Abstract
Mitochondria play a central role in the aging process. Studies in model organisms have started to integrate mitochondrial effects on aging with the maintenance of protein homeostasis. These findings center on the mitochondrial unfolded protein response (UPRmt), which has been implicated in lifespan extension in worms, flies and mice, suggesting a conserved role for in the long-term maintenance of cellular homeostasis. Here, we review current knowledge of the UPRmt and discuss its integration with cellular pathways known to regulate lifespan. We highlight how insight into the UPRmt is revolutionizing our understanding of mitochondrial lifespan extension and of the aging process.
Keywords: mitochondria, unfolded protein response, proteostasis, aging, mtUPR
Introduction
Aging is a complex process of cellular and physiological decline that is caused by a combination of intrinsic and extrinsic factors. Strikingly, the study of single-gene mutations that extend lifespan has shown that the rate of aging is plastic and can be modulated by a defined number of biological pathways (For overview see e.g. (Kenyon, 2005)). Such discoveries have provided fertile ground for a still-growing field investigating the basic mechanisms behind aging, which in turn has provided new insight into basic physiological regulation.
Mitochondrial function has been studied as a key influence on aging for four decades, yet the processes and mechanisms governing this role remain controversial. Mitochondria were initially seen as a source of molecular damage on account of their production of reactive oxygen species (ROS); However, it has since become apparent that mitochondria play a more active and defined role in regulating both cellular and physiological function, often using ROS as signaling molecules (Finkel, 2011).
Of the roughly 1500 proteins known to act within human mitochondria, only 13 subunits of the electron transport chain (ETC) are encoded by mitochondrial DNA (Anderson et al., 1981). Thus, constant inter-organelle communication is required to regulate expression of the mitochondrial proteome from nuclear DNA, and specialized machinery must transport nascent proteins past the two mitochondrial membranes. Nuclear-encoded mitochondrial gene expression can be modified by cellular signals (e.g. nitrous oxide), cell non-autonomous cues (e.g. exercise-induced hormones), as well as by cues from mitochondria themselves (Murakami et al., 1998; Nisoli and Carruba, 2006). Mediators of such mitochondria-initiated ‘retrograde’ signal transduction include byproducts of mitochondrial metabolism, calcium ions (for which mitochondria serve as active reservoirs), as well as modulation of mitochondrial protein import (Finley and Haigis, 2009). These signals allow adjusting mitochondrial activity to cellular needs, and can be initiated by external conditions, such as when a cold environment promotes uncoupling and thereby thermogenesis (Golozoubova et al., 2006), or by general dysfunction of the mitochondria themselves. An example of the latter is loss of mitochondrial protein homeostasis (proteostasis), which can occur when expression of its mitochondria- and nuclear-encoded components are not coordinated, when protein import or folding is impaired, or when unnecessary or aggregated proteins cannot be disposed of.
Loss of proteostasis has been proposed to play an important role in age-related decline, as impaired protein quality control and accumulation of misfolded and unfolded proteins has been identified as a hallmark of aging organisms, and as promoting proteostasis by increasing molecular chaperone expression, limiting translation, or increasing protein turnover has been implicated in lifespan extension through various molecular perturbations (for review see (Taylor and Dillin, 2011)). Recent studies have implicated a mitochondrial stress response, the mitochondrial unfolded protein response (UPRmt) as a link between mitochondrial proteostasis and aging in various organisms.
The UPRmt is a vigorous transcriptional response that has been proposed to allay proteostatic stress in mitochondria by promoting folding, limiting import, and reducing translation of mitochondrial proteins (Haynes and Ron, 2010; Haynes et al., 2013). Its role in longevity regulation has emerged from studies in worms focusing on the paradoxical lifespan extension of mutants in which mitochondrial activity is impaired (Dillin et al., 2002; Feng et al., 2001) and has now been supported by findings in flies and mice (Houtkooper et al., 2013; Owusu-Ansah et al., 2013). The UPRmt can be regarded as a hormetic mechanism that extends lifespan in spite of mitochondrial dysfunction. Supporting this view, and highlighting the importance of a measured response to stress, transient activation of the UPRmt during development can produce lasting protective effects and extend lifespan, while chronic or overly powerful induction reduces lifespan (Owusu-Ansah et al., 2013; Pimenta de Castro et al., 2012; Rea et al., 2007).
Similar effects have been observed for activation of the endoplasmic reticulum- and cytoplasm-specific unfolded protein responses, classically known as the unfolded protein response in the ER (UPRER) and heat-shock response (HSR), respectively. In both cases mild transient stimulation of the response, or overexpression of the transcription factors mediating the response, can extend lifespan, while excessive or chronic activation can have deleterious effects (Cohen et al., 2006; Hsu et al., 2003; Labunskyy et al., 2014).
How these proteostatic pathways tie into other processes known to regulate aging is only beginning to be understood. This question is particularly relevant for the UPRmt, given that mitochondria have previously been implicated in a large number of processes influencing longevity (Dillin et al., 2002; Fridell et al., 2005; Rera et al., 2011; Schmeisser et al., 2013; Schriner et al., 2005; Trifunovic and Wredenberg, 2004; Zarse et al., 2012). Does the UPRmt act to either regulate or affect these pathways, or is it a largely independent protective mechanism that alleviates specific cellular dysfunctions during aging by improving mitochondrial function? In this review we survey current knowledge about the activation, transduction and consequences of the UPRmt, and discuss open questions. We then address how the UPRmt ties into current models of longevity regulation.
The UPRmt: a definition
The UPRmt was first described as transcriptional activation of mitochondria-specific chaperones in mitochondrial DNA deficient (ρ0) rat hepatoma cells (Martinus et al., 1996), and mitochondrial chaperone expression remains the gold standard for measuring UPRmt activity. This transcriptional program is selective for specific chaperones, but can also partially overlap with the HSR and the UPRER, and the precise group of chaperones induced by either of these responses seems to vary by study and model organism.
The UPRmt has been studied most extensively in C. elegans, where it leads to upregulation of hsp-6 and hsp-60 (Durieux et al., 2011; Houtkooper et al., 2013; Mouchiroud et al., 2013; Yoneda et al., 2004). Both chaperones are nuclear-encoded but localize to the mitochondrial matrix; hsp-6 is homologous to mtHsp70 in mammals, while hsp-60 is Hsp60 in mammals. To distinguish this response from other proteostatic stress responses, hsp-4 and hsp-16/hsp-70/hsp-90 have been used as markers of UPRER and the HSR, respectively (Durieux et al., 2011; Houtkooper et al., 2013; Mouchiroud et al., 2013; Yoneda et al., 2004). Although mild activation of hsp-6 has been observed when the UPRER and HSR are activated with tunicamycin and heat respectively (Yoneda et al., 2004), the three unfolded protein responses, if defined by induction of the aforementioned chaperones, appear to be mostly selective. However, a comprehensive characterization of the full complement of chaperones induced by each response remains to be performed.
The Drosophila genome encodes a greater number of mitochondrial chaperones, complicating such studies somewhat: While there is only one known homologue for hsp-6/mtHsp70 (Hsc70-5), Hsp10 is represented by two (CG11267 and CG9920), and hsp-60/Hsp60 by four (Hsp60 & Hsp60B-D) genes. In addition, the small heat shock protein Hsp22 also localizes to mitochondria (Morrow et al., 2000). Some activation of hsc70-5 is seen at the transcriptional level during ETC dysfunction or overexpression of misfolding ornithine transcarbamylase (dOTC) in mitochondria (Owusu-Ansah et al., 2013; Pimenta de Castro et al., 2012), but changes in protein expression have not been reported thus far. Hsp60 RNA levels are also only mildly higher under these conditions, but the protein is strongly up-regulated (Pimenta de Castro et al., 2012). Hsp60 and Hsp60C (but not B or D) are required for lifespan extension by electron transport chain perturbation, and their overexpression produces a phenotype similar to activation of the UPRmt (Owusu-Ansah et al., 2013). Changes in gene expression of each of these Hsp60 variants in response to the UPRmt have not been reported, however.
In mammalian cells, the UPRmt response has only been characterized in a few studies (Houtkooper et al., 2013; Martinus et al., 1996; Zhao et al., 2002). As in the worm and fly, Hsp60 is upregulated in mouse, rat and human cells treated with mitochondrial stress-inducing compounds (ethidium bromide (EtBr) or doxycycline), or by over-expression of dOTC. These treatments also induce the mitochondrial chaperones Hsp10 and mtDnaJ. Conversely, increased mtHsp70 was not observed, suggesting increased selectivity and specificity of the UPRmt in mammals, and highlighting the need for further studies to characterize the vertebrate UPRmt.
Expression of mitochondrial chaperones is thus our current best measure of UPRmt activity, but a refinement and clarification of the definition of the UPRmt will be required and possible as more information from diverse model organisms becomes available. Critically, it will be necessary to look beyond chaperone expression in the cellular response to mitochondrial dysfunction to understand the connection between the UPRmt and longevity.
Conditions triggering the UPRmt
Using mitochondrial chaperone induction as the benchmark, a number of studies have explored the signals triggering the UPRmt. As discussed above, increased mitochondrial protein stress is such a signal: Overexpressing folding-deficient mutant proteins targeted to mitochondria is sufficient to elicit a vigorous UPRmt response, as demonstrated by over-expressing dOTC in worms, flies, or mammalian cells (Jin and Youle, 2013; Pimenta de Castro et al., 2012; Rath et al., 2012; Zhao et al., 2002). Similarly, knockdown of mitochondrial chaperones and proteases (which is expected to disrupt mitochondrial proteostasis) activates the UPRmt (Benedetti et al., 2006; Yoneda et al., 2004).
Early studies further suggested that the UPRmt is triggered not only by loss of mitochondrial proteostasis, but also when the complement of nuclear and mitochondrially encoded mitochondrial proteins present in the cell is not synchronized (Martinus et al., 1996; Yoneda et al., 2004). This concept was further developed by the Auwerx lab, who showed that a range of conditions that produced either an abundance or scarcity of mitochondrial ETC components triggered the UPRmt (Houtkooper et al., 2013). Accordingly, reducing the production of proteins from mtDNA can trigger the UPRmt. This has been accomplished using EtBr, which impairs transcription preferentially in mitochondria (Yoneda et al., 2004). Extended EtBr treatment can completely eliminate mitochondrial DNA, and in rat hepatoma cells this increases the steady state level of mitochondrial chaperones (Martinus et al., 1996). The UPRmt is also activated by inhibiting mitochondrial translation directly, either by treatment with doxycycline or by knockdown of mRpS5 and other mitochondrial ribosomal subunits (Houtkooper et al., 2013; Yoneda et al., 2004). Conversely, the UPRmt is activated by knockdown of several nuclear-encoded protein subunits of the ETC: The complex IV subunit Vb/COX4, complex I subunit ND75, and complex III subunit ISP-1 have been studied in in a variety of systems (Durieux et al., 2011; Houtkooper et al., 2013; Owusu-Ansah et al., 2013), but several other ETC components have been implicated (Runkel et al., 2014; Yoneda et al., 2004).
Supporting a general role for the UPRmt in the cellular response to mitonuclear imbalance, UPRmt activation has paradoxically also been reported for genetic and pharmacological interventions that should decrease protein stress in mitochondria by either increasing mitochondrial biogenesis (Houtkooper et al., 2013; Mouchiroud et al., 2013) or by impairing the mitochondrial import machinery (Nargund et al., 2012; Rainbolt et al., 2013). In the ‘mitonuclear imbalance’ model it is proposed that the imbalance (in either direction) of mitochondria- and nuclear-encoded subunits of the ETC leads to accumulation of ‘orphaned’ proteins and thereby activates the UPRmt (Houtkooper et al., 2013; Yoneda et al., 2004).
Perturbing mitochondrial physiology more generally is a third way of activating the UPRmt. Mutations in clk-1 (encoding a coenzyme Q biosynthetic enzyme) interfere with ubiquinone synthesis, affecting both the ETC and the cellular redox environment, and lead to UPRmt activation in worms (Baker et al., 2012; Durieux et al., 2011; Yang and Hekimi, 2010a). Compounds that interfere with ETC function (rotenone and antimycin A) also induce hsp-6 in C. elegans (Runkel et al., 2013). Along these lines, treatment with paraquat to increase mitochondrial ROS production induces UPRmt chaperones in worms, while non-mitochondrial ROS production caused by acrylamide does not (Runkel et al., 2013; Yoneda et al., 2004). However, it cannot be discounted that the activation of the UPRmt in these conditions is a consequence of local protein damage triggered by ROS production, rather than a direct ROS-mediated signaling event.
The UPRmt is thus emerging as an evolutionarily conserved surveillance system that is generally engaged when mitochondrial homeostasis has to be restored.
UPRmt signal transduction
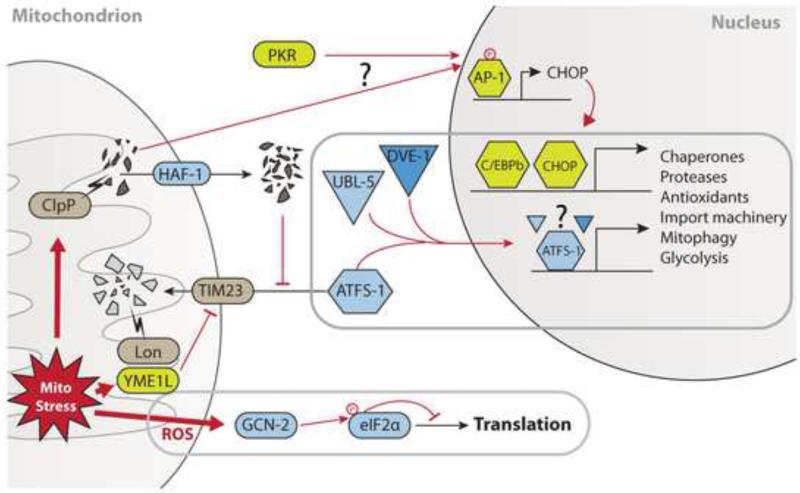
Our understanding of UPRmt signal transduction stems overwhelmingly from C. elegans, with additional insight gained from mammalian studies. To date, the evolutionary conservation of the pathways identified in worms remains unclear, and the exact mechanism(s) leading to the coordinated and selective transcriptional induction of mitochondrial chaperones have not been determined exhaustingly. Nevertheless, genetic and biochemical studies in various models have started to assemble a working model for UPRmt signal transduction, as illustrated in Figure 1.
Figure 1. UPRmt signal transduction.
Current model for UPRmt signal transduction based on studies in worms and mammalian cells. Signaling components identified in worms are shaded blue and signaling components identified in mammalian cells are shaded green. See text for details.
The ATFS-1 pathway
At least two separate pathways have been proposed to link mitochondrial stress to the proteostatic response in worms: a response dependent on the bZip transcription factor ATFS-1 (previously known by its gene name ZC376.7) (Haynes et al., 2010), and a pathway involving the kinase GCN-2 (Baker et al., 2012).
ATFS-1 contains a nuclear localization signal, but also nuclear export and mitochondrial localization signals. Consequently, it is normally excluded from the nucleus and imported into mitochondria where it is degraded by the protease Lon (Nargund et al., 2012). When the import of ATFS-1 is inhibited by mitochondrial stress the transcription factor enters the nucleus to activate the UPRmt. The transcriptional induction of UPRmt response genes by ATFS-1 depends on the ubiquitin-like protein UBL-5 and the homeobox protein DVE-1: mitochondrial stress increases UBL-5 expression and promotes its nuclear translocation, where it dimerizes with DVE-1 (Benedetti et al., 2006; Haynes et al., 2007). While both ATFS-1 and the UBL-DVE complex are required to induce mitochondrial chaperones, a direct interaction between them has not been described thus far.
Mitochondrial localization of ATFS-1 in basal conditions depends on several factors, which seem to converge on the mitochondrial import machinery. Reducing import efficiency of the TIM23 complex in C. elegans and human cells, by RNAi of either TIM17 or TIM23, activates the UPRmt in an ATFS-1 dependent manner (Nargund et al., 2012; Rainbolt et al., 2013). Mitochondrial import is also sensitive to the electrochemical potential across the inner membrane (Martin et al., 1991), suggesting a mechanism whereby ETC dysfunction activates the UPRmt. However, direct membrane depolarization using dinitrophenol does not activate the UPRmt (Yoneda et al., 2004). On the other hand, import inhibition by mitochondrial oxidative stress has been reported (Wright et al., 2001), and paraquat treatment induces mitochondrial chaperones (Nargund et al., 2012).
The nuclear relocalization of ATFS-1 during stress requires the inner membrane ABC transporter HAF-1, and it has been proposed that an efflux of degraded peptides through HAF-1 prevents mitochondrial import of ATFS-1 (Haynes et al., 2010; Nargund et al., 2012). While it has not been demonstrated that HAF-1 inhibits import in this manner, indirect evidence is provided by the fact that HAF-1 is not required for UPRmt activation by methods that directly interfere with mitochondrial import, e.g. RNAi of inner or outer membrane translocases, paraquat treatment and ETC dysfunction (Nargund et al., 2012). Instead, HAF-1 is required for UBL-5 up-regulation in a heat-sensitive model of UPRmt (Haynes et al., 2010). Furthermore, both HAF-1 and the protease CLPP-1 were found to be required for nuclear localization of GFP-tagged ATFS-1 when the UPRmt was induced by knockdown of hsp-6, and CLPP-1 promotes HAF-1 dependent protein efflux from mitochondria (Haynes et al., 2010). However, any epistatic relationship between the two proteins has yet to be clearly demonstrated.
The GCN-2 pathway
Recently, it was reported that several ETC mutations (clk-1 and isp-1) activate the second branch of the UPRmt (Baker et al., 2012). Rather than on ATFS-1, this branch depends on phosphorylation of eIF2α by the kinase GCN-2, which increases under conditions of mitochondrial and ER stress. eIF2α phosphorylation reduces cytosolic translation, presumably to reduce the load on the mitochondrial proteostatic machinery. GCN-2 is stimulated by ROS, and lifespan extension in clk-1(qm30) and isp-1(qm150) mutant worms is dependent on increased superoxide levels (Yang and Hekimi, 2010b).
Knockdown of GCN-2 (and thus exacerbated protein load in mitochondria) leads to greater induction of mitochondrial chaperones during stress, while knockdown of HAF-1 or ATFS-1 correspondingly increases eIF2α phosphorylation exclusively in stressed conditions. Furthermore, obstructing both these pathways has cumulative effects on the developmental phenotype of spg-7, clk-1 and isp-1 mutant worms, suggesting that they play complementary roles in the UPRmt (Baker et al., 2012). A clear partition of these pathways into a ROS-dependent and a protein stress-dependent branch is not possible, however: paraquat has been shown to induce chaperones via ATFS-1 (Pimenta de Castro et al., 2012; Runkel et al., 2013), while eIF2α phosphorylation in murine cells is ClpP dependent (Rath et al., 2012). Further work will be needed to clarify the relative contribution and regulation of transcriptional induction of mitochondrial chaperones versus reduced translation during the UPRmt, as well as the role of UBL-5 and DVE-1 (if any) in the latter.
Proteases in the UPRmt
As mentioned above, inhibition of mitochondrial proteases can induce the UPRmt, presumably by increasing the load of misfolded proteins. The most extensively studied example is SPG-7, a subunit of the m-AAA protease that is orthologous to human paraplegin (Benedetti et al., 2006; Haynes et al., 2010; Yoneda et al., 2004). In yeast this protease forms a supercomplex with prohibitin 1 and 2 (Steglich et al., 1999) and RNAi of prohibitin 2 has also been shown to induce the UPRmt in C. elegans, though not extending lifespan (Schleit et al., 2013).
Other proteases have been placed in the signaling cascade leading to UPRmt induction, most notably orthologues of the bacterial protease ClpP: The worm orthologue CLPP-1 localizes to mitochondria and is required for hsp-6 and hsp-60 induction (Haynes et al., 2007). It interacts with either of the two homologues of the bacterial AAA+ ATPase ClpX, and this interaction is required for its protease activity and for triggering the UPRmt (Haynes et al., 2010). It has been suggested that ClpP activity is required because it produces the peptides that are transported through HAF-1 and inhibit the mitochondrial import of ATFS-1 through an unresolved mechanism (Haynes and Ron, 2010; Haynes et al., 2007, 2010). RNA interference (RNAi) of the Drosophila ClpX homologue (CG4538) is synthetic lethal with mitochondrial mutations (Owusu-Ansah et al., 2013), suggesting an evolutionarily conserved role of the CLPP-1/ClpX complex. In human cells, mitochondrial ClpP is transcriptionally induced by, and immunoprecipitates with, unfolded proteins in mitochondria (Zhao et al., 2002). The role of human ClpX in activating the UPRmt has not yet been studied.
The metalloprotease YME1L has also been implicated in triggering the UPRmt in human cells. It is induced by mitochondrial stress (Aldridge et al., 2007), and, consistent with a role in mitochondrial proteostasis, its absence leads to accumulation of non-assembled respiratory chain subunits, disrupting mitochondrial morphology (Stiburek et al., 2012). YME1L is essential for degradation of the TIM17A subunit of the inner membrane translocase (TIM23) upon eIF2α phosphorylation, thus reducing mitochondrial import (Rainbolt et al., 2013).
Transcriptional induction of the UPRmt in vertebrates
In mammalian cells, it has been proposed that the up-regulation of mitochondrial chaperones, proteases and import machinery is mediated by heterodimers of the CHOP and C/EBPβ transcription factors (Aldridge et al., 2007; Horibe and Hoogenraad, 2007; Zhao et al., 2002). These transcription factors also play a role in the UPRER, and it has been proposed that mitochondrial specificity is achieved by selective transcriptional induction of CHOP through activator protein-1 (AP-1) elements in the chop promoter (Horibe and Hoogenraad, 2007). The c-Jun subunit of AP-1 is phosphorylated in intestinal epithelial cells over-expressing dOTC (Rath et al., 2012) by a ClpP and protein kinase R (PKR)-dependent mechanism (Rath et al., 2012). How the specificity of the AP-1 transcriptional output is determined during the UPRmt response, however, remains unclear.
Open questions
A wide range of treatments has been used to induce the UPRmt, and there is some variation both in the chosen readout for activation and in the observed effects. For example, induction of the UPRmt by spg-7 RNAi was initially reported to be HAF-1 dependent (Haynes et al., 2010) while a later study using the same reporter showed HAF-1 independent activation (Nargund et al., 2012). Moreover, qualitatively different transcriptional responses have been reported when comparing RNAi of ETC subunits with point mutations in the same protein (Yang and Hekimi, 2010a). Only RNAi approaches led to induced chaperones and reduced ATP levels, while mutations decreased oxygen consumption and increased antioxidant protein levels. Both types of perturbation extended lifespan, and the effects of knocking down one subunit was additive with mutation in another (whereas double mutants did not live longer than single mutants). Some of this complexity may be a consequence of different levels of mitochondrial perturbation in the different loss of function conditions (see Rea et al., 2007), but it also brings to mind the two branches of UPRmt, and could imply that what we currently describe as the UPRmt is really a set of interrelated responses to more specific mitochondrial stressors. An important consideration for future efforts will thus be to effectively and clearly distinguish between effectors and regulators of the UPRmt, and to obtain more detailed insight into the signaling kinetics involved.
Crosstalk between the UPRmt and other stress signaling pathways
Attenuated translation through eIF2α phosphorylation also occurs in response to ER stress, and as in the UPRmt, this response occurs in parallel with a separate transcriptional program, which for the UPRER is activated by IRE-1 and XBP-1 (Shen et al., 2001). The UPRER thus converges with the UPRmt on the regulation of eIF2α, which is phosphorylated by PEK-1 (PERK in mammals) in response to ER stress, but by GCN-2 upon mitochondrial stress. Loss of either kinase stimulates and exacerbates the corresponding organelle-specific stress response (Baker et al., 2012). Mechanisms that ensure selectivity of these responses remain incompletely understood.
Another pathway that potentially overlaps with the UPRmt is the mitochondrial retrograde response, which has been particularly well studied in S. cerevisiae (see e.g. Liu and Butow, 2006). Although the pivotal trigger seems to be perturbation of glutamate/glutamine levels, the pathway can be activated by a variety of disturbances in membrane potential and ATP synthesis. The transcriptional response is aimed at restoring the transcarboxylic cycle while providing alternative sources of nitrogen for biosynthesis. A retrograde response has also been reported in mammalian cells, where signal transduction occurs via release of Ca2+ to the cytosol (Biswas et al., 1999). Since the retrograde response regulates longevity in yeast (Kirchman et al., 1999) and, like the UPRmt, can be activated through ETC dysfunction and in ρ0 cells, crosstalk between these pathways is worth considering. Ca2+ did not appear to play a role in UPRmt activation in murine cells (Rath et al., 2012), but most studies have not examined the activation of the retrograde and UPRmt responses simultaneously. Further, transcriptional activation through the retrograde response varies somewhat depending on the activating stimulus, and not all transcriptional changes in ρ0 cells are mediated by the retrograde response (Epstein et al., 2001).
Spatiotemporal coordination of the UPRmt
A particularly intriguing feature of the UPRmt is the fact that it can be activated by and can elicit secondary UPRmt responses distal to the tissue experiencing stress. This non-autonomy of the UPRmt is significant for the effects of the UPRmt on lifespan, presumably because it allows coordinating cellular stress responses to improve viability systemically whenever proteostatic stress is experienced locally (Durieux et al., 2011). A related aspect of the UPRmt is its perdurance, where induction of the UPRmt during development is ‘remembered’ by increased chaperone expression in adult animals (Durieux et al., 2011). Current knowledge of the molecular mechanisms mediating these responses remains limited, however:
Spatial coordination
The non-autonomous nature of the UPRmt was initially identified in worms, where RNAi knockdown of ETC components in neurons induces chaperone expression in the gut to the same level as cell-autonomous RNAi in the gut (Durieux et al., 2011). Neuronal hsp-6::GFP induction is not observed even for neuron-specific knockdown, and it is therefore unclear whether the non-autonomous communication is mono- or bi-directional. It has been proposed that such communication is mediated by secreted molecules (dubbed mitokines) during mitochondrial stress (Durieux et al., 2011), although the nature and receptors of such mitokines are not known. It is noteworthy that while systemic UBL-5 knockdown prevents lifespan extension in ETC loss of function conditions, gut-specific knockdown of UBL-5 does not prevent UPRmt activation in conditions of neuronal ETC dysfunction. This suggests a UBL-5 independent pathway of mitochondrial chaperone induction in the gut in response to neuronal UPRmt activation, the nature of which remains unclear (Durieux et al., 2011). Recent data suggest that the non-autonomous coordination of proteostatic stress responses in worms is not limited to the UPRmt, as activation of the UPRER in neurons or the intestine can also induce the UPRER in other tissues (Taylor and Dillin, 2013).
The spatial coordination of the UPRmt seems to be conserved in Drosophila, where RNAi knockdown of some ETC components in the brain is sufficient to extend lifespan (Copeland et al., 2009), and where knockdown of the ETC component ND75 in the flight muscle is sufficient to induce chaperone expression and extend lifespan (Owusu-Ansah et al., 2013). At the same time, overexpression of Hsp60 or Hsp60C in muscle is sufficient to extend mean lifespan, suggesting that mitochondrial homeostasis in this tissue is limiting for fly lifespan (Owusu-Ansah et al., 2013). The muscle UPRmt was found to trigger systemic activation of the insulin/IGF signaling (IIS)-responsive transcription factor Foxo, a well-known mediator of systemic stress responses and regulator of lifespan (Karpac and Jasper, 2009; Kenyon, 2010): ND75 knockdown in the muscle lead to increased expression of the insulin-like peptide binding protein ImpL2 (an orthologue of human IGFBP7), resulting in systemic inhibition of IIS activity and consequent Foxo activation in peripheral tissues (Owusu-Ansah et al., 2013). Accordingly, overexpression of ImpL2 in the muscle phenocopies growth defects caused by ND75 knockdown and is sufficient to extend lifespan. Furthermore, knockdown of ImpL2 reduces the lifespan of ND75 RNAi flies, suggesting that ImpL2 acts downstream of the local UPRmt to mediate the systemic response. It was not tested, however, whether inhibiting UPRmt signaling components prevents ImpL2 upregulation under conditions of impaired mitochondrial electron transport. It thus remains unclear whether the UPRmt is involved in the induction of ImpL2 in ND75 loss of function conditions, or whether ImpL2 is induced by alternative stress responses. Importantly, the C.elegans Foxo homologue DAF16 is not required for lifespan extension by ETC disruption in worms (Dillin et al., 2002), indicating that an involvement of systemic IIS perturbation in the UPRmt-mediated longevity in flies may be a species-specific effect.
Temporal coordination
Another interesting observation from (Durieux et al., 2011) is that ETC perturbations in larval stages are sufficient to increase chaperone expression at least into early adulthood, while perturbation after the L3 larval stage did not induce chaperone expression. This accords with previous observations that L3/L4 is a critical stage for ETC-mediated longevity in worms (Dillin et al., 2002; Rea et al., 2007), and that lifelong UPRmt induction most strongly induces chaperones in L4 and early adulthood (Yoneda et al., 2004). Most studies linking the UPRmt to longevity are carried out using mutant animals or with RNAi throughout development, but one later study corroborated that UPRmt activation after the L3 stage does not extend lifespan or even induce chaperone expression (Houtkooper et al., 2013). On the other hand, the GCN-2 branch of the UPRmt can be activated in adult worms (Baker et al., 2012), and low-dose paraquat treatment can extend lifespan even when initiated in the adult (Lee et al., 2010). The importance of the L3 stage has been proposed to result from a dramatic mitochondrial expansion at this point (Tsang and Lemire, 2002), and it is possible that the peak of mitochondrial proteostatic stress experienced here sets a baseline that modifies chaperone expression in adults. It is tempting to speculate that this regulation may be mediated by nuclear epigenetic modifications, as has recently been demonstrated for mitochondrial ROS-induced longevity in yeast (Schroeder et al., 2013).
In Drosophila, RNAi of certain ETC components in the adult extends lifespan, although other components only extend lifespan when removed through development (Copeland et al., 2009). In the flight muscle, adult-onset RNAi of ND75 is sufficient to extend lifespan, and to preserve mitochondrial function, motility and muscle structure in aging animals (Owusu-Ansah et al., 2013). This difference between the temporal requirement of UPRmt activation for lifespan extension in worms and flies may result from a more temporally diffuse program of mitochondrial biogenesis throughout fly development, commencing with a significant expansion in oocytes (Tourmente et al., 1990).
The UPRmt and aging: physiological consequences of UPRmt activation
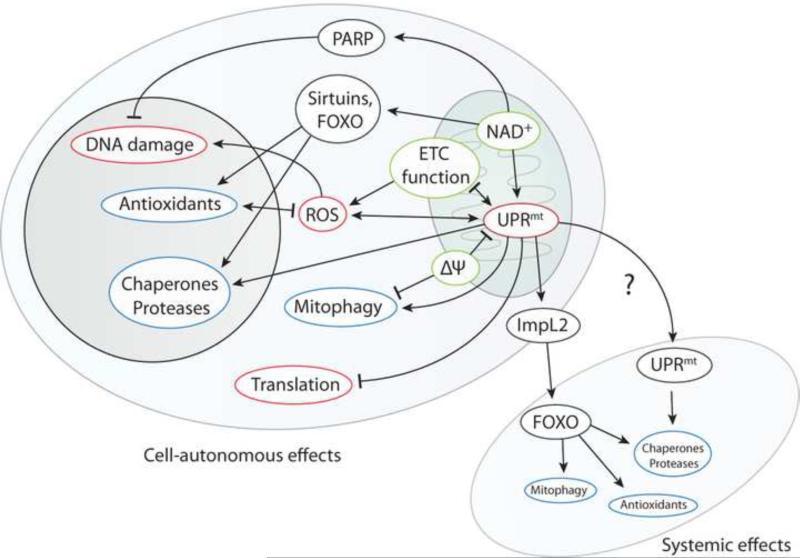
The lifespan extension observed after activation of the UPRmt is accompanied by pleiotropic physiological changes in the animal. Mitochondrial stressors that activate the UPRmt in C. elegans, for example, typically slow development and reduce both the size and motility of adult worms, while compromising the integrity of the body wall muscle in freshly molted adults. Blocking UPRmt signaling compounds this phenotype (Baker et al., 2012; Haynes et al., 2007, 2010; Houtkooper et al., 2013). In Drosophila, reduced size and perturbed flight muscle structure are also observed when the UPRmt is activated in development (Owusu-Ansah et al., 2013; Pimenta de Castro et al., 2012). On the other hand, the age-related decline in muscle structure and function is mitigated in worms and flies with chronically activated UPRmt (Houtkooper et al., 2013; Mouchiroud et al., 2013; Owusu-Ansah et al., 2013). It seems pertinent that, to understand the lifespan extension elicited by the UPRmt mechanistically, a more comprehensive insight into the cellular and physiological consequences of UPRmt activation has to be achieved. The following sections, summarized in Figure 2, will provide an overview of our current understanding.
Figure 2. Cellular and systemic outcomes of UPRmt activation.
Cell autonomous interactions between the UPRmt and processes ensuring cellular homeostasis, as well as non-autonomous interactions between local UPRmt activation and peripheral cells are depicted. Individual interactions have been identified in different model systems, and detailed characterization of these interactions is needed to obtain a comprehensive model for UPRmt responses in specific systems / tissues. See text for details.
Consequences of UPRmt activation for mitochondrial function
Activation of the UPRmt is accompanied by changes in mitochondrial structure and function. The mitochondrial network often becomes more fragmented (Houtkooper et al., 2013; Runkel et al., 2013), potentially aiding autophagy of mitochondria (mitophagy) (Jin and Youle, 2013; Papa and Germain, 2013; Pimenta de Castro et al., 2012). Cellular oxygen consumption is commonly decreased (Baker et al., 2012; Haynes et al., 2010; Houtkooper et al., 2013; Yang and Hekimi, 2010a), and ATP production is compromised (Copeland et al., 2009; Houtkooper et al., 2013). While reduced oxygen consumption is not surprising in response to ETC perturbations, these phenotypes may constitute a regulated response to mitochondrial stress, suspending respiration to allow for mitochondrial repair, and may thus promote homeostasis and cell survival, increasing lifespan. Consistent with this idea, one study has reported the induction of several glycolysis genes by ATFS-1 in worms, suggesting a regulated shift from oxidative phosphorylation to glycolysis (Nargund et al., 2012).
Changes in cellular ATP levels do not always correlate with changes in oxygen consumption following UPRmt activation (Copeland et al., 2009; Houtkooper et al., 2013; Yang and Hekimi, 2010a), although they do echo changes in mitochondrial content (Houtkooper et al., 2013; Mouchiroud et al., 2013; Owusu-Ansah et al., 2013). This variance could reflect the response of multiple metabolic regulators to mitochondrial dysfunction. A simpler explanation, however, would be that the UPRmt can affect mitochondrial coupling (and thereby the ratio of heat/ATP production). Alternatively the UPRmt could affect ATP consumption in the cell, which would impact steady-state levels of ATP independently of its production. Further functional characterization of the UPRmt should help clarify the situation.
It seems clear that the UPRmt links ETC dysfunction with the well-established longevity phenotype of animals with perturbed ETC function, but a UPRmt-independent role for ETC dysfunction in lifespan extension cannot yet be discounted. Mitochondrial efficiency has been linked to longevity outside the context of ETC-deficient mutants, an example of which is lifespan extension by neuronal overexpression of uncoupling protein 2 (Fridell et al., 2005). In genetic screens, transducers of ETC-mediated longevity have also been identified that are not obviously associated with the UPRmt (Walter et al., 2011). Moreover, genetic induction of mitochondrial biogenesis can increase longevity (Rera et al., 2011) and increased mitochondrial DNA mutations produce an aged phenotype independent of ROS production (Trifunovic and Wredenberg, 2004; Trifunovic et al., 2005). Detailed mechanistic dissection of the effects of increased mitochondrial chaperone production on mitochondrial biochemistry and bioenergetics is thus required.
Mitochondrial metabolic byproducts in UPRmt-mediated longevity
Evidence is emerging that changes to the production of other mitochondrial metabolic byproducts wrought by the UPRmt also have important physiological effects. Among these, ROS likely elicit cellular stress responses and thus influence homeostasis and longevity (Ristow and Schmeisser, 2011). Others include the cholesterol-producing mevalonate pathway, relevant as the target of statins (Rauthan et al., 2013), and NAD+, a substrate for both poly(ADP-ribose) polymerases (PARPs) and the sirtuin deacetylases (Mouchiroud et al., 2013; Papa and Germain, 2013), and a major emerging regulator of longevity (Gomes et al., 2013; Mouchiroud et al., 2013; Schmeisser et al., 2013).
Excessive ROS can be damaging to cellular components, but the relationship between ROS production and lifespan is complicated (Cohen et al., 2009; Copeland et al., 2009; Lee et al., 2010; Ristow and Schmeisser, 2011; Sena and Chandel, 2012). Studies in both C. elegans and Drosophila suggest that reducing ROS through overexpressed or supplemented antioxidants blocks both lifespan extension and transcriptional responses induced by ETC dysfunction (Lee et al., 2010; Owusu-Ansah et al., 2013; Rea et al., 2007; Yang and Hekimi, 2010b). Other reports, however, have observed activation of the UPRmt and lifespan extension in the presence of antioxidants (Durieux et al., 2011; Houtkooper et al., 2013). This dichotomy extends to the resulting redox state and response: MnSOD is not induced with the UPRmt in human cells (Zhao et al., 2002), but elevated levels of both ROS and antioxidants have been reported in the worm (Nargund et al., 2012; Owusu-Ansah et al., 2013; Yang and Hekimi, 2010a). These discrepancies may arise from qualitatively different responses being activated under specific conditions; phosphorylation of eIF2α by GCN-2 is ROS dependent (Baker et al., 2012) and phosphorylation of eIF2α by other kinases also induces oxidative stress resistance (Harding et al., 2003), suggesting the possibility that this branch of the UPRmt is particularly responsive to oxidative stress.
An intriguing possibility is that the UPRmt promotes longevity through its impact on NAD+ metabolism. NAD+ has a critical role in cellular energy metabolism, and is emerging as a central metabolite affecting aging (Jasper, 2013). The role of NAD+ in aging has first been explored due to its function as co-factor and substrate of NAD-dependent histone deacetylases (sirtuins). Overexpression of sirtuins has been reported to extend lifespan in multiple organisms, including mammals (Kanfi et al., 2012; Rogina and Helfand, 2004; Tissenbaum and Guarente, 2001), although the robustness of this effect has been called into question (Burnett et al., 2011). Sirtuins regulate energy metabolism, and their function in aging intersects with the response to calorie restriction (Guarente, 2013), but also a broader range of processes including control of cell death (Araki et al., 2004) and circadian rhythms (Chang and Guarente, 2013). Another group of NAD+ consuming enzymes, PARPs, have recently been implicated in longevity regulation, a function that is most likely a consequence of their effects on cellular NAD metabolism (Bai et al., 2011). PARPs were initially recognized as part of the DNA damage response, signaling damage through poly ADP-ribosylation (Durkacz et al., 1980), but also play a role in normal cell cycle progression (Augustin and Spene, 2003) and transcription (Meisterernst et al., 1997). They have been implicated in aging both through investigations of DNA repair and in cross-sectional studies of longevity (Kobbe et al., 2003; Muiras et al., 1998). PARP activity rapidly consumes ADP-ribose monomers, and can thereby deplete their precursor NAD+ and induce cell death (Yang et al., 2007).
NAD+ can be biosynthesized from nicotinamide (NAM), which can also be methylated to form 1-methylnicotinamide (MNA); supplementation of NAM and/or increased conversion to MNA, as well as PARP inhibition, was recently shown to extend lifespan in the worm, independently of sirtuins (Schmeisser et al., 2013). NAD+ levels naturally decline with age in worms and mice, causing a loss of mitochondrial (but not of nuclear) encoded ETC subunits (Gomes et al., 2013), and depleting NAD+ levels genetically or chemically accelerates mortality (Gomes et al., 2013; Mouchiroud et al., 2013). In mice, Sirt1 knockout phenocopies this age-dependent decline of mitochondrial function, and elevating NAD+ levels in old animals restores mitochondrial function in a SirT1-dependent manner. The authors propose that loss of NAD+ or Sirt1 results in a pseudohypoxic state that includes accumulation of HIF-1α and Warburg-like reprogramming of cellular metabolism (Gomes et al., 2013). Despite the associated imbalance in mitochondrial and nuclear encoded ETC subunits, the authors do not report activation of the UPRmt. However, analogous results were found in worms and a mouse cell line, where increased NAD+ levels increased the ratio between mitochondrial and nuclear ETC subunits (Mouchiroud et al., 2013). In these cases, mitochondrial chaperones were rapidly induced, and an antioxidant response followed with some delay. sir-2.1 over-expression was sufficient to induce this UPRmt, and both ubl-5 and sir-2.1 were required for lifespan extension. In line with this, SirT3 has been reported to activate branches of the UPRmt in human cells (Papa and Germain, 2013). It remains to be seen whether the UPRmt is a critical ingredient of sirtuin-mediated longevity, or whether a more complex interplay is responsible.
Insulin/IGF signaling
The observation that the UPRmt affects insulin signaling in flies provides another possible route for its effect on longevity. Reduced Insulin/IGF signaling (IIS) is among the best understood conditions that extend lifespan (For review see (Kenyon, 2010)). Impaired IIS activity has been linked to a number of protective mechanisms, including activation of heat shock factor 1 (HSF-1), which acts alongside DAF-16 to increase expression of small heat shock proteins, and thus increases lifespan in worms (Hsu et al., 2003), and regulation of the UPRER (Henis-Korenblit et al., 2010). Furthermore, DAF-16 is required for longevity resulting from loss of hypoxia-induced factor 1 (HIF-1) (Zhang et al., 2009), and HIF-1 has been implicated in the longevity phenotype of ETC mutants (Lee et al., 2010). On the other hand, activation of HIF-1 also reduces protein aggregation and extends lifespan in an IIS-independent but proteasome-dependent manner (Mehta et al., 2009; Zhang et al., 2009), and the absence of a requirement for DAF-16 in ETC-mediated lifespan extension suggests that IIS regulation is not required for lifespan extension by the UPRmt, at least in worms (Dillin et al., 2002). Nevertheless, recent studies report that impaired IIS extends lifespan by inducing a transient ROS signal through mitochondrial metabolism (Zarse et al., 2012). Such a signal is consistent with one method of activating the UPRmt and inducing longevity, although no causal relationship has been reported thus far (Runkel et al., 2013; Yang and Hekimi, 2010a, 2010b). Further studies revisiting and characterizing the interaction between IIS components and the UPRmt in the worm in detail thus seem warranted.
In flies, FOXO preserves proteostasis in aging muscle, triggering a systemic proteostatic response that increases lifespan (Demontis and Perrimon, 2010). Combined with the recently identified systemic inhibition of IIS activity by muscle-specific mitochondrial perturbation (Owusu-Ansah et al., 2013), it seems likely that local UPRmt activation triggers a systemic Foxo-mediated improvement of proteostasis, and thereby extends lifespan even while the activating tissue is adversely affected.
TOR signaling
Signaling through Target of Rapamycin (TOR) Kinase, which constitutes a second signaling branch regulating lifespan downstream of IIS, has also been implicated in UPRmt signaling. TOR responds to nutrient availability by inducing an anabolic state and promoting translation and growth, and both pharmacological and genetic studies have repeatedly implicated the TOR pathway in aging and age-related diseases (for a thorough review see (Johnson et al., 2013)). The consequences of TOR inhibition mimic the consequences of UPRmt activation on several levels, including an increase in autophagy and reduction of translation, and TOR also regulates mitochondrial biogenesis (Johnson et al., 2013). TOR is regulated by the GTP-binding protein Rheb, and a worm homologue of Rheb was identified in an RNAi screen for proteins involved in the UPRmt (Haynes et al., 2007). Despite these indications, it is not yet clear whether TOR pathway regulation is part of the longevity effects of the UPRmt.
Hormetic cellular maintenance
Even though the studies we have presented thus far seem to place the UPRmt in a complex web of interactions, it is possible that the longevity it promotes is simply the result of improved cellular housekeeping. Loss of proteostasis plays an important role in age-related decline: maintaining proteostasis is an essential part of proper physiological function, but aging brings an impaired protein quality control and accumulation of misfolded proteins (for thorough reviews see: (Morimoto, 2008; Taylor and Dillin, 2011)). As might be expected from the nature of the response, the effects of the UPRmt help restore protein quality control: increased chaperone expression assists correct protein folding, but also allows refolding of incorrect products and can sequester aggregated protein in less cytotoxic states (Behrends et al., 2006); higher protease activity helps degrade misfolded and aggregated proteins, while reduced translation and import increases the ratio between chaperones and unfolded proteins. Improved antioxidant defenses prevents oxidative damage from interfering with protein folding, as well as from damaging DNA and lipids, while a shift towards glycolysis serves to ensure the supply of ATP during a crisis. There is also evidence that UPRmt activation increases mitophagy (Jin and Youle, 2013; Owusu-Ansah et al., 2013; Papa and Germain, 2013), although it is possible that this is a last resort that occurs when mitochondrial proteotoxic stress cannot be alleviated by other branches of the UPRmt.
One model of cellular proteostasis is that the steady-state level of chaperones is only barely able to maintain proteostasis (Morimoto, 2008). This would predict stochastic failure of protein folding, even when fluctuating conditions are not severe enough to cause stress responses. This in turn suggests that the activity of stress responses like the UPRmt could improve proteostasis beyond what ‘normal’ cellular function begets, and thereby extend lifespan. This is essentially the idea of hormesis, where mild stress induces a protective response that more than compensates for the stressor's negative effect. Under this hypothesis activation of the UPRmt would remedy and/or augment the baseline proteostatic machinery, which is not inconceivable based on the lasting effects seen when the UPRmt is activated during development (and that this persistence in many cases correlates with longevity). In line with this supposition, it has been shown that increased expression of chaperones (Morrow et al., 2004; Yokoyama et al., 2002), proteases (Luce and Osiewacz, 2009), increased mitophagy (Rana et al., 2013), and reduced translation (Hansen et al., 2007) can extend lifespan.
Conclusions
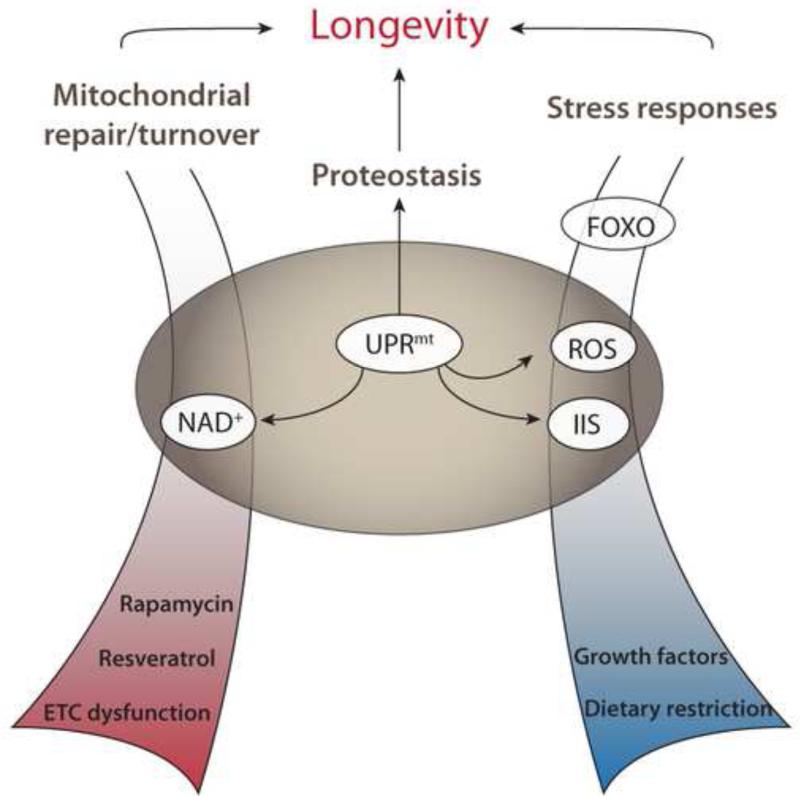
Within a short period, the UPRmt has assumed a conspicuous position in the study of longevity. Being involved in numerous biological pathways that can modulate lifespan (Figure 3), it holds the potential to provide a more unified conception of how longevity is achieved in concrete terms. While this rapid development is tremendously exciting, the pace has meant that some ambiguity has arisen regarding central elements of this response: Mitochondrial chaperone expression is currently the gold standard for activation of what we call the UPRmt, but in some cases aspects of the UPRmt phenotype occur without increased chaperone expression. Nor is UPRmt activation the only way to induce mitochondrial chaperones, or at least there is not a single, well-defined UPRmt responsible for all such induction. It will likely be beneficial to narrow down more specific definitions for the subjects of study, and to develop functional readouts that go beyond expression of single reporters. Thus, when describing the phenotypes of UPRmt activation it is important to keep in mind that these observations encompass both the imbalance that activates the UPRmt, the cellular and physiological consequences of this imbalance, and the protective response it provokes. Placing a particular phenotype in one of these categories can be difficult, as in the case of reduced ETC activity, but this will be critical in understanding the protective role of the UPRmt.
Figure 3. Integration of the UPRmt with processes influencing lifespan.
The UPRmt intersects with most processes influencing lifespan known to date, suggesting that interventions that preserve mitochondrial function or that trigger mitoprotective responses are promising candidates for therapies extending lifespan.
In a similar vein, not every activation of the UPRmt extends lifespan, nor does it seem that longevity always results from the same mechanism: the UPRmt now links the dysfunction of the ETC with the paradoxical extension of lifespan this can produce. But only some types of dysfunction extend lifespan, and only in some cases does this correlate with specific types of stress resistance (e.g. Copeland et al., 2009). Likewise, both the severity and duration of UPRmt activation determines whether lifespan is extended or shortened (Owusu-Ansah et al., 2013; Rea et al., 2007). Furthermore, induction of mitochondrial chaperones does not correlate directly with longevity, and in some cases the two physiological outcomes can be dissociated (Bennett et al., 2014). With this perspective, it is clear that it will be important not only to delineate different aspects of the UPRmt, but also to determine which of these represent the triggering cellular/organellar states, resulting adaptations and protective mechanisms with comprehensible effects.
Along these lines, the discrepant requirements and effects observed in different species brings up questions regarding the origin of lifespan extension by the UPRmt: does the requirement for activation during mitochondrial expansion in the worm imply lifespan extension by a single (chaperone mediated) mechanism, while another/multiple mechanism(s) are active in the fly? It is noteworthy that although the GCN-2 and HAF-1/ATFS-1 pathways have been described as complementary, disruption of either is sufficient to eliminate the lifespan extension conferred by UPRmt activation in worms (Baker et al., 2012; Houtkooper et al., 2013). The timing requirement could mean that the protective effect comes from a transient signal that alters the baseline function of cells rather than from the UPRmt itself. In line with this, constitutive activation of the HAF-1/ATFS-1 pathway by a nuclear-localizing ATFS-1 mutant did not improve mitochondrial stress resistance (Nargund et al., 2012), and atfs-1 gain-of-function mutant worms show reduced lifespan (Bennett et al., 2014; Rauthan et al., 2013). On the other hand, constitutive activation of the UPRER was recently reported to extend lifespan (Labunskyy et al., 2014).
Finally, it will be gratifying to discover which role the UPRmt plays in more complex organisms. The clause of developmental activation in the worm already provides one example of species-specific features, but most likely not the last. Further work in Drosophila and mice is likely to improve our understanding of how the UPRmt acts in and between different tissues, and of course demonstrating protective effects of the UPRmt in mammals will be an important step towards therapeutic applications.
Research Highlights.
Mitochondria possess a distinct response to the presence of unfolded proteins
This response is implicated in life-extending mitochondrial perturbations
The signaling pathways and consequences of this response are now being clarified
Its impact on mitochondrial metabolism could help explain its effect on longevity
Acknowledgements
Work in Dr. Jasper's lab is supported by the National Institute on Aging (RO1 AG028127), the National Institute on General Medical Sciences (RO1 GM100196), and the National Eye Institute (R01 EY018177). Dr. Borch Jensen is supported by the Alfred Benzon Foundation. The authors would like to thank Dr. Jason Karpac for helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One. 2007;2:e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Augustin A, Spene PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J. Cell Sci. 2003;116:1551–1562. doi: 10.1242/jcs.00341. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C, Brunyánszki A, Huber A, Szántó M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, et al. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 2011;13:450–460. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Langer C. a, Boteva R, Böttcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol. Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat. Commun. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, Adebanjo O. a, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvári M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H-C, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153:1448–1460. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139:1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr. Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu A-L, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain- mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- Epstein CB, Waddle J. a, Hale W, Davé V, Thornton J, Macatee TL, Garner HR, Butow R. a. Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussière F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Finkel T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LWS, Haigis MC. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res. Rev. 2009;8:173–188. doi: 10.1016/j.arr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell Y-WC, Sánchez-Blanco A, Silvia B. a, Helfand SL. Targeted expression of the human uncoupling protein 2 (hUCP2) to adult neurons extends life span in the fly. Cell Metab. 2005;1:145–152. doi: 10.1016/j.cmet.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Golozoubova V, Cannon B, Nedergaard J. UCP1 is essential for adaptive adrenergic nonshivering thermogenesis. Am. J. Physiol. Endocrinol. Metab. 2006;291:350–357. doi: 10.1152/ajpendo.00387.2005. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJY, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD+ Induces a Pseudohypoxic State Disrupting Nuclear-Mitochondrial Communication during Aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Calorie restriction and sirtuins revisited. Genes Dev. 2013;27:2072–2085. doi: 10.1101/gad.227439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee S-J, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Harding H, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J. Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert T. a, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Fiorese CJ, Lin Y-F. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol. 2013;23:311–318. doi: 10.1016/j.tcb.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit S, Zhang P, Hansen M, McCormick M, Lee S-J, Cary M, Kenyon C. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9730–9735. doi: 10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horibe T, Hoogenraad NJ. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS One. 2007;2:e835. doi: 10.1371/journal.pone.0000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Jasper H. Longevity focuses on NAD+. Nat. Chem. Biol. 2013;9:2–4. doi: 10.1038/nchembio.1369. [DOI] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- Karpac J, Jasper H. Insulin and JNK: optimizing metabolic homeostasis and lifespan. Trends Endocrinol. Metab. 2009;20:100–106. doi: 10.1016/j.tem.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobbe C, Von, Harrigan JA, May A, Opresko PL, Dawut L, Cheng W-H, Bohr VA. Central role for the Werner syndrome protein/poly (ADP-ribose) polymerase 1 complex in the poly (ADP-ribosyl) ation pathway after DNA damage. Mol. Cell. Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labunskyy VM, Gerashchenko MV, Delaney JR, Kaya A, Kennedy BK, Kaeberlein M, Gladyshev VN. Lifespan extension conferred by endoplasmic reticulum secretory pathway deficiency requires induction of the unfolded protein response. PLoS Genet. 2014;10:e1004019. doi: 10.1371/journal.pgen.1004019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow R. a. Mitochondrial retrograde signaling. Annu. Rev. Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Luce K, Osiewacz HD. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat. Cell Biol. 2009;11:852–858. doi: 10.1038/ncb1893. [DOI] [PubMed] [Google Scholar]
- Martin J, Mahlke K, Pfanners N. Role of an Energized Inner Membrane in Mitochondrial Protein Import. J. Biol. Chem. 1991;266:18051–18057. [PubMed] [Google Scholar]
- Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Høj PB, Hoogenraad NJ. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur. J. Biochem. 1996;240:98–103. doi: 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- Mehta R, Steinkraus K. a, Sutphin GL, Ramos FJ, Shamieh LS, Huh A, Davis C, Chandler-Brown D, Kaeberlein M. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 2009;324:1196–1198. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisterernst M, Stelzer G, Roeder RG. Poly(ADP-ribose) polymerase enhances activator-dependent transcription in vitro. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Inaguma Y, Kato K, Tanguay RM. The small heat shock protein Hsp22 of Drosophila melanogaster is a mitochondrial protein displaying oligomeric organization. J. Biol. Chem. 2000;275:31204–31210. doi: 10.1074/jbc.M002960200. [DOI] [PubMed] [Google Scholar]
- Morrow G, Samson M, Michaud S, Tanguay RM. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Cantó C, Mottis A, Jo Y-S, Viswanathan M, Schoonjans K, et al. The NAD+/sirtuin pathway controls longevity through the mitochondrial UPR and daf-16-dependent ROS defense. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiras ML, Müller M, Schächter F, Bürkle a. Increased poly(ADP-ribose) polymerase activity in lymphoblastoid cell lines from centenarians. J. Mol. Med. (Berl) 1998;76:346–354. doi: 10.1007/s001090050226. [DOI] [PubMed] [Google Scholar]
- Murakami T, Shimomura Y, Yoshimura A, Sokabe M, Fujitsuka N. Induction of nuclear respiratory factor-1 expression by an acute bout of exercise in rat muscle. Biochim. Biophys. Acta -Gen. Subj. 1998;1381:113–122. doi: 10.1016/s0304-4165(98)00018-x. [DOI] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E, Carruba MO. Nitric oxide and mitochondrial biogenesis. J. Cell Sci. 2006;119:2855–2862. doi: 10.1242/jcs.03062. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle Mitohormesis Promotes Longevity via Systemic Repression of Insulin Signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa L, Germain D. SirT3 regulates a novel arm of the mitochondrial unfolded protein response. Mol. Cell. Biol. 2013;34:699–710. doi: 10.1128/MCB.01337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta de Castro I, Costa AC, Lam D, Tufi R, Fedele V, Moisoi N, Dinsdale D, Deas E, Loh SHY, Martins LM. Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Cell Death Differ. 2012;19:1308–1316. doi: 10.1038/cdd.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbolt TK, Atanassova N, Genereux JC, Wiseman RL. Stress-Regulated Translational Attenuation Adapts Mitochondrial Protein Import through Tim17A Degradation. Cell Metab. 2013;18:908–919. doi: 10.1016/j.cmet.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath E, Berger E, Messlik A, Nunes T, Liu B, Kim SC, Hoogenraad N, Sans M, Sartor RB, Haller D. Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation. Gut. 2012;61:1269–1278. doi: 10.1136/gutjnl-2011-300767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauthan M, Ranji P, Aguilera Pradenas N, Pitot C, Pilon M. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5981–5986. doi: 10.1073/pnas.1218778110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Schmeisser S. Extending life span by increasing oxidative stress. Free Radic. Biol. Med. 2011;51:327–336. doi: 10.1016/j.freeradbiomed.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel ED, Liu S, Baumeister R, Schulze E. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS Genet. 2013;9:e1003346. doi: 10.1371/journal.pgen.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel ED, Baumeister R, Schulze E. Mitochondrial stress: Balancing friend and foe. Exp. Gerontol. 2014 doi: 10.1016/j.exger.2014.02.013. DOI: 10.1016/j.exger.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Schleit J, Johnson SC, Bennett CF, Simko M, Trongtham N, Castanza A, Hsieh EJ, Moller RM, Wasko BM, Delaney JR, et al. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell. 2013;12:1050–1061. doi: 10.1111/acel.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser K, Mansfeld J, Kuhlow D, Weimer S, Priebe S, Heiland I, Birringer M, Groth M, Segref A, Kanfi Y, et al. Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat. Chem. Biol. 2013;9:693–700. doi: 10.1038/nchembio.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- Schroeder E, Raimundo N, Shadel G. Epigenetic Silencing Mediates Mitochondria Stress-Induced Longevity. Cell Metab. 2013;17:954–964. doi: 10.1016/j.cmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena L. a, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon a, Yoshida H, Morimoto R, Kurnit DM, Mori K, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Steglich G, Neupert W, Langer T. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 1999;19:3435–3442. doi: 10.1128/mcb.19.5.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiburek L, Cesnekova J, Kostkova O, Fornuskova D, Vinsova K, Wenchich L, Houstek J, Zeman J. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol. Biol. Cell. 2012;23:1010–1023. doi: 10.1091/mbc.E11-08-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 2011;3:1–17. doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. XBP-1 Is a Cell-Nonautonomous Regulator of Stress Resistance and Longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tourmente S, Lecher P, Degroote F, Renaud M. Mitochondrial development during Drosophila oogenesis: distribution, density and in situ RNA hybridizations. Biol. Cell. 1990;68:119–127. doi: 10.1016/0248-4900(90)90296-f. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, Larsson N-G. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17993–17998. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Lemire BD. Mitochondrial genome content is regulated during nematode development. Biochem. Biophys. Res. Commun. 2002;291:8–16. doi: 10.1006/bbrc.2002.6394. [DOI] [PubMed] [Google Scholar]
- Walter L, Baruah A, Chang H-W, Pace HM, Lee SS. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS Biol. 2011;9:e1001084. doi: 10.1371/journal.pbio.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G, Terada K, Yano M, Sergeev I, Mori M. Oxidative Stress Inhibits the Mitochondrial Import of Preproteins and Leads to Their Degradation. Exp. Cell Res. 2001;263:107–117. doi: 10.1006/excr.2000.5096. [DOI] [PubMed] [Google Scholar]
- Yang W, Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell. 2010a;9:433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010b;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur J. a, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr V. a, Rosenzweig A, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Fukumoto K, Murakami T, Harada S, Hosono R, Wadhwa R, Mitsui Y, Ohkuma S. Extended longevity of Caenorhabditis elegans by knocking in extra copies of hsp70F, a homolog of mot-2 (mortalin)/mthsp70/Grp75. FEBS Lett. 2002;516:53–57. doi: 10.1016/s0014-5793(02)02470-5. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M. Impaired Insulin/IGF1 Signaling Extends Life Span by Promoting Mitochondrial L-Proline Catabolism to Induce a Transient ROS Signal. Cell Metab. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS One. 2009;4:e6348. doi: 10.1371/journal.pone.0006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]