Abstract
The standard approach to analyzing 16S tag sequence data, which relies on clustering reads by sequence similarity into Operational Taxonomic Units (OTUs), underexploits the accuracy of modern sequencing technology. We present a clustering-free approach to multi-sample Illumina data sets that can identify independent bacterial subpopulations regardless of the similarity of their 16S tag sequences. Using published data from a longitudinal time-series study of human tongue microbiota, we are able to resolve within standard 97% similarity OTUs up to 20 distinct subpopulations, all ecologically distinct but with 16S tags differing by as little as one nucleotide (99.2% similarity). A comparative analysis of oral communities of two cohabiting individuals reveals that most such subpopulations are shared between the two communities at 100% sequence identity, and that dynamical similarity between subpopulations in one host is strongly predictive of dynamical similarity between the same subpopulations in the other host. Our method can also be applied to samples collected in cross-sectional studies and can be used with the 454 sequencing platform. We discuss how the sub-OTU resolution of our approach can provide new insight into factors shaping community assembly.
Introduction
Host-associated microbial communities are known to be of tremendous importance for host fitness, improving nutrient uptake, training the immune system and resisting invasion by pathogens (see, for example, Brestoff and Artis, 2013; Fredricks, 2013; Kamada et al., 2013). Our understanding of these communities, however, remains remarkably poor. The origin, maintenance and importance of community diversity (Fierer and Lennon, 2011), the factors determining community stability and resilience (Shade et al., 2012) and the mechanisms of community assembly (Costello et al., 2012) are only some of the questions driving this rapidly expanding field.
Although most microorganisms cannot be cultured in a laboratory setting, advances in genome-sequencing technology now allow organisms to be probed in their natural environments. In particular, the 16S ribosomal RNA tag-sequencing approach identifies community members using fragments of DNA from the hypervariable regions of the ribosomal 16S gene. The development of this technique and the decreasing cost of high-throughput sequencing have prompted a large number of tag-sequencing experiments, including such large-scale efforts as the Human Microbiome Project or the Earth Microbiome Project. The amount of collected data is growing exponentially. However, our ability to interpret this data still has important limitations.
The de facto standard approach to 16S data analysis begins by clustering reads by sequence similarity into ‘Operational Taxonomic Units' (OTUs); see Figure 1a (Quince et al., 2009; Kunin et al., 2010; Huse et al., 2010). A variety of clustering techniques have been developed and are widely used in popular software tools or packages (Hunt et al., 2008; Schloss et al., 2009; Edgar, 2010; Huang et al., 2010; Edgar et al., 2011; Quince et al., 2011; Schloss et al., 2011; Sul et al., 2011; Caporaso et al., 2012; Zheng et al., 2012; Morgan et al., 2013; Youngblut et al., 2013). Despite significant progress in the development of such software, all clustering-based approaches suffer from a major shortcoming (Prosser et al., 2007; Hamady and Knight, 2009; Schloss and Westcott, 2011). Although an OTU is a useful concept for coarse-graining sequencing data, its definition is not biologically motivated, but as its name acknowledges is purely operational. Sequences assigned to a particular OTU are generally presumed to be close phylogenetic relatives and therefore likely to derive from ecologically similar bacterial subpopulations. However, the assumption that 16S sequence similarity is a good proxy for ecological similarity is notoriously problematic (Prosser et al., 2007; Preheim et al., 2013). Moreover, OTU assignments are not definitive but depend on both the clustering algorithm and the random seed chosen (Schloss and Westcott, 2011).
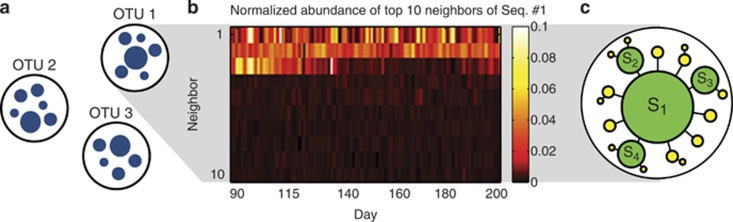
Figure 1.
Clustering reads into OTUs underexploits the quality of modern sequence data. (a) Cartoon illustrating OTU-based noise filtering. Due to sequencing errors, PCR errors or natural intra-strain variability, each bacterial ‘species' generates a cloud of similar 16S sequences (blue circles; the radius of a circle represents the abundance of a given 16S sequence in a sample, and spacing represents distance in sequence space). Clustering reads into OTUs by sequence similarity is a standard approach to filter this noise. (b) Heat map of the abundance, for 100 consecutive samples, of the 10 highest-abundance direct neighbors (Hamming distance=1) of Seq. #1, normalized for each sample to the abundance of Seq. #1 (4600 counts per day on average). Three specific direct neighbors are strongly and consistently overrepresented and exhibit distinct dynamics. (c) Cartoon based on b of the expected structure of an ‘error cloud'. Each circle is a unique sequence, with size representing abundance in a sample. True biological sequences (S1–S4; green circles) generate ‘daughter' variants due to substitution errors (yellow circles). Black lines denote Hamming distance=1 in sequence space.
Several approaches have been proposed to improve the resolution of 16S data analysis beyond the standard 97%-similarity OTUs. Denoising algorithms exploit the predictable structure of certain error types to attempt to reassign or eliminate noisy reads (Huse et al., 2010; Quince et al., 2011; Rosen et al., 2012). These algorithms are widely used for identifying low-abundance (‘rare') species against a noisy background, often with the aim of improving estimates of ecological diversity. These objectives, however, remain very challenging due to issues that no denoiser can fully address. Any error model is necessarily approximate, and no denoising algorithm can deal with errors that are not adequately described by its error model; when calling low-abundance species this issue becomes particularly problematic. An alternative approach termed Distribution-Based Clustering (Preheim et al., 2013) aims to circumvent the limitations of conventional denoisers by using cross-sample comparisons, that is, supplementing sequence information by ecological information (distribution of abundance across multiple biological samples). However, Distribution-Based Clustering as an OTU clustering algorithm also has important limitations: for low-count sequences, cross-sample comparisons necessarily become unreliable, and the execution time is prohibitively long even for moderately-sized data sets.
Here, we build on the above methods to address a distinct question. Rather than trying to further improve the existing approaches to OTU clustering and rare species identification, we combine error-model-based denoising and systematic cross-sample comparisons to resolve the fine (sub-OTU) structure of moderate-to-high-abundance community members in 16S Illumina data. Importantly, our method does not rely on clustering similar sequences together. In this regard, our method is similar to oligotyping (Eren et al., 2013), but our approach does not require manual supervision and applies to an entire community rather than an isolated OTU. Using published data from a longitudinal study where the tongue community of two human individuals was sampled almost daily for several months (Caporaso et al., 2011), we demonstrate that sequence similarity is a very poor predictor of ecological similarity, which we quantify for two bacteria as the correlation of their abundance time traces (‘dynamical similarity'). Thus, most clustering-based approaches would erroneously group together bacterial subpopulations of high ecological diversity for this data set. However, a comparative analysis of the tongue communities of the two individuals also shows that when a pair of 16S tags is observed in both individuals, the dynamical similarity of the pair as measured independently in the two individuals is highly correlated. This correlation falls off substantially when sequences differing by 1 nucleotide (nt) out of 130 are compared. In other words, the exact sequence of the 16S tag carried by a bacterial subpopulation is predictive of its ecology, while even 99.2% similarity between tags of different subpopulations is generally not predictive of dynamical similarity, as defined above. Our results lend support to the recent idea that even a purely 16S-based study can provide insight into functional relatedness of community members (cf. PiCRUST, Langille et al., 2013), while also exhibiting and beginning to quantify the limitations of such methods. We demonstrate the applicability of our approach to a broad range of data set types (host-associated longitudinal; environmental cross-sectional; mock community), providing examples when highly similar sequences were found to exhibit ecologically significant distinctions. Finally, we discuss how the single-nt sub-OTU resolution of our method can provide new insights into factors shaping community assembly.
Materials and methods
Data selection and quality filtering
We used the raw data from a published long-term longitudinal sampling from four body sites (gut: feces, right and left palm, and tongue) of one male and one female individual (Caporaso et al., 2011). In this study, the hypervariable region V4 of the bacterial 16S ribosomal RNA gene was amplified and sequenced with Illumina GA-IIx (Illumina, Inc., San Diego, CA, USA). For details on collection and sequencing see the original reference (Caporaso et al., 2011). Quality-filtered data published with that study is available at MG-RAST:4457768.3-4459735.3 and is sufficient to reproduce our results using provided analysis scripts (see Supplementary Information). However, to investigate the performance of our filtering approach at different quality filtering settings, for this work we used the demultiplexed, but not quality-filtered FastQ data, kindly provided to us by the study authors. We split this data into per-sample FastQ files using a custom MatLab script (Mathworks, Inc., Natick, MA, USA) and subjected it to minimal quality filtering using USEARCH v.7.0.1090 (Edgar, 2010), truncating reads at Phred quality score 2 (other thresholds were also evaluated; see Supplementary Figure S4), trimming to a fixed length of 130 nt and eliminating reads with ambiguous characters (N). In addition, we removed reads with expected number of base call errors exceeding 1 (maxEE parameter in USEARCH). This criterion only eliminated 1% of trimmed reads. Notably, our approach does not rely on assumptions about a maximum number of errors in a read. Finally, to facilitate cross-sample comparisons, we compiled a library of all 1.4M unique reads ever observed and a global table listing the abundances of each of these reads across samples. This was done using a custom Perl script (mergeSeqs.pl). This script and others referenced in bold below are freely available at https://github.com/hepcat72/CFF. Finally, the abundance table was normalized to 2.4 × 104 total reads per sample, to correct for varying sample size.
Read quality varied across lanes, so the number of reads after quality filtering was highest in a subset of tongue and fecal samples. In this work, we focused primarily on the tongue samples, as these come closest to probing the internal dynamics of a community living in a well-defined location on the body; however, the analysis of fecal samples supports the same conclusions and is presented in Supplementary Figure S11.
Tongue samples were distributed over two lanes. The lane 6 samples from the male subject from day 65 onwards (314 consecutive samples covering a period of 355 days, 2.4±0.4 × 104 reads in quality-filtered samples before normalization) had approximately fourfold more reads than those from the female subject and from days 1–64 of the male subject (all on lane 5). Consequently, the analysis below uses the data from the male subject from day 65 onwards, and, for the comparative analysis of the two individuals, also the 135 samples collected from the female subject. The early samples from the male subject (days 1–64) are only used for illustrative purposes (Figure 3d).
To demonstrate the broad applicability of our method we also employed other published data (Supplementary Figures S7 and S11); the data is described in the corresponding legends.
Cluster-free filtering
Clustering can be a useful strategy to coarse-grain 16S data while also reducing noise, but if sequencing noise is low enough, such coarse-graining may not be necessary. At low noise, each community member is predominantly represented by the same 16S sequence, surrounded by a cloud of low-abundance error sequences with the structure of the cloud determined by reproducible error rates. Prior work has described such error clouds in the data (Quince et al., 2009; Edgar, 2013), and the assumption that high-abundance sequences are likely to be error-free is used in several rank-based denoising and chimera-checking algorithms (Single-linkage preclustering, Perseus, Uchime de novo, Uparse and AbundantOTU).
The treatment of reads that are very similar to high-abundance sequences is different across existing algorithms. For example, single-linkage preclustering (Huse et al., 2010) would consider any read differing by a single nt from a higher-abundance sequence (its ‘direct neighbor' in sequence space) as an error. However, some of these reads may actually represent true community members (Preheim et al., 2013). A more nuanced treatment can accept a sequence as likely to be real if its observed abundance is highly unlikely to have arisen in error, given some assumptions about error rates. This idea is at the foundation of error-model-based denoising. It was used in AmpliconNoise (Quince et al., 2011), and its recent implementation in DADA (Rosen et al., 2012) makes DADA, to our knowledge, the best denoiser currently available.
However, no error model is perfect, and for all denoisers, errors not explicitly described by their model are labeled as true sequences. Thus a denoising algorithm alone is insufficient for achieving sub-OTU resolution: if two close sequences that would fall within a single OTU are both identified as ‘probably real', one of these could still be an error. In the context of a single sample, confidently resolving close sequences as ‘independently real' requires a different experimental technique (Faith et al., 2013) or a complete, high-quality reference database of all bacteria in the sample, which in practice is available only for mock communities.
It is possible to resolve this problem in the framework of standard 16S experiments through a comparison of multiple samples, either longitudinal or cross-sectional (Preheim et al., 2013). As an example, Figure 1b shows the abundances of the 10 highest-abundance direct neighbors of the overall top sequence of the tongue community, Seq.#1, for a representative set of 100 consecutive samples. We see that three specific direct neighbors are strongly and consistently overrepresented compared with the other neighboring sequences and, more importantly, exhibit a dynamical behavior of their own (consider, for example, the third most abundant neighbor). This has a clear interpretation (Figure 1c): these three sequences must belong to other, fairly abundant bacterial subpopulations, possibly related to Seq.#1, but distinct and with their own dynamics.
To achieve sub-OTU resolution, we adopt precisely this strategy, namely a cross-sample correlation analysis of individually denoised samples. Which denoiser should we use? DADA would be an excellent option; however, its estimated execution time on the tongue data set used here is 2.3 × 105 s (see Supplementary Information). This is largely due to its exact treatment of probabilities, critically important for the processing of sequences with an abundance of just a few counts. However, for such sequences the imperfections of the error model become non-negligible and cannot be controlled, since cross-sample comparisons are interpretable only for sequences with sufficient abundance. We therefore designed a new, simplified denoiser. Our algorithm, described below, takes two orders of magnitude less time to execute, yet for sequences of moderate abundance considered here achieves performance equal to DADA, as demonstrated using mock community data (Supplementary Table S2).
Cluster-free filtering: the denoiser
For 16S data obtained using the Illumina platform, the main sources of errors are PCR substitutions, PCR chimeras and substitution errors due to Illumina base call errors. Of these, the substitution errors are responsible for generating the largest number of unique sequences (Supplementary Figure S2; see also Edgar, 2013) and have the most predictable structure: their rates can be estimated directly from the data. To do so, we considered the error clouds around the top 10 sequences by overall abundance (in all tongue samples combined). Assuming that most of these sequences are in fact errors, we determined the rates of specific one-nt substitutions (errorRates.pl with z-score threshold of 2; see Supplementary Information). These inferred rates were consistent across error clouds observed in the data (Supplementary Figure S3), with the average error rate of only 0.10% per nt (Supplementary Table 1; compare with Quince et al., 2011; Supplementary Table 2). We then used these error rates to predict the expected abundance of any given sequence if its presence were entirely due to independently generated sequencing errors of its more abundant neighbors (the ‘null model' Supplementary Figure S5; nZeros.pl). Sequences whose abundance exceeded a threshold of 10 counts and the null-model prediction by at least 10-fold (very conservative filtering parameters), were marked as ‘candidates' their presence cannot be explained as an error within a substitution-only error model (getCandidates.pl). Candidate sequences include true biological 16S sequences, but also sequences that arose through a different type of error, most notably PCR chimeras. Chimeric sequences were identified using UCHIME de novo (Edgar et al., 2011) on the pooled data from all samples. Most such sequences were already eliminated by the abundance threshold requirement: if we relax the abundance threshold to 2 (excluding singletons only), we find that the chimeras detected by UCHIME, when present in a sample, have abundance under 10 counts in 95% of cases. However, chimeras of highly abundant parents reproducibly occur at higher abundances (Haas et al., 2011) and are filtered at this step.
Candidate sequences that remained after filtering chimeras were labeled ‘real'. Our highly conservative filtering criteria allow us to assume that this list contains only true biological sequences, that is, there are no false positives (cf. Supplementary Table S2), except possibly those due to some exceptionally frequent errors not described by our error model (see Supplementary Information). This stringency comes at the expense of low-abundance false negatives (true biological sequences labeled as ‘possible noise'). Our strategy is to retain all sequences marked ‘real' in two or more samples (out of 507; getReals.pl). This makes our denoiser specifically adapted to multi-sample analysis: in each sample, only high-confidence detections are identified, which is very fast, and then a liberal criterion applied across samples retains all sequences that ever generated a high-confidence detection, except sample singletons. In particular, we stress that our detection threshold of 10 counts is not equivalent to removing all sequences with abundance below 10; the only sequences excluded from consideration are those that never rise to 10 counts in the entire set of 507 tongue samples, or do so only once. For such sequences, the measured counts are dominated by detection and counting noise.
In the interest of speed, and to ensure the robustness of reported sequence-abundance values with respect to the details of the error model, we did not attempt to remap noisy reads to their most probable source. Our approach relies on the accuracy of measurement of relative abundances of true sequences. The error remapping process modifies sequence counts in a way that depends on the assumptions of the error model, distorting the relative abundance values whenever neighboring sequences are incorrectly classified as ‘reals' or ‘errors'. In contrast, discarding noisy reads leaves the relative abundances intact, as long as the probability of making zero errors is approximately constant across all sequences. This assumption is much weaker than adopting a particular error model. We estimate the zero-error probability at ≈85% (see Supplementary Information); in other words, discarding noisy reads leads only to a ≈15% loss of sequencing depth. If read remapping is desired, the analysis described below can be applied to DADA denoiser output.
Since non-identical reads are never clustered together, ours is a single-nt resolution approach. The complete workflow of cluster-free filtering is outlined in Supplementary Figure S6 and detailed in the Supplementary Information. The code is freely available at https://github.com/hepcat72/CFF.
Results
The starting point for our analysis is a global sequence-abundance table listing the abundances of each unique 16S sequence across samples. We retained the 307 sequences that passed the multi-sample filtering criteria described in Methods, and thus putatively belong to bacteria present in the population at least part of the time. We denote these sequences by their overall abundance rank: Seq.#1, #2 and so on. In this list, 184 pairs of sequences were direct neighbors in sequence space (Hamming distance 1). These pairs had 99.2% sequence similarity but were resolved by our criteria as independently present in the community. The population of bacteria sharing the exact same sequenced fragment of the 16S gene (at 100% identity) is the smallest taxonomic unit resolvable by 16S analysis. For notational convenience, throughout this work we call it the ‘subpopulation' identified by a sequence.
Sequence similarity need not imply ecological similarity, and vice versa
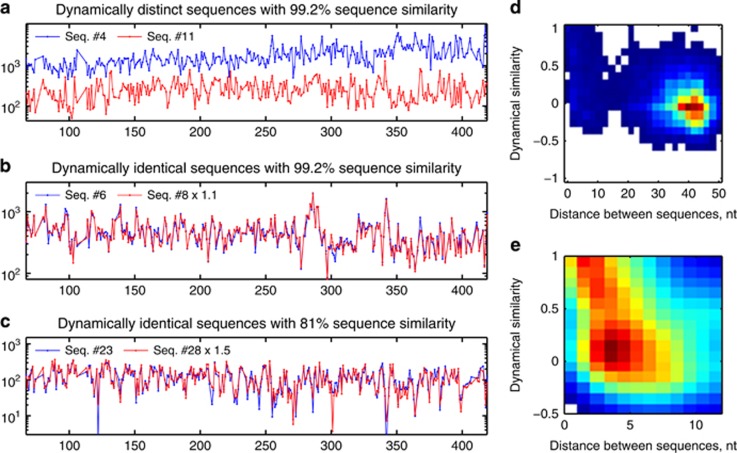
In the standard approach to tag-sequencing data, it is assumed that sequence similarity of 16S hypervariable regions can be used as a proxy for phylogenetic, and therefore ecological, relatedness. Our new filtering method, applied to time-series data, allows us to bypass this assumption and assess ecological relatedness independently, based on the similarity of time traces, since each distinct subpopulation will respond in its own way to variation in environmental conditions (Youngblut et al., 2013), causing the abundance time traces to be more or less correlated (or possibly anticorrelated; see Supplementary Figure S8). Figures 2a–c illustrate this by showing time traces (normalized counts versus observation day) for three examples of sequence pairs. We find that sequences differing by as little as one nt (99.2% similarity) can be ecologically distinct as evidenced by their very different time series (Figure 2a); see also VandeWalle et al., 2012. For comparison, Figure 2b shows another pair of sequences, also with 99.2% sequence similarity but whose abundance time traces appear indistinguishable. The remarkable correlation between these two traces provides an internal control and demonstrates that the much lower correlation of traces in Figure 2a cannot be explained by measurement error but reflects a true ecological difference. Note that the abundances of the two sequences shown in Figure 2b are not equal, but occur with a highly stable ratio. This could reflect a stable difference in abundance of the bacteria they represent, but is more likely caused by differential amplification efficiency of these sequences by the PCR primers (Turnbaugh et al., 2010; Klindworth et al., 2013) and/or a different number of genomic 16S copies per cell (Tourova, 2003). Panels a and b show that sequence similarity need not imply ecological similarity. Finally, Figure 2c illustrates that the converse is also true: sequences exhibiting identical time dependence may have as little as 81% sequence identity.
Figure 2.
Sequence similarity need not imply dynamical similarity, and vice versa. Panels show sequence counts versus observation day, for days 65–420. (a) Seq. #4 and #11, despite 99.2% sequence similarity, display significant differences in time dependence, indicating that these 16S tags belong to ecologically distinct bacterial subpopulations. (b) For Seq. #6 and #8, 99.2% sequence similarity (one-nt difference) is mirrored by near perfect correlation of time series. Red trace renormalized for best overlap. (c) Seq. #23 and #28, with only 81% sequence similarity, nevertheless display near perfect correlation. Red trace renormalized for best overlap. (d) Two-dimensional histogram of dynamical similarity (Pearson correlation of abundance traces, normalized by maximum expected correlation cmax, see text) versus distance in sequence space (nt), for all pairs of the top 200 sequences (19 900 data points). (e) Zoom-in of d (1321 sequence pairs), showing the most similar sequences. Histogram smoothed for clarity.
To quantify the generality of these examples, it is useful to define a measure of the ecological similarity of the bacterial subpopulations represented by two sequences. A natural candidate metric is the Pearson correlation of the measured abundance traces. Note, however, that the maximum correlation one can expect between the time traces of two sequences depends on their abundance: for low-abundance sequences Poisson sampling noise becomes non-negligible and sets an upper bound on the correlation coefficient. We therefore define the ‘dynamical similarity' of two traces as the Pearson correlation of their abundance, normalized by their maximum possible correlation cmax, computed as the correlation of the higher-abundance time trace with a Poisson-downsampled version of itself (see Supplementary Information). For sequence distance, we use the Hamming distance between sequences after pairwise alignment (see Supplementary Information). With these definitions, we can present a two-dimensional histogram of dynamical similarity versus distance in sequence space for all sequence pairs constructed from the top 200 real sequences (Figure 2d). As expected, most sequence pairs exhibit no significant dynamical similarity and are also far apart in sequence space, but a subset of closely similar sequences appears to display some degree of anticorrelation between the two measures. Zooming in on this region (Figure 2e) makes this anticorrelation more apparent; however, even when restricted to the subset shown in Figure 2e, the correlation coefficient remains weak (R=–0.3). Sequences separated by up to six or seven nt (95% sequence similarity) tend to be dynamically similar, the effect increasing for smaller distances, but this general trend is very loose and is not a reliable predictor of similarity for any particular pair. This result was not unexpected, and is frequently used in arguments against over-reliance on the 16S gene sequence (see, for example, Prosser et al., 2007), in favor of methods providing functional information, such as shotgun metagenomics. The novelty of Figure 2e lies in the fact that it was obtained entirely within the framework of 16S tag-sequencing methodology.
Cluster-free filtering can resolve distinct subpopulations with high dynamical similarity
As explained in the previous section, 16S tags with low dynamical similarity clearly derive from distinct bacterial subpopulations, even if the sequences are themselves highly similar. We now consider pairs of sequences with highly correlated time traces such as observed in Figures 2b and c. Such correlated pairs could derive from the same bacterial cells (as multiple genomic copies of the 16S gene, or as exceptionally common PCR errors not included in our model). Alternatively, they could derive from distinct bacterial subpopulations that either occupy the same ecological niche or engage in a strong obligate symbiosis. Such pairs are thus of significant ecological interest, provided it can be shown that the sequences actually derive from different bacterial cells. In this section, we demonstrate that cross-sample correlation analysis can, in some cases, successfully make this subtle distinction between same-cell or different-cell sources.
To draw this distinction, we make use of the following observation. The abundance ratio of two sequences that derive from the same bacterium is set by some sample-independent parameter (for example, involving differential amplification efficiency, 16S copy number, and/or PCR error rate); therefore, any fluctuation in their abundance ratio is due to measurement noise, and must be uncorrelated between samples. Any statistically significant time (or location; see Supplementary Information) correlation of abundance ratio fluctuations, for example, in consecutive (or proximate) samples, is therefore strong evidence that the two sequences are at least partially contributed by physically distinct subpopulations.
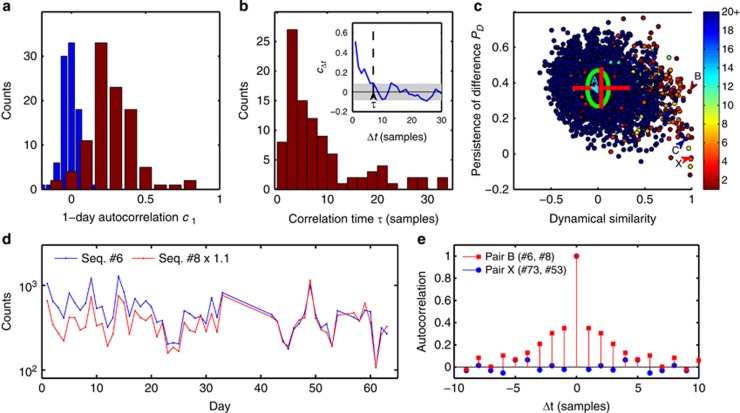
For this approach to succeed, the dynamics of individual subpopulations must be slow enough to allow correlations between consecutive samples to be observed. We therefore began by computing, for each of the top 100 sequences, the autocorrelation function cΔt, defined as the correlation between abundance fluctuations in samples separated by Δt time points, and normalized so that c0=1 (for simplicity, we treat samples as though they were equally spaced in time, which is approximately correct; the mean separation between samples was 1.1 days). The environment experienced by tongue microorganisms changes frequently, and one might have expected that daily sampling would probe the space of possible community states, but provide little information about community dynamics as these would occur on a faster time scale. Surprisingly, we found the time dependence of most sequences in the top 100 to have a significant autocorrelation despite the relatively low sampling rate (Figure 3a). Although conditions on the tongue make fast abundance changes possible, as evidenced by the large, rapid fluctuations in Figures 2a–c, we found the correlation time for the top 100 sequences to be surprisingly long, typically 2–4 days but often longer (Figure 3b), sometimes exceeding a month (Supplementary Figure S10).
Figure 3.
Dynamical similarity versus 16S similarity. (a) 100 most abundant sequences of the population exhibit significant autocorrelation. Histogram of autocorrelation coefficients of sequence abundance for consecutive samples (red), and after randomly permuting sample labels (blue). (b) Histogram of autocorrelation times of 100 most abundant sequences. We define the autocorrelation time τ as the time shift Δt at which the autocorrelation function cΔt falls below the threshold of statistical significance as illustrated in the inset (see Supplementary Information). For 19 sequences the autocorrelation time exceeds 35 days (not shown). (c) Persistence of difference PD for all pairs of sequences from the top 100, plotted against the correlation of their abundances (normalized by maximum expected correlation cmax). Green ellipse indicates mean and standard deviation for the null model obtained by reversing in all pairs the time order for one of the sequences. Most pairs are consistent with the null model, except for a broadening of the correlation coefficient distribution (mean and standard deviations indicated by the red cross). Pairs to the right of the plot are dynamically similar (strong abundance correlation), often accompanied by high sequence similarity (color code indicates Hamming distance between aligned sequences in the pair; see Supplementary Information). Of these, a subset (bottom right) also exhibit weak or negligible persistence of difference. These pairs, such as pair ‘X', most likely correspond to genomic 16S variants found within a single bacterium. Letters A–C identify pairs shown in Figures 2a–c. The large persistence of difference identifies pair B as coming from distinct bacterial cells. (d) Sequence counts versus observation day for early samples of Seq. #6 and #8 (99.2% similarity), normalized as in Figure 2b but excluded there due to relatively poor sequencing depth. The clear separation observed prior to day 40 confirms that these two sequences are contributed at least in part by distinct bacterial subpopulations. (e) Autocorrelation functions of the relative difference Δ(t) for two pairs identified in c: pair ‘B' (red squares; high PD indicative of distinct bacterial cells) and pair ‘X' (blue circles; low PD indicative of 16S variants found within a single bacterium).
These multi-day autocorrelations make it plausible that for physically distinct subpopulations, the fluctuations of their abundances relative to each other could be slow enough to be detectable even if their ecology is similar. Consider two sequences A and B whose abundance time traces are highly correlated. Denote by nA(t), nB(t) the two traces renormalized to the same mean for best overlap, as in Figures 2b and c, and let Δ(t) be their fractional difference in a given sample (a quantity more robust to noise than the naïve abundance ratio):
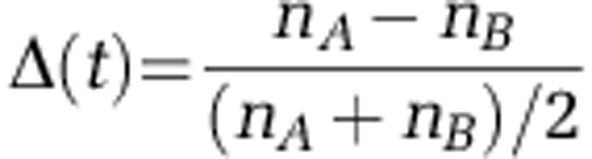 |
If nA,B (t) reflects abundances of two distinct subpopulations, then Δ(t) can be expected to exhibit an autocorrelation on par with that observed for the individual sequences. Intuitively, if on day 1, subpopulation A is, say, 10% more abundant than B, and the dynamics of both are slow, then A is likely to maintain its lead on day 2. In contrast, if the two sequences are genomic variants contained within the same bacterium, then any difference between nA (t) and nB (t) must be due to measurement noise, and Δ(t) will be uncorrelated between samples. We therefore introduce the persistence of difference PD as the 1-day autocorrelation coefficient of Δ(t):
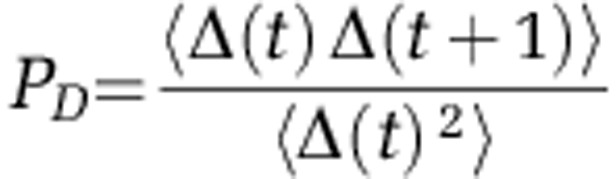 |
where angular brackets denote averaging over time. PD characterizes the persistence of abundance fluctuations of two sequences relative to each other. For sequences arising from the same cells, PD must vanish. Any pair of sequences exhibiting a statistically significant PD must be contributed, at least in part, by two physically distinct bacterial subpopulations. Note that the absolute abundance of a sequence may change dramatically between days (for example, more favorable conditions can cause both subpopulations to proliferate quickly), but the normalization of Δ(t) makes PD insensitive to such overall correlated behavior.
Summarizing the above, we have the following expectation for PD: for a randomly chosen pair of sequences, with insignificant dynamical similarity, PD should be significantly non-zero (due to the slow dynamics of the individual subpopulations; see Supplementary Information), and form a unimodal distribution consistent with the null model of unrelated subpopulations. In contrast, pairs displaying high dynamical similarity come in two types, and the persistence of difference PD should display a bimodal distribution: pairs of sequences found within the same bacterial cell will have vanishing or insignificant PD, while pairs belonging to distinct subpopulations will likely exhibit a persistence of difference comparable with the null-model prediction.
This is precisely what we observe. Figure 3c shows, for all sequence pairs constructed from the top 100 sequences, a scatter plot of their persistence of difference PD versus dynamical similarity as defined previously (the normalized Pearson correlation of their abundances). The mean and standard deviations of the distribution predicted by the null model (unrelated subpopulations) are indicated by the green ellipse, and were computed directly from the data by reversing in all pairs the time order for one of the sequences. The mean and standard deviations of the actual data are indicated by the red cross. We find, as expected, that the PD score of dynamically dissimilar sequence pairs is unimodal and consistent with the null-model prediction. In contrast, the PD score of dynamically similar pairs exhibits the predicted bimodality (right side of the plot), with a subset exhibiting weak or negligible persistence of difference (bottom right). As explained above, we interpret these low-PD pairs as corresponding to genomic 16S variants found within a single bacterium. Letters A–C identify pairs shown on Figures 2a–c. Note that the strong persistence of difference identifies the pair ‘B' as being contributed, at least in part, by distinct bacterial cells, despite 99.2% sequence similarity and an almost perfect correlation of abundances (Figure 2b). Conversely, the low-PD pair ‘C' (with only 81% sequence similarity) likely corresponds to an example of two dissimilar 16S genes contained within a single bacterium. Note the enrichment of pairs with high sequence similarity among the dynamically similar pairs, as indicated by the color code (compare with Figure 2d).
Remarkably, in the case of pair ‘B', the conclusion of distinct bacterial subpopulations drawn from Figure 3c can be confirmed directly. Panel d shows the time traces of this pair for days 1–64 (normalization as in Figure 2b). Due to the relatively poor sequencing depth in these early samples, they were not included in Figure 2b. The clear separation observed prior to day 40 provides an independent confirmation of our conclusion. We stress that these data were not used in the analysis presented in Figure 3c, but the sensitivity of the autocorrelation method was sufficient to identify these sequences as deriving from physically distinct cells based solely on the data shown in Figure 2b. The autocorrelation function of the fractional difference Δ(t) for this pair is shown in Figure 3e. We have verified that the persistence of difference for this pair does not change significantly if any window of 100 consecutive samples is used instead of the full time series (data not shown).
Clustering reads into OTUs vastly underestimates ecological richness
Figure 2a; Supplementary Figures S7, S10, and S11 provide examples of some fine features that standard OTU-based methods would fail to detect, but which become accessible with cluster-free filtering. We now ask whether such cases are the exception or the rule. For a given sequence similarity threshold, we can define, for each of the most abundant sequences, its would-be OTU, namely the ensemble {Si } of all ‘real' sequences within the chosen similarity threshold. We construct the time trace of the abundance of this OTU as the sum of the abundances of all its members. We can now ask: how representative is this time trace of the true behavior of the member sequences? Let {ci} be the correlation coefficients between time traces of individual members and the OTU itself, normalized to the maximum expected correlation as before. We define unweighted and weighted OTU quality scores Qu and Qw as, respectively, the simple average of {ci}, and an average weighted by the abundance of the member:
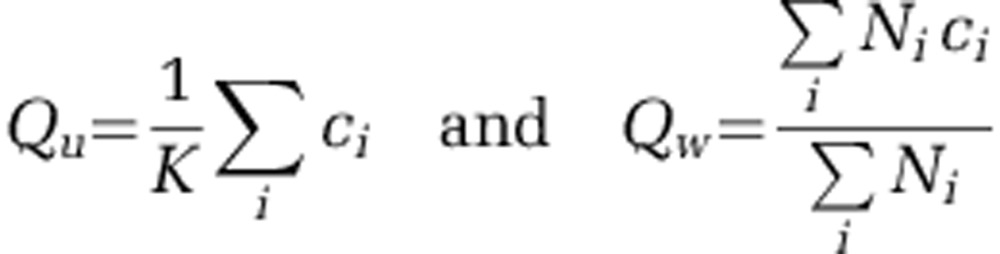 |
Here K is the number of subpopulations in the OTU and Ni is the average abundance of member i. The weighted quality score Qw is always larger, because the most abundant sequence dominates the sum and so is better correlated with the OTU trace. Thus Qw tells us how representative the OTU is of its most abundant member. The unweighted quality score Qu tells us how diverse is the group of subpopulations lumped together into an OTU. If the sequences grouped into an OTU are all dynamically identical (are Poisson-resampled versions of each other at different abundances), both quality scores will be close to 1. If the OTU is dominated by one subpopulation, with other members dynamically different but very low in abundance, we will have Qw≈1, but Qu≪ 1. Finally, if the OTU contains several dynamically distinct subpopulations at comparable abundances, both quality scores will be low.
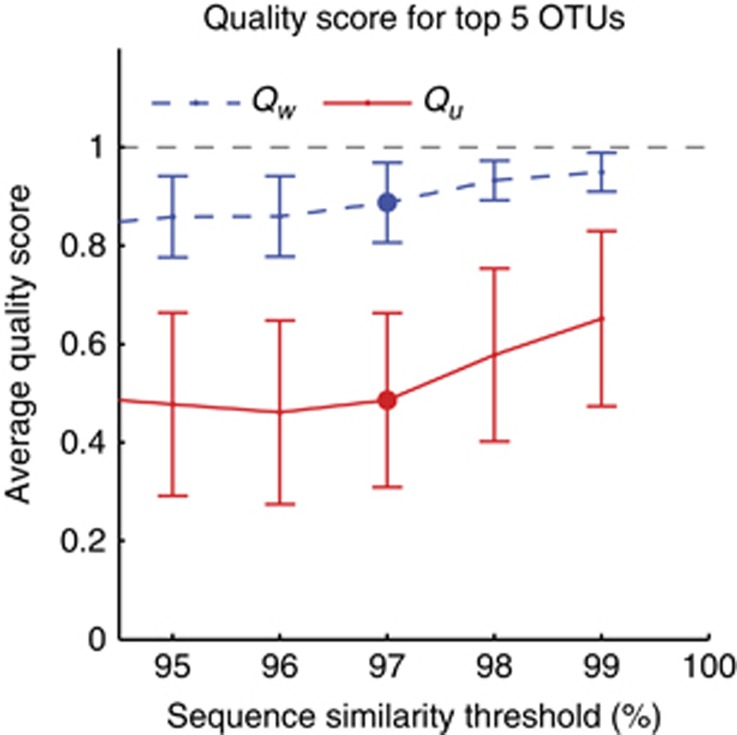
The average quality scores for OTUs assembled around the top 5 sequences are presented in Figure 4 as a function of sequence similarity threshold. The high weighted quality score Qw means that an OTU time trace is, on an average, fairly representative of its most abundant member. The unweighted score Qu is, however, dramatically lower, indicating that the OTUs group together sequences from subpopulations with high dynamical diversity.
Figure 4.
Clustering reads into OTUs vastly underestimates dynamical diversity. Average quality score for OTUs assembled around the top five sequences (defined as the ensemble of ‘real' sequences within a given sequence similarity threshold), as a function of similarity threshold. Error bars are standard deviations across five considered OTUs. Weighted quality score Qw (dashed line; see text) is high, indicating that the OTU time traces are representative of the time traces of their most abundant members. However, the unweighted score Qu (solid line) is dramatically lower, indicating that the OTUs group together sequences with very different time traces. Thus OTUs combine sequences with high dynamical diversity. The commonly used ‘species-level' similarity threshold of 97% is highlighted.
These quality scores rely on abundance time-trace correlations, which become contaminated with noise for low-abundance sequences. For the purposes of Figure 4, to apply these definitions conservatively, we therefore restricted our attention only to high-abundance members of the OTU, considering only sequences from the top 200 by overall abundance. Further, our cluster-free filtering method also has finite resolution, as the sequences we analyze are only 130 nt long and may derive from distinct 16S genes, implying some unresolved diversity. This limited resolution leads to an artificial inflation of OTU quality scores as the similarity threshold approaches 100%. For both these reasons the true quality scores of OTUs are likely even lower (see Supplementary Information).
Exact tag sequence identity is substantially more predictive of subpopulation dynamics than 99.2% sequence similarity
The fact that tag sequence similarity within the 16S gene is only loosely correlated with dynamical similarity (Figure 2e) was not unexpected (see, for example, Prosser et al., 2007 and references therein). At a neutral mutation rate of order 10−9 per base pair per generation (Ochman, 2003), an average difference of a single nt out of 100 would already require divergence for millions of generations. A more precise estimate of divergence time should take into account the possibility of horizontal gene transfer, whose rate in an ecologically relevant setting is hard to assess. However, it is clear that, generically, two bacteria that differ by even one nt in a particular hypervariable region of the 16S gene likely diverged a long time ago. These bacteria are likely to also differ elsewhere in their 16S gene, and to carry even more significant differences in functional parts of their genome.
In contrast, what if we consider two bacteria whose sequenced portions of their 16S genes are identical? Since the length of the sequenced fragment is small (typically ∼100 nt) and the mutation rate is low, these bacteria could still have diverged a very long time ago (Lukjancenko et al., 2010). However, depending on circumstances, the actual time since the last common ancestor may be much shorter. For example, consider two communities that frequently exchange members. If two bacteria drawn from two such communities are 100% identical in their 16S tags, a likely explanation for this identity is a recent exchange event, in which case the entire genomes of these bacteria may be close to identical. We conclude that in the presence of strain exchange between communities, exact sequence identity and near-identity may have fundamentally different implications. The study of Caporaso et al. (2011) sampled the tongue microbiota of two cohabiting individuals (Rob Knight, personal communication), and so strain exchange is likely to be a highly significant factor (Song et al., 2013). We hypothesized, therefore, that these communities would share some non-negligible number of subpopulations at 100% sequence identity, and that these common subpopulations might have similar ecology in both communities.
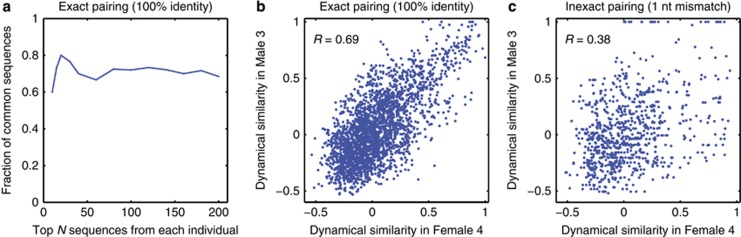
We began by identifying the fraction of common 16S sequences in the list of the top N for each individual (at 100% sequence identity). Based on our strain exchange hypothesis, we expected to find some matches, but were still surprised to find this fraction to be as high as 75% (Figure 5a). Such a high proportion of perfect matches provides strong evidence that the identical sequences found in these two communities most likely diverged from a common ancestor more recently than any pair of close, but non-identical sequences within the same community. The same conclusion is supported by the analysis of fecal samples from the two individuals (Supplementary Figure S11).
Figure 5.
Comparative analysis at 100% sequence identity of oral community composition in two cohabiting individuals reveals shared subpopulations. (a) Fraction of shared 16S sequences, defined as the fraction of common tags (at 100% sequence identity) among the most abundant N sequences in each of the two individuals, plotted as a function of N. (b) Scatter plot of the dynamical similarity of pairs of common sequences, as measured independently in the two individuals, for all possible pairs among the 73 common sequences shared within the top N=100. (c) Same as b, but with intentionally inexact pairing of sequences across individuals (each sequence is mapped to a partner differing by exactly one nt). Despite 99.2% sequence similarity of such pairs, allowing the one-nt mismatch significantly decreases the degree to which dynamical similarity as observed in the two individuals is correlated.
We then considered the 73 sequences that were found among the top 100 of both individuals and asked whether the behavior of these subpopulations was predominantly shaped by their presumed common origin (causing them to be similar) or by local adaptation (causing them to diverge while leaving the 16S region intact; see Lukjancenko et al., 2010). To this end, for each pair of sequences (i, j) drawn from this list, we measured their dynamical similarity independently in the two data sets; SijM for the male and SijF for the female. If the effect of local adaptation were dominant, then the exactness of a match of 16S sequences would not carry much information: the ecologies and genomes would be no more similar between 100%-identical partners in the two communities than between any other sequences within the same bacterial ‘species' (OTU); this scenario is implicitly assumed by taxonomy-based methods. Alternatively, if the ecology were determined primarily by the shared recent ancestor, then identical 16S tag sequences in the two communities would correspond to bacterial subpopulations with almost identical genomes. In this scenario, provided local adaptation did not modify the ecology of a subpopulation significantly, SijM and SijF should be strongly correlated, and unlike the first scenario, this correlation would be noticeably degraded for any less than 100% sequence identity. The latter is indeed what we observe (Figures 5b and c). Figure 5b demonstrates that subpopulations identified by the exact same 16S tags in the two individuals are dynamically similar; see also Supplementary Figures S11D and S12. To obtain Figure 5c, we constructed an ‘inexact pairing' of sequences between individuals, whereupon each sequence from the top 100 in the female individual was matched to the highest-abundance sequence from the top 100 in the male individual that differed from it by exactly one nt, when such a match existed. This matching corresponds to 99.2% sequence identity, yet already substantially degrades the correlation between SijM and SijF (Figure 5c). We conclude that 100% identity of tag sequences has qualitatively different implications from even 99.2% near-identity.
Discussion
In this work, we have demonstrated that cross-sample correlation analysis of denoised 16S data can be exploited to achieve sub-OTU resolution. The cluster-free filtering approach we presented reliably identified up to 20 distinct subpopulations within standard 97% similarity OTUs, and a comparative analysis of oral communities of two cohabiting individuals demonstrates that most such subpopulations are shared between the two communities. Furthermore, subpopulations identified by the exact same 16S tags in the two individuals are dynamically similar, whereas even a single-nt mismatch is enough to degrade this similarity. Overall, our analysis shows that coarse-graining sequence data into OTUs is not essential for ecological applications of 16S tag-sequencing methodology.
Our approach combines two novelties. First and foremost, we do not cluster similar sequences together. Regrettably, in the literature the term ‘clustering' has multiple meanings. Most denoising algorithms aim to assign erroneous reads to their most likely source, to make the abundance estimates of true sequences more accurate. The same term ‘clustering' is used both for this read remapping and for merging multiple true sequences into a single OTU. However, these two practices are fundamentally different. Read remapping constitutes data denoising; as such, it is always advantageous, can be done in a principled way, and can be evaluated against an objective standard of performance. Adding it to our approach would likely somewhat improve the results. In contrast, OTU clustering is a form of data coarse-graining, and the optimal degree of coarse-graining is necessarily application-dependent. Importantly, for some applications it may not be necessary or desirable. When studying coarse features of community composition and dynamics, e.g., comparing communities across habitats (Costello et al., 2009; Huttenhower et al., 2012), coarse-graining is appropriate. For example, metrics of community comparison such as UniFrac (Lozupone and Knight, 2005) are widely used precisely because, by construction, they are not sensitive to OTU sub-structure. However, when studying subtle differences between broadly similar communities, e.g., samples from similar habitats or repeated sampling of the same habitat, the sub-OTU structure becomes a valuable source of insight. This is the intended application for our approach. Although we focused on longitudinal Illumina data, the denoising algorithm we developed does not assume short read length or low error rate and is directly applicable to a wide range of data set types (see examples in Supplementary Figures S7 and S11), provided the error structure is consistent across samples (Preheim et al., 2013). We expect our approach to be useful for investigating the structure and dynamics of discrete community subtypes such as those observed in the vaginal community (Huttenhower et al., 2012).
Our second novelty is to exploit the quantitative advantage offered by multi-sample (time course or cross-sectional) data. Since the copy number of the 16S gene carried by a bacterium is typically unknown (Tourova, 2003), and the PCR amplification bias among different 16S fragments can sometimes reach orders of magnitude (Turnbaugh et al., 2010; Klindworth et al., 2013), the 16S data from a single sample carries very little quantitative information about community composition. In contrast, the ratios of sequence abundance are highly informative and can be measured very precisely, as demonstrated in Figures 2b and c. Recently, time-course data collection has been gaining popularity, as it was recognized that such experiments can offer valuable insight into community dynamics (Shade et al., 2013 and references therein). However, another major advantage of such data sets, namely that changes in sequence-abundance ratios can be measured much more accurately than absolute abundances, is only beginning to be explored. For us, time-series data provides a context where sub-OTU resolution acquires its full power. Specifically, we have shown that cross-sample comparisons enable us to decouple sequence similarity from dynamical similarity while remaining fully within the framework of 16S tag sequencing. High-quality reference databases can complement our approach to facilitate paralog identification. The basic methodology described here should also be extendable to other marker genes.
The new approach described in this work is not a replacement for OTU clustering; it discards low-abundance sequences and so is unsuitable for studies of population-level alpha or beta diversity. However, the novel statistical and computational techniques we present allow full utilization of the quantitative information carried by sequences with a moderate-to-high abundance. This has promising applications for the study of factors affecting community assembly. As discussed above, sub-OTU resolution can provide insight into the prevalence of strain exchange between communities, invasion/extinction dynamics of OTU subpopulations (Supplementary Figure S10), and the time scale of ecological divergence relative to sequence divergence. In addition, the dynamics of individual-specific subpopulations could help characterize the role of host genetics or the host immune system on shaping the community, particularly in the context of highly controlled experiments with germ-free animals.
Acknowledgments
We thank W Bialek, AF Bitbol, CP Broedersz, DS Fisher, R Knight and members of his lab; SA Levin, Y Meir, SW Pacala, SP Preheim and MJ Rosen for helpful discussions; G Caporaso for providing the data and R Edgar for USEARCH support. This work was partially supported under the DARPA Biochronicity program, grant D12AP00025, and the National Science Foundation grants PHY-0957573, PHY-1305525 and CCF-0939370.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, et al. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013;4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439–1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Lennon JT. The generation and maintenance of diversity in microbial communities. Am J Bot. 2011;98:439–448. doi: 10.3732/ajb.1000498. [DOI] [PubMed] [Google Scholar]
- Fredricks DN. The Human Microbiota: How Microbial Communities Affect Health and Disease. Wiley-Blackwell: Hoboken, NJ, USA; 2013. [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;3:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu BF, Gao Y, Fu LM, Li WZ. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukjancenko O, Wassenaar TM, Ussery DW. Comparison of 61 sequenced Escherichia coli genomes. Microbial Ecol. 2010;60:708–720. doi: 10.1007/s00248-010-9717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MJ, Chariton AA, Hartley DM, Court LN, Hardy CM. Improved inference of taxonomic richness from environmental DNA. PLOS One. 2013;8:e71974. doi: 10.1371/journal.pone.0071974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H. Neutral mutations and neutral substitutions in bacterial genomes. Mol Biol Evol. 2003;20:2091–2096. doi: 10.1093/molbev/msg229. [DOI] [PubMed] [Google Scholar]
- Preheim SP, Perrotta AR, Martin-Platero AM, Gupta A, Alm EJ. Distribution-based clustering: using ecology to refine the operational taxonomic unit. Appl Environ Microbiol. 2013;79:6593–6603. doi: 10.1128/AEM.00342-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser JI, Bohannan BJM, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, et al. Essay—the role of ecological theory in Microbial Ecol. Nat Rev Microbiol. 2007;5:384–392. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–U627. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen MJ, Callahan BJ, Fisher DS, Holmes SP. Denoising PCR-amplified metagenome data. BMC Bioinformatics. 2012;13:283. doi: 10.1186/1471-2105-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol. 2011;77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Peter H, Allison SD, Baho D, Berga M, Buergmann H, et al. Fundamentals of microbial community resistance and resilience. Front Microbiol. 2012;3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Caporaso JG, Handelsman J, Knight R, Fierer N. A meta-analysis of changes in bacterial and archaeal communities with time. ISME J. 2013;7:1493–1506. doi: 10.1038/ismej.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sul WJ, Cole JR, Jesus ED, Wang Q, Farris RJ, Fish JA, et al. Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc Natl Acad Sci USA. 2011;108:14637–14642. doi: 10.1073/pnas.1111435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourova TP. Copy number of ribosomal operons in prokaryotes and its effect on phylogenetic analyses. Microbiology. 2003;72:389–402. [PubMed] [Google Scholar]
- Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeWalle JL, Goetz GW, Huse SM, Morrison HG, Sogin ML, Hoffmann RG, et al. Acinetobacter, Aeromonas and Trichococcus populations dominate the microbial community within urban sewer infrastructure. Environ Microbiol. 2012;14:2538–2552. doi: 10.1111/j.1462-2920.2012.02757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblut ND, Shade A, Read JS, McMahon KD, Whitaker RJ. Lineage-specific responses of microbial communities to environmental change. Appl Environ Microbiol. 2013;79:39–47. doi: 10.1128/AEM.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZJ, Kramer S, Schmidt B. DySC: software for greedy clustering of 16S rRNA reads. Bioinformatics. 2012;28:2182–2183. doi: 10.1093/bioinformatics/bts355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.