Interactions between ankyrin-B and both dynactin and phosphatidylinositol 3-phosphate lipids promote fast axonal transport of organelles.
Abstract
Axon growth requires long-range transport of organelles, but how these cargoes recruit their motors and how their traffic is regulated are not fully resolved. In this paper, we identify a new pathway based on the class III PI3-kinase (PIK3C3), ankyrin-B (AnkB), and dynactin, which promotes fast axonal transport of synaptic vesicles, mitochondria, endosomes, and lysosomes. We show that dynactin associates with cargo through AnkB interactions with both the dynactin subunit p62 and phosphatidylinositol 3-phosphate (PtdIns(3)P) lipids generated by PIK3C3. AnkB knockout resulted in shortened axon tracts and marked reduction in membrane association of dynactin and dynein, whereas it did not affect the organization of spectrin–actin axonal rings imaged by 3D-STORM. Loss of AnkB or of its linkages to either p62 or PtdIns(3)P or loss of PIK3C3 all impaired organelle transport and particularly retrograde transport in hippocampal neurons. Our results establish new functional relationships between PIK3C3, dynactin, and AnkB that together promote axonal transport of organelles and are required for normal axon length.
Introduction
Long-range axonal transport of organelles mediated by microtubule-based motors is essential for axonal morphogenesis and maintenance (Hirokawa et al., 2010). Moreover, defects in axonal transport contribute to the pathogenesis of neurodegenerative diseases (Hirokawa et al., 2010; Goldstein, 2012; Millecamps and Julien, 2013). Despite remarkable progress in the elucidation of how motor proteins target to their cargoes in neurons, a comprehensive explanation of the mechanisms that dictate motor–cargo specificity remains elusive, and the motor–cargo interactions involving cytoplasmic dynein are the least well understood (Akhmanova and Hammer, 2010). Although multiple kinesin motors and associated factors provide the basis for both cargo specificity and redundancy in motor recruitment for anterograde transport, a single dynein is responsible for the majority of minus end–directed motion (Kardon and Vale, 2009). In a few cases, dynein selectively binds cargoes through direct association of some of its subunits with endomembranes (Tai et al., 1999; Fejtova et al., 2009; Tan et al., 2011). More commonly, dynein associates with membranes through adaptors (Cai et al., 2010; Horgan et al., 2010; Splinter et al., 2012; Drerup and Nechiporuk, 2013).
The dynactin complex is one of the best studied dynein cargo adaptors, and it is an essential partner in organelle transport as well as other cellular functions (Schroer and Sheetz, 1991; Holleran et al., 1996; Waterman-Storer et al., 1997; Muresan et al., 2001; Schroer, 2004; Johansson et al., 2007; van Spronsen et al., 2013). The pointed-end module of dynactin containing Arp11, p62 (also known as Dyn4), p25, and p27 subunits facilitates dynactin–dynein recruitment to cargoes (Zhang et al., 2011; Yeh et al., 2012). However, the mechanism for the coupling of these dynactin subunits to membranes remains to be determined. A current view is that multiple adaptor proteins and cofactors facilitate the coupling of the dynein–dynactin retrograde motor complex to specific membrane cargoes (Akhmanova and Hammer, 2010). However, cargo–motor recognition systems based predominantly on adaptors do not explain how the adaptors themselves are targeted to membrane organelles.
Ankyrin-B (AnkB) is a member of the ankyrin family of adaptors that contribute to the assembly of specialized plasma membrane domains (Bennett and Lorenzo, 2013). AnkB, in contrast to other ankyrins, can associate with intracellular membranes as a result of autoinhibition of its plasma membrane interactions (He et al., 2013). In striated muscle, AnkB simultaneously interacts with dystrophin, the p62 subunit of the dynactin complex, and microtubules and is required for the association of these proteins and the β-dystroglycan complex with costameres (Ayalon et al., 2008, 2011). Acute depletion of either AnkB or p62 in adult mouse skeletal muscle, or expression of mutations of AnkB and of dynactin-4 that selectively impair binding between these proteins destabilizes costamere-associated microtubules and causes muscle fragility (Ayalon et al., 2008, 2011).
Here, we report that AnkB is a general physiological adaptor for dynactin in the nervous system, linking this complex to multiple axonally transported organelles through direct binding to p62, and that this interaction is required for efficient cargo transport and for normal axonal elongation. We further find that AnkB associates with intracellular membranes by binding to phosphoinositide (PtdIns) 3-phosphate (PtdIns(3)P) lipids through an as-yet-uncharacterized basic pocket in its ZU5 domain. Moreover, we determined that the PtdIns(3)P lipids responsible for dynactin-AnkB membrane attachment in neurons are produced by class III PI3 kinase (PIK3C3). Together, these experiments provide evidence for an AnkB-based pathway required for normal axon length that links dynactin–dynein-based microtubule transport with PtdIns’s.
Results
Formation of long axon tracts and axonal elongation require AnkB
AnkB-null (AnkB−/−) mice lacking expression of 440- and 220-kD AnkB isoforms in the brain (Fig. S1, A and B) exhibit severe malformation of multiple brain structures involving long axon tracts and experience early neonatal mortality (Scotland et al., 1998). To better characterize these morphological defects, we visualized fiber tracts in postnatal day (PND) 2 control (AnkB+/+) and AnkB−/− brains using diffusion tensor imaging (DTI), a magnetic resonance imaging-based technique that measures the displacement of water molecules on the micrometer scale around white matter tracts as a function of spatial location (Assaf and Pasternak, 2008). DTI analysis allows delineation of white matter structures in the brain and 3D reconstruction of white matter bundles and fiber connectivity (Mori et al., 1999; Jiang and Johnson, 2011). Fractional anisotropy (FA) maps derived from DTI images show the absence of long axonal projections, including the corpus callosum (Fig. 1 A, arrowheads) and anterior commissure (Fig. 1 A, asterisks) in AnkB−/− animals (Fig. 1 A and Fig. S1 C). Analysis of 3D-reconstructed fibers showed a major reduction in the length and the number of white matter tracts in AnkB−/− brains (Fig. 1 B and Fig. S1 D). Additionally, a time course of axonal growth in neonatal cultures of control and AnkB−/− hippocampal neurons revealed that day in vitro (DIV) 3 AnkB−/− neurons specified axons normally as shown by the localization of ankyrin-G to axon initial segments but failed to elongate their axons to the same length as control neurons (Fig. 1, C and D), whereas their dendritic length was unaffected (Fig. 1 E). These results suggest that AnkB is not required for the initial stages of neuronal polarization but rather plays a role in the growth and/or the stability of axons.
Figure 1.
AnkB is required for axonal growth in mouse brains and cultured hippocampal neurons. (A) FA maps of mouse brains and 3D images of white matter tracts colored by FA values. Positions of the corpus callosum (arrowheads, red boxes) and anterior commissures (asterisks, white boxes) are indicated. (B) Size distribution of axonal tracts in control and AnkB−/− brains (n = 4 mice). The data shown represent the mean of four independent DTI scans per each genotype performed as part of one single experiment. (C) Time course of axonal growth in hippocampal neurons. Axon initial segments were labeled by ankyrin-G staining (AnkG). Axons and dendrites were labeled by neurofilament and MAP2 staining, respectively. (D and E) Quantification of axonal length at different DIVs (D) and dendritic branches length at DIV8 (E). Data were compiled from a single representative experiment out of three repeats. For the experiment shown, data represent means ± SEM; n = 40 cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001, two-tailed t test.
Unperturbed organization of the axonal cytoskeleton and microtubule dynamics in AnkB-null neurons
We initially hypothesized that the reduced axon length observed in AnkB−/− brains and cultured neurons was caused by impaired assembly of the axonal membrane skeleton, composed of actin, spectrin, and adducin, recently imaged in axons using 3D stochastic optical reconstruction microscopy (STORM; Xu et al., 2013). However, 3D-STORM images showed an unperturbed arrangement of spectrin, with a 180-nm periodic spacing in both control and AnkB−/−axons (Fig. 2, A and B). Similarly, we did not detect changes in axon initial segments of DIV8 AnkB−/− neurons or in expression of major axonal cytoskeleton proteins (Fig. 2, C and D). Moreover, microtubule dynamics, which are proposed to be essential for axonal elongation (Conde and Cáceres, 2009), were preserved upon AnkB loss, with no change in mean number and mean velocity of EB1 as moving comets on microtubule plus ends (Fig. 3, A–C).
Figure 2.
Normal axonal cytoskeleton structure in AnkB-deficient neurons. (A and B, top) 3D-STORM images of β2-spectrin structure in DIV10 AnkB+/+ (A) and AnkB−/− (B) axons. Violet to red color-coded z position is shown, representing closest to and farthest from the substratum, respectively. (middle) Graphs show the distribution of molecules along the axon long axis. (bottom) Fourier transform of the 1D localization distributions shows a fundamental frequency of 180 nm−1 and an overtone. Data are representative of axonal STORM images and analysis from one experiment out of three repeats. For the experiment shown, n = 10. (C) Expression and localization of total and 440-kD AnkB, β2-spectrin, and ankyrin-G proteins (AnkG) and MAP2 in DIV8 AnkB+/+ and AnkB−/− hippocampal neurons. Arrowheads indicate axon initial segments. Color arrows represent the direction of coordinate planes in the rotated 3D image. (D) Immunoblots show ankyrin-G (all neuronal isoforms), β2-spectrin, β4-spectrin, and actin expression in PND2 brains. All experiments were repeated three times.
Figure 3.
Normal microtubule dynamics and stability in AnkB-deficient neurons. (A) Kymographs illustrate the movement of EB1-GFP comets in AnkB+/+ and AnkB−/− hippocampal axons. (B and C) Quantification of the mean number (B) and mean velocity (C) of EB1-GFP comets in AnkB+/+ (n = 25) and AnkB−/− (n = 29) axons from five independent preparations. For all graphs, data represent means ± SEM and were obtained from one experiment out of three repeats. (D) Representative images show immunofluorescent staining of DIV10 control and AnkB−/− neurons with antibodies to either acetylated (left), detyrosinated (center), or polyglutamylated (right) tubulin subunits.
Similarly, we observed no differences in microtubules’ posttranslational modifications assessed by the levels and localization of acetylated, detyrosinated, and glutamylated tubulin between control and AnkB−/− axons (Fig. 3 D). These results indicate that the axonal growth defects in AnkB−/− neurons are not caused by changes in the dynamics and stability of microtubules or the overall organization of the spectrin–actin skeleton.
AnkB regulates axonal transport of multiple cargoes
Neurons rely on transport of organelles to build and maintain dendrites and axons (Hollenbeck and Bray, 1987; Martenson et al., 1993; Zhou et al., 2012; van Spronsen et al., 2013). Given that AnkB binds to the p62 subunit of the dynactin complex and that AnkB also associates with intracellular membranes (Ayalon et al., 2008, 2011; He et al., 2013), we evaluated whether the axonal growth phenotypes in AnkB−/− mice were associated with impaired cargo motility in cultured hippocampal neurons using time-lapse video microscopy. In control axons, the synaptic vesicle proteins synaptophysin-YFP and synaptotagmin-GFP moved bidirectionally with high processivity (Fig. 4 A, Fig. S2 A, and Video 1). In contrast, AnkB−/− axons exhibited a significant increase in the percentage of synaptic vesicles that were either stationary or trapped within axonal swellings all along the axons (Fig. 4 A, arrowheads; and Video 1). Those vesicles that retained motility also reversed direction frequently (Fig. 4 A and Fig. S2, A and D). Slower velocities and shorter distances traveled by both synaptophysin-YFP and synaptotagmin-GFP vesicles compared with controls provided additional evidence for impaired bidirectional transport of synaptic vesicles in AnkB−/− axons (Fig. 4, B and C; and Fig. S2, B and C). Similarly, we observed that AnkB deficiency specifically affected the retrograde motility of dsRed mitochondria, Rab5-RFP endosomes, and LAMP1-GFP lysosomes in axons. These effects were indicated by a significant decrease in the percentage of dynamic organelles and the presence of numerous cargo-filled swellings that were present all along the axon but significantly concentrated at the axonal terminal (Fig. S2 E, Fig. S3, and Videos 3, 5, and 6). Furthermore, the retrograde velocity and run length of all these cargoes decreased significantly (Fig. 4, B and C; Fig. S2, F and G; and Fig. S3, C, D, H, and I). Thus, the fast axonal transport of multiple classes of organelle cargoes depends on AnkB.
Figure 4.
AnkB interaction with p62 regulates the motility of multiple axonal cargoes and growth of axons. (A) For each genotype the top represents the distribution of synaptophysin-YFP vesicles and 220-kD WT or DD1320AA AnkB-mCherry in DIV7 axons. Arrowheads indicate large axonal aggregates. (middle) Kymograph analysis of the motion of all particles. (bottom) Corresponding color-coded trajectories for a subset of YFP-positive particles show static vesicles (blue) and anterograde (green)- and retrograde (red)-moving vesicles. (B and C) Axonal velocity (B) and run length (C) for the indicated cargoes. The data shown are from a single representative experiment out of five repeats. For the experiment shown, data were computed from n = 10 axons from five independent neuronal preparations (for synaptophysin-YFP, anterograde [n = 175] and retrograde [n = 150] YFP-particles; for other organelles, see supplemental figure legends). (D) Representative images of DIV8 hippocampal neurons expressing dsRed alone or in combination with WT or DD1320AA AnkB-GFP (arrowheads indicate axons). (E) Quantification of axonal length at DIV8 (n = 30 cells). The data shown are from a single representative experiment out of three repeats. Data represent means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, one-way ANOVA with Tukey’s post-test.
To assess whether AnkB itself is axonally transported, we imaged the dynamic behavior of 220-kD AnkB expressed in AnkB−/− neurons. We observed that axonal AnkB-mCherry and AnkB-GFP proteins were motile (Videos 2 and 4–7, top). Initially, we tried to identify the subcellular compartment that AnkB associates with. However, fluorescently tagged AnkB did not completely localize to one particular organelle and instead was associated and cotransported with all the aforementioned cargoes (Videos 2 and 4–7, bottom). In the case of mitochondria, we observed that instead of decorating the entire mitochondrial surface, AnkB localization was limited to a few points of contact (Video 4). Importantly, AnkB’s motility and its association with axonal cargoes were independent of the fluorescent tag used to label and track these proteins (Video 7). Using rescue experiments, we evaluated whether the reported association between AnkB and the dynactin subunit p62 played a role in axonal transport (Ayalon et al., 2008, 2011). The DD1320AA AnkB-mCherry mutant, which binds β2-spectrin but is unable to associate with p62 (Ayalon et al., 2011; Fig. 5, C and F), also failed to rescue transport deficits of various cargoes in AnkB−/− axons, whereas wild-type (WT) AnkB was active (Fig. 4, B and C; Fig. S2; Fig. S3; and Videos 2 and 4–7, bottom).
Figure 5.
AnkB interaction with the dynactin subunit p62 through DD1320 residues recruits dynein–dynactin to organelle membranes. (A and B) Immunoprecipitation from adult mouse brain homogenates show 220-kD AnkB association with DIC and the dynactin subunits p50 and p62. (C) Coimmunoprecipitation experiments from COS7 cells expressing p62-HA and WT or DD1320AA GFP-AnkB proteins. (D) The amounts of membrane-associated AnkB, β2-spectrin, dynein, dynactin, and kinesin were evaluated by immunoblotting after subcellular fractionation of brain homogenates on a sucrose step gradient. Rab5, LAMP1, and synaptophysin are membrane controls. GAPDH is a loading control. (E) Time-lapse images and kymographs of p62-GFP dynamics in axons of control or AnkB-null neurons. Data also show results of rescue experiments in which WT or mutant DD1320AA AnkB-mCherry was coexpressed with p62-GFP in axons. (F) Immunoprecipitation from COS7 cells expressing GFP–β2-spectrin and AnkB-HA proteins. IP, immunoprecipitation; IB, immunoblotting.
To further examine whether transport deficits contributed to axonal growth defects, we determined whether WT and DD1320AA AnkB could rescue the reduced axonal length of AnkB−/− neurons. Consistent with previous observations, DIV 8 control axons were on average markedly longer (207 ± 3 µm) than AnkB−/− axons (110 ± 3 µm). WT 220-kD AnkB partially rescued lengths to 178 ± 2 µm, whereas DD1320AA AnkB-mCherry failed (117 ± 5 µm) to restore axonal length (Fig. 4, D and E). These results demonstrate that AnkB requires p62 binding activity to promote axonal organelle transport and normal axonal growth in cultured neurons.
Because 220-kD AnkB also localizes to dendrites (Fig. 2 C and Fig. 4 D), we assessed whether dendritic cargoes require AnkB. The bidirectional motility of dendritic transferrin receptor (TfR; TfR-YFP) and TGN38-GFP (not depicted) along dendritic shafts and spines was similar in both AnkB−/− and control neurons (Fig. S4). Thus, AnkB is necessary for membrane transport in axons but not for these particular cargoes in dendrites. Interestingly, 440-kD AnkB, the major splice variant in the embryonic and premyelinated brain, contains a single additional giant exon and is exclusively localized to axons (Fig. S1 A and Fig. 2 C; Kunimoto, 1995; Scotland et al., 1998). This large AnkB isoform shares all the functional domains of the 220-kD polypeptide, including the dynactin binding site. In vivo, 440-kD AnkB targets to axons through yet to be identified determinants in its giant exon, and it could specifically promote axonal transport. However, neurons did not express the GFP-tagged full-length 440-kD AnkB cDNA at sufficient levels for live imaging, which precluded further evaluation of the dynamic behavior of this polypeptide.
AnkB recruits dynactin and dynein to membrane cargoes in neurons
AnkB interacts with the dynactin subunit p62 in yeast two-hybrid assays as well as in binding assays using purified proteins (Ayalon et al., 2008, 2011). We verified a p62–AnkB interaction in neurons by coimmunoprecipitation of p62 with an antibody against AnkB from brain homogenates (Fig. 5 A). Similarly, AnkB was coimmunoprecipitated using antibodies against p62 (Fig. 5, A and B) as well as with antibodies against dynactin p50 and dynein intermediate chain (DIC; Fig. 5 B). Coimmunoprecipitation using lysates from transfected COS7 cells expressing both AnkB-GFP and p62-HA showed binding of AnkB to p62. An anti-HA antibody coimmunoprecipitated WT AnkB-GFP with p62-HA but failed to pull down GFP-tagged AnkB harboring the DD1320AA mutation (Fig. 5 C). However, the DD1320AA mutation that eliminated AnkB binding to p62-HA did not affect interaction with GFP–β2-spectrin (Fig. 5 F). Despite these interactions, AnkB deletion did not noticeably affect expression levels of p62, p150Glued, p50, Arp1, or dynein heavy chain (DHC) proteins based on immunoblot analysis of brain homogenates of AnkB−/− and control littermates (Fig. 5 D, postnuclear supernatant [PNS] lanes).
To directly evaluate whether AnkB interaction with p62 contributes to the recruitment of dynactin and dynein to organelles, we isolated total membrane fractions from brain homogenates from PND1 control and AnkB−/− mice by upward flotation in a sucrose step gradient. AnkB primarily associates with brain membranes under these conditions (Fig. 5 D). Strikingly, multiple dynactin subunits, including p50, p62, Arp1, and p150Glued, as well as DHC, were markedly reduced in membrane fractions isolated from AnkB−/− compared with WT brains (Fig. 5 D). In contrast, other membrane-associated proteins, including synaptophysin, Rab5, and LAMP1, as well as β2-spectrin and kinesin heavy chain, remained normally associated with membrane fractions from AnkB-null brains (Fig. 5 D).
We further evaluated the significance of the AnkB interaction with p62 for dynactin recruitment to membrane cargo by assessing the dynamic localization of p62-GFP in WT and AnkB−/− axons. Although p62-GFP was preferentially associated with retrograde-directed cargoes in control axons, it exhibited a marked loss of its association with particulate structures in the absence of AnkB (Fig. 5 E and Video 8). Strong colocalization of 220-kD AnkB-mCherry with p62-GFP in motile organelles (Pearson’s colocalization coefficient = 0.76) indicates that AnkB cotransports with p62 in the same dynamic compartments (Video 8). Moreover, WT AnkB, but not DD1320AA AnkB lacking p62-binding activity, effectively restored p62-GFP association with motile cargoes (Fig. 5 E and Video 8). The minor p62-GFP population associated with cargo in axons expressing DD1320AA AnkB suggests that other AnkB interactions, including those with β2-spectrin, which remains associated with membranes in the absence of AnkB (Fig. 5 D), may provide an alternative mechanism for coupling dynactin to intracellular membranes (Holleran et al., 1996; Muresan et al., 2001). The distinct location of binding sites for β2-spectrin in the ZU5N domain and for p62 in the urokinase-type plasminogen activator (UPA) domain in the atomic structure of AnkB’s ZU5N-ZU5C-UPA supramodule suggests that these proteins bind AnkB independently (Wang et al., 2012). The colocalization and cotransport of AnkB and p62 on particulate structures suggest a novel mechanism for targeting dynactin to different subcellular membranes.
AnkB binds axonal cargoes through a basic pocket on its ZU5C domain
We next sought to identify the cargo-interacting site of AnkB. We began by evaluating association of synaptophysin-dsRed with either the N-terminal ankyrin repeats or the C-terminal domains (ZU5N-ZU5C-UPA-death domain [DD]-Ct) tagged with GFP. Although full-length 220-kD AnkB associated with motile synaptic vesicles in axons, strikingly, the ankyrin repeats, which bind to multiple membrane proteins (Bennett and Lorenzo, 2013), associated poorly (Fig. S5 A). In contrast, the ZU5N-ZU5C-UPA-DD-Ct-GFP portion of AnkB (Fig. S5 A) or the same protein lacking the DD (not depicted) bound and cotransported with cargoes. Because the cargo-binding region in AnkB does not involve ankyrin repeats, we postulated that its coupling to membranes occurs through membrane components other than membrane-spanning proteins that are shared across cargoes. We initially evaluated AnkB binding to membrane lipids using a protein–lipid overlay assay. His-tagged C-terminal domains of AnkB (WT ZU5-DD) bound strongly and specifically to PtdIns(3)P and weakly to PtdIns 4-phosphate (PtdIns(4)P; Fig. 6, A and C; and Fig. S5, B–F) but not to other PtdIns’s. Binding essays using polyPIPosomes and PtdIns phosphate (PtdInsP)–coated beads (Fig. 6 D and Fig. S5, G and H) confirmed binding to PtdIns(3)P as well as a weaker interaction with PtdIns(4)P.
Figure 6.
AnkB’s ZU5C domain interacts with membrane PtdIns(3)P. (A) Coomassie blue staining and immunoblots of purified AnkB proteins. (B) Image shows the known crystal structure of the ZU5N-ZU5C-UPA-DD supermodule of AnkB and highlights a positively charged surface (yellow) within the ZU5C domain. High magnification view of the boxed region depicts all residues mutated for analysis, including those found to be essential for AnkB binding to PtdIns(3)P lipids (red dotted lines). (C) Protein–lipid overlay assay showing affinity of the ZU5-DD region for PtdIns(3)P. IB, immunoblot; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PA, phosphatidic acid; PS, phosphatidylserine; S1P, sphingosine-1-phosphate. (D) Binding of WT 220-kD AnkB, WT ZU5N-ZU5C-UPA-DD, and the ZU5C mutant proteins to polyPIPosomes bearing the indicated PtdInsPs measured through ELISA. The data shown are for one experiment out of three repeats. Data represent mean ± SEM. RU, relative unit. (E) Time-lapse image and kymographs of AnkB-mCherry proteins and synaptophysin-YFP localization and dynamics in axons.
Although AnkB lacks a known PtdInsP-binding domain (Lemmon, 2008), its ZU5N-ZU5C-UPA domains are folded into a supermodule with a cluster of highly conserved positively charged residues within the ZU5C domain (Fig. 6 B, yellow contour; Wang et al., 2012). We tested whether these basic residues are required to bind to PtdInsPs. His-tagged ZU5N-ZU5C-UPA-DD with K1157E, R1158E, R1160E, R1193E, R1194E, and K1195E amino acid substitutions (polybasic motif ZU5C mutant) showed nearly complete loss of binding to PtdIns(3)P and PtdIns(4)P (Fig. 6 D and Fig. S5, B–F and H) but bound normally to p62 and β2-spectrin (Fig. S5, I and J). In addition, we found that individual R1194E or K1195E substitutions (Fig. 6 B, red dotted circles in inset) were sufficient to impair AnkB association with PtdIns(3)P (Fig. 6, C and D). Moreover, both the polybasic motif ZU5C mutant and the single R1194E mutation in either full-length or ZU5N-ZU5C-UPA-DD-Ct AnkB abolished their association with synaptic vesicles in live imaging (Fig. 6 E).
Post facto molecular docking simulations are consistent with the interaction of PtdIns(3)P with the proposed ZU5C binding sites of native AnkB (see Fig. 10). The lowest energy pose generated by a Lamarckian genetic algorithm depicted the putative binding of the PtdIns(3)P’s inositol 1,3-bisphosphate head group to K1157, R1194, and K1195 as well as to T1156 and H1197 (see Fig. 10, purple contour). Modeling thus supports binding disruption from abrogation of multiple hydrogen bonds and salt bridges of the mutated residues. Stable binding of PtdIns(3)P in biological membranes with most of its effectors typically occurs through association with multiple sites or avidity effects (Schink et al., 2013). Our results suggest that simultaneous interactions of PtdIns(3)P with multiple residues within the positively charged pocket in the ZU5C domain are essential for AnkB recruitment to membranes.
Figure 10.
Model for AnkB interactions with dynactin and axonal cargoes. (left) Proposed model for AnkB coupling of the dynein–dynactin motor complex to cargoes at approximate scale. (center) Homology model of AnkB’s ZU5N-ZU5C-UPA-DD region highlighting binding sites (spheres) for the dynactin subunit p62 and for PtdIns(3)P lipids in membrane cargoes. (right) Molecular docking simulation of PtdIns(3)P binding to ZU5C (dotted lines show interacting residues). Mutated (yellow contour) and additional potential interacting (purple contour) residues are highlighted.
Deletion of PIK3C3 disrupts AnkB association with cargoes and affects long-range multiorganelle transport in axons
PtdIns(3)P is enriched in membranes of the endolysosomal–autophagosomal pathway, where it recruits effector protein complexes to restricted membrane domains in a reversible manner and regulates membrane dynamics, vesicular trafficking, and cell signaling (Di Paolo and De Camilli, 2006; Devereaux et al., 2013). However, coexpression of the PtdIns(3)P biosensor GFP-2×FYVEEEA1 (Schink et al., 2013) with markers for axonal cargoes revealed colocalization of GFP-2×FYVEEEA1 not only with endosomes and lysosomes but also with synaptic vesicles (Fig. 7, A–C, arrowheads) and, to a lesser extent, with mitochondria in which the association was restricted to a few points of contact (Fig. 7 D, arrowheads). Although present at different levels, these data show that PtdIns(3)P is an intrinsic component of various axonal organelle membranes, consistent with a previous study (Gillooly et al., 2000).
Figure 7.
PtdIns(3)P lipids are found in multiple organelle membranes. (A–D) The GFP-2×FYVE probe was used as a marker for the localization of cellular PtdIns(3)P lipids. Representative images show GFP-2×FYVE localization to PtdIns(3)P-containing membranes of synaptic vesicles labeled by synaptophysin-dsRed (A), lysosomes labeled by LAMP1-GFP (B), early endosomes labeled by Rab5-RFP (C), and mitochondria labeled by mito-dsRed (D). Insets show high magnification views of the boxed regions. Arrowheads indicate areas of colocalization. Data shown are for one experiment out of three repeats.
Cellular PtdIns(3)P is generated by three PtdIns kinases by either direct phosphorylation of PtdIns precursors or production of PtdIns(3,4)P2 and PtdIns(3,4, 5)P3 species, which are subsequently modified by PtdIns phosphatases (Schink et al., 2013). Interestingly, neuronal-specific knockdown of the class III PI3K (PIK3C3, also known as Vps34), which directly yields PtdIns(3)P, results in both the loss and the shortening of axons and in rapid neuronal degeneration, which the authors attributed to defective endolysosomal pathways (Zhou et al., 2010; Wang et al., 2011).
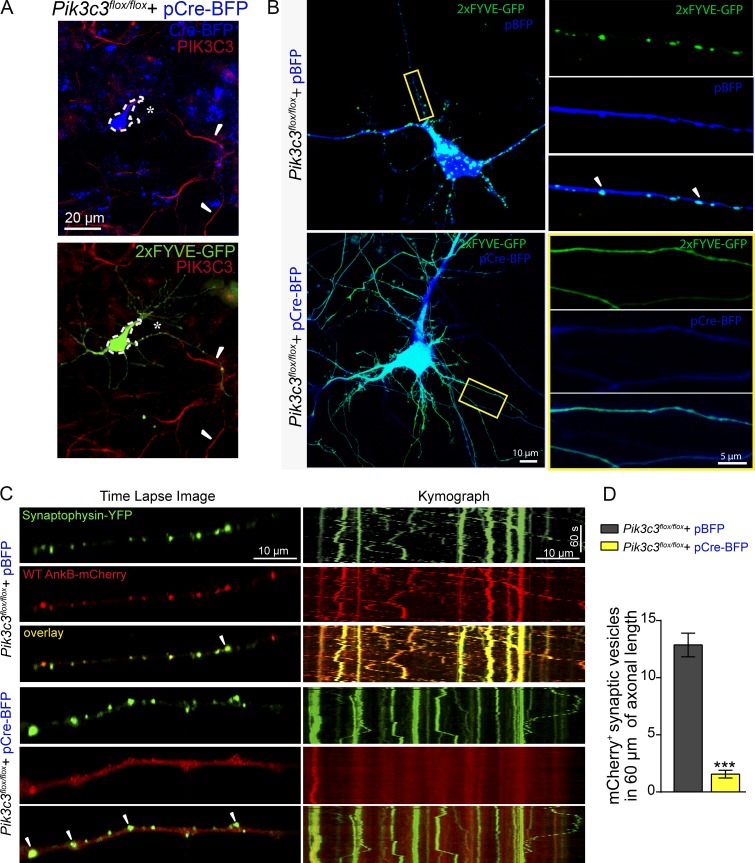
To directly evaluate the role of PIK3C3 in AnkB’s association with membrane cargo and overall axonal organelle motility, we transfected hippocampal neurons from Pik3c3flox/flox mice with either a pCAG-Cre-BFP plasmid encoding the Cre-recombinase enzyme (to induce PIK3C3 knockdown) or control pCAG-BFP. Expression of the Cre-BFP fusion protein caused a marked reduction in PIK3C3 labeling in neuronal processes (Fig. 8 A, asterisks) when compared with untransfected neurons (Fig. 8 A, arrowheads). An efficient depletion of PtdIns(3)P in Cre-BFP–expressing neurons was demonstrated by a major loss of GFP-2×FYVEEEA1 signal from punctate structures in the cell body and processes and a switch to a diffuse pattern (Fig. 8, A and B). Consistent with our hypothesis, reduced levels of neuronal PtdIns(3)P abolished AnkB association with synaptic vesicles (Fig. 8, C and D; and Video 10). Moreover, knockout of PI3KC3 also disrupted the axonal transport of synaptophysin-YFP vesicles, which in turn resulted in a significant increase in both the number of axonal swellings (Fig. 8 C and Fig. 9, A [arrowheads] and G) and stationary vesicles (Fig. 9 F) as well as a decrease in the velocity and run length of YFP-positive vesicles (Fig. 9, D and E; and Video 9).
Figure 8.
PtdIns(3)P lipids are required for AnkB association with membrane cargo in axons. (A) Images show loss of PIK3C3 expression and diffuse staining for the PtdIns(3)P biosensor 2×FYVE in Pik3c3flox/flox neurons expressing Cre-BFP (dotted lines, asterisks) when compared with untransfected cells (arrowheads). (B) Detection of PtdIns(3)P distribution using the 2×FYVE-GFP probe in Pik3c3flox/flox neurons expressing BFP or Cre-BFP proteins. Insets are enlarged views of boxed axonal regions. Arrowheads indicate areas of localization of GFP signal to vesicular structures. (C) Time-lapse image and kymographs of synaptophysin-YFP and WT 220-kD AnkB-mCherry localization and dynamics in Pik3c3flox/flox axons transfected with BFP or Cre-BFP. Arrowheads indicate large axonal swellings. (D) Quantification of AnkB-mCherry localization to YFP-positive synaptic vesicles (n = 10 axons, 5 independent preparations; n = 100 YFP vesicles). Data represent means ± SEM and correspond to one experiment out of three repeats. ***, P < 0.001, two-tailed t test.
Figure 9.
Neuronal PIK3C3 and PtdIns(3)P lipids regulate the transport dynamics of multiple axonal cargoes. (A–C) Distribution of synaptophysin-YFP (A), LAMP1-GFP (B), and Rab5-RFP (C) in Pik3c3flox/flox axons expressing BFP or Cre-BFP proteins. Arrowheads indicate large axonal aggregates. Kymographs illustrate synaptophysin-YFP, LAMP1-GFP, and Rab5-RFP motility in axons. For each kymograph, both grayscale images showing the motility of all particles and color-coded trajectories for a small subset of particles are provided. (D and E) Axonal velocity (D) and run length (E) for synaptophysin-YFP, LAMP1-GFP, and Rab5-RFP cargoes. (F) Relative motility of axonal synaptophysin-YFP, LAMP1-GFP, and Rab5-RFP in Pik3c3flox/flox neurons expressing BFP or Cre-BFP proteins. (G) Quantification of axonal swellings. For all graphs, the data shown are for one experiment out of three repeats. For the experiment shown, motility parameters were quantified from a total of 10 axons, 5 independent preparations, and 250 total particles for each organelle per genotype tested. Data represent means ± SEM. *, P < 0.05; ***, P < 0.001, two-tailed t test.
We further assessed a requirement for PIK3C3 and neuronal PtdIns(3)P lipids for the regulation of axonal transport dynamics of other organelles by imaging the motility of LAMP1-GFP and Rab5-RFP cargoes in control (Pik3c3flox/flox + pBFP) and PIK3C3-deficient (Pik3c33flox/flox + pCre-BFP) axons. Similar to the results seen in AnkB−/− neurons, reduction in PtdIns(3)P levels resulted in an increase in the percentage of stationary LAMP1-GFP and Rab5-RFP cargoes (Fig. 9, B, C, and F; and Video 10), with a major impact on the retrograde movement of cargoes. Furthermore, PtdIns(3)P depletion caused an increase in the frequency of axonal swellings (Fig. 9, B, C, and G, arrowheads) and a significant reduction in both the retrograde velocity and run length of these motile organelles (Fig. 9, D and E).
Discussion
We report the discovery of a new pathway required for fast long-range transport of multiple organelles in axons and normal axonal elongation (Fig. 10). We demonstrate that AnkB recruits the dynein–dynactin retrograde motor complex to diverse membrane cargoes by coupling both the p62 subunit of dynactin and membrane PtdIns(3)P lipids, predominantly generated by PIK3C3. Moreover, we show that loss of AnkB or of its binding to either p62 or PtdIns(3)P lipids, or loss of neuronal PIK3C3, impaired retrograde movement of synaptic vesicles, endosomes, mitochondria, and lysosomes. AnkB knockout in mice also impaired the formation of multiple brain structures involving long axon tracts, and resulted in shorter axons both in vivo and in cultured neurons.
Our results establish as of yet unsuspected functional relationships between PIK3C3, a known player in autophagy and endosomal dynamics, the dynactin complex, and AnkB, previously implicated in assembly of multiprotein complexes in specialized plasma membrane domains, which together promote the transport of multiple organelles over long distances in axons. Although motor–cargo coupling and transport regulation via PtdIns have been reported previously for specific motor–cargo pairs (Klopfenstein et al., 2002; Hoepfner et al., 2005; Niu et al., 2013), to our knowledge, this is the first observation of multiple organelles using a shared membrane docking molecule.
Loss of AnkB or of its linkages to either cargo or p62, in addition to having effects on retrograde transport of multiple organelles, also exclusively impaired rapid anterograde axonal transport of synaptic vesicles (Fig. 4, A–C; and Fig. S2, A–D). This disruption in the bidirectional motility of synaptic vesicles phenocopies transport deficits, resulting from inhibition of dynein interaction with membrane-bound dynactin (Waterman-Storer et al., 1997). Coordination of the bidirectional motion of vesicles has been observed in a variety of biological systems and can operate through the direct interaction of dynein and dynactin with kinesin, and it can also be regulated by factors that bind both motor complexes (Martin et al., 1999; Barkus et al., 2008; Fu and Holzbaur, 2013). In addition to direct interactions with kinesin motors, dynein and dynactin have been proposed to help organelles reverse the direction of transport as they bypass obstacles encountered along the crowded microtubule tracks (Jolly and Gelfand, 2011). This ability for quick reversal in transport direction is likely to be especially important for synaptic vesicles in axons because of their small caliber and strong dependence on long-range transport.
Although loss of AnkB impaired axonal transport of multiple cargoes, it did not entirely eliminate cargo motility. Disruption of the linkage between specific motor–adaptor pairs results in similar partial effects (Cai et al., 2010; van Spronsen et al., 2013). The observed residual transport may operate through alternative pathways, such as those of the known organelle-specific effectors, including the coupling of dynactin to intracellular membranes through β-spectrin proteins (Holleran et al., 1996; Muresan et al., 2001). This supports the hypothesis of the existence of numerous mechanisms for mediating dynein attachment to a particular type of cargo (Akhmanova and Hammer, 2010). Axons, however, are apparently especially vulnerable to transport defects in spite of these redundant pathways. Although our findings are consistent with the idea of the existence of various adaptors for the same type of cargo, presumably to assure redundancy, our discoveries also represent a departure from the commonly observed theme of adaptors being specific to only one type of organelle in two distinctive ways: (1) AnkB connects different organelles to the same axonal retrograde machinery, and (2) it does so using the same membrane coupling mechanism across endomembranes.
The identified PtdIns(3)P binding site within the ZU5C domain in AnkB is independent of ankyrin repeats, which mediate interactions of ankyrins with membrane-spanning proteins (Bennett and Lorenzo, 2013). The highly conserved, positively charged residues of this site confer interaction specificity and are essential for AnkB recruitment to intracellular membranes. Interestingly, the residues comprising this PtdIns binding site of AnkB are conserved among all bilaterian ankyrins, suggesting other ankyrins may also be PtdIns effectors.
Intriguingly, we find that AnkB, as well as the PtdIns(3)P biosensor GFP-2×FYVEEEA1, associates not only with early endosomes, a PtdIns(3)P-enriched organelle, but also with other membranes where PtdIns(3)P is presumably not the predominant PtdIns species, such as synaptic vesicles. Thus, our findings uncover a novel and central role for PtdIns(3)P lipids and neuronal PIK3C3 in regulating dynein targeting to cargoes and in long-range organelle motility in axons and axonal growth. Our results suggest that defects in these functions contribute to the loss of axons and rapid neurodegeneration observed in PIK3C3-null mouse brains (Zhou et al., 2010; Wang et al., 2011). However, we cannot rule out contributions from potential impairments in the function of other PI3K effectors, such as AKT and GSK-3, with known roles in axonal transport and growth (Polleux and Snider, 2010; Weaver et al., 2013; Dajas-Bailador et al., 2014). Interestingly, AnkB and PtdIns(3)P localization to mitochondria is not as pronounced as the one observed with other organelles and occurs at only a few points of contact. Nevertheless, our results clearly demonstrate that the retrograde transport of mitochondria is impaired in AnkB−/− axons and is restored by expression of WT AnkB but not the DD1320AA AnkB mutant unable to associate with the dynactin complex. It is likely that only a limited degree of contact between AnkB and mitochondria is required to mediate its transport. Moreover, it is possible that other well-characterized mechanisms for axonal mitochondrial transport, such as the one mediated by the TRAK (trafficking kinesin protein)/Milton motor–adaptor proteins (van Spronsen et al., 2013), facilitate mitochondrial motion in AnkB-deficient neurons.
Our findings establish AnkB’s role as an essential transport adaptor and expand the general functional relevance of ankyrin proteins beyond their “static” role as membrane organizers. Ankyrin-G, in contrast to AnkB, primarily associates with plasma membranes rather than intracellular organelles (He et al., 2013) and lacks dynactin-binding activity (unpublished data). Ankyrin-G and AnkB shared a common ancestral gene that was duplicated early in vertebrate evolution (Cai and Zhang, 2006). AnkB, thus, either gained dynactin-binding activity, or alternatively, ankyrin-G may have lost this function. In either case, dual functions in intracellular membrane trafficking and plasma membrane stabilization help explain roles of ankyrins in coordinating assembly of plasma membrane domains such as costameres (Ayalon et al., 2008, 2011). Our results also reveal versatile modes of coupling dynactin to membranes through AnkB, which can either function to stabilize membrane-associated microtubule populations, as observed with costamere-associated microtubules (Ayalon et al., 2008, 2011) or to promote organelle transport along microtubules in axons.
In summary, we present a novel pathway responsible for the transport of multiple organelles in axons and for normal axonal elongation involving AnkB as a novel multiorganelle adaptor of the dynactin–dynein motor complex. We also show the essential role of PIK3C3 in this mechanism, which via the production of membrane PtdIns(3)P, enables AnkB binding to cargoes. Consequently, we present evidence showing that deficiencies in AnkB or PIK3C3 lead to impaired transport and axonal growth abnormalities.
Materials and methods
Mouse lines and animal care
All animal care and procedures were approved by the Institutional Animal Care and Use Committee of Duke University. AnkB-null mice (Scotland et al., 1998) were generated by targeted disruption of the endogenous AnkB (Ank2) gene by homologous recombination. In brief, a clone containing 17 kb of the AnkB gene isolated from a 129SVJ genomic λ DNA library was modified to introduce a NotI site-flanked cassette containing a neomycin resistance gene, an in-frame HA epitope, and a stop codon within an exon in the spectrin-binding domain of AnkB. Male C57BL/6 chimeras containing the targeting construct were crossed to C57BL/6 females to assure germline transmission. Heterozygous carriers (AnkB+/−) were maintained in a mixed 129SVJ/C57BL/6 genetic background and used to generate WT (AnkB+/+) and AnkB-null mice (AnkB−/−) littermates. Pik3c3 conditional knockout (Pik3c3flox/flox) mice (gift from F. Wang and Q.-J. Li, Duke University, Durham, NC; Zhou et al., 2010; Devereaux et al., 2013) were generated by homologous recombination using a targeting construct that contained exons 15–19 of the mouse Pik3c3 gene, corresponding to the ATP binding domain, flanked by LoxP sites and a flippase recognition target–flanked neomycin-resistant gene cassette.
Constructs
Plasmids used in transfection experiments included synaptophysin-YFP and synaptophysin-DsRed (gifts from A.M. Craig, University of British Columbia, Vancouver, British Columbia, Canada), which were cloned as full-length sequences in the HindIII and AgeI sites of peYFP-N1 and pDsRed-N1 backbones (Takara Bio Inc.), respectively; DsRed2-mito containing the mitochondrial targeting sequence (mito) cloned into the NheI and BamHI sites of pDsRed2 (Takara Bio Inc.), synaptotagmin-I–GFP (gift from F. Valtorta, Università Vita-Salute San Raffaele, Milano, Italy) containing the synaptotagmin-I coding sequence cloned into the HindIII and BamHI sites of pEGFP-N3, and GFP-2×FYVEEEA1 (gift from P. De Camilli, Yale University, New Haven, CT). TfR-YFP (gift from A.M.J. VanDongen, Duke-NUS [National University of Singapore], Singapore) was generated by insertion of the full-length coding sequence of human TfR into the AgeI and BsrGI sites of peYFP-N1. Full-length EB1-GFP (cloned as an XhoI–BamHI fragment in pEGFP-N1), LAMP1-GFP and LAMP1-dsRed (cloned as an EcoRI–SalI fragment in either pEGFP-N3 or pDsRed vectors), and Rab5-RFP (cloned as a HindIII–BamHI fragment in pmRFP-C3) were purchased from Addgene.
The p62-GFP construct (Ayalon et al., 2008, 2011) was generated by cloning into the EcoRI and XhoI sites of pEGFP-N1. For the preparation of p62-3HA, GFP was first cut out of pEGFP-N1 vector between HindIII and NotI, and in its place, we introduced a triple HA (3×-YPYDVPDYA) sequence that was synthesized by PCR and ligated into the vector. The full-length p62 cDNA sequence was inserted into the XhoI and HindIII sites of the modified 3×HA-containing vector.
HA-AnkB was prepared based on an AnkB-GFP template in a pEGFP-N1 vector. GFP was cut out between the PmeI and the NotI sites and replaced with a 2×HA tag–encoding sequence with a PmeI site on its 5′ end and a stop codon followed by a NotI site on the 3′ end. For the preparation of DD1320AA-AnkB-HA and DD1320AA-AnkB-GFP, the DD1320AA double mutation was introduced into the tagged AnkB constructs by site-directed mutagenesis using a site-directed mutagenesis kit (QuikChange II XL; Agilent Technologies).
mCherry derivatives of p62-GFP and AnkB-GFP constructs were made by respective EcoRI–XhoI and EcoRI–AgeI restriction enzyme cloning into pmCherry-N1 (Takara Bio Inc.). GFP-tagged truncation constructs of the ankyrin repeat domain (ANK-GFP) of AnkB and the rest of the molecule (ZU5N-ZU5C-UPA-DD-Ct-GFP; He et al., 2013) were cloned into pEGFP-N1 using EcoRI and SalI restriction sites.
The maltose binding protein (MBP)–ZU5N-ZU5C-UPA-DD-7×-His AnkB construct was generated by PCR amplification of a BamHI–XhoI fragment comprising amino acids S966 to T1535 of human AnkB using the sense, 5′-GGGGGATCCTCAGGTTTCCTGGTTAGTTTTATGGTGGATGCC-3′, and antisense, 5′-GGGCTCGAGTGTGTTGGTCTCCATGAGATGAACAATATCCAT-3′, oligonucleotides (oligos). This product was cloned into a modified version of the pGEX6P-1 vector (GE Healthcare) that includes a C-terminal 6×-His tag. DNA constructs were verified by sequencing and transformed into BL21 (DE3) pLysS competent cells. The MBP-ZU5N-ZU5C-UPA-DD-7×-His and 220-kD AnkB-GFP and AnkB-mCherry constructs were further modified using the site-directed mutagenesis kit (QuikChange II XL) to generate the polybasic motif ZU5C mutant constructs carrying the K1157E, R1158E, R1160E, R1193E, R1194E, and K1195E amino acid substitutions as well as the R1194E and K1195E mutants. Sense oligos used for mutagenesis include KRIR1157EEIE, 5′-CCAGAGGGGGCACTCACCGAAGAGATCCGCGTAGGCCTGCAGGCTCAAC-3′; RK1193EE, 5′-CACTTTGGAACCTAGAGAAGAAGAGTTCCACAAACCAATTAC-3′; R1194E, 5′-GTCCACTTTGGAACCTAGAAGAGAGAAATTCCACAAACCAATTACC-3′; K1195E, 5′-CTTTGGAACCTAGAAGAAGAGAGTTCCACAAACCAATTACCATG-3′.
Antibodies
Antiserum against 440-kD AnkB was generated by immunization of rabbits with a purified peptide containing amino acids 2,427–2,617 of 440-kD AnkB. Rabbit affinity-purified antibodies against total AnkB (C-terminal domain), ankyrin-G (C-terminal domain), β2-spectrin (spectrin repeats 4–9), GFP, and β4-spectrin (amino acids 800–1,094 of mouse β4-spectrin σ 6 isoform) were all generated in our laboratory (Ayalon et al., 2008, 2011). Other antibodies included rabbit anti-MAP2 and rabbit anti-Tau (Synaptic Systems), goat anti-p62 and rabbit anti-DHC (Santa Cruz Biotechnology, Inc.), mouse anti-neurofilament (Covance), rabbit anti-Arp1, mouse anti-synaptophysin, mouse anti–acetylated tubulin, and mouse anti–detyrosinated tubulin (Sigma-Aldrich), mouse anti-p50 and anti-p150Glued (BD), mouse anti-DIC, anti–kinesin heavy chain, and anti-GAPDH (EMD Millipore), mouse anti-mCherry (Abcam), mouse anti-HA and chicken anti-GFP (Aves Labs), rabbit anti–β-actin and anti-Rab5 (Cell Signaling Technology), rabbit anti-PIK3C3 (Abnova), mouse anti-LAMP1 (Developmental Studies Hybridoma Bank), mouse anti-His (QIAGEN), and mouse anti–polyglutamylated tubulin (Adipogen). For β2-spectrin labeling of axons in STORM images, we used mouse anti–β2-spectrin (BD), which labels a position close to the middle of each α2-β2 spectrin tetramer (Bennett and Baines, 2001; Baines, 2010).
Secondary antibodies used for ELISA included HRP-conjugated goat anti–rabbit IgG (EMD Millipore) and goat anti–mouse IgG (Sigma-Aldrich). Secondary antibodies used for conventional fluorescence imaging of neurons were donkey and goat labeled with Alexa Fluor 488, 555, and 568 (Invitrogen). For single-color STORM imaging, secondary antibodies were custom labeled with a photoswitchable reporter dye (Alexa Fluor 647) and an activator dye (Alexa Fluor 405), which facilitates the photoswitching of the reporter dye (Bates et al., 2007). In brief, donkey anti–mouse and donkey anti–rabbit secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) were each labeled with a mixture of amine-reactive activator and reporter dyes in a one-step reaction. In some experiments, commercial Alexa Fluor 647–conjugated donkey anti–mouse, anti–rabbit, or anti–goat secondary antibodies (Invitrogen) were used.
Hippocampal neuronal culture
Primary hippocampal neuronal cultures were established from PND0 AnkB-null and WT mice or from Pik3c3flox/flox mice. Hippocampi were dissected in Hibernate E (BrainBits) and digested with 0.05% trypsin-EDTA (Invitrogen) for 15 min at 37°C. Tissue was washed three times with Hibernate E and dissociated in Neurobasal-A medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), B27 supplement (Invitrogen), 2 mM glutamine (Invitrogen), and penicillin/streptomycin (Invitrogen) and gently triturated through a glass pipette with a fire-polished tip. Dissociated cells were filtered through a 100-µm cell strainer (VWR International) to remove any residual nondissociated tissue and plated onto poly-d-lysine– and laminin-coated dishes (MatTek Corporation) for transfection and time-lapse microscopy imaging. For STORM imaging, neurons were plated onto poly-l-lysine– and laminin-coated 12-mm coverslips (BD) or poly-l-lysine–coated 8-well chambers. Media were replaced 16 h after plating with serum-free Neurobasal-A medium containing B27 and glutamine (culture medium). 5 µM cytosine-d-arabinofuranoside (Sigma-Aldrich) was added to the culture medium to inhibit the growth of glial cells 3 d after plating. Neurons were fed twice a week with freshly made culture medium until use.
Neuronal transfection
For time-lapse imaging experiments, DIV5 hippocampal neurons were transfected with 1 µg of each plasmid DNA after a modified Ca2+-phosphate transfection protocol (Jiang and Chen, 2006) using the CalPhos Mammalian Transfection kit (Takara Bio Inc.) and imaged 48 h after transfection. For experiments that evaluate axonal length DIV2 AnkB−/− and control, neurons were transfected with 300 ng of pdsRed-N1 (Takara Bio Inc.) alone or in combination with 500 ng AnkB-GFP rescue plasmids using a similar Ca2+-phosphate transfection protocol. Neurons were processed for immunofluorescence 6 d after transfection. For experiments that evaluate PIK3C3 knockdown, Pik3c3flox/flox hippocampal neurons were also transfected using the Ca2+-phosphate method. In this case, DIV2 neurons were transfected with 1 µg of either pCAG-BFP or pCAG-Cre-BFP plasmids in combination with 500 ng of each of the other plasmids and imaged at DIV8.
Fluorescence labeling of neurons
Neuronal cultures were washed with cold PBS, fixed for 15 min at room temperature in a 4% paraformaldehyde, 4% sucrose solution in PBS, and permeabilized with 0.2% vol/vol Triton X-100 in PBS at room temperature for 10 min. Samples were then blocked for 30 min in blocking buffer (4% BSA in PBS) and stained with primary antibodies in blocking buffer overnight at 4°C. Cells were washed three times with PBS, incubated with fluorescent-labeled secondary antibody conjugates in blocking buffer for 1 h at room temperature, washed five times with PBS, and mounted in Vectashield mounting media (Vector Laboratories).
For STORM imaging, DIV10 cultured neuron were initially fixed and extracted for 1 min in a solution of 0.3% (vol/vol) glutaraldehyde and 0.25% (vol/vol) Triton X-100 in cytoskeleton buffer (10 mM MES, pH 6.1, 150 mM NaCl, 5 mM EGTA, 5 mM glucose, and 5 mM MgCl2). Samples were postfixed for 15 min in 2% (vol/vol) glutaraldehyde in cytoskeleton buffer and treated with freshly prepared 0.1% (wt/vol) sodium borohydride for 7 min to reduce background fluorescence caused by glutaraldehyde fixation.
For labeling of axonal microtubules, DIV10 cultured neurons were quickly rinsed once with PBS at 37°C and treated with PEM buffer (100 mM Pipes-KOH, pH 6.9, 1 mM MgCl2, and 1 mM EGTA) containing 1% Triton X-100, 2% polyethylene glycol (molecular weight of 35,000), 2 µM phalloidin, and 2 µM taxol for 3 min at room temperature. Cells were then rinsed three times with PBS and fixed at room temperature with 0.2% glutaraldehyde in PBS for 20 min. Fixed samples were quenched twice with 2 mg/ml NaBH4 for 5 min, washed in PBS, and then blocked with 10% goat serum for 30 min and stained overnight with antibodies against different tubulin posttranslational modifications.
STORM imaging of fixed neuronal preparations
STORM imaging acquisition was conducted in PBS containing 100 mM cysteamine, 5% glucose, 0.8 mg/ml glucose oxidase (Sigma-Aldrich), and 40 µg/ml catalase (Roche). To image the samples from 12-mm coverslips, ∼4 µl of imaging buffer was dropped at the center of a freshly cleaned, #1.5 rectangular coverslip (22 × 60 mm), and the sample on the 12-mm coverslip was mounted on the rectangular coverslip and sealed with nail polish or Cytoseal. To image samples from 8-well chambers, 400 µl of imaging buffer was added to the imaging chamber.
The STORM setup was based on an inverted optical microscope (IX-71; Olympus) in which 405-nm (CUBE 405-50C; Coherent), 460-nm (Sapphire 460–10; Coherent), 532-nm (GCL-200-I; CrystaLaser), and 657-nm (RCL-300-656; CrystaLaser) lasers were introduced into the sample through the back focal plane of the microscope. A translation stage allowed the laser beams to be shifted toward the edge of the objective so that the emerging light reached the sample at incidence angles slightly smaller than the critical angle of the glass–water interface, thus illuminating only the fluorophores within a few micrometers of the coverslip surface. A T660LPXR (Chroma Technology Corp.) was used as the dichroic mirror, and a band pass filter (ET705/72M; Chroma Technology Corp.) was used as the emission filter. All laser lines were combined and coupled into an optical fiber (OZ Optics), and the fiber output was collimated and focused on the back focal plane of a high- NA oil-immersion objective (100× U Plan S Apochromat, NA 1.4; Olympus). The fluorescence emission was collected by the same objective and imaged onto a back-illuminated electron-multiplying charge-coupled device (EMCCD) camera (iXon DU-860 or iXon DU-897; Andor Technology). To stabilize the focus during data acquisition, an 830-nm fiber-coupled diode laser (LPS-830-FC; Thorlabs) was introduced into the microscope in a separate objective-type total internal reflection path. The reflected infrared beam from the coverglass–water interface was directed to a quadrant photodiode. The position readout of the quadrant photodiode provided feedback to a piezo objective positioner (Nano-F100; Mad City Labs), keeping the focal drift to <20 nm.
For single-color STORM imaging of Alexa Fluor 647–labeled samples, the 647-nm laser was used to excite fluorescence from Alexa Fluor 647 molecules and switch them into the dark state. A pair of 647-nm notch filters (NF01-543/647; Semrock), which reject the 647-nm excitation light, were inserted into the imaging paths of the two objectives. After passing through these filters, fluorescence from Alexa Fluor 647 collected by the two objectives were each focused by a 20-cm achromatic lens, cropped by a slit at the focal plane, and then separately projected onto two different areas of the same EMCCD camera (iXon DU-897) using two pairs of relay lenses. A single-band band pass filter (ET700/75m; Chroma Technology Corp.) was placed in front of the camera. Before acquiring STORM images, we first used relatively weak 647-nm light (∼0.05 W/cm2) to illuminate the sample and recorded the conventional fluorescence image before a substantial fraction of the dye molecules were switched off. We then increased the 647-nm light intensity (to ∼2 kW/cm2) to rapidly switch the dyes off for STORM imaging. The 405-nm laser (when Alexa Fluor 405 was used as the activator dye or no activator dye was used) or 532-nm laser (when Cy3 was used as the activator dye) was used to reactivate the fluorophores from the dark state back to the emitting state. The power of the activation lasers (typical range of 0–1 W/cm2) was adjusted during image acquisition so that at any given instant, only a small, optically resolvable fraction of the fluorophores in the sample was activated. STORM images were acquired from both objectives simultaneously at a frame rate of 60 Hz using an EMCCD camera and the free-access STORM-Control acquisition software developed by the Zhuang laboratory.
A typical STORM image was generated from a sequence of ∼30,000–60,000 image frames at a frame rate of 60 Hz. The recorded STORM video was analyzed according to previously described methods (Huang et al., 2008; Dani et al., 2010). In brief, images of individual molecules were identified and fit to an elliptical Gaussian function to obtain the centroid position coordinates (x0 and y0) and the Gaussian widths (dx and dy). The lateral position of the molecule was determined as x0 and y0, and the z position was calculated from dx and dy using a calibration curve independently determined by imaging a 100-nm fluorescent bead (TetraSpeck; Invitrogen) on a coverglass while scanning in the z direction. The z position was then corrected for refractive index mismatch between glass and the imaging medium. Super-resolution images were reconstructed from the molecular coordinates by depicting each location as a 2D Gaussian peak. Localization precisions were ∼10 nm for lateral (xy) and ∼22 nm for axial (z) directions measured in SD or ∼25 nm for lateral and ∼50 nm for axial directions measured in full width at half-maximum. Quantitative measurements of the structure of β2-spectrin in neurons were obtained by performing Fourier transform analysis (Xu et al., 2013).
DTI analysis
Brain specimens from PND2 AnkB−/− mice (n = 4) and WT littermates (n = 4) were actively stained by transcardial perfusion following a reported protocol (Johnson et al., 2002). In detail, mice were first perfused with 0.9% PBS followed by a mixture of 10% buffered formalin and the contrast agent gadoteridol (10:1, vol/vol; ProHance; Bracco Imaging). Heads were stored in formalin overnight and moved to a ProHance/PBS (1:200) solution. Brains were kept in the skull for the entire procedure, including during the magnetic resonance scanning to avoid physical distortion and were immersed in perfluorocarbon (Solvay Solexis) to minimize susceptibility artifacts and to limit specimen dehydration. Images were acquired using a 9.4 T vertical bore magnet interfaced to a magnetic resonance imaging console (GE EXCITE running version 12M5; Epic 12.4X; GE Healthcare) equipped with shielded coils (Resonance Research, Inc.) that provide gradients of up to 2,000 mT/m. We used in house–developed solenoid coils to acquire 3D volume images with a field of view of 22 × 11 × 11, a matrix size of 512 × 256 × 256, and 43-µm isotropic resolutions. The diffusion-weighted spin–echo pulse sequences used repetition time of 100 ms, echo time of 11.83 ms, and number of excitations of 2, with one baseline image (b0) and six diffusion encoding directions (diffusion pulse width of 1.3 ms, b = ∼1.5 × 103 s/mm2, separation of 6.4 ms, and gradient amplitude of 1,600 mT/m), as in Jiang and Johnson (2011). The six diffusion-weighted acquisitions were aligned to b0 to compensate for Eddy current distortions.
Diffusion-weighted image processing was performed using an automated DTI processing pipeline written in Perl and MATLAB (MathWorks) as previously described (Johnson et al., 2012). Diffusion Toolkit (TrackVis) command line tools were used to calculate the diffusion tensor and other DTI parameters and to generate the apparent diffusion coefficient map, isotropic diffusion-weighted image, longitudinal diffusivity map, radial diffusivity map, FA map, and directionally encoded FA color map. Automated skull stripping of the diffusion-weighted image was performed using a modified version of the skull-stripping algorithm previously described (Badea et al., 2007), and the mask was propagated to all other coregistered image sets. Finally, the mask was used as a region of interest for whole-brain fiber tractography using the Diffusion Toolkit implementation of the FACT (fiber assignment by continuous tracking) algorithm (Mori et al., 1999) with an FA threshold of 0.1 and an angle threshold of 45°. All tractography data were visualized and analyzed using the TrackVis interactive visualization environment.
Fluorescence image acquisition
Fluorescent antibody labeling of fixed neuronal preparations was visualized at room temperature using an inverted laser-scanning confocal microscope (LSM 780; Carl Zeiss) equipped with a digital microscope camera (AxioCam; Carl Zeiss) and a GaAsP detector (Nikon). Z stacks with optical sections of 0.5-µm intervals were collected using a 40×, 1.3 NA, and 60× oil, 1.4 NA, objective lenses and the Zen 2011 acquisition and imaging software (Zen Black 2011; Carl Zeiss). 3D rendering of confocal z stacks was performed using the Volocity software (PerkinElmer) with the 3D rendering option set to opacity.
Time-lapse video microscopy and video analyses
Live microscopy of neuronal cultures was performed using a laser inverted scanning confocal microscope (LSM 780) equipped with a digital microscope AxioCam camera, a GaAsP detector, and a temperature- and CO2-controlled incubation chamber. Time-lapse images were collected from neuronal preparation incubated at 37°C and 5% CO2 in phenol red-free Hibernate E and captured in the midaxon, ∼60 µm away from the soma, with a 40× oil objective, 1.3 NA, at a rate of 1 frame/s for time intervals ranging from 60 to 300 s using the zoom and definite focus functions of the Zen 2011 acquisition and imaging software (Zen Black 2011).
Time-lapse images were exported as 15 frames/s video files using the Zen 2011 acquisition and imaging software (Zen Black 2011) that were processed and analyzed using ImageJ (National Institutes of Health). Kymographs were obtained using the KymoToolBox plugin for ImageJ (Zala et al., 2013). In detail, space (x axis in micrometers)- and time (y axis in seconds)-calibrated kymographs were generated from video files. In addition, the KymoToolBox plugin was used to manually follow a subset of particles from each kymograph and reported the tracked particles on the original kymograph and video files using a color code for movement directionality (red for anterograde, green for retrograde, and blue for stationary particles).
Quantitative analyses were performed manually by following the trajectories of individual particles to calculate dynamic parameters, including net and directional velocities and net and directional run length, as well as time of pause or movement in a particular direction of transport. Anterograde and retrograde motile vesicles were defined as particles showing a net displacement >3 µm in one particular direction, and stationary vesicles were defined as particles with a net displacement <2 µm.
Immunoprecipitation
Total protein homogenates from mouse brains were prepared in PBS containing 150 mM NaCl, 0.32 M sucrose, 2 mM EDTA, 0.1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and protease inhibitors (10 µg/ml AEBSF, 30 µg/ml benzamidine, 10 µg/ml pepstatin, and 10 µg/ml leupeptin; EMD Millipore). Samples were centrifuged at 100,000 g for 30 min, and the supernatants were precleared with protein G Dynabeads (Invitrogen) and subjected to immunoprecipitation using antibodies against AnkB, p62, p50, DIC, or control IgG. Immunoprecipitation samples were resolved by SDS-PAGE and Western blotting, and antibodies were detected using I125-labeled protein A/G (Thermo Fisher Scientific).
For coimmunoprecipitation experiments, 6 × 106 COS7 cells were plated in 10-cm dishes and transfected with 4 µg of each plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were harvested 48 h after transfection and lysed in 0.5% Triton X-100 in lysis buffer (10 mM sodium phosphate, 0.32 M sucrose, 2 mM EDTA, and protease inhibitors). Cell lysates were centrifuged at 100,000 g for 30 min, and the soluble fraction was collected and precleared by incubation with protein G Dynabeads. Coimmunoprecipitation experiments were performed using protein G Dynabeads and mouse anti-HA or rabbit anti-GFP antibodies. Immunoprecipitation samples were resolved by SDS-PAGE and Western blot as described in the previous paragraph.
Membrane fractionation assay
Membrane isolation from brain homogenates was conducted as previously reported (Yeh et al., 2012). In brief, four brains from PND1 control or AnkB−/− mice were homogenized in TMEE buffer (50 mM Tris, pH 7.4, 5 mM MgSO4, 1 mM EGTA, 0.5 mM EDTA, and protease inhibitors). Nuclei were pelleted by centrifugation at 1,600 rpm for 10 min at 4°C. The PNS was fractionated by centrifugation at 44,700 rpm for 30 min at 4°C (SW 55 Ti rotor; Beckman Coulter) into supernatant (soluble fraction) and pellet (total membrane fraction). The pellet was then brought to 2 M sucrose in TMEE (1:4 vol/vol) and overlaid stepwise with 1.2 and 0.2 M sucrose in TMEE. The gradient was centrifuged at 44,700 rpm for 60 min at 4°C (SW55Ti rotor), and floated membranes were collected from the 1.2/0.2 M sucrose interface. Membranes were diluted to 700 µl in TMEE and repelleted at 44,700 rpm for 30 min at 4°C. The final pellet was dissolved in 5× PAGE, and equal amounts of protein for each fraction (PNS, soluble, and membranes) were solved by SDS-PAGE and Western blotting.
Protein expression and purification
Recombinant WT and ZU5C mutant (MBP-ZU5N-ZU5C-UPA-DD-7×-His) AnkB proteins were expressed in BL21 (DE3) pLysS Escherichia coli cells. Dual-tagged MBP-His fusion proteins were first isolated on Ni-Sepharose beads (High Performance Ni-Sepharose; GE Healthcare), washed with NiNTA column buffer (50 mM phosphate buffer, pH 8.0, 0.3 M sodium bromide, 0.2 mM EDTA, 0.2 mM DTT, and 20 mM imidazole) and eluted with same buffer containing 300 mM imidazole. Proteins eluted from the Ni-Sepharose beads were diluted 1:3 vol/vol in amylose column buffer (20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, and 1 mM NaN3) and immediately loaded onto an amylose resin (New England Biolabs, Inc.). The column was washed extensively, and bound proteins were eluted with amylose column buffer containing 10 mM maltose.
Histidine-tagged 220-kD AnkB was expressed using a baculovirus protein expression system (BacPAK; Takara Bio Inc.) and purified as previously reported (Abdi et al., 2006). In brief, a plasmid encoding 220-kD WT AnkB-7×-His was cotransfected into Sf21 insect cells along with viral plasmid pBakPak6 (Takara Bio Inc.) to generate recombinant virus. Single viral clones were isolated and amplified for expression. Cells were lysed in 50 mM phosphate buffer, 0.5 M NaBr, 20 mM imidazole, and 0.2 mM β-mercaptoethanol and coupled batchwise to High Performance Ni-Sepharose. Sepharose was washed with binding buffer, and proteins were finally eluted in buffer containing 50 mM phosphate buffer, 0.3 M NaBr, and 70 mM imidazole.
β2-spectrin binding assay
Purified His-tagged WT 220-kD (200 nM) and WT or mutant His-tagged ZU5N-ZU5C-UPA-DD (1 µM) AnkB proteins were incubated with glutathione-Sepharose beads (GE Healthcare), which were preloaded with either β2-spectrin (repeats 14–16)-GST or GST-alone purified proteins. Beads and proteins were incubated for 1 h at 25°C in binding buffer (10 mM Tris, pH 7.8, 100 mM NaCl, 1 mg/ml BSA, and 1 mM sodium azide). Beads were layered onto 20% glycerol barrier gradients and centrifuged at 4,000 rpm for 10 min. The tubes were frozen, and the tips were cut off into 5× PAGE buffer. SDS-PAGE and Western blotting were performed as described in the previous paragraph to detect bound proteins using an anti-His monoclonal antibody.
Protein–lipid overlay assay
The protein–lipid binding assay was conducted according to the manufacturer’s protocol. In brief, PtdIns strips or PtdIns arrays (Echelon Biosciences) were blocked for 1 h at room temperature in blocking buffer (10 mM Tris, pH 7.8, 100 mM NaCl, 0.05% Tween 20, and 2 mg/ml fatty acid–free BSA) followed by a 2-h incubation with 200 nM of purified His-tagged WT 220-kD or WT or mutant His-tagged ZU5N-ZU5C-UPA-DD AnkB proteins in blocking buffer. Membranes were then washed three times in wash buffer (10 mM Tris, pH 7.8, 100 mM NaCl, and 0.05% Tween 20) and sequentially incubated for 1 h at room temperature with mouse anti-His antibody, rabbit anti–mouse IgG, and I125-labeled protein A/G. Densitometric analysis was used to quantify the relative binding of AnkB proteins to various PtdInsP.
PtdIns specificity ELISA
The specificity of AnkB binding to PtdIns lipids was also assessed by ELISA. Cova PtdInsP specificity 96-well plates (Echelon Biosciences) were blocked with PBS containing 0.05% Tween 20, 1% normal goat serum, and 0.5 mg/ml fatty acid–free BSA (binding buffer) for 30 min at room temperature. 200 nM of purified His-tagged WT 220-kD or WT or mutant His-tagged ZU5N-ZU5C-UPA-DD AnkB proteins in binding buffer was added to the wells, incubated for 1 h, and washed three times with PBS containing 0.05% Tween 20 (PBST). Plates were sequentially incubated with mouse anti-His antibody and HRP-conjugated goat anti–mouse IgG. A colorimetric reaction was initiated by addition of 100 µl of the HRP’s substrate TMB (3, 3′, 5, 5″-tetramethylbenzidine; Sigma-Aldrich) and quenched with 1 M H2SO4. Absorbance was read at 450 nm.
Lipid–protein binding assays
The binding of AnkB to PtdIns-coated beads was conducted according to the manufacturer’s protocol. In brief, 20-µl PtdIns-coated beads (Echelon Biosciences) were blocked for 1 h at room temperature in binding buffer (10 mM Tris, pH 7.8, 100 mM NaCl, and 0.25% IGEPAL) supplemented with 1 mg/ml fatty acid–free BSA (blocking buffer) and 1% normal goat serum. Beads were further incubated for 1 h with 200 nM of purified His-tagged WT 220 kD in blocking buffer at room temperature. Samples were layered onto 20% glycerol barrier gradients and centrifuged at 4,000 rpm for 10 min. The tubes were frozen, and the tips were cut off into 5× PAGE buffer. SDS-PAGE and Western blotting were performed as described previously to detect bound proteins using an anti-His monoclonal antibody.
Binding of AnkB proteins to polyPIPosomes was assessed by protein pull-down. Biotinylated PtdIns(3)P, PtdIns(4)P, or control PolyPIPosomes (Echelon Biosciences) were incubated with 200 nM of purified His-tagged WT 220-kD or WT or mutant His-tagged ZU5N-ZU5C-UPA-DD AnkB proteins in 200 µl of binding buffer (20 mM Hepes, pH 7.4, 120 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1 mg/ml BSA, 0.2 mM CaCl2, and 5 mM KCl) for 1 h at 4°C. Samples were centrifuged for 10 min at 13,000 rpm and 4°C and washed three times in binding buffer with 10-min centrifugations at 13,000 rpm in-between washes. Pellets and initial supernatants were resolved by SDS-PAGE and Western blotting as described previously.
Binding of AnkB proteins to polyPIPosomes was also assessed using an ELISA format developed by Echelon Biosciences. In brief, 96-well streptavidin-coated plates (Thermo Fisher Scientific) were incubated for 1 h at room temperature with 5 µM of biotinylated PtdIns(3)P, PtdIns(4)P, or control PolyPIPosomes in binding buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, and 0.5 mg/ml BSA) and washed with PBST. Plates were incubated for 1 h at room temperature with 200 nM of purified His-tagged WT 220-kD or WT or mutant His-tagged ZU5N-ZU5C-UPA-DD AnkB proteins in 100 µl of binding buffer followed by PBST washes and 1-h sequential incubations with mouse anti-His antibody and HRP-conjugated goat anti–mouse IgG. A colorimetric reaction was initiated by addition of 100 µl of the HRP’s substrate TMB and quenched with 1 M H2SO4. Absorbance was read at 450 nm.
Docking simulation
The AutoDock Vina program was used for the docking experiments (Trott and Olson, 2010). To simplify the simulation, only PtdIns(3)P’s head group (inositol 1,3-bisphosphate), singly methylated (methyl group not depicted) at the phosphate 1 position was used as ligand in the simulation. The ligand molecule was created using ChemBio3D (CambrigeSoft) and the ZU5N-ZU5C-UPA-DD tandem receptor (PDB accession no. 4D8O). The hydrogen positions and Gasteiger charges were calculated by AutoDockTools (ADT 1.5.6; Morris et al., 2009). The ligand was allowed torsional flexibility, whereas the macromolecule was kept rigid. The grid box was centered at the proposed binding region. The program was run using default parameters, and the energy optimization was performed using the Lamarckian genetic algorithm as described previously (Morris et al., 1998). The lowest mean energy was selected as a plausible model for the binding of the inositol 1,3-bisphosphate head group to the protein. The schematic diagrams of protein–ligand interactions were visualized with the PyMOL program (Schrödinger, LLC).
Statistical analysis
Prism (GraphPad Software) was used for statistical analysis. Two groups of measurements were compared by unpaired t test. Multiple groups were compared by repeated measures one-way analysis of variance (ANOVA) followed by a Tukey’s multiple comparisons test. The criterion for statistical significance was set at *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Online supplemental material
Fig. S1 shows reduced number of axonal tracts and loss of long axonal projections in AnkB−/− brains. Fig. S2 shows that AnkB promotes the bidirectional transport of synaptic vesicles and the retrograde transport of mitochondria. Fig. S3 shows that AnkB regulates the retrograde transport of early endosomes and lysosomes in axons. Fig. S4 shows that AnkB is not required for the transport dynamics of TfR-containing vesicles in dendrites. Fig. S5 shows that AnkB binds PtdInsP lipids through a polybasic motif in the ZU5C domain. Video 1 shows that AnkB regulates the bidirectional transport of synaptic vesicles in axons. Video 2 illustrate a requirement for AnkB’s interaction with p62 for the regulation of the bidirectional transport of synaptic vesicles in axons. Video 3 shows that AnkB regulates the retrograde transport of axonal mitochondria. Video 4 shows that AnkB association with p62 regulates the retrograde motion of axonal mitochondria. Videos 5 and 6 show that AnkB association with p62 regulates the retrograde motion of early endosomes and lysosomes, respectively. Video 7 illustrates that GFP-tagged AnkB also regulates the retrograde motion of lysosomes through is interaction with p62. Video 8 shows that AnkB association with p62 through DD1320 residues recruits dynactin to axonal membrane cargoes. Video 9 shows that knockdown of PIK3C3 disrupts AnkB association with axonal cargo and the motility of synaptic vesicles. Video 10 illustrates that knockdown of PIK3C3 disrupts the axonal motility of early endosomes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201407063/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201407063.dv.
Supplementary Material
Acknowledgments
We thank Dr. A.M. Craig for the gift of synaptophysin-YFP and synaptophysin-dsRed plasmids, Dr. F. Valtorta for synaptotagmin-GFP, Dr. A.M.J. Van Dongen for TGN38-GFP and TfR-YFP, and Dr. P. De Camilli for the GFP-2×FYVEEEA1 construct. We also thank Drs. F. Cordelières and F. Saudou for providing us with the KymoToolBox plugin for ImageJ, Drs. F. Wang and Q. J. Li for the Pik3c3flox/flox mice, and Y. Qi, M. Sherrier, and Dr. G.A. Johnson at the Duke Center for In Vivo Microscopy (National Institutes of Health/National Institute of Biomedical Imaging and Bioengineering National Biomedical Technology Resource Center; 41 EB015897) for help with DTI experiments.
A. Badea is supported by an NIA-K01 AG041211 grant. X. Zhuang and V. Bennett are investigators in the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AnkB
- ankyrin-B
- ANOVA
- analysis of variance
- DD
- death domain
- DHC
- dynein heavy chain
- DIC
- dynein intermediate chain
- DIV
- day in vitro
- DTI
- diffusion tensor imaging
- EMCCD
- electron-multiplying charge-coupled device
- FA
- fractional anisotropy
- MBP
- maltose-binding protein
- PND
- postnatal day
- PNS
- postnuclear supernatant
- PtdIns
- phosphoinositide
- PtdIns(3)P
- PtdIns 3-phosphate
- PtdIns(4)P
- PtdIns 4-phosphate
- PtdInsP
- PtdIns phosphate
- STORM
- stochastic optical reconstruction microscopy
- TfR
- transferrin receptor
- UPA
- urokinase-type plasminogen activator
- WT
- wild type
References
- Abdi K.M., Mohler P.J., Davis J.Q., and Bennett V.. 2006. Isoform specificity of ankyrin-B: a site in the divergent C-terminal domain is required for intramolecular association. J. Biol. Chem. 281:5741–5749 10.1074/jbc.M506697200 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., and Hammer J.A. III. 2010. Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 22:479–487 10.1016/j.ceb.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf Y., and Pasternak O.. 2008. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: a review. J. Mol. Neurosci. 34:51–61 10.1007/s12031-007-0029-0 [DOI] [PubMed] [Google Scholar]
- Ayalon G., Davis J.Q., Scotland P.B., and Bennett V.. 2008. An ankyrin-based mechanism for functional organization of dystrophin and dystroglycan. Cell. 135:1189–1200 10.1016/j.cell.2008.10.018 [DOI] [PubMed] [Google Scholar]
- Ayalon G., Hostettler J.D., Hoffman J., Kizhatil K., Davis J.Q., and Bennett V.. 2011. Ankyrin-B interactions with spectrin and dynactin-4 are required for dystrophin-based protection of skeletal muscle from exercise injury. J. Biol. Chem. 286:7370–7378 10.1074/jbc.M110.187831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea A., Ali-Sharief A.A., and Johnson G.A.. 2007. Morphometric analysis of the C57BL/6J mouse brain. Neuroimage. 37:683–693 10.1016/j.neuroimage.2007.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines A.J.2010. The spectrin-ankyrin-4.1-adducin membrane skeleton: adapting eukaryotic cells to the demands of animal life. Protoplasma. 244:99–131 10.1007/s00709-010-0181-1 [DOI] [PubMed] [Google Scholar]
- Barkus R.V., Klyachko O., Horiuchi D., Dickson B.J., and Saxton W.M.. 2008. Identification of an axonal kinesin-3 motor for fast anterograde vesicle transport that facilitates retrograde transport of neuropeptides. Mol. Biol. Cell. 19:274–283 10.1091/mbc.E07-03-0261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M., Huang B., Dempsey G.T., and Zhuang X.. 2007. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 317:1749–1753 10.1126/science.1146598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V., and Baines A.J.. 2001. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol. Rev. 81:1353–1392. [DOI] [PubMed] [Google Scholar]
- Bennett V., and Lorenzo D.N.. 2013. Spectrin- and ankyrin-based membrane domains and the evolution of vertebrates. Curr Top Membr. 72:1–37 10.1016/B978-0-12-417027-8.00001-5 [DOI] [PubMed] [Google Scholar]
- Cai Q., Lu L., Tian J.H., Zhu Y.B., Qiao H., and Sheng Z.H.. 2010. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 68:73–86 10.1016/j.neuron.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., and Zhang Y.. 2006. Molecular evolution of the ankyrin gene family. Mol. Biol. Evol. 23:550–558 10.1093/molbev/msj056 [DOI] [PubMed] [Google Scholar]
- Conde C., and Cáceres A.. 2009. Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 10:319–332 10.1038/nrn2631 [DOI] [PubMed] [Google Scholar]
- Dajas-Bailador F., Bantounas I., Jones E.V., and Whitmarsh A.J.. 2014. Regulation of axon growth by the JIP1-AKT axis. J. Cell Sci. 127:230–239 10.1242/jcs.137208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani A., Huang B., Bergan J., Dulac C., and Zhuang X.. 2010. Superresolution imaging of chemical synapses in the brain. Neuron. 68:843–856 10.1016/j.neuron.2010.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereaux K., Dall’Armi C., Alcazar-Roman A., Ogasawara Y., Zhou X., Wang F., Yamamoto A., De Camilli P., and Di Paolo G.. 2013. Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS ONE. 8:e76405 10.1371/journal.pone.0076405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., and De Camilli P.. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Drerup C.M., and Nechiporuk A.V.. 2013. JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet. 9:e1003303 10.1371/journal.pgen.1003303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejtova A., Davydova D., Bischof F., Lazarevic V., Altrock W.D., Romorini S., Schöne C., Zuschratter W., Kreutz M.R., Garner C.C., et al. . 2009. Dynein light chain regulates axonal trafficking and synaptic levels of Bassoon. J. Cell Biol. 185:341–355 10.1083/jcb.200807155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M.M., and Holzbaur E.L.. 2013. JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J. Cell Biol. 202:495–508 10.1083/jcb.201302078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., and Stenmark H.. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L.S.2012. Axonal transport and neurodegenerative disease: can we see the elephant? Prog. Neurobiol. 99:186–190 10.1016/j.pneurobio.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Tseng W.C., and Bennett V.. 2013. A single divergent exon inhibits ankyrin-B association with the plasma membrane. J. Biol. Chem. 288:14769–14779 10.1074/jbc.M113.465328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Niwa S., and Tanaka Y.. 2010. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 68:610–638 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- Hoepfner S., Severin F., Cabezas A., Habermann B., Runge A., Gillooly D., Stenmark H., and Zerial M.. 2005. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 121:437–450 10.1016/j.cell.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Hollenbeck P.J., and Bray D.. 1987. Rapidly transported organelles containing membrane and cytoskeletal components: their relation to axonal growth. J. Cell Biol. 105:2827–2835 10.1083/jcb.105.6.2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran E.A., Tokito M.K., Karki S., and Holzbaur E.L.. 1996. Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J. Cell Biol. 135:1815–1829 10.1083/jcb.135.6.1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan C.P., Hanscom S.R., Jolly R.S., Futter C.E., and McCaffrey M.W.. 2010. Rab11-FIP3 links the Rab11 GTPase and cytoplasmic dynein to mediate transport to the endosomal-recycling compartment. J. Cell Sci. 123:181–191 10.1242/jcs.052670 [DOI] [PubMed] [Google Scholar]
- Huang B., Wang W., Bates M., and Zhuang X.. 2008. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 319:810–813 10.1126/science.1153529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., and Chen G.. 2006. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat. Protoc. 1:695–700 10.1038/nprot.2006.86 [DOI] [PubMed] [Google Scholar]
- Jiang Y., and Johnson G.A.. 2011. Microscopic diffusion tensor atlas of the mouse brain. Neuroimage. 56:1235–1243 10.1016/j.neuroimage.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V.M., and Neefjes J.. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7–RILP–p150Glued, ORP1L, and the receptor βlll spectrin. J. Cell Biol. 176:459–471 10.1083/jcb.200606077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G.A., Cofer G.P., Gewalt S.L., and Hedlund L.W.. 2002. Morphologic phenotyping with MR microscopy: the visible mouse. Radiology. 222:789–793 10.1148/radiol.2223010531 [DOI] [PubMed] [Google Scholar]
- Johnson G.A., Calabrese E., Badea A., Paxinos G., and Watson C.. 2012. A multidimensional magnetic resonance histology atlas of the Wistar rat brain. Neuroimage. 62:1848–1856 10.1016/j.neuroimage.2012.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly A.L., and Gelfand V.I.. 2011. Bidirectional intracellular transport: utility and mechanism. Biochem. Soc. Trans. 39:1126–1130 10.1042/BST0391126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon J.R., and Vale R.D.. 2009. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 10:854–865 10.1038/nrm2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein D.R., Tomishige M., Stuurman N., and Vale R.D.. 2002. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 109:347–358 10.1016/S0092-8674(02)00708-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto M.1995. A neuron-specific isoform of brain ankyrin, 440-kD ankyrinB, is targeted to the axons of rat cerebellar neurons. J. Cell Biol. 131:1821–1829 10.1083/jcb.131.6.1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M.A.2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9:99–111 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- Martenson C., Stone K., Reedy M., and Sheetz M.. 1993. Fast axonal transport is required for growth cone advance. Nature. 366:66–69 10.1038/366066a0 [DOI] [PubMed] [Google Scholar]
- Martin M., Iyadurai S.J., Gassman A., Gindhart J.G. Jr, Hays T.S., and Saxton W.M.. 1999. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol. Biol. Cell. 10:3717–3728 10.1091/mbc.10.11.3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S., and Julien J.P.. 2013. Axonal transport deficits and neurodegenerative diseases. Nat. Rev. Neurosci. 14:161–176 10.1038/nrn3380 [DOI] [PubMed] [Google Scholar]
- Mori S., Crain B.J., Chacko V.P., and van Zijl P.C.. 1999. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann. Neurol. 45:265–269 [DOI] [PubMed] [Google Scholar]
- Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., and Olson A.J.. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19:1639–1662 [DOI] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., and Olson A.J.. 2009. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30:2785–2791 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V., Stankewich M.C., Steffen W., Morrow J.S., Holzbaur E.L., and Schnapp B.J.. 2001. Dynactin-dependent, dynein-driven vesicle transport in the absence of membrane proteins: a role for spectrin and acidic phospholipids. Mol. Cell. 7:173–183 10.1016/S1097-2765(01)00165-4 [DOI] [PubMed] [Google Scholar]
- Niu Y., Zhang C., Sun Z., Hong Z., Li K., Sun D., Yang Y., Tian C., Gong W., and Liu J.J.. 2013. PtdIns(4)P regulates retromer-motor interaction to facilitate dynein-cargo dissociation at the trans-Golgi network. Nat. Cell Biol. 15:417–429 10.1038/ncb2710 [DOI] [PubMed] [Google Scholar]
- Polleux F., and Snider W.. 2010. Initiating and growing an axon. Cold Spring Harb. Perspect. Biol. 2:a001925 10.1101/cshperspect.a001925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink K.O., Raiborg C., and Stenmark H.. 2013. Phosphatidylinositol 3-phosphate, a lipid that regulates membrane dynamics, protein sorting and cell signalling. BioEssays. 35:900–912. [DOI] [PubMed] [Google Scholar]
- Schroer T.A.2004. Dynactin. Annu. Rev. Cell Dev. Biol. 20:759–779 10.1146/annurev.cellbio.20.012103.094623 [DOI] [PubMed] [Google Scholar]
- Schroer T.A., and Sheetz M.P.. 1991. Two activators of microtubule-based vesicle transport. J. Cell Biol. 115:1309–1318 10.1083/jcb.115.5.1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotland P., Zhou D., Benveniste H., and Bennett V.. 1998. Nervous system defects of AnkyrinB (−/−) mice suggest functional overlap between the cell adhesion molecule L1 and 440-kD AnkyrinB in premyelinated axons. J. Cell Biol. 143:1305–1315 10.1083/jcb.143.5.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D., Razafsky D.S., Schlager M.A., Serra-Marques A., Grigoriev I., Demmers J., Keijzer N., Jiang K., Poser I., Hyman A.A., et al. . 2012. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol. Biol. Cell. 23:4226–4241 10.1091/mbc.E12-03-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai A.W., Chuang J.Z., Bode C., Wolfrum U., and Sung C.H.. 1999. Rhodopsin’s carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 97:877–887 10.1016/S0092-8674(00)80800-4 [DOI] [PubMed] [Google Scholar]
- Tan S.C.J.S., Scherer J., and Vallee R.B.. 2011. Recruitment of dynein to late endosomes and lysosomes through light intermediate chains. Mol. Biol. Cell. 22:467–477 10.1091/mbc.E10-02-0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., and Olson A.J.. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen M., Mikhaylova M., Lipka J., Schlager M.A., van den Heuvel D.J., Kuijpers M., Wulf P.S., Keijzer N., Demmers J., Kapitein L.C., et al. . 2013. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron. 77:485–502 10.1016/j.neuron.2012.11.027 [DOI] [PubMed] [Google Scholar]
- Wang C., Yu C., Ye F., Wei Z., and Zhang M.. 2012. Structure of the ZU5-ZU5-UPA-DD tandem of ankyrin-B reveals interaction surfaces necessary for ankyrin function. Proc. Natl. Acad. Sci. USA. 109:4822–4827 10.1073/pnas.1200613109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Budolfson K., and Wang F.. 2011. Pik3c3 deletion in pyramidal neurons results in loss of synapses, extensive gliosis and progressive neurodegeneration. Neuroscience. 172:427–442 10.1016/j.neuroscience.2010.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Karki S.B., Kuznetsov S.A., Tabb J.S., Weiss D.G., Langford G.M., and Holzbaur E.L.. 1997. The interaction between cytoplasmic dynein and dynactin is required for fast axonal transport. Proc. Natl. Acad. Sci. USA. 94:12180–12185 10.1073/pnas.94.22.12180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver C., Leidel C., Szpankowski L., Farley N.M., Shubeita G.T., and Goldstein L.S.. 2013. Endogenous GSK-3/shaggy regulates bidirectional axonal transport of the amyloid precursor protein. Traffic. 14:295–308 10.1111/tra.12037 [DOI] [PubMed] [Google Scholar]
- Xu K., Zhong G., and Zhuang X.. 2013. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 339:452–456 10.1126/science.1232251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T.Y., Quintyne N.J., Scipioni B.R., Eckley D.M., and Schroer T.A.. 2012. Dynactin’s pointed-end complex is a cargo-targeting module. Mol. Biol. Cell. 23:3827–3837 10.1091/mbc.E12-07-0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zala D., Hinckelmann M.V., Yu H., Lyra da Cunha M.M., Liot G., Cordelières F.P., Marco S., and Saudou F.. 2013. Vesicular glycolysis provides on-board energy for fast axonal transport. Cell. 152:479–491 10.1016/j.cell.2012.12.029 [DOI] [PubMed] [Google Scholar]
- Zhang J., Yao X., Fischer L., Abenza J.F., Peñalva M.A., and Xiang X.. 2011. The p25 subunit of the dynactin complex is required for dynein–early endosome interaction. J. Cell Biol. 193:1245–1255 10.1083/jcb.201011022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Cai Q., Xie Y., and Sheng Z.H.. 2012. Snapin recruits dynein to BDNF-TrkB signaling endosomes for retrograde axonal transport and is essential for dendrite growth of cortical neurons. Cell Reports. 2:42–51 10.1016/j.celrep.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang L., Hasegawa H., Amin P., Han B.X., Kaneko S., He Y., and Wang F.. 2010. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc. Natl. Acad. Sci. USA. 107:9424–9429 10.1073/pnas.0914725107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.