Abstract
Environmental stimuli, including elevated carbon dioxide levels, regulate stomatal development1–3; however, the key mechanisms mediating the perception and relay of the CO2 signal to the stomatal development machinery remain elusive. To adapt CO2 intake to water loss, plants regulate the development of stomatal gas exchange pores in the aerial epidermis. A diverse range of plant species show a decrease in stomatal density in response to the continuing rise in atmospheric CO2 (ref. 4). To date, one mutant that exhibits deregulation of this CO2-controlled stomatal development response, hic (which is defective in cell-wall wax biosynthesis, ref. 5), has been identified. Here we show that recently isolated Arabidopsis thaliana β-carbonic anhydrase double mutants (ca1 ca4)6 exhibit aninversion in their response to elevated CO2, showing increased stomatal development at elevated CO2 levels. We characterized the mechanisms mediating this response and identified an extracellular signalling pathway involved in the regulation of CO2-controlled stomatal development by carbonic anhydrases. RNA-seq analyses of transcripts show that the extracellular pro-peptide-encoding gene EPIDERMAL PATTERNING FACTOR 2 (EPF2)7,8, but not EPF1 (ref. 9), is induced in wild-type leaves but not inca1 ca4 mutant leaves at elevated CO2 levels. Moreover, EPF2 is essential for CO2 control of stomatal development. Using cell-wall proteomic analyses and CO2-dependent transcriptomic analyses, we identified a novel CO2-induced extracellular protease, CRSP (CO2 RESPONSE SECRETED PROTEASE), as a mediator of CO2-controlled stomatal development. Our results identify mechanisms and genes that function in the repression of stomatal development in leaves during atmospheric CO2 elevation, including the carbonic-anhydrase-encoding genes CA1 and CA4 and the secreted protease CRSP, which cleaves the pro-peptide EPF2, in turn repressing stomatal development. Elucidation of these mechanisms advances the understanding of how plants perceive and relay the elevated CO2 signal and provides a framework to guide future research into how environmental challenges can modulate gas exchange in plants.
CO2 exchange between plants and the atmosphere, and water loss from plants to the atmosphere, depends on the density and the aperture size of plant stomata, and plants have evolved sophisticated mechanisms to control this flux1–3,10,11. Ecophysiological studies have highlighted the importance of stomatal density in the context of global ecology and climate change12. Plants adapt to the continuing rise in atmospheric CO2 concentration by reducing their stomatal density4 (that is, the number of stomata per unit of epidermal surface area). This change causes the leaf temperature to rise because of a decrease in the plant’s evapotranspirative cooling ability, while simultaneously increasing the transpiration efficiency of plants13. These phenomena, combined with the increasing scarcity of fresh water for agriculture, are predicted to dramatically impact on plant health12,14,15.
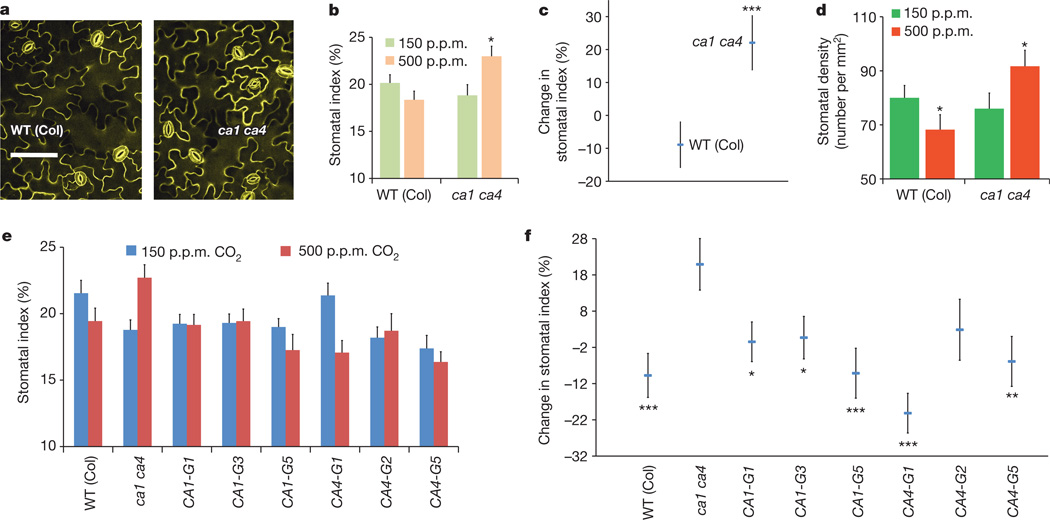
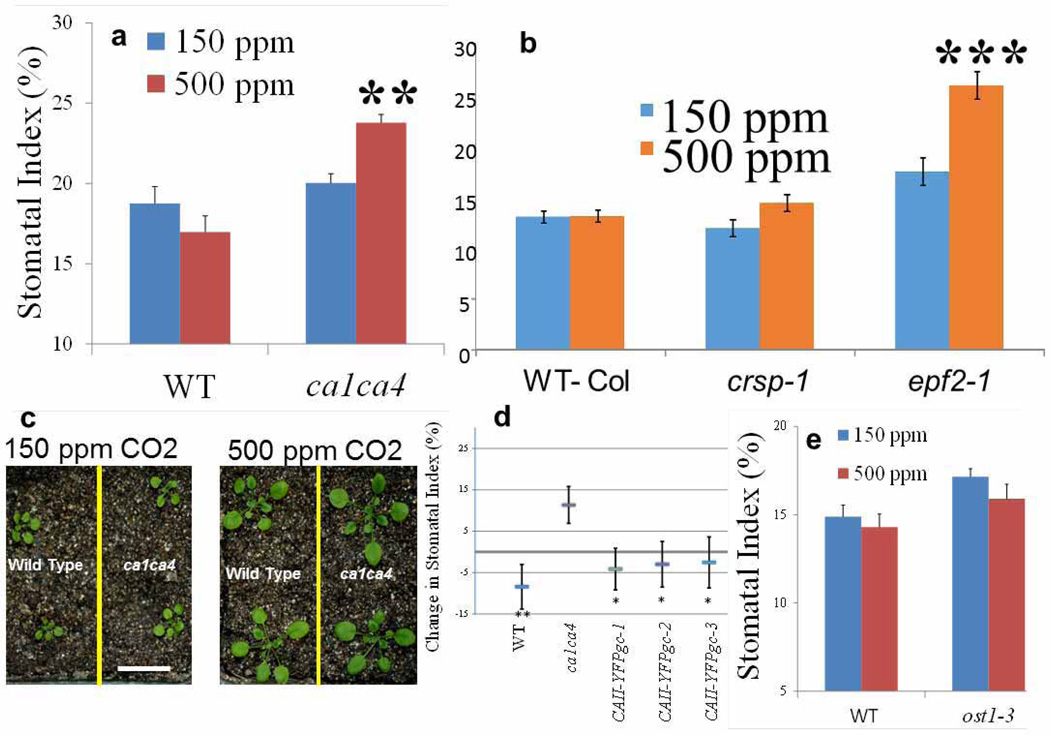
In recent research, we identified mutations in the A. thaliana βcarbonic anhydrase genesCA1 (At3g01500) andCA4 (At1g70410) that impair the rapid, short-term CO2-induced stomatal movement response6. Although ca1 ca4 (double mutant) plants show a higher stomatal density than wild-type plants, it remains unknown whether CO2 control of stomatal development is affected in these plants6.We investigated whether the long-term CO2 control of stomatal development is altered in ca1 ca4 plants. We analysed the stomatal index of wild-type (WT) and ca1 ca4 plants grown at low(150 p.p.m.) and elevated (500 p.p.m.) CO2 concentrations. For WT plants (Columbia (Col)), growth at the elevated CO2 concentration resulted in, on average,8%fewer stomata than growth at the low CO2 concentration (Fig. 1a–c and Extended Data Fig. 1). The ca1 ca4 mutant did not show an elevated CO2-induced repression of the stomatal index; however, interestingly, ca1 ca4 plants grown at the elevated CO2 concentration showed an average 22% increase in the stomatal index in their cotyledons (P<0.024; Fig. 1b, c) compared with ca1 ca4 plants grown at the low CO2 concentration. Similar results were obtained when stomatal density measurements were analysed (Fig. 1d). The mature rosette leaf phenotype in ca1 ca4 mutants also showed an increase in the stomatal index at the elevated CO2 concentration, which is consistent with the observations in the cotyledons (Extended Data Fig. 1a; stomatal indices rather than densities were analysed for accuracy; see Methods and Extended Data Fig. 1c legend).
Figure 1. The carbonic anhydrases CA1 and CA4 are required for repression of stomatal development at elevated CO2 concentrations.
a, Confocal images of the abaxial cotyledon epidermis of 10-day-old ca1 ca4 and WT (Col) seedlings grown at 500 p.p.m. CO2. Scale bar, 100 µm. b, Stomatal index of WT and ca1 ca4 seedlings grown at 150 and 500 p.p.m. CO2, showing an inverted stomatal development response to elevated CO2 by the mutant. c, Elevated CO2-induced changes in the stomatal index (data from b) shown as percentage changes in the stomatal index at 500 p.p.m.CO2 relative to 150 p.p.m. CO2. d, Stomatal density (data from c) for WT and ca1 ca4 seedlings. e, Stomatal index for six independent complementation lines of ca1 ca4 transformed with genomic copies of either A. thaliana CA1 (CA1-G) or A. thaliana CA4 (CA4-G). f, Elevated CO2-induced changes in stomatal development (data from e). b–f, Statistical comparisons were made between CO2 treatments (b and d) or were compared with the WT (c) or the ca1 ca4 data (f). Stomatal density and index measurements were conducted on 10-day-old seedlings. Error bars show mean ± s.e.m., n = 20 for b–f. ***, P<0.00005; **, P<0.005; *, P<0.05, using analysis of variance (ANOVA) and Tukey’s post-hoc test.
We transformed the ca1 ca4 mutant with genomic constructs expressing either CA1 or CA4 and investigated complementation of their stomatal development responses to CO2. Five of six independent transformant lines for either the CA1 or CA4 gene showed a significant suppression of the elevated CO2-induced inversion in the stomatal index found in ca1 ca4 plants (Fig. 1e, f). By contrast, ca1 ca4 leaves showed an average of 20% more stomata than WT leaves at the elevated CO2 concentration. The complementation lines showed varying levels of suppression of the inverted stomatal development phenotype of ca1 ca4 plants (Fig. 1e, f).
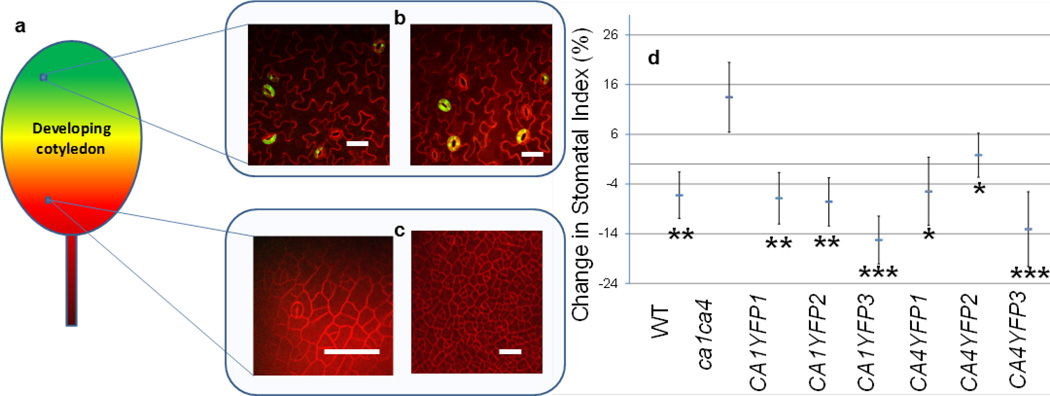
We tested the effects of preferential expression of these native A. thaliana carbonic anhydrases in mature guard cells6,16, as yellow fluorescent protein (YFP) fusion proteins (Extended Data Fig. 2a–c). These cell-type-specific complementation analyses showed that the enhanced stomatal development in ca1 ca4 plants at the elevated CO2 concentration can be suppressed by preferential expression of either CA1 or CA4 in mature guard cells (Extended Data Fig. 2b–d). This result provides initial evidence for extracellular signalling in the CO2 response mediated by these carbonic anhydrases during protodermal cell fate specification in developing cotyledons. It also indicates that the catalytic activity of the carbonic anhydrases may be required for CO2 control of stomatal development (see Extended Data Fig. 1d for data on complementation analyses with an unrelated, human, carbonic anhydrase, CA-II). We note that although we can complement the ca1 ca4 mutant phenotype with mature-guard-cell-targeted carbonic anhydrase overexpression, this finding does not exclude the possibility that expression in other cell types could function in this process. For example, in addition to being highly expressed in mature guard cells, CA1 and CA4 are also highly expressed in meristemoids, pavement cells and mesophyll cells6,16,17. Experiments analysing CO2 control of stomatal development in the open stomata 1 mutant ost1-3 show a divergence in the CO2-mediated signalling pathways controlling stomatal movements18 and stomatal development (Extended Data Fig. 1e).
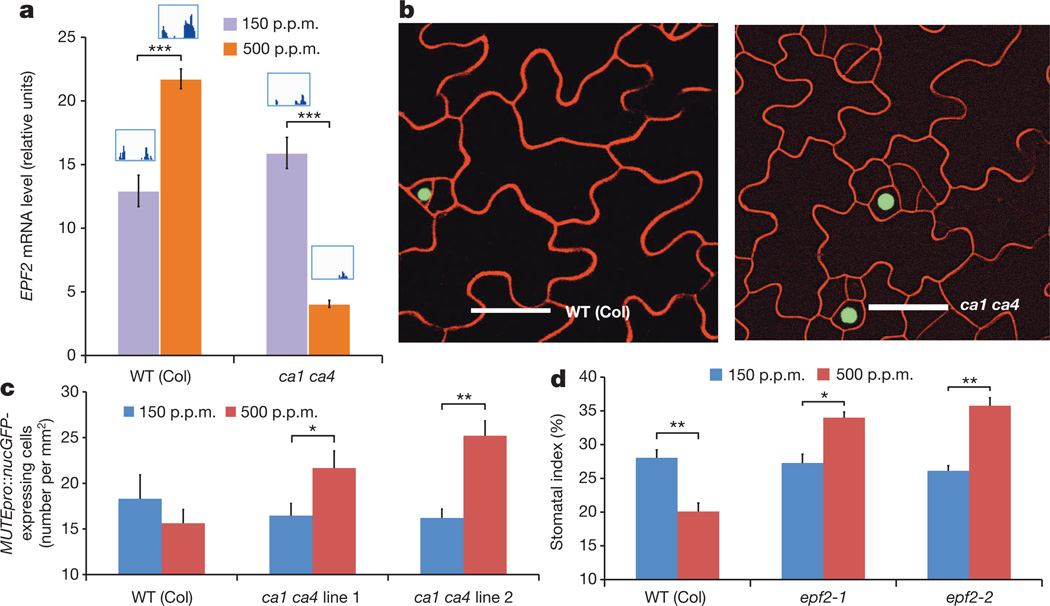
To gain initial insight into the regulatory mechanisms by which signalling in response to an elevated CO2 concentration exerts CA1- and CA4-dependent repression of stomatal development, we conducted high-throughput RNA-seq transcriptomics on immature aerial tissues of A. thaliana seedlings grown at the low and elevated CO2 concentrations. These analyses and independent single gene quantitative PCR (qPCR) studies of developing cotyledons showed that elevated CO2 induced upregulation of transcripts of EPF2 (which encodes an extracellular pro-peptide ligand)7,8 in WT plants but not ca1 ca4 plants (Fig. 2a). Our mature guard cell complementation analyses support a role for extracellular signalling in the elevated CO2-mediated repression of stomatal development (Extended Data Figs 1d and 2).
Figure 2. EPF2 expression is regulated by CO2 concentration and is essential for CO2 control of stomatal development.
a, EPF2 messenger RNA levels in developing 5 DAG (days after germination) cotyledons of WT and ca1 ca4 seedlings, showing induction, at the elevated CO2 concentration in the WT but not ca1 ca4. Levels were normalized to those of the CLATHRIN gene. The insets show the normalized RNA-seq expression of EPF2 exons from an RNA-seq experiment (5 DAG). b–d, MUTE expression correlates with the stomatal development phenotype of the ca1 ca4 mutant. Confocal images showing MUTEpro::nucGFP expression (green) in developing (5 DAG) cotyledons of WT and ca1 ca4 plants (b). Scale bars, 100 µm. Quantitation of MUTEpro::nucGFP-expressing cells in the WT and two independent lines in the ca1 ca4 background, at low and elevated CO2 concentrations (c). d, Stomatal index in WT plants and plants carrying either of two independent mutant alleles of epf2, at low and elevated CO2 concentrations, demonstrating that epf2 mutants show an inversion of the elevated CO2-mediated control of stomatal development. Error bars, mean ± s.e.m., n = 10 in a and n = 20 in c and d. ***, P<0.00005; **, P<0.005; *, P<0.05, using ANOVA and Tukey’s post-hoc test.
EPF2 is an early mediator of protodermal cell fate specification and controls cell entry to the stomatal lineage by limiting asymmetric divisions7,8.MUTE19,20 expression is a reliable indicator of cells that are committed to the stomatal lineage19,20. We transformed and examined WT and ca1 ca4 plants harbouring aMUTEpro::nucGFPconstruct19 (which allows expression of green fluorescent protein localized to the nucleus). Compared with WT plants, ca1 ca4 plants expressedMUTE pro::nuc GFP in 33%more cells, on average, at the elevated CO2 concentration but not the low CO2 concentration (Fig. 2b, c). TheMUTE pro::nuc GFP expression data provide an independent measure of the effect of ca1 ca4 on the CO2 response and are correlated with the increased stomatal index of ca1 ca4 leaves that is found at the elevated CO2 concentration (Fig. 1b). These data suggest that the increased stomatal development in ca1 ca4 plants at the elevated CO2 concentration progresses via components upstream of MUTE.
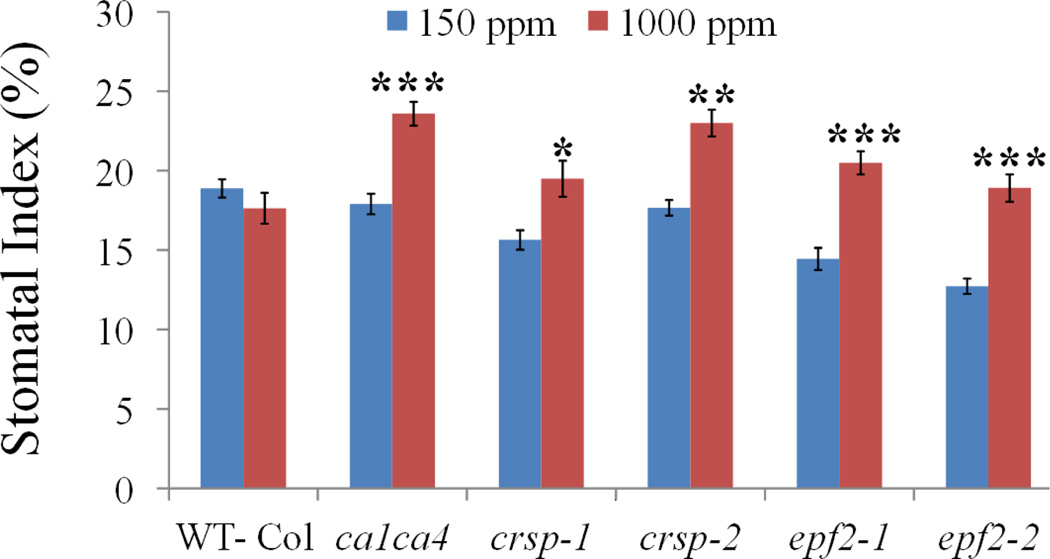
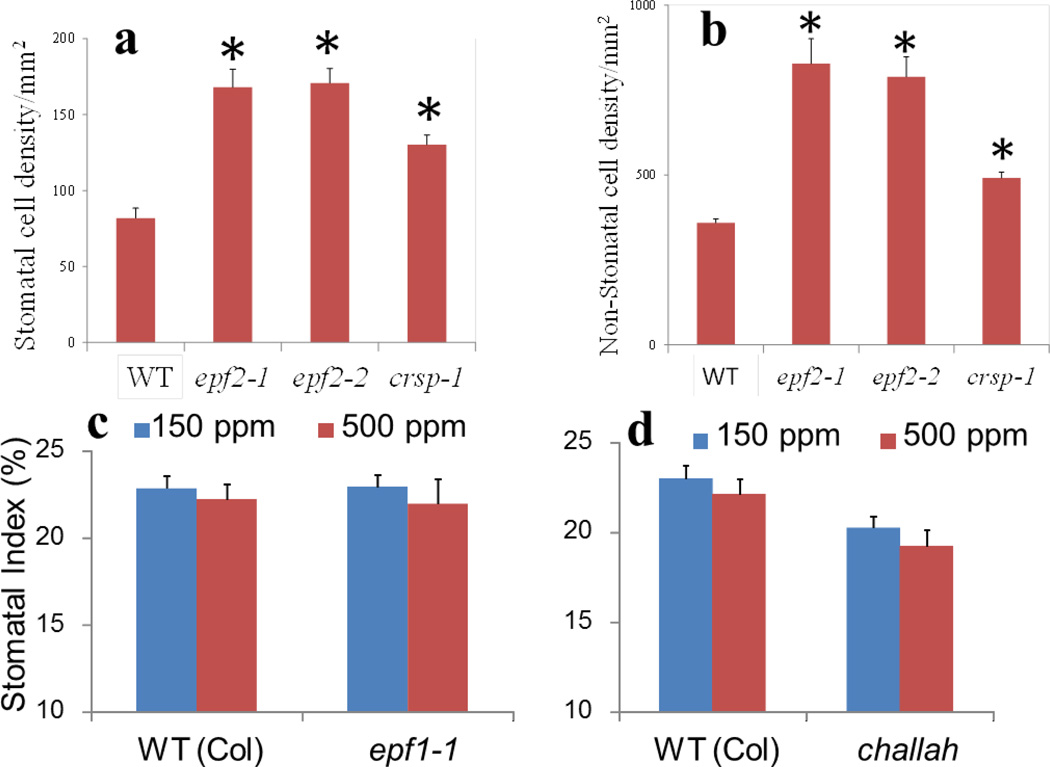
We analysed whether genetic perturbation of EPF2 results in an abnormal stomatal development response to CO2 concentration. Remarkably, plants carrying either of two independent mutant epf2 alleles showed a clear inversion in CO2 control of stomatal development (Fig. 2d and Extended Data Fig. 1b), with an average of 23% more stomata at the elevated CO2 concentration than at the low concentration. We also tested the effects of a very high (1,000 p.p.m.) CO2 concentration and found a similar inversion in the stomatal index of epf2-1 and epf2-2 plants (Extended Data Fig. 3). The epf2 mutant epidermis has been shown to have more non-stomatal cells than WT plants7,8. The epf2 mutants also had more non-stomatal cells at the elevated CO2 concentration than WT plants (Extended Data Fig. 4a, b). Conversely, plants with a mutation in the related negative-regulatory secreted peptides EPF1 (ref. 9) or EPFL6 (also known as CHALLAH)21, which also have roles in stomatal development, did not show an inversion of the CO2-controlled stomatal development response to the elevated CO2 concentration (Extended Data Fig. 4c, d).
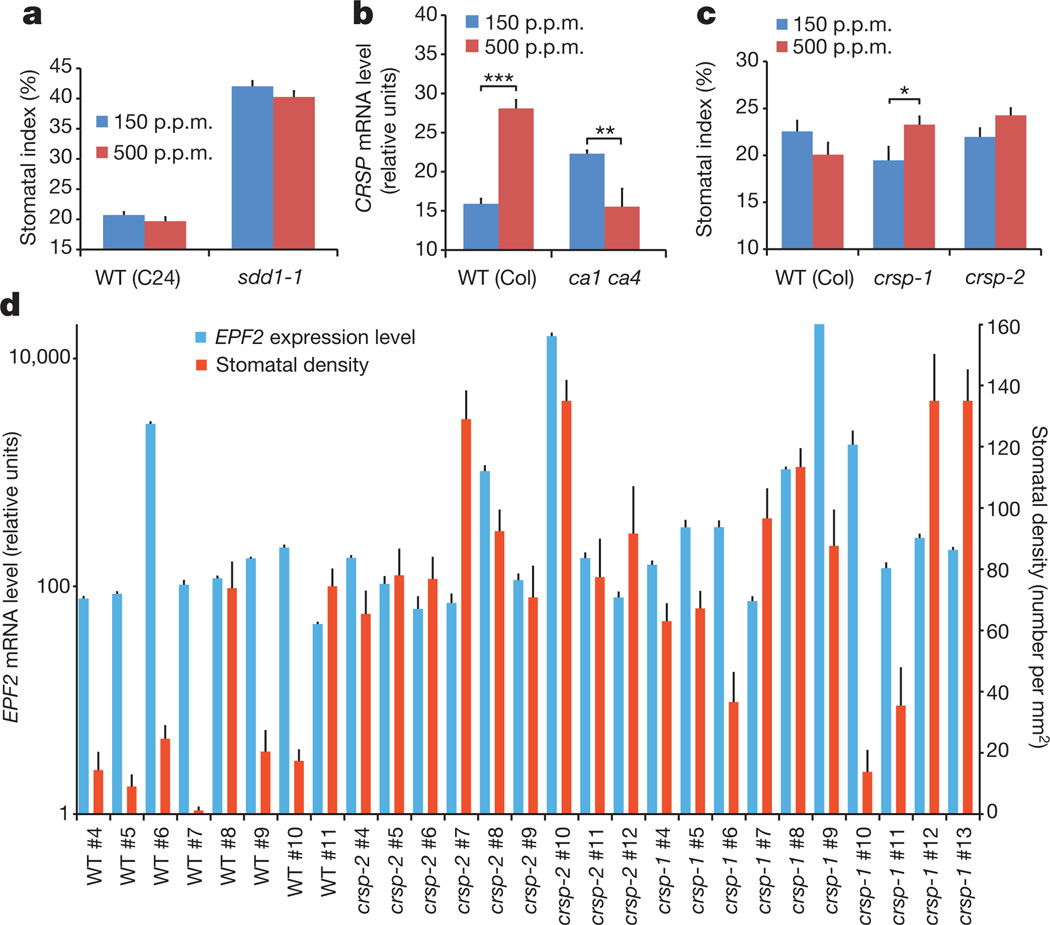
EPF2 belongs to a family of 11EPF and EPFL peptide proteins, which are predicted to be converted to an active peptide ligand isoform upon cleavage22–25. Hence, we tested plants with mutated SDD1, which has been shown to be a negative regulator of stomatal development and which encodes an extracellular subtilisin-like serine protease26. The stomatal index of the sdd1-1 mutant was much higher than that of the corresponding C24 WT accession at both the low and elevated CO2 concentrations (Fig. 3a). The sdd1-1 mutant showed, on average, a 4% decrease in the stomatal index at the elevated CO2 concentration compared with the low concentration, similar to the C24 WT background line (Fig. 3a). This result indicates that the protease SDD1 is not, alone, essential for CO2 control of stomatal development, consistent with studies suggesting that SDD1 does not function in the same pathway as EPF2 (refs 7, 8) and that extracellular proteases that function in the EPF1, EPF2 and STOMAGEN (also known as EPFL9 (refs 23, 24, 27), a positive-regulatory peptide related to EPF1 and EPF2) pathways remain unknown. At present, no environmental signals that clearly mediate the control of stomatal development via the extracellular pro-peptides EPF1, EPF2 and EPFL9 or the protease SDD1 have been identified.
Figure 3. A CO2-regulated, secreted subtilisin-like serine protease, CRSP, is a mediator of elevated CO2 repression of stomatal development.
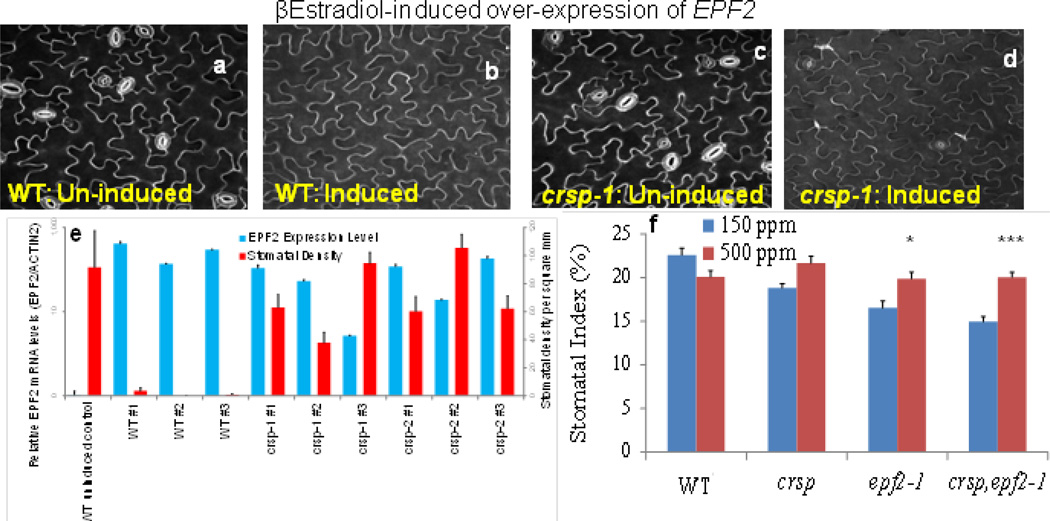
a, Stomatal index of the WT(C24) and the sdd1-1 mutant grown at the low and elevated CO2 concentrations. b, CO2 control of CRSP (SBT5.2) mRNA levels in developing (5 DAG) cotyledons of WT (Col) and ca1 ca4 seedlings grown at low and elevated CO2 concentrations (qPCR data, with cDNA levels normalized to CLATHRIN (At4G24550) expression). c, Stomatal index of WT cotyledons and those carrying either of two independent crsp alleles at low and elevated CO2 concentrations. d, Quantitation of the effects of EPF2 transcript levels on the stomatal density of 5 DAG cotyledons in 27 independent lines harbouring the β-oestradiol-inducible EPF2 overexpression construct in the WT(Col), crsp-1 and crsp-2 mutant backgrounds (normalized to ACTIN 2 expression). For each line, 20 images from 10 cotyledons (2 images per cotyledon; 10 separate seedlings used) were analysed, and RNA was extracted from 10 separate seedlings (see Methods and Extended Data Fig. 7e). Error bars, mean ± s.e.m., n = 20 in a, c and d and n = 10 in b. b, c, ***, P<0.00005; **, P<0.005; *, P<0.05, using ANOVA and Tukey’s post-hoc test.
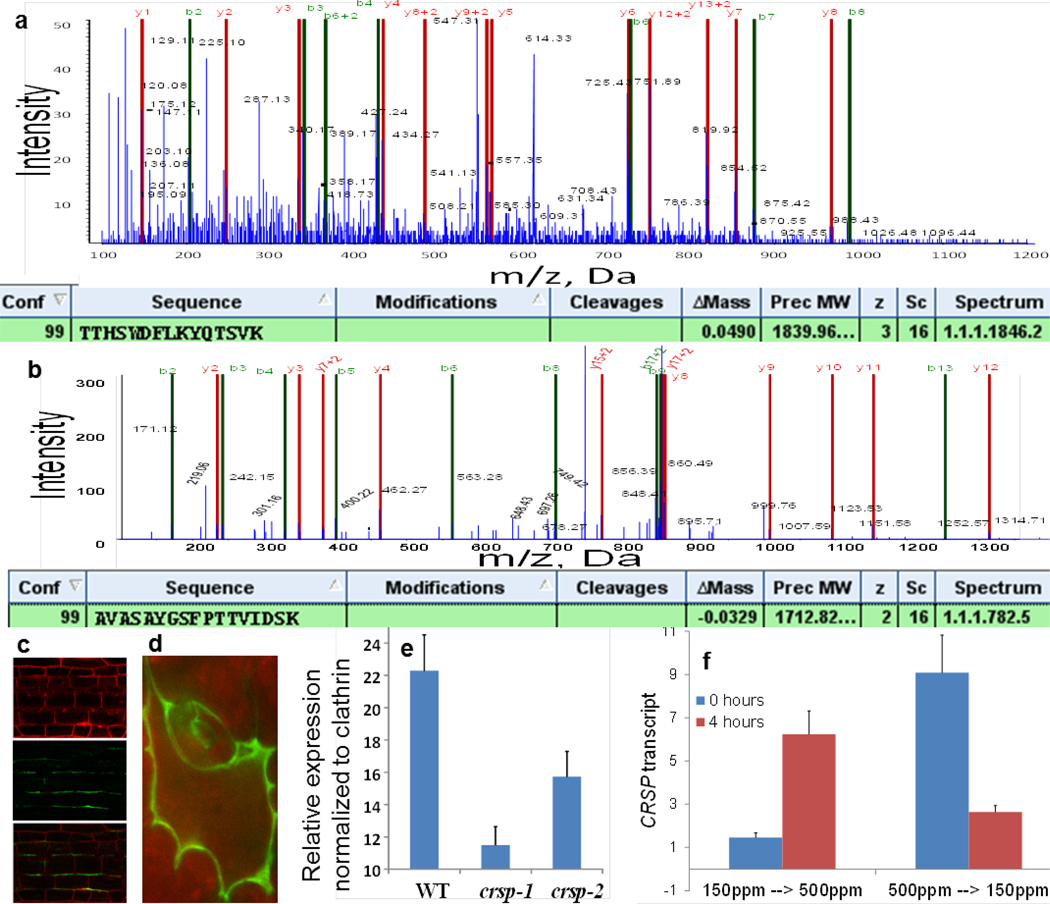
We hypothesized that there is a distinct extracellular protease(s) that mediates CO2 control of stomatal development. SDD1 belongs to a 56-member subtilisin-like serine protease family (subtilases). Therefore, we pursued proteomic analyses of apoplast proteins in leaves and identified four abundant subtilases (SBT1.7 (also known as ARA12), SBT1.8 (At2g05920), SBT3.13 (At4g21650) and SBT5.2; Extended Data Fig. 5). Because SBT1.7 has been shown to be required for seed mucilage release28 and SBT3.13 was detected in two of five experiments, we focused on SBT5.2 rather than SBT3.13, SBT1.7 or its closest homologue, SBT1.8. Interestingly, qPCR data from developing cotyledons showed an increase in the abundance of SBT5.2 transcripts in WT plants after both long term (5 days; Fig. 3b) and short term (4 h; Extended Data Fig. 5f) exposure to the elevated CO2 concentration. By contrast, the ca1 ca4 plants failed to show this increase in SBT5.2 transcript abundance at the elevated CO2 concentration (Fig. 3b). We named SBT5.2 as CRSP (CO2 RESPONSE SECRETED PROTEASE). CRSP is widely expressed in guard cells and meristemoid- and pavement-cell-enriched samples, as well as in other plant tissues, including high expression in roots17,29. Our experiments with a CRSP–VENUS construct showed that CRSP is targeted to the cell wall (Extended Data Fig. 5c, d). We tested the effect on CO2 control of stomatal development of two T-DNA insertion alleles encoding mutated forms of this extracellular protease (Fig. 3c and Extended Data Figs 1b, 3, 4 and 5e). Interestingly, the two distinct crspmutant alleles (Extended Data Fig. 5e) conferred, on average, deregulation of stomatal development, with more stomata at the elevated CO2 concentration than at the low concentration (Fig. 3c and Extended Data Figs 1b and 3). Furthermore, when epidermal cell types were analysed individually, the crsp-1 mutant had more stomata and non-stomatal cells than the WT, which is a similar phenotype to (but not as severe as) the epf2 mutant (Extended Data Fig. 4a, b), implicating the functions of additional proteases. It should be noted that, similar to ERECTA, the wide expression pattern of CRSP indicates that the CRSP protein could have additional roles in plant growth and development.
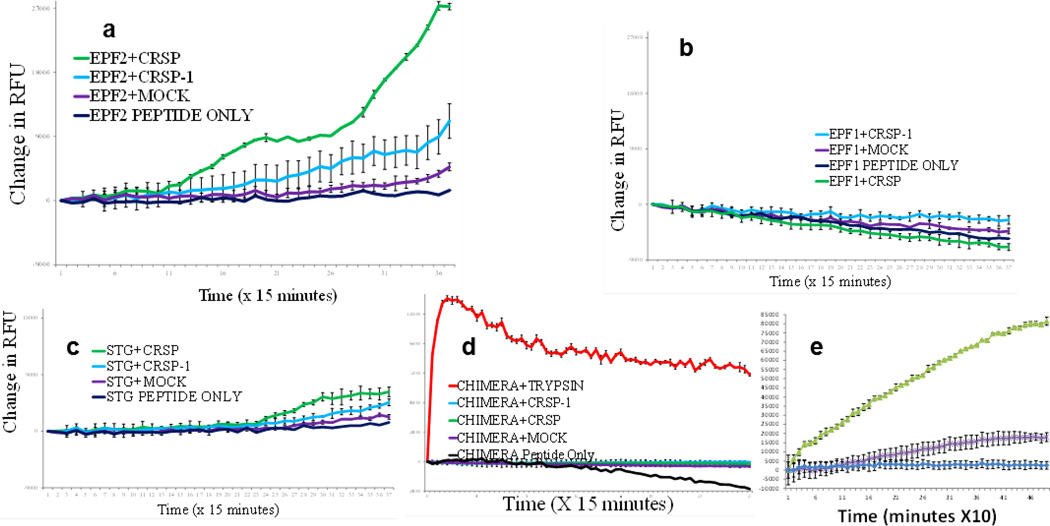
To determine whether the EPF2 pro-peptide can be cleaved by CRSP, we constructed two synthetic peptides spanning the predicted EPF2 cleavage site. We subjected these peptides to in vitro proteolytic analyses using in vitro-synthesized CRSP protein. CRSP showed robust cleavage of both synthetic EPF2 (synEPF2) peptides in vitro, and this cleavage was greatly reduced by the inclusion of protease inhibitors or the mutant form of the CRSP protein (CRSP-1) in the reaction (Extended Data Fig. 6a, e). To test the specificity of CRSP-mediated cleavage, we generated an EPF2 mutant peptide sequence with 7 residue substitutions to mimic a 12-residue sequence that surrounds the cleavage site in STOMAGEN; this mutant was not cleaved by CRSP (Extended Data Fig. 6d). We also tested the synthetic EPF1 and STOMAGEN peptides, and both of these control peptides showed negligible cleavage in vitro in the presence of either CRSP or the mutant CRSP-1 (Extended Data Fig. 6b, c). These data support the function of CRSP in the modulation of EPF2 activity.
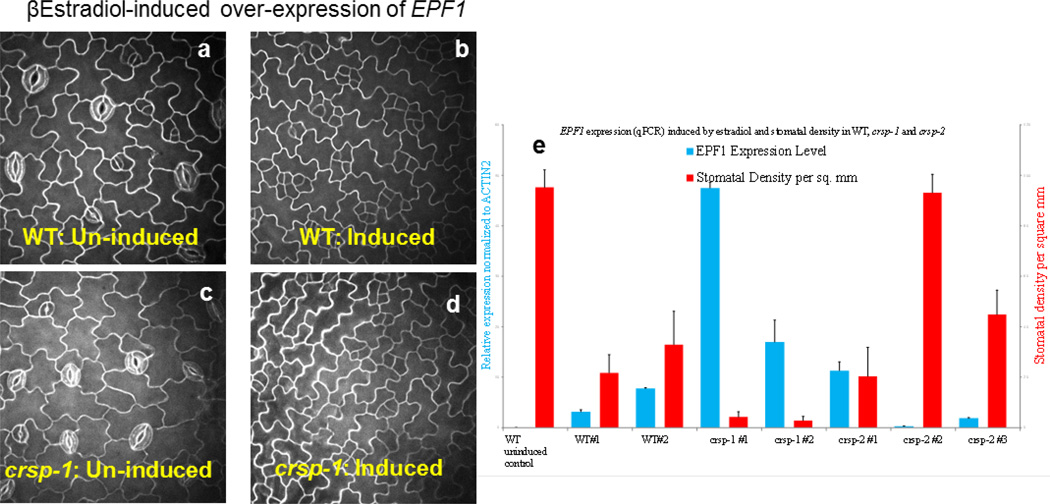
Several proteomic approaches were unsuccessful at detecting low-abundance EPF1 and EPF2 peptides in cell-wall extracts (see Methods). To further analyse whether EPF2 and CRSP function in the same pathway, we conducted epistasis analyses by generating crsp epf2 double mutant lines. Double mutant plants did not show clearly additive mutant phenotypes (Extended Data Fig. 7f). We then overexpressed EPF2 in the WT and crsp mutant backgrounds using an oestradiol-inducible system. Analysis of 36 independent lines showed that equivalent quantified levels of EPF2 overexpression repressed stomatal development to a lesser degree in the crsp background than in the WT (Fig. 3d and Extended Data Fig. 7a–e). The partial repression of stomatal density in high-EPF2-expressing crsp lines, the epistasis analysis and the non-stomatal cell densities implicate the function of additional proteases in EPF2 activation (Extended Data Figs 3, 8 and 9). These data also do not exclude a possible role for CRSP in other stomatal responses. Controls using inducible EPF1 overexpression showed similar effects on stomatal development in the WT and crsp backgrounds (Extended Data Fig. 8).
We have uncovered key elements in a long-sought pathway by which elevated CO2 concentrations control cell fate and the stomatal development machinery4. The results of our study identify new players in CO2 control of stomatal development:CA1,CA4,CRSP and EPF2.Together, the present findings point to the extracellular protease CRSP, identified here as functioning in the CO2-controlled stomatal development response, and further suggest that the activity of the negative regulator EPF2 is modulated by CRSP.EPF2 peptides are predicted to be activated by cleavage, thus signalling the repression of stomatal development7,8,22. CRSP can cleave EPF2 (Extended Data Fig. 6a, e), and our data provide evidence that CRSP functions in EPF2 signalling to mediate the repression of stomatal development (Fig. 3d and Extended Data Figs 6–8). An inverted CO2-dependent stomatal development response in erecta plants potentially correlates with the preferential binding ofEPF2 to the receptor kinase ERECTA22 (Extended Data Fig. 9).
The finding that the stomatal index is similar in ca1 ca4 and WT plants at a low CO2 concentration indicates that additional regulatory mechanisms exist and that CO2 control is not entirely disrupted in ca1 ca4 plants. In the absence of the elevated CO2-mediated modulation of CRSP and EPF2, competing extracellular signals that promote stomatal development (for example, the STOMAGEN peptide23,24,27) might contribute to the inverted CO2 control of stomatal development found here in the ca1 ca4, epf2 and crsp mutants (Figs 1–3). The mechanisms reported here may also aid in understanding the natural variation in stomatal developmental responses to elevated CO2 concentrations that has been observed in A. thaliana and other plant species30. Globally, as plants grow and respond to the continuing rise in atmospheric CO2 concentrations, an understanding of the key genetic players that mediate the CO2-controlled plant developmental response could become critical for agriculturally relevant efforts aimed at improving water use efficiency or plant heat resistance.
METHODS
Statistical analyses
In all figures, statistical analyses were conducted using the OriginPro 8.6 software package, and comparisons were made for individual genotypes between CO2 treatments or with the WT data or with the ca1 ca4 mutant data using analysis of variance (ANOVA) and Tukey’s post-hoc test. ***,P<0.00005; **, P<0.005; *, P<0.05. For all figures, n = 20 images derived from10 independent seedlings were analysed per genotype and CO2 treatment; error bars, mean ± s.e.m.
Plant growth
WT(Col and C24 accessions) and individual mutant seedlings were grown in plant growth chambers (Percival) under identical conditions of light (16 h light:8 h dark cycles; 100 µ molm−1 s−1), humidity (80–90%) and temperature (21 °C), with only the CO2 concentration being varied (low = 150 p.p.m. and elevated = 500 p.p.m. (or 1,000 p.p.m. where noted)).
Stomatal development analyses
The T-DNA insertion alleles used were: SALK_ 132812C = crsp-1; SALK_099861C = crsp-2; SALK_102777 = epf2-1; and GK-673E01 = epf2-2.Theca1 ca4 carbonic anhydrase double mutant has been described previously6. Seedlings were grown for 10 days, at which point the abaxial epidermal surfaces of mature cotyledons from 10 independent seedlings were imaged using propidium iodide staining and a confocal microscope (two non-overlapping images per cotyledon for a total n = 20 per genotype per CO2 treatment). Images were acquired from the centre of the cotyledon, away from the margin and midrib. Imaging for seedlings harbouring the MUTEpro::nucGFP19 construct was also conducted with a confocal microscope. Epidermal cells were counted, and the stomatal density and index were quantitated using the Image J software. Stomatal density = number of stomata per mm2; stomatal index = percentage of epidermal cells that are stomata, as calculated by stomatal index = 100× (number of stomata)/(number of stomata + number of pavement cells). Multiple environmental stimuli can influence stomatal development and control the baseline stomatal density or indices (which can vary slightly from experiment to experiment, similar to the findings of previous studies5); therefore, for all experiments, WT controls were grown side by side (in parallel), and the data from within each experiment were analysed in comparison with the corresponding mutants. Furthermore, all experiments were repeated at least three times, and blinded experiments were conducted, in which either the genotype, or both the genotype and the CO2 concentration (double blind), were unknown to the experimenter until after the data quantitation had been completed for the experiments.
RNA-seq and qPCR analyses
Hypocotyls and cotyledons of developing seedlings (5 DAG; WT and ca1 ca4 mutant plants; n>1,000 per sample) grown in the low and elevated CO2 concentrations were used as source tissue to extract total RNA and conduct RNA-seq experiments using the HiSeq 2000 platform (Illumina). The raw data from three independent biological replicates (experiments) have been deposited in the BioProject database under accession number PRJNA218542. qPCR experiments were conducted on cDNA synthesized from total RNA extracted from 500 pooled 5 DAG seedlings from the indicated CO2 treatments. Three biological replicates were conducted, and candidate gene expression was normalized to that of the CLATHRIN gene.
Primer sequences
The primer sequences used for qPCR were as follows: EPF2. For 5′-CGCCGCGTGTTCTTTGGTCG-3′, EPF2.Rev 5′-CGGCGTTTTTCTTT TCTCCGCCA-3′; CLATHRIN(AT4G24550). For 5′-ATACGCGCTGAGTTCCC -3′, CLATHRIN(AT4G24550). Rev 5′-CTGACTGGCCCTGCTT-3′; andCRSP.For 5′-ATGGCAGCTCCTCATGTTTCAGC-3′, CRSP.Rev 5′-CGTTGTTTGTTTG AGTCGCTGTTG-3′. MultiSite Gateway cloning was used to generate a full-length CRSP translational fusion with VENUS. The primer sequences for the CRSP–VENUS fusion protein were: CRSPpro For 5′-GGGGACAACTTTGTATAGAAAAGTTG GATAGACCTTTCTCG-3′, CRSPpro Rev 5′-GGGGACTGCTTTTTTGTACAA ACTTGTACATACCTCAACTCAAG-3′; CRSPcds For 5′-GGGGACAAGTTTG TACAAAAAAGCAGGCTTAATGAAAGGCATTACATTCTTC-3′, CRSPcds Rev 5′-GGGGACAGCTTTCTTGTACAAAGTGGGATTTTTCAAATTGAGGATG AGACCAGGAGCCGCCGCCGCCGTTTGTGCGGCTACTCTCGC-3′; andVENUS cds For 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAGTGAGCAAGGG CGAGGAG-3′,VENUScdsRev 5′-GGGGACAACTTTGTATAATAAAGTTGTA TTACTTGTACAGCTCGTCCATGCCG-3′. (We amplified a 2,000-basepair genomic region directly upstream of the first ATG of CRSP to drive CRSP–VENUS expression.)
In vitro cleavage of synthetic EPF peptides
All synthetic EPF peptides were manufactured and purified to a purity >97%by LifeTein. Peptides were conjugated at the carboxy and amino termini to fluorophore and quencher moieties, respectively. The 30-residue (synEPF2-Short) or the 69-residue (synEPF2-Long) EPF2 peptides included the predicted cleavage site. The peptide sequences used were as follows: EPF2-Short, Dabcyl-SKNGGVEMEMYPTGSSLPDCSYACGACSPC-E- (EDANS); EPF2-Long, Dabcyl-HKKEISKNGGVEMEMYPTGSSLPDCSYACGA CSPCKRVMISFECSVAESCSVIYRCTCRGRYYHVPSRA-HHHHHH-E-(EDANS); EPF1, Dabcyl-KRQRRRPDTVQVAGSRLPDCSHACGSCSPC-E-(EDANS); STOMAGEN, Dabcyl-LLPQVHLLNSRRRHMIGSTAPTCTYNECRG-E-(EDANS); and CHIMERA, Dabcyl-SKNGGVEMEMYPIGSTAPTCTYNEGACSPC-E-(EDANS).
The synthetic EPF2-Long peptide (69 residues) does not inhibit stomatal development, possibly owing to misfolding or another missing post-translational modification(s) compared with the native EPF2 peptide. STREP II-tagged CRSP and mutatedCRSP-1 proteases were synthesized using the TNT SP6 High-Yield Wheat Germ Protein Expression System (Promega) and purified using the Strep-Tactin MacroPrep resin (IBA). In vitro cleavage reactions (100 µl) in 1× PBS were incubated at 30 °C in a Mithras LB 940 96-well plate reader (Berthold Technologies), and fluorescence readings were acquired every 10 min after shaking the plate for 1 s. A final concentration of 30 µM synthetic peptide and approximately 10 pmol wheat-germ-synthesized protease were used in the reactions. Inclusion of a 1:20 dilution of plant Protease Inhibitor Cocktail (Sigma) and peptide or CRSP protease only were used as controls. The fluorescence data were normalized for background fluorescence using buffer only controls, and the change in relative fluorescence was calculated by subtracting the initial fluorescence measurement for each sample. Mean values are shown, and error bars represent s.e.m. In independent experiments under different concentrations of protease (20 pmol) and synEPF2 (50 µM), similar results were obtained.
Oestradiol induction of EPF1 and EPF2
T2 transgenic seeds of hygromycin-resistant lines harbouring the previously published22 EPF1 (pTK-102) or EPF2 (pTK-103) inducible overexpression constructs were germinated on 0.5×MSplates (pH5.7) containing 10 µM β-oestradiol (Sigma), and images of the epidermis of the cotyledons were captured using propidiumiodide staining and confocal microscopy. To attempt to detect EPF peptides in planta, mature rosette leaves of lines harbouring the EPF1 or EPF2 inducible overexpression constructs were sprayed with 10 µM β-oestradiol, and apoplast proteomes were extracted and analysed (see below) 16 h and 72 h later in two separate experiments.
Apoplast and secreted protein isolation
Rosettes of 10 soil grown plants (8 weeks old, or in the case of cotyledon apoplast extraction, cotyledons and hypocotyls from 5-day-old seedlings) were vacuum-infiltrated with 0.3 M mannitol for 2 min at room temperature, after which leaves were centrifuged at 200g in a swinging bucket rotor at 4 °C for 15 min. The same leaves were re-infiltrated with 0.2M CaCl2 in 0.3Mmannitol for 3min under vacuumat room temperature, after which the leaves were centrifuged at 200g in a swinging bucket rotor at 4 °C for 20 min. The pH of this extraction buffer was varied between 4 and 9 to maximize the capture of proteins based on their predicted pKa values. The centrifugation step produced 19 ml apoplast fluid, which was separated on an Amicon Ultra-15 filter column (15 ml capacity) in a swinging bucket rotor at 4,100 r.p.m. and 4 °C. The flow-through was passed through the column three times, resulting in a final volume of 300 µl in the filter cup. Protease Inhibitor Cocktail (Sigma, 30 µl) was added to the 300 µl protein sample. The 300 µl protein sample was then acidifiedwith1%trifluoroacetic acid (TFA) to a final concentration of 0.1% TFA. ZipTip pipette tips (Millipore) were used according to the manufacturer’s protocols, and protein samples were eluted in an acetonitrile dilution series as follows: 5, 10, 20, 30, 40 and 50% acetonitrile in 0.1% TFA. The samples were desiccated and re-dissolved in 0.1% TFA and 5% acetonitrile. The peptides were then extracted and desalted using Aspire RP30 desalting columns (Thermo Scientific). For the isolation of secreted cysteinerich peptides, two separate experiments including WT and ca1 ca4 seedlings or WT and crsp-1 seedlings were cultured in 0.5×MSliquid medium under constant agitation and light for 10 days. Secreted proteins from the liquid growth medium were size-fractionated to isolate peptides of 3–10 kDa using Amicon Ultra-15 filter columns. Cysteine-rich peptides were purified on Thiopropyl Sepharose 6B (Sigma) with and without a dithiothreitol pre-reduction step. The eluted and flow-through samples were analysed as described below. We attempted several proteomic approaches (including 35S promoter-driven EPF1 and EPF2 overexpression, inducible oestradiol-mediated overexpression of EPF2, liquid culture of seedlings followed by enrichment of cysteine-rich secreted peptides, and analysing the apoplast proteomes of 5-day-old cotyledons and hypocotyls) and did not detect these low abundance EPF peptides from in planta samples.
Sample trypsinization
As described previously31, samples were diluted in TNE buffer (50mMTris, pH8.0, 100mMNaCl and 1mMEDTA). RapiGest SF(Waters) was added to the mixture to a final concentration of 0.1%, and the samples were boiled for 5 min. Tris-(2-carboxyethyl) phosphine (TCEP) was added to a final concentration of 1mM, and the samples were incubated at 37 °C for 30 min. Next, the samples were carboxymethylated with 0.5 mg ml−1 iodoacetamide for 30 min at 37 °C, followed by neutralization with 2mMTCEP (final concentration). The protein samples prepared above were digested with trypsin (trypsin:protein ratio = 1:50) overnight at 37 °C. The RapiGest SF was degraded and removed by treating the samples with 250mM HCl at 37 °C for 1 h, followed by centrifugation at 15,800g for 30 min at 4 °C. The soluble fraction was transferred to a new tube, and the peptides were extracted and desalted using Aspire RP30 desalting columns. Trypsin-digested peptides and directly extracted peptides were analysed by high pressure liquid chromatography (HPLC) coupled with tandem mass spectroscopy (LC-MS/MS) using nanospray ionization, as described previously32 with the following changes: the nanospray ionization experiments were performed using a QSTAR-Elite hybrid mass spectrometer (AB SCIEX) interfaced with a nanoscale reversed-phase HPLC system (Tempo) using a 10-cm, 100-µm internal diameter glass capillary packed with 5-µm C18 ZORBAX beads (Agilent Technologies). Peptides were eluted from the C18 column into the mass spectrometer using a linear gradient (5–60%) of acetonitrile at a flow rate of 400 µl min−1 for 1 h. The buffers used to create the acetonitrile gradient were: Buffer A (97.795% H2O, 2% acetonitrile, 0.2% formic acid and 0.005% TFA) and Buffer B (99.795% acetonitrile, 0.2% formic acid, and 0.005% TFA). MS/MS data were acquired in a data dependent manner in which the MS1 data were acquired from m/z 400 Da to 1,800 Da and the MS/MS data were acquired fromm/z 50 Da to 2,000 Da. The MS/MS data were analysed using the software ProteinPilot 4.0 (AB SCIEX) for peptide identification.
In an alternative protocol, protein samples were prepared for SDS–PAGE using the protocol described previously33. Briefly, proteins from apoplast fluid were extracted by addition of an equal volume of Tris-buffered phenol. After centrifugation at 10,000g for 10 min, the aqueous phase was removed, and the proteins in the organic phase were precipitated by adding five volumes of 0.1 M ammonium acetate in methanol. After overnight incubation at −20 °C, the samples were centrifuged at 10,000g for 5 min, and the pelleted proteins were washed twice with 80% acetone. The protein pellets in 80% acetone were air-dried, resuspended in SDS– PAGE loading buffer, and separated (50 µg) by SDS–PAGE in a 10% gel. Proteins were visualized by Coomassie blue G-250 staining, and each sample lane was cut into 10 separate gel slices. Reduction, alkylation and in-gel trypsin digestion of the individual gel slices were performed as described previously34. Tryptic peptides were extracted by sequential addition and removal of 100 µl 1% TFA, 50% acetonitrile and 0.5%TFA twice, then 100%acetonitrile. For each sample, the solutions containing the extracted peptides were pooled in a fresh tube and lyophilized overnight. The lyophilized peptides were dissolved in 1.0% formic acid and 5% acetonitrile, applied to a 12-cm, 150-µminternal diameter silica column packed in-house with MagicC18 medium (Michrom) and eluted into the nanoelectrospray ion source of an LTQ-Orbitrap LC-MS/MS mass spectrometer (ThermoElectron) controlled by the software Xcalibur version 2.2.1. A fully automated chromatography run using 0.1% formic acid (Buffer A) and 99.9% acetonitrile and 0.1% formic acid (Buffer B) was performed with the following settings: increase from 0 to 40% Buffer B over 70 min and then increase to 80% Buffer B in 1 min and hold at 80% Buffer B for 5min.Mass spectrometer settings were as described previously35. The MS/MS spectra were extracted by Mascot Distiller version 2.3.1 (Matrix Science). Mascot (server version 2.3,Matrix Science) and X! Tandem (The GPM; version 2010.12.01.1) were used to analyse the MS/MS spectra by searching an in-house A. thaliana TAIR10 protein database assuming the digestion enzyme was trypsin. Searches were performed with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 0.80Da.Oxidation of methionine and the iodoacetamide derivative of cysteine were specified as variable modifications. Scaffold (version Scaffold_3.6.4, Proteome Software) was used to validate MS/MS-based peptide and protein identifications with identifications accepted if they could be established at greater than 99.0%probability and contained at least two identified peptides. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. The mass spectrometry proteomics data have been deposited in the PRIDE database under the accessions PXD000692, PXD000693 and PXD000956
Extended Data
Extended Data Figure 1. Mutations in CA1, CA4, CRSP and EPF2 affect CO2 control of stomatal development in mature rosette leaves.
a, WT (Col) and ca1 ca4 double mutants. b, WT (Col) and crsp-1 and epf2-1 single mutants grown for 6 weeks at low (150 p.p.m.; blue) and high (500 p.p.m.; orange) CO2 concentrations. Small cell clusters (SLGCs) are not included in these stomatal index (SI) calculations. Abaxial stomatal indices (that is, the percentage of epidermal cells that are stomata: 100×[number of stomata]/[number of stomata + number of pavement cells]) for mature rosette leaves (seventh and eighth leaves). c, Twenty-one-day-old ca1 ca4 double mutant and WT plants grown at 150 p.p.m. and 500 p.p.m. CO2. Scale bar, 2 cm. ca1 ca4 mutant and WT plants were morphologically indistinguishable under the imposed growth conditions. No obvious aberrations in stomatal shape or size were found in the ca1 ca4 mutant (Fig. 1a). Examination of the epidermis of ca1 ca4 mutant plants revealed that adjacent stomata had at least one epidermal cell between them with no stomatal pairing or clusters (unlike what is observed in epf1 mutants9), indicating that spacing divisions were enforced in the mutant during stomatal lineage establishment (Fig. 1a). The WT and ca1 ca4 plants grown at 150 p.p.m. CO2 were smaller than their 500-p.p.m.-grown counterparts; the cotyledons and leaves of the WT and the ca1 ca4 mutant were similar in size and shape at each CO2 concentration. Because seedlings grown at 150 p.p.m. CO2 have smaller leaf areas, such size differences may generate artefacts when analysing stomatal density. Hence, in this study, we employed stomatal index analyses as a reliable measure of comparing stomatal developmental changes between CO2 treatments. d, CO2-induced change in stomatal index (500 p.p.m. versus 150 p.p.m.) of three independent lines of the ca1 ca4 mutant complemented with guard cell preferential overexpression of a YFP fusion of the human carbonic anhydrase II (CA-II). The significance of suppression was analysed relative to ca1 ca4. We interrogated whether carbonic anhydrase enzyme activity or the specific structure of CA1 and CA4 are important for mediating CO2 control of stomatal development. We transformed the ca1 ca4 mutant with the unrelated gene human CA2 (ref. 6) as a YFP fusion protein under the control of the mature guard cell preferential promoter (pGC1; Extended Data Fig. 2b,c). Human CA-II has low protein sequence identity to A. thaliana CA1 (9%) and CA4 (12%)6 and, as such, is an ideal candidate for these studies. In all three independent transformant lines tested, the elevated CO2-induced inversion in the stomatal index of ca1 ca4 mutant plants was partially suppressed by mature-guard-cell-targeted expression of the human carbonic anhydrase gene. This result suggests that the catalytic activity of the carbonic anhydrases may be required for CO2 control of stomatal development. The requirement for catalytic carbonic anhydrase activity for this CO2 response would be consistent with a background CO2 response rate even in ca1 ca4 mutant plants, owing to spontaneous CO2 hydration. e, Altering rapid CO2-induced stomatal movements and transpiration efficiency did not invert the elevated-CO2-mediated control of stomatal development. The stomatal index in the WT (Col) and in the OPEN STOMATA 1 mutant ost1-3 at low and elevated CO2 concentrations is shown. Leaf transpiration rates control stomatal development36. As CO2 levels affect transpiration by regulating stomatal movements6,10,12, we examined whether the processes governing transpiration and CO2-induced stomatal movements are distinct from CO2 regulation of stomatal development. We chose a mutant of the protein-kinase-encoding gene OST1 for these studies as OST1 is an upstream regulator of abscisic acid-induced stomatal closure and mutations in this gene result in plants with a higher transpiration rate37. Furthermore, OST1 is a key mediator of CO2-induced stomatal closure18, and whether CO2 control of stomatal development requires OST1 is unknown. Thus, we investigated whether ost1-3 mutant plants also show an inversion of the CO2-controlled stomatal development response. We found that ost1-3 mutant plants grown at the elevated CO2 concentration showed an average 7% reduction in the stomatal index. Furthermore, ost1-3 mutant leaves had slightly larger average stomatal indices than WT leaves at low and elevated CO2 concentrations (P = 0.097 at 150 p.p.m.). Hence, we conclude that disrupted stomatal movements and increased transpiration do not cause the CO2-induced inverted stomatal development response in ost1 mutants. This finding is in contrast to that for ca1 ca4 leaves, which have an increased stomatal conductance and an inverted stomatal development response to the elevated CO2 concentration. For a, b, d and e, n = 20. ***, P<0.00005; **, P<0.005; *, P<0.05, using ANOVA and Tukey’s post-hoc test. Error bars, mean ± s.e.m. in a, b, d and e.
Extended Data Figure 2. Mature-guard-cell-targeted carbonic anhydrase catalytic activity suppresses stomatal development via extracellular signalling in ca1 ca4 mutants.
a, Cartoon showing epidermal cell differentiation in an immature cotyledon. Green indicates differentiated epidermis with stomata (shown in b); red indicates epidermal cells that have entered the stomatal lineage (shown in c). b, c, Confocal images of mature (b) or developing (c) stomata in cotyledons at 5 DAG for lines expressing the human CA-II–YFP construct driven by the mature guard cell preferential promoter pGC1 (ref. 16), illustrating mature guard cell targeting of pGC1::CA-II-YFP. Two representative images of the distal end of the cotyledon epidermis, where stomatal differentiation has already occurred (b). Two representative images (the image on the left is at higher magnification) of the proximal end of the cotyledon, where stomatal differentiation has not yet taken place (c). d, The stomatal index of six independent complementation lines of the ca1 ca4 mutant transformed with either CA1–YFP or CA4–YFP (significance of suppression was determined relative to ca1 ca4). All scale bars, 20 µm. Error bars, mean ± s.e.m.; n = 20. ***, P<0.00005; **, P<0.005; *, P<0.05, using ANOVA and Tukey’s post-hoc test.
Extended Data Figure 3. CO2 regulation of stomatal indices at 1,000 p.p.m. CO2 in mature leaves.
Abaxial stomatal indices (that is, the percentage of epidermal cells that are stomata) for mature cotyledons (10 DAG) of WT (Col), the epf2-1, epf2-2, crsp-1 and crsp-2 single mutant alleles and the ca1 ca4 double mutant grown at 150 and 1,000 p.p.m. CO2. Small cell clusters are included in the calculations for the epf2 mutants. Error bars, mean ± s.e.m., n = 20. ***, P<0.00005; **, P<0.005; *, P<0.05, using ANOVA and Tukey’s post-hoc test.
Extended Data Figure 4. Numbers of stomatal and non-stomatal cells in WT, epf2-1, epf2-2 and crsp-1 mutants at the elevated CO2 concentration, and mutations in the negative-regulatory extracellular signals of stomatal development.
The secreted EPF signalling pro-peptides have been identified as extracellular pro-peptide ligands that mediate the repression of stomatal development via extracellular signalling7–9,22–24,27. Abaxial cell densities for stomatal cells (a) and non-stomatal cells (b; all epidermal pavement and SLGC cells except guard cells) (per mm2) in mature cotyledons (10 DAG) of WT, epf2-1, epf2-2 and crsp-1 mutants grown at 500 p.p.m. CO2. Note that the stomatal density effects in epf2 mutants are larger than those on stomatal index (see the main text, Methods and Extended Data Fig. 1c legend). *, P<0.05 for comparisons with WT. c, d, Seedlings carrying mutations in the negative-regulatory extracellular signals of stomatal development, EPF1 and CHALLAH (EPFL6), did not exhibit inverted CO2 control of stomatal development in cotyledons. Stomatal indices of 10-day-old WT (Col), epf1-1 single mutant7 (c) and challah single mutant21 (d) seedlings grown at low (150 p.p.m.) and elevated (500 p.p.m.) CO2 concentrations are shown. In all panels, error bars, mean ± s.e.m., n = 20. *, P<0.05, using ANOVA and Tukey’s post-hoc test.
Extended Data Figure 5. Tandem mass spectrometry (MS/MS) spectra identifying the protease CRSP in the apoplast proteome, CRSP localization, qPCR for T-DNA insertion alleles in CRSP and the effects of short term exposure to step changes in the CO2 concentration on CRSP mRNA levels.
Leaf apoplast proteomic experiments identified the following: SBT1.7 (also known as ARA12; identified in four out of five experiments), SBT1.8 (the closest homologue of ARA12; identified in three out of five experiments), SBT5.2 (At1g20160; identified in four out of five independent apoplast proteomic experiments) and SBT3.13 (identified in two out of five independent apoplast proteomic experiments). SDD1 is distantly related to SBT5.2 and has been shown to function independently of EPF1 and EPF2. It belongs to the SBT1 clade of the subtilisin-like serine proteases. a, Example product ion spectrum for the native peptide TTHSWDFLKYQTSVK of CRSP, which was recovered directly from the apoplast extract before trypsin digestion. The product ion spectrum for the parent ion of m/z = 614.33 (+ 3) is shown. Apoplast proteins were isolated, purified and subjected to MS/MS as described in the Methods. b, The product ion spectrum for the peptide AVASAYGSFPTTVIDSK of CRSP, which was identified from trypsin digestion of the apoplast extract. The product ion spectrum for the parent ion of m/z = 857.44 (+ 2) is shown. The product ion spectra are annotated for y, y + 2, b and b + 2, using the Paragon algorithm (ProteinPilot 4.0 AB SCIEX). The tables show the identification results for the peptides using ProteinPilot 4.0. Conf. denotes the percent confidence (99%) score for the identified peptide. Cleavages means any potential mis-cleavage. Delta Mass is the theoretical mass – the measured mass. Z is the charge state. c, d, A translational fusion of the CRSP protease with VENUS (driven by the 5′ promoter fragment comprising the 2,000 basepairs of genomic sequence directly upstream of the first ATG of CRSP) localizes to the cell wall in A. thaliana plants. Hypocotyl (c) and sixth leaf epidermal cells (d) of 10-day-old seedlings are shown. Hypocotyl samples were counter-stained with propidium iodide (top panel) and imaged for VENUS fluorescence (middle panel); the bottom panel shows the merged image. Pending detailed characterization of the sites of CRSP protein expression and localization, it is not known whether the biological activity of CRSP’s modulation of stomatal development in response to an elevated CO2 stimulus originates either from stomatal precursor stem cells or from other cell types such as mature stomata. e, qPCR analyses of 10-day-old seedlings were conducted for WT, crsp-1 (SALK_132812C) and crsp-2 (SALK_099861C) seedlings. Twenty seedlings were pooled, and the RNA was isolated for cDNA synthesis and subsequent qPCR. The expression levels were normalized to those of the CLATHRIN gene. qPCR results suggest approximately 55% reduction in CRSP transcript abundance in seedlings carrying the crsp-1 mutant allele upstream of the T-DNA insertion site. Note that the CRSP-1 translated protein exhibits reduced cleavage of EPF2 (Extended Data Fig. 6a). The crsp-2 mutant has a T-DNA insertion at the 3′ end of the last (ninth) exon and shows partially reduced CRSP transcript levels. Primer sequences 5′ of the T-DNA insertion sites amplified CRSP transcripts (Methods, for primer sequences). f, qPCR analyses of 10-day-old WT seedlings were conducted for plants grown at 150 p.p.m. (left) or 500 p.p.m. (right) CO2. After 10 days of growth at these conditions, the plants were transferred to the opposite CO2 growth conditions for 4 h. CRSP transcripts were quantified via qPCR in cotyledons (ACTIN 2 was used as the housekeeping gene with which to normalize cDNA levels) before (0 h; blue bars) and after (4 h; red bars) the step change in CO2 concentration. n = 10 in e and n = 20 in f. Error bars, mean ± s.e.m. in e and f.
Extended Data Figure 6. CRSP cleaves synEPF2 in vitro.
In vitro cleavage reactions over time of synthetic EPF family peptides incubated with CRSP, (mutated) CRSP-1 and negative control (mock, wheat germ extract only) proteases. The synEPF peptides are flanked by fluorophore and quencher moieties, and fluorescence can be measured when the quencher–fluorophore interaction is disrupted by cleavage of the synEPF peptide. EPF2 (a); EPF1 (b); STOMAGEN (STG) (c); a chimaeric peptide of EPF2, including seven amino acid substitutions corresponding to STOMAGEN in the region of the cleavage (d). The EPF2 peptide that was used comprises the 69 carboxyterminal amino acids of the native EPF2 peptide and includes the predicted cleavage site. This peptide lacks the 51 amino-terminal amino acids of the native EPF2 peptide. We mapped an in vitro cleavage site of the synthetic EPF2 peptide using MS/MS analyses, and our results show predominant cleavage at the site in bold: SKNGGVEMEMYPTGSSLPD|CSYACGACSPC. When aligned with the STOMAGEN protein sequence, this in vitro cleavage site of EPF2 by CRSP is within seven residues of the native STOMAGEN peptide cleavage site23,27. It remains to be determined whether an EPF2 cleavage site corresponding to the STOMAGEN cleavage site23,27 occurs in vivo. The CHIMERA peptide was also cleaved by trypsin to demonstrate the functionality of the synthetic fluorogenic peptide (the EPF1 and STOMAGEN peptides also showed a robust fluorescence signal when cleaved with trypsin). To test the specificity of CRSP-mediated EPF2 cleavage, we conducted cleavage experiments with a re-designed EPF2–STOMAGEN chimaeric peptide. This peptide included 7 amino acid substitutions in the EPF2 sequence, converting a stretch of 12 EPF2 residues into the aligned STOMAGEN sequence (the 12 residue stretch spans the LPD|CS site). The modified EPF2 cleavage site containing the STOMAGEN sequence is SKNGGVEMEMYPIGSTA PTCTYNEGACSPC. We changed the D (in the LPD|CS site) to a T since this corresponds to the sequence of STOMAGEN and EPFL4, a negative regulatory peptide related to EPF2. The modified sequence contained the STOMAGEN-specific TTNE motif. These experiments show that CRSP mediated cleavage is abolished in this chimaeric EPF2–STOMAGEN peptide. Fluorescence data were normalized for background fluorescence by using buffer only controls, and the change in the relative fluorescence was calculated by subtracting the initial fluorescence measurement for each sample. e, The change in the relative fluorescence emitted over time on cleavage of the synthetic EPF2 peptide (synEPF2) by CRSP in the presence or absence of protease inhibitors is shown (Methods). In all panels n = 3. Error bars, mean ± s.e.m.
Extended Data Figure 7. CRSP is required for EPF2 function in planta and CO2 control of stomatal development in crsp epf2 double mutant plants.
a–d, WT and crsp mutant seedlings harbouring an oestradiol-inducible EPF2 construct were germinated in the absence (uninduced; a and c) or presence (induced; b and d) of β-oestradiol. The cotyledon epidermis of 5-day-old seedlings was imaged using a confocal microscope and propidium iodide staining. e, Quantitation of the effects of EPF2 transcript levels on 5-day-old cotyledon stomatal density (number of stomata per mm2) in nine independent lines harbouring the β-oestradiol-inducible EPF2 overexpression construct in the WT, crsp-1 or crsp-2 mutant backgrounds and the WT control (uninduced). For each line, 20 images from 10 cotyledons (2 images per cotyledon; 10 separate seedlings used) were analysed, and RNA was extracted from 10 separate seedlings (see Methods). f, Abaxial stomatal indices for mature cotyledons (10 DAG) of WT (Col), the crsp-1 and epf2-1 single mutants, and the crsp-1 epf2-1 double mutant plants grown at low (blue) and high (red) CO2 concentrations. SLGCs are included in these stomatal index (SI) calculations. n = 20 in e and f. In f, ***, P< 0.00005;*, P < 0.05, using ANOVA and Tukey’s post-hoc test. Error bars, mean ± s.e.m.
Extended Data Figure 8. CRSP is not clearly required for EPF1 function in planta.
a–d, WT and crsp mutant seedlings harbouring an oestradiol inducible EPF1 construct were germinated in the absence (uninduced; a and c) or presence (induced; b and d) of β-oestradiol. The cotyledon epidermis of 5-day-old seedlings was imaged using a confocal microscope and propidium iodide staining. e, Quantitation of the effects of EPF1 transcript levels on 5-dayold cotyledon stomatal density (number of stomata per mm2) in independent lines harbouring the oestradiol-inducible EPF2 overexpression construct in the WT, crsp-1 and crsp-2 mutant backgrounds. n = 20 in e. Error bars, mean ± s.e.m.
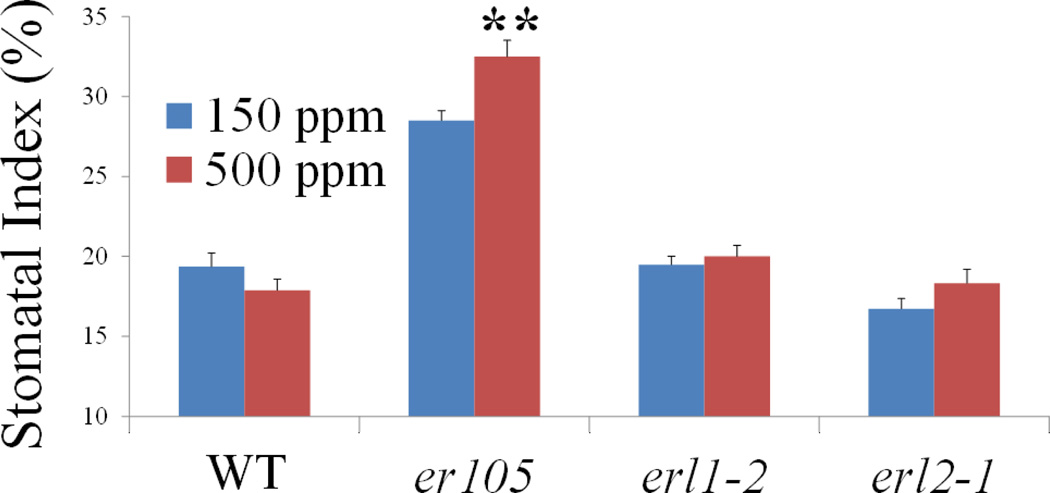
Extended Data Figure 9. erecta mutant exhibits impaired CO2 control of stomatal development.
It has previously been shown that EPF2 binds to the receptor ERECTA22,38, and it has been shown that the mitogen-activated protein (MAP) kinase kinase kinase YODA39 represses stomatal development. Hence, we tested the effects of the elevated CO2 concentration on stomatal development in plants carrying an erecta mutant or erecta like 1 (erl1) or erl2 mutant alleles: er-105, erl1-2 and erl2-1 (ref. 40). The er-105 mutant showed an inversion of CO2 control of stomatal development, and the erl2-1 single mutant showed a possible increase in the stomatal index at elevated CO2 concentration but weaker than that for er-105 Abaxial stomatal indices of WT (Col) and the er-105, erl1-2 and erl2-1 single mutants grown at low (150 p.p.m.; blue) and high (500 p.p.m.; red) CO2 concentrations. SLGCs are excluded from these stomatal index (SI) calculations. n = 20.**, P < 0.005, using ANOVA and Tukey’s post-hoc test. Error bars, mean ± s.e.m.
Acknowledgements
We thank K. Knepper for conducting independent CO2-dependent stomatal development analyses. We thank A. Ries for help with generating the CA–YFP-fusion complementation lines. We thank D. Bergmann for providing the epfl6 mutant line and DNA constructs for MUTEpro::nucGFP expression; K. Torii for providing DNA constructs for MUTEpro::MUTE-GFP expression, erecta mutants and the oestradiol-inducible EPF constructs; T. Altmann for providing the sdd1-1mutant; and M. Estelle, Y. Zhao, A. Stephan and M. Facette for comments on the manuscript. This project was funded by grants from the National Science Foundation (MCB0918220 and MCB1414339 to J.I.S. and IOS-1025837 to S.C.P.) and the National Institutes of Health (GM060396-ES010337 to J.I.S.), a BAYER-UC Discovery grant (J.I.S.) and a seed grant from the UCSD-SDCSB (GM085764) Systems Biology Center (C.B.E.). A grant from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy (DE-FG02-03ER15449) to J.I.S. funded complementation and localization analyses.
Footnotes
Author Contributions J.I.S. and C.B.E. designed experiments. C.B.E. conducted experiments (except for mass spectrometry analyses). C.B.E. generated GFP reporter expression and EPF overexpression plant lines. H.H. generated genetic constructs and transgenic plant lines for ca1 ca4mutant YFP-fusion lines. Mass spectrometry analyses were conducted by M.G., J.C.A. and S.C.P. J.I.S. proposed the project. The manuscript was written by C.B.E. and J.I.S.
The raw data from three independent biological replicates in RNA-seq experiments have been deposited in the BioProject database under accession number PRJNA218542. The mass spectrometry proteomics data have been deposited in the Proteomics Identification Database (PRIDE) under accession numbers PXD000692, PXD000693 and PXD000956.
The authors declare no competing financial interests.
References
- 1.Bergmann DC, Sack FD. Stomatal development. Annu. Rev. Plant Biol. 2007;58:163–181. doi: 10.1146/annurev.arplant.58.032806.104023. [DOI] [PubMed] [Google Scholar]
- 2.Pillitteri LJ, Torii KU. Mechanisms of stomatal development. Annu. Rev. Plant Biol. 2012;63:591–614. doi: 10.1146/annurev-arplant-042811-105451. [DOI] [PubMed] [Google Scholar]
- 3.Nadeau JA, Sack FD. Control of stomatal distribution on the Arabidopsis leaf surface. Science. 2002;296:1697–1700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- 4.Woodward FI. Stomatal numbers are sensitive to increases in CO2 from pre-industrial levels. Nature. 1987;327:617–618. [Google Scholar]
- 5.Gray JE, et al. The HIC signalling pathway links CO2 perception to stomatal development. Nature. 2000;408:713–716. doi: 10.1038/35047071. [DOI] [PubMed] [Google Scholar]
- 6.Hu H, et al. Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nature Cell Biol. 2010;12:87–93. doi: 10.1038/ncb2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hara K, et al. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009;50:1019–1031. doi: 10.1093/pcp/pcp068. [DOI] [PubMed] [Google Scholar]
- 8.Hunt L, Gray JE. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 2009;19:864–869. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- 9.Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim TH, Bohmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodward FI. Potential impacts of global elevated CO2 concentrations on plants. Curr. Opin. Plant Biol. 2002;5:207–211. doi: 10.1016/s1369-5266(02)00253-4. [DOI] [PubMed] [Google Scholar]
- 12.Hetherington AM, Woodward FI. The role of stomata in sensing and driving environmental change. Nature. 2003;424:901–908. doi: 10.1038/nature01843. [DOI] [PubMed] [Google Scholar]
- 13.Masle J, Gilmore SR, Farquhar GD. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature. 2005;436:866–870. doi: 10.1038/nature03835. [DOI] [PubMed] [Google Scholar]
- 14.Sellers PJ. Modeling the exchanges of energy, water, and carbon between continents and the atmosphere. Science. 1997;275:502–509. doi: 10.1126/science.275.5299.502. [DOI] [PubMed] [Google Scholar]
- 15.Battisti DS, Naylor RL. Historical warnings of future food insecurity with unprecedented seasonal heat. Science. 2009;323:240–244. doi: 10.1126/science.1164363. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. Isolation of a strong Arabidopsis guard cell promoter and its potential role as a research tool. Plant Methods. 2008;4:1. doi: 10.1186/1746-4811-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pillitteri LJ, Peterson KM, Horst RJ, Torii KU. Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell. 2011;23:3260–3275. doi: 10.1105/tpc.111.088583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue S, et al. Central functions of bicarbonate in S-type anion channel activation and OST1 protein kinase in CO2 signal transduction in guard cell. EMBO J. 2011;30:1645–1658. doi: 10.1038/emboj.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- 20.Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- 21.Abrash EB, Bergmann DC. Regional specification of stomatal production by the putative ligand CHALLAH. Development. 2010;137:447–455. doi: 10.1242/dev.040931. [DOI] [PubMed] [Google Scholar]
- 22.Lee JS, et al. Direct interaction of ligand–receptor pairs specifying stomatal patterning. Genes Dev. 2012;26:126–136. doi: 10.1101/gad.179895.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugano SS, et al. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463:241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- 24.Kondo T, et al. Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 2010;51:1–8. doi: 10.1093/pcp/pcp180. [DOI] [PubMed] [Google Scholar]
- 25.Uchida N, Tasaka M. Regulation of plant vascular stem cells by endodermis-derived EPFL-family peptide hormones and phloem-expressed ERECTA-family receptor kinases. J. Exp. Bot. 2013;64:5335–5343. doi: 10.1093/jxb/ert196. [DOI] [PubMed] [Google Scholar]
- 26.Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- 27.Ohki S, Takeuchi M, Mori M. The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nature Commun. 2011;2:512. doi: 10.1038/ncomms1520. [DOI] [PubMed] [Google Scholar]
- 28.Rautengarten C, et al. A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 2008;54:466–480. doi: 10.1111/j.1365-313X.2008.03437.x. [DOI] [PubMed] [Google Scholar]
- 29.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nature Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 30.Woodward FI, Lake JA, Quick WP. Stomatal development and CO2: ecological consequences. New Phytol. 2002;153:477–484. doi: 10.1046/j.0028-646X.2001.00338.x. [DOI] [PubMed] [Google Scholar]
- 31.Guttman M, et al. Interactions of the NPXY microdomains of the low density lipoprotein receptor-related protein 1. Proteomics. 2009;9:5016–5028. doi: 10.1002/pmic.200900457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCormack AL, et al. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal. Chem. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 33.Anderson JC, Peck SC. A simple and rapid technique for detecting protein phosphorylation using one-dimensional isoelectric focusing gels and immunoblot analysis. Plant J. 2008;55:881–885. doi: 10.1111/j.1365-313X.2008.03550.x. [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols. 2007;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 35.Niehl A, Zhang ZJ, Kuiper M, Peck SC, Heinlein M. Label-free quantitative proteomic analysis of systemic responses to local wounding and virus infection in Arabidopsis thaliana. J. Proteome Res. 2013;12:2491–2503. doi: 10.1021/pr3010698. [DOI] [PubMed] [Google Scholar]
- 36.Lake JA, Woodward FI. Response of stomatal numbers to CO2 and humidity: control by transpiration rate and abscisic acid. New Phytol. 2008;179:397–404. doi: 10.1111/j.1469-8137.2008.02485.x. [DOI] [PubMed] [Google Scholar]
- 37.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torii KU, et al. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergmann DC, Lukowitz W, Somerville CR. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304:1494–1497. doi: 10.1126/science.1096014. [DOI] [PubMed] [Google Scholar]
- 40.Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]