Abstract
Autophagy is a conserved lysosomal self-digestion process used for the breakdown of long-lived proteins and damaged organelles, and it is associated with a number of pathological processes, including cancer. Phospholipase D (PLD) isozymes are dysregulated in various cancers. Recently, we reported that PLD1 is a new regulator of autophagy and is a potential target for cancer therapy. Here, we investigated whether PLD2 is involved in the regulation of autophagy. A PLD2-specific inhibitor and siRNA directed against PLD2 were used to treat HT29 and HCT116 colorectal cancer cells, and both inhibition and genetic knockdown of PLD2 in these cells significantly induced autophagy, as demonstrated by the visualization of light chain 3 (LC3) puncta and autophagic vacuoles as well as by determining the LC3-II protein level. Furthermore, PLD2 inhibition promoted autophagic flux via the canonical Atg5-, Atg7- and AMPK-Ulk1-mediated pathways. Taken together, these results suggest that PLD2 might have a role in autophagy and that its inhibition might provide a new therapeutic basis for targeting autophagy.
Introduction
Autophagy is an essential lysosomal degradation pathway that eliminates protein aggregates and damaged organelles from the cytoplasm to maintain cytoplasmic quality and homeostasis.1 Autophagy is promoted in response to physiological conditions, such as starvation, and in response to diverse pathological stresses, including the formation of protein aggregates or infection, thus allowing cells to adapt to environmental and developmental changes. Autophagy is initiated by the formation of double membrane-bound vesicles, called autophagosomes, which sequester cytoplasmic material in a non-degradative compartment.2 The final stage of autophagy, the fusion of autophagosomes with lysosomes, allows the degradation of autophagic cargo and the subsequent recycling of nutrients and membranes. Autophagy may be dysregulated in several disorders, including metabolic diseases, neurodegenerative disorders, infectious diseases and cancer. The prevailing view is that autophagy functions both as a tumor suppressor pathway that prevents tumor initiation and as a pro-survival pathway that helps tumor cells endure metabolic stress and resist death triggered by chemotherapeutic agents.3 Certain agents that are used as a preventive form of cancer therapy induce autophagy.4, 5 Several signaling molecules and cascades modulate autophagy in response to numerous cellular and environmental cues. The best-characterized modulator of autophagy is mammalian target of rapamycin (mTOR), which negatively regulates autophagy by inhibiting the ULK1 complex through direct phosphorylation and is inhibited by rapamycin, which induces autophagy.6 Activation of mTOR requires the lipid second messenger phosphatidic acid (PA), which binds to mTOR.7, 8 Phospholipase D (PLD) hydrolyzes phospholipids to generate PA, which is known to provide survival signals.9 Phosphatidylcholine-specific PLD1 and PLD2 are the classic mammalian isoforms of PLD,10, 11 and elevated expression and activity of PLDs have been detected in various human cancers.9 Recently, we reported PLD1 as a new regulator of autophagy.12 PLD1 suppresses autophagy, and inhibition of PLD1 promotes autophagy.12 Furthermore, the PLD2 expression level is correlated with tumor size and survival of patients with colorectal carcinoma, indicating that it might be a prognostic marker in colorectal cancers.13 In addition, elevated expression of PLD2 in low invasive breast cancer cells has been shown to induce a highly aggressive phenotype, with primary tumors that form following xenotransplantation being larger, growing faster and developing lung metastases more readily.14 In the present study, we demonstrate that PLD2 inhibition induces autophagy.
Materials and methods
Cell culture and reagents
HT29 and HCT116 human colorectal cancer cells were maintained in DMEM with 10% fetal bovine serum and were incubated at 37 °C in a humidified atmosphere of 5% CO2. Atg5+/+ and Atg5−/− RasV12/T-large antigen mouse embryo fibroblasts (MEFs) were provided by Dr G. Velasco (School of Biology, Complutense University); Atg7+/+ and Atg7−/− MEFs were provided by Dr M Komatsu (Tokyo Metropolitan Institute of Medical Science); and Ulk1+/+ and Ulk1−/− MEFs were provided by Dr M Kundu (St Jude Children's Hospital). A PLD2 inhibitor (VU0285655-1) was purchased from Cayman Chemical (Ann Arbor, MI, USA), and bafilomycin A1 (BafA1) was obtained from Sigma (Beverly, MA, USA). An siRNA corresponding to the PLD2 sequence (5′-AAGAGGUGGCUGGUGGUGAAGdTdT-3′) was used.
Western blot analysis
Cell lysates were analyzed by immunoblotting, and enhanced chemiluminescence was used for signal detection. Antibodies against the following proteins were used: tubulin (Sigma), green fluorescent protein (GFP) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), phospho-AMPK (Thr172, Cell Signaling, Bray, Ireland), AMPK (Cell Signaling), LC3B (Cell Signaling), phospho-ULK1 (Ser555/Ser757, Cell Signaling), ULK1 (Cell Signaling), ATG5 (Santa Cruz Biotechnology) and ATG7 (Abcam, Cambridge, MA, USA). Rabbit polyclonal anti-PLD antibody, which recognizes both PLD1 and PLD2, was generated as previously described.12
Autophagy analysis by light chain 3 monitoring
To monitor the formation of light chain 3 (LC3) puncta, cells were transiently transfected with red fluorescent protein (RFP)-LC3 and then cultured under nutrient starvation conditions such as on HBSS (Hank's Buffered Salt Solution; amino acid-free) medium. The cells were then fixed with 4% paraformaldehyde for fluorescence microscopy and visualized, and the images were collected using a fluorescence microscope (Axiovert200 M, Zeiss, Wetzlar, Germany). Quantification of autophagic vacuoles was analyzed by calculating the numbers of LC3 puncta (endogenous LC3, RFP-LC3, or RFP+/GFP-LC3 from some fields containing more than 3–5 randomly selected cells in the microscopy-captured images). RFP-LC3 was provided by Dr Colombo (Universidad Nacional de Cuyo, Mendoza, Argentina), and monomeric RFP (mRFP)-GFP-LC3 was provided by Dr T Yoshimori (Research Institute for Microbial Diseases, Osaka University).
Electron microscopy
The sample preparation for electron microscopy was performed as described previously.12 The sample was pre-fixed with 2.5% glutaraldehyde (4 °C, phosphate buffer, pH 7.2) and post-fixed with 1% osmium tetroxide in the same buffer. The sample was then dehydrated with a series of graded ethyl alcohol and embedded in epoxy resin (Epon 812 mixture). Thick sections (1 μm) were stained with 1% toluidine blue for light microscopy. In addition, thin sections (50–60 nm) were prepared using an ultramicrotome (Leica, Reichert SuperNova, Heidelberg, Germany), double stained with uranyl acetate and lead citrate and then examined using a transmission electron microscope (JEM1200EX-II, JEOL, Tokyo, Japan).
Data analysis
The results are expressed as the means±s.d. of the determinations. The statistical significance of the differences was determined by ANOVA, and significance was accepted when the P-value was lower than 0.05.
Results
Knockdown of PLD2 induces autophagy
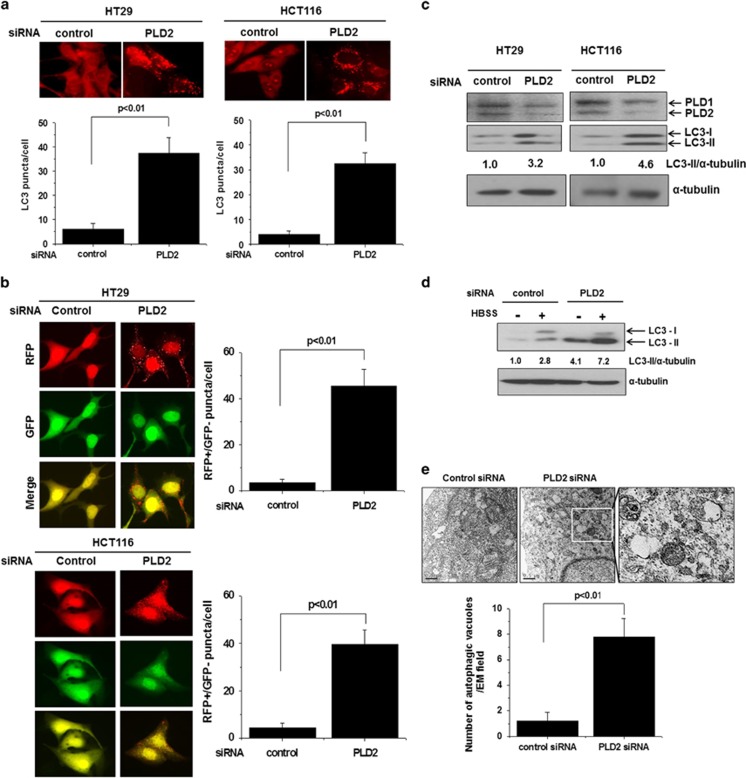
To investigate whether PLD2 is involved in the regulation of autophagy, we used RNA interference against PLD2. Autophagy mediates the bulk degradation of cytoplasmic components through the delivery of these components to lysosomes via autophagosomes. Microtubule-associated 1 LC3 localizes to autophagosomal membranes and is a specific marker for autophagosomes,15, 16 and the development of autophagy is frequently assessed by the number and intensity of LC3 vesicles.17, 18 We first assessed the effect of PLD2 depletion on autophagy. Depletion of PLD1 significantly induced autophagy in HT29 and HCT116 colorectal cancer cells, as indicated by punctate dots of endogenous LC3 (Figure 1a). Moreover, the maturation process through which autophagosomes are converted into autolysosomes can be monitored by the mRFP-GFP tfLC3 (tandem fluorescent-tagged LC3) method.19 The GFP-LC3 punctate feature of the mRFP-GFP tandem autophagy flux reporter was not detectable in the acidic pH lysosomal environment by lysosomal hydrolysis, whereas the mRFP-LC3 punctate feature was still detectable in this compartment. Depletion of PLD2 significantly increased the transition of RFP-GFP/LC3-positive autophagosomes to RFP-positive, GFP-negative autolysosomes in RFP-GFP/LC3-transfected cells (Figure 1b), suggesting that knockdown of PLD1 promotes autophagic flux. The C-terminal fragment of LC3 is cleaved immediately following its synthesis to yield a cytosolic form called LC3-І (18 kDa),17, 18 and a sub-population of LC3-І is further converted into an autophagosome-associating form, LC3-II (16 kDa),16, 20 which localizes to autophagosomal membranes and delivers degraded cytoplasmic components to lysosomes. To examine whether PLD2 knockdown induces autophagy in HT29 and HCT116 cells, we investigated the conversion of LC3 from I to II, a known autophagosome marker. Depletion of PLD2 in HT29 and HCT116 cells increased the LC-II levels and reduced the expression of PLD2, as analyzed by immunoblotting (Figure 1c). In addition, depletion of PLD2 increased the level of LC3-II induced by amino acid and serum starvation conditions (HBSS media) for 6 h (Figure 1d), suggesting that silencing of PLD2 increases both basal and starvation-induced autophagy. Moreover, PLD2 depletion revealed a significant increase in the number of autophagic vacuoles, as analyzed by transmission electron microscopy (Figure 1e). Taken together, these results suggest that depletion of PLD2 induces autophagy.
Figure 1.
Depletion of PLD2 induces autophagy. (a) HT29 and HCT116 cells were transfected with an siRNA directed against PLD2 and then immunostained with antibody specific for LC3. Endogenous LC3 punctate dots were observed by fluorescence microscopy, and the number of puncta per cell was quantified (7–10 cells were assessed). The data are representative of three independent experiments. (b) The cells were co-transfected with mRFP-GFP-LC3 and a PLD2-directed siRNA for 24 h, and the total number of RFP-positive/GFP-negative puncta per cell was counted. The data are representative of three independent experiments. (c) The cells were transfected with an siRNA directed against PLD2, and the lysates were immunoblotted with the indicated antibodies. The levels of LC3-II compared with that of α-tubulin were quantified using densitometer analysis. (d) HT29 cells were transfected with an siRNA directed against PLD2 and then cultured under amino acid and serum starvation conditions (HBSS media) for 6 h, after which the lysates were immunoblotted with the indicated antibodies. The levels of LC3-II compared with that of α-tubulin were quantified by densitometer analysis. (e) HT29 cells were transfected with the indicated siRNAs and then fixed and examined by transmission electron microscopy to detect autophagic vacuoles, which were counted in the field of view. Arrows indicate autophagic vacuoles. The values are the means±s.d. of three independent experiments.
Inhibition of PLD2 activity induces autophagy
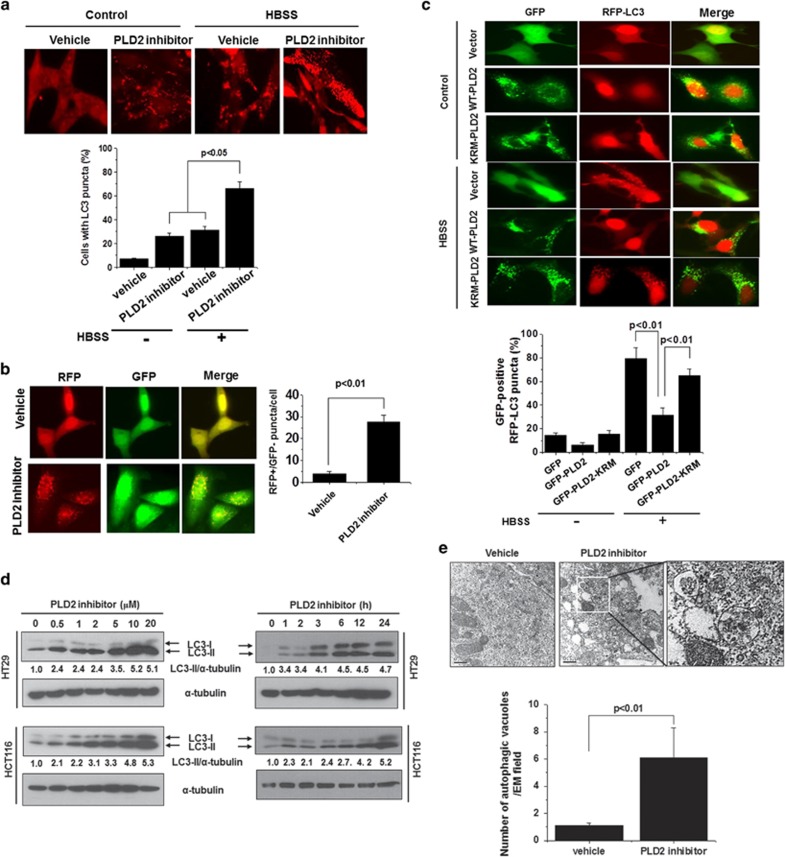
The PLD2 inhibitor VU0285655-1 is known to selectively inhibit PLD2.21 PLD2 inhibition significantly increased autophagy under basal conditions and starvation in HT29 cells, as indicated by punctate dots of endogenous LC3 (Figure 1a). We further examined the effect of VU0285655-1 on autophagy. HT29 cells were transfected with mRFP-GFP tandem LC3 and then treated with VU0285655-1. PLD2 inhibition significantly increased autophagy, as analyzed by mRFP-GFP tandem LC3 puncta (RFP-positive, GFP-negative puncta) formation (Figure 2b), suggesting that PLD2 inhibition promotes the on-rate of autophagy, at least in vitro, in cultured cells. To further examine whether PLD activity is involved in autophagy, we co-transfected RFP-LC3 with wild-type GFP-PLD2 or a catalytically inactive mutant form of PLD2, GFP-PLD2K758R(KRM-PLD2), in HT29 cells. Expression of PLD2wt, but not of PLD2K758R, significantly inhibited GFP-positive RFP-LC3 puncta formation induced by starvation (Figure 2c), suggesting the involvement of PLD2 activity in the regulation of autophagy. Under basal conditions, PLD2 inhibition increased the level of the LC3-II protein in a dose- and time-dependent manner in HT29 and HCT116 cells (Figure 2d). Furthermore, PLD2 inhibition significantly increased the formation of autophagic vacuoles, as analyzed by transmission electron microscopy (Figure 2e), suggesting that PLD2 inhibition induces autophagy.
Figure 2.
Inhibition of PLD2 activity induces autophagy. (a) HT29 cells were pre-treated with or without PLD2 inhibitor (10 μM) for 1 h and then cultured in normal or HBSS medium for 6 h. The cells were then fixed, permeabilized and stained with a Texas Red-conjugated antibody specific for endogenous LC3, and the number of LC3 puncta per cell was counted (7–10 cells were assessed). The data are representative of three independent experiments. (b) HT29 cells were transfected with mRFP-GFP-LC3 and then pre-treated with or without PLD2 inhibitor (10 μM) for 1 h, after which the cells were cultured in normal or HBSS medium for 6 h. Autolysosomes were quantified by counting the RFP-positive/GFP-negative puncta per cell (7–10 cells were counted). (c) HT29 cells were co-transfected with GFP vector, GFP-WT-PLD2 or GFP-KRM-PLD2 and RFP-LC3 and then cultured in HBSS or normal medium for 6 h, after which the percentage of RFP punctate cells relative to the percentage of GFP-expressing cells was determined. The data are representative of three independent experiments. (d) HT29 and HCT116 cells were treated with the indicated concentrations of PLD2 inhibitor for 12 h or with 10 μM of PLD2 inhibitor for the indicated time, and the lysates were immunoblotted with the indicated antibody. The level of LC3-II compared with that of α-tubulin was quantified using densitometer analysis. The data are representative of three independent experiments. (e) HT29 cells were treated with PLD2 inhibitor (10 μM) for 12 h and then fixed and examined by transmission electron microscopy to visualize autophagic vacuoles. The quantification was based on counting the autophagic vacuoles in the field of view. Arrows indicate autophagic vacuoles. The values are the means±s.d. of three independent experiments.
PLD2 inhibition induces autophagy via Atg5-, Atg7- and AMPK-Ulk1-mediated pathways
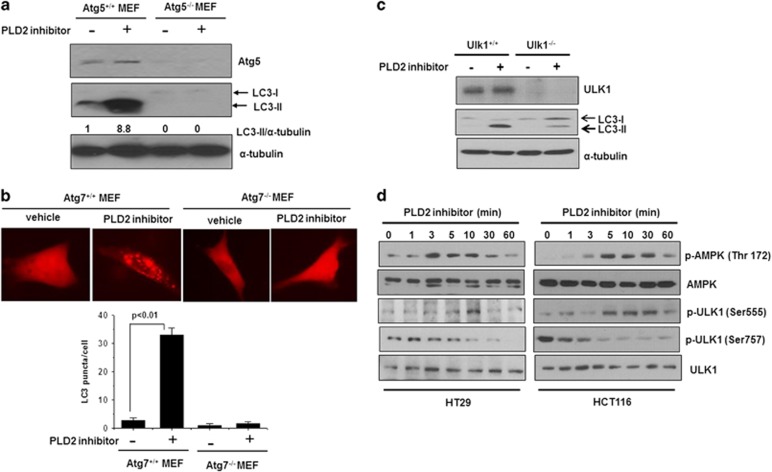
To investigate the regulation of autophagy pathways by PLD1, we examined whether PLD2 inhibition is dependent on Atg genes. A series of protein complexes composed of autophagy-related gene (Atg gene) products coordinate the formation of autophagosomes. In these experiments, we used isogenic MEFs, deficient in the essential autophagy genes, Atg5 or Atg7; these cells are completely defective for autophagy.22, 23 PLD2 inhibition increased the LC3-II protein level and the number of LC3 puncta in Atg5 and Atg7 MEFs, but the LC3 level and number of LC3 puncta induced by PLD2 inhibition were dramatically suppressed in Atg5 and Atg7 null MEFs (Figures 3a and b). The ATG1/ULK1 complex (Atg1 in yeast and ULK1 in mammals) is an essential positive regulator of autophagosome formation,24 and PLD1 inhibition dramatically suppressed the level of LC3-II in Ulk1 null MEFs (Figure 3c). Autophagy is controlled by several kinases, including mTOR, a negative regulator of autophagy,25 and monophosphate-activated protein kinase (AMPK), a positive regulator of autophagy.26 mTOR inhibits autophagy initiation by phosphorylating ULK1Ser75,6 and the cellular energy-sensing pathway that is controlled by AMPK is upstream of mTOR.27 Under starvation conditions, activated AMPK inhibits mTOR to relieve the phosphorylation of ULK1 on Ser757, leading to the ULK1-AMPK interaction.6 AMPK then phosphorylates ULK1 on Ser555, Ser317 and Ser777, activates ULK1 kinase, and eventually leads to the induction of autophagy.6 PLD2 inhibition increased both the phosphorylation of AMPK (Thr172) and the phosphorylation of ULK1 Ser555, which is mediated by AMPK, in a time-dependent manner, whereas PLD1 inhibition suppressed the phosphorylation of ULK1 Ser757 mediated by mTOR (Figure 3d). Collectively, these results suggest that PLD2 inhibition induces autophagy via canonical Atg5-, Atg7- and AMPK-Ulk1-mediated pathways.
Figure 3.
PLD2 inhibition induces autophagy via Atg5-, Atg7- and Ulk1-mediated pathways. (a) Atg5 MEFs were treated with or without PLD2 inhibitor (10 μM) for 6 h, and the lysates were immunoblotted with the indicated antibodies. The level of LC3-II compared with that of α-tubulin was quantified by densitometer analysis. The data are representative of three independent experiments. (b) Atg7+/+ and Atg7−/− MEFs were transfected with RFP-LC3 for 24 h and then treated with PLD2 inhibitor (10 μM) for 6 h. RFP-LC3 punctate dot cells were observed by fluorescence microscopy, and the number of RFP punctate cells was quantified. The values are the means±s.d. of three independent experiments. (c) Two isogenic MEF cell lines (Ulk1+/+ and Ulk1-null) were treated with PLD2 inhibitor (10 μM) for 6 h, and the lysates were immunoblotted with the indicated antibodies. The data are representative of three independent experiments. (d) HT29 and HCT116 cells were treated with PLD2 inhibitor (10 μM) for the indicated time, and the lysates were immunoblotted with the indicated antibodies. The data are representative of three independent experiments.
PLD2 inhibition increases the on-rate of autophagy
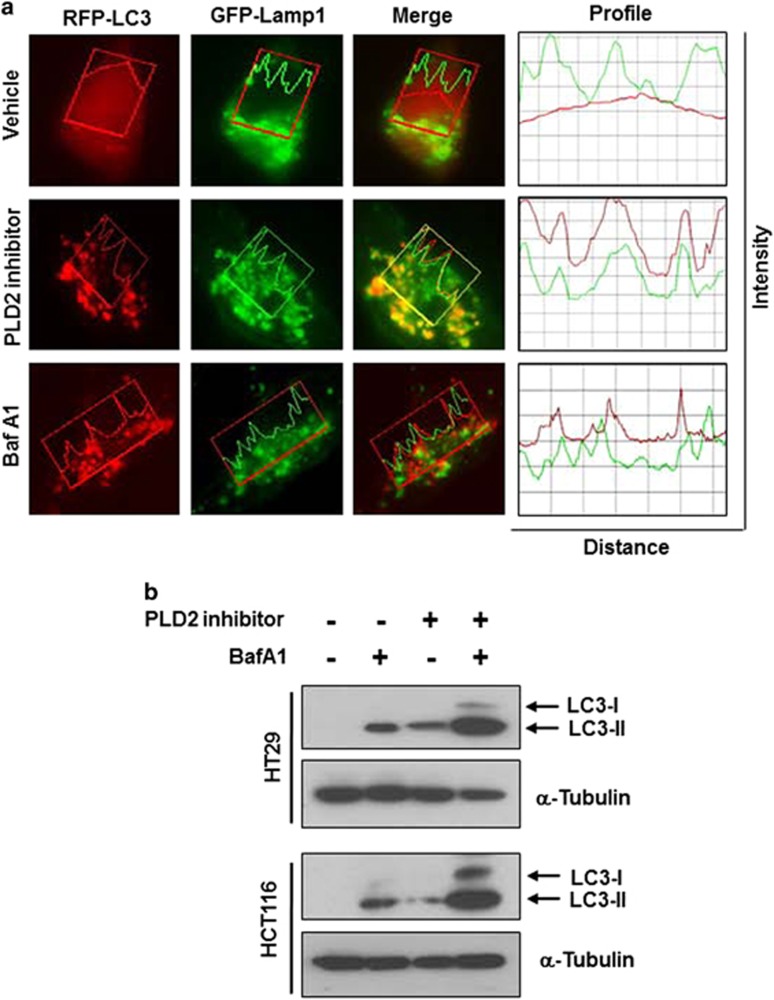
The accumulation of autophagosomes and autolysosomes by inhibiting or depleting PLD2 could involve enhanced autophagic sequestration (on-rate) or reduced degradation of autophagic material by fusion with lysosomes (off-rate). To distinguish between these possibilities, we assessed PLD2 inhibitor-induced autophagic vacuolization by monitoring the colocalization of an autophagic marker, GFP-LC3, with a lysosomal marker, Lamp-1, in the presence or absence of BafA1, which is known to inhibit autophagosome–lysosome fusion.28 The PLD2 inhibitor, but not BafA1, induced colocalization between Lamp-1 and LC3, suggesting that the autophagosome might encounter the lysosome in PLD2 inhibitor-treated HT29 cells (Figure 4a). Moreover, BafA1 further increased the PLD2 inhibitor-triggered induction of LC3-II in HT29 and HCT116 cells (Figure 4b). These results suggest that PLD2 inhibition promotes autophagic flux.
Figure 4.
PLD2 inhibition increases the on-rate of autophagy. (a) HT29 cells were co-transfected with RFP-LC3 and GFP-Lamp1 and then pre-treated with BafA1 (100 nM) or PLD2 inhibitor (10 μM) for 10 h, and the colocalization of RFP-LC3 and GFP-Lamp-1 was analyzed. Representative fluorescence microphotographs are shown together with the profiles of colocalization. (b) The cells were treated with PLD2 inhibitor and/or BafA1 for 10 h, and the lysates were immunoblotted with the indicated antibody. The data are representative of three independent experiments.
Discussion
In the present study, we demonstrate that PLD2 modulates autophagy; specifically, overexpression of PLD2 suppresses autophagy, and inhibition of PLD2 induces autophagy. Overexpression of PLD1 or PLD2 has been reported to protect cancer cells from apoptosis,29 and aberrant expression of PLDs has been widely implicated in a variety of cancers.9 Furthermore, ablation and pharmacological inhibition of PLD suppresses tumor growth, invasion and metastasis.14, 30, 31 Autophagy is dysregulated in a wide spectrum of human cancers, and some of these abnormalities have been shown to correlate with clinicopathological parameters and disease outcomes such as overall survival in cancer patients. Thus, it is suggested that autophagy has a pivotal role in tumorigenesis. The role of autophagy in cancer and treatment responsiveness is complicated; the double-edged sword function of autophagy, both as a tumor suppressor and as a protector of cancer cell survival, likely impacts anticancer treatment efficacy in opposing ways. The exploitation of the functional autophagy status in tumors and pharmacological autophagy modulation for cancer treatment provide opportunities for cancer management. PLD2 inhibition increased the number of LC3 punctate dots and the level of the LC3-II protein. Recently, we reported that PLD1 suppresses autophagy and that PLD1 inhibition augments the efficacy of anticancer regimens through promoting autophagic pathways.12 PLD2 inhibition significantly increased both basal autophagy and nutrient starvation-induced autophagy in colorectal cancer cells, and PLD2 activity is involved in suppressing autophagy. We found that PA, the product of PLD activity, suppressed starvation-induced autophagy and thus has a negative role in autophagy.12 We have reported that PLD1 may be a critical regulator of autophagy by coordinating the major player of autophagy, the AMPK–mTOR–ULK1 complex.12 In this study, we demonstrated that PLD2 inhibition induced the canonical autophagic pathways through Atg5, Atg7 and Ulk1 (Atg1). Moreover, PLD2 inhibition positively regulates autophagy by modulating the differential phosphorylation of Ulk1 by AMPK and mTOR. Because LC3-II formation can be altered by perturbation, we examined its level in the presence of BafA1, which inhibits LC3-II degradation by blocking autophagosome–lysosome fusion. PLD2 inhibition enhanced autophagic flux, similar to PLD1 inhibition.12 In contrast, it has been reported that PLD inhibition suppressed autophagic flux in HeLa cells and glioma cells,32, 33 and p53 has been reported to have a dual role in autophagy regulation.34, 35 Thus, PLD1 may function as both a positive and a negative modulator of the autophagy pathway, depending on the cell type and the cellular context. Further studies will be required to understand the mechanisms underlying the dual role of PLD1 in autophagy. Collectively, the pharmacological and genetic inhibition of PLD2 provides a new therapeutic basis for targeting the autophagic process.
Acknowledgments
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST; No. 2012002009) and a Translational Research Center for Protein Function Control Grant (NSF 2009-0092960). We thank Dr G Velasco (Complutense University), M Komatsu (Tokyo Metropolitan Institute of Medical Science) and M Kundu (St Jude Children's Hospital) for materials.
The authors declare no conflict of interest.
References
- Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- Cuzick J, DeCensi A, Arun B, Brown PH, Castiglione M, Dunn B, et al. Preventive therapy for breast cancer: a consensus statement. Lancet Oncol. 2011;12:496–503. doi: 10.1016/S1470-2045(11)70030-4. [DOI] [PubMed] [Google Scholar]
- Bursch W, Ellinger A, Kienzl H, Török L, Pandey S, Sikorska M, et al. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis. 1996;17:1595–1607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DA. Regulation of mTOR by phosphatidic acid. Cancer Res. 2007;67:1–4. doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- Sun Y, Chen J. mTOR signaling: PLD takes center stage. Cell Cycle. 2008;7:3118–3123. doi: 10.4161/cc.7.20.6881. [DOI] [PubMed] [Google Scholar]
- Kang DW, Choi KY, Min DS. Functional regulation of phospholipase D expression in cancer and inflammation. J Biol Chem. 2014;289:22575–22582. doi: 10.1074/jbc.R114.569822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Altshuller YM, Sung TC, Rudge SA, Rose K, Engebrecht J, et al. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J Biol Chem. 1995;270:29640–29643. doi: 10.1074/jbc.270.50.29640. [DOI] [PubMed] [Google Scholar]
- Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, et al. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol. 1997;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- Jang YH, Choi KY, Min DS. Phospholipase D-mediated autophagic regulation is a potential target for cancer therapy. Cell Death Differ. 2014;21:533–546. doi: 10.1038/cdd.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Iwadate M, Higashimoto M, Ono K, Takebayashi Y, Takenoshita S. Expression of phospholipase D2 in human colorectal carcinoma. Oncol Rep. 2007;18:1329–1334. [PubMed] [Google Scholar]
- Henkels KM, Boivin GP, Dudley ES, Berberich SJ, Gomez-Cambronero J. Phospholipase D (PLD) drives cell invasion, tumor growth and metastasis in a human breast cancer xenograph model. Oncogene. 2013;32:5551–5562. doi: 10.1038/onc.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Atg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya N, Mizushima T, Ueno A, Yamamoto T, Kirisako T, Noda E, et al. LC3, a mammalian homologue of yeast Atg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, et al. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2007;581:2623–2629. doi: 10.1016/j.febslet.2007.05.061. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–690. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5:671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, et al. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol. 2010;22:169–176. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee YH, Kwon TK, Chang JS, Chung KC, Min DS. Phospholipase D prevents etoposide-induced apoptosis by inhibiting the expression of early growth response-1 and phosphatase and tensin homologue deleted on chromosome 10. Cancer Res. 2006;66:784–793. doi: 10.1158/0008-5472.CAN-05-1316. [DOI] [PubMed] [Google Scholar]
- Chen Q, Hongu T, Sato T, Zhang Y, Ali W, Cavallo JA, et al. Key roles for the lipid signaling enzyme phospholipase D1 in the tumor microenvironment during tumor angiogenesis and metastasis. Sci Signal. 2012;5:ra79. doi: 10.1126/scisignal.2003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, et al. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Armi C, Hurtado-Lorenzo A, Tian H, Morel E, Nezu A, Chan R, et al. The phospholipase D1 pathway modulates macroautophagy. Nat Commun. 2010;1:142–150. doi: 10.1038/ncomms1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruntz RC, Taylor HE, Lindsley CW, Brown HA. Phospholipase D2 mediates survival signaling through direct regulation of Akt in glioblastoma cells. J Biol Chem. 2014;289:600–616. doi: 10.1074/jbc.M113.532978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]