Abstract
The efficacy of inoculation of single pure bacterial cultures into complex microbiomes, for example, in order to achieve increased pollutant degradation rates in contaminated material (that is, bioaugmentation), has been frustrated by insufficient knowledge on the behaviour of the inoculated bacteria under the specific abiotic and biotic boundary conditions. Here we present a comprehensive analysis of genome-wide gene expression of the bacterium Sphingomonas wittichii RW1 in contaminated non-sterile sand, compared with regular suspended batch growth in liquid culture. RW1 is a well-known bacterium capable of mineralizing dibenzodioxins and dibenzofurans. We tested the reactions of the cells both during the immediate transition phase from liquid culture to sand with or without dibenzofuran, as well as during growth and stationary phase in sand. Cells during transition show stationary phase characteristics, evidence for stress and for nutrient scavenging, and adjust their primary metabolism if they were not precultured on the same contaminant as found in the soil. Cells growing and surviving in sand degrade dibenzofuran but display a very different transcriptome signature as in liquid or in liquid culture exposed to chemicals inducing drought stress, and we obtain evidence for numerous ‘soil-specific' expressed genes. Studies focusing on inoculation efficacy should test behaviour under conditions as closely as possible mimicking the intended microbiome conditions.
Introduction
Environmental pollution is one of the most relevant challenges of our time in terms of potential harmful effects on biodiversity and human health (Kulkarni et al., 2008; Camilli et al., 2010; Hazen et al., 2010; Megharaj et al., 2011). Many pollutants are spontaneously transformed and removed from the environment by microbial activity (Medina-Bellver et al., 2005; Camilli et al., 2010; Hazen et al., 2010; Mrozika and Piotrowska-Segetb, 2010). As a consequence, there has been considerable interest in understanding the capacity and the roles of bacteria for degradation of pollutants (de Lorenzo, 2001; Andreoni and Gianfreda, 2007; de Lorenzo, 2009; Haritash and Kaushik, 2009; de Lorenzo et al., 2013). This has resulted in isolation of a wide variety of specific strains capable to degrade particular contaminants and in characterization of their catabolic activity under laboratory conditions. It has been proposed that inoculation of preenriched strains or pure culture isolates with biodegradative properties may be beneficial for enhancing the degradation rates of organic pollutants at contaminated sites or for achieving degradation of one or more specific organic pollutants for which no ‘inherent' capacity exists at a site (de Lorenzo, 2001; de Lorenzo, 2009; Tyagi et al., 2011; de Lorenzo et al., 2013). The success of such bioaugmentation, however, is mostly anecdotic and the activity of inoculated pure culture isolates to degrade pollutants in the environment is still relatively unpredictable (Tchelet et al., 1999; Mrozika and Piotrowska-Segetb, 2010; Megharaj et al., 2011; Jeon and Madsen, 2013). It is clear that we do not understand sufficiently well how introduced pure culture isolates behave under the environmental conditions and within a native microbiome. Strain behaviour in a complex system in first instance will depend on its ability to survive and/or grow to a sizeable population. Several factors have been implicated in survival of introduced bacterial strains in the environment, such as water availability, pH or temperature (Megharaj et al., 1997; Halden et al., 1999; Backman and Jansson, 2004). In second instance, even when surviving and growing, the activity of introduced bacteria for degradation depends on the accessibility of the target pollutant to the cells, the presence of other metabolizable substrates and, more generally, available nutrients. Autoecological properties such as adaptation to a broad range of environments and carbon sources, biofilm formation, motility, biosurfactant production (Cunliffe and Kertesz, 2006) and genome structure have been implicated as well (Mongodin et al., 2006), but the general molecular and functional response of cells to a given environment has been poorly explored (Desai et al., 2010; Puglisi et al., 2010; Wang et al., 2011).
The overarching goal of this work is thus to improve our understanding of the environmental behaviour of bacterial strains degrading aromatic hydrocarbons and of the factors that determine their activity. In particular, we were interested to study the global reactions of bacteria with biodegradative properties under near-environmental conditions as compared with laboratory culture conditions. Global reactions can be deduced from analysing recorded genome-wide expression profiles under different growth conditions, which we hypothesize will highlight factors and pathways specifically needed under those conditions.
The microorganism we chose to work with is Sphingomonas wittichii RW1, which is capable to degrade dibenzo-p-dioxins and dibenzofurans (DBF), and to cometabolize some of their chlorinated substitutes (Wittich et al., 1992; Wilkes et al., 1996; Yabuuchi et al., 2001; Hong et al., 2002; Nam et al., 2012). S. wittichii RW1 has been proposed as a candidate pure culture isolate to achieve targeted degradation of dioxins and DBFs and has been used in several studies. Megharaj et al. (1997) showed that preadaptation of RW1 to soil before inoculation enhanced its survival and increased biodegradation rates of DBF and dibenzodioxins. Halden et al. (1999) revealed the negative influence of organic matter on the kinetics of biodegradation of 2-chlorodibenzo-p-dioxin, whereas Nam et al. (2005) showed survival of RW1 in minimal medium with fly ash from solid waste incinerators and demonstrated that RW1 can act as a sorbent for dioxins. The complete genome of RW1 has been sequenced and was recently published (Miller et al., 2010). Following up on this, we analysed gene expression in RW1 upon growth on salicylate, DBF and phenylalanine and showed how likely several parallel ‘lower' pathways operate in DBF metabolism after the initial unique angular dioxygenase attack (Coronado et al., 2012). We further examined genome-wide expression of RW1 in response to laboratory condition-induced water stress (Johnson et al., 2011). Finally, we performed a genome-wide transposon scanning of RW1 to identify putative functions important for survival under drought stress and in soil (Roggo et al., 2013).
In an attempt to better understand the strategies that RW1 displays once it is introduced into a (non-lab) environment, we compared here the genome-wide responses of RW1 between regular laboratory batch growth on the aromatic substrates DBF and salicylate with growth in sand with or without the same aromatic compounds. We analysed the cellular reactions immediately after introduction into the sand, during early and late growth phases, all in carefully controlled and replicated experimental conditions. DBF degradation by the inoculated RW1 population in the sand was measured. Genome-wide transcriptome changes were recorded by micro-array hybridizations of purified and reverse-transcribed labelled RW1 cDNA, as previously described (Johnson et al., 2011). We find that global reactions of RW1 are extremely different to liquid batch cultures and soil batch incubations with the same major carbon substrate, even though the specific growth rates are not very different in the two situations.
Materials and methods
Culture conditions
S. wittichii RW1 was cultured in phosphate-buffered mineral medium (medium DSM457 from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) with salicylate (5 mM) or DBF as the sole carbon source. Liquid batch cultures were incubated at 30 °C and with 180 r.p.m. rotary shaking. DBF was dosed as crystals (1.6 g l−1) to sterilized medium.
Preparation of sandy soil
Sand was collected in Spring 2011 on a beach of Lausanne near Lake Leman (46.5079741 N, 6.545103 E). The sand was sieved through a 2-mm mesh, dried at 80 °C for 16 h and stored at room temperature (18–22 °C). pH-H2O of the sand was 7.14±0.02, its CaCO3 content is between 25–55%, whereas the percentage of organic matter content is 0.028±0.005. Dry sand was contaminated with DBF by spraying 10 ml of a dichloromethane/DBF solution (100 mg DBF per ml dichloromethane) on 500 g of sand on aluminium foil. Dichloromethane was allowed to evaporate for 16 h under a chemical hood after which aliquots of 2 g were placed in a 15-ml Greiner tube and homogenized by rotary mixing. Volumes of 25, 50, 250 and 500 μl of minimal medium were added per 1 g of sand to produce microcosms with gravimetric water content (GWC) of 2.4, 4.8, 20 and 33% (visible saturation), respectively.
Survival in sand
To measure the survival of RW1 in sand after inoculation and as a function of GWC and physiological state, we used cells from salicylate-grown cultures. For exponentially growing cells, we added 1 ml, 0.5 ml, 100 μl or 50 μl of culture with turbidity (optical density (OD), at 600 nm) of 0.3 into 2 g dry sand in 15-ml tubes to produce 33, 20, 4.8 and 2.4% GWC, respectively. For stationary phase, the same volumes were added but sampled at a culture turbidity of 0.9. Cells were maintained in the sandy microcosm for 1 h at 24–26 °C, after which they were extracted by adding 5 ml of saline solution (0.9%) to the tube and vortexing for 30 s. Larger particles were allowed briefly to settle, after which the supernatant was 10-fold serially diluted in sterile saline solution. Fifty microlitres of aliquots were plated on DSM457 agar medium with 5 mM salicylate to calculate the number of colony-forming units (CFU) per g of soil. The inoculum was also serially diluted to count the number of CFU per ml. Survival was calculated as the percentage of total CFU recovered after 1 h from the soil compared with the inoculated CFU.
The recovered cell suspension from sand was also stained using Live/Dead staining (Life Technologies, Carlsbad, CA, USA). Volumes of 5 μl of SYBR Green and 0.1 μl of propidium iodide were added per aliquot of 500 μl of suspension, mixed by vortexing and incubated for 10 min in the dark at room temperature. After that, the sample was filtered over a black 0.2-μm isopore membrane filter (Merck Millipore AG, Zug, Switzerland). The proportion of red and green fluorescent cells was counted by epifluorescence microscopy (Zeiss Axioplan II imaging microscope, Carl Zeiss, Jena, Germany).
Growth in sand
To follow growth of RW1 in the sand contaminated or not with DBF, we inoculated ∼2.5 × 105 cells per g soil in triplicate microcosm series (50 ml Greiner tube with 10 g soil). RW1 cells in this case were precultured on DBF as the sole carbon and energy source in liquid medium, harvested at a culture turbidity of 0.3 and diluted to 5 × 106 per ml in sterile minimal medium without added carbon source. A volume of 500 μl of this diluted cell suspension was added to 10 g sand to produce 4.8% GWC at the start of incubation. At regular time intervals, three replicate microcosms were killed to extract and dilute cells from the sand as described above. To count the number of CFU per g soil, serial dilutions were plated on selective plates containing minimal media agar with salicylate as carbon source, streptomycin (50 μg ml−1) to inhibit growth of other bacteria (RW1 is spontaneously resistant to streptomycin) and cyclohexamide (100 μg ml−1, Chemie Brunschwig, Basel, Switzerland) to limit fungal growth. The growth rate in soil was calculated from the increase of the log CFU over time in the triplicate assays, which was compared with that measured in liquid batch culture on DBF.
DBF degradation by RW1 in sand
The DBF content in the sand was measured over time in parallel incubated triplicate sand microcosms, inoculated or not with RW1 as described above. For every time point, three microcosms were killed and extracted by adding 5 ml of a 80:20 (v/v) mixture of hexane:acetone per g sand. Extraction was allowed to proceed for 5 min in an ultrasound bath (35 kHz), after which the organic phase was recovered by decanting. This was followed by twice an extraction with 5 ml dichloromethane per g sand and ultrasound treatment (5 min, 35 kHz). The organic phases were pooled into a single 20-ml amber glass vial (Infochroma AG, Zug, Switzerland; G075B-27/057). For analysis, 0.2 ml of the organic phase was diluted in 20 ml isooctane, of which 0.2 ml was mixed in 1 ml of isooctane containing 200 mg ml−1 13C-DBF (Cambridge Isotope Laboratories, Inc., Tewksbury, MA, USA; 50 μg ml−1, CLM-1561-1.2). Volumes of 2 μl were injected on a Thermo Scientific GC Trace 1310-ISQ gas chromatograph with mass spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) (GC–MS) equipped with a 60 m × 0.25 mm × 0.25 μm ZB-5MS column. The GC–MS was operated under constant flow (1.5 ml min−1) of He gas with a temperature programme of 80 °C for 0.5 min, an increase of 50 °C min−1 until 150 °C, and then 10.0 °C min−1 until 250 °C, followed by 3 min at 250 °C. Mass analysis was carried out at 250 °C, and DBF in sand was identified by comparison of the parent and fragment masses to an authentic DBF standard (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland; 23,637-3).
Genome-wide responses of RW1 to inoculation in sand
To measure the immediate genome-wide reaction of RW1 to sand with or without DBF, we inoculated ∼108 RW1 cells taken from exponentially growing liquid batch culture per g sand at 4.8% GWC, which had resulted in 80% survival after 1-h incubation. Cells for inoculation were harvested from a 500-ml culture in exponential phase either on salicylate or on DBF by centrifugation for 5 min at 4000 r.p.m. Cells were then resuspended in 1.5 ml leftover medium to obtain a minimum of 12 replicas of 100 μl, of which four replicas each were used per treatment. The following treatments were produced: (i) cells harvested from salicylate-growing cultures inoculated to sand without (SAL-NOTH) or (ii) with DBF (SAL-DBF); (iii) cells harvested from DBF-growing cultures inoculated to sand with DBF (DBF-DBF); (iv) as (i) but not inoculated (control SAL); (v) as (iii) but not inoculated (control DBF).
Aliquots of 100-μl cell suspension were added to 2 g dry sand in a 15-ml Greiner tube to start the incubation and produce 4.8% GWC. All tubes were incubated on a tube roller at 50 r.p.m. min−1 and 24–26 °C. After 1 h, the cells were extracted from the sand by adding 5 ml of sterile saline solution (0.9%) to the tube and vortexing for 30 s, after which the suspension was filtered over a cell strainer of 70-μm pore size to remove sand grains. The filtrate was then immediately filtered over a 0.22-μm Durapore membrane filter (Merck Millipore AG) by vacuum suction. The filter with the cells was removed, placed in a 2-ml tube containing 0.5 g of acid-washed glass beads (Sigma-Aldrich Chemie GmbH), frozen in liquid nitrogen and stored at −80 °C for RNA isolation. For the control treatments, the cells were maintained in the resuspension solution (100 μl) for 1 h at 24–26 °C. Subsequently, each of the 100-μl control cell suspensions was added to 2 g of dry sand, after which immediately 5 ml saline solution was added to the mixture and vortexed. Cells were re-extracted immediately from the sand as described above and stored for RNA isolation.
Genome-wide response of RW1 during growth in sand
To measure genome-wide gene expression of RW1 during growth in sand, we inoculated microcosms contaminated with DBF (see above) with ∼2.5 × 105 CFU of cells pregrown on DBF (see above) per g sand. In order to obtain enough cells to extract RNA, we started with 80 microcosms, each with 10 g sand. After 16 h, 64 microcosms were killed, cells were extracted by adding 15 ml of saline solution, vortexing and filtering as described above. Cells from 16 microcosms were pooled to obtain four replicates for the ‘exponential phase' in sand (SAND-DBF-EXPO). Forty hours after inoculation, we killed the remaining 16 microcosms, extracted cells as before and pooled cells from four microcosms together to produce four replicates of ‘stationary phase' in sand (SAND-DBF-STAT). As controls for growth in sand, we inoculated fourfold replicate batch cultures with RW in liquid medium with DBF crystals. Cells were recovered by filtration at an OD of 0.2–0.3 (LIQ-DBF-EXPO) and at an OD of 0.9–1 (LIQ-DBF-STAT).
RNA isolation
For extraction of RNA from cells recovered after 1 h (that is, treatments SAL-NOTH, SAL-DBF, DBF-DBF, control SAL and control DBF), we used a modified acid-phenol method (Johnson et al., 2008); (Supplementary Methods). For extraction of RNA from growing cells in sand, it was not possible to use the hot phenol procedure because of the large quantity of soil. Therefore, for the treatments SAND-DBF-EXPO, SAND-DBF-STAT, LIQ-DBF-EXPO and LIQ-DBF-STAT, we used the RNA PowerSoil Total RNA Isolation Kit (Mobio Laboratories, Carlsbad, CA, USA). The replicates were maintained at −80 °C until extraction, and filters still frozen were broken inside the tube with RNAse-free tweezers to reduce the size of the filters, before following the protocol indicated by the manufacturer. The RNA pellet was resuspended in a final volume of 20 μl to obtain concentrations >500 ng μl−1 necessary for subsequent labelling.
RNA quality in the purified solutions was verified by quantification of the A260/A280 and A260/A230 ratios using a NanoDrop spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA) and by electrophoresis on an Agilent Bioanalyser (Agilent Technologies, Santa Clara, CA, USA) to detect intact 16S and 23S rRNA. If ratio values in NanoDrop were below 1.9 or if rRNA bands were visually degraded on the Bioanalyser diagram, the RNA was discarded, and the experiment was repeated. RNA was stored at −80 °C before cDNA labelling.
Labelling and microarray hybridization
cDNAs were produced by reverse transcription using cyanine-3-labelled (Cy-3)-dCTP as described previously (Johnson et al., 2011). The labelled cDNA was purified with a MinElute PCR Purification Kit (Qiagen, Hilden, Germany), and the quantity of Cy-3-dCTP was calculated by the MICROARRAY function of the NanoDrop spectrophotometer. The criteria for sufficient labelling were a ratio of absorbances at 260/280 nm >1.6 and an absorbance at 553 nm >0.01. If these criteria were not met, the labelling was repeated.
The volume of the different samples was adjusted for the hybridization in order to have at least 2 pmol of labelled Cy-3-cDNA per array on the slide. Slides consisted of a custom 8 × 15 K array format (Agilent Technologies), which have 8 replicate 15 000 feature arrays per slide, including on average 3–4 probes per gene (Johnson et al., 2011). Slides were hybridized at 65 °C for 17 h, after which they were washed according to the procedures by Agilent and scanned. The AGILENT FEATURE EXTRACTION SOFTWARE (vs 10.7.1.1, Agilent Technologies) was used to extract the signal intensities of the probes from the scanned images. The text data file obtained was used as input in GeneSpring GX (vs 12, Agilent Technologies). Data were 2log transformed, normalized by quantile and scaled with the baseline to the median of all samples. Genes with a signal intensity above the 20th percentile in at least one of the samples were retained for analysis and comparisons between treatments. The RW1 transcriptomes described here were deposited in the GEO database under accession numbers GSE54814-54816.
Data analysis
A Welch's T-test with unequal variances was used to calculate P-values on triplicate or quadruplate probe signal intensity comparisons. P-values were converted into false-discovery rates with the Benjamini and Hochberg procedure for multiple hypotheses testing. Genes were considered statistically significantly differentially expressed between two conditions at a false-discovery rate of <0.05 and a fold-difference between the average grouped probe-per-gene hybridization intensity of >2. Reproducibility of samples (genes and conditions) was examined in GeneSpring by hierarchical clustering of normalized data, which were filtered on expression levels, after which the distance and similarity were calculated by using Euclidean distance and linkage rule average. Data were further examined by Principal Component Analysis (GeneSpring). Transcriptome data from the inoculation experiments in soil vs suspended batch liquid culture were further analysed using a two-way analysis of variance (ANOVA) with interpretation groups ‘environment' and ‘growth phase' (GeneSpring, Table 1).
Table 1. Two-way ANOVA comparison groups of transcriptome data.
| Transcriptome |
Comparison group |
|
|---|---|---|
| Environment | Growth phase | |
| CTRL DBF | Liquid with DBF | LAG |
| WS-DBF | Liquid with DBF | EXPO |
| LIQ-DBF-STAT | Liquid with DBF | STAT |
| DBF-DBF | Sand with DBF | LAG |
| SAND-DBF-EXPO | Sand with DBF | EXPO |
| SAND-DBF-STAT | Sand with DBF | STAT |
Abbreviations: ANOVA, analysis of variance; DBF, dibenzofuran.
Genes statistically different between two or more different treatments were subsequently interpreted by using Gene Ontology (GO) terminology (GOConsortium et al., 2000). GO terms of all RW1 genes or a series of differentially expressed genes between comparisons were extracted using the program DAVID (Huang da et al., 2009). The web-based bioinformatics tool GOEAST (Zheng and Wang, 2008) was then used to analyse GO data sets of statistically significantly differentially expressed genes in each pair-wise treatment comparison, under implementation of the Alexa's algorithm (Alexa et al., 2006).
Heat maps of normalized expression of genes common to specific comparison sets were produced by using the web-based program Matrix2png (Pavlidis and Noble, 2003). Thresholded and 2log transformed expression data from GeneSpring were used as input in Matrix2png under the parameters: normalize rows and preset map 17. The average of the expression data per gene (in 10log) was used further as input in Matrix2png to produce an expression scale (that is, log AVG, with the minimum value displayed in white and the maximum value in black).
A network for RW1 metabolism of DBF and other aromatic compounds as predicted in earlier work (Coronado et al., 2012) was manually created in Cytoscape (version 2.8.3), with nodes representing metabolites and edges gene expression data. Thickness of the edge line width (linear scale) was used to represent expression values of the relevant RW1 genes under the different experimental conditions, normalized per gene to the highest expression (100%, line width=15).
Results
Survival and growth of S. wittichii RW1 in sand contaminated with DBF
In order to compare the global behaviour of S. wittichii RW1 between contaminated sand and liquid batch culture, we first ensured that cells were surviving after transfer from liquid batch and were growing in the sand (that is, detectable exponential growth and stationary phase).
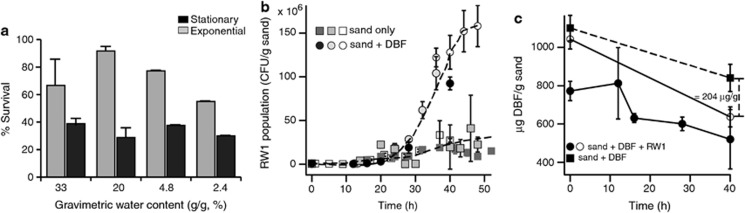
First, we established the conditions under which at least 70% of RW1 cells survived a transition from culture flask to sand (equivalent to a lag phase). These conditions would then allow us to examine the immediate RW1 sand-exposed transcriptome compared with cells remaining in liquid suspension after batch growth. We inoculated approximately 108 RW1 cells per gram of sand taken from exponential or stationary phase in liquid culture and measured survival (as CFU) and percentage of live cells (by staining with SYBR Green) after 1 h in sand with different GWC. The best survival was found for cells taken from exponential phase and in sand with 20% GWC (Figure 1a). A threshold of 70% survival was also attained for exponential phase cells inoculated in sand with 4.8% GWC. Concomitant SYBR Green-staining of recovered cells revealed 80–90% live cells. In contrast, inoculation of exponential phase cells into sand with 2.4% GWC diminished the surviving fraction to below 70% (Figure 1a). Finally, not more than 30–40% of RW1 cells inoculated from stationary phase survived in the sand, irrespective of the water content. Because of the necessity to have sufficient live cells, but considering that soils may often be drier than 20% GWC, we decided to carry out transcriptome analysis in sand at 4.8% GWC and using exponential phase cells for inoculation.
Figure 1.
Survival (a), growth of (b) and degradation of DBF by (c) Sphingomonas wittichii RW1 in sand supplemented or not with DBF. (a) Percentage of survival of RW1 cells taken from exponential and stationary phase cultures on salicylate, inoculated in sand at different GWCs during 1 h. (b) Sampling points represent the average number of CFU per g sand from four independent microcosm replicates. Data are combined from three independent inoculation series (open, grey and black symbols) in sand with (circles) or without added DBF (squares). (c) DBF degradation was measured in two independent triplicate microcosm series inoculated with RW1 (circles) or not (squares).
Next, in order to measure their capacity to actually grow in the sand, we inoculated a low number of RW1 cells (∼2.5 × 105 cells g−1) in sand at 4.8% GWC contaminated with or without 2 mg DBF g−1. RW1 growth in sand without added DBF reached a population size of 2.5 × 107 CFU g−1 (Figure 1b). Trace amounts of DBF (0.8–1.6 μg g−1) were present in non-amended sand. The RW1 population in sand with added DBF increased after 2 days to up to 1.5 × 108 CFU g−1 sand, which is evidence for active growth on DBF in the sand (Figure 1b). RW1 cells used on average 0.2 mg DBF g−1 sand during the first 40 h of incubation (Figure 1c), which is sufficient to sustain the net development of an RW1 population of 108 cells (assuming 0.4 pg C per cell and a yield on DBF of 20%). Population development of RW1 during the early phase in sand with DBF (12–28 h) was best represented by exponential growth (Supplementary Figure S1), with a calculated growth rate of 0.24±0.06 h−1. This is similar to the growth rate observed in liquid cultures with DBF crystals (0.23±0.01 h−1). Although population growth on poorly water-soluble substances like DBF is often controlled by their dissolution rates and is therefore better described by pseudolinear rather than exponential growth (Wick et al., 2001), for simplicity we refer to the early phase (12–28 h) in sand as ‘exponential growth phase' and the later phase (28–48 h) as ‘stationary phase'. We concluded that the RW1 transcriptomes in exponential and stationary phases under both conditions could be compared without being compromised by growth rate effects alone.
Analysis of the immediate response of RW1 after inoculation into sand
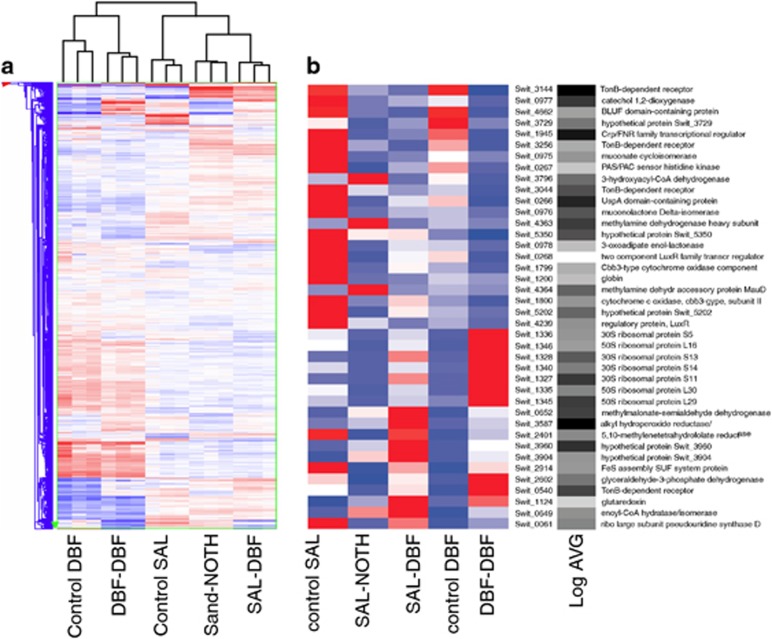
Next we asked the question whether and how RW1 cells would respond to the transition into sand after having been cultured in liquid batch with a specific aromatic carbon source (similar to the ‘inoculation' phase in a bioaugmentation process). In order to study this, we examined genome-wide gene expression of RW1 cells among five experimental conditions: (i) cells pregrown in liquid culture on DBF and inoculated for 1 h in sand (at 4.8% GWC) with DBF (DBF-DBF); (ii) cells grown on salicylate but introduced for 1 h in sand with DBF (SAL-DBF), or (iii) in sand without any addition of carbon substrate (SAL-NOTH); (iv) cells grown on DBF or (v) on salicylate but not incubated in sand (yet otherwise treated similarly—see Materials and methods section; named: control DBF and control SAL). Microarray analysis showed good replica clustering (Figure 2a), and replicas grouped closely together in the Principal Component Analysis (not shown). What becomes evident from the clustering analysis in Figure 2a is that cells pregrown on DBF or with salicylate display globally very different transcriptomes. In contrast, a transition from cells grown in DBF to sand with DBF is globally speaking less of a change than the difference between growth on DBF and salicylate. Cells grown on salicylate but introduced to sand with DBF or without any C-source addition maintain a global ‘salicylate' signature (Figure 2a).
Figure 2.
Hierarchical clustering of expression from (a) all RW1 genes (single blue or red lines) in microarray analysis or (b) of a set of 40 commonly differentially expressed genes. (a) Transcriptomes of RW1 cells introduced for 1 h in sand without or with DBF, compared with controls. Gene groups cluster in Y-direction; samples in X-direction. Colours represent genes exhibiting high (red), global average (white) or low (blue) intensity values of normalized signals in the comparison group. Sample designations, see main text. (b) Normalized gene expression of 40 RW1 genes commonly differentially expressed in all pair-wise comparisons of 1-h inoculation in sand with DBF (see Figure 3a). Colours indicate high (red), median (white) and low relative (blue) signal intensities. Log AVG, 10log of the average absolute hybridization intensity per indicated gene, on a scale from low (white) to high (black). Heat map generated by Matrix2png (Pavlidis and Noble 2003).
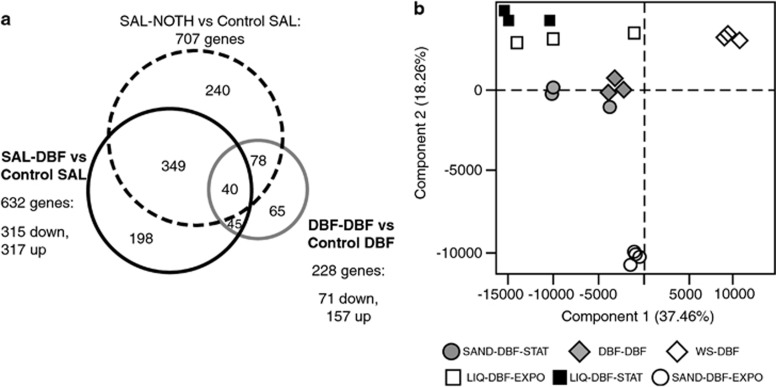
The expression of between 4% and 13% of all genes in the RW1 genome was affected by a 1 h presence in sand (Figure 3a). The largest number of (statistically significantly) differentially expressed genes was found in cells taken from salicylate culture and inoculated in sand without any further carbon source (SAL-NOTH vs control SAL: 707 genes). In all, 11% of all genes (632 genes) were differentially expressed in cells passed from salicylate culture to sand with DBF (SAL-DBF vs control SAL). The lowest number of differentially expressed genes (228) occurred among cells taken from DBF culture and introduced in sand contaminated with DBF (DBF-DBF vs control DBF). A common group of 45 genes were similarly differentially expressed (24 higher and 21 lower than in the controls) upon inoculation in sand with DBF, irrespective of preculturing of the cells with salicylate or DBF. A total of 40 differentially expressed genes were common to all sand transitions (Figures 2b and 3a). Of these, 19 were always lower expressed than in the controls, 6 were always higher expressed and 15 were either lower or higher expressed than in the controls (Table 2). These genes might thus represent the core reaction of RW1 to a transition in sand (under the used aromatic growth substrates).
Figure 3.
Group clustering of transcriptome data. (a) Venn diagram of the numbers of differentially expressed RW1 genes in pair-wise comparisons between 1-h treatments. SAL-NOTH, cells inoculated in bare sand from salicylate cultures; SAL-DBF, cells from salicylate cultures inoculated in sand contaminated with DBF; DBF-DBF, cells from DBF liquid cultures inoculated in sand contaminated with DBF. (b) Two-dimensional Principal Component Analysis of Sphingomonas wittichii transcriptomes under different growth conditions. Global patterns of RW1 cells growing in sand in exponential (SAND-DBF-EXPO) or stationary phase (SAND-DBF-STAT) vs liquid cultures with crystals of dibenzofuran in exponential (LIQ-DBF-EXPO) or stationary phase (LIQ-DBF-STAT). DBF-DBF, transcriptome from cells grown on DBF inoculated during 1 h in sand with DBF. WS-DBF exponential growth in liquid with saturated amounts of DBF (WS-DBF).
Table 2. Differentially expressed genes of S. wittichii RW1 common to pair-wise comparisons in short contact sand experiments.
| Gene | Annotation |
Fold change in comparisona |
|||||
|---|---|---|---|---|---|---|---|
| SAL-NOTHb vs ctrl SAL | SAL-DBF vs ctrl SAL | DBF-DBF vs ctrl DBF |
Two-way ANOVAc |
||||
| 1 h | EXPO | STAT | |||||
| Swit_0061 | Ribosomal large subunit pseudouridine synthase D | −5.3 | −3.3 | 2 | 2.6 | −1.9 | 1.0 |
| Swit_0266 | UspA domain-containing protein | −4.7 | −5.7 | −4.6 | −1.7 | −3.2 | −1.1 |
| Swit_0540 | TonB-dependent receptor | −2.6 | −2.3 | 3.5 | 2.6 | −1.4 | 1.1 |
| Swit_0652 | Methylmalonate-semialdehyde dehydrogenase | 4.2 | 4.5 | 6.5 | 1.1 | 6.2 | 1.9 |
| Swit_0975 | Muconate cycloisomerase | −21.7 | −21.1 | −6.3 | 1.5 | −1.9 | −1.1 |
| Swit_0976 | Muconolactone Delta-isomerase | −12.9 | −11.9 | −4.5 | 1.7 | −5.4 | 1.2 |
| Swit_0977 | Catechol 1,2-dioxygenase | −11.4 | −12.9 | −9.8 | −1.1 | −2.6 | 1.6 |
| Swit_1124 | Glutaredoxin | 3.7 | 4.3 | 2.7 | 1.8 | 2.2 | 33.9 |
| Swit_1200 | Globin | −21.8 | −22.1 | −3.3 | −4.6 | −1.3 | −1.3 |
| Swit_1327 | 30S ribosomal protein S11 | −2.3 | 2.5 | 10.5 | 2.2 | 1.4 | 2.6 |
| Swit_1328 | 30S ribosomal protein S13 | −2.1 | 2.8 | 14.1 | 4.9 | 1.5 | 2.4 |
| Swit_1336 | 30S ribosomal protein S5 | −6.3 | −2.3 | 35.2 | 7.7 | −1.4 | 1.2 |
| Swit_1346 | 50S ribosomal protein L16 | −5.6 | −2.2 | 22.2 | 4.6 | −1.3 | −1.0 |
| Swit_1799 | Cbb3-type cytochrome oxidase component | −4.4 | −4.5 | −3.5 | −1.5 | −2.8 | −1.7 |
| Swit_1800 | Cytochrome c oxidase, cbb3-type, subunit II | −3.3 | −3.9 | −2.8 | 1.2 | −2.4 | −1.4 |
| Swit_2401 | 5,10-Methylenetetrahydrofolate reductase | −14.3 | −2.4 | 4.6 | 3.0 | −3.1 | −1.5 |
| Swit_2914 | FeS assembly SUF system protein | −6.2 | −3.1 | 3.7 | 2.3 | −1.1 | 1.4 |
| Swit_3044 | TonB-dependent receptor | −3.1 | −8.8 | −4.6 | 2.7 | −4.0 | −18.7 |
| Swit_3144 | TonB-dependent receptor | −3.1 | −8.2 | −17.8 | −1.0 | −1.4 | 3.4 |
| Swit_3256 | TonB-dependent receptor | −2.5 | −8.2 | −6.5 | 3.7 | −1.6 | 1.6 |
| Swit_3587 | Alkyl hydroperoxide reductase | 2.5 | 6.3 | 5.4 | 49.3 | −1.1 | 7.4 |
| Swit_3729 | Hypothetical protein Swit_3729 | −3 | −3.9 | −8.2 | −1.2 | −1.8 | 5.3 |
| Swit_3904 | Hypothetical protein Swit_3904 | 11.7 | 9.8 | 3.8 | 51.7 | 2.4 | 31.3 |
| Swit_3960 | Hypothetical protein Swit_3960 | 2.5 | 4.1 | 4.4 | 1.7 | 1.6 | 5.0 |
| Swit_4364 | Methylamine dehydrogenase accessory protein MauD | 9 | −2.2 | −3.1 | 1.6 | −1.1 | −2.5 |
| Swit_4662 | BLUF domain-containing protein | −2.8 | −7.9 | −9.2 | 1.0 | −3.1 | 1.8 |
| Swit_5202 | Hypothetical protein Swit_5202 | −8.7 | −5.6 | −2.6 | 1.2 | 1.4 | −2.5 |
Abbreviation: ANOVA, analysis of variance.
Fold change (absolute) in condition compared with control, negative values indicate decreased expression in soil.
Sample indications: Bare sand vs control cells growing in salicylate (SAL-NOTH vs ctrl SAL), sand contaminated with DBF vs control cells growing in salicylate (SAL-DBF vs ctrl SAL), and sand contaminated with DBF vs control cells growing in dibenzofuran (DBF-DBF vs ctrl DBF).
Two-way ANOVA in the condition ‘growth phase' 1 h: DBF-DBF vs ctrl DBF, EXPO: SAND-DBF-EXPO vs wsDBF, and STAT: SAND-DBF-STAT vs LIQ-DBF-STAT.
Because a Venn representation is only informative for the identification of genes that are statistically significantly differentially expressed between treatments, but not on their expression levels, we examined more precisely the normalized and averaged expression levels of those 40 genes shared between all sand transitions and their controls. Figure 2b clearly shows how two opposing (control SAL and control DBF, vs SAL-DBF and DBF-DBF) and one intermediate expression pattern arise (SAL-NOTH) among the 40 shared differentially expressed genes. Functions that tend to be lower expressed after transition to sand with DBF (both SAL-DBF and DBF-DBF) include a group of genes (Swit_0975 to Swit_0978) previously implicated in degradation of salicylate (Coronado et al., 2012), three genes (Swit_3144, Swit_3256 and Swit_3044) for TonB-dependent receptors, which may be associated with uptake of scarce resources such as vitamins, trace metals or heme (Lim, 2010), a small gene cluster encoding a cbb3-type cytochrome oxidase (Swit_1799 and Swit_1800) and an UspA stress domain-containing protein (Swit_0266). Furthermore, a number of potential regulatory proteins are consistently lower expressed in sand with DBF, namely, a Crp/FNR family transcriptional regulator (Swit_1945), a LuxR-type two component system (Swit_0267 and Swit_0268) and another LuxR-type regulator (Swit_4239). In contrast, there is a very clear increase of expression of two gene clusters for ribosomal proteins (Swit_1327-1328, and Swit_1335-1346) in sand with DBF (Figure 2b). Furthermore, genes for another putative TonB-dependent receptor (Swit_0540) and for a putative glutaredoxin (Swit_1124) are higher expressed in sand with DBF. Genes specifically higher expressed in sand without extra added carbon include Swit_4363/_4364, part of a cluster putatively encoding methylamine degradation, and a 3-hydroxyacyl-CoA dehydrogenase (Swit_3796).
GO interpretation of the changes upon inoculation in sand
In order to further interpret the transcriptome changes and cellular reactions during the different transitions, we used GO terminology analysis (http://omicslab.genetics.ac.cn/GOEAST/). A complete detailed list of enriched GO terms for each comparison is presented in Supplementary Information (Supplementary Tables S1–S3), but reactions can be summarized as follows. From the GO category ‘Biological processes', it became evident that cells taken from exponential phase on salicylate and introduced into sand without DBF completely interrupt their metabolism (deficient GO terms: respiration, oxidative phosphorylation, glycolysis, methionine biosynthetic process; Supplementary Table S1), try to scavenge nutrients and maintain cell survival (enriched GO terms: regulation of nitrogen compound metabolic process, methylamine and valine metabolic process, DNA repair, cellular homeostasis). This response is dramatically changed in cells introduced into sand supplemented with DBF and different for cells coming from cultures grown on salicylate or DBF. Cells grown on salicylate change their metabolism in order to adapt to the new carbon source (enriched GO terms: aromatic compound and catechol-containing compound catabolic process, Supplementary Table S2), and adapt to life in sand (GO terms: polysaccharide, lipopolysaccharide and glutamate biosynthetic process). When the cells are already adapted to the carbon source present in the sand (that is, DBF), they continue without major changes in their metabolism. In the DBF-DBF vs control DBF comparison, enriched GO terms relate to oxidative phosphorylation, respiration and translation, indicative for growing and active cells (Supplementary Table S3). Interestingly, in all cases the transition to sand resulted in a decrease of expression of functions implicated in oxygen transport and binding, which may be due to the higher provision rate of oxygen in sand at 4.8% GWC (for example, thin water films) than in liquid culture.
Of further interest are a number of discernable stress functions that RW1 displays under different transition conditions (Supplementary Table S4). RW1 growing on DBF transited to sand with DBF turned off the expression of various two-component response regulators. A total of 21 genes distributed across 6 different GO terms related to stress is higher expressed in sand without DBF in comparison to the control on salicylate (Supplementary Table S4). Interesting among those is Swit_3927, predicted to code for an EcnAB entercidin (58.6-fold increase), which is part of a family of proteins called the entericidin antidote/toxin peptides. These proteins are activated in Escherichia coli in stationary phase under high osmolarity (Bishop et al., 1998).
Genome-wide expression differences between RW1 cells growing in sand contaminated with DBF vs liquid culture
After having examined the specific expression differences during a transition from liquid culture to sand at 4.8% GWC, we analysed the RW1 genome-wide gene expression under four new experimental conditions: (i) RW1 growing in sand on DBF, sampled in early phase (16 h, SAND-DBF-EXPO) and (ii) in late phase (40 h, SAND-DBF-STAT); (iii) cells growing in liquid batch culture on DBF crystals, sampled in early phase (LIQ-DBF-EXPO), and (iv) sampled during late phase (LIQ-DBF-STAT). In addition, we included previously established transcriptomes of RW1 during early exponential phase growth with water-saturated amounts of DBF (WS-DBF, Table 1) (Coronado et al., 2012).
Principal Component Analysis indicated that RW1 genome-wide gene expression during early growth in sand with DBF (open circles, Figure 3b) is very different from late growth phase in sand (filled circles), which on its turn resembles more the growth in liquid culture with DBF (open squares; both components explaining 59% of the variation). Genome-wide expression during early (OD=0.3) and late phase (OD=0.9) in liquid culture with DBF crystals varied quite a bit between replicates but was not statistically significantly different between them (Figure 3b). The reason for this may be that cells growing with DBF crystals in liquid culture actually display pseudolinear growth, in which case the growth rate is governed by the crystal dissolution rate (Wick et al., 2001). For this reason, we used the transcriptome data set WS-DBF instead of LIQ-DBF-EXPO in the two-way ANOVA presented below, which better represents RW1 exponential growth on DBF in liquid culture (Coronado et al., 2012). The genome-wide expression of RW1 cells 1 h after transition to sand with DBF (DBF-DBF, see above) was very different from both the liquid and the sand exponential growth phases (Figure 3b, filled diamonds) but more similar as in late growth phase in sand plus DBF (SAND-DBF-STAT, two components explaining 53% of variation; Figure 3b).
Microarray data were grouped and further analysed in a two-way ANOVA examining the effect of ‘Environment' (that is, Sand or Liquid) and ‘Growth Phase' (lag phase of 1-h contact, ‘exponential' or ‘stationary phase', Table 1). A total of 1418 genes were identified, whose expression reacted statistically significantly different to the condition ‘Environment' (P<0.001). Two thousand and thirty-six genes were identified, which showed statistically significant interaction between the two terms (‘Environment' and ‘Growth Phase', P<0.001), of which 728 were shared with condition ‘Environment' alone. Gene functions differentially expressed to the condition ‘Environment' are implicated, among others, in cellular homeostasis, response to stress, protein secretion, a variety of biosynthetic and metabolic processes, inorganic ion scavenging or glutamine synthesis (Supplementary Table S5).
Compared with 1h- contact in sand with DBF, RW1 cells growing exponentially in sand (early phase: 12–28 h, Supplementary Figure S1) preferentially expressed genes related to energy generation (GO terms: ATP synthesis and oxidative phosphorylation) and biosynthesis of phospholipids, cell wall, ribonucleoproteins and fatty acids (Supplementary Table S6). In contrast, a wide range of genes decreased their expression in sand during exponential phase vs 1-h contact, which may have been particularly important for the first adaptation step after inoculation. GO analysis suggests that these are primarily functions in nutrient scavenging, transport, cellular homeostasis and oxidative damage repair (for example, antioxidant and electron carrier activity; Supplementary Table S6).
In comparison to cells growing exponentially in liquid suspended culture (WS-DBF) and at a relatively conservative cutoff level of more than fourfold expression difference, 142 genes were higher and 90 were lower expressed in sand (Supplementary Table S7). Among those are numerous genes for putative TonB-dependent receptors, which are indicative for scavenging of substrates, minerals and recycling of nitrogen from organo-N compounds (Lim, 2010). Interestingly, some genes putatively associated with adhesion were much more expressed among exponentially growing cells in soil. This includes the 85-fold more highly expressed gene Swit_0615 (annotated as Flp/Fap pilin component) and the 38-fold more highly expressed Swit_0163 (annotated as Type IV secretory pathway TrbD component-like protein). The expression of a range of other genes is diminished among cells growing in sand with DBF compared with liquid culture (Supplementary Tables S7 and S8). This affects notably genes involved in stress response, in flagellar biosynthesis and a range of (two-component) regulatory systems (Supplementary Table S8).
RW1 cells sampled in late phase in sand with DBF (SAND-DBF-STAT) diminished expression of energy generation processes, biosynthesis and cellular metabolism and cell wall production (Supplementary Tables S9 and S10) but increased expression of functions implicated in oxidative stress response, cellular homeostasis, nutrient scavenging, turnover of proteins, polysaccharide production and transport and production of osmoprotectants (glutamate biosynthetic process, Supplementary Table S9). Interestingly, also expression of genes for DNA-modifying enzymes increased (Supplementary Table S9).
In comparison to RW1 cells growing in liquid suspension with DBF crystals (LIQ-DBF-STAT), cells in sand with DBF in late phase have a quite drastically different gene expression pattern (167 genes with higher and 161 with lower expression than in liquid), which can be interpreted by a variety of enriched GO terms (Supplementary Tables S11 and S12). As an example, cells in sand with DBF in late phase (SAND-DBF-STAT) showed consistently higher expression of genes involved in cellular homeostasis, stress response, glutamate biosynthesis and ‘compound' binding (for example, GTP binding, NADP binding), suggestive for specific protection needed in the sand and osmotic balancing (Supplementary Table S11). On the other hand, genes involved in aromatic compound metabolism and flagellar synthesis decreased expression in soil. Frequently, differentially expressed genes also clustered in a number of potential co-transcribed regions, suggesting concerted regulation to specific environmental signals (Supplementary Table S12).
Aromatic compound metabolism
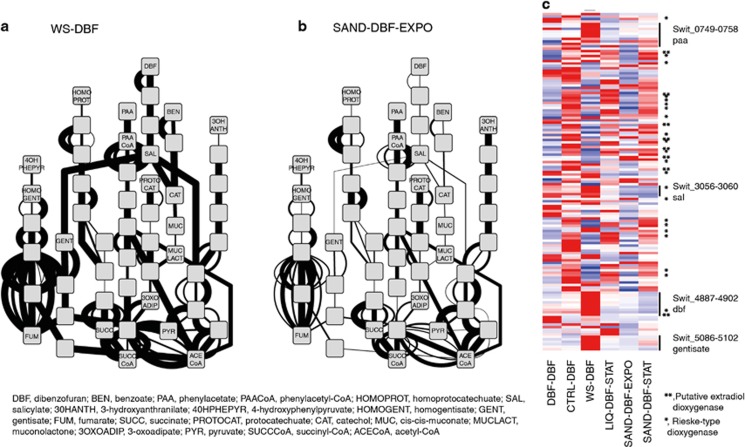
In order to analyse more specifically expression changes of RW1 genes potentially implicated in DBF and aromatic compound metabolism among the different growth conditions, we constructed a network with metabolites as nodes and known or predicted RW1 gene functions as edges (Supplementary Figure S2), based on previous analyses (Coronado et al., 2012). Compared with exponential growth in liquid medium with DBF (WS-DBF, Figure 4a), cells in early growth phase in sand with DBF (SAND-DBF-EXPO) clearly changed the expression of genes implicated in DBF and aromatic compound metabolism (Figure 4b). Interestingly, expression of many of the known genes for DBF metabolism diminished in this phase in sand, whereas that of other genes with predicted function in aromatic compound metabolism but no known specificity increased (Figure 4c, Supplementary Figure S3, for example, Rieske-type dioxygenases). Expression of the known DBF network genes partially returned in later phase in the sand (Supplementary Figure S4), but overall, the expression of genes potentially implicated in aromatic compound metabolism in RW1 was strikingly different between exponential growth in liquid culture with DBF (WS-DBF) vs the other conditions (Figure 4c). Furthermore, expression of genes for aromatic compound metabolism was markedly different between the transition (DBF-DBF) and early phase in sand (SAND-DBF-EXPO) and the other conditions (CTRL-DBF, LIQ-DBF-STAT and SAND-DBF-STAT, Figure 4c; Supplementary Figure S3).
Figure 4.
Network analysis of DBF and aromatic compound metabolism by S. wittichii RW1 under different growth conditions. (a, b) Inferred DBF and aromatic compound metabolic network in liquid cultures growing exponentially with DBF (WS-DBF) or in sand (SAND-DBF-EXPO), respectively. Nodes represent substrates and metabolites converted in RW1-predicted enzyme reactions (edges). Edge line width is a linear-scale representation of normalized expression of the gene coding for the particular enzyme. For a complete overview of all node names and edges, see Supplementary Figure S2, whereas Supplementary Figure S4 shows the expression networks under all six growth conditions. (c) Heat map of normalized expression of all RW1 genes predicted to be implicated in aromatic compound metabolism (including those of unknown specificity and not represented in the metabolic network) under the different growth conditions. Colours and heat map generation as in Figure 2b. For a full-scale version with all gene designations and predicted functions, see Supplementary Figure S3.
Discussion
The work presented here shows a comprehensive analysis of the global reactions of bacteria inoculated and growing in (contaminated) sand, compared with suspended batch growth in regular liquid culture. The types of global transcriptome global transcriptome reactions uncovered here may be representative for those which such cells would undergo when being deployed for targeted bioaugmentation processes of contaminated sites. We chose here to work with S. wittichii RW1 as an example of a strain that can degrade a number of pertinent aromatic hydrocarbons (DBF and dioxin) and which as such has been proposed in the past as a realistic strain to be inoculated with the purpose of achieving increased DBF and dioxin biodegradation rates in the environment (Wittich et al., 1992; Wilkes et al., 1996; Megharaj et al., 1997; Halden et al., 1999; Miller et al., 2010). Globally speaking, and despite being mostly descriptive, we believe our results help to explain a number of common trivial practical observations, from which various crucial measures may be suggested to avoid immediate failure of future strain inoculation efforts.
We are aware that some compromises had to be made in our experimental setup in order to allow proper analysis and comparison of RW1 genome-wide transcriptional responses. For example, we used regular sand which was contaminated with DBF, rather than a more complex soil with more clay minerals and organic matter. This was crucial in order to obtain sufficient high-quality RW1 RNA but certainly has given strain RW1 some competitive advantage given the lower background of native microbiome in this sand compared with soils with higher organic matter. Notwithstanding the use of sand, other groups have shown that inoculated sphingomonads do grow in more complex soils and at the expense of polycyclic aromatic hydrocarbons (Megharaj et al., 1997; Halden et al., 1999; Fida et al., 2013). In addition, in order to sample sufficient RNA for the immediate response of RW1 cells to a soil transition (the 1-h response), we had to inoculate 108 cells per g of material, which may have exacerbated the lack of nutrients and minerals on a per cell basis. Inoculation of high cell densities is not uncommon in bioaugmentation efforts, which, however, impedes such cells from actually growing and establishing themselves at the expense of their unique target substrate in the soil (here: DBF). At much lower starting cell concentrations (2 × 105 per g), we did observe specific growth in sand at the expense of DBF (Figures 1b and c) with similar growth rates as in liquid culture. Finally, by including a wide range of control conditions for each of the examined steps in the inoculation process (that is, lag transition, exponential growth and stationary phase), we are confident that our comparative analyses of the RW1 genome-wide responses correctly highlight the respective transcriptome changes.
Our observations during the first step of inoculation (1-h contact, lag phase) show that cells that have been precultured on the same carbon substrate as their target contaminant (here, as example, DBF) display the least amount of gene expression differences, compared with those that have been precultured on a different (aromatic) substrate (here: salicylate). This was somewhat surprising, given that salicylate is an intermediate of DBF metabolism (Wittich et al., 1992). Cells prepared and inoculated under such circumstances do not have to readjust their primary metabolism, although we can still see evidence of increased scavenging reactions for nutrients. Interestingly, even inoculated cells precultured on DBF after 1 h in sand with DBF show a ‘stationary phase' signature (Figure 3b), indicating they are going through a period of cell growth arrest after inoculation. Cells that had been pregrown on the DBF-related aromatic substrate salicylate showed major transitions of carbon metabolism and osmotic adaptations (Supplementary Table S2), which in a field situation might mean that their capacity for substrate competition is diminished compared with native bacteria. In extremo, when cells are inoculated into sand which does not contain DBF, they display extreme carbon and nutrient shortage stress, even though they do grow to some extent (Figure 1b). This indicates that when the intended target substrate is not sufficiently bioavailable, the inoculation is likely to fail. We found some 40 genes commonly statistically significantly expressed during sand transitions compared with the controls, the majority of which also change expression during growth and stationary phase in sand with DBF (Table 2), which might therefore constitute essential elements governing this transition state.
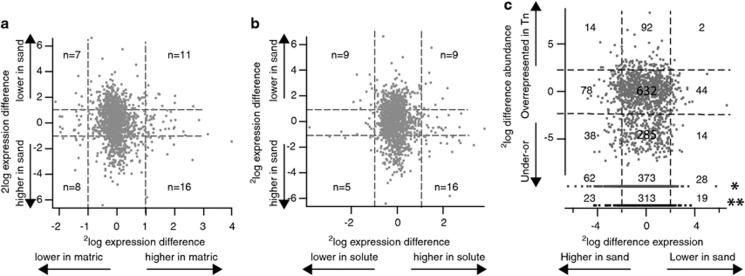
One of the interesting basic questions our work may help to address is whether cells ‘realise' that they are in a ‘soil' rather than in liquid culture. Clearly, RW1 gene expression during the transition phase and during exponential growth in sand with DBF was very different from that in all liquid cultivations (Figures 2 and 4), whereas late growth phase in sand resembled more stationary phase and slow growth in liquid culture. This resemblance was also reflected in expression of genes potentially implicated in aromatic compound metabolism (Figure 4c). It has previously been suggested that behaviour in dry soil might be experimentally induced by lowering of the water potential in liquid culture through the addition of salt (solute potential (SP)) or of swelling agents, such as polyethylene glycol (matric potential (MP)) (Roberson and Firestone, 1992; Halverson and Firestone, 2000; Johnson et al., 2011; van de Mortel and Halverson, 2004). When we compare, however, the RW1 differentially expressed genes during exponential growth in sand at 4.8% GWC with previous data on the differentially expressed genes in RW1 induced upon SP or MP stress (both not reducing the growth rate by >20%) (Johnson et al., 2011), we find very little overall similarities (Figures 5a and b, Supplementary Table S13). For most of these genes, the response to MP and SP stress is mutually exclusive, but the expression of several genes for motility is consistently diminished and genes for polysaccharide biosynthesis are consistently more highly expressed under sand, MP and SP stress (Supplementary Table S13). Interesting is a gene for a putative glutathione-dependent formaldehyde-activating protein suspected in formaldehyde detoxification (Vorholt, 2002) (Swit_1412), which showed 56-fold higher expression in cells growing exponentially in sand with DBF compared with liquid, compared with fivefold under SP stress and twofold under MP stress (Supplementary Table S13). We conclude that, although MP and SP stress each produce a number of useful expression signatures related to that in sand, they are not quite representative for sand behaviour. The gene expression programme displayed in exponentially growing RW1 cells in sand at 4.8% GWC with DBF must, therefore, be a specific reaction to the sand physico-chemical environment.
Figure 5.
Correlation between genome-wide gene expression of S. wittichii RW1 growing exponentially in sand with DBF and (a) matric stress, (b) solute stress or (c) relative fitness cost of gene interruption on survival and growth in sand. Each dot is the average 2log expression difference from triplicate treatments vs controls (for example, matric stress vs regular growth, expression in sand vs liquid culture). Numbers represent the number of genes in the respective quadrant. Dottes lines indicate twofold cutoff. Dots in panel (c) marked with a single (*) or a double asterisk (**) indicate genes judged to be essential in sand or under all growth conditions, respectively. Matric and solute stress transcriptome data from Johnson et al. (2011); fitness cost data taken from the transposon scanning approach in Roggo et al. (2013).
As an extension to this question, it is interesting to determine whether there are specific gene functions that seem to be very important for the survival or growth of RW1 under sand conditions. As transcriptome data as recorded here remain essentially descriptive, we compared the genome-wide gene expression data with previous transposon scanning of RW1 for survival functions (Roggo et al. 2013). In Figure 5c, we plot hereto the 2log difference of normalized expression of genes appearing in the ANOVA interaction terms against the 2log ratio of the abundance of transposon insertions in exponentially growing RW1 populations in sand (vs the control of the transposon abundance distribution in the starter transposon library), with gene lists appearing in Supplementary Tables S14 and S15. The transposon scanning approach is based on the premise that any transposon insertion in a gene affecting growth under a specific set of growth conditions will lead to a lower abundance (or even disappearance) of that transposon mutant population in the metapopulation of all mutants (Gawronski et al., 2009; van Opijnen et al., 2009; Roggo et al., 2013). Under exclusion of essential genes and only focussing on those genes with higher expression in sand, one can conclude from GO classification that a wide range of metabolic processes is specific for growth in soil (Table 3), and a further range of gene functions is implicated in specific survival in soil upon cessation of growth (cellular homeostasis, nutrient scavenging, stress response, Table 3). The list of gene functions is too long to discuss in detail, but a few genes are worth highlighting given previous discussions (Johnson et al., 2011; Roggo et al., 2013). In contrast to previous conclusions on the role of a putative trehalose synthesis cluster (Swit_4523-4533), these genes are not particularly differentially expressed in sand; neither are genes for putative polysaccharide biosynthesis (Swit_3608-3613), which were both higher expressed during matric and solute stress (Johnson et al., 2011) and disruption of which caused drastic fitness loss in sand (Roggo et al., 2013). Rather, it seems that glutamate biosynthesis is used to compensate for osmolarity differences in sand (Swit_0656-0659; Supplementary Table S16). On the other hand, previously mentioned genes involved in fatty acid metabolism (Swit_3903-3908), for a 17-kDa ‘surface antigen' (Swit_1507-1509), and for a cell wall hydrolase SleB (Swit_3463) are higher expressed in sand, as well as under solute or matric stress, and transposon insertion causes fitness loss (Supplementary Tables S14 and S15). Also, pertinently higher expressed in sand is a gene cluster for arsenic detoxification (Swit_2243-2244), interruption of which causes strong fitness loss (Supplementary Table S15). The cluster around Swit_0163 (type IV secretory pathway TrbD component-like protein) is also intriguing, because it is essential for growth in sand and is up to 38-fold higher expressed (Supplementary Table S16). A few further putative operons seem specific for sand growth, such as putative RND efflux systems (Swit_1152-1154; Swit_3230-3231), or alkyl hydroperoxidase activity (Swit_3585-3587) (Supplementary Table S16). Interestingly and similar as under solute or matric stress, expression of the flagellar cluster Swit_1260-1293 is decreased in soil, although insertions in this gene region mostly cause fitness loss (Supplementary Table S14). From recent work on Pseudomonas putida, it has been suggested (Martinez-Garcia et al., 2014) that flagella allow bacteria to explore the environment searching for nutrients and help escaping from predators or adverse conditions. But mutants without flagellar machinery are actually more resistant to oxidative stress and ultraviolet exposure (Martinez-Garcia et al., 2014). In addition, cells without flagella have more metabolic energy in the form of ATP and reducing power in the form of NADPH, which could potentially be used to better cope with environmental stresses. Behaviour in soil may thus require low or temporarily restricted expression of flagellar systems, in order to optimize metabolic energy for survival and yet allow migration, when necessary. It is further interesting to note that a number of plasmid functions are differentially expressed in liquid or soil and have important fitness effects. For example, important for sand fitness and higher expressed in liquid in stationary phase are Swit_5005-5010 (a putative type IV secretion system encoded on pSWIT02) and Swit_5364-5467 (putative type IV functions encoded on pSWIT01; Supplementary Table S16). On the other hand, a clear polycistronic unit on pSWIT01 is higher expressed in exponentially growing cells in sand (Swit_5192-5196, mostly hypothetical function). Even though the molecular mechanisms of these effects are not immediately trivial, this underscores that natural plasmids can have important roles in general survival or growth in the environment.
Table 3. Gene ontology interpretation of gene functions important for fitness in sand and higher expressed in sand than in liquid.
| Growth phase | GOID | Ontology | Term | No. of probes | Total No. of probes | Log odds ratio | P-value |
|---|---|---|---|---|---|---|---|
| Exponential | GO:0008152 | BP | Metabolic process | 111 | 2082 | 0.1 | 0.0678 |
| GO:0055114 | BP | Oxidation–reduction process | 38 | 573 | 0.4 | 0.0219 | |
| GO:0006573 | BP | Valine metabolic process | 2 | 7 | 2.5 | 0.0424 | |
| GO:0009276 | CC | Gram-negative-bacterium-type cell wall | 2 | 10 | 2.0 | 0.0403 | |
| GO:0003995 | MF | Acyl-CoA dehydrogenase activity | 5 | 47 | 1.1 | 0.0819 | |
| GO:0004491 | MF | Methylmalonate-semialdehyde dehydrogenase (acylating) activity | 2 | 5 | 3.0 | 0.0223 | |
| GO:0016811 | MF | Hydrolase activity, acting on carbon–nitrogen (but not peptide) bonds, in linear amides | 5 | 35 | 1.5 | 0.0278 | |
| Stationary | GO:0019725 | BP | Cellular homeostasis | 8 | 43 | 2.7 | 0.0002 |
| GO:0006950 | BP | Response to stress | 7 | 79 | 1.7 | 0.0014 | |
| GO:0030163 | BP | Protein catabolic process | 2 | 5 | 3.8 | 0.0081 | |
| GO:0006508 | BP | Proteolysis | 9 | 96 | 1.7 | 0.0017 | |
| GO:0006096 | BP | Glycolysis | 2 | 9 | 2.9 | 0.0270 | |
| GO:0009306 | BP | Protein secretion | 4 | 32 | 2.1 | 0.0135 | |
| GO:0006879 | BP | Cellular iron ion homeostasis | 2 | 10 | 2.8 | 0.0332 | |
| GO:0043231 | CC | Intracellular membrane-bounded organelle | 2 | 12 | 2.5 | 0.0372 | |
| GO:0008270 | MF | Zinc ion binding | 6 | 84 | 1.3 | 0.0336 | |
| GO:0008233 | MF | Peptidase activity | 11 | 112 | 1.8 | 0.0429 | |
| GO:0016624 | MF | Oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulphide as acceptor | 2 | 14 | 2.3 | 0.0604 | |
| GO:0008289 | MF | Lipid binding | 3 | 24 | 2.1 | 0.0386 | |
| GO:0008236 | MF | Serine-type peptidase activity | 5 | 28 | 2.6 | 0.0031 | |
| GO:0016668 | MF | Oxidoreductase activity, acting on a sulphur group of donors, NAD(P) as acceptor | 3 | 7 | 3.9 | 0.0008 | |
| GO:0008565 | MF | Protein transporter activity | 4 | 38 | 1.9 | 0.0231 | |
| GO:0004175 | MF | Endopeptidase activity | 4 | 42 | 1.7 | 0.0321 | |
| GO:0016209 | MF | Antioxidant activity | 4 | 27 | 2.4 | 0.0070 | |
| GO:0008199 | MF | Ferric iron binding | 2 | 11 | 2.7 | 0.0386 |
Not only did introduced RW1 cells survive in sand but they also actively degraded DBF in the early and late phases (Figure 1c). Expression analysis of RW1 genes predicted to be implicated in aromatic compound metabolism showed that introduced cells adjust their metabolic network immediately after transition into sand with DBF, during the early (exponential) growth phase in sand and in later growth phases (Figure 4; Supplementary Figures S3 and S4). Interestingly, expression of the predicted aromatic compound network is quite dissimilar to that obtained during exponential growth on DBF in liquid culture (Figure 4). Although it cannot be excluded that RW1 in the early growth phase in sand profits from other available organic compounds, it is also possible that the strain deploys other unknown metabolic branches for DBF degradation, dependent on the growth environment (Supplementary Figure S3).
In conclusion, our results demonstrate for the first time the specific cellular reactions of a typical bacterial strain intended for environmental inoculation (S. wittichii RW1) to a contaminated environment (sand). We further conclude that such cellular reactions are mostly different from typical water stress achieved with SP or MP change in liquid cultures and, therefore, that sand or soil inoculations themselves should be used to understand cellular reactions to these environmental changes. Such experiments are necessary not only for the practise of bioaugmentation but also will help to more generally understand how single strains inoculated into complex microbiomes (for example, plant leaf surfaces, intestinal tract) are behaving. In the long term, this may help to better design and predict the success of inoculation efforts.
Acknowledgments
This work was supported by grant KBBE-211684 from the European Commission within the FP7 Framework Programme. We thank Dominique Grandjean and Felippe Dealencastro from the Central Environmental Laboratory of the Ecole Polytechnique de Lausanne for their help in the analysis of DBF content in contaminated sand and Aline Buri for help in determining sand characteristics.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Andreoni V, Gianfreda L. Bioremediation and monitoring of aromatic-polluted habitats. Appl Microbiol Biotechnol. 2007;76:287–308. doi: 10.1007/s00253-007-1018-5. [DOI] [PubMed] [Google Scholar]
- Backman A, Jansson JK. Degradation of 4-chlorophenol at low temperature and during extreme temperature fluctuations by Arthrobacter chlorophenolicus A6. Microb Ecol. 2004;48:246–253. doi: 10.1007/s00248-003-2026-3. [DOI] [PubMed] [Google Scholar]
- Bishop RE, Leskiw BK, Hodges RS, Kay CM, Weiner JH. The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. J Mol Biol. 1998;280:583–596. doi: 10.1006/jmbi.1998.1894. [DOI] [PubMed] [Google Scholar]
- Camilli R, Reddy CM, Yoerger DR, Van Mooy BA, Jakuba MV, Kinsey JC, et al. Tracking hydrocarbon plume transport and biodegradation at Deepwater Horizon. Science. 2010;330:201–204. doi: 10.1126/science.1195223. [DOI] [PubMed] [Google Scholar]
- Coronado E, Roggo C, Johnson DR, van der Meer JR. Genome-wide analysis of salicylate and dibenzofuran metabolism in Sphingomonas wittichii RW1. Front Microbiol. 2012;3:300. doi: 10.3389/fmicb.2012.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe M, Kertesz MA. Autecological properties of soil sphingomonads involved in the degradation of polycyclic aromatic hydrocarbons. Appl Microbiol Biotechnol. 2006;72:1083–1089. doi: 10.1007/s00253-006-0374-x. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V. Cleaning up behind us. The potential of genetically modified bacteria to break down toxic pollutants in the environment. EMBO Rep. 2001;2:357–359. doi: 10.1093/embo-reports/kve100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V. Recombinant bacteria for environmental release: what went wrong and what we have learnt from it. Clin Microbiol Infect. 2009;15:63–65. doi: 10.1111/j.1469-0691.2008.02683.x. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V, Pieper DH, Ramos JL. From the test tube to the environment - and back. Environ Microbiol. 2013;15:6–11. doi: 10.1111/j.1462-2920.2012.02896.x. [DOI] [PubMed] [Google Scholar]
- Desai C, Pathak H, Madamwar D. Advances in molecular and ‘-omics' technologies to gauge microbial communities and bioremediation at xenobiotic/anthropogen contaminated sites. Bioresour Technol. 2010;101:1558–1569. doi: 10.1016/j.biortech.2009.10.080. [DOI] [PubMed] [Google Scholar]
- Fida TT, Moreno-Forero SK, Heipieper HJ, Springael D. Physiology and transcriptome of the polycyclic aromatic hydrocarbon-degrading Sphingomonas sp. LH128 after long-term starvation. Microbiology. 2013;159:1807–1817. doi: 10.1099/mic.0.065870-0. [DOI] [PubMed] [Google Scholar]
- Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOConsortium. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halden RU, Halden BG, Dwyer DF. Removal of dibenzofuran, dibenzo-p-dioxin, and 2-chlorodibenzo-p-dioxin from soils inoculated with Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1999;65:2246–2249. doi: 10.1128/aem.65.5.2246-2249.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson LJ, Firestone MK. Differential effects of permeating and nonpermeating solutes on the fatty acid composition of Pseudomonas putida. Appl Environ Microbiol. 2000;66:2414–2421. doi: 10.1128/aem.66.6.2414-2421.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritash AK, Kaushik CP. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater. 2009;169:1–15. doi: 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- Hong HB, Chang YS, Nam IH, Fortnagel P, Schmidt S. Biotransformation of 2,7-dichloro- and 1,2,3,4-tetrachlorodibenzo-p-dioxin by Sphingomonas wittichii RW1. Appl Environ Microb. 2002;68:2584–2588. doi: 10.1128/AEM.68.5.2584-2588.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jeon CO, Madsen EL. In situ microbial metabolism of aromatic-hydrocarbon environmental pollutants. Curr Opin Biotechnol. 2013;24:474–481. doi: 10.1016/j.copbio.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Johnson DR, Brodie EL, Hubbard AE, Andersen GL, Zinder SH, Alvarez-Cohen L. Temporal transcriptomic microarray analysis of ‘Dehalococcoides ethenogenes' strain 195 during the transition into stationary phase. Appl Environ Microbiol. 2008;74:2864–2872. doi: 10.1128/AEM.02208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DR, Coronado E, Moreno-Forero SK, Heipieper HJ, van der Meer JR. Transcriptome and membrane fatty acid analyses reveal different strategies for responding to permeating and non-permeating solutes in the bacterium Sphingomonas wittichii. BMC Microbiol. 2011;11:250. doi: 10.1186/1471-2180-11-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni PS, Crespo JG, Afonso CA. Dioxins sources and current remediation technologies—a review. Environ Int. 2008;34:139–153. doi: 10.1016/j.envint.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Lim BL. TonB-dependent receptors in nitrogen-fixing nodulating bacteria. Microbes Environ. 2010;25:67–74. doi: 10.1264/jsme2.me10102. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia E, Nikel PI, Chavarria M, de Lorenzo V. The metabolic cost of flagellar motion in Pseudomonas putida KT2440. Environ Microbiol. 2014;16:291–303. doi: 10.1111/1462-2920.12309. [DOI] [PubMed] [Google Scholar]
- Medina-Bellver JI, Marin P, Delgado A, Rodriguez-Sanchez A, Reyes E, Ramos JL, et al. Evidence for in situ crude oil biodegradation after the Prestige oil spill. Environ Microbiol. 2005;7:773–779. doi: 10.1111/j.1462-2920.2005.00742.x. [DOI] [PubMed] [Google Scholar]
- Megharaj M, Wittich RM, Blasco R, Pieper DH, KN T. Superior survival and degradation of dibenzo-p-dioxin and dibenzofuran in soil by soil-adapted Sphingomonas sp. strain RW1. Appl Microbiol Biotechnol. 1997;48:109–114. [Google Scholar]
- Megharaj M, Ramakrishnan B, Venkateswarlu K, Sethunathan N, Naidu R. Bioremediation approaches for organic pollutants: a critical perspective. Environ Int. 2011;37:1362–1375. doi: 10.1016/j.envint.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Miller TR, Delcher AL, Salzberg SL, Saunders E, Detter JC, Halden RU. Genome sequence of the dioxin-mineralizing bacterium Sphingomonas wittichii RW1. J Bacteriol. 2010;192:6101–6102. doi: 10.1128/JB.01030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongodin EF, Shapir N, Daugherty SC, DeBoy RT, Emerson JB, Shvartzbeyn A, et al. Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1. PLoS Genet. 2006;2:e214. doi: 10.1371/journal.pgen.0020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrozika A, Piotrowska-Segetb Z. Bioaugmentation as a strategy for cleaning up of soils contaminated with aromatic compounds. Microbiol Res. 2010;165:363–375. doi: 10.1016/j.micres.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Nam IH, Hong HB, Kim Y-M, Kim BH, Murugesan K, Chang YS. Biological removal of polychlorinated dibenzo-p-dioxins from incinerator fly ash by Sphingomonas wittichii RW1. Water Res. 2005;39:4651–4660. doi: 10.1016/j.watres.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Nam IH, Kim Y-M, Murugesan K, Chang YS. A catabolic activity of Sphingomonas wittichii RW1 in the biotransformation of carbazole. Water Air Soil Pollut. 2012;223:943–949. [Google Scholar]
- Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- Puglisi E, Cahill MJ, Lessard PA, Capri E, Sinskey AJ, Archer JA, et al. Transcriptional response of Rhodococcus aetherivorans I24 to polychlorinated biphenyl-contaminated sediments. Microb Ecol. 2010;60:505–515. doi: 10.1007/s00248-010-9650-5. [DOI] [PubMed] [Google Scholar]
- Roberson EB, Firestone MK. Relationship between desiccation and exopolysaccharide production in a soil Pseudomonas sp. Appl Environ Microbiol. 1992;58:1284–1291. doi: 10.1128/aem.58.4.1284-1291.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggo C, Coronado E, Moreno-Forero SK, Harshman K, Weber J, van der Meer JR. Genome-wide transposon insertion scanning of environmental survival functions in the polycyclic aromatic hydrocarbon degrading bacterium Sphingomonas wittichii RW1. Environ Microbiol. 2013;15:2681–2695. doi: 10.1111/1462-2920.12125. [DOI] [PubMed] [Google Scholar]
- Tchelet R, Meckenstock R, Steinle P, van der Meer JR. Population dynamics of an introduced bacterium degrading chlorinated benzenes in a soil column and in sewage sludge. Biodegradation. 1999;10:113–125. doi: 10.1023/a:1008368006917. [DOI] [PubMed] [Google Scholar]
- Tyagi M, da Fonseca MMR, de Carvalho CCCR. Bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation processes. Biodegradation. 2011;22:231–241. doi: 10.1007/s10532-010-9394-4. [DOI] [PubMed] [Google Scholar]
- van de Mortel M, Halverson LJ. Cell envelope components contributing to biofilm growth and survival of Pseudomonas putida in low-water-content habitats. Mol Microbiol. 2004;52:735–750. doi: 10.1111/j.1365-2958.2004.04008.x. [DOI] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorholt JA. Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch Microbiol. 2002;178:239–249. doi: 10.1007/s00203-002-0450-2. [DOI] [PubMed] [Google Scholar]
- Wang Y, Morimoto S, Ogawa N, Fujii T. A survey of the cellular responses in Pseudomonas putida KT2440 growing in sterilized soil by microarray analysis. FEMS Microbiol Ecol. 2011;78:220–232. doi: 10.1111/j.1574-6941.2011.01146.x. [DOI] [PubMed] [Google Scholar]
- Wick LY, Colangelo T, Harms H. Kinetics of mass transfer-limited bacterial growth on solid PAHs. Environ Sci Technol. 2001;35:354–361. doi: 10.1021/es001384w. [DOI] [PubMed] [Google Scholar]
- Wilkes H, Wittich RM, Timmis KN, Fortnagel P, Francke W. Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by Sphingomonas sp strain RW1. Appl Environ Microbiol. 1996;62:367–371. doi: 10.1128/aem.62.2.367-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittich R-M, Wilkes H, Sinnwell V, Francke W, Fortnagel P. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1992;58:1005–1010. doi: 10.1128/aem.58.3.1005-1010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuuchi E, Yamamoto H, Terakubo S, Okamura N, Naka T, Fujiwara N, et al. Proposal of Sphingomonas wittichii sp. nov. for strain RW1T, known as a dibenzo-p-dioxin metabolizer. Int J Syst Evol Microbiol. 2001;51:281–292. doi: 10.1099/00207713-51-2-281. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36:358–363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.