Abstract
Systemic lupus erythematosus (SLE) is a prototypic systemic autoimmune disorder. Considerable progress has been made to delineate the genetic control of this complex disorder. In this review, selected aspects of human and mouse genetics related to SLE are reviewed with emphasis on genes that contribute to both innate and adaptive immunity and to genes that contribute directly to susceptibility to end organ damage. It is concluded that the interactions among these two major pathways will provide further insight into the pathogenesis of SLE. An interactive model of the two major pathways is proposed without emphasis on the importance of breaking tolerance to autoantigens.
Systemic lupus erythematosus (SLE) is considered to be a prototype of systemic autoimmune disorder. It is characterized by the presence of autoantibodies with complex specificities and protean clinical presentations at the initial diagnosis and relapses [1]. Both genetic and environmental factors play significant roles in its pathogenesis. The high heritability of human SLE and a higher disease concordance rate in monozygotic twins support a strong genetic contribution to the development of SLE [2•]. Most genetic studies have focused on genes affecting immune responsiveness. End organ responses to immune effectors have rarely been considered. In this review, the recent advances in the genetics of SLE will be reviewed. Emphasis will be made on the recent observation that end organ resistance to damage may be crucial to the clinical manifestation of SLE.
Overview of human SLE genetics
The initial approaches, including linkage analysis and candidate gene association studies, identified and confirmed a limited number of SLE-associated loci (e.g. HLA-DR2/DR3). The genome-wide association study (GWAS) approach to screen hundreds of thousands of single nucleotide polymorphisms (SNPs) across the genome in a hypothesis-free manner has advanced our understanding of genetic basis of SLE. Since 2007, eight GWAS in SLE (four in European-derived [3–6] and four in Asian populations [7•,8–10]) and subsequent meta-analysis and large-scale replication studies have revealed a growing number of risk loci exceeding the genome-wide significance level (P < 5 × 10−8). Fine mapping and functional characterization of GWAS signals have localized candidate causative variants, identification of target gene(s) directly influenced by the associated variants and elucidation of pathogenic mechanisms to understand how SLE susceptibility genes affect disease risk. Few of disease-associated variants affect gene coding sequences altering functions of the encoded proteins, whereas most fall in the noncoding regions regulating gene expression through transcriptional and/or posttranscriptional mechanisms. The majority of the established SLE susceptibility genes encode products involved in innate and adaptive immunity, particularly the three key immunological pathways contributing to the pathogenesis of SLE: firstly, clearance of apoptotic cells and immune complexes (ICs); secondly, activation of toll-like receptor (TLR), type I interferon (IFN) and NF-κB signaling; and finally, multiple dysfunctions in T/B cell signaling (Table 1). Two recent reviews have appeared to deal with gene-function studies in SLE genetics [11,12]. Only selected areas of interest will be discussed.
Table 1.
SLE-associated genes in the disease pathwaysa
| Function | Position | Gene | OR | Population |
|---|---|---|---|---|
| Innate immune response | ||||
| Clearance of apoptotic cells and Immune Complexes | 1p36 | C1Q | Rare, complete deficiency | |
| 1q23 | FCGR2A | 1.3–1.4 | EU,EA,AA,AS | |
| FCGR3A | 1.2–1.5 | EU,AA | ||
| FCGR2B | 1.3–2.5 | AS | ||
| FCGR3B | 1.7–2.3 | EU,AA | ||
| 3p21.31 | TREX1 | Rare mutation | ||
| 6p21.3 | C4A/4B, C2 | Rare, complete deficiency | ||
| 12p13 | C1R/C1S | Rare, complete deficiency | ||
| 16p13.3 | DNASE1 | Rare mutation | ||
| 6q21 | ATG5 | 1.2–1.3 | EU,AS | |
| 16p11.2 | ITGAM | 1.3–2.1 | EA,EU,AA,AS,HS | |
| Type I IFN pathway | 2q24 | IFIH1 | 1.1–1.4 | EA,AA |
| 2q32 | STAT4 | 1.4–1.8 | EU,EA,AS,HS,AA | |
| 5q34 | miR146a | 1.2–1.3 | AS | |
| 7q32 | IRF5 | 1.3–1.9 | EU,EA,AA,AS,HS | |
| 11p15 | IRF7 | 1.3–2.0 | EU,EA,AA,AS | |
| 12q24.32 | SLC15A4 | 1.1–1.3 | EA,AS | |
| 16q24 | IRF8 | 1.2–1.3 | EU,EA | |
| 19p13 | TYK2 | 1.3 | EA | |
| Xp22 | TLR7 | 1.2–2.3 | AS,EA,AA,HS | |
| NFκB pathway | 5q33.1 | TNIP1 | 1.2–1.4 | EA,AS |
| 6q23 | TNFAIP3 | 1.7–2.3 | EU,EA,AS | |
| 22q11.21 | UBE2L3 | 1.2–1.4 | EU,EA,AS | |
| Xq28 | IRAK1 | 1.1–1.6 | EA,AS,AA,HS | |
| Adaptive immune response | ||||
| Antigen presentation T & B cell signaling | 6p21.3 | HLA region | 1.5–2.5 | EU, AS |
| 1p13.2 | PTPN22 | 1.4–2.4 | EU,HS | |
| 1q25 | TNFSF4 | 1.2–1.5 | EU,EA,AS,AA,HS | |
| 1q31-q32 | IL10 | 1.2–1.3 | EU,EA | |
| 2p25-p24 | RASGRP3 | 1.2–1.4 | AS,EU | |
| 3q13 | CD80 | 1.3 | AS | |
| 4q21 | AFF1 | 1.2 | AS | |
| 4q24 | BANK1 | 1.2–1.4 | EU,EA,AS,AA | |
| 4q26-q27 | IL21 | 1.1–1.6 | EA,AA | |
| 6q21 | PRDM1 | 1.2 | EA | |
| 7p12.2 | IKZF1 | 1.2–1.4 | EU, AS | |
| 8p23 | BLK | 1.2–1.6 | EU,EA,AS,AA | |
| 8q13 | LYN | 1.2–1.3 | EU | |
| 10q21 | ARID5B | 1.2 | AS | |
| 11p13 | PDHX/CD44 | 1.2–1.4 | EA,AA,AS | |
| 11q23.3 | ETS1 | 1.2–1.4 | AS,EU | |
| 13q13 | ELF1 | 1.3 | AS | |
| 15q24.1 | CSK | 1.3 | EU | |
| 16p11.2 | PRKCB | 1.2 | AS | |
| 17q21 | IKZF3 | 1.2–1.9 | EU,AA | |
| Others | ||||
| ROS generation | 1q25 | NCF2 | 1.2–2.8 | EA,AA,HS |
| DNA methylation | Xq28 | MECP2 | 1.1–1.6 | EA,AS,AA,HS |
| Cell adhesion | 19p13 | ICAM1/4/5 | 1.2 | EU,AA,HS,AS |
| Unknown | 2p13 | TET3 | 1.3 | AS |
| 3p14.3 | PXK | 1.2–1.3 | EU,EA | |
| 3q13.33 | TMEM39A | 1.2–1.4 | EU,AA,AS | |
| 5q35 | PTTG1 | 1.2 | EU | |
| 6p21 | UHRF1BP1 | 1.2–1.5 | EU,AS | |
| 7p15.2 | JAZF1 | 1.2 | EA | |
| 8p23.1 | XKR6 | 1.2–1.3 | EU | |
| 10q11.23 | WDFY4 | 1.2–1.3 | AS | |
| 12p13 | CDKN1B | 1.2 | AS | |
| 12q23 | DRAM1 | 1.2 | AS | |
| 16p13.13 | CLEC16A | 1.2 | EU,AS |
Abbreviation AA, African American; AS, Asian; EA, European American; EU, European; HS, Hispanic. ARID5B, AT rich interactive domain 5B; AFF1, AF4/FMR2 family member 1; ATG5, autophagy related 5; BANK1, B-cell scaffold protein with ankyrin repeats 1; BLK, B lymphoid tyrosine kinase; C1Q, complement component 1, q subcomponent; C1R, complement component 1, r subcomponent; C1S, complement component 1, s subcomponent; C2, complement component 2; C4A/4B, complement component 4A/4B; CD44, CD44 molecule; CD80, CD80 molecule; CDKN1B, cyclin-dependent kinase inhibitor 1B; CLEC16A, C-type lectin domain family 16 member A; CSK, c-src tyrosine kinase; DNASE1, deoxyribonuclease I; DRAM1, DNA-damage regulated autophagy modulator 1; ELF1, E74-like factor 1; ETS1, v-ets avian erythroblastosis virus E26 oncogene homolog 1; FCGR2A, Fc fragment of IgG low affinity IIa receptor; FCGR3A, Fc fragment of IgG low affinity IIIa receptor; FCGR2B, Fc fragment of IgG low affinity IIb receptor; FCGR3B, Fc fragment of IgG low affinity IIIb receptor; ICAM1, intercellular adhesion molecule 1; ICAM4, intercellular adhesion molecule 4; ICAM5, intercellular adhesion molecule 5; IFIH1, interferon induced with helicase C domain 1; IKZF1, IKAROS family zinc finger 1; IKZF3, IKAROS family zinc finger 3; IL10, interleukin 10; IL21, interleukin 21; IRF5, interferon regulatory factor 5; IRF7, interferon regulatory factor 7; IRF8, interferon regulatory factor 8; IRAK1, interleukin-1 receptor-associated kinase 1; ITGAM, integrin alpha M; JAZF1, JAZF zinc finger 1; LYN, v-yes-1 Yamaguchi sarcoma viral related oncogene homolog; MECP2, methyl CpG binding protein 2; miR146a, microRNA146a; NCF2, neutrophil cytosolic factor 2; PDHX, pyruvate dehydrogenase complex component X; PRDM1, PR domain containing 1 with ZNF domain; PRKCB, protein kinase C beta; PTPN22, protein tyrosine phosphatase non-receptor type 22; PTTG1, pituitary tumor-transforming 1; PXK, PX domain containing serine/threonine kinase; RASGRP3, RAS guanyl releasing protein 3; SLC15A4, solute carrier family 15 (oligopeptide transporter) member 4; STAT4, signal transducer and activator of transcription 4; TET3, tet methylcytosine dioxygenase 3; TLR7, toll-like receptor 7; TMEM39A, transmembrane protein 39A; TNFAIP3, tumor necrosis factor alpha-induced protein 3; TNIP1, TNFAIP3 interacting protein 1; TNFSF4, tumor necrosis factor superfamily member 4; TREX1, three prime repair exonuclease 1; TYK2, tyrosine kinase 2; UBE2L3, ubiquitin-conjugating enzyme E2L3; UHRF1BP1, UHRF1 binding protein 1; WDFY4, WDFY family member 4; XKR6, XK Kell blood group complex subunit-related family member 6.
Most of these loci have been reviewed in the reference [13], and those that have causative variants are highlighted in the text.
SLE susceptibility genes in innate immune responses
Clearance of apoptotic cells and ICs
Inefficient clearance of apoptotic cells and ICs that may result in autoantigens accumulation promote initiation and maintenance of autoimmune responses in SLE. Deficiencies or polymorphisms in genes encoding components required for this process confer susceptibility to SLE (reviewed in [13]). For example, ITGAM encodes the α-chain of αMβ2 integrin that functions in phagocytosis of complement-coated particles and ICs as well as regulation of leukocyte apoptosis, adhesion and migration via interaction with a range of ligands. The SLE-associated missense ITGAM variant confers impaired phagocytosis of complement-opsonized targets by monocytes, neutrophils and macrophages, which might alter IC clearance and deposition, resulting in tissue damage [14]. This is supported by the finding that patients carrying the ITGAM risk variant show an increased risk in development of lupus nephritis [15,16].
Type I IFN pathway
Dysregulation of type I IFN is considered as one of the central drivers of SLE pathogenesis. More than half of the identified SLE susceptibility genes encode proteins that can be directly or indirectly linked to this pathway. TLRs (e.g. TLR7) or other cytosolic sensors (e.g. IFIH1) is a major trigger of type I IFN production in SLE. SLE-associated variants of TLR7 that bind cellular and microbial nucleic acids or IFIH, a viral nucleic acid sensor confer functional effects on affecting gene transcription or structure, and patients carrying the risk alleles show increased IFN signature [17•,18••,19,20••]. IRF5, IRF7 and IRF8, a family of transcription factors downstream of endosomal TLRs, are required for activating transcription of type I IFN genes. SLE-associated variants of IRF5 and IRF7 have functional impacts on increased serum IFN-α and such impacts depend on the presence of specific autoantibodies [21]. MiR-146a has been recognized as a negative regulator of immune activation by inhibiting key signaling proteins in the type I IFN pathway [22]. The SLE-risk miR146a promoter variant confers lower levels of its transcript due to decreased binding of transcription factor Ets-1 [23].
NFκB pathway
SLE risk genes that function in the NFκB pathway downstream of TLR engagement include TNFAIP3, TNIP1, UBE2L3 and IRAK1. TNFAIP3 encodes a deubiquitinating enzyme (A20) participating in the termination of NFκB signaling. Development of lupus-like phenotypes in mice with full or conditional A20 deficiency indicates that reduced A20 expression predisposes to autoimmunity [24]. Consistently, the SLE-associated TNFAIP3 variant with inefficient delivery of nuclear protein complex to the TNFAIP3 promoter attenuates A20 expression, resulting in enhanced NFκB pathway activity and predisposition to SLE [25••]. The X-linked IRAK1 gene encodes a kinase acting with MyD88 to regulate NFκB activation. The missense variant in IRAK1 which confers increased NFκB activity captures a risk haplotype associated with SLE in multiple ancestries [26••].
SLE susceptibility genes in adaptive immune responses
T cell signaling and function
Genes involved in T cell functions have been associated with SLE. The importance of highly conserved and extended haplotypes bearing the class II HLA-DR2 and HLA-DR3 alleles in SLE susceptibility has been well recognized [13]. HLA class II molecules are critical in mediating host defense responses through antigen presentation and immune tolerance by self/non-self recognition. Given of their roles in T-cell dependent antibody and effector responses, cumulative evidence supports association of class II alleles with autoantibody production, especially HLA-DR3 with anti-Ro/La antibodies [27•]. PTPN22 encodes a tyrosine phosphatase that regulates immune signaling. Humans carrying the SLE-risk 620W allele and knockin mice expressing the analogous 619W mutation show altered TCR and BCR signaling with enhanced B cell autoreactivity (reviewed in [28••]). In myeloid cells [29], the risk R620W allele is associated with diminished TLR-induced type I IFN production, but with enhanced functions in neutrophils including increased transendothelial migration, Ca2+ release and ROS (reactive oxygen species) production [30].
B cell signaling and function
B cells are critical players in SLE pathogenesis through both antibody-dependent and independent functions (reviewed in [31]). Multiple genes encoding kinases, adaptor molecules and cytokines associated with B cell functions confer risk for SLE. CSK encoding c-Src tyrosine kinase regulates Src kinases (e.g. Lyn) activation by interaction with tyrosine phosphatases in lymphocytes. The SLE-risk CSK variant is associated with high CSK expression, increased phosphorylation of Lyn, enhanced BCR-mediated activation of mature B cells and expansion of transitional B cells, suggesting CSK is involved in multiple developmental stages of B cells [32]. BLK encodes a member of the Src family kinases functioning in intracellular signaling and regulation of B cell differentiation and tolerance. SLE-risk alleles of the BLK promoter variants confer reduced promoter activity in B cell lines representing different B cell developmental stages, implicating that decreased BLK expression may affect B cell development and impair functional responses in B cells [33••]. BANK1, encoding an adaptor/scaffold protein, facilitates intracellular calcium release and alters the B cell activation threshold. The three SLE-associated BANK1 variants contribute to sustained BCR signaling and B cell hyperactivity characteristic of SLE [4]. IL-10 is a pivotal cytokine with immunosuppressive and immunostimulatory properties. The IL10 risk allele confers increased IL10 expression by preferentially binding to the activated transcription factor Elk-1 associated with disease activity in B cells of SLE patients [34••]. Other SLE-associated genes with roles in T/B cell functions (Table 1) await delineation of functional mechanisms by which the associated variants affect disease risk.
SLE susceptibility genes with other functions
In addition to immune signaling, several SLE susceptibility genes with functions linked to the other pathways contribute to the pathogenesis of SLE. For example, NCF2 encodes a subunit of NADPH (nicotinamide adenine dinucleotide phosphate) oxidase complex involved in the ROS generation. Overproduction of ROS may lead to oxidative stress that stimulates autoimmune responses. A surprising finding of the SLE-risk variant in human NCF2 gene leading to low NADPH oxidase activity and reduced ROS production provides new insight of an antiinflammatory role for ROS in autoimmune disease [35•]. MECP2 encodes a transcriptional regulator to modulate expression of methylation-sensitive genes. Healthy individuals carrying the SLE-risk IRAK1-MECP2 haplotype with increased levels of a specific MECP2 transcript isoform in stimulated T cells showed DNA hypomethylation of IFN-regulated genes [36••]. Other SLE-associated loci (Table 1) with unknown immune function require fine mapping and characterization of functional pathways relevant to the development of SLE.
Emergence of genes controlling susceptibility to end organ damage
Since non-muscle myosin heavy chain 9 (MYH9) has been implicated in focal segmental glomerulosclerosis, HIV-associated nephropathy, hypertensive end-stage renal disease (ESRD) and diabetes and non-diabetic ESRD, its role in lupus nephritis was investigated together with the closely-linked gene APOL1 [37]. No significant association was found in EA, AA, Asians, Amerindians or Hispanics. However, a recent study published in Arthritis & Rheumatology by Freedman et al. indicates that APOL1 G1/G2 alleles strongly impact the risk of lupus nephritis-ESRD in African Americas, as well as the time of progression to ESRD [38••]. This suggests that APOL1 is associated with renal susceptibility to damage. This interpretation is supported by the clinical observation that kidneys from diseased donors with two APOL1 nephropathy alleles fail more rapidly than those from diseased donors with 0 or 1 risk allele [39]. This is the first human gene to be associated with end organ damage in SLE.
Lesions learned from New Zealand Mixed (NZM) models of lupus
NZM2410
The contributions to our understanding of the pathogenesis of SLE by studies in animal models have been extensively reviewed by Hahn and Kono [40]. The recent studies on the NZM strains have enhanced our understanding of the complexity of lupus genetics. The two most studied strains are NZM2410 and NZM2328. L. Morel has summarized the studies on NZM2410 [41••]. Because of the approach taken by her and her colleagues by generating congenic lines with Sle1-3 on B6 background and the lack of glomerulonephritis (GN) in the single congenic lines, the genes in these loci contributing to lupus GN which is the phenotype in the initial mapping studies have proven to be elusive. Nevertheless, the genetic dissection of the genes in NZM2410 has provided us further understanding of genes that control autoimmune responses and in some case provided us novel insights in the immune system. In addition, a suppressor gene Sles1 has been identified, further illustrating the complexity of lupus genetics.
NZM2328
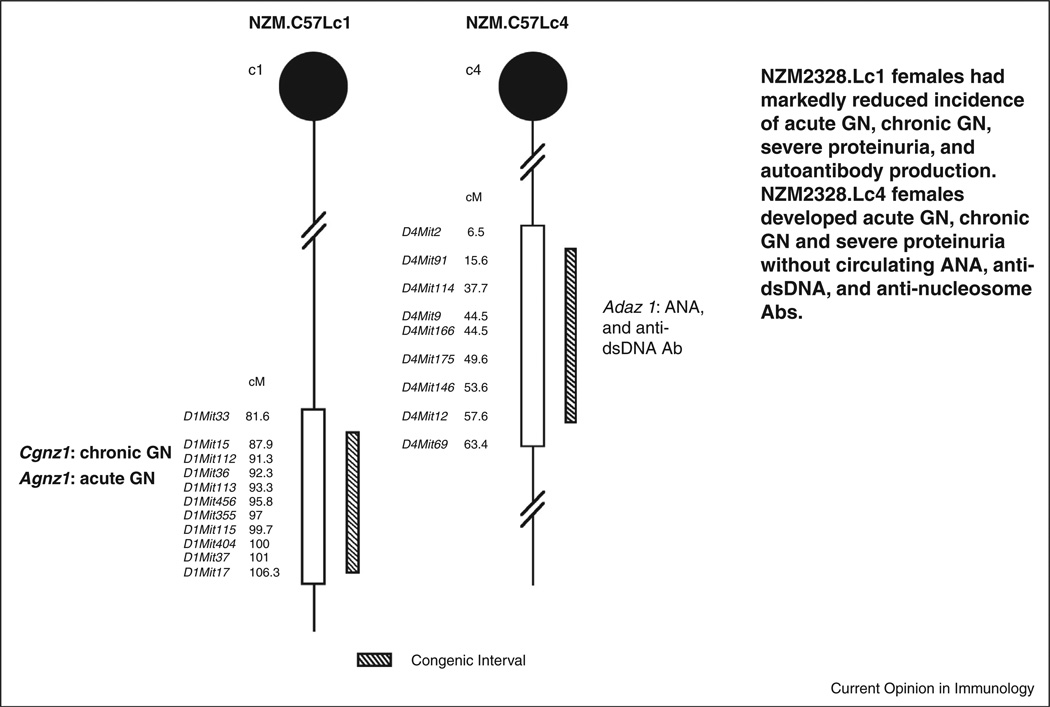
The genetic dissection of lupus pathogenesis was carried out in Fu’s laboratory. NZM2328 was chosen because its GN resembles that of human proliferative GN with severe proteinuria and ESRD observed predominantly in female mice [42•]. Analysis of a cohort of female mice of the (NZM2328XC57L/J) F1XNZM2328 identified a locus Cgnz1 on chromosome 1 that contributes to the development of severe proteinuria and ESRD, a locus Adaz1 on chromosome 4 for ANA and anti-dsDNA antibody production and three loci, Agnz1 on chromosome 1, and H2-Tnf complex and Agnz2 on chromosome 17 for acute GN with IC deposition and complement fixation without tubular damage and interstitial fibrosis. Two congenic strains, NZM.C57Lc1 and NZM.C57Lc4 (Figure 1) were generated by transgressing genetic regions from C57L/J containing the loci of Cgnz1 and Agnz1 on chromosome 1 and Adaz1 on chromosome 4 respectively [43••]. NZM.Lc1 female mice had markedly reduced incidence of severe proteinuria and ESRD. NZM.Lc4 female mice developed acute GN with IC deposition and complement activation that progressed to chronic GN and ESRD with the same kinetics as NZM2328 female mice without ANA and anti-dsDNA antibodies. NZM.Lc1 provided us the opportunity to further dissect the gene(s) associated with Cgnz1. NZM.Lc4 provided us the evidence that ANA and anti-dsDNA antibodies are dispensable in the induction of lupus GN suggesting to us that a less ANA-and-anti-dsDNA antibody-centric hypothesis is warranted.
Figure 1.
NZM.C57/Lc1 (Lc1) and NZM.C57Lc4 (Lc4) congenic lines were derived by replacing the genetic intervals in NZM2328 with those from C57L/J (hatched bar). The genetic intervals with SLE susceptibility genes in NZM2328 delineated by informative microsatellite markers are shown (open bars). Chromosome intervals are drawn to scale. The genes involved in Lc1 and Lc4 congenic lines are Cgnz1 and Agnz1, and Adaz1 respectively. The characteristics of these two congenic lines are included (modified from [43••]).
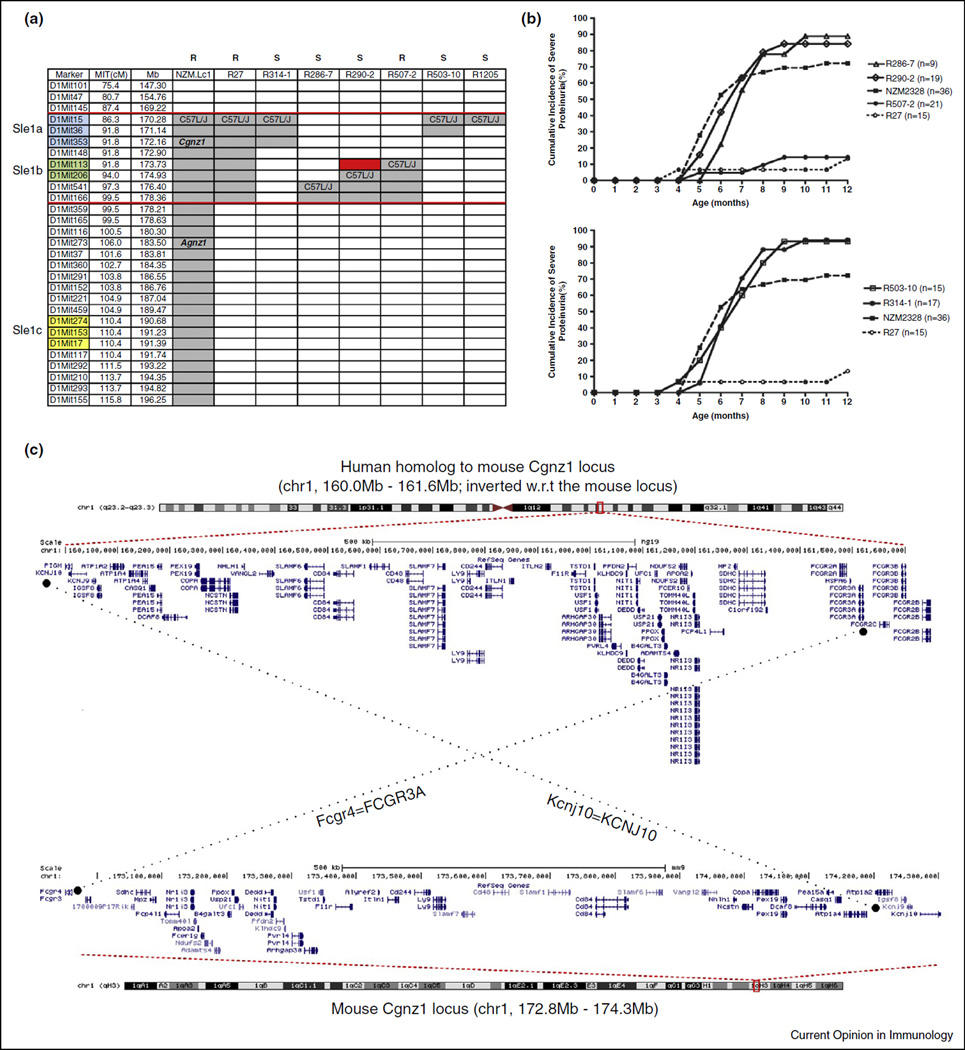
Recently we published our characterization of the genetic interval associated with chronic GN and ESRD [44••]. The results are summarized in Figure 2a. Cgnz1 has been localized to a 1.34 Mb region on chromosome 1. The region contains 45 genes including many of the Slam family members and those controlling metabolism, apoptosis, autophagy and other biological processes. This region mapped within Sle1b indicating Cgnz1 is identical to Sle1. The human homologous region is also depicted in Figure 2c. The human homologous region is on chromosome 1q23 from 160.0 to 161.1 Mb. With the exception of LOC10041098, each of the mouse genes in the Cgnz1 locus has human counterparts in the homologous region and the exact order is preserved between these two species. The conservative nature of the gene order indicates that this region is likely to be important in human lupus nephritis.
Figure 2.
Fine mapping of Cgnz1 region on mouse chromosome 1 and the human homologous region. (a) Six intrachromosomal recombinant lines, R314-1,R286-7,R290-2, R507-2, R503-10 and R1205 were generated by generation of heterozygous mice from (R27×NZM2328)F1 × (R27×NZM2328)F1 and the heterozygous mice were bred to generate homozygous lines. R27 has a c1 segment covering both Sle1a and Sle1b from C57L/Jc1. 314-1 has the whole region of Sle1a from C57L/Jc1. R503-10 has only part of the Sle1a from C57L/Jc1. R286-7 lacks both Sle1a and Sle1b from C57L/Jc1. R290-2 contains part of the Sle1b from C57L/Jc1. R507-2 has the whole Sle1b region from C57L/Jc1. R1205 is a newly generated line with early mortality and it contains 2–19 genes depending on further delineation of the recombinant sites. The red rectangle depicts the region of 1.34 Mb where Cgnz1 locates. The gray areas denote the intervals in NZM2328 that are replaced with that of C57L/J (modified from [44••]). (b) Five intrachromosomal recombinant lines, R314-1, R286-7, R290-2, R507-2 and R503-10 were generated and a female cohort of these lines was followed for 12 months. All but R507-2 were susceptible to cGN. These results provide a genetic region located between microsatellite markers Mit148 and Mit206. This region is within the Sle1b region. The data show that the other two Sle1 subregions, that is, Sle1a and Sle1c, are not important for cGN. Female mice of R314-1, R286-7, R290-2, and R503-10 develop cGN in larger percentages in comparison with the parental line NZM2328. Almost 100% of R314-1 and R503-10 develop severe proteinuria by seven months of age (modified from [44••]). (c) The mouse Cgnz1 locus is nearly perfectly homologous to a 1.6 Mb locus on human chromosome 1. The assembled human locus is inverted with respect to the mouse locus; nonetheless the exact gene order is preserved. Dotted lines connect the human and mouse homologs at the boundaries of the mouse and human Cgnz1 loci.
The phenotypes of intra-chromosomal 1 recombinant congenic lines as shown in Figure 2b suggest that there are significant interactions among the genes in the c1 interval. Female mice of NZM.Lc1 R314-1 and NZM.Lc1 R503-10 have accelerated development of severe proteinuria and ESRD, suggesting that a gene within the 169.22–172.16 Mb in NZM2328 has suppressor function. A new recombinant line NZM.Lc1R1205 appears to have similar phenotypes as those of R314-1 and R503-10. Further phenotypic and genetic characterization of congenic lines in this region indicate that this suppressive gene is located between 169.22 Mb and 171.14 Mb. Similarly there appears to be a suppressor gene within 174.93 Mb and 178.21 Mb in NZM2328. This complexity in gene-gene interaction on chromosome 1 mirrors that in NZM2410.
Future directions
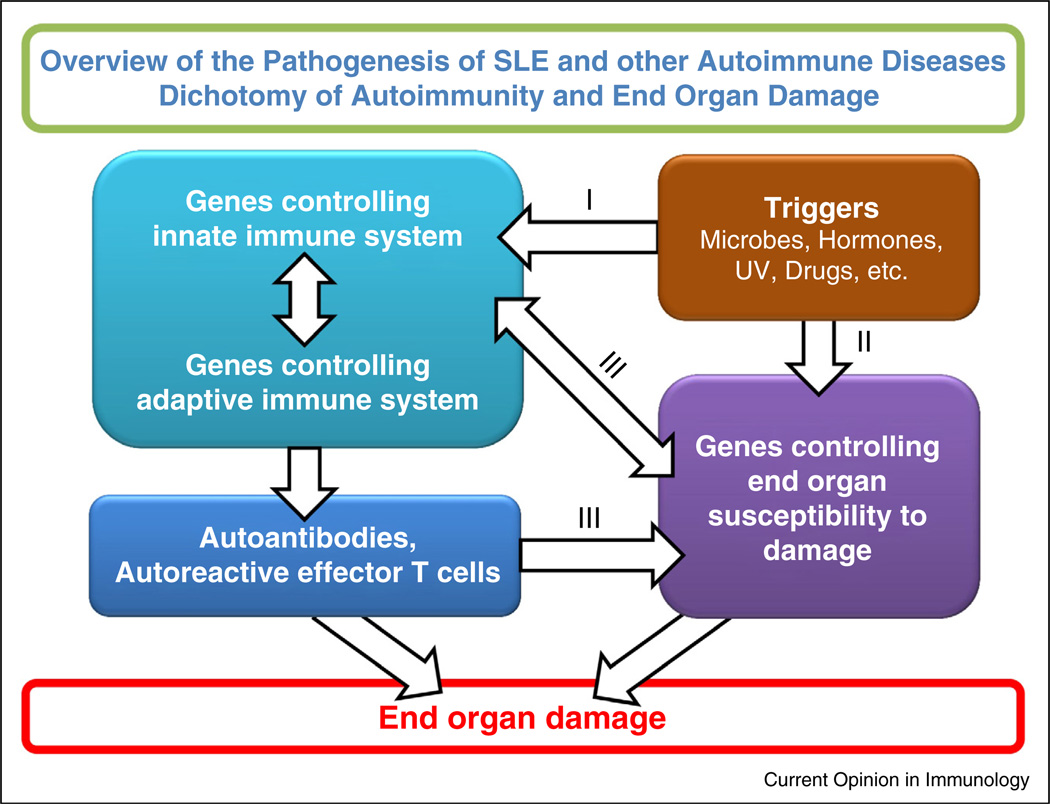
The implication of our findings in NZM2328 has been discussed in two review papers [45••,46]. The findings reviewed here provide us a frame-work for future investigation on the pathogenesis of SLE. Figure 3 summarizes our working model for the pathogenesis of SLE. It is important to note that this model has a general application to other autoimmune disorders such as rheumatoid arthritis, ankylosing spondylitis, type I diabetes mellitus, etc.
Figure 3.
Interactive model for the pathogenesis of SLE. This model makes the assumption that environmental triggers act on susceptible hosts. The triggers act on both genes controlling immune responsiveness and genes for end organ damage. These are two independent yet interactive pathways. Pathway I leads to the generation of autoantibodies and autoreactive effector T cells. Pathway II provides autoantigens and/or soluble mediators that influence immune responsiveness. Pathways I and II interact at several levels as indicated by III. These interactions can lead to end organ damage.
Future challenges in understanding SLE genetics include discovery of novel associations responsible for the missing heritability, identification of causal variants with functional characterization at each locus, and implication of new pathogenic pathways contributing to SLE development. Next-generation sequencing based on whole-exome/genome investigation may accelerate the identification of novel, particularly rare variants, which are functionally important, penetrant and harbor large effect sizes for SLE risk. The emergence of genomic and epigenomic data sets, such as eQTL (expression quantitative-trait loci) and ENCODE (Encyclopedia of DNA Elements), have revolutionized functional annotation of many GWAS-identified variants with cis/trans-regulatory effects in the relevant cell types, which help to predict the most likely causal variants and target genes in SLE. Pathway-based and network-based analyses are another useful strategy to prioritize disease target genes. New technologies, like CRISPR/Cas-based methods to generate isogenic cell lines or heterozygous allele modifications in animal models, will expand our ability to validate the functional mechanisms. Continuing efforts should be made to utilize mouse models to identify human homologous genes that can be explored further. We expect that these approaches will enable rapid progress from genetic studies to biological knowledge, provide a better understanding of SLE molecular pathogenesis and guide the development of therapeutic interventions.
Acknowledgements
This work is supported by grants from NIH P50-AR04522, R01-AR047988, and R01-AR049449 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, R01-AI079621 from the National Institute of Allergy and Infectious Diseases and grants from Alliance for Lupus Research to SMF, a grant from National Human Genome Research Institute, R01-HG006693 to AQ and grants from NIH R01-AR043814 and R21-AR065626 and the Alliance for Lupus Research to BPT.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2. Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, Walker A, Mack TM. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992;35:311–318. doi: 10.1002/art.1780350310. This remains the best study on twins in SLE providing an estimate of environmental influences on its pathogenesis.
- 3.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 4.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, Gunnarsson I, Svenungsson E, Sturfelt G, Jonsen A, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 5.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, Burtt NP, Guiducci C, Parkin M, Gates C, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. This study shows importance of racial composition of large cohort GWA studies.
- 8.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, Hirankarn N, Ying D, Pan HF, Mok CC, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okada Y, Shimane K, Kochi Y, Tahira T, Suzuki A, Higasa K, Takahashi A, Horita T, Atsumi T, Ishii T, et al. A genome-wide association study identified AFF1 as a susceptibility locus for systemic lupus eyrthematosus in Japanese. PLoS Genet. 2012;8:e1002455. doi: 10.1371/journal.pgen.1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee HS, Kim T, Bang SY, Na YJ, Kim I, Kim K, Kim JH, Chung YJ, Shin HD, Kang YM, et al. Ethnic specificity of lupus-associated loci identified in a genome-wide association study in Korean women. Ann Rheum Dis. 2014;73:1240–1245. doi: 10.1136/annrheumdis-2012-202675. [DOI] [PubMed] [Google Scholar]
- 11.Guerra SG, Vyse TJ, Graham DSC. The genetics of lupus: a functional perspective. Arthritis Res Ther. 2012;14:211. doi: 10.1186/ar3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crispin JC, Hedrich CM, Tsokos GC. Gene-function studies in systemic lupus erythematosus. Nat Rev Rheumatol. 2012;9:476–484. doi: 10.1038/nrrheum.2013.78. [DOI] [PubMed] [Google Scholar]
- 13.Deng Y, Tsao BP. Genetics of human SLE. In: Wallace DJ, Hahn BH, editors. Dubois’ Lupus Erythematosus and Related Syndromes. Elsevier Health Sciences; 2012. pp. 35–45. [Google Scholar]
- 14.Fossati-Jimack L, Ling GS, Cortini A, Szajna M, Malik TH, McDonald JU, Pickering MC, Cook HT, Taylor PR, Botto M. Phagocytosis is the main CR3-mediated function affected by the lupus-associated variant of CD11b in human myeloid cells. PLoS ONE. 2013;8:e57082. doi: 10.1371/journal.pone.0057082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W, Zhao M, Hirankarn N, Lau CS, Mok CC, Chan TM, Wong RW, Lee KW, Mok MY, Wong SN, et al. ITGAM is associated with disease susceptibility and renal nephritis of systemic lupus erythematosus in Hong Kong Chinese and Thai. Hum Mol Genet. 2009;18:2063–2070. doi: 10.1093/hmg/ddp118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim-Howard X, Maiti AK, Anaya JM, Bruner GR, Brown E, Merrill JT, Edberg JC, Petri MA, Reveille JD, Ramsey-Goldman R, et al. ITGAM coding variant (rs1143679) influences the risk of renal disease, discoid rash and immunological manifestations in patients with systemic lupus erythematosus with European ancestry. Ann Rheum Dis. 2010;69:1329–1332. doi: 10.1136/ard.2009.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen N, Fu Q, Deng Y, Qian X, Zhao J, Kaufman KM, Wu YL, Yu CY, Tang Y, Chen JY, et al. Sex-specific association of X-linked Toll-like receptor 7 (TLR7) with male systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2010;107:15838–15843. doi: 10.1073/pnas.1001337107. This is a well-designed investigation on the role of TLR7 in male lupus patients.
- 18. Deng Y, Zhao J, Sakurai D, Kaufman KM, Edberg JC, Kimberly RP, Kamen DL, Gilkeson GS, Jacob CO, Scofield RH, et al. MicroRNA-3148 modulates allelic expression of toll-like receptor 7 variant associated with systemic lupus erythematosus. PLoS Genet. 2013;9:e1003336. doi: 10.1371/journal.pgen.1003336. This study establishes the TLR7 3′UTR SNP as a causal variant associated with SLE in multiple ancestries, which confers allelic effect on transcript turnover via differential binding to the epigenetic factor miR-3148.
- 19.Robinson T, Kariuki SN, Franek BS, Kumabe M, Kumar AA, Badaracco M, Mikolaitis RA, Guerrero G, Utset TO, Drevlow BE, et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. J Immunol. 2011;187:1298–1303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Molineros JE, Maiti AK, Sun C, Looger LL, Han S, Kim-Howard X, Glenn S, Adler A, Kelly JA, Niewold TB, et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS Genet. 2013;9:e1003222. doi: 10.1371/journal.pgen.1003222. This genome-wide admixture mapping study reveals three independently associated variants in the IFIH1 gene and explains their molecular basis for SLE pathogenesis.
- 21.Salloum R, Niewold TB. Interferon regulatory factors in human lupus pathogenesis. Transl Res. 2011;157:326–331. doi: 10.1016/j.trsl.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen N, Liang D, Tang Y, de Vries N, Tak PP. MicroRNAs — novel regulators of systemic lupus erythematosus pathogenesis. Nat Rev Rheumatol. 2012;8:701–709. doi: 10.1038/nrrheum.2012.142. [DOI] [PubMed] [Google Scholar]
- 23.Luo X, Yang W, Ye DQ, Cui H, Zhang Y, Hirankarn N, Qian X, Tang Y, Lau YL, de Vries N, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011;7:e1002128. doi: 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vereecke L, Beyaert R, van Loo G. Genetic relationships between A20/TNFAIP3, chronic inflammation and autoimmune disease. Biochem Soc Trans. 2011;39:1086–1091. doi: 10.1042/BST0391086. [DOI] [PubMed] [Google Scholar]
- 25. Wang S, Wen F, Wiley GB, Kinter MT, Gaffney PM. An enhancer element harboring variants associated with systemic lupus erythematosus engages the TNFAIP3 promoter to influence A20 expression. PLoS Genet. 2013;9:e1003750. doi: 10.1371/journal.pgen.1003750. This study provides functional evidence supporting a causal role for TT > A risk variants in the genetic predisposition to SLE.
- 26. Kaufman KM, Zhao J, Kelly JA, Hughes T, Adler A, Sanchez E, Ojwang JO, Langefeld CD, Ziegler JT, Williams AH, et al. Fine mapping of Xq28: both MECP2 and IRAK1 contribute to risk for systemic lupus erythematosus in multiple ancestral groups. Ann Rheum Dis. 2013;72:437–444. doi: 10.1136/annrheumdis-2012-201851. This fine mapping study identifies a risk haplotype tagged by rs1059702 associated with SLE in four major ancestries. This SNP leads to an amino acid change in IRAK1 with known function of increasing NFκB activity and is associated with low MECP2 expression levels in PBMCs, suggesting both IRAK1 and MECP2 contribute to SLE susceptibility.
- 27. Morris DL, Fernando MM, Taylor KE, Chung SA, Nititham J, Alarcón-Riquelme ME, Barcellos LF, Behrens TW, Cotsapas C, Gaffney PM, et al. MHC associations with clinical and autoantibody manifestations in European SLE. Genes Immun. 2014;15:210–217. doi: 10.1038/gene.2014.6. This study provides evidence for a multilevel risk model for HLA-DRB1*03:01 in SLE, where the association with anti-Ro/La antibody-positive SLE is much stronger than SLE without these autoantibodies.
- 28. Bottini N, Peterson EJ. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu Rev Immunol. 2014;32:83–119. doi: 10.1146/annurev-immunol-032713-120249. This review summarizes current insights into functions of PTPN22 and its autoimmunity-associated variant in regulation of immune cell signaling and the mechanisms of action in autoimmune disease susceptibility.
- 29.Wang Y, Shaked I, Stanford SM, Zhou W, Curtsinger JM, Mikulski Z, Shaheen ZR, Cheng G, Sawatzke K, Campbell AM, et al. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity. 2013;39:111–122. doi: 10.1016/j.immuni.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayley R, Kite KA, McGettrick HM, Smith JP, Kitas GD, Buckley CD, Young SP. The autoimmune-associated genetic variant PTPN22 R620W enhances neutrophil activation and function in patients with rheumatoid arthritis and healthy individuals. Ann Rheum Dis. 2014 doi: 10.1136/annrheumdis-2013-204796. http://dx.doi.org/10.1136/annrheumdis-2013-204796. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 31.Anolik JH. B cell biology: implications for treatment of systemic lupus erythematosus. Lupus. 2013;22:342–349. doi: 10.1177/0961203312471576. [DOI] [PubMed] [Google Scholar]
- 32.Manjarrez-Orduno N, Marasco E, Chung SA, Katz MS, Kiridly JF, Simpfendorfer KR, Freudenberg J, Ballard DH, Nashi E, Hopkins TJ, et al. CSK regulatory polymorphism is associated with systemic lupus erythematosus and influences B-cell signaling and activation. Nat Genet. 2012;44:1227–1230. doi: 10.1038/ng.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guthridge JM, Lu R, Sun H, Sun C, Wiley GB, Dominguez N, Macwana SR, Lessard CJ, Kim-Howard X, Cobb BL, et al. Two functional lupus-associated BLK promoter variants control cell-type- and developmental-stage-specific transcription. Am J Hum Genet. 2014;94:586–598. doi: 10.1016/j.ajhg.2014.03.008. This study identifies two causal variants within the BLK promoter associated with SLE susceptibility by modulating BLK transcription in B cells and altering immune responses.
- 34. Sakurai D, Zhao J, Deng Y, Kelly JA, Brown EE, Harley JB, Bae SC, Alarcomicronn-Riquelme ME, Edberg JC, Kimberly RP, et al. Preferential binding to Elk-1 by SLE-associated risk allele upregulates expression. PLoS Genet. 2013;9:e1003870. doi: 10.1371/journal.pgen.1003870. This study identifies the IL10 upstream SNP rs3122605 as the likely causal variant responsible for the genetic association with SLE in European Americans. The risk allele preferentially binds to the activated Elk-1 conferring elevated IL10 expression.
- 35. Jacob CO, Eisenstein M, Dinauer MC, Ming W, Liu Q, John S, Quismorio FP, Jr, Reiff A, Myones BL, Kaufman KM, et al. Lupus-associated causal mutation in neutrophil cytosolic factor 2 (NCF2) brings unique insights to the structure and function of NADPH oxidase. Proc Natl Acad Sci U S A. 2012;109:E59–E67. doi: 10.1073/pnas.1113251108. This study identifies NCF2 as a candidate gene for childhood lupus and links the mutation to specific molecular function.
- 36. Koelsch KA, Webb R, Jeffries M, Dozmorov MG, Frank MB, Guthridge JM, James JA, Wren JD, Sawalha AH. Functional characterization of the MECP2/IRAK1 lupus risk haplotype in human T cells and a human MECP2 transgenic mouse. J Autoimmun. 2013;41:168–174. doi: 10.1016/j.jaut.2012.12.012. This study identifies that the SLE-risk haplotype confers increased expression level of MECP2 transcript isoform 2 in stimulated T cells and is associated with DNA methylation changes in multiple genetic loci, providing evidence for genetic-epigenetic interaction in SLE.
- 37.Lin CP, Adrianto I, Lessard CJ, Kelly JA, Kaufman KM, Guthridge JM, Freedman BI, Anaya JM, Alarcon-Riquelme ME, Pons-Estel BA, et al. Role of MYH9 and APOL1 in African and non-African populations with lupus nephritis. Genes Immun. 2012;13:232–238. doi: 10.1038/gene.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE, Birmingham DJ, Hebert LA, Hicks PJ, Segal MS, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;66:390–396. doi: 10.1002/art.38220. This study identifies the apolipoprotein L1 nephropathy risk alleles G1/G2 that strongly impact the risk and the time of end-stage renal disease in African Americans with lupus nephritis.
- 39.Reeves-Daniel AM, DePalma JA, Bleyer AJ, Rocco MV, Murea M, Adams PL, et al. The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant. 2011;11:1025–1030. doi: 10.1111/j.1600-6143.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn BH, Kono K. Animal models of SLE. In: Wallace DJ, Hahn BH, editors. Dubois’ Lupus Erythematosus and Related Syndromes. Elsevier Health Sciences; 2012. pp. 190–237. [Google Scholar]
- 41. Morel L. Mapping lupus susceptibility genes in the NZM2410 mouse model. Adv Immunol. 2012;115:113–139. doi: 10.1016/B978-0-12-394299-9.00004-7. This review summarizes all the work on NZM2410 with references to other investigations on lupus mouse models.
- 42. Waters ST, Fu SM, Gaskin F, Deshmukh US, Sung S-SJ, Kannapell CC, Tung KSK, McEwen SB, McDuffie M. NZM2328: a new mouse model of systemic lupus erythematosus with unique genetic susceptibility loci. Clin Immunol. 2001;100:372–383. doi: 10.1006/clim.2001.5079. This study identifies genes that are associated with two stages of lupus glomerulonephritis.
- 43. Waters ST, McDuffie M, Bagavant H, Deshmukh US, Gaskin F, Jiang C, Tung KSK, Fu SM. Breaking tolerance to double stranded DNA, nucleosome, and other antigens is not required for the pathogenesis of lupus glomerulonephritis. J Exp Med. 2004;199:255–264. doi: 10.1084/jem.20031519. This paper provides the evidence for the independent nature of genetic control of autoimmunity and susceptibility to end organ damage. It provides evidence that anti-dsDNA antibodies and ANA are not indispensable for the development of chronic GN and ESRD.
- 44. Ge Y, Jiang C, Sung S-SJ, Bagavant H, Wang H, Dai C, Cathro HP, Gaskin F, Fu SM. Cgnz1 allele confers kidney resistance to damage preventing progression of immune complex-mediated acute lupus glomerulonephritis. J Exp Med. 2013;210:2387–2401. doi: 10.1084/jem.20130731. This paper provides genetic evidence that acute GN is an autoimmune response and its progression to chronic GN is dependent on susceptibility of kidneys to damage.
- 45. Fu SM, Deshmukh US, Gaskin F. Pathogenesis of systemic lupus erythematosus revisited 2011: End organ resistance to damage, autoantibody initiation and diversification, and HLA-DR. J Autoimmun. 2011;37:104–112. doi: 10.1016/j.jaut.2011.05.004. This review summarizes the authors’ opinion on the pathogenesis of SLE.
- 46.Lewis JE, Fu SM, Gaskin F. Autoimmunity, end organ damage and the origin of autoantibodies and autoreactive T cells in systemic lupus erythematosus. Discov Med. 2013;15:85–92. [PMC free article] [PubMed] [Google Scholar]