Abstract
Objectives:
Patient outcomes for esophageal adenocarcinoma (EAC) have not improved despite huge advances in endoscopic therapy because cancers are being diagnosed late. Barrett's esophagus (BE) is the primary precursor lesion for EAC, and thus the non-endoscopic molecular diagnosis of BE can be an important approach to improve EAC outcomes if robust biomarkers for timely diagnosis are identified. MicroRNAs (miRNAs) are tissue-specific novel biomarkers that regulate gene expression and may satisfy this requirement.
Methods:
Patients with gastroesophageal reflux disease (GERD) and BE were selected from an ongoing tissue and serum repository. BE was defined by the presence of intestinal metaplasia. Previously published miRNA sequencing profiles of GERD and BE patients allowed us to select three miRNAs, miR-192-5p, -215-5p, and -194-5p, for further testing in a discovery cohort and an independent validation cohort. Receiver operating curves were generated to calculate the diagnostic accuracy of these miRNAs for BE diagnosis. To test specificity, the miRNA signature was compared with those of the gastric cardia epithelium and the non-intestinal-type columnar epithelium (another definition of BE). In addition, to gain insights into BE origin (intestinal vs non-intestinal), global BE miRNA profiles were compared with the published miRNA profiles of other columnar epithelia in the gastrointestinal tract, that is, normal stomach and small and large intestine.
Results:
The discovery cohort included 67 white male patients (40 with GERD and 27 with BE). The validation cohort included 28 patients (19 with GERD and 11 with BE). In the discovery cohort, the sensitivity, specificity and area under the curve (AUC) of the three mRNAs for BE diagnosis were 92–100%, 94–95%, and 0.96–0.97, respectively. During validation, the sensitivity and specificity of miRNAs for BE diagnosis were as follows: miR-192-5p, 92% and 94%, AUC 0.94 (0.80–0.99, P=0.0004); miR-215-5p, 100% and 94%, AUC 0.98 (0.84–1, P=0.0004); and miR-194-5p, 91% and 94%, AUC 0.96 (0.80–0.99, P=0.0001), respectively. The tested miRNAs identified all BE patients in both the discovery and the validation cohorts. When compared with non intestinal-type columnar and gastric cardia epithelia, the miRNA signature was specific to the intestinal-type columnar epithelium. Comparisons of BE miRNA sequencing data to published data sets for the normal stomach, small intestine and large intestine confirmed that two of the three miRNAs (miR-215-5p and -194-5p) were specific to the intestinal-type epithelium.
Conclusions:
MicroRNAs are highly accurate for detecting intestinal-type BE epithelia and should be tested further for the non-endoscopic molecular diagnosis of BE.
Introduction
Esophageal adenocarcinoma (EAC) is a cancer of significant importance to public health because of rapidly increasing incidence1 and poor survival.2 The major determinant of the outcome of EAC is the stage at diagnosis.2, 3 Early-stage cancer can be treated with endoscopic therapies to achieve high cure rates.4 Although EAC progresses stepwise from the premalignant lesion of Barrett's esophagus (BE), >90% of cancer cases arise in patients without a prior diagnosis of BE, which can result in delayed cancer detection.5 Because current methods have failed to improve the outcomes for EAC, newer approaches are needed.6 The American Gastroenterological Association7 and the American College of Gastroenterology8 guidelines suggest an earlier diagnosis of BE as a strategy to halt the significant increase in the rate of esophageal cancer. An important yet underappreciated fact is that ~70% of EACs are prevalent cancers that can be detected by index endoscopy.9 Thus, expedient BE diagnosis will be beneficial.
Currently, upper endoscopy is the standard for BE diagnosis and is typically recommended for patients with chronic gastroesophageal reflux disease (GERD).7, 8 However, upper endoscopy is not logistically feasible for testing the 10 million individuals with GERD who are at risk for BE. Recently, investigators from England demonstrated the feasibility of testing BE-specific molecular markers in non-endoscopically obtained cytology specimens as a practical alternative for BE diagnosis.10 This approach could benefit from the establishment of novel markers with high accuracy. MicroRNAs (miRNAs) are small RNA molecules that regulate gene expression and have shown promise as highly specific tissue biomarkers in multiple premalignant and malignant conditions.11, 12, 13, 14 In a landmark paper, miRNAs were more specific than messenger RNA profiles for diagnosis in multiple cancer types.12 In addition, miRNAs were found to be useful in determining the developmental lineage of various cancers.12 These data led us to hypothesize that miRNA expression could accurately discriminate between GERD and BE epithelia. Previous studies found candidate miRNAs to be specific for BE epithelium15, 16, 17, 18, 19 but did not systematically evaluate their diagnostic performance. Prior to larger trials to test the clinical utility of miRNA for BE diagnosis, their performance characteristics need to be defined to facilitate study design. We previously sequenced the miRNA transcriptome in patients with GERD and BE to identify miRNAs associated with BE pathogenesis, and we identified 10 miRNAs that were differentially expressed.20 In this study, we had two aims: (1) to determine the accuracy of select candidate miRNAs for differentiating between patients with GERD and BE, and (2) to gain insights into the tissue of BE origin (intestinal vs non-intestinal epithelium) based on the miRNA expression patterns. For aim 1, we compared miRNA expression in discovery and validation cohorts between independent patients with GERD and BE to determine their diagnostic accuracy, and for aim 2, we correlated the miRNA profiles of BE with the published miRNA profiles of other columnar epithelia in the gastrointestinal tract, that is, the normal stomach,21 small intestine,22 and large intestine.22
Methods
Subject selection, definitions, and histologic evaluation
Patients with BE and GERD were selected from an ongoing prospective tissue and serum repository at the Veterans Affairs Medical Center, Kansas City, MO, USA (clinical trials #NCT00574327). The protocol was initially approved on 25 April 2005 and has been annually reviewed and renewed by the local Institutional Review Board. Details of the repository have been published previously.20 Patients referred for upper endoscopy were invited to participate and contribute specimens for molecular research related to GERD and BE. All patients signed an informed consent document prior to participation.
GERD was defined on the basis of a validated GERD questionnaire.23 Based on endoscopy, GERD patients were further subdivided into those with non-erosive reflux disease (NERD) and erosive esophagitis (EE). The presence of EE was graded according to the Los Angeles Classification. BE was defined by the presence of columnar lined esophagus at least 1 cm in length and the presence of intestinal metaplasia on the basis of goblet cells. Biopsies were not obtained from patients with active ulcerations in the BE segment. The gastroesophageal junction was identified by the top of gastric folds. BE lengths were determined according to the Prague criteria.24 Standard-of-care clinical biopsies were obtained in a four-quadrant manner every 1–2 cm in all BE patients using Radial Jaw 4 biopsy forceps (Boston Scientific, Quincy, MA, USA). Additional research biopsies were obtained every 2 cm in BE patients. In patients with GERD, only research biopsies were obtained from the squamous epithelium 1 cm above the gastroesophageal junction. The gastric cardia biopsies were provided by Dr Krishnadath of the Academic Medical Center, Amsterdam, Netherlands under an approved protocol (study number: MEC 01/288#08.17.1042). Paired gastric cardia biopsies were obtained for molecular testing and routine histopathological evaluation within 2 cm below the gastric folds. All BE biopsies were reviewed by an experienced gastrointestinal pathologist (SCM) according to the standardized criteria described by Montgomery et al.25 If there was concern for high-grade dysplasia and/or cancer, the biopsies were reviewed by a second pathologist. All discrepant diagnoses were solved by consensus. For this proposal, we only included BE patients without dysplasia.
The inclusion criteria for the current feasibility study were as follows:
BE lengths longer than 2 cm. We chose this criterion to include only well-defined BE patients for this pilot study.
BE without dysplasia in both clinical and research biopsies. We excluded patients with dysplasia to minimize variability in miRNA expression based on the presence of dysplasia.
Cases, controls, and selection of candidate miRNAs
Cases were defined as patients with BE. The control group was composed of patients with GERD (without BE). Both the cases and the controls were selected randomly from the repository. We chose GERD patients as the control group because this is the group at the highest risk for BE and endorsed for BE screening by the American Gastroenterological Association7 and was included as the control group in previous studies on molecular diagnosis of BE.10, 26
To discover miRNAs associated with BE pathogenesis, we previously sequenced the entire miRNA transcriptome of GERD and BE patients followed by verification of the sequencing results by quantitative real-time polymerase chain reaction (qRT-PCR) in 67 independent patients.20 These 67 patients formed the discovery cohort for the current study—40 with GERD (20 with EE and 20 with NERD) and 27 with BE.20 We identified 10 miRNAs (three up- and seven downregulated) that were differentially expressed between squamous and intestinal-type columnar epithelia.20 Select miRNAs were further evaluated for BE diagnosis in the current study. The miRNA selection criteria were as follows: (a) upregulation in BE compared with GERD, (b) high expression in BE tissues by next-generation sequencing and (c) high degree of differential expression between GERD and BE tissues. We restricted the analysis to the upregulated miRNAs because for non-endoscopic molecular methods, upregulated miRNAs are more likely to be useful for BE. We finally selected the following miRNAs for this study: miR-192-5p (240 611 reads per million (RPM) in BE, fold change 7.9 vs GERD), miR-215-5p (69250 RPM in BE, fold change 9.6 vs GERD), and miR-194-5p (8 209 RPM in BE, fold change 6.5 vs GERD), respectively.
We first calculated the sensitivity and specificity of the above miRNAs for BE diagnosis in the initial discovery cohort of 67 patients and then confirmed the results in an independent validation cohort.
Additional comparisons to test specificity of the BE miRNA signature
To test specificity, we also tested miR-192-5p, -215-5p, and -194-5p in two additional epithelia:
Columnar lined epithelium in the distal esophagus but without intestinal metaplasia (columnar lined esophagus-no IM). These patients were selected from the aforementioned repository. All of these patients underwent four-quadrant standard-of-care clinical biopsies every 1–2 cm. Additional research biopsies were obtained. Columnar lined esophagus-no IM patients were defined by the lack of intestinal metaplasia in both clinical and research biopsies.
Gastric cardia epithelium. Given that esophageal sampling devices may obtain cells from gastric cardia, we also evaluated gastric cardia biopsies to test the specificity of miRNA markers for BE epithelium. Mucous columnar cells without goblet cells confirmed gastric cardia epithelium in the H&E staining of biopsies taken adjacent to the research biopsies.
Quantitative real-time polymerase chain reaction
RNA isolation was performed by the TRIzol method (Sigma, St Louis, MO, USA). Quantitative reverse transcription polymerase chain reactions were performed as previously described20 by research associate (XH) blinded to the clinical diagnosis. In brief 50 ng total RNA was reverse-transcribed using the Universal RT primer system (Exiqon, Denmark) or stem-loop primers from Applied Biosystems (Grand Island, NY, USA). An RT-negative control was included in each reaction. Each RT reaction was then run in triplicate qPCR reactions, and the average threshold cycle for that sample (if variation was <0.3 cycles) was used in all subsequent calculations. If qPCR variation exceeded 0.3 Ct, then the sample was re-analyzed. Small nucleolar RNAU6B was used to normalize for RT and RNA quantification differences. Relative fold changes were determined by the delta Ct method using the formula 2−dCt (Ct miRNA-Ct RNAU6B). Calibrator samples were included across all PCR reactions, and all values were adjusted based on the Ct values for the calibrator samples across PCR experiments. In two patients, PCR reactions did not amplify the template for miR-192-5p and -215-5p in any of the triplicate reactions and were excluded from the final analysis.
Statistical analysis
Receiver operating curves were generated to first calculate the sensitivity and specificity of the above miRNAs for BE diagnosis in the initial discovery cohort of 67 patients followed by confirmation in an independent validation cohort. Receiver operating curves with 95% confidence intervals were computed by logistic regression models using fold changes. Interactions between miRNA panels were also tested to improve diagnostic accuracy. To test reliability, bootstrap analysis was performed, drawing 1000 random samples with replacement, and receiver operating curves values with 95% confidence intervals were recalculated.27, 28 Bootstrapping is a technique used to estimate variance in studies with small sample sizes by resampling the original population multiple times and calculating the distribution of means. BE miRNA profiles were compared with the published profiles of normal human stomach (n=5),21 normal mouse small intestine (n=7),22 and normal mouse large intestine (n=6),22 which is reasonable because miRNA expression is highly conserved across species.29 Because the published studies used different miRBase versions (13.0–18.0), the analysis was restricted to those miRNA included in all of the versions. MicroRNA expression (RPM) was log transformed. Pearson's correlation coefficients with 95% confidence intervals were calculated.
Results
The discovery cohort included 40 GERD (20 NERD and 20 EE) and 27 BE patients as described.20 The validation cohort included 28 white male patients. For validation, GERD was defined by the presence of EE. All GERD patients had EE grades B or higher. The mean age for patients with GERD (n=17) was 59±7 years and for patients with BE (n=11) was 61±6 years. The average BE length was Prague C2.7M4.5 cm (range C0-6 M2-9 cm). The characteristics of GERD and BE patients in the discovery and validation cohorts are compared in Supplementary Table 1 online.
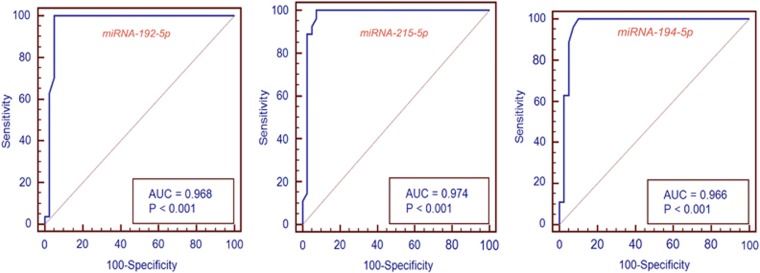
In the discovery cohort, the sensitivity and specificity of miRNAs for BE diagnosis were as follows: miR-192-5p, 100 and 95%, P=0.0004; miR-215-5p, 100 and 93%, P=0.0004; and miR-194-5p, 97 and 92%, P=0.0001) (Figure 1). Bootstrap analysis did not substantially alter the results (Table 1). Individual comparisons of the EE and NERD groups with the BE group revealed a similar accuracy for BE diagnosis (Table 2 and Supplementary Figure 1). In the validation cohort, the sensitivity and specificity were as follows: miR-192-5p, 92 and 94%, area under the curve (AUC) 0.94 (0.80–0.99, P=0.0004); miR-215-5p, 100 and 94%, AUC 0.98 (0.84–1, P=0.0004); and miR-194-5p, 91 and 94%, AUC 0.96 (0.80–0.99, P=0.0001) (Figure 2). Combining miRNAs into panels did not further improve accuracy in the discovery (AUC 0.97–0.99) or the validation cohorts (AUC 0.96–0.99).
Figure 1.
Receiver operating curves for miRNAs -192-5p, -215-5p, and -194-5p in the discovery cohort. The graphs show a near complete area under the curve for BE diagnosis for all the three miRNAs.
Table 1. Bootstrap analysis of the ROC curves in the discovery cohort.
| miRNA | ROC | ROC bootstrap |
|---|---|---|
| miR-192-5p | 0.97 (0.89–1) | 0.97 (0.91–1) |
| miR-215-5p | 0.97 (0.90–0.99) | 0.98 (0.92–1) |
| miR-194-5p | 0.96 (0.89–0.99) | 0.97 (0.91–1) |
ROC, receiver operating curve.
Table 2. Performance characteristics of the miRNAs for BE vs EE and NERD groups.
| miRNA | BE vs EE | BE vs NERD |
|---|---|---|
| miR-192-5p | ||
| Sensitivity | 100% | 97% |
| Specificity | 95% | 95% |
| AUC (95% CI) | 0.98 (0.89–1.0) | 0.95 (0.85–0.99) |
| miR-215-5p | ||
| Sensitivity | 100% | 100% |
| Specificity | 100% | 85% |
| AUC (95% CI) | 1.0 (0.93–1.0) | 0.95 (0.84–0.99) |
| miR-194-5p | ||
| Sensitivity | 95% | 96% |
| Specificity | 95% | 90% |
| AUC (95% CI) | 0.98 (0.89–1.0) | 0.95 (0.85–0.99) |
BE, Barrett's esophagus; CI, confidence interval; EE, erosive esophagitis; NERD, non-erosive reflux disease.
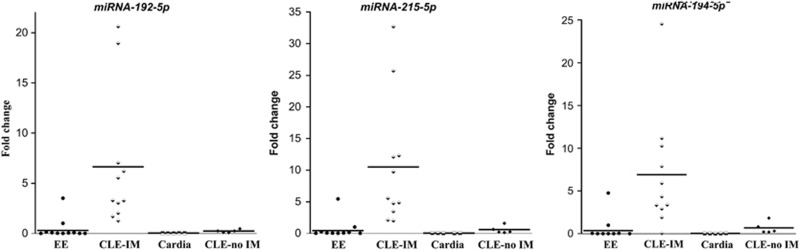
Figure 2.
Scatter plots for miRNAs -192-5p, -215-5p, and -194-5p in the validation cohort. Four different types of epithelia were compared: squamous mucosa from GERD patients with erosive esophagitis (EE), columnar lined esophagus with and without intestinal metaplasia (CLE-IM and CLE-no IM that represent two definitions of BE) and gastric cardia epithelium. All miRNAs show nice separation of BE defined by intestinal metaplasia from other epithelia with miRNA-215-5p detecting all BE patients. The solid line represents median values.
To test specificity, we compared the miRNA expression of intestinal-type columnar epithelium that defined BE in the current study with (a) gastric cardia epithelium (all white males, aged 61±4 years) and (b) non intestinal-type columnar epithelium from columnar lined esophagus-no IM patients (all white males, aged 59±6 years, columnar lined esophagus length 1.3±0.5 cm) (Figure 2). We found that miR-192-5p, -194-5p, and -215-5p effectively distinguished gastric cardia epithelium and non-intestinal-type columnar epithelium from intestinal-type columnar epithelium (Figure 2).
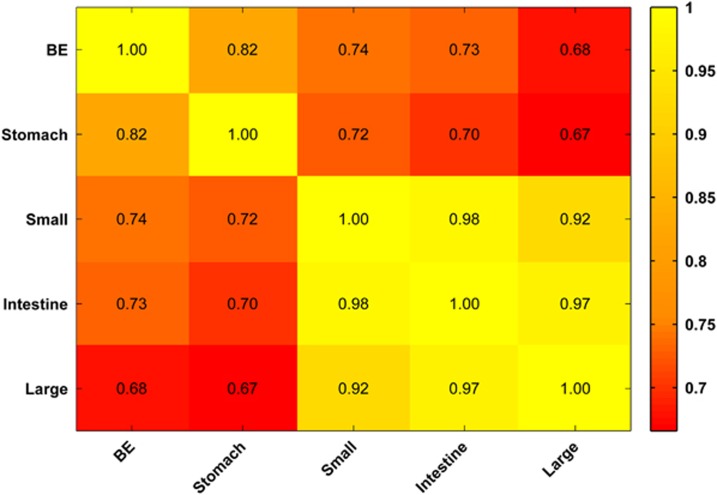
Controversy continues regarding the requirement of intestinal versus non-intestinal-type columnar epithelium (commonalities with gastric epithelium) to define BE.30, 31 To gain insights into the origin of BE, we compared the global miRNA sequencing profiles of BE mucosa20 with the published miRNA sequencing profiles of other columnar epithelia in the gastrointestinal tract—normal stomach,21 small intestine22 and large intestine.22 Globally, there was an incremental reduction in the correlation of BE miRNA expression with miRNA expression in the stomach, small intestine and large intestine (r=0.82>r=0.74>r=0.68, respectively), with a gradient in miRNA expression following proximity to the esophagus (Figure 3). When we compared the three miRNAs of interest, we found that miR-192-5p was highly expressed across all the tissue types (>40 000 RPM). MicroRNA-215-5p was highly abundant in BE (69 250 RPM) and was also abundant in the small and large intestines (>50 000 RPM in both) but was minimally expressed in the stomach (200 RPM). MicroRNA-194-5p was expressed similarly among BE, small intestine and large intestine (8209, 5225, and 4256, respectively) but was minimally expressed in the stomach (178 RPM). Thus, miR-215-5p and -194-5p were specific to intestinal-type epithelia.
Figure 3.
Heatmap displays the correlation coefficients of global miRNA sequencing profiles of BE epithelia compared with those of normal gastric, small intestinal and large intestinal epithelia. Under 'intestine', the miRNA profiles of the small and the large intestine were combined for analysis. Although there is a gradient in correlation as a function of the proximity of the organs, global miRNA profiles of various columnar epithelia are quite similar.
Discussion
We previously profiled miRNA expression in GERD and BE epithelia to identify the miRNAs with a potential role in BE pathogenesis.20 The current study focused on the clinical utility of specific miRNAs for the expedient diagnosis of BE. For several reasons, non-endoscopic molecular screening of BE32 with accurate biomarkers is feasible and could become a novel, effective approach to control the rapidly increasing rates of EAC. First, BE is the primary premalignant condition that predisposes patients to EAC and clearly identifies the at risk population, and second, two-thirds of all EACs are diagnosed by the index procedure.9 Thus, timely BE diagnosis could have the added value of EAC detection. Third, database studies suggest that if there is no BE on the initial endoscopy, it is unlikely to develop subsequently.33 Therefore, a single test at an appropriate age may suffice. In well-defined patients, we found multiple miRNAs, miR-192-5p, -194-5p, 215-5p to have an excellent accuracy for BE diagnosis in the discovery and validation cohorts (91–100% sensitivity and >90% specificity). We also found that the BE miRNA signature was specific to the intestinal-type columnar epithelium compared with the non-intestinal-type columnar epithelium and gastric cardia epithelium. The results were similar when NERD and EE groups were individually compared with BE, which suggests that miRNA expression is a strong indicator of epithelial type in the esophagus and is unlikely to be confounded by the presence of EE in the GERD group. In addition, comparisons of BE miRNA sequencing data with published data sets of other columnar epithelia in the gastrointestinal tract confirmed the specificity of two of the three miRNAs (-215-5p, -194-5p) to the intestinal-type epithelium. The results of this feasibility study strongly support the inclusion of miRNAs as candidate biomarkers in future larger-scale studies for the molecular diagnosis of BE. Our findings can be directly translated to non-invasive screening by using non-endoscopic cytology sponges that are highly effective at acquiring cells.10 Combination of highly specific markers with esophageal sampling techniques that do not require endoscopy can be applied in the primary care and community settings and will provide a practical, convenient and accurate method of BE diagnosis.
Other investigators have evaluated miRNA differences between squamous and Barrett's epithelia.15, 16, 17, 18, 19 Our results are consistent with the study by Fassan et al., who found miRs -192, -215, and -194 to be progressively upregulated along the spectrum of squamous/gastric metaplasia/intestinal metaplasia. We present additional comparisons with normal gastric cardia and small and large intestinal epithelium, and we confirmed the signature to be specific for intestinal-type BE. In additional, miR-192 was associated with Barrett's metaplasia in another study.16 The miRNAs described here are different than the study by Garman et al.18 possibly due to differences in methodology (microarray vs sequencing). Future studies should comprehensively compare the multiple miRNAs identified by various studies. Importantly, none of the studies quantitatively describe the diagnostic utility of miRNAs for BE, sometimes due to a lack of independent groups. In the absence of quantitative data, it is difficult to recognize the clinical utility of miRNA for BE diagnosis. We present quantitative data on the ability of miRNAs to diagnose BE; these data are needed to facilitate the design and sample size calculations of future prospective clinical trials and to move the field of BE molecular diagnosis forward. Another application of our results would be the reduction of diagnostic confusion that occurs with shorter lengths of BE. Because goblet cell distribution is patchy and can be missed, especially with shorter lengths, our results argue that miRNA expression might be used to molecularly confirm intestinal-type BE. Further confirmation is needed.
The feasibility of the molecular diagnosis of BE on cytology specimens was recently demonstrated by testing for Trefoil Factor 3, with a sensitivity and specificity of 73% and 94%, respectively for BE longer than 1 cm.10 The investigators used immunohistochemistry, a widely available technique in clinical laboratories. Given that miRNAs are disease specific34 and that the miRNAs tested in the current study identified all BE patients both in the discovery and validation cohorts and were specific to intestinal-type columnar epithelium, miR-192-5p, -215-5p, -194-5p could improve upon these accuracy rates and should be further validated in cytology specimens. Other studies that performed genome-wide methylation analysis showed the methylation signature to discriminate well between squamous and columnar (BE) epithelia.35 Prospective trials are needed to compare the accuracy, variability and reproducibility of miRNA versus methylation signatures for BE detection. An important question is how these results can be applied in clinical practice. PCR-based detection has revolutionized the rapid diagnosis of Clostridium Difficile and is being used widely.36 We evaluated miRNA expression based on relative fold changes. However, we envision that future advancements in PCR technology such as digital PCR37 will provide an absolute copy number (rather than relative fold change) for the miRNA of interest, which could then be used to develop normative values similarly to clinically used laboratory tests, for example, liver function tests.
Histopathologically, the BE epithelium is comprised of multiple cell types with resemblance to both gastric and intestinal epithelia.30 A recent study by Lavery et al. using histochemical and mitochondrial DNA mutational analysis argued that Barrett's glands resemble pyloric-type gastric glands with intestinal differentiation over time.31 Other data suggest that the differentiation into non-specialized columnar metaplasia vs intestinal metaplasia is modulated by interactions between multiple molecular factors, including pSMAD/CDX2.38 Analysis in the current study showed the global miRNA profiles of BE mucosa overlapped with those of normal gastric, small intestinal and large intestinal mucosa. However, the expression of specific miRNAs such as -215-5p and -194-5p was restricted to the intestinal epithelium and may be an important determinant of the epithelial phenotype; further study is warranted. MicroRNAs -192, -215, -194 coordinately regulate important targets in cell cycle regulation such as Smad interacting protein 1, ZEB2,39 and p5340, 41 and may help us better understand the molecular pathways driving the 'intestinalization' of columnar epithelia in BE with a role in BE development. Other miRNAs may have a role in BE carcinogenesis. MicroRNAs -221 and -222 were recently shown to be upregulated in EAC and were activated by FXR, a ligand for bile acids.42 These miRNAs promoted degradation of cdx2 that is important for intestinal differentiation and their inhibition could have a role in management of BE and EAC. MicroRNAs can be therapeutically modulated relatively easily with limited toxicity.43 Thus, miRNA-based therapy could have a role in chemoprevention and cancer treatment in BE and EAC, respectively.44
Our study has several limitations but made strong and novel observations. We included patients with GERD as the control group instead of healthy volunteers. This study design is clinically relevant because GERD identifies the population at highest risk for BE, and the American Gastroenterological Association guidelines recommend that this population undergo BE screening.7 For practical purposes, molecular testing for BE may need to begin with patients at the highest risk for BE, that is, those with GERD, prior to widespread population testing. Also, our design is consistent with previous studies on molecular diagnosis of BE, which defined controls based on the history of reflux.10, 26 Multiple questionnaires are available to define GERD.23, 45, 46, 47 We used a validated questionnaire that we have previously used48, 49 to define GERD. This questionnaire has been shown to be reliable on test-retest procedure with a median k statistic for the symptom items of 0.71 (interquartile range, 0.63–0.81).23 We included only well-defined BE patients with BE lengths greater than 2 cm, and thus the results may be different for shorter lengths of BE. The sample sizes were relatively small, but the intent was to conduct a feasibility study prior to large-scale studies. We performed bootstrap analysis to statistically test the impact of small sample sizes on our results. The narrow distribution of confidence intervals after bootstrapping underscores the reliability of our results (Table 1). A comparison of the BE miRNA signature with the columnar epithelia of multiple gastrointestinal organs confirmed specificity and is a strength of the study. Given that the results were compelling and consistent between discovery and validation cohorts, this study provides a strong rationale to conduct prospective studies to confirm these observations in cytology specimens.
In conclusion, the BE miRNA signature has shown significant promise and should be evaluated further as a specific marker for the presence of intestinal-type BE; this signature has the potential to improve our understanding of BE pathogenesis.
Study Highlights
Acknowledgments
We would like to acknowledge the meticulous management of the tissue and serum repository by Tracy Shipe, MS.
Guarantors of the article: Ajay Bansal, MD and Lane K. Christenson, PhD.
Specific author contributions: A.B. conceived of the study, participated in its design and coordination and drafted the manuscript, X.H. performed the RNA isolation and polymerase chain reaction (PCR) experiments and has extensive experience with these techniques, I.H.L. performed the sequence alignment, K.K.K. contributed gastric cardia biopsies and critically reviewed the manuscript, S.C.M. reviewed all the biopsies to confirm the diagnosis of Barrett's esophagus and provided important feedback, S.G. did the statistical analysis, A.R. contributed to the repository and provided important feedback, P.S. made significant contributions to the repository and critically reviewed the manuscript to improve the composition of the manuscript, L.K.C. participated in the design of the study, supervised the PCR experiments and helped to draft the manuscript. All authors read and approved the final manuscript.
Financial support: The current work was supported by a pilot grant from the American Cancer Society (A.B. and L.K.C.), the American College of Gastroenterology Junior Faculty Development Award (A.B.) and grants from Hall Family Foundation (L.K.C.) and Kansas IDeA Network of Biomedical Research Excellence (A.B., L.K.C.). None of the funding bodies had any role in design, in the collection, analysis and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
Potential competing interests: A provisional patent application on using microRNA for BE diagnosis (A.B., P.S., L.K.C.) has been filed. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Supplementary Material
References
- Shaheen NJ. What is behind the remarkable increase in esophageal adenocarcinoma. Am J Gastroenterol. 2014;109:345–347. doi: 10.1038/ajg.2014.35. [DOI] [PubMed] [Google Scholar]
- Dubecz A, Gall I, Solymosi N, et al. Temporal trends in long-term survival and cure rates in esophageal cancer: a SEER database analysis. J Thorac Oncol. 2012;7:443–447. doi: 10.1097/JTO.0b013e3182397751. [DOI] [PubMed] [Google Scholar]
- Cen P, Banki F, Cheng L, et al. Changes in age, stage distribution, and survival of patients with esophageal adenocarcinoma over three decades in the United States. Ann Surg Oncol. 2012;19:1685–1691. doi: 10.1245/s10434-011-2141-1. [DOI] [PubMed] [Google Scholar]
- Prasad GA, Wu TT, Wigle DA, et al. Endoscopic and surgical treatment of mucosal (T1a) esophageal adenocarcinoma in Barrett's esophagus. Gastroenterology. 2009;137:815–823. doi: 10.1053/j.gastro.2009.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley DA, Levin TR, Habel LA, et al. Surveillance and survival in Barrett's adenocarcinomas: a population-based study. Gastroenterology. 2002;122:633–640. doi: 10.1053/gast.2002.31879. [DOI] [PubMed] [Google Scholar]
- Shaheen NJ, Hur C. Garlic, silver bullets, and surveillance upper endoscopy for Barrett's esophagus. Gastroenterology. 2013;145:273–276. doi: 10.1053/j.gastro.2013.06.028. [DOI] [PubMed] [Google Scholar]
- American Gastroenterological Association. Spechler SJ, Sharma P, Souza RF, Inadomi JM, Shaheen NJ. American Gastroenterological Association Medical Position Statement on the Management of Barrett's Esophagus. Gastroenterology. 2011;140:1084–1091. doi: 10.1053/j.gastro.2011.01.030. [DOI] [PubMed] [Google Scholar]
- Wang KK, Sampliner RE. Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett's esophagus. Am J Gastroenterol. 2008;103:788–797. doi: 10.1111/j.1572-0241.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- Sharma P, Falk GW, Weston AP, et al. Dysplasia and cancer in a large multicenter cohort of patients with Barrett's esophagus. Clin Gastroenterol Hepatol. 2006;4:566–572. doi: 10.1016/j.cgh.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Kadri SR, Lao-Sirieix P, O'Donovan M, et al. Acceptability and accuracy of a non-endoscopic screening test for Barrett's oesophagus in primary care: cohort study. BMJ. 2010;341:c4372. doi: 10.1136/bmj.c4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Chen NY, Cui RX, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13:633–641. doi: 10.1016/S1470-2045(12)70102-X. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang L, Bayaxi N, et al. A microRNA panel to discriminate carcinomas from high-grade intraepithelial neoplasms in colonoscopy biopsy tissue. Gut. 2013;62:280–289. doi: 10.1136/gutjnl-2011-301554. [DOI] [PubMed] [Google Scholar]
- Cortez MA, Bueso-Ramos C, Ferdin J, et al. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nature reviews. Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassan M, Volinia S, Palatini J, et al. MicroRNA Expression Profiling in the Histological Subtypes of Barrett's Metaplasia. Clin Transl Gastroenterol. 2013;4:e34. doi: 10.1038/ctg.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzna P, Gregar J, Uberall I, et al. Changes of microRNAs-192, 196a and 203 correlate with Barrett's esophagus diagnosis and its progression compared to normal healthy individuals. Diagn Pathol. 2011;6:114. doi: 10.1186/1746-1596-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streppel MM, Pai S, Campbell NR, et al. MicroRNA 223 is upregulated in the multistep progression of Barrett's esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin Cancer Res. 2013;19:4067–4078. doi: 10.1158/1078-0432.CCR-13-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garman KS, Owzar K, Hauser ER, et al. MicroRNA expression differentiates squamous epithelium from Barrett's esophagus and esophageal cancer. Dig Dis Sci. 2013;58:3178–3188. doi: 10.1007/s10620-013-2806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidner RS, Ravi L, Leahy P, et al. The microRNAs, MiR-31 and MiR-375, as candidate markers in Barrett's esophageal carcinogenesis. Genes Chromosomes Cancer. 2012;51:473–479. doi: 10.1002/gcc.21934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A, Lee IH, Hong X, et al. Discovery and validation of Barrett's esophagus MicroRNA transcriptome by next generation sequencing. PLoS One. 2013;8:e54240. doi: 10.1371/journal.pone.0054240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-dos-Santos A, Khayat AS, Silva A, et al. Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach. PLoS One. 2010;5:e13205. doi: 10.1371/journal.pone.0013205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna LB, Schug J, Vourekas A, et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function Gastroenterology 20101391654–1664.1664 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke GR, Talley NJ, Weaver AL, et al. A new questionnaire for gastroesophageal reflux disease. Mayo Clin Proc. 1994;69:539–547. doi: 10.1016/s0025-6196(12)62245-9. [DOI] [PubMed] [Google Scholar]
- Sharma P, Dent J, Armstrong D, et al. The development and validation of an endoscopic grading system for Barrett's esophagus: the Prague C & M criteria. Gastroenterology. 2006;131:1392–1399. doi: 10.1053/j.gastro.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Montgomery E, Bronner MP, Goldblum JR, et al. Reproducibility of the diagnosis of dysplasia in Barrett esophagus: a reaffirmation. Hum Pathol. 2001;32:368–378. doi: 10.1053/hupa.2001.23510. [DOI] [PubMed] [Google Scholar]
- Lao-Sirieix P, Boussioutas A, Kadri SR, et al. Non-endoscopic screening biomarkers for Barrett's oesophagus: from microarray analysis to the clinic. Gut. 2009;58:1451–1459. doi: 10.1136/gut.2009.180281. [DOI] [PubMed] [Google Scholar]
- Efron BT, Tibshirani RJ. An Introduction to the Bootstrap. Chapman & Hall, Inc: New York; 1993. [Google Scholar]
- Steyerberg EW, Harrell FE, Jr, Borsboom GJ, et al. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J Clin Epidemiol. 2001;54:774–781. doi: 10.1016/s0895-4356(01)00341-9. [DOI] [PubMed] [Google Scholar]
- Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- Odze R, Goldblum JR. Surgical Pathology of the GI Tract, Liver, Biliary Tract and Pancreas. 2nd edition. Philadelphia; Elsiever; 2009. [Google Scholar]
- Lavery DL, Nicholson AM, Poulsom R, et al. The stem cell organisation, and the proliferative and gene expression profile of Barrett's epithelium, replicates pyloric-type gastric glands. Gut. 2014;63:1854–1863. doi: 10.1136/gutjnl-2013-306508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao-Sirieix P, Fitzgerald RC. Screening for oesophageal cancer. Nature reviews. Clin Oncol. 2012;9:278–287. doi: 10.1038/nrclinonc.2012.35. [DOI] [PubMed] [Google Scholar]
- Stoltey J, Reeba H, Ullah N, et al. Does Barrett's oesophagus develop over time in patients with chronic gastro-oesophageal reflux disease. Aliment Pharmacol Ther. 2007;25:83–91. doi: 10.1111/j.1365-2036.2006.03138.x. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu E, Gu J, Hawk ET, et al. Genome-wide methylation analysis shows similar patterns in Barrett's esophagus and esophageal adenocarcinoma. Carcinogenesis. 2013;34:2750–2756. doi: 10.1093/carcin/bgt286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont HL.Diagnosis and management of Clostridium difficile infection Clin Gastroenterol Hepatol 2013111216–1223.quiz e73. [DOI] [PubMed] [Google Scholar]
- Bizouarn F. Clinical applications using digital PCR. Methods Mol Biol. 2014;1160:189–214. doi: 10.1007/978-1-4939-0733-5_16. [DOI] [PubMed] [Google Scholar]
- Mari L, Milano F, Parikh K, et al. A pSMAD/CDX2 complex is essential for the intestinalization of epithelial metaplasia. Cell Rep. 2014;7:1197–1210. doi: 10.1016/j.celrep.2014.03.074. [DOI] [PubMed] [Google Scholar]
- Khella HW, Bakhet M, Allo G, et al. miR-192, miR-194 and miR-215: a convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis. 2013;34:2231–2239. doi: 10.1093/carcin/bgt184. [DOI] [PubMed] [Google Scholar]
- Braun CJ, Zhang X, Savelyeva I, et al. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichiorri F, Suh SS, Rocci A, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18:367–381. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Matsuzaki J, Suzuki H, Tsugawa H, et al. Bile acids increase levels of microRNAs 221 and 222, leading to degradation of CDX2 during esophageal carcinogenesis. Gastroenterology. 2013;145:1300–1311. doi: 10.1053/j.gastro.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Bostrom P, Frisen J. New cells in old hearts. N Engl J Med. 2013;368:1358–1360. doi: 10.1056/NEJMcibr1300157. [DOI] [PubMed] [Google Scholar]
- Matsuzaki J, Suzuki H. MicroRNAs in Barrett's esophagus: future prospects. Front Genet. 2014;5:69. doi: 10.3389/fgene.2014.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R, Junghard O, Dent J, et al. Development of the GerdQ, a tool for the diagnosis and management of gastro-oesophageal reflux disease in primary care. Aliment Pharmacol Ther. 2009;30:1030–1038. doi: 10.1111/j.1365-2036.2009.04142.x. [DOI] [PubMed] [Google Scholar]
- Shaw MJ, Talley NJ, Beebe TJ, et al. Initial validation of a diagnostic questionnaire for gastroesophageal reflux disease. Am J Gastroenterol. 2001;96:52–57. doi: 10.1111/j.1572-0241.2001.03451.x. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Matsuzaki J, Okada S, et al. Validation of the GerdQ questionnaire for the management of gastro-oesophageal reflux disease in Japan. United European Gastroenterol J. 2013;1:175–183. doi: 10.1177/2050640613485238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Wani S, Bansal A, et al. A feasibility trial of narrow band imaging endoscopy in patients with gastroesophageal reflux disease Gastroenterology 2007133454–464.quiz 674. [DOI] [PubMed] [Google Scholar]
- Balasubramanian G, Singh M, Gupta N, et al. Prevalence and predictors of columnar lined esophagus in gastroesophageal reflux disease (GERD) patients undergoing upper endoscopy. Am J Gastroenterol. 2012;107:1655–1661. doi: 10.1038/ajg.2012.299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.