Abstract
The assembly of nucleic acid nanostructures with controlled size and shape has large impact in the fields of nanotechnology, nanomedicine and synthetic biology. The directed arrangement of nanostructures at interfaces is important for many applications. In spite of this, the use of laterally mobile lipid bilayers to control RNA three-dimensional nanostructure formation on surfaces remains largely unexplored. Here, we direct the self-assembly of RNA building blocks into three-dimensional structures of RNA on fluid lipid bilayers composed of cationic 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) or mixtures of zwitterionic 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) and cationic sphingosine. We demonstrate the stepwise supramolecular assembly of discrete building blocks through specific and selective RNA-RNA interactions, based on results from quartz crystal microbalance with dissipation (QCM-D), ellipsometry, fluorescence recovery after photobleaching (FRAP) and total internal reflection fluorescence microscopy (TIRF) experiments. The assembly can be controlled to give a densely packed single layer of RNA polyhedrons at the fluid lipid bilayer surface. We show that assembly of the 3D structure can be modulated by sequence specific interactions, surface charge and changes in the salt composition and concentration. In addition, the tertiary structure of the RNA polyhedron can be controllably switched from an extended structure to one that is dense and compact. The versatile approach to building up three-dimensional structures of RNA does not require modification of the surface or the RNA molecules, and can be used as a bottom-up means of nanofabrication of functionalized bio-mimicking surfaces.
Introduction
Nucleic acid architectures can be used to construct self-assembling, programmable objects with well-defined properties for applications such as therapeutics, diagnostic tools, and biomimetic systems in biophysical and biochemical studies.1–6 In RNA and DNA self-assembly strategies, the specificity of the interactions between complementary bases enables directed, selective self-assembly of nanoscale objects.7–16 So-called RNA architectonics offers the possibility to design and assemble RNA into specific shapes, such as RNA polyhedrons,14,17 RNA fibers,18,19 squares and nanorings,20,21 as well as programmable arrays and nanogrids.9,21 Three-dimensional (3D) RNA nanostructures can also be used as scaffolding to direct high precision assembly of nano-sized materials, such as colloidal particles and membrane proteins, into objects with the desired spacing, shape and organisation.1,22,23
So far, most self-assembly strategies of RNA or DNA 3D nanostructures are performed in bulk solution after which the pre-assembled structures are deposited at a solid support when so desired.9,14,19,24 The potential of mediating the assembly of nucleic acid objects on a surface has recently been exploited by using chemically modified nucleic acids that are bound or grafted to a solid surface or a bilayer.25–34 This opens up new possibilities to use biomolecules to construct functional nanostructured surfaces with applications in, for example, bio analytical and diagnostic devices. However, previous studies mainly involve hybridization of short modified DNA or RNA oligomers, e.g. by coupling a DNA strand to a biotin-modified bilayer using streptavidin as a molecular link. One drawback of these methods is the need for chemical modification of the RNA or/DNA molecules.
In the present study, we propose a bottom-up approach to guide 3D RNA self-assembly at a model lipid membrane without chemical modification of the RNA building blocks. By exploiting the attractive electrostatic interactions between RNA polyanions and cationic bilayers, we first direct the adsorption of RNA nanostructures on a lipid bilayer scaffold before promoting further RNA self-assembly through selective RNA-RNA interactions (Figure 1). Taking advantage of an experimental flow-system set-up, RNA self-assembly can be controlled in-situ by sequential changes of the solution composition. More specifically, we use RNA polyhedrons that we previously built by stepwise assembly in solution from eight tRNA building blocks (Figures 1 and S1).14 First, two different sets of four tRNA units assemble into two distinct square-shaped RNA nanostructures, named tectosquares TS3 and TS4 (Table S1). AFM experiments confirm the formation of RNA tectosquares (Figure S2). These tectosquares can then assemble further into RNA polyhedrons (TO3-4) by means of complementary base pairings formed between single stranded tails appended to the corners of each square (Figure 1a and S1).14 The non-uniform square antiprism shape adopted by these polyhedrons was characterized by cryo-EM imaging with single particle reconstruction (Figure S1).14 To provide fundamental insight on the balance of forces that control the process of adsorption and self-assembly of these RNA nanostructures at deposited lipid bilayers, we have investigated their stepwise assembly directly on the bilayers (Figure 1b) and explored the effects of various solution conditions (e.g. counterion concentration) and surface properties (e.g. changing the bilayer composition) on this process.
Figure 1.
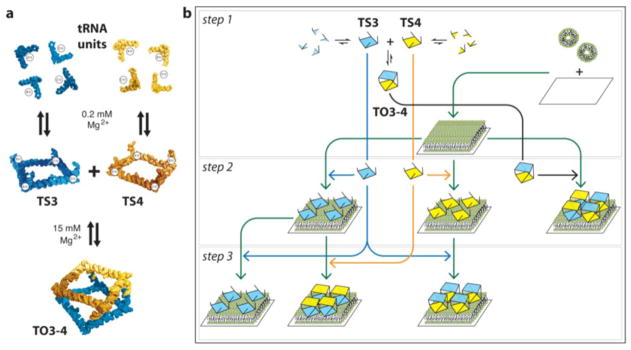
RNA self-assembly though loop-loop interactions at 0.2 mM Mg2+. The TO3-4 polyhedron is formed by assembly of TS3 with TS4 through selective tail-tail interactions: this is favoured at higher magnesium concentrations. The shape of the polyhedron is the one of an RNA square antiprism according to Severcan et al.14 (b) Multistep assembly processes studied in this paper.
Results and Discussion
The main goal of this work is to demonstrate that fluid supported bilayers can be used as a scaffold for the sequential building of complex 3D RNA polyhedron nanostructures, and to explore how RNA self-assembly can be controlled by regulating the properties of the aqueous solution and deposited bilayers. We monitored the assembly of 3D RNA polyhedrons on lipid bilayers using four main complementary surface techniques: quartz crystal microbalance with dissipation (QCM-D), null ellipsometry, fluorescence recovery after photobleaching (FRAP), and total internal reflection fluorescence microscopy (TIRF). The reason for choosing QCM-D and ellipsometry as the main techniques was that they do not rely on labelling for quantifying the interaction and they also complement each other; the QCM-D method is very sensitive to the extent by which assemblies protrude from the surface into the bulk solution, while ellipsometry allows the direct quantification of the mass adsorbed without the coupled water.
RNA 3D assemblies readily adsorb to bilayers
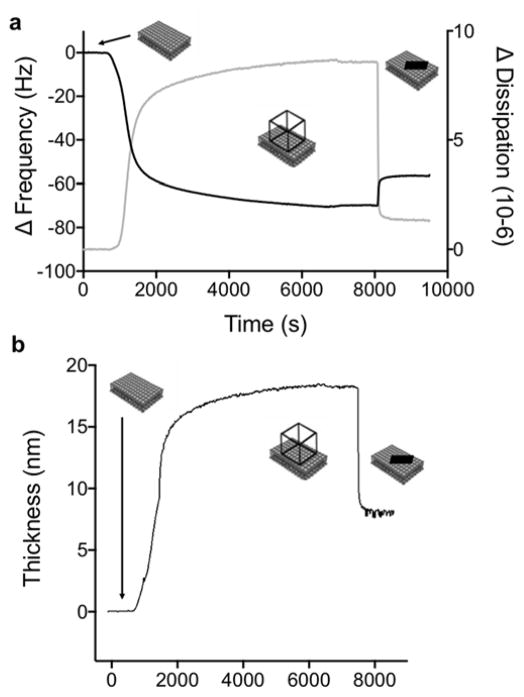
We first investigated the adsorption of fully assembled RNA TO3-4 polyhedrons, pre-formed in solution in the presence of magnesium as previously described14 (Figure S1,S2), onto bilayer surfaces. Under the same solution conditions as those in this study, TO3-4 assemblies were previously demonstrated to form 3D antiprisms by cryo-EM (Figures 1 and S1).14 A bilayer comprised of the cationic lipid, 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), was formed on a silica surface via the vesicle fusion technique, and monitored by simultaneous QCM-D and ellipsometry measurements (Figure 2). Ellipsometry data showed an increase in the amount of lipid on the surface after fusion of vesicles. To probe the structure of the bilayer at the surface, we simultaneously measured the adsorption with QCM-D, which provides information on the total amount of material (including the coupled solvent) attached to the surface from the change in frequency (ΔF) as well as the viscoelastic properties of the attached layer from the dissipation (ΔD). The temporal changes in ΔF and ΔD observed upon vesicle fusion are consistent with a DOTAP bilayer of high coverage, as described in detail in the Methods section.
Figure 2.
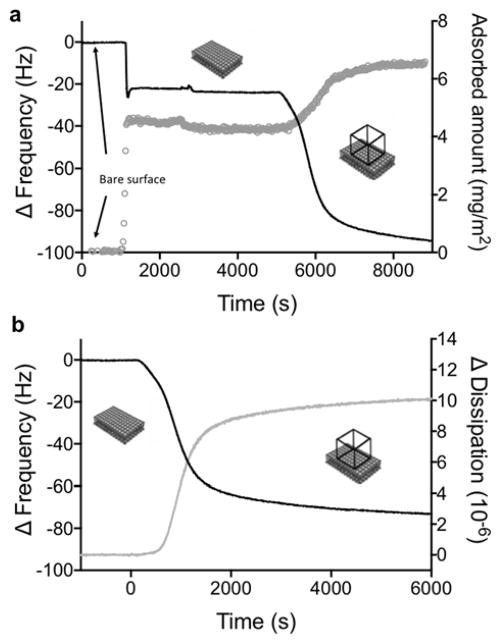
Adsorption of TO3-4 polyhedrons to a deposited bilayer, as simultaneously recorded by means of QCM-D (overtone 7) and ellipsometry. (a) A large decrease in frequency, as measured by QCM-D (black curve, left axis) is observed at the same time as the adsorbed amount (grey curve, right axis) increases. The adsorbed amount is calculated from the ellipsometry measurements. (b) The change in frequency (black curve, left axis) coincides with a large increase in dissipation (grey curve, right axis) as measured by QCM-D.
After ensuring the quality of the bilayer, a dispersion of negatively charged TO3-4 polyhedrons, quality-checked by native PAGE (Figure S2) was flowed over the DOTAP bilayer and shown to readily adsorb to the cationic bilayer surface. This is demonstrated by the large increase in adsorbed mass observed with ellipsometry, which coincided with a decrease in frequency in the QCM-D measurements (Figure 2a). Measured ΔD for adsorbed TO3-4 polyhedrons compared to those from tectosquare TS3 and RNA9-10 (Figures 2b, S3 and S4), indicates that the adsorbed 3D TO3-4 protrudes further into solution than the flat and less structured RNA9-10 and the smaller TS3 tectosquare, and therefore contains a substantial mass of coupled solvent (see below).
RNA adsorption on DOTAP bilayers was further confirmed by TIRF after addition of the fluorescent nucleic acid stain, GelStar™ to the RNA solution (Figure S5a). Pre-formed RNA TO3-4 polyhedrons adsorb to the bilayer as indicated by an increase of fluorescence at the bilayer over the time course of the experiment. By contrast, no fluorescence signal is observed at the bilayer when GelStar™ is added alone. Taken together, the QCM-D, ellipsometry and TIRF results indicate that pre-formed RNA TO3-4 polyhedrons are readily adsorbed at the cationic bilayer surface to form a single layer of RNA polyhedrons that protrudes as a 3D structure from the surface.
QCM-D probes the protrusion of RNA nanostructures from the bilayer surface
We first compared the adsorption of the TO3-4 polyhedrons to the adsorption of TS3 and TS4 (Figure 1): TS3 and TS4 are designed to assemble into the TO3-4 polyhedron when mixed together in association buffer (Figures 1 and S1).17 Figure 3 and S4 show the adsorption of the TS3 and TS4 (Figure 1b, step 2)14 compared to the adsorption of TO3-4. Adsorption of a very different RNA complex, RNA9-10 formed of RNA9 and RNA10, and known to assemble into flat networks on lipid layers (Figure S3),35 was also investigated (Figures 3 and S4).
Figure 3.
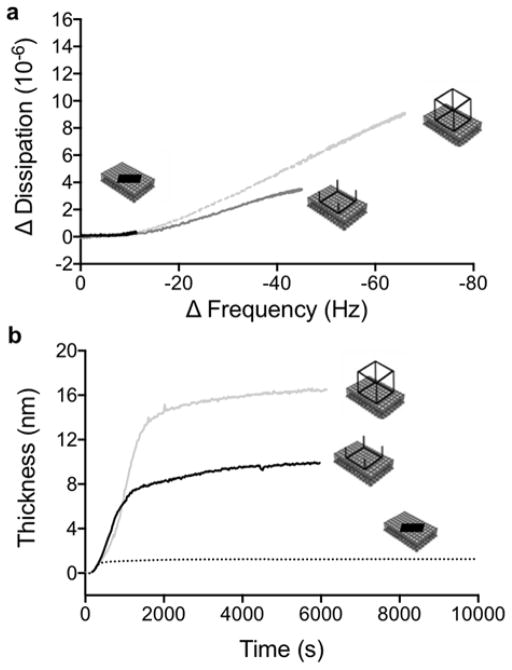
(a) A correlation plot of ΔD as a function of ΔF for RNA9-10 (black curve), RNA TS3 square (dark grey curve) and RNA polyhedron (light grey curve). (b) The change in thickness of the adsorbed layer after adsorption of RNA polyhedrons (light grey curve), tectosquares (dark grey curve) and RNA9-10 (dotted black curve) to a deposited DOTAP bilayer, as calculated from the fitted QCM data. (Overtone 7)
The correlation between the changes in the dissipation ΔD, and the frequency ΔF (Figure 3a), reveals valuable information about structural changes at the lipid bilayer during the adsorption process of these RNA structures. The small shift in both ΔF and ΔD observed for the RNA9-10, indicates that the oligomer complexes adsorb to form a flat and thin layer at the surface, consistent with previous observations of adsorbed layers of other linear RNA and DNA.27,36 In contrast, the large changes in both ΔF and ΔD observed during adsorption of the TO3-4 polyhedrons and the TS3 and TS4 RNA tectosquares onto the bilayer, imply that there is a significant amount of coupled water associated with the RNA structures, and that the adsorbed structures retain their 3D shapes. Thus, the RNA assemblies are not an integral part of the bilayer but rather form a protruding layer. The overall change in ΔF and ΔD was significantly larger for TO3-4 compared to TS3 (Figure 3a) indicating that the tectosquares form a layer that has lower mass and is less protruding into the bulk solution compared to the TO3-4 polyhedron.
Interestingly, the ΔD versus ΔF data show that the initial adsorption regime (the low frequency region) was similar for all three RNA systems investigated (Figures 3a and S4). At low coverage of RNA, the decrease in ΔF led to a minor change in ΔD, which implies formation of a compact RNA film with little coupled solvent (ΔF <10Hz) on the surface. This regime corresponds to the overall response observed for the RNA9-10 oligomers, and the adsorbed amount RNA9-10 was estimated to be 0.2 mg/m2 using Eq 2. For the RNA polyhedrons and tectosquares, a second adsorption regime can be distinguished (with ΔF> 10 Hz), where the continued decrease in ΔF coincides with a large increase in ΔD. For TO3-4 and TS3, these QCM-D data directly demonstrate the formation of a protruding non-rigid film with a substantial amount of coupled solvent. The two-regime behaviour observed for the adsorption of TO3-4 and TS3 indicates that, at low surface coverage, these RNA structures adsorb as a thin layer comparable to those observed for linear RNA molecules (e.g. RNA9-10 and previous studies36). When the number of RNA structures at the surface increases, the surface layer starts to protrude into the bulk and there is room for more RNA assemblies at the surface. This interpretation is further supported by the dramatic reduction in shear viscosity versus time obtained from the viscoelastic modelling of the QCM-D data obtained for both TS3 and the TO3-4 polyhedrons (Figure S6).
Next, the adsorption was quantified, in terms of amount and the thickness of the layer. As discussed above, the data implies that the viscoelastic properties of the layer depend on the species that is adsorbing. The modelling of the QCM data is therefore challenging, as different models have to be applied for different systems. The Sauerbrey equation (explained in the Methods section) can be used to calculate the thickness of the deposited lipid bilayer and the adsorbed layer of RNA9-10. However, this equation is not a valid model for the extended layers of adsorbed RNA tectosquares and polyhedrons. For these systems, we used an extended Voigt-based viscoelastic representation of the film to quantify the coupled mass, the thickness of adsorbed layer and its viscoelastic properties.37 In this modelling, the frequency dependence of the effective viscosity and shear modulus was included.38,39
As calculated from the modelling of QCM data using a Voigt-based viscoelastic representation of the film, the adsorbed layer thickness for TO3-4 polyhedrons and TS3 tectosquares was estimated to be 16.5 nm and 8 nm, respectively (Figure 3b). In contrast, the adsorbed layer of the RNA oligomer complex, RNA9-10, was much thinner and was determined to be 1 nm using the Sauerbrey expression with a layer density of 1.6 kg/m3. The thickness of the adsorbed layer of TO3-4 polyhedrons is in the size range, although a bit larger compared to the expected size for these nanostructures, which have been characterized as non-uniform square antiprisms with dimensions 14×14×8 nm.14 The differences in the measured thickness obtained from the QCM measurements and the dimensions obtained by cryo-EM characterization (Figure S114) can be explained by that the QCM measurements sense a layer that also includes water associated with the RNA polyhedron layer, which is coupled to the movement of the RNA polyhedrons (i.e. the coupled solvent). The modelled thickness of the adsorbed layer may thus be better described as a hydrodynamic thickness of the adsorbed layer rather than the atomic dimension of the individual polyhedron, and these values can therefore not be directly compared. We also conclude that the RNA polyhedrons are tightly packed at the bilayer surface. The adsorbed amount was determined by ellipsometry to be 2.2±0.1 mg/m2 (Figure 2), which implies that the effective area per RNA polyhedron is 240 nm2 (Mw(TO3-4 polyhedron)=317 520 g/mol). This value should be compared to the estimated area of 110–200 nm2 for each polyhedron (depending on which side of the polyhedron faces the bilayer). Such high packing at the surface is only possible on a fluid substrate like the DOTAP bilayer. In this system, the bilayer fluidity together with the reduction of the charge repulsion between poly-anionic polyhedrons as a consequence of the interaction with the oppositely charged lipid bilayer and the relatively high ionic strength together allows efficient packing of the polyhedrons. This is analogous to previous observations that streptavidin coupled to biotinylated lipids in a fluid supported bilayer can form crystalline layers due to the fluidity of the bilayer.40–42
Building RNA polyhedrons on a bilayer scaffold
We next investigated whether it is possible to assemble the TO3-4 polyhedron on the surface of the lipid bilayer in a stepwise manner from its constituent tectosquares, as shown in Figure 1. Tectosquares can readily adsorb on the cationic bilayer. Because of the 3D shape of these nanostructures (Figures 1 and S1), they are most likely adsorbing on their flatter side, with their single strand tail regions extending out into the bulk solution as suggested by QCM-D data. The tails of one adsorbed tectosquare are thus accessible to base pairings with the complementary single strands from another tectosquare present in the bulk solution. As shown Figures 1b, it is therefore possible to envision the stepwise assembly of TO3-4 polyhedrons from TS3 and TS4 tectosquares on a deposited DOTAP bilayer.
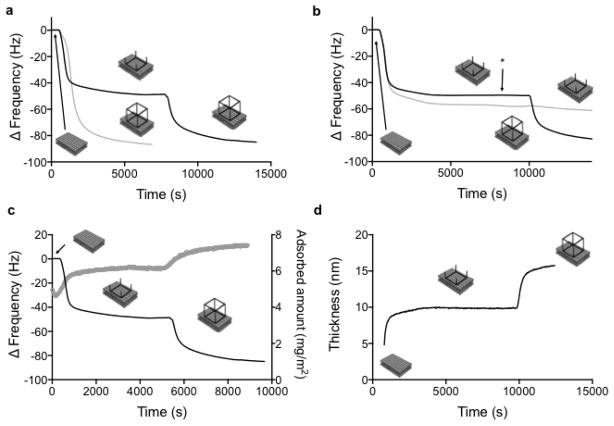
The two-step assembly of the RNA polyhedron was performed by the sequential addition of two complementary RNA squares (Figure 4a). It should be noted that the experiments were performed in the same medium as the previous step-wise assembly by Severcan et al.14, that is in a dilute (<100 nM) aqueous solution. TS3 was first adsorbed to the bilayer resulting in a smaller ΔF (Figure 4a, black line) than the one observed for adsorption of the pre-assembled TO3-4 (Figure 4a, grey line). Next, the bulk solution was replaced with association buffer containing 15 mM Mg2+ and the complementary TS4 was added. Figure 4a shows that the final change in ΔF (Figure 4a, black line) is very similar to the one measured for adsorption of the pre-assembled TO3-4 (Figure 4a, grey line). The RNA layer formed on the bilayer surface by the two-step assembly has the same thickness (Figure 4d) as the adsorbed layer of pre-assembled TO3-4 (Figure 2), as obtained by fitting the Voigt model to the QCM-D data. Therefore, these results strongly support the interpretation that the layer of RNA polyhedrons that are assembled at the bilayer is very similar to the layer of adsorbed pre-assembled RNA polyhedrons. The same results were obtained when the order of addition of the complementary squares was reversed, i.e. when TS3 was added to the pre-adsorbed TS4. By contrast, no further RNA assembly was detected in the negative control experiments, when the adsorption of the TS3 was followed by addition of a solution with the same non-complementary TS3 tectosquare (Figure 4b, grey line). This demonstrates that the increase in thickness of the RNA layer upon addition of TS4 to adsorbed TS3 is mediated by specific complementary tail-tail interactions.
Figure 4.
(a) QCM-D frequency data (overtone 7) for the stepwise assembly of the TO3-4 RNA polyhedron to a DOTAP bilayer. TS3 tectosquare is adsorbed first. TS4 tectosquare is added second (black curve). The assembly at the surface is compared with adsorption of the pre-assembled RNA polyhedron (grey curve). (b) QCM-D frequency data for polyhedron assembly (TS3 + TS4, black curve) together with one negative control (TS3 + TS3, grey curve). The rinsing step is denoted with an arrow. (c) Combined QCM-D and ellipsometry measurements for the stepwise polyhedron assembly (TS3 + TS4), showing that the frequency shift measured with QCM (black curve, left axis) coincides with the increase in adsorbed amount (grey curve, right axis) as calculated from the ellipsometry data. (d) Modelled QCM-D data for the thickness of the adsorbed layer during the building of the RNA polyhedron at a DOTAP bilayer.
Quantitative measurements of the amount of RNA adsorbed onto the bilayer during stepwise assembly were obtained by ellipsometry, simultaneously performed with QCM-D (Figure 4c). The amount of pre-adsorbed TS3 (1.2 mg/m2) was nearly equal to that of the complementary TS4 adsorbed in the second step of the assembly (1.1 mg/m2) (Figure 4c). This equality is consistent with the expected 1:1 stoichiometry for the assembly of a polyhedron from TS3 and TS4 (Figure S7). It is also noteworthy that the amount adsorbed for the two-step assembly of the polyhedron on the bilayer surface (2.3±0.1 mg/m2) as determined by ellipsometry was very close to the corresponding value after adsorption of the pre-assembled polyhedron (2.2±0.1 mg/m2). This indicates that the pre-programmed RNA polyhedron is correctly assembled via the two-step assembly, and that the bilayer can be used as a scaffold for assembly. Previous self-assembly experiments with tectosquares in solution have clearly established that the correct assembly into polyhedrons is highly constrained by the geometry and orientation of the tail-tail interactions.14 As such, tetrameric TS3 and TS4 are pre-programmed to assemble into closed octameric polyhedrons with high yields.14 Considering the high density of the adsorbed layer and the close to 1:1 stoichiometry in the assembly reaction (Figure 4c), it is therefore highly unlikely that the adsorbed layer consists of a random network of coupled TS3 and TS4 units rather than discrete RNA polyhedrons (Figure S7).
Reversibility of RNA nanostructure self-assembly on the bilayer scaffold
We next explored different solution conditions to switch on and off the assembly of the RNA polyhedron on the bilayer scaffold. The assembly of the tectosquares into 3D nanostructures can be controlled by the concentration of divalent cations such as magnesium, which are needed for promoting the formation of the sequence specific tail-tail interactions between complementary tectosquares.14 Removal of the divalent cations may thus lead to desorption or dissociation of the adsorbed RNA assemblies.
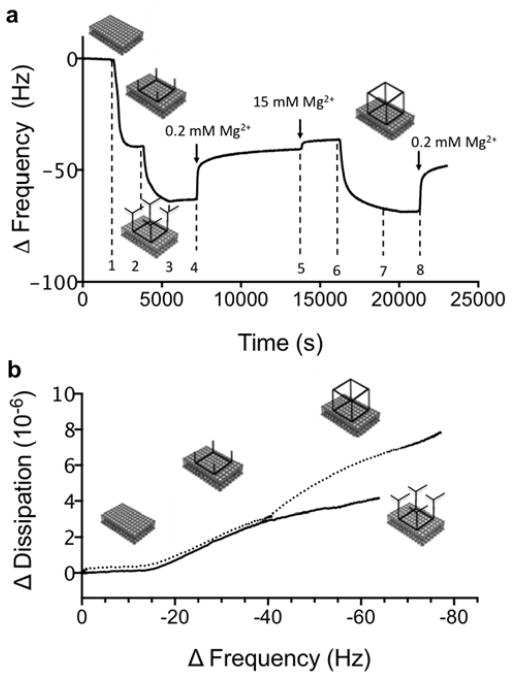
Figures 5 and S8 show the QCM-D data from experiments with sequential changes in solution conditions that can be used to control the assembly of RNA structures of different sizes at the bilayer surfaces. After adsorption of TS3 tectosquares to the DOTAP bilayer, the solution was first replaced with association buffer containing 15mM Mg2+, and then followed by the addition of one complementary tRNA unit (At1′) (Figure 1a). The At1′ tRNA is one of the building blocks comprising the TS4 tectosquare (Figure S1c), and it contains one single stranded tail that is complementary to all four tails of the TS3 tectosquare. It is therefore possible for four At1′ tRNA to bind to each TS3 tectosquare when At1′ is present in excess in buffer with 15mM Mg2+ (saturation binding is reached, see Figure S9). The QCM-D data show that the addition of At1′ tRNA leads to a decrease in ΔF that was smaller than the corresponding decrease in ΔF observed when the full TS4 tectosquare was added. The decrease in ΔF was accompanied by a small increase in ΔD, which was much smaller than the corresponding increase upon addition of TS4 (Figure 5b). This implies a structure that is less protruding out into the solution. In the quantitative comparison of these data, it is important to point out that for soft layers that extend into the solution, the measured ΔF is not proportional to the change in mass, and it has to be analysed in relation to the change in dissipation, ΔD.37 Based on these experiments we can confirm that the RNA assembly at the bilayer scaffold is specific and directed through tail-tail interactions to form the pre-programmed nanostructure. It is also shown that binding of At1′ to the pre-adsorbed TS3 RNA square leads to an assembled structure that is less protruding than the RNA polyhedron.
Figure 5.
a) QCM frequency data (overtone 7) for a sequential experiment. (step 1) TS3 is first adsorbed on a DOTAP bilayer. After rinsing in buffer (15 mM Mg2+); (step 2) the At1′ tRNA unit is added, leading to a decrease in ΔF, i.e. increase in mass; (step 3) Rinsing in buffer with 15 mM Mg2+ does not affect the adsorption, while (step 4) rinsing in buffer with 0.2 mM Mg2+ lead to an increase in ΔF, indicating desorption. Quantitative comparisons can only be done between situations where the buffer is the same, i.e., after the (step 5) subsequent change to the buffer with 15 mM Mg2+. The magnitude of the overall increase in ΔF after the rinsing steps (steps 3–5) is almost equal to the increase in ΔF upon At1′ adsorption (step 2); (step 6) The full TS4 RNA square is then added. This leads to significant decrease in ΔF, which implies that the tail regions of the pre-adsorbed TS3 tectosquare are again available for base pairings with those from TS4. Finally, (step 7) rinsing in buffer with 15 mM Mg2+ does not lead to desorption, while (step 8) rinsing in buffer with 0.2 mM Mg2+ leads partial desorption of TS3. (b) A correlation plot of ΔD as a function of ΔF (on=7) for the surface assembly of At1′ tRNA units to the pre-adsorbed TS3 (solid line) and for the addition of TS4 to pre-adsorbed TS3 (dotted line).
In the next step of the experiment in Figure 5a, the Mg2+ concentration was reduced from 15 mM to 0.2 mM in order to weaken the strength of the complementary tail-tail interactions. This led to an increase in ΔF, which is an indication of dissociation of the assembly. A minor complication in the interpretation of these QCM-D data results from the fact that both ΔF and ΔD are slightly affected by changing the buffer, and quantitative comparisons can therefore only be done between situations where the buffer is the same. In the present experiment, comparison is possible after changing back to the buffer with 15 mM Mg2+. It is here noteworthy that the change in ΔF after the addition of At1′ almost equals the opposite change in ΔF when replacing the RNA solution for neat buffer in 2 steps. This suggests removal of the At1′ during rinsing, and that in this case tail-tail interactions are reversible and controlled by the presence of Mg2+. From the results for the RNA 3D assembly in Figure 4, together with the control experiments with the smaller tRNA building blocks in Figure 5a, we conclude that the RNA assembly occurs only based on complementary RNA tail-tail interactions. Moreover, our data imply that tectosquares are adsorbed at the surface through their flatter side with their single strand tail regions protruding out into the bulk solution so that their RNA tails are accessible for interactions with complementary strands. This further supports our hypothesis that the RNA 3D assemblies are formed at the bilayer surface, which acts as a scaffold. Note that removal of pre-adsorbed tectosquares from the bilayer has not been detected for our experimental conditions and DOTAP bilayers.
In order to check whether the pre-adsorbed TS3 is still accessible for base pairing after the removal of the At1′ tRNA units, TS4 tectosquare was then added. The measured ΔF and ΔD were almost identical to the corresponding changes measured when TS4 RNA square was added directly to the pre-adsorbed TS3 RNA squares without intermediate steps (compare Figure 5a to Figure 4a). This confirms that TO3-4 polyhedrons can be generated after dissociation of At1′ tRNA units from the pre-adsorbed tectosquare at low concentrations of divalent cations. Finally, the layer composed of surface-assembled RNA polyhedrons was rinsed in buffer solution with 0.2 mM Mg2+, which led to partial release of TS4. However, the ΔD was still high, which is consistent with protruding 3D structures at the bilayer surface. From these experiments we conclude that the assembly between the TS3 RNA square and four At1′ tRNA units is significantly weaker than that of the assembly of two complementary squares. In conclusion, these experiments (Figures 4 and 5) show that full reversibility of the base pairing at the bilayer scaffold depends on the number of interactions and the size of the RNA nanostructures. Finally, when pre-assembled RNA polyhedrons are adsorbed to a bilayer and then rinsed in buffer with low Mg2+ concentration, there is no observed dissociation (not shown). This suggests that the RNA polyhedron assembled in bulk solution is slightly more stable than the one assembled at the bilayer scaffold, which might be due to remaining divalent ions strongly associated with the pre-assembled polyhedron, making it more stable against lowering the ionic strength.
FRAP study of lipids and RNA polyhedrons
The mobility within the lipid bilayer is important for the buildup of the RNA layer. Therefore we investigated the lateral diffusion in the adsorbed layer by means of fluorescence recovery after photobleaching (FRAP). FRAP experiments on RNA polyhedrons labelled with Gelstar™ at the DOTAP bilayer surface showed no recovery of fluorescence signal from Gelstar™ during the course of the experiment (60 s) (Figure S5b), indicating that the adsorbed RNA polyhedrons are immobile on this time scale. The slow diffusion is consistent with the presence of tightly packed, large RNA polyhedron assemblies anchored to a bilayer by strong attractive electrostatic interactions with several lipid molecules. The number of DOTAP molecules that can associate with each polyhedron depends on the size of the polyhedron plane that faces the bilayer (either 14×8 nm or 14×14 nm), and is estimated to be in the order of 150 or 270 cationic DOTAP molecules, assuming that the area per DOTAP headgroup does not change upon association. In the high ionic strength buffer used here, long-range electrostatic attractions are screened and the positive charges from DOTAP are balanced with those negative charges from the RNA polyhedron that are sufficiently close to the bilayer. This mainly includes the parallel strands, ca. 110 or 200 negative charges, depending on the orientation of the polyhedron.
Influence of RNA polyhedrons on the mobility in the fluid bilayer
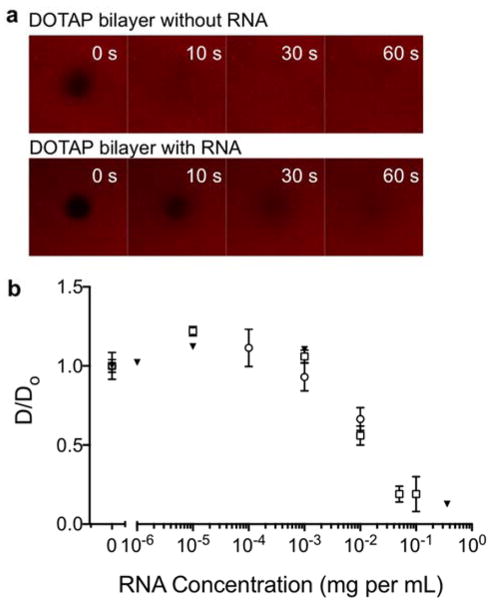
The influence of the adsorbed RNA polyhedron on the mobility of lipids and RNA in the plane of the fluid bilayer was investigated by means of FRAP using two different fluorescent lipid analogues, Rh-PE or NBD-DOTAP, as tracers in the DOTAP bilayer. The measurements were performed before and after incubation with RNA polyhedrons at different concentrations (Figure 6). Separate QCM-D measurements performed in similar conditions further confirm that there is no detectable desorption from the surface upon rinsing, implying very strong RNA association (Figure 4b). There was no detectable change in the lateral diffusion coefficient for any of the lipid analogues at the lower RNA concentration (Figure 6d). However, for RNA polyhedron bulk concentrations above ca. 0.01 mg mL−1, a dramatic decrease in the diffusion coefficient was measured. When RNA polyhedron bulk concentrations were further increased, the diffusion coefficients became close to zero.
Figure 6.
(a) Representative FRAP data for bilayers of DOTAP labeled with Rhodamine-PE (0.5 mol%) performed before and after exposure to TO3-4 RNA polyhedron (0.1 mg mL−1). The 0 s images show each bilayer immediately after bleaching the spot. Each image shown is 40 × 40 μm. (b) The normalized diffusion coefficients for fluorescent probes in DOTAP bilayers after exposure to increasing RNA polyhedron concentrations. The RNA concentration refers to the bulk polyhedron concentration used for incubation. FRAP measurements were performed after rinsing with buffer. The diffusion coefficient was obtained from FRAP experiments performed using a scanning confocal microscope on bilayers of DOTAP labeled with NBD-DOTAP (open circles) and Rhodamine-PE (open squares), or using a TIRF microscope on bilayers of DOTAP labeled with Rhodamine-PE (solid triangles). The diffusion coefficient was normalized for each series by the diffusion coefficient determined for the bilayer without RNA.
Our interpretation of the combined FRAP, QCM and ellipsometry data is that the bilayer scaffold is saturated with RNA polyhedrons that form a single and dense layer at bulk concentrations above 0.01 mg mL−1. At lower bulk RNA concentrations, the saturation adsorption is not reached, as also confirmed by QCM (Figure S9). Before discussing the mobility data in greater detail it is important to consider the lateral distribution of the two different fluorescent lipid analogues. First we note that there is a strong electrostatic attraction between the adsorbed RNA polyhedrons and the oppositely charged DOTAP bilayer. The Rh-PE is effectively negatively charged, and the NBD-DOTAP carries the bulky fluorescent probe in the acyl-chain, making them less likely to partition into the RNA-DOTAP complexes. One therefore expects that the fluorescent lipid analogues are present in the surrounding fluid bilayer and not in the more condensed RNA polyhedron-DOTAP complexes. Similar specific association with certain lipid/surfactant species have previously also been shown in complexes containing DNA.43 The FRAP measurements in Figure 6 and Figure S5 provide a measure of the 2D diffusion of the fluorescent lipid analogues in a bilayer containing small and virtually immobile obstacles, that are the RNA polyhedrons and the associated DOTAP molecules. When the surface coverage of RNA polyhedrons is low, the presence of few obstacles does not significantly influence the diffusion of the excluded dye molecules in the plane. However, at higher coverage, the obstacle density is so high that the surrounding fluid bilayer regions are disconnected or the diffusion route is highly tortuous. The sharp decrease in diffusion coefficient therefore indicates the percolation threshold. The lateral diffusion can then not occur over large distances, and this is manifested as strongly reduced fluorescence recovery. The reduction in mobility can be quantified based on a first order approximation44 as
| (1) |
where p is the fraction of the bilayer surface in which the fluorescent lipid analogues cannot move, Dobs is the diffusivity observed after RNA addition and Dintial is observed diffusivity in the bilayer prior to RNA polyhedron binding. The present data show that p is close to zero at low RNA polyhedron coverage, and that p increases to close to unity at the percolation threshold (Figure S5d). Taken together, the diffusion behaviour, as determined by FRAP, and the adsorption data from the QCM-D and ellipsometry, imply a single and densely packed layer of RNA polyhedrons at the bilayer scaffold surface. Furthermore, the reduction in diffusivity close to zero above the percolation threshold implies coupling between the two monolayers in the deposited bilayer scaffold. This is also consistent with previous diffraction studies on deposited 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) lipid bilayers in the gel state, where it was found that the lipids in the two leaflet always become coupled even if they are deposited on the supporting surface independently.45
Collapse of RNA polyhedrons with dendrimers as compacting agents
An efficient way to control nucleic acid conformation is to add compacting agents, like cationic surfactant or dendrimers.46,47,48 The ability to collapse the RNA nanostructure layer on the lipid surface can be seen as additional indirect evidence for the extended, “wire-framed” nano-architecture of the RNA polyhedrons layer. The collapse of the adsorbed layer of RNA polyhedrons on lipid bilayers was triggered by highly charged cationic dendrimers (PAMAM, generation 4). QCM-D data show that ΔD decreased to values close to zero upon the addition of the dendrimers, accompanied by an increase in ΔF (Figure 7a). These changes in ΔD and ΔF allow us to readily distinguish between a 3D structure and compact layer. The modelling of the QCM data showed that thickness of the adsorbed layer decreased from 16.5 nm to 7 nm upon addition of dendrimers (Figure 7b). Beyond showing that RNA can be reduced in size on the bilayer surface, these data show that the RNA polyhedron can be switched from an extended 3D structure to a more compact and dense layer at the surface. This in turn can potentially be used to trigger release of active components enclosed in the polyhedron. Here we note that we recently showed that a highly charged polyelectrolyte, heparin, can be used to release nucleic acids from complexes with dendrimers.49
Figure 7.
(a) QCM-D frequency (black, left axis) and dissipation data (grey, right axis) (overtone 7) for the adsorption of TO3-4 polyhedron to a DOTAP bilayer, followed by the collapse of the polyhedron after the addition of dendrimers (b) Modeled QCM data for the thickness of the adsorbed RNA-dendrimer layer.
Adsorption of 3D RNA assemblies to bilayers with different lipid composition
In the previous sections we have described the adsorption and assembly of RNA polyhedrons on supported bilayers composed of a cationic lipid analogue, DOTAP. We also explored the corresponding processes utilizing more biologically relevant lipid systems that are composed of two lipid species, a natural cationic lipid, sphingosine (Sph), and a zwitterionic phospholipid, DOPC. By varying the composition of these two lipids from 10 mole % Sph to 50 mole % Sph, we can reveal the effect of the bilayer charge density on RNA adsorption and directed self-assembly. Table 1 summarizes measured values of ΔD and ΔF together with the calculated thickness of the adsorbed layer of pre-assembled RNA polyhedrons as well as TS3 and TS4 RNA squares at deposited bilayers with different compositions. It is clear that the charged RNA polyhedrons and RNA squares not only adsorb to the deposited bilayer composed of 100% cationic surfactant, but to all positively charged lipid and surfactant bilayers investigated. The observed changes were similar for all (completely or partly) cationic bilayer systems studied, and even at the lowest concentration of charged lipid (10 mol% charged lipids), we observed significant adsorption. From these experiments we conclude that a relatively low bilayer charge density is sufficient in order for the RNA polyhedrons and RNA squares to adsorb, and that the adsorbed RNA polyhedra retain similar 3D shape after adsorption onto bilayers with either high or low charge density. Furthermore, the adsorption did not differ much between the DOTAP and the DOPC/sphingosine bilayer systems. However, the bilayer charge density made a difference in how tightly the adsorbed RNA layer is associated with the bilayer, as implied from the extent of desorption when the bulk RNA solution was exchanged with neat buffer. In fact, desorption of the RNA nanostructures was observed for all mixed DOPC/sphingosine bilayers, and the only bilayer system where desorption was insignificant was for the completely cationic DOTAP bilayers. Adsorption of RNA polyhedrons and TS3 RNA squares was also detected to bilayers composed of only zwitterionic DOPC bilayers. However, the adsorbed amount was very small and no further experiments using pure DOPC bilayers were performed.
Table 1. 3D RNA assemblies at bilayers with different lipid composition.
ΔF and ΔD (overtone 7) for the adsorption of RNA tectosquare and polyhedron to bilayers with different composition (Mixed DOPC/Sphingosine bilayers and pure DOTAP bilayers). Modelled data for the thickness of the adsorbed RNA layer are also shown.
| Lipid bilayer | Charged lipids (mol%) | RNA | Δ D (*10−6) | Δ F (Hz) | Δ h (nm) |
|---|---|---|---|---|---|
| DOPC Sph | 10 % | TS3 | 4.5±0.4 | −32.9±3 | 8.8±0.9 |
| DOPC Sph | 50 % | TS3 | 3.9±0.4 | −38.6±3 | 8.9±0.9 |
| DOTAP | 100 % | TS3 | 4.1±0.4 | −53.3±4 | 10.3±0.9 |
| DOPC Sph | 10 % | TO3-4 | 12.7* | −73.2* | 20.1* |
| DOPC Sph | 30 % | TO3-4 | 9.1±0.8 | −67.4±6 | 15.2±1.3 |
| DOPC Sph | 50 % | TO3-4 | 8.2±0.8 | −61.8±6 | 13.4±1.3 |
| DOTAP | 100 % | TO3-4 | 8.9±0.8 | −71.7±7 | 16.7±1.3 |
indicates that only one replicate was done for this lipid composition.
Materials and Methods
Chemicals
DOPC (1,2-dioleoyl-sn-glycero-3-phosphatidylcholine), D-erythro-Sphingosine (18:1), DOTAP (18:1 TAP, 1,2-dioleoyl-3-trimethylammonium-propane) and Liss Rhod PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) ) with 99% purity were purchased from Avanti polar lipids Inc. (Alabaster, USA). Chloroform and methanol (both 99.8% purity) were purchased from Merck (Darmstadt, Germany). GelStar® nucleic acid gel stain (10 000x concentrate) was purchased from Lonza Bioscience. Tris(hydroxymethyl)aminomethane (Tris) of ultra pure grade was purchased from Fischer Scientific and used without further purification. The poly(amido amine) (PAMAM) dendrimers of generation 2 and generation 4 and with ethylenediamine cores were purchased from Sigma. The PAMAM dendrimers were dissolved in methanol and were dried over night in a vacuum oven and then left to hydrate at 4 °C for 1–2 days prior to use. Boric acid (approx. 99% purity), Mg(OAc)2 (99% purity) and 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES) (≥99.5% purity) were purchased from Sigma Aldrich. KCl (≥99.5% purity) was purchased from Fluka.
RNA synthesis
The eight RNA segments used for assembling the two squares and the polyhedron were designed and produced according to protocol from Severcan et al.14 The design characteristics and the tRNA sequences are shown in Figure S1 and Table S1. In brief, the preparation of the eight RNA building blocks that assemble into a polyhedron was done as follows: DNA templates of each of the eight segments were put in separate test tubes and amplified using PCR with 25 cycles (94°C for 75 seconds cool to 54°C for 75 seconds and heat again to 72°C for another 75 seconds). The DNA produced in the PCR was purified and collected using the Quiagen PCR purification kit. After purification the DNA was then transcribed to RNA, and after the transcription the RNA was purified using a 10% bisacrylamide (19:1)/8M Urea gel. RNA was detected with UV light, and the RNA containing bands were cut out and the RNA was extracted from the gel using a “crush and soak” buffer (200 mM NaCl, 10 mM Tris, pH 7.5, 0.5 mM EDTA). RNA was than precipitated using ethanol and finally dried. The TS3, TS4 RNA squares and the TO3-4 polyhedron were characterized by native polyacrylamide gel electrophoresis in 15 mM Mg(OAc)2 and at 10°C (Figure S2 a) as well as AFM imaging of the square (Figure S2 b). RNA molecules were used without further purification. All RNA buffer solutions were prepared using ultrapure water from a Milli-Q® Ultrapure water purification system from Millipore (Massachusetts, USA). All glassware was soaked in 10% hydrochloric acid for several hours and then washed 10 times with Milli-Q water. As RNA is sensitive to nuclease degradation, all glassware used was sterilized at 200°C for 8 hours and all solutions were autoclaved (121°C for 20 minutes).
Self-assembly of RNA squares and polyhedron
The RNA used in this study was self-assembled square shaped and polyhedron shaped RNA structures. Each RNA square (TS3 or TS4) was assembled in solution by mixing 4 RNA components constituting that square (as shown in Figure S1 c). TS3 and TS4 are designed to assemble into the RNA polyhedron when mixed.14 To assemble the squares, a solution containing the four RNA sequences was heated to 90°C for three minutes then cooled on ice for three minutes. After cooling, the solution was kept at 30°C for 3 minutes, and then mixed with the association buffer with 0.2 mM Mg2+ (10 mM Tris-borate, 50 mM KCl and 0.2 mM Mg(OAc)2). The solution containing the segments for assembly of TS3 and of TS4 was thereafter incubated for another 30 minutes. The RNA polyhedron was assembled in equimolar mixtures of TS3 and TS4 using association buffer with 15 mM Mg2+. The solution was heated to 60°C for 3 minutes and then slowly cooled down to 10°C.14
AFM imaging
RNA tectosquares were deposited on freshly cleaved mica surface (muscovite mica grade V-1) that was pre-treated with the deposition buffer (10 mM Tris-borate, 50 mM KCl and 15 mM Mg(OAc)2), incubated on the surface for 5 minutes, rinsed with Milli-Q water and dried under nitrogen. The topography was imaged in air by non-contact mode AFM (XE-100 Park system) using PPP-NCHR cantilevers (spring constant: k=42 N/m, resonance frequency: f=330 kHz). Sample tilt was corrected during image deposition with Park system software.
Lipid vesicle preparation
Deposition of the supported bilayers was done by the vesicle fusion technique.50 Stock solutions of the lipid mixtures used were prepared by dissolving the lipids/surfactants in chloroform/methanol 2/1 (vol ratio). Smaller aliquots from the stock solutions were taken out and dried under a stream of air until the solvent was evaporated and a thin lipid film remained. The lipid film was suspended in buffer/aqueous solution and left above the melting point of the lipids for at least 30 min before sonication. The lipid dispersion was sonicated using 30 second pulses at 30% amplitude (Vibracell tip sonicator, Sonics & materials Ltd, USA) until a clear dispersion of small unilamellar vesicles (SUVs) was obtained, a process that usually took less than 10 minutes. Care was taken not to overheat the sample.
QCM-D measurements
Quartz crystal microbalance with dissipation (QCM-D) measurements were performed using a Q Sense E4 system from Q Sense (Gothenburg, Sweden) with four temperature controlled flow modules. In each flow module one sensor crystal was placed. The crystals used for the present experiments were quartz crystals with a fundamental frequency of 4.95MHz covered by a thin gold surface layer connected to the two electrodes. The gold surface was coated with a 50 nm thick SiO2 layer (QSX 303, Q Sense). The flow of liquid through the modules was controlled by means of an external peristaltic pump (Ismatec IPC-N 4) to assure constant flow throughout the experiment. Prior to use the sensor crystals were cleaned and stored in 2% SDS solution for a minimum of 1 hour, washed with MQ water, rinsed with ethanol, dried under nitrogen and thereafter treated in a plasma cleaner from Harrick Scientific (New York, USA) for 5 minutes. Directly after plasma cleaning the crystals were inserted into the QCM-D cells. Before each measurement, the crystals were allowed to equilibrate in water until a stable baseline was reached. The QCM crystals were then equilibrated with 10 mM HEPES buffer at pH7 prior to deposition of the bilayer by pumping the lipid SUV solution through the cell at a flow rate of 100 μl/min. The formation of a stable bilayer was fast and already after 3–5 minutes it was possible to start rinsing the surfaces with 10 mM HEPES buffer. The bilayers formed with a high surface coverage (close to 100%).51 After exchanging vesicular dispersion with neat buffer, i.e. thoroughly rinsing the bilayer, the buffer solution was changed either to association buffer with 0.2 mM Mg2+ or to association buffer with 15 mM Mg2+. Solutions of freshly prepared RNA polyhedrons or squares were added when a constant signal for bilayer was reached. A successful approach relies on robust and reproducible model system, e.g. the deposited bilayer has high surface coverage, and strong adsorption of the RNA building block, where the RNA remains attached the bilayer even if the RNA solution is replaced with pure buffer. Minimal RNA desorption (i.e., not significant within errors of the measurements) upon rinsing with buffer was observed for the bilayer systems composed of only cationic DOTAP. The bilayers were deposited on a SiO2 surface, which is slightly negatively charged at neutral pH. In separate experiments, we show that the RNA molecules do not adsorb to the bare SiO2 surface. The RNA concentration was in all cases 10 μg/ml and the nucleic acids were added continuously with a flow of 10 μL/min. In order for the nucleic acids to faster reach the cell and thus reduce the lag time, the pump speed during the first 1 min 15 sec was 100 μL/min. All experiments were performed at 25 °C.
The wet mass of the lipid film adsorbed to the silica was calculated according to the Sauerbrey expression
| (2) |
where C ≈ 17.7 ng Hz−1 cm−2 for a 5 MHz crystal and on is the overtone number.52 The validity of applying this equation to non-rigid biomolecular systems has been addressed by Höök et al.,37 concluding that the Sauerbrey expression is a good approximation for thin, acoustically rigid and evenly distributed adsorbed films. However, also an acoustically rigid film may trap solvent, which becomes sensed as an additional mass. Hence, only in cases when the amount of coupled water is low, the “Sauerbrey mass” correspond well to the adsorbed molecular (lipid) mass. Planar supported lipid bilayers constitute one such example, as verified with other methods such as ellipsometry.50 For the present experiments, the mass of the deposited bilayers were calculated to 410–440 ng/cm2 for the DOPC/sphingosine bilayers and 380–400 ng/cm2 for the DOTAP bilayers, using the Sauerbrey expression.
When the change in dissipation becomes large, as observed for, e.g. the RNA polyhedrons, the Sauerbrey expression is not applicable to calculate the adsorbed mass for two reasons.37 First, the adsorbed material may extend into the solution in such a way that the amount of hydrodynamically coupled water becomes significant. Secondly, if the induced dissipation is significant, the conversion of ΔF into wet mass fails. While the former condition leads to an overestimation of the adsorbed molecular mass using the Sauerbrey expression, the latter yields an underestimation. Hence, when a non-rigid extended layer is deposited at the surface, a viscoelastic representation of the adsorbed film needs to be used for the data analysis. The model used here for analysing the data obtained for adsorption of RNA polyhedron and squares was an extended viscoelastic model, where an explicit frequency dependence of the viscoelastic parameters has been taken into account. Data was modelled using a Voigt-based model for one adsorbed layer, representing the properties of the viscoelastic film, using experimental data from the 3, 5 and 7th overtone. The parameters that were kept fixed in the modelling were the fluid density (1000 kg/m3) and the fluid viscosity (0.001 kg/ms). The best fit was obtained using a fixed layer density of 1600 kg/m3. The layer thickness, viscosity and shear modulus were fitting parameters, but kept within the following boundaries; layer thickness (10−10–10−7m), viscosity (0.001–0.01 kg/ms) and shear modulus (1000–108 kg/(ms2)). Note that in QCM-D experiments the change of buffer also gives rise to a change in both measured frequency and dissipation, due to slight changes in density and viscosity. The most pronounced effect was observed when changing to the buffer containing 15 mM Mg2+, and this has to be taken into account in the analysis of the data. Each of experiments presented here were reproduced at least two times with good agreement between the different experiments. One example to demonstrate the high reproducibility is shown in Figure S10.
Combined QCM-D and ellipsometry module
Ellipsometry is based on measurements of changes in the polarization in terms of the amplitude difference, Ψ, and the phase shift, Δ when light is reflected on a surface.51 From these data we can obtained information about the thickness and refractive index of the film formed at the surface using an iterative process,51,52 provided accurate determination of the substrate can be obtained. Modeling of the optical properties of the QCM-D crystals for unambiguous determination of thickness of the adsorbed layer with ellipsometry is challenging as the surface consists of a 1000 Å thick layer of titanium, covered by about 5 Å thick layer titanium oxide that is covered by about 800 Å thick layer of silica.53 The main purpose of using ellipsometry here was however to determine the “dry” mass of adsorbed material. In order to do so, we can instead determine an effective refractive index of the substrate (in this study about N=1.7–2.5i, depending on the crystal used), which takes into account the oxide layers and the substrate. In this way the adsorbed amount can be calculated accurately, although thickness and refractive index cannot be resolved.52 The validity of the mass calculation was confirmed by test experiments using C12E5, giving an adsorbed amount of 1.27±0.08 mg/m2,52 as well as the fact that the amount adsorbed lipid bilayer determined by QCM-D and ellipsometry is the same. For calculation of adsorbed amount (Γ) we used the approach of de Feijter et al:54
| (3) |
where df is film thickness, nf is the film refractive index and n0 is the refractive index of the solvent. Here we used a refractive index increment of dn/dc = 0.148 cm3/g, which is the value for the lipid55 and for the RNA adsorbed to the lipid bilayer dn/dc = 0.168 cm3/g was used as previously determined for RNA.56 The lipid layer was assumed to have same refractive index and thickness as before adding RNA and the refractive index of the RNA layer was calculated assuming that the measured nf is the average of refractive index for the lipid and RNA layers scaled by the determined thickness of the layers.
An Optrel Multiskop ellipsometer (Optrel, Berlin, Germany, www.optrel.de) with a Nd:YAG, λ =5320 Å laser, operating as a null-elipsometer was used in this study.57 The Q-sense ellipsometry module (QELM 401) was used for the combined QCM-D and ellipsometry measurements. The module has a fixed angle of incidence of 65°. The QCM-D ellipsometry module is connected to an external peristaltic pump (Ismatec IPC-N 4), the cell has one inlet for fluid and three outlets, to assure constant flow in the whole cell during the experiment. The substrate used for the experiments was a quartz crystal covered with a thin layer of titanium that is further coated with silicon dioxide (QSX335, Q-Sense). The outer surface layer is as for the standard QCM SiO2 coated quartz crystals gold (QSX303, Q-Sense), and the fundamental frequency is also here 4.95 MHz. DOTAP bilayers formed in the ellipsometry module were done in the same way as in the standard QCM-D module except that the bilayers were rinsed first with water before the solution was changed to the correct buffer.
FRAP and TIRF measurements
Fluorescence microscopy images were recorded using either a confocal laser scanning microscope (CLSM) or a total internal reflection fluorescence (TIRF) microscope. For the confocal microscopy, a Leica CLSM SP5 was operated in the inverted mode (D60001) with a 60x glycerol-immersion objective. HeNe (543 nm) and Ar ion (488 nm) lasers were used to excite the rhodamine-labeled (543 nm) and NBD-labeled (463 nm) lipids, respectively. Fluorescence recovery after photobleaching (FRAP) experiments were performed by bleaching a spot with a diameter of 10 μm with the argon ion laser set to a tube intensity of 80%. Subsequently, images were rcorded every 1 s to monitor the fluorescence recovery. The field of imaging was 40 μm × 40 μm (512 × 512 pixels). The TIRF microscopy was performed on an inverted Eclipse TE 2000 microscope (Nikon) equipped with a high-pressure mercury lamp, an Apo TIRF 60× oil objective (NA 1.49) and a CCD camera (1002 × 1004 pixel in binning mode). Filter sets and dichroic mirrors in the filter cubes were chosen to match the excitation and emission properties of the selected fluorophores. Images (190 μm × 190 μm) were recorded using acquisition times of 200 ms. In the TIRF set-up, FRAP experiments were performed by directing a focused (diameter: 20 – 30 μm) beam of a blue (wavelength: 473 nm) or green (wavelength 532 nm) solid state laser on the center of the microscopy image. The bleaching process was continued for a time span of 10 s – 20 s reducing the fluorescence emission intensity in this area by 30% – 50%. Right after the bleaching step, time lapse TIRF images were recorded in short time intervals of 10s to monitor the fluorescence recovery at an appropriate time resolution.
For both types of microscopy, cover glasses were cleaned by soaking for 15 minutes in 5 parts H2O: 1 part 27% NH4OH: 1 part 30% H2O2 at 70 °C followed by 5 minutes of treatment in reduced air plasma of 0.02 mbar (Harrick Scientific, New York, USA) or 20 minutes of treatment in UV ozone. DOTAP vesicles were prepared as described in the previous section, except that 0.5 mol% Liss Rhod PE was included as the fluorescent probe. Bilayers were self-assembled on the glass surface by flowing vesicle dispersions over the cleaned cover glasses, sealed in disposable flow cells (sticky-Slide I0.6 Luer, ibidi GmbH, Martinsried, Germany). Bilayers were imaged and FRAP experiments were performed on the bilayers alone. Afterwards, the bilayer was incubated with a solution of RNA polyhedrons in association buffer for 30 minutes. The bilayer was subsequently rinsed by flowing copious amounts of buffer through the flow cell (at least 10 x the volume of the flow cell). FRAP measurements were performed on the bilayers which had been incubated with various concentrations of RNA polyhedrons and subsequently rinsed. The fraction of immobile molecules was negligible at low RNA polyhedron concentrations (Figure S5c). Diffusion constants of the bleached lipid bilayer were calculated according to Jönsson et al.44 At least three measurements were made on different locations of each sample.
Conclusions
We have demonstrated that it is possible to adsorb and assemble complex RNA polyhedron structures on a supported lipid fluid bilayer as a scaffold. The use of a bottom-up approach allowed us to direct RNA self-assembly at a model membrane by exploiting the strong attractive electrostatic interactions between RNA polyanions and cationic bilayer. Quantitative in-situ information on this directed RNA assembly reaction at the bilayer upon sequential changes of the solution composition was obtained using a flow-system set-up. The presence of a scaffold membrane surface was found to favour the nucleation of the assembly after adsorption of only a few molecules on the surface. Therefore, the surface induced process required less material compared to the corresponding bulk self-assembly reaction.
Our results imply that the adsorbed RNA polyhedrons retain their 3D structure when adsorbed. RNA polyhedron adsorption affected the mobility of the fluid lipid bilayer with loss of mobility above a percolation threshold. The RNA polyhedrons as well as TS3 and TS4 tectosquares adsorb readily to different charged bilayer systems, largely independent on the surface charge density. We also show that the 3D structure of the RNA polyhedron can be collapsed by addition of highly charged cationic dendrimers. By changing the solution conditions, we show that assembly of the pre-programmed 3D structure can be modulated by sequence specific interactions, surface charge and changes in the salt environment. In addition, the tertiary structure of the RNA polyhedron can be controllably switched from an extended structure to one that is dense and compact.
Previously, it was demonstrated that in vitro selected RNA molecules could interact with lipid membranes by forming RNA networks that could affect membrane permeation.35,58 There are also recent studies showing synthetic membrane-spanning nanochannels formed by synthetic DNA with cholesterol or porphyrin based bilayer anchors covalently attached to the DNA strands.33,34 Our results provide evidence that complex and programmable RNA and DNA self-assembly processes can be controlled on lipid membranes without impacting the structural integrity and stability of the immobilized three-dimensional RNA nanostructures or of the membranes themselves. Furthermore, the fact that our RNA polyhedron can assemble on model lipid membranes composed of phospholipids and sphingosine, which could be of biological relevance, is particularly encouraging for developing novel therapeutic all-RNA nanoparticles that might directly interact with natural cellular membranes. Indeed, RNA-based nanoparticles such as the RNA polyhedron could readily be used as scaffolds for aptamers, ribozymes or therapeutics siRNAs.14,15,17,59 As such, a combination of rational design and in vitro selection approaches could lead to the development of novel classes of RNA-based nanoparticles that could facilitate the interaction and passage of therapeutics RNAs through a lipid membrane. Additionally, our data suggests the possibility of creating self-assembling RNA (or DNA) nanostructures acting as “rafts” on lipid membranes. For instance, by contrast to static mineral surfaces, lipid membranes offer the advantage of more fluid, dynamical surfaces on which RNA nanostructures could undergo structural rearrangements upon modulation of the membrane fluidity (e.g. in function of temperature, hydration or solution conditions). The molecular organization and dynamics in the single layer of RNA nanostructures can the thereby be controlled by external conditions in a rather easy and accessible way. Recently, it has been shown that DNA (or RNA) could complex with surfactant (like DDAB) to form self-standing films able to undergo major structural rearrangement upon hydration and variation of temperature.9,60–62 Therefore, in conjunction with the data presented herein, responsive RNA/lipid based films could undergo complex rearrangement leading to the self-assembly of complex RNA nanostructures on the membranes upon hydration. While some of these ideas are still speculative, it is already apparent that our present results open up promising new routes to functionalize bio-mimicking surfaces with applications in nanotechnology, nanomedicine and synthetic biology.12,63,64
Supplementary Material
Acknowledgments
Peter Jönsson is acknowledged for valuable discussions about the analysis of the FRAP experiments and D. Csontos for reviewing our manuscript prior to submission. LJ wishes to dedicate this paper to Saint Birgitta of Sweden. The Swedish Research Council (VR) is gratefully acknowledged for financial support both through regular grants and the Linnaeus Center of Excellence “Organizing molecular matter” (ES, TN). E.S. acknowledges The Swedish Foundation for Strategic Research (SSF) for financial support and A. D. and T. N. acknowledge support from the Nanometer Structure Consortium at Lund University. Knut and Alice Wallenberg’s foundation funded the acquisition of the QCM-D equipment and the confocal microscope. This work was partially funded by the National Institutes of Health (R01-GM079604 to LJ).
Footnotes
Electronic Supplementary Information (ESI) available: Table with sequences of tRNA units used in this study; schematic structures of the RNA polyhedron and its building blocks; gel electrophoresis characterization of the RNA polyhedron and squares; AFM characterization of RNA tectosquare; schematic structures of RNA-9 and RNA-10 and their association with lipid bilayers; QCM-D frequency and dissipation data (as function of time) for adsorption of RNA polyhedrons, RNA squares and RNA9-10; TIRF images of RNA with Gelstar after photobleaching with analysis; Correlation plot in change of shear viscosity for TS3 and TO3-4; Models for the stoichiometry of TS; QCM-D dissipation data for the sequential experiment in Figure 5a; QCM-D and for the assembly of building blocks at the bilayer scaffold at varying bulk concentrations; QCM-D of adsorption of TS3. See DOI: 10.1039/b000000x/
Notes and references
- 1.Schreiber R, Do J, Roller EM, Zhang T, Schuller VJ, Nickels PC, Feldmann J, Liedl T. Nature Nanotechnol. 2013;9:74. doi: 10.1038/nnano.2013.253. [DOI] [PubMed] [Google Scholar]
- 2.Pinheiro AV, Han D, Shih VM, Yan H. Nature Nanotechnol. 2011;6:763. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aagaard L, Rossi JJ. Adv Drug Deliver Rev. 2007;59:75. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros V, Chen XM. Development. 2007;134:1635. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 5.Garzon R, Calin GA, Croce CM. Ann Rev Med. 2009;60:167. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 6.Zhao YX, Shaw A, Zeng X, Benson E, Nyström AM, Högberg B. ACS Nano. 2012;6:8684. doi: 10.1021/nn3022662. [DOI] [PubMed] [Google Scholar]
- 7.Rothemund PWK. Nature. 2006;440:297. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 8.Nangreave J, Han DR, Liu Y, Yan H. Curr Op Chem Biol. 2010;14:608. doi: 10.1016/j.cbpa.2010.06.182. [DOI] [PubMed] [Google Scholar]
- 9.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 10.Jaeger L, Chworos A. Curr Op Struct Biol. 2006;16:531. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Liedl T, Högberg B, Tytell J, Ingber DE, Shih WM. Nature Nanotechnol. 2010;5:520. doi: 10.1038/nnano.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabow WW, Jaeger LIpPoM. Account Chem Res. 2014;2014 doi: 10.1021/ar500076k. In press. [DOI] [PubMed] [Google Scholar]
- 13.Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Nature. 2009;459:414. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. Nature Chem. 2010;2:772. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo P. Nature Nanotechnol. 2010;5:833. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu YM, Zeng DD, Chao J, Jin YQ, Zhang Z, Liu HJ, Li D, Ma HW, Huang Q, Gothelf KV, Fan CH. J Am Chem Soc. 2013:135. doi: 10.1021/ja3076692. [DOI] [PubMed] [Google Scholar]
- 17.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. Nature Nanotechnol. 2010;5:676. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasalean L, Baudrey S, Leontis NB, Jaeger L. Nucleic Acids Res. 34:1381. doi: 10.1093/nar/gkl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geary C, Chworos A, Jaeger L. Nucleic Acids Res. 2011;39:1066. doi: 10.1093/nar/gkq748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L. Nano Letters. 2011;11:878. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Severcan I, Geary C, Verzemnieks E, Chworos A, Jaeger L. Nano Letters. 2009;9:1270. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellot G, McClintock MA, Chou JJ, Shih WM. Nature Protocol. 2013;8:755. doi: 10.1038/nprot.2013.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juul S, Iacovelli F, Falconi M, Kragh SL, Christensen B, Frohlich R, Franch O, Kristoffersen EL, Stougaard M, Leong KW, Ho YP, Sorensen ES, Birkedal V, Desideri A, Knudsen BR. ACS Nano. 2013;7:9724. doi: 10.1021/nn4030543. [DOI] [PubMed] [Google Scholar]
- 24.Aldaye FA, Sleiman HF. J Am Chem Soc. 2007;129:13376. doi: 10.1021/ja075966q. [DOI] [PubMed] [Google Scholar]
- 25.Wittung-Stafshede P, Rodahl M, Kasemo B, Nielsen P, Nordén B. Coll Surf A. 2000;174:269. [Google Scholar]
- 26.Börjesson K, Lundberg EP, Woller JG, Norden B, Albinsson B. Angew Chemie. 2011:50. doi: 10.1002/anie.201103338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundberg EP, Feng BB, Mohammadi AS, Wilhelmsson LM, Norden B. Langmuir. 2013;29:285. doi: 10.1021/la304178f. [DOI] [PubMed] [Google Scholar]
- 28.Höök F, Ray A, Nordén B, Kasemo B. Langmuir. 2001;17:8305. [Google Scholar]
- 29.Larsson C, Rodahl M, Hook F. Anal Chem. 2003;75:5080. doi: 10.1021/ac034269n. [DOI] [PubMed] [Google Scholar]
- 30.Kufer SK, Puchner EM, Gumpp H, Liedl T, Gaub HE. Science. 2008;319:594. doi: 10.1126/science.1151424. [DOI] [PubMed] [Google Scholar]
- 31.Kufer SK, Strackharn M, Stahl SW, Gumpp H, Puchner EM, Gaub HE. Nature Nanotechnol. 2009;4:45. doi: 10.1038/nnano.2008.333. [DOI] [PubMed] [Google Scholar]
- 32.Frykholm K, Bombelli FB, Norden B, Westerlund F. Febs J. 2010;277:234. [Google Scholar]
- 33.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. Science. 2012;338:932. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burns JR, Göpfrich K, Wood JW, Thacker VV, Stulz E, Keyser UF, HS Angew Chem Int Ed. 2013;52:12069. doi: 10.1002/anie.201305765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vlassov A, Khvorova A, Yarus M. Proc Natl Acad Sci USA. 2001;98:7706. doi: 10.1073/pnas.141041098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michanek A, Björklund M, Nylander T, Sparr E. Soft Matter. 2012;8:10428. [Google Scholar]
- 37.Höök F, Kasemo B, Nylander T, Fant C, Sott K, Elwing H. Anal Chem. 2001;73:5796. doi: 10.1021/ac0106501. [DOI] [PubMed] [Google Scholar]
- 38.Eisele NB, Frey S, Piehler J, Gorlich D, Richter RP. EMBO Rep. 2010;11:366. doi: 10.1038/embor.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reviakine I. D Johannsmann and RP Richter. 2011;83:8838. doi: 10.1021/ac201778h. [DOI] [PubMed] [Google Scholar]
- 40.Horton MR, Reich C, Gast AP, Rädler JO, Nickel B. Langmuir. 2007;23:6263. doi: 10.1021/la063690e. [DOI] [PubMed] [Google Scholar]
- 41.Reviakine I, Brisson A. Langmuir. 2001;17:8293. [Google Scholar]
- 42.Brisson A, Olofsson A, Ringler P. M Schmutz and S Stoylova, Biol Cell. 1994;80:221. [PubMed] [Google Scholar]
- 43.Leal C, Sandström D, Nevsten P, Topgaard D. Biochim Biophys Acta. 2008;1778:214. doi: 10.1016/j.bbamem.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 44.Jönsson P, Jonsson MP, Tegenfeld JO, Höök F. Biophys J. 2008;95:5334. doi: 10.1529/biophysj.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watkins EB, Miller CE, Mulder DJ, Kuhl TL, Majewski J. Phys Rev Lett. 2009;102:238101. doi: 10.1103/PhysRevLett.102.238101. [DOI] [PubMed] [Google Scholar]
- 46.Cardenas M, Braem A, Nylander T, Lindman B. Langmuir. 2003;19:7712. [Google Scholar]
- 47.Dias R, Rosa M, Pais AC, Miguel M, Lindman B. J Chin Chem Soc. 2004;51:447. [Google Scholar]
- 48.Carnerup AM, Ainalem ML, Alfredsson V, Nylander T. Soft Matter. 2011;7:760. [Google Scholar]
- 49.Ainalem ML, Bartles A, Muck J, Dias RS, Carnerup AM, Zink D, Nylander T. PLOS ONE. 2014;9:e92692. doi: 10.1371/journal.pone.0092692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandoolaeghe P, Rennie AR, Campbell RA, Thomas RK, Hook F, Fragneto G, Tiberg F, Nylander T. Soft Matter. 2008;4:2267. [Google Scholar]
- 51.Azzam RMA, Bashara NM. Ellipsometry and polarized light. Elsevier Science BV; Amsterdam: 1996. [Google Scholar]
- 52.Tiberg F, Landgren M. Langmuir. 1993;9:927. [Google Scholar]
- 53.Carton I, Brisson AR, Richter RP. Anal Chem. 2010;82:9275. doi: 10.1021/ac102495q. [DOI] [PubMed] [Google Scholar]
- 54.de Feijter JA, Benjamins J, Veer FA. Biopolymers. 1978;17:1759. [Google Scholar]
- 55.Ainalem ML, Kristen N, Edler KJ, Hook F, Sparr E, Nylander T. Langmuir. 2010;26:4965. doi: 10.1021/la9036327. [DOI] [PubMed] [Google Scholar]
- 56.Nieuwenhuysen P, De Voeght F, Clauwaert J. Biochem J. 1981;197:689. doi: 10.1042/bj1970689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harke M, Teppner R, Schulz O, Motschmann H, Orendi H. Rev Sci Instrum. 1997;68:3130. [Google Scholar]
- 58.Janas T, Janas T, Yarus M. Nucleic Acids Res. 2006;34:2128. doi: 10.1093/nar/gkl220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, Shapiro BA, Jaeger L. Nat Protocol. 2011;6:2022. doi: 10.1038/nprot.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smitthipong W, Neumann T, Gajria S, Li Y, Chworos A, Jaeger L, Tirrell M. Biomacromolecules. 2008;10:221. doi: 10.1021/bm800701a. [DOI] [PubMed] [Google Scholar]
- 61.Neumann T, Gajria S, Bouxsein NF, Jaeger L, Tirrell M. J Am Chem Soc. 2010;132:7025. doi: 10.1021/ja909514j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neumann T, Gajria S, Tirrell M, Jaeger L. J Am Chem Soc. 2009;131:3440. doi: 10.1021/ja809349m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grabow WW, Jaeger L. F1000Prime Reports. 2013;5:46. doi: 10.12703/P5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hess H, Jaeger L. Curr Op Biotech. 2010;21:373. doi: 10.1016/j.copbio.2010.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.