGCCUCUGAGAACCUUCCGUU-3′; RNF20 A, 5′-CCGUGUCCCAGAUUGUGACUGUUUA-3′; RNF20 B, 5′-CAGUCACAGUUCUCCGUCUUGUAUA-3′; ACF1 A, 5′-CAAGUAUAAAGUGCAACCCACUAAA-3′; ACF1 B, 5′-UCAAGAUCCUCAGGUAUCCACUAAA-3′; CHD3 A, 5′-GGGCCAUCAUUCGUGAGAAUGAAUU-3′; CHD3 B, 5′-AGGCACAGGUGAAGUUCCAUGUUCU-3′; RNF8 A, 5′-GGACAAUUAUGGACAACAA-3′; RNF8 B, 5′-UGCGGAGUAUGAAUAUGAA-3′; and 53BP1, 5′-AGAACGAGGAGACGGUAAUAGUGGG-3′.
For plasmid expression, adherent cells were transfected with 2 µg plasmid 48 h after siRNA treatment using Metafectene Pro according to methods described in Goodarzi et al. (2008, 2011) and Noon et al. (2010) and, 16–24 h later, were irradiated for analysis. For pEGFP-N1–SNF2H expression constructs (SNF2HGFP), full human SNF2H cDNA were cloned into the pEGFP-N1 backbone under a cytomegalovirus (CMV) promoter (Takara Bio Inc.; plasmid backbone information found at Addgene). For the SNF2HFLAG construct, full human SNF2H cDNA were fused with FLAG cDNA (followed by a stop codon) cloned into pEGFPN3 (Takara Bio Inc.) backbone, under a CMV promoter with the stop codon preventing C-terminal GFP expression. Silent point mutations (T2715C + T2718C + T2721C) were introduced into SNF2HFLAG and SNF2HGFP expression constructs to generate siRNA B–resistant SNF2H cDNA, using a mutagenesis kit (QuikChange XL; Agilent Technologies; this kit was used for all point mutagenesis). Impacting point mutations were then introduced: A1093G→K211Q, G3154T→E898X, and G3487T→L1007X. To generate siRNA A–resistant RNF20HA expression constructs, (G651A + T654A + G657A), silent point mutations were introduced into full-length RNF20 cDNA (constructs obtained from Y. Shiloh, University of Tel Aviv, Tel Aviv, Israel), which was cloned originally into pcDNA3.0 under a CMV promoter, as described in Moyal et al. (2011). Impacting point mutations were then introduced: T604G→S172A and T1747G→S553A.
For expression of ACF1 fused at the C terminus to GFP, the ACF1.1 (GenBank accession no. NM_013448) coding sequence was amplified by PCR and cloned in frame into pEGFP-N1 (Takara Bio Inc.) at the Kpn1 and BamHI sites, under a CMV promoter. Silent point mutations (T3127C + G3130A + A3133T) were introduced into pEGFP-N1–ACF1 expression constructs to generate siRNA B–resistant ACF1 cDNA, using mutagenesis kit (QuikChange XL). To create ACF1.1ΔBAZ and ACF1.2 (GenBank accession no. NM_182648), bases 2,003–2,797 and 1,511–1,606 were deleted, respectively, from the ACF1.1 coding sequence by overlap extension PCR. PHD finger mutations were introduced as follows: (C3520G + A3521C + T3529G + G3530C→H1174A + C1177A = Zn-3 mutation) + (T3574G + G3575C + T3583G + G3584C→C1192A + C1195A = Zn-4 mutation) to generate a combined H1174A + C1177A + C1192A + C1195A mutant for both Zn-3 and Zn-4 (referred to as ACF1.1ΔPHD).
Wild-type and K767Q siRNA-resistant CHD3.1FLAG constructs were as in Goodarzi et al. (2011) and represent full-length CHD3.1 cDNA cloned into pCIneoB-3FLAG plasmid under a CMV promoter. To generate PHD finger mutants, the following point mutations were introduced: PHD finger 1, (C1354G + A1355C + T1363G + G1364C→H402A + C405A = Zn-3 mutation) + (T1408G + G1409C + T1317G + G1318C→C420A + C423A = Zn-4 mutation) to generate a combined H402A + C405A + C420A + C423A for both Zn-3 and Zn-4 (referred to as CHD3.1ΔPHD1); and PHD finger 2, (T1525G + G1526C + T1334G + G1335C→C459A + C462A = Zn-1 mutation) + (T1561G + G1562C + T1570G + G1571C→C471A + C474A = Zn-2 mutation) to generate a combined C459A + C462A + C471A + C474A for both Zn-1 and Zn-2 (referred to as CHD3.1ΔPHD2). The PHD finger 1 and 2 mutations described were combined to generate CHD3.1ΔPHD1+2.
Antibodies and IF
The following Abcam antibodies were used (note, r, rabbit host; m, mouse host; g, goat host): anti–KAP-1 ab10484(r), anti-CHD3.1 ab84528(r), anti-H3K9me3 ab8898(r), anti-H3K9me2+3 ab71604(m), anti-H4K20me3 ab9053(r), anti-H3K4me3 ab8580(r), anti-H4K8ac ab15823(r), anti-H2B ab1790(r), anti-SNF2H ab72499(r), anti-ACF1 ab94749(r), anti-RNF20 ab32629(r), anti-53BP1 ab21083(r) and ab36823(r), anti-GFP ab290(r), anti-Myc ab9106(r), and anti-FLAG ab1240(g). We also used anti-Myc 9E10(m) obtained from Santa Cruz Biotechnology, Inc., anti-HA clone 7 H3663(m) obtained from Sigma-Aldrich, anti-ATMS1981p 2152–1(r) obtained from Epitomics, and anti–γ-H2AX clone JBW301(m) obtained from EMD Millipore. Custom anti–KAP-1S824p (rabbit polyclonal, raised to phosphorylated S824 C-terminal peptide) and anti-ATM 4BA (rabbit polyclonal, raised against aa 2,323–2,740 of human ATM) are as described in Noon et al. (2010). Identical results were achieved using commercially available anti–KAP-1S824p A300-767A(r) purchased from Bethyl Laboratories, Inc. All IF was performed as in Goodarzi et al. (2008, 2011) and Noon et al. (2010); in brief, washed (in PBS) cells were fixed in 3% (wt/vol) PFA + 2% (wt/vol) sucrose for 10 min, permeabilized for 3 min in 0.2% (vol/vol) Triton X-100 (in PBS), and immunostained for 1 h with primary antibody (diluted in 2% [wt/vol] BSA in PBS) the 30 min with 1:200 dilutions of secondary antibodies (also in 2% BSA as before). All secondary antibodies for IF were as follows: anti–mouse or anti–rabbit IgG coupled to Alexa Fluor 488 or 594 (Invitrogen/Molecular Probes). Antibodies for immunoblotting were as follows: anti–mouse/rabbit/goat-HRP obtained from Sigma-Aldrich. Cells were cultured in a quiescent state for 1–2 wk for observing changes in CHD3.1 detergent extractability using the method described in Goodarzi et al. (2011). In brief, cells were extracted first in PBS + 0.2% (vol/vol) Triton X-100 for 3 min and then fixed and immunostained as described previously in this section; poor results were obtained with logarithmically dividing cells. Where indicated, cells were counterstained with 0.1 µg/ml DAPI to visualize nuclei and were mounted using Polymount G. Samples were imaged with a platform microscope (Axio Observer.Z1; Carl Zeiss), with a Plan Apochromat 20×/0.8 NA, an EC Plan Neofluar 40×/0.75 NA, or a Plan Apochromat 63×/1.4 NA (oil immersion) objective and a camera (AxioCam MRm Rev.3; Carl Zeiss). Acquisition and analysis software used was Zen Pro (Carl Zeiss).
Heterochromatic IRIF overlap microscopy analysis
Highly resolved z stacks were captured by a camera (AxioCam MRm) using a confocal microscope (LSM 510 meta; Carl Zeiss) with an oil immersion 100× objective used at room temperature. Subsequently, 3D rendering was performed to convert the 3D z stacks into a 2D image. The overlap between γ-H2AX foci and heterochromatin markers was quantified by ImageJ software (National Institutes of Health) with the Colocalization Analysis plugin. Mean values represent 15–20 cells in each of the three experiments for each condition.
MNase digestion assay
Nucleosome relaxation assays were performed as in Ziv et al. (2006) and Goodarzi et al. (2011). For each sample, prepare two T25 flasks of 100% confluent cells (4 ml media per flask). To treat with NCS, remove all but 2 ml of the media, turn off lights, and add NCS to a final concentration of 200 ng/ml. 0.5 h later, harvest cells by trypsinization, wash once with 10 ml PBS, and count the cells at this point, to ensure an equal number between samples. Resuspend cell pellets in 1 ml PBS and transfer to a 1.5-ml tube on ice. Centrifuge down cells again and remove PBS. Note the packed cell volume (PCV). Generally, two T25 flasks of confluent HeLa cells should give ∼50 µl packed cells; primary cells will give ∼25 µl PCVs. Ensure that PCVs are equivalent between samples. Keep all samples on ice. Use cold microcentrifuge for every spin step. Resuspend cells with 10 volumes (0.5 ml if PCV = 50 µl) ice-cold hypotonic buffer with spermidine and spermine (HBSS buffer = 340 mM sucrose, 15 mM Tris, pH 7.5, 15 mM NaCl, 60 mM KCl, 10 mM DTT, 0.15 mM spermine, 0.5 mM spermidine, and 0.5% [vol/vol] Triton X-100). Make sure cells are completely resuspended and incubate on ice for ≤10 min with periodic vortexing. Centrifuge cells at 11,000 rpm for 5 min at 4°C. Discard supernatant and resuspend in 0.25 ml (5× the original PCV) HBSS. Generally, the PCV should have reduced by 50% if cells are efficiently lysed. Rinse the nuclei in another 0.25 ml HBSS (5× the original PCV), vortex, centrifuge cells at 11,000 rpm for 5 min, and discard supernatant. Resuspend nuclei in 0.25 ml of 1:1 HBSS/glycerol and store at −20°C overnight. If desired, freeze cells in liquid nitrogen and continue with the next step immediately. Thaw the nuclei and remove 75 µl to a fresh tube (rest can be frozen). Centrifuge at 11,000 rpm for 5 min and resuspend in 75 µl MNase digestion buffer (250 mM sucrose, 15 mM Tris, pH 7.5, 15 mM NaCl, 60 mM KCl, 0.5 mM DTT, and 1 mM CaCl2), ensuring that they are completely resuspended and that there are no lumps. Prepare a fresh dilution of 1 U/µl MNase (Nuclease S7; catalog no. 107921; Roche), from a 10-U/µl stock solution dissolved in 5 mM Tris, pH 7.5, containing 25 mM CaCl2. Collect aliquot and store at −80°C. Heat a water bath to 25°C. Ensure that the 0.5 M EDTA is close at hand. Using a timer to accurately measure time of addition between samples, add 1.7 µl MNase to 75 µl of resuspended nuclei, mix well, and put in the 25°C water bath. 3 min after the addition of MNase, add 1.5 µl of 0.5-M EDTA to stop the reaction and put the sample on ice. Add 8 µl of 5% (wt/vol) SDS + 1 mg/ml Proteinase K (Sigma-Aldrich), vortex, and incubate at 37°C for 0.5 h. Dilute each proteinase-digested sample to a final volume of 250 µl with MNase buffer and vortex to mix. Add 250 µl of Tris-buffered phenol/chloroform/isoamyl-alcohol. Vortex and centrifuge at 14,000 rpm for 2 min. Transfer the clear (or mostly clear) supernatant to a new tube, avoiding the white precipitate found at the interface with phenol. Add 1 ml of water-saturated diethyl-ether to each sample, vortex, and spin at 14,000 rpm for 1 min. Discard the supernatant and repeat the ether wash step. Add 25 µl of 3 M sodium acetate, pH 5.8, mix by vortexing, and add 750 µl of cold 100% ethanol, vortex, and put at −20°C overnight. The next day, centrifuge samples at 14,000 rpm for 20 min to pellet precipitated DNA. Discard the supernatant and wash pellets with 0.5 ml of 70% ethanol. Air dry pellets (no more than 1 h) and resuspend in 40 µl of water. Allow 5–10 min for DNA to fully dissolve. Measure [DNA] and resolve 2.5 µg of each sample by agarose gel electrophoresis (1.2% agarose gels in TAE [Tris acetate EDTA] buffer).
Chromatin segregation assay
This assay was adapted from the NIH3T3 cell chromatin segregation procedure performed as in Goodarzi et al. (2008). Confluent NIH3T3 cells were transfected with siRNA using Metafectene Pro as outlined in the methods section entitled Transient knockdown of protein expression and siRNA-resistant construct expression. 107 cells were washed with PBS and once with 1 ml of low salt buffer (LSB). Pelleted cells were resuspended in 6× the PCV of LSB (10 mM Hepes, pH 7.4, 25 mM KCl, 10 mM NaCl, 1 mM MgCl2, and 0.1 mM EDTA) + 0.1 µM microcystin (MC)-LR and protease inhibitor cocktail and snap frozen in liquid nitrogen. Samples were quick thawed and immediately centrifuged for 10 min at 10,000 rpm (supernatant = S10). The pellet was gently resuspended (by tapping, but not pipetting to prevent chromatin decondensation) in 1 vol of high salt buffer (HSB; 50 mM Tris-HCl, pH 8.0, 5% [vol/vol] glycerol, 1 mM EDTA, 10 mM MgCl2, 400 mM KCl, protease inhibitors, and 0.1 µM MC-LR), which was equal to 0.25 vol LSB used to lyse the cells. Samples were immediately centrifuged for 10 min at 10,000 rpm (supernatant = P10). The pellet was then resuspended in nucleosome preparation buffer (NPB; 10 mM Hepes, pH 7.9, 10 mM KCl, 1.0 mM CaCl2, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, and 0.1% [vol/vol] Triton X-100; same volume as HSB) containing 5 U/ml MNase and incubated at 37°C for 5 min. To this, 0.25 vol of nucleosome solubilization buffer (NSB; NPB + 2% [vol/vol] NP-40, 2% [vol/vol] Triton X-100, and 600 mM NaCl) was added, and samples were then centrifuged for 5 min at 10,000 rpm (supernatant = C1). The pellet was resuspended in NPB (volume same as used for HSB) containing 250 U/ml MNase and was incubated at 37°C for 10 min before 0.5 vol of NSB was added. Samples were vortexed briefly and centrifuged for 5 min at 10,000 rpm (supernatant = C2). The pellet was resuspended in NPB (volume same as used for HSB) containing 100 U/ml MNase and was incubated at 37°C for 20 min before 1 vol of NSB was added. Samples were vortexed briefly and centrifuged for 5 min at 10,000 rpm (supernatant = C3). The remaining pellet was resuspended in NSB buffer (0.5 vol as HSB) and an equal volume of nucleosome denaturing buffer (50 mM Tris, pH 6.8, 1% [vol/vol] SDS, 100 mM DTT, and 10% glycerol) before centrifugation at 10,000 rpm for 5 min (supernatant = C4). The remaining pellet was resuspended in nucleosome denaturing buffer (same volume as HSB) before sonication (2× 5 s), boiling at 100°C for 5 min, and centrifuging at 10,000 rpm for 5 min (supernatant = C5).
The fractionation of chromatin using the methods shown in Fig. 4 (B and C) do not produce perfectly “clean” euchromatic- versus heterochromatic-typical nucleosome pools. As in the cell, the separation of euchromatin and heterochromatin is more of a gradient of enrichment for certain histone marks than a clear-cut total segregation. In this case, we were able to separate a very nuclease-resistant fraction of nucleosomes (fraction C5), which was enriched for H3K9me3 and relatively depleted for H3K4me3 or H4K8ac—suggesting this fraction was largely sourced from very dense heterochromatin. Earlier fractions show relatively equal abundance of all three marks, indicative of a mixture of nucleosome subtypes. The utility of this assay was the observed mobility of nucleosomes away from the C5 fraction after irradiation and the effect that either SNF2H or RNF20 depletion had on attenuating this.
IP and immunoblotting
Cells expressing CHD3.1, ACF1, SNF2H, RNF20, and/or RNF40 were resuspended in 3× PCV of NPB (10 mM Hepes, pH 7.9, 10 mM KCl, 1.0 mM CaCl2, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, and 0.1% [vol/vol] Triton X-100) containing protease inhibitors (Sigma-Aldrich), 0.5 µM MC-LR, 1 µM Wortmannin, 10 mM N-ethyl maleimide, 0.5 µM Trichostatin A, and 100 U/ml MNase (note: MC-LR is required to block protein phosphatase activity, Wortmannin is required to block in vitro DNA-dependent protein kinase/ATM activation, and Trichostatin A prevents in vitro histone deacetylation). Resuspended cells were incubated for 30 min at 37°C. An equal volume (i.e., 3× original PCV) of NSB (NPB + 600 mM NaCl, 2% [vol/vol] Triton X-100, and 2% [vol/vol] NP-40) was then added followed by sonication (1× 5 s) and clarification by centrifugation (10,000 g) for 10 min. 50 µg of extract was used for all immunoblotting. For IP of GFP/FLAG fusion proteins, 500 µg of extract was incubated with 10 µl bead-conjugated anti-GFP antibody (Sepharose; ab69314) or anti-DDDDK (FLAG) antibody (agarose; ab1240) for 4 h at 4°C with rotation. Immunoprecipitates were washed 3× with 1 ml ice-cold 1:1 NPB + NSB and resuspended in 2× SDS sample buffer. For histone modification immunoprecipitates, 2.5 µl of chromatin IP–grade antibody was incubated with 400 µg of extract for 4 h at 4°C with rotation.
Targeted LacO array heterochromatin relaxation assay
The RNF20 cDNA was amplified using 5′-CCGCTCGAGGAATGTCAGGAATTGGAAATAAAAGAG-3′ and 5′-CGGGATCCTCAACCAATGTAGATGCGATGAAAATC-3′ and inserted into mCherry-LacR-C1 (as in Coppotelli et al., 2013), with a CMV promoter and pmCherry-C1 as a backbone (Takara Bio Inc.; plasmid information found at Addgene). siRNA oligonucleotides (Thermo Fisher Scientific) were synthesized to Luciferase (5′-CGUACGCGGAAUACUUCGA-3′) or human SNF2H (pool of 5′-GGAUUAAACUGGCUCAUUU-3′ + 5′-GAGGAGAUGUAAUACCUUAUU-3′ + 5′-GGAAUGGUAUACUCGGAUA-3′ + 5′-GGGCAAAUAGAUUCGAGUA-3′) as described previously (Smeenk et al., 2013). GFP-LacR-stop was generated by replacing mCherry for GFP. Fusions of SNF2H-GFP-LacR were generated by exchanging GFP with GFP-LacR-stop in SNF2HWT-GFP and SNF2HK211R-GFP. LacR-tagged versions of CHD3 were generated by inserting CHD3WT or CHD3K767Q taken from FLAG-CHD3 plasmids in mCherry-LacR-C1. All constructs were verified by sequencing. U2OS 2-6-3 cells were transfected twice with 40 nM siRNA using RNAiMAX (Invitrogen). The next day, cells were cotransfected with RNF40Myc and either LacRmCherry or LacR-RNF20mCherry using Lipofectamine 2000 according to the manufacturer’s instructions. Cells were typically analyzed 60 h after the first transfection. Immunofluorescent labeling of cells was performed as outlined in the Materials and methods section Antibodies and IF, with specific details in Luijsterburg et al. (2012). Images of fixed samples were acquired on a wide-field fluorescence microscope (Axio Imager.M2; Carl Zeiss) equipped with a 100× Plan Apochromat (1.4 NA) oil-immersion objective (Carl Zeiss) and a metal-halide lamp (HXP 120) used for excitation. Images were captured using a camera (AxioCam MRm Rev.3; Carl Zeiss), recorded using ZEN 2012 software, and images were quantified using ImageJ software. In brief, 8-bit grayscale images were converted into colored images using a lookup table based on the pixel intensities in the grayscale image. The lookup table is shown next to the images and utilizes colors ranging from black to white to represent pixel intensities ranging from 0 (black) to 256 (white). A segmentation tool was used to measure the surface of the LacO array (based on the mCherry-LacR/GFP-LacR image), which was divided over the surface of the nucleus (based on the DAPI image) to express the relative size of the LacO array (Luijsterburg et al., 2012; Coppotelli et al., 2013). The mean LacO array size reflects the quantification of ∼100 cells from three independent experiments.
Targeted LacO array heterochromatin DSB-induction assay
siRNA oligonucleotides targeting Luciferase (5′-CGUACGCGGAAUACUUCGA-3′) or RNF20 (5′-CCAAUGAAAUCAAGUCUAA-3′) were purchased from Thermo Fisher Scientific. U2OS 2-6-3 cells stably expressing estrogen receptor (ER)–Fok1-mCherry-LacR–destabilization domain (DD; (obtained from R. Greenberg, University of Pennsylvania, Philadelphia, PA; Tang et al., 2013) were induced with 300 nM 4-OHT and 1 µM Shield-I for 5 h. Subsequently, cells were preextracted using 0.25% Triton X-100 in cytoskeletal buffer (10 mM Hepes, 300 mM sucrose, 100 mM NaCl, and 3 mM MgCl2) for 10 min, fixed with formaldehyde, and immunostained with the indicated antibodies. Expression of RNF20 or the ubiquitylation status of H2B was analyzed by immunoblotting with rabbit anti-RNF20 (A300-714A; Bethyl Laboratories, Inc.) and mouse anti-uH2B (MM-0029; NRO3; MediMabs) followed by secondary antibodies donkey anti–rabbit 700CW at 1:20,000 and donkey anti–mouse 800CW at 1:20,000 (Sigma-Aldrich) and detection using the infrared imaging system (Odyssey; LI-COR Biosciences).
Laser microirradiation
HeLa cells transfected with constructs (as outlined in the Materials and methods section Transient knockdown of protein expression and siRNA-resistant construct expression) were incubated with 10 µM BrdU for 16–24 h before laser microirradiation. DSB tracks were induced in live-imaged cells (kept at 37°C in a humidified environment at 5% CO2) using a 355-nm, 5-mW self-aligning solid-state diode laser at 33% power projected through an EC Plan Neofluor 100×/1.3 NA oil immersion objective, via a laser microdissection module (PALM MicroBeam; Carl Zeiss) on an Axio Observer.Z1 platform. Images were captured on a camera (AxioCam MRm Rev.3). Laser irradiation was controlled by RoboSoftware 4.5 (Carl Zeiss). Acquisition and analysis software used was Zen Pro. DNA damage equivalent at the site of the laser track is estimated to be approximately equivalent to 8–10 Gy IR, with doses estimated using the same methodology described in Bekker-Jensen et al. (2006). TIFF image files were captured every 30 s for ≤4.5 min and subsequently analyzed using ImageJ to quantify relative fluorescence intensity at tracks. 15–20 cells were irradiated and tracked over time per condition.
For 405-nm UV laser irradiation, experiments were performed as described by Kruhlak et al. (2006) using photosensitization with Hoechst dye before irradiation. In brief, HeLa cells pretreated with 2 µM Hoechst 33342 (Sigma-Aldrich) for 5 min were imaged at 37°C using a custom-built microscope (Cell Observer; Carl Zeiss/Intelligent Imaging Innovations), equipped with a heated CO2 incubator, diode-based lasers (405, 488, 561, and 633 nm), and a spinning-disk confocal scanning unit (CSU-X1; Yokogawa Electric Corporation) using a 40×, 1.4 NA immersion oil objective lens. UV laser damage was induced by a 100-mW, 405-nM diode laser using a Vector Scan Unit (Intelligent Imaging Innovations), where the effective light output was measured as ∼8 mW at the objective when using 100% power. A single line scan of the 405-nm laser at 70% power was sufficient to generate DNA DSBs as demonstrated by the rapid recruitment of KU70 (Andrin et al., 2012), which was estimated to be equivalent to ∼40–60 Gy cellular dose by the aforementioned method and in Bekker-Jensen et al. (2006). Images were captured every 10 s for 6 min and analyzed using an electron-multiplying charge-coupled device camera (Evolve; Photometrics) and SlideBook 5.5 software (Intelligent Imaging Innovations), respectively. A minimum of 25 cells were irradiated and tracked over time per condition.
Online supplemental material
Fig. S1 shows controls for SNF2H, ACF1, and RNF20 siRNA efficacy, additional DSB repair data for ATM, Artemis, and RNF168 mutated cells, methodology for heterochromatic DSB repair analysis, and the impact of SNF2H depletion of γ-H2AX foci size. Fig. S2 shows full controls for the (lack of) impact of SNF2H depletion on 53BP1 foci, controls for SNF2H, CHD3.1, ACF1, and RNF20 plasmid expression, and nuclear localization and the efficacy of plasmid siRNA resistance. Fig. S3 shows controls for LacR expression constructs, siRNA controls for CHD3.1 and KAP-1 knockdown, representative gels of MNase assays using primary fibroblasts, IP experiments using anti-H3K9me3 and anti-H4K8ac, alignments of the PHD fingers of ACF1 with CHD3.1, CHD4, and BPTF, and 405-nm laser microirradiation data with expression construct controls. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201405077/DC1.
Supplementary Material
Acknowledgments
We thank Mr. Guilherme Rodrigues (University of Calgary) for ACF1siRNAresistance and RNF20S(172/553)A mutagenesis, Dr. Yosef Shiloh for wild-type RNF20 and RNF40 constructs, Dr. Susan M. Janicki for U2OS 2-6-3 cells, and Dr. Roger A. Greenberg for U2OS 2-6-3 cells stably expressing ER-Fok1-mCherry-LacR-DD.
G. Dellaire is a Senior Scientist of the Beatrice Hunter Cancer Research Institute, and his laboratories are funded by a Discovery Grant (RGPIN 386049) from the Natural Science and Engineering Research Council. J.B. Pinder was supported by Beatrice Hunter Cancer Research Institute with funds provided by Harvey Graham Cancer Research Fund as part of The Terry Fox Strategic Health Research Training Program in Cancer Research at Canadian Institute of Health Research. The H. van Attikum laboratory is supported by a European Research Council Consolidator grant to H. van Attikum and a VENI grant from the Netherlands Organization for Scientific Research to M.S. Luijsterburg. The A.A. Goodarzi laboratory is supported by the Canadian Institutes of Health Research and Alberta Cancer Foundation. K. Klement was supported by a postdoctoral fellowship from the Engineered Air Chair in Cancer Research. A.A. Goodarzi is currently the Canada Research Chair for Genome Damage and Instability Disease, and this work was undertaken, in part, thanks to funding from the Canada Research Chairs program.
The authors declare no competing financial interests.
Author contributions: A.A. Goodarzi conceived and designed the study and provided Figs. 1 (A, D, and E), 2 (B–D and F–H), 3, 4, 6, 8, 10, S1 (A–F and H), S2 (A–F), and S3 (B–E). K. Klement and V. Del Nero developed and performed the analysis shown in Figs. 1 C and S1 G. K. Klement, V. Del Nero, C.S. Cena, C.M. Wintersinger, and A.A. Goodarzi contributed experimental data to means shown in Figs. 1 (B and F), 2 (A and E), and 9 (C and D) and prepared/validated the SNF2H, ACF1, and/or RNF20 expression constructs. K. Klement developed the system and provided the data in Fig. 9 (A and B). M.S. Luijsterburg and H. van Attikum provided Figs. 5, 7, and S3 A, constructed RNF20-LacR, SNF2H-LacR, and CHD3.1-LacR vectors, and provided the original SNF2H construct. J.B. Pinder and G. Dellaire provided data in Fig. S3 (F and G) and constructed ACF1 vectors. K. Klement, M.S. Luijsterburg, J.B. Pinder, G. Dellaire, H. van Attikum, and A.A. Goodarzi all contributed to manuscript preparation.
Footnotes
Abbreviations used in this paper:
- AID
- ACF1-interacting domain
- ATM
- ataxia telangiectasia mutated
- BPTF
- bromodomain PHD finger transcription factor
- CMV
- cytomegalovirus
- DD
- destabilization domain
- DSB
- double-strand break
- ER
- estrogen receptor
- HR
- homologous recombination
- HSB
- high salt buffer
- IF
- immunofluorescence
- IP
- immunoprecipitation
- IR
- ionizing radiation
- IRIF
- IR-induced foci
- LSB
- low salt buffer
- MC
- microcystin
- MNase
- micrococcal nuclease
- NCS
- neocarzinostatin
- NHEJ
- nonhomologous end joining
- NPB
- nucleosome preparation buffer
- NSB
- nucleosome solubilization buffer
- PCV
- packed cell volume
- PHD
- plant homeodomain
- SUMO
- small ubiquitin-like modifier
- WSTF
- Williams syndrome transcription factor
References
- Andrin C., McDonald D., Attwood K.M., Rodrigue A., Ghosh S., Mirzayans R., Masson J.Y., Dellaire G., and Hendzel M.J.. 2012. A requirement for polymerized actin in DNA double-strand break repair. Nucleus. 3:384–395 10.4161/nucl.21055 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., and Lukas J.. 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173:195–206 10.1083/jcb.200510130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher A., Birraux J., Tchouandong L., Barton O., Shibata A., Conrad S., Goodarzi A.A., Krempler A., Jeggo P.A., and Löbrich M.. 2009. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 28:3413–3427 10.1038/emboj.2009.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A., Längst G., Kehle J., Clapier C.R., Imhof A., Eberharter A., Müller J., and Becker P.B.. 2000. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 19:4332–4341 10.1093/emboj/19.16.4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunton H., Goodarzi A.A., Noon A.T., Shrikhande A., Hansen R.S., Jeggo P.A., and Shibata A.. 2011. Analysis of human syndromes with disordered chromatin reveals the impact of heterochromatin on the efficacy of ATM-dependent G2/M checkpoint arrest. Mol. Cell. Biol. 31:4022–4035 10.1128/MCB.05289-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N., Poot R.A., Kukimoto I., García-Jiménez C., Dellaire G., and Varga-Weisz P.D.. 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32:627–632 10.1038/ng1046 [DOI] [PubMed] [Google Scholar]
- Coppotelli G., Mughal N., Callegari S., Sompallae R., Caja L., Luijsterburg M.S., Dantuma N.P., Moustakas A., and Masucci M.G.. 2013. The Epstein-Barr virus nuclear antigen-1 reprograms transcription by mimicry of high mobility group A proteins. Nucleic Acids Res. 41:2950–2962 10.1093/nar/gkt032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher D., and Wellinger R.J.. 2010. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 6:e1001082 10.1371/journal.pgen.1001082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi A.A., and Jeggo P.A.. 2012a. The heterochromatic barrier to DNA double strand break repair: how to get the entry visa. Int. J. Mol. Sci. 13:11844–11860 10.3390/ijms130911844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi A.A., and Jeggo P.A.. 2012b. Irradiation induced foci (IRIF) as a biomarker for radiosensitivity. Mutat. Res. 736:39–47 10.1016/j.mrfmmm.2011.05.017 [DOI] [PubMed] [Google Scholar]
- Goodarzi A.A., and Jeggo P.A.. 2013. The repair and signaling responses to DNA double-strand breaks. Adv. Genet. 82:1–45 10.1016/B978-0-12-407676-1.00001-9 [DOI] [PubMed] [Google Scholar]
- Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., and Jeggo P.A.. 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 31:167–177 10.1016/j.molcel.2008.05.017 [DOI] [PubMed] [Google Scholar]
- Goodarzi A.A., Noon A.T., and Jeggo P.A.. 2009. The impact of heterochromatin on DSB repair. Biochem. Soc. Trans. 37:569–576 10.1042/BST0370569 [DOI] [PubMed] [Google Scholar]
- Goodarzi A.A., Kurka T., and Jeggo P.A.. 2011. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 18:831–839 10.1038/nsmb.2077 [DOI] [PubMed] [Google Scholar]
- Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., and Spector D.L.. 2004. From silencing to gene expression: real-time analysis in single cells. Cell. 116:683–698 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Guermah M., McGinty R.K., Lee J.S., Tang Z., Milne T.A., Shilatifard A., Muir T.W., and Roeder R.G.. 2009. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 137:459–471 10.1016/j.cell.2009.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M.J., Celeste A., and Nussenzweig A.. 2006. Spatio-temporal dynamics of chromatin containing DNA breaks. Cell Cycle. 5:1910–1912 10.4161/cc.5.17.3169 [DOI] [PubMed] [Google Scholar]
- Lan L., Ui A., Nakajima S., Hatakeyama K., Hoshi M., Watanabe R., Janicki S.M., Ogiwara H., Kohno T., Kanno S., and Yasui A.. 2010. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell. 40:976–987 10.1016/j.molcel.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Lee D.-H., Goodarzi A.A., Adelmant G.O., Pan Y., Jeggo P.A., Marto J.A., and Chowdhury D.. 2012. Phosphoproteomic analysis reveals that PP4 dephosphorylates KAP-1 impacting the DNA damage response. EMBO J. 31:2403–2415 10.1038/emboj.2012.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löbrich M., Shibata A., Beucher A., Fisher A., Ensminger M., Goodarzi A.A., Barton O., and Jeggo P.A.. 2010. γH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 9:662–669 10.4161/cc.9.4.10764 [DOI] [PubMed] [Google Scholar]
- Luijsterburg M.S., Acs K., Ackermann L., Wiegant W.W., Bekker-Jensen S., Larsen D.H., Khanna K.K., van Attikum H., Mailand N., and Dantuma N.P.. 2012. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J. 31:2511–2527 10.1038/emboj.2012.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Bailly D., Roche D., Montes de Oca R., Probst A.V., Vassias I., Dingli F., Lombard B., Loew D., Quivy J.P., and Almouzni G.. 2011. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat. Genet. 43:220–227 10.1038/ng.765 [DOI] [PubMed] [Google Scholar]
- Mansfield R.E., Musselman C.A., Kwan A.H., Oliver S.S., Garske A.L., Davrazou F., Denu J.M., Kutateladze T.G., and Mackay J.P.. 2011. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J. Biol. Chem. 286:11779–11791 10.1074/jbc.M110.208207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S.A., Baek S., Sung M.H., John S., Wiench M., Johnson T.A., Schiltz R.L., and Hager G.L.. 2014. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat. Struct. Mol. Biol. 21:73–81 10.1038/nsmb.2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin Y.M., Chalkley G.E., Kan T.W., Reddy B.A., Ozgur Z., van Ijcken W.F., Dekkers D.H., Demmers J.A., Travers A.A., and Verrijzer C.P.. 2012. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol. Cell. Biol. 32:675–688 10.1128/MCB.06365-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyal L., Lerenthal Y., Gana-Weisz M., Mass G., So S., Wang S.Y., Eppink B., Chung Y.M., Shalev G., Shema E., et al. 2011. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell. 41:529–542 10.1016/j.molcel.2011.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund A., Schubert T., Staege H., Kinkley S., Reumann K., Kriegs M., Fritsch L., Battisti V., Ait-Si-Ali S., Hoffbeck A.S., et al. 2012. SPOC1 modulates DNA repair by regulating key determinants of chromatin compaction and DNA damage response. Nucleic Acids Res. 40:11363–11379 10.1093/nar/gks868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Kato A., Kobayashi J., Yanagihara H., Sakamoto S., Oliveira D.V., Shimada M., Tauchi H., Suzuki H., Tashiro S., et al. 2011. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell. 41:515–528 10.1016/j.molcel.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Noon A.T., Shibata A., Rief N., Löbrich M., Stewart G.S., Jeggo P.A., and Goodarzi A.A.. 2010. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol. 12:177–184 10.1038/ncb2017 [DOI] [PubMed] [Google Scholar]
- Oliveira D.V., Kato A., Nakamura K., Ikura T., Okada M., Kobayashi J., Yanagihara H., Saito Y., Tauchi H., and Komatsu K.. 2014. Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J. Cell Sci. 127:763–772 10.1242/jcs.135855 [DOI] [PubMed] [Google Scholar]
- Pessina F., and Lowndes N.F.. 2014. The RSF1 histone-remodelling factor facilitates DNA double-strand break repair by recruiting centromeric and Fanconi Anaemia proteins. PLoS Biol. 12:e1001856 10.1371/journal.pbio.1001856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price B.D., and D’Andrea A.D.. 2013. Chromatin remodeling at DNA double-strand breaks. Cell. 152:1344–1354 10.1016/j.cell.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E., Kühne M., Rief N., Doherty A., Smith G.C., Recio M.J., Reis C., Dahm K., Fricke A., Krempler A., et al. 2004. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to γ-H2AX foci. Mol. Cell. 16:715–724 10.1016/j.molcel.2004.10.029 [DOI] [PubMed] [Google Scholar]
- Rowbotham S.P., Barki L., Neves-Costa A., Santos F., Dean W., Hawkes N., Choudhary P., Will W.R., Webster J., Oxley D., et al. 2011. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell. 42:285–296 10.1016/j.molcel.2011.02.036 [DOI] [PubMed] [Google Scholar]
- Ruthenburg A.J., Li H., Milne T.A., Dewell S., McGinty R.K., Yuen M., Ueberheide B., Dou Y., Muir T.W., Patel D.J., and Allis C.D.. 2011. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 145:692–706 10.1016/j.cell.2011.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D.C., Friedman J.R., and Rauscher F.J. III. 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15:428–443 10.1101/gad.869501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D.C., Ayyanathan K., Negorev D., Maul G.G., and Rauscher F.J. III. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16:919–932 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Böckler B., and Lehner B.. 2012. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 488:504–507 10.1038/nature11273 [DOI] [PubMed] [Google Scholar]
- Smeenk G., Wiegant W.W., Marteijn J.A., Luijsterburg M.S., Sroczynski N., Costelloe T., Romeijn R.J., Pastink A., Mailand N., Vermeulen W., and van Attikum H.. 2013. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J. Cell Sci. 126:889–903 10.1242/jcs.109413 [DOI] [PubMed] [Google Scholar]
- Soria G., Polo S.E., and Almouzni G.. 2012. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol. Cell. 46:722–734 10.1016/j.molcel.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Stanley F.K., Moore S., and Goodarzi A.A.. 2013. CHD chromatin remodelling enzymes and the DNA damage response. Mutat. Res. 750:31–44 10.1016/j.mrfmmm.2013.07.008 [DOI] [PubMed] [Google Scholar]
- Tang J., Cho N.W., Cui G., Manion E.M., Shanbhag N.M., Botuyan M.V., Mer G., and Greenberg R.A.. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20:317–325 10.1038/nsmb.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S.D., Li H., Ruthenburg A.J., Allis C.D., and Patel D.J.. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14:1025–1040 10.1038/nsmb1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjeertes J.V., Miller K.M., and Jackson S.P.. 2009. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28:1878–1889 10.1038/emboj.2009.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D., Erdel F., Bouazoune K., Silberman D.M., Zhong L., Mulligan P., Sebastian C., Cosentino C., Martinez-Pastor B., Giacosa S., et al. 2013. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell. 51:454–468 10.1016/j.molcel.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbine L., Brunton H., Goodarzi A.A., Shibata A., and Jeggo P.A.. 2011. Endogenously induced DNA double strand breaks arise in heterochromatic DNA regions and require ataxia telangiectasia mutated and Artemis for their repair. Nucleic Acids Res. 39:6986–6997 10.1093/nar/gkr331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Li H., Shechter D., Ahn S.H., Fabrizio L.A., Erdjument-Bromage H., Ishibe-Murakami S., Wang B., Tempst P., Hofmann K., et al. 2009. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 457:57–62 10.1038/nature07668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., and Shiloh Y.. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8:870–876 10.1038/ncb1446 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.