Abstract
The microbiota of multi-pond solar salterns around the world has been analyzed using a variety of culture-dependent and molecular techniques. However, studies addressing the dynamic nature of these systems are very scarce. Here we have characterized the temporal variation during 1 year of the microbiota of five ponds with increasing salinity (from 18% to >40%), by means of CARD-FISH and DGGE. Microbial community structure was statistically correlated with several environmental parameters, including ionic composition and meteorological factors, indicating that the microbial community was dynamic as specific phylotypes appeared only at certain times of the year. In addition to total salinity, microbial composition was strongly influenced by temperature and specific ionic composition. Remarkably, DGGE analyses unveiled the presence of most phylotypes previously detected in hypersaline systems using metagenomics and other molecular techniques, such as the very abundant Haloquadratum and Salinibacter representatives or the recently described low GC Actinobacteria and Nanohaloarchaeota. In addition, an uncultured group of Bacteroidetes was present along the whole range of salinity. Database searches indicated a previously unrecognized widespread distribution of this phylotype. Single-cell genome analysis of five members of this group suggested a set of metabolic characteristics that could provide competitive advantages in hypersaline environments, such as polymer degradation capabilities, the presence of retinal-binding light-activated proton pumps and arsenate reduction potential. In addition, the fairly high metagenomic fragment recruitment obtained for these single cells in both the intermediate and hypersaline ponds further confirm the DGGE data and point to the generalist lifestyle of this new Bacteroidetes group.
Introduction
Multi-pond solar salterns consist of a series of interconnected ponds with increasing salinity, from seawater level to sodium chloride saturation (Benlloch et al., 2002; Pašić et al., 2005; Oh et al., 2010; Boujelben et al., 2012). In continuously operated salterns, such as the Santa Pola (Spain) facilities studied here, seawater is pumped through a set of shallow ponds where sequential precipitation of calcium carbonate, calcium sulfate and sodium chloride occurs. This salinity gradient is accompanied by a succession of different groups of microorganisms, from marine to extremely halophilic microbiota, each adapted to a salinity range.
Saltern ponds have been traditionally considered ‘steady-state' systems because of the stability of both salinity and microbial composition of the different ponds (Guixa-Boixereu et al., 1996; Ghai et al., 2011). Although it is generally assumed that the composition of saltern microbial communities is ‘fairly stable over time even when subjected to environmental perturbations' (Gasol et al., 2004; Ghai et al., 2011), most studies do not include different time points and provide only snapshots of the microbial diversity without addressing in situ temporal dynamics. However, some studies hint at a more dynamic picture of these systems. A metagenomic analysis of the San Diego salterns community (Rodriguez-Brito et al., 2010) using samples harvested at various time points separated by 1 day to >1 year showed that most abundant microbial ‘species' persisted, but their relative abundances and microdiversity changed over time. Notwithstanding, this study only included two time points (01 July 2004 and 28 November 2005) for ponds of medium high salinities (above 18%), and thus its conclusions cannot be extrapolated to wider temporal ranges for this kind of environments. More recently, a study of the Tunisian solar salterns of Sfax (Boujelben et al., 2012) also found a temporal succession of phylotypes in medium salinity (around 20%) ponds, with some specific bacterial sequences appearing only at some time points along the year.
In order to gain insight into the changes experienced by microbial communities in saltern ponds we have undertaken a comprehensive seasonal study of five ponds of medium and high salinity (from 18% to 40% total salts) from Santa Pola solar salterns. The prokaryotic community of these saltern ponds has been studied for decades using several approaches, depending on the available tools, from classical cultivation to culture-independent techniques. The lower-medium (15–25%) salinity ponds are inhabited by members of the Proteobacteria, Cyanobacteria, Bacteroidetes or haloarchaea-like Halosarcina or Halorubrum, whereas crystallizers (ponds where NaCl precipitates) are dominated by high numbers of the archaeon Haloquadratum walsbyi and extremely halophilic Bacteroidetes, such as Salinibacter ruber, that account for up to 75% and 18% of the total community, respectively (Antón et al., 1999; Benlloch et al. 2002; Burns et al. 2004; Estrada et al., 2004; Ghai et al., 2011; Narasingarao et al., 2012; Zhaxybayeva et al., 2013). Recently, the use of Next-Generation Sequencing (NGS) metagenomics and single-cell genomics has led to the discovery of new abundant groups (Ghai et al., 2011; Narasingarao et al., 2012), such as a group of low GC Actinobacteria in low and intermediate salinities, and a new archaeal linage of Nanohaloarchaea in crystallizers.
Our results indicate that saltern microbiota is dynamic and within a given pond microbial composition can change considerably along the year. In addition, total salinity is not the only factor structuring microbial assemblages in hypersaline environments as ponds of the same salinity harbored different microbial communities. Some microbial phylotypes were strongly associated with specific environmental conditions while others were very widely distributed both spatially and temporally. Especially noteworthy was the case of an uncultured Bacteroidetes phylotype that was present along the whole salinity gradient and could thus constitute a generalist strategist. Here, we have further characterized this bacterial group by means of database search and single-cell genome analyses.
Materials and methods
Sample collection
Five contiguous ponds of Bras del Port salterns (Santa Pola, Spain, 38° 12′ N, 0° 36′ W) were analyzed in this study (Figure 1): two medium concentrators (CM1 and CM2), a brine concentrator (CCAB), and two crystallizers (CR30 and CR41). Samples were collected on the following dates in 2006: 23 January (JN), 7 March (MR), 26 April (AP), 6 June (J1), 27 June (J2), 25 July (JL), 5 September (SP), 2 October (OC) and 28 November (NV).
Figure 1.
Aerial photograph of Bras del Port saltern ponds, Santa Pola, Alicante. Water is pumped from the sea to preconcentrators, medium concentrators, a brine concentrator and finally to the crystallizers. The ponds analyzed in this study are framed with dotted white lines: CM1 and CM2 (medium concentrators), CCAB (brine concentrator) and CR30 and CR41 (crystallizers). Arrows indicate the water circuit.
Environmental parameters
Temperature, pH and salinity were measured in situ while solar radiation and rainfall data were provided by the meteorological station of Bras del Port. Chemical analyses (sulfate, chloride, carbonate, phosphate, sodium, potassium, calcium, magnesium and ammonium) were carried out by atomic absorption spectrometry (Unicam SOLAAR 969, Unicam Ltd., Cambridge, UK) at the Department of Agrochemistry and Environment (University Miguel Hernández de Elche, Spain).
CARD-FISH
Archaeal and bacterial counts were measured following an optimized CARD-FISH protocol modified from Pernthaler et al. (2004). Samples were fixed with formaldehyde (7% final concentration; Sigma, St Louis, MO, USA) at 4 °C for 16 h, diluted (1/1000, 1/500 and 1/200) in 1X phosphate-buffered saline (8 g NaCl, 1.44 g Na2HPO4, 2 g KCl, 2 g KH2PO4, pH 7.0) and filtered through 0.2-μm pore diameter GTTP Isopore filters (Millipore, Madrid, Spain). The filters were stored at −20 °C until used. Cell permeabilization was carried out with proteinase K (150 μg ml−1 final concentration; Promega, Madrid, Spain) in Tris-HCl 20 mM pH 8.0 at 37 °C for 5 min, followed by incubation with lysozyme (3 mg ml−1 final concentration; Sigma) in TE buffer pH 8.0 at 37 °C for 60 min. Peroxidase inactivation was performed in 0.1 M HCl at room temperature for 1 min. Probes Eub338-I-III and Arc915 were used for Bacteria (Amann et al., 1995; Daims et al., 1999) and Archaea (Massana et al., 1997), respectively. Probe Non338 was used to check for non-specific hybridization (Wallner et al., 1993). Hybridization was performed in filters for 150 min at 35 °C in an isotonically humid chamber. For signal amplification, filters were incubated with fluorescein–tyramides conjugates at 37 °C for 30 min. Finally, filters were stained with DAPI (4′,6-diamidino-2-phenylindole-dihydrochloride; 1 μg ml−1; Sigma) for 1 min and observed using a Leica DMLA microscope (Leica Microsystems, Barcelona, Spain).
Nucleic acids extraction
Samples (500 ml) were centrifuged in a Beckman Coulter Avanti J-25centrifuge (Beckman Instruments, GmbH, Munich, Germany), with a JLA-16 rotor at 38 000 g, for 15 min at 4 °C. Cell pellets were resuspended in 600 μl of extraction buffer (100 mM Tris-HCl pH 8.0, 100 mM EDTA, pH 8.0) in 2 ml phenol-resistant tubes. Lysozyme (3 mg ml−1 final concentration) was added, and samples were incubated for 15 min at 37 °C with agitation (180 r.p.m.). Then proteinase K (150 μg ml−1 final concentration) and sodium dodecyl sulfate 10% (6 μg ml−1 final concentration) were added, and the samples were incubated for 30 min at 37 °C (180 r.p.m.); 120 μl of 5 M NaCl and 120 μl of hexadecyltrimethylammonium bromide (10% CTAB, 0.7 M NaCl) were added to the samples and incubated at 65 °C for 10 min, followed by three cycles of freezing in liquid nitrogen and thawing in a water bath at 65 °C. Finally, 900 μl of phenol:chloroform:isoamyl alcohol (25:24:1) were added to each sample and then centrifuged for 5 min at 16 000 g (4 °C). After centrifugation, the aqueous phases were transferred to clean tubes, and the phenol extraction was repeated until the interphase between the aqueous and organic phases was completely clean (two or three times approximately). Nucleic acids were precipitated by adding 0.1 volumes of sodium acetate 3 M (pH 4.8) and 0.6 volumes of cold isopropanol (−20 °C) and centrifuged for 30 min at 16 000 g (4 °C). Nucleic acids were then washed with 70% ethanol and centrifuged for 15 min at 16 000 g (4 °C). Pellets were dried for 30 min to evaporate residual ethanol and resuspended in 50–100 μl of milli-Q water. The extracted nucleic acids were analyzed by electrophoresis in 0.8% agarose gels (LE, FMC Bioproducts, Rockland, ME, USA) and visualized under ultraviolet light after staining with ethidium bromide (100 μg ml−1).
PCR amplification and DGGE analysis of 16S rRNA genes
Archaeal and bacterial 16S rRNA genes were partially amplified with forward primers 341f-GC and 344F-GC, respectively, and the reverse universal primer 907R (Muyzer et al., 1996; Schäfer et al., 2001). Each PCR reaction contained MgCl2 1.5 mM, Tris-HCl 10 mM pH 9.0, KCl 50 mM, 200 μM of each dNTP, 0.2 M of each primer, 1 U Taq DNA polymerase recombinant (Invitrogen, Prat de Llobregat, Spain) and approximately 100 ng of template DNA. The PCR reactions were performed in a PTC-100 (Peltier-Effect Cycling). The PCR program for Bacteria was: 94 °C for 5 min, 65 °C for 1 min, 72 °C for 3 min and 9 touchdown cycles of: 94 °C for 1 min, 65 °C (with a decreasing of 1 °C in each cycle) for 1 min, 72 °C for 3 min, followed by 20 cycles of: 94 °C for 1 min, 55 °C for 1 min, 72 °C for 3 min. The PCR program for Archaea was: 94 °C for 5 min and 30 cycles of: 94 °C for 30 s, 56 °C for 45 s, 72 °C for 2 min. To minimize double-band formation, both programs included a final extension of 30 min at 72 °C (Janse et al., 2004). A reconditioning PCR was performed to avoid heteroduplex formation using a 10-fold dilution of the original PCR product (Thompson et al., 2002). Reconditioning PCR conditions were: 94 °C for 3 min, 3 cycles of 94 °C for 15 s, 55 °C for 30 s, 72 °C for 2 min and finally one cycle of 7 min at 72 °C. PCR products were concentrated and purified with Gel Band Purification Kit (GE Healthcare, Madrid, Spain), eluted in 20–30 μl milli-Q water and quantified using NanoDrop ND-1000 spectrophotometer (Thermo, Wilmington, MA, USA).
DGGE was performed by using the D-Code System (Bio-Rad, Madrid, Spain). PCR products were loaded onto 6% (w/v) polyacrylamide (acrylamide:bis-acrylamide gel stock solution 37.5:1; Bio-Rad), with 45–65% denaturing gradient (where 100% of denaturant contains 7 M urea and 40% formamide) in 1X TAE buffer (40 mM Tris, pH 8.0; 20 mM acetic acid; 1 mM EDTA) and subjected to 18 h of electrophoresis at 60 °C and 70 V. After electrophoresis, the gels were stained with SYBR Green (1:10 000; Fluka, Madrid, Spain) for 15 min, washed in TAE 1X for 30 min and visualized using a computer image analyzer Typhoon 9410 (Amersham Biosciences, Cerdanyola del Valles, Spain). Bands were cut, transferred to sterile 1.5-ml tubes with 20 μl milli-Q water and incubated overnight at 4 °C. Two microliters (approximately 100 ng) of each band were then re-amplified with the same primer pairs. Purity of amplification products was checked by DGGE and, finally, products yielding a single band were sequenced in the Technical Services Unit of the University of Alicante, using a GeneAmp PCR System 2400 and an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Alcobendas, Spain). PCR products were sequenced with primer 907R in an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). For an initial affiliation, sequences were checked at the GenBank nucleotide database using the BLAST software at the National Centre of Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) (Altschul et al., 1997). The tree reconstruction was performed by using the ARB software package (Ludwig et al., 2004), with the SILVA REF 111 database (Pruesse et al., 2007). The tree reconstruction was made using the neighbor-joining algorithm using a 40% conservational filter as implemented in the ARB package. To stabilize the tree, a selection of about 700 almost complete high-quality sequences representing a wide range of phyla within Bacteria and Archaea were included. For the final tree, only sequences of Chlorobi were kept as outgroup. Partial sequences were aligned against the same database using the SINA aligner, and the aligned sequences were inserted in the preexisting neighbor-joining tree by using the parsimony tool, all implemented in the ARB package. The DGGE band sequences obtained here have been deposited in the GenBank database (accession nos. FJ648083-FJ648198 and KF679127-KF679133).
SAG (single amplified genome) genomic sequencing, assembly and annotation
A 50-μl water sample taken from CR30 in June 2011 was used for single-cell genome analysis. At that time, the pond had a salinity of 37.2% and harbored 1.74 × 107 cells ml−1. Single-cell sorting was performed at the Single Cell Genomics Center (Bigelow Laboratory, Bigelow, ME, USA) as described elsewhere (Martínez-García et al., 2012a, 2012b). In brief, prokaryotic community was DNA-stained with SYTO-9, and single cells with high nucleic acid content were sorted with a MoFlo (Beckman Coulter) flow cytometer. Then, single cells deposited in 384-well plates were lysed with KOH, and their genomes were amplified by a multiple displacement amplification (Raghunathan et al., 2005). These generated SAGs were PCR screened for 16S rRNA genes by using the primer sets 27F (5′-AGRGTTYGATYMTGGCTCAG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) for Bacteria and Arc-344F (5′-ACGGGGYGCAGCAGGCGCGA-3′) and Arch-915R (5′-GTGCTCCCCCGCCAATTCCT-3′) for Archaea. Sixteen S rRNA gene sequences of Bacteroidetes SAGs have been deposited in the GenBank database (accession nos. KF679134–KF679143). Finally, five SAGs belonging to the uncultured BC3 group were selected, and their genomes were sequenced by using paired-end Hi-Seq 2000 Illumina Technology (Solexa, San Diego, CA, USA) according to the manufacturer's protocol at the Center for Genomic Regulation (Barcelona, Spain).
Reads were 96 bases long and had an average insert size of 550 bases. Raw Illumina reads, which have been deposited in SRA (BioSample accessions: SAMN02434673, SAMN02434674, SAMN02434675, SAMN02434676, SAMN02434677) were preprocessed before assembly to trim either at the first undetermined base or at the first base having a PHRED quality below 20. The pairs having at least one read shorter than 30 (or 20 for SAG AB578_D06) bases after trimming were excluded from the assembly process (see Supplementary Table S1). SOAPdenovo2 (Luo et al., 2012) was used to assemble paired-end reads into contigs. In order to ensure the minimum number of contigs with the highest N50 and N90 values, different combinations of kmer sizes were tested (range 21–41, in steps of 2) and merging rounds (from 0 to 3) allowing up to three mismatches during the merging process. Final assembly statistics are shown in Supplementary Table S2.
Genomic annotation was performed at the US Department of Energy Joint Genome Institute (JGI-DOE) with the pipeline annotation Prodigal (Hyatt et al., 2010). These sequence data annotations were produced by the JGI-DOE (http://www.jgi.doe.gov/) in collaboration with the user community, and the corresponding SAGs were deposited and made accessible at the Integrated Microbial Genomes of JGI with the following IMG genome Taxon ID numbers: AB577-O10 (2522125006), AB578-D06 (2522125009), AB578-K19 (2522125007), AB578-L11 (2522125008) and AB577-P23 (3300002078). Analyses of average nucleotide identities values were performed by using JSpecies (Richter and Rosselló-Móra, 2009).
Metagenomic analyses
The basic approach of Rusch et al. (2007) was used to estimate the abundance of SAGs in the previously described metagenomes SS19 and CR30 (Ghai et al., 2011). BLAST+ v2.2.22 was used to recruit metagenome sequences to each SAG assembly using the following parameters: −evalue 0.0001 –perc_identity 60 –outfmt 6. Then, BLAST output was parsed according to the selected cutoff identities, and the percentage of unique reads for each metagenome–genome pair was determined at 100, 95, 90, 80 and 70% identity thresholds. Finally, these hits were normalized according to the respective genome and metagenome sizes. To determine the presence of the previously characterized genes involved in arsenic resistance (Morgante and González-Pastor, personal communication) in the CR30 metagenome and the resulting Bacteroidetes SAGs assembly, a custom database was built with these genes, and BLAST searches were performed with an e-value threshold of 0.00001 by using the stand-alone blast version BLAST+ v2.2.22.
Multivariate analysis
Dimension reduction of environmental parameters (salinity, ionic composition, PAR, temperature, solar radiation and hours of light) was carried out by principal component analysis using the SPSS software (IBM, Chicago, IL, USA) (Business Editors/High-Tech Writers, 2002). Relationships between environmental and biological variables (cell numbers and Shannon index diversity, calculated using the PAST program (Hammer et al., 2001)) were analyzed by the Redundancy Analysis (RDA) lineal ordination method. The Canonical Correspondence Analysis (CCA) unimodal ordination method was used to correlate environmental variables with prokaryotic phylotypes, while environmental and biological parameters were correlated using RDA. In both cases, Monte Carlo tests with 499 permutations were carried out to ensure the significance of canonical axes. CCA and RDA were carried out using the CANOCO 4.5 software package (Microcomputer Power, Ithaca, NY, USA) with the tool CANODRAW for biplot visualization (ter Braak and Smilauer, 2002).
Shannon index (H) was calculated according to the following formula where pi is the proportion of phylotype ‘i' relative to the total number of phylotypes:
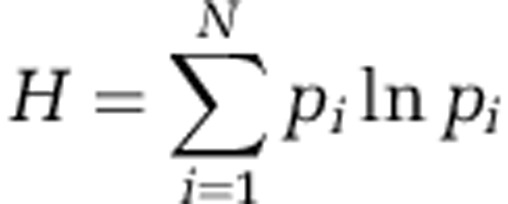 |
Analysis of Similarity (ANOSIM) test was used to compare the variation in phylotypes abundance and composition among temporal samples of each pond. In addition, Similarity Percentage tool was used to identify the contribution of phylotypes to similarities bewteen different samples from each pond Supplementary Table S6). ANOSIM analysis was carried out with the Bray-Curtis distance and 999 permutations by means of the PRIMER 6.0 software (Plymouth Marine Laboratories, Plymouth, UK) computer program (Clarke, 1993).
Results and discussion
Description of samples: spatial and temporal ‘environments'
Samples were taken from five hypersaline (Figure 1) ponds at nine time points during the year 2006, yielding a total of 45 water samples. For each of them, salinity, pH, photosynthetically active radiation, ionic composition (four cations and five anions) and temperature were measured; radiation, rainfall and hours of light were also recorded for every sampling time (Supplementary Table S3). These sixteen ‘environmental' parameters will be referred to as the independent variables. Total cell counts, Bacteria and Archaea numbers and diversity (Shannon indices) will be considered as the dependent variables (Supplementary Table S4). In order to reduce the number of independent variables and make them amenable for subsequent analyses, a principal component analysis was carried out and three components (C1–C3), which explained 82% of environmental data variance, were obtained (Supplementary Table S5). Component 1 was mainly determined by salinity, pH and calcium concentration. Hours of light, solar radiation and rainfall dominated C2, while C3 was defined by sodium, chloride and magnesium concentrations. When the 45 samples were arranged based on their three component values, they were grouped according to their pond of origin (medium concentrators, CM1 and CM2; brine concentrator, CCAB; and crystallizers, CR30 and CR41) and the sampling season. The corresponding spatial and temporal ‘environments' are marked with discontinuous lines in Supplementary Figure S1.
Archaeal and bacterial diversity
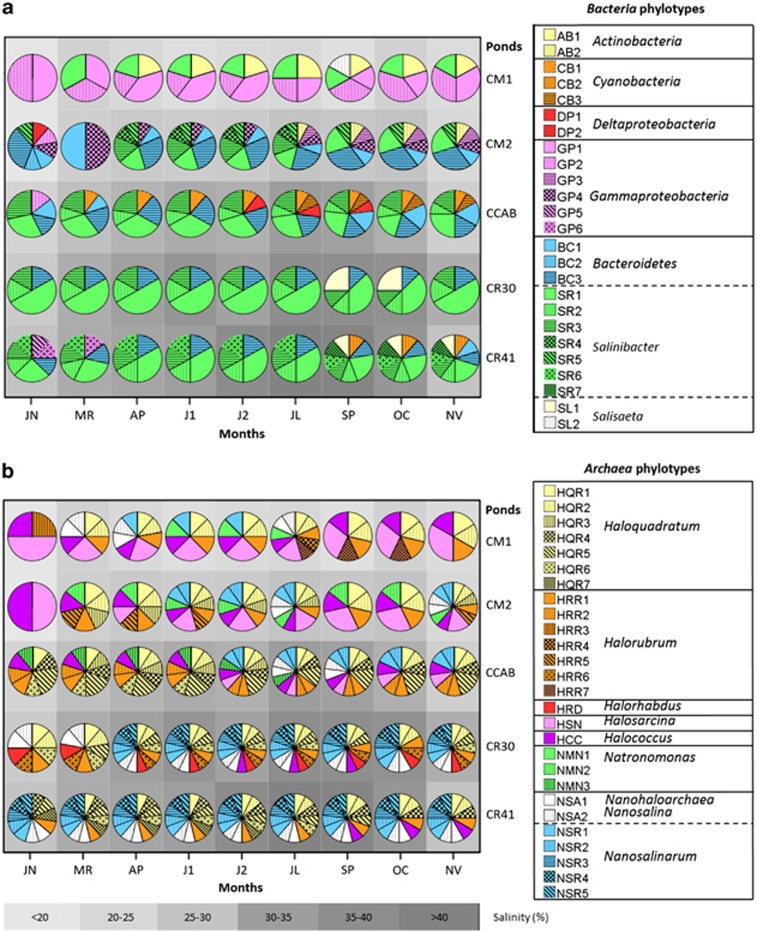
Bacteria and Archaea assemblages were analyzed by DGGE (Supplementary Figure S2), which yielded a specific band pattern for each sample. Bands were recovered from each gel, sequenced and the corresponding partial 16S rRNA gene sequences grouped into phylotypes (that is, a group of partial 16S rRNA gene sequences sharing ⩾97% similarity). We take this value as a conservative boundary that would embrace species or very closely related species of the same genus according to Stackebrandt and Goebel (1994). A total of 45 phylotypes were detected (25 bacterial and 20 archaeal, Tables 1 and 2, respectively) whose distribution changed throughout the year in the different ponds (Figure 2). For labeling purposes, these groups of sequences were named as the closest cultured genera, although it must be noticed that this does not necessarily mean that the phylotypes belonged to these genera.
Table 1. Bacteria phylotypes retrieved during the course of this study.
| Phylotypes | Uncultured closest relativea Cultured closest relativea | Similarity | Pondsb | Months | Salinity |
|---|---|---|---|---|---|
| Phylum | ACTINOBACTERIA (AB) | ||||
| Genus | Candidatus Flaviluna | ||||
| AB1 | Uncultured marine Bacteria clone MAS85 (AM747379) (1) | 96% | CM2 | ANNUAL (except JN) | 20–30% |
| Candidatus Flaviluna lacus partial, strain MWH-Creno3D3 (AM939567.1) | 94% | ||||
| Genus | Candidatus Rhodoluna | ||||
| AB2 | Uncultured Actinobacteria, clone lagoaC01 (EF598921.1) (3) | 98% | CM1 | JL-NV | 20–30% |
| Candidatus Rhodoluna planktonica, strain MWH-Dar1 (AJ565415.1) | 96% | ||||
| Phylum | CYANOBACTERIA (CD) | ||||
| Genus | Dactylococopsis | ||||
| CB1 | Dactylococopsis sp. strain PCC 8305 (AJ000711.1) | 99% | CCAB | ANNUAL | 20–40% |
| Genus | Euhalothece | ||||
| CB2 | Euhalothece sp. strain MPI 95AH10 (AJ000709.1) | 97% | CCAB | ANNUAL (except JN) | 20–40% |
| Genus | Cyanothece | ||||
| CB3 | Cyanothece sp. 115 (DQ243690.1) | 79% | CR41 | SP, OC, and NV | 20–40% |
| Phylum | DELTAPROTEOBACTERIA (DP) | ||||
| Genus | Desulfuromonas | ||||
| DP1 | Uncultured Proteobacteria, clone At12OctA12 (AY862790.1) | 94% | CM2 | J2, JL and SP | 20–30% |
| Desulfuromonas alkaliphilus strain Z-0531 (DQ309326.1) | 86% | ||||
| Genus | Desulfosarcina | ||||
| DP2 | Uncultured Bacteria clone P9_LJL_0205_030 (EU677285.1) | 99% | CCAB | JN | 20–30% |
| Desulfosarcina sp. clone SB4_53 (AY177791.1) | 96% | ||||
| Phylum | GAMMAPROTEOBACTERIA (GP) | ||||
| Genus | Alkalilimnicola | ||||
| GP1 | Uncultured Bacteria clone 185ZF11 (CU467460.1) | 99% | CM1 | ANNUAL | 19–30% |
| Alkalilimnicola ehrlichei MLHE-1 (CP000453.1) | 94% | ||||
| Genus | Alkalispirillum | ||||
| GP2 | Uncultured Bacteria clone 185ZE06 (CU467472.1) | 99% | CM1 | ANNUAL | 18–40% |
| Alkalispirillum sp. Z 7008 (DQ217920.1) | 99% | CM2 | JN | ||
| CCAB | JN | ||||
| Genus | Nitrococcus | ||||
| GP3 | Uncultured Bacteria clone 185ZE06 (CU467472.1) | 4% | CM2 | JL-NV | 20–30% |
| Nitrococcus mobilis (ATCC 25380) | 88% | ||||
| Genus | Halospina | ||||
| GP4 | Halospina denitrificans strain 30–40%D 1-3 (DQ072719.1) | 98% | CM2 | ANNUAL | 18–30% |
| Genus | Halovibrio | ||||
| GP5 | Uncultured Bacteria clone SN144 (EU735693.1) Halovibrio denitrificans strain C4-2 (EU868859.1) | 83 | CR41 | JN | 20–30% |
| GP6 | Pseudomonas sp., clone 2B4 (AY987840.1) | 95% | CR41 | JN, MR | 20–30% |
| Halovibrio denitrificans, strain C4-2 (EU868859.1) | 94% | ||||
| Phylum | BACTEROIDETES (BC) | ||||
| Genus | Psychroflexus | ||||
| BC1 | Uncultured Bacteria, clone E4aA01 (DQ103640.1) | 98% | CM2 CCAB CR41 | ANNUAL | 20–40% |
| Psychroflexus sp. sp.gap-d-25 (DQ530461.1) | 98% | ||||
| Genus | Owenweeksia | ||||
| BC2 | Owenweeksia hongkongensis (DQ103638.1) | 98% | CM2 | JN | 18% |
| Genus | Cytophaga | ||||
| BC3 | Uncultured Bacteria, clone LCKS000B34 (EF201719.1) | 98% | All except CM1 | ANNUAL | 18–>40% |
| Cytophaga sp. Dex80-37 (AB239011.1) | 87% | ||||
| Genus | Salinibacter | ||||
| SR1 | Salinibacter ruber DSM 13855 (CP000159.1) | 99% | CMs | ANNUAL (except JN) | 20–>40% |
| CCAB and CRs | ANNUAL | ||||
| SR2 | Uncultured Bacteria, clone SFC1D061 (AM981340.1) | 98–99% | CCAB | ANNUAL (except JN) | 30–>40% |
| Salinibacter ruber DSM 13855 (CP000159.1) | 93% | CRs | ANNUAL | ||
| SR3 | Uncultured Bacteria, clone SFA1H111 (CU467389.1) | 99% | CM2 | AB—JL | 30–>40% |
| Salinibacter ruber DSM 13855 (CP000159.1) | 92% | CCAB | ANNUAL | ||
| SR4 | Uncultured Bacteria, clone SFC1D061 (AM981343.1) | 78% | CM2 | AB—JL | 20–30% |
| Salinibacter ruber DSM 13855 (CP000159.1) | 71% | ||||
| SR5 | Uncultured Bacteria, clone SFA1H111 (CU467389.1) | 93% | CM2 | AB—NV | 20–30% |
| Salinibacter ruber DSM 13855 (CP000159.1) | 88% | ||||
| SR6 | Salinibacter sp. clone SFD1E121 (FN393500.1) | 93% | CR41 | ANNUAL | 20–>40% |
| Salinibacter ruber DSM 13855 (CP000159.1) | 89% | ||||
| SR7 | Uncultured Bacteria, clone SFA1C121 (CU467418.1) | 99% | CR41 | SP, OC and NV | 20–>40% |
| Salinibacter ruber DSM 13855 (CP000159.1) | 95% | ||||
| Genus | Salisaeta | ||||
| SL1 | Salinibacter sp. clone salH12 (EF598986.1) | 98% | CR30 | SP and OC | 20–>40% |
| Salisaeta longa strain S4-4 (EU426570.2) | 93% | CR41 | SP, OC and NV | ||
| SL2 | Uncultured Bacteria, clone XB27 (FM211011.1) | 96% | CM1 | SP | 20–30% |
| Salisaeta longa strain S4-4 (EU426570.2) | 93,5% | ||||
Pubmed access number.
Ponds: Medium concentrators CMs: CM1 and CM2; Brine concentrator: CCAB; and Crystallizers: CRs: CR30 and CR41.
Table 2. Archaea phylotypes retrieved during the course of this study.
| Phylotypes | Uncultured closest relativea Cultured closest relativea | Similarity | Pondsb | Months | Salinity |
|---|---|---|---|---|---|
| Genus | Haloquadratum | ||||
| HQR1 | Haloquadratum walsbyi DSM 16790 (AM180088.1) | 98% | CMs | ANNUAL (except JN) | 20–>40% |
| CCAB | ANNUAL | ||||
| CR30 | ANNUAL | ||||
| CR41 | ANNUAL (except JN) | ||||
| HQR2 | Uncultured Haloarchaea clone Cry7_clone27 (GQ374968.1) | 99% | CMs | ANNUAL (except JN) | 20–>40% |
| Haloquadratum walsbyi C23 (NR_028207.1) | 92% | CCAB | AB-NV | ||
| CR30 | ANNUAL | ||||
| CR41 | J2-NV | ||||
| HQR3 | Uncultured Haloarchaea, clone Cry7_clone20 (GQ374960.1) | 95% | CM2 | MR-JL and NV | 20–40% |
| Haloquadratum walsbyi DSM 16790 (AM180088.1) | 95% | CCAB | MR-J2 | ||
| HQR4 | Haloquadratum walsbyi DSM 16790 (AM180088.1) | 98–99% | CCAB | ANNUAL | 20–>40% |
| Haloquadratum walsbyi DSM 16790 (AM180088.1) | CR41 | ANNUAL (except JN) | |||
| HQR5 | 99% | CCAB | ANNUAL | 20–>40% | |
| CR30 | MR, SP and NV | ||||
| HQR6 | Haloquadratum walsbyi DSM 16790 (AM180088.1) | 99% | CCAB | EN-J1 | 20–>40% |
| CR30 | ANNUAL(except SP and OC) | ||||
| CR41 | MR-J2 | ||||
| HQR7 | Uncultured Archaea, clone 186ZH05 (CU467175.1) | 96% | CR41 | JN-J2 | 20–>40% |
| Haloquadratum walsbyi DSM 16790 (AM180088.1) | 96% | ||||
| Genus | Halorubrum | ||||
| HRR1 | Uncultured Archaea clone Baj_clon46 (GQ374995.1) Halorubrum orientale strain EJ-52T (AM235786.1) | 99–100% | CMs CCAB CR30-CR41 | ANNUAL (except JN from CM2) | 20–>40% |
| HRR2 | Uncultured Archaea clone Cry7_clone60 (GQ375002.1) | 98% | CCAB | ANNUAL | 20–>40% |
| Halorubrum sp. DV427 (FJ492047.1) | 92% | ||||
| HRR3 | Uncultured Archaea clone SFH1B111 (FN391236.2) | 94% | CM1 | JN | 19% |
| Halorubrum sp strain DW6 (EF194098.1) | 92% | ||||
| HRR4 | Halorubrum sp. clone SFH1A082 (FN391273.2) Halorubrum sp. strain EN-2 (AM268115.1) | 85% | CM1 | JL | 20–30% |
| HRR5 | Uncultured Archaea clone Baj_clone46 (GQ374995.1) | 94% | CM2 | ANNUAL | 20–30% |
| Halorubrum orientale, strain EJ-52T (AM235786.1) | 94% | ||||
| HRR6 | Uncultured Archaea clone Cry7_clone81 | 99% | CR30 | ANNUAL | 20–>40% |
| Halorubrum alkaliphilum strain DZ-1 (NR_028206.) | 98% | ||||
| HRR7 | Uncultured Haloarchaea clone SFG1H031 (AM947489.1) | 85% | CM1 | JL, SP and OC | 20–30% |
| Halorubrum sp. EN-2 (AM268115.1) | 85% | ||||
| Genus | Halorhabdus | ||||
| HRD | Uncultured Haloarchaea clone 186ZF04 (CU467268.1) | 99% | CR30 | ANNUAL | 20–>40% |
| Halorhabdus utahensis AX-2 (NR_028172.1) | 91% | ||||
| Genus | Halosarcina | ||||
| HSN | Uncultured Haloarchaea clone 3a-PML 22% pond (AF477932.1) | 99% | CMs | ANNUAL | 18–40% |
| Halosarcina pallida (EF055454.1) | 94% | CCAB | J2-NV | ||
| Genus | Halococcus | ||||
| HCC | Halococcus hamelinensis (NR_028168.1) | 92% | CMs | ANNUAL | 18–>40% |
| CCAB | ANNUAL | ||||
| CR30 | J2- JL | ||||
| CR41 | SP-NV | ||||
| Genus | Natronomonas | ||||
| NMN | Natronomonas sp. DV462A (FJ492048.1) | 93% | CM1 | De J1 a JL | 20–30% |
| NMN2 | Natronomonas moolapensis 8.8.11 strain CSW8.8.11 (AY498645.2) | 93% | CM2 | ANNUAL (-JN) | 20–30% |
| NMN3 | Natronomonas sp. DV462A (FJ492048.1) | 94% | CCAB | JN-JL | 30–40% |
| Linage | Nanohaloarchaea | ||||
| NSA1 | Candidatus Nanosalinarum (Clade J0756AB) | 97–99% | CM1 | MR, AB and JL | 20–>40% |
| CM2 | JL and NV | ||||
| CCAB | JL and SP | ||||
| CRs | ANNUAL | ||||
| NSA2 | CM1 | MR, AB and JL | |||
| NSR1 | Candidatus Nanosalina (Clade J07AB43) | 97–99% | CM1 | AP, J1 and J2 | 20–>40% |
| CM2 | J1, J2, JL and NV | ||||
| CCAB | J2-NV | ||||
| CRs | AP-NV | ||||
| NSR2 | CM1 | AP, J1 and J2 | |||
| CM2 | J1, J2, JL and NV | ||||
| NSR3 | CCAB | J2-NV | 20–>40% | ||
| NSR4 | CRs | AP-NV | |||
| NSR5 | CRs | AP-NV | |||
Pubmed access number.
Ponds: Medium concentrators CMs: CM1 and CM2; Brine concentrator: CCAB; and Crystallizers: CRs: CR30 and CR41.
Figure 2.
Temporal distribution of relative abundance of bacterial (a) and archaeal (b) phylotypes in the five analyzed ponds (medium concentrators CM1 and CM2, brine concentrator CCAB, and crystallizers CR30 and CR41). A complete description of the phylotypes is provided in Tables 1 and 2.
As in previous studies (Benlloch et al., 2002), Actinobacteria, Gammaproteobacteria related to Alkalilimnicola and Nitrococcus and Cyanobacteria were detected in lower and medium salinity ponds (CMs and CCAB) together with Euryarchaea related to the genera Halosarcina, Halococcus and Halorubrum. The relative recovery frequency of these groups decreased along the salinity gradient, at the same time that the proportion of phylotypes related to Bacteroidetes, in particular with S. ruber, and Archaea, such as Hqr. walsbyi, increased over 25% salinity (Figure 2, and Supplementary Table S6, in which a list of the phylotypes found in each pond at all sampling times is shown). These two groups are frequently dominant in close-to-saturation ponds and will be discussed in more detail later. In addition, phylotypes related to Nanohaloarchaea were detected in the different ponds. As mentioned above, this new group had been discovered by using 'modern' molecular ecology techniques, such as NGS metagenomics and single-ell genomics (Ghai et al., 2011; Narasingarao et al., 2012). The detection of nanohaloarchaeal phylotypes in our samples reinforces the use of the 16S rRNA gene DGGE approach despite the fact that some authors considered it to be obsolete and very limited for environmental studies (Legault et al., 2006), as it can only retrieve phylotypes that account for >0.4% of the community (Casamayor et al., 2000). Indeed, around 92% of the phylotypes detected in June 2008 in CR30 using a NGS metagenomic approach (Ghai et al., 2011) have been detected at different time points in the different samples studied here. In addition, many phlotypes described in this work were not present in the NGS study.
A total of seven phylotypes related to the genus Salinibacter (see Table 1 and Figure 2) were recovered in this study. In 1998, the presence of extremely halophilic bacteria (later described as ‘Salinibacter') was discovered in the crystallizer CR30 (Antón et al., 2000). However, at that time, only two phylotypes were detected, and only one of them (named EHB-1, from ‘extremely halophilic bacterium' 1) has been brought to pure culture and classified as S. ruber (Antón et al., 2002). Currently, all Salinibacter sp. isolates recovered from different hypersaline environments around the world (Antón et al., 2008, 2013) correspond to EHB-1. Here, in samples taken 8 years after the first description of Salinibacter in these salterns, a total of seven different phylotypes related to this genus have been detected. Only one of them (SR1) had a similarity >97% with S. ruber isolates, providing a new example of the biases imposed by cultivation even in ‘culturable' species. Other Salinibacter-related phylotypes (see Table 1) presented the highest identities with environmental clones from medium salinity ponds in Tunisian salterns (Baati et al., 2008, 2010, 2011). Remarkably, some Salinibacter phylotypes, like SR4 and SR7, were detected only at specific temporal points from CM2 and CR41, respectively, underscoring the dynamic nature of the microbial communities in the salterns.
Although highly divergent Salinibacter-related phylotypes have been frequently retrieved from hypersaline environments (Mutlu et al., 2008), most Haloquadratum sequences previously detected were very closely related to each other. For instance, Hqr. walsbyi cultured strains have <1.8% divergence in their 16S rRNA gene sequences (Legault et al., 2006) while environmental phylotypes recovered from highly distant saltern ponds in Spain (Bolhuis et al., 2006; Legault et al., 2006), Israel (Soerensen et al., 2005), Turkey (Mutlu et al., 2008), Tunisia (Baati et al., 2008), Peru (Maturrano et al., 2006) and Australia (Oh et al., 2010) normally present <1% divergence. In contrast, some Haloquadratum sequences recovered in this work (Table 2 and Figure 2) showed a higher divergence (up to 5%) in the analyzed 16S rRNA gene fragment. In particular, phylotype HQR3 was detected only at some time points in low and medium salinity ponds while HQR7 was detected only in the crystallizer CR41 that had not been sampled previously. These examples, again, underscore the relevance of taking samples at different time points and from different crystallizers.
A main outcome of this study is that, despite the fact that two analyzed crystallizers displayed very similar salinities (Figure 2, Tables 1 and 2), remarkable differences in their microbial community structures were found (28.73% dissimilarity as calculated with ANOSIM, Supplementary Table S7). This fact is intriguing if we consider that both ponds are fed with the same water from CCAB. Although a common portion of CCAB community remained in both crystallizers, there were also crystallizer-specific phylotypes, such as Halorhabdus-related phylotypes that were detected at all sampling times in CR30 or HQR7 Haloquadratum and Salinibacter phylotype SR6 present only in CR41 samples (HQR7 from January to June and SR6 in all samples). In addition, CR41 harbored Cyanobacteria from September to November, which is noteworthy as normally this group of prokaryotes is absent from close-to-saturation hypersaline waters being the green alga Dunaliella the only photoautotroph (Estrada et al., 2004; Gasol et al., 2004).
Prokaryotic community dynamics and their relation to environmental parameters
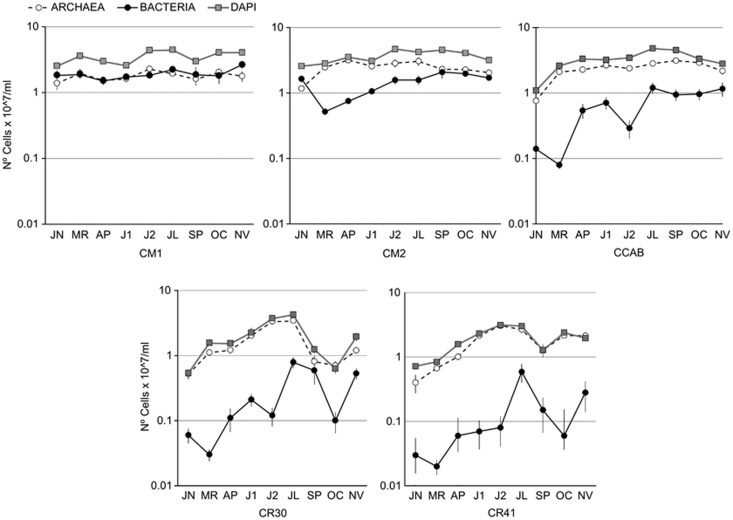
Total microbial counts were in the range previously observed in solar salterns (Guixa-Boixereu et al., 1996; Antón et al. 1999, 2000; Benlloch et al. 2002; Oren, 2002; Estrada et al., 2004; Burns et al., 2007; Mutlu et al. 2008; Eulloumi et al., 2009) ranging from 6.3 × 106 to 4.79 × 107 cells ml−1 in CR30 (OC) and CCAB (JL), respectively (Figure 3). Although throughout the concentration process from seawater to saturated brines there is an overall increase of cell density, at concentrations >20% this trend is not always observed (Guixa-Boixereu et al., 1996; Boujelben et al., 2012), as shown here. Total counts changed in the different ponds throughout the year, with lower salinity ponds being more stable than crystallizers. As expected, the highest counts in each pond were reached in summer samples while samples taken in colder months harbored less dense communities. This is in agreement with the low microbial numbers (around 5 × 106 cells ml−1) detected in Andean Maras salterns (Maturrano et al., 2006), with an annual average temperature <20 °C, which present the lowest cell concentration reported from crystallizer ponds.
Figure 3.
Temporal variation of total counts (grey symbols), Archaea (white symbols) and Bacteria (black symbols) in the different ponds.
Bacterial and archaeal numbers (determined by CARD-FISH) also changed with salinity and in every pond throughout the year. As expected for hypersaline systems (Antón et al., 2000, Maturrano et al., 2006; Mutlu et al., 2008) Bacteria were less abundant than Archaea in almost all the samples, except in a few CM samples (Figure 3).
Along with the changes in cell numbers, there was also a temporal succession of different phylotypes in each pond (Figure 2). Changes in phylotype composition mainly occurred during winter–spring and summer–autumn transitions and were more drastic in CMs and CCAB than in crystallizers (in fact, CR30 was the steadiest pond). Microbial diversity in the environment can be explained in the frame of the ‘core-seed bank' model proposed by Pedrós-Alió (2006), according to which ‘core' (most abundant) microorganisms would be maintained through active growth, while minority species (‘seed' bank) would be below the detection threshold of molecular techniques, such as DGGE. However, apart from differences among environments, temporal variations should also be taken into account as, depending on the time of sampling, a given phylotype could be part of either the core or the seed bank. For instance, that would be the case of Salinibacter SR4 phylotype that was only detected from AP to JL samples (CM2), while was below the DGGE detection limit (that is, part of the seed bank) the rest of the year.
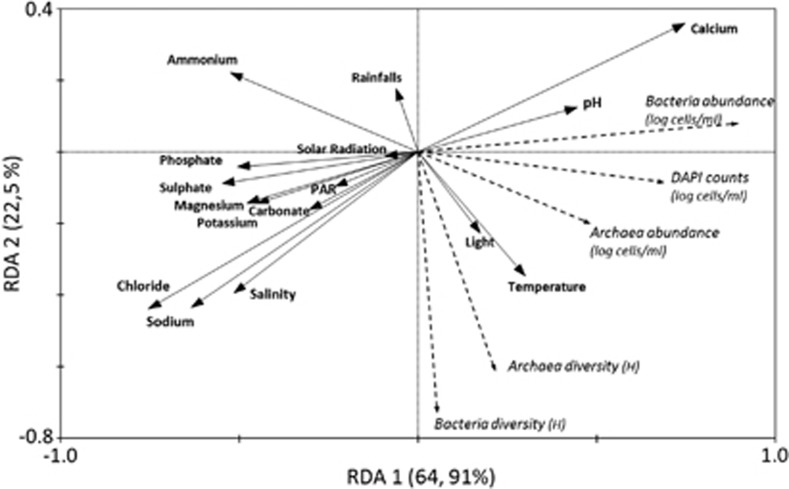
Relationships among environmental parameters and prokaryotic numbers and diversity were investigated by redundant detrended analysis (RDA; Figure 4; see Supplementary Tables S8–11). In this analysis, RDA1 and RDA2 ordination axes explained 87.5% total variance data. Salinity was positively correlated with most ions and negatively with calcium, although ammonium concentration was independent of salinity. Numbers of Bacteria and total cell counts showed a negative dependence with salinity, while Archaea was rather independent of this variable (Supplementary Tables S12 and S13). This may seem contradictory with previous results (Guixa-Boixereu et al., 1996; Antón et al., 2000; Pedrós-Alió, 2005) that indicated that archaeal numbers increased with salinity. This discrepancy is explained, because the correlation between archaeal numbers and salinity is only circumscribed to some months. However, this correlation disappears when the whole data set is considered. In addition, if ponds of lower salinity (that is, from marine to around 15%) had been included in the analysis, correlation between salinity and archaeal number would probably be more apparent. Both Archaea and Bacteria diversity were highly influenced by salinity and by meteorological factors (Supplementary Table S8). In particular, archaeal diversity was strongly correlated with hours of light and temperature. Temperature had also been found to have a strong influence on the prokaryotic community structure in ∼20% salinity ponds from Sfax solar salterns (Boujelben et al., 2012).
Figure 4.
Redundance detrented analysis (RDA) canonical ordination biplot of environmental and biological parameters. Environmental parameters are represented by black arrows and biological parameters by dotted arrows. DAPI, Archaea and Bacteria abundances are represented by the logarithm of the number of cells per milliliter and the diversity of Archaea and Bacteria by Shannon diversity indices (H). RDA yielded two synthetic canonical axes (RDA1 and RDA2) that explained 89.2% of data variance (Supplementary Table S7).
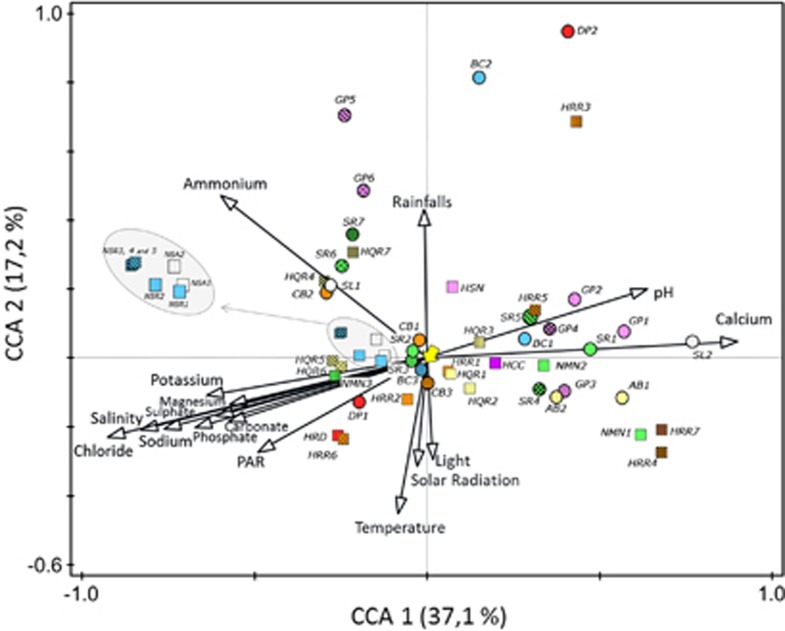
The different phylotypes found in the analyzed ponds were arranged by canonical correspondence analysis, CCA, according to their optimum environmental factors (Figure 5). CCA axes 1 and 2 explained 54.3% total variance data (Supplementary Table S11). As in the RDA analysis, the first canonical axis (CCA1) was highly correlated with salinity and ion concentrations (except ammonium) and the second one (CCA2) with meteorological factors. One of the main outcomes of this analysis was that phylotypes belonging to the same genus could have very different environmental optima (for instance, this is the case of SR1 and SR6, both related to Salinibacter sp. but located in very different regions of CCA space; see Figure 5). In addition, ammonium concentration, which was less correlated with the rest of ions, influenced the distribution of some phylotypes, such as SR7 or HQR7 (related to S. ruber and Hqr. walsbyi, respectively). Nanohaloarchaea-related phylotypes showed a relatively narrow range of environmentally optimum conditions (circled in Figure 5) with Candidatus Nanosalinarum phylotypes (except NRS1) being more ‘halophilic' than Candidatus Nanosalina phylotypes.
Figure 5.
Canonical correspondence analysis (CCA) ordination biplot of phylotypes (symbols) and environmental parameters (arrows). Zone A (right) is defined by low salinity, high calcium concentration and high pH values; zone B (center) represents medium values (and non-determining environmental factor) and zone C (left) high salinity, low calcium concentration, and low pH. Archaeal (squares) and bacterial (circles) phylotypes are named as in Tables 1 and 2. Symbol colors are as in Figure 2. The yellow arrow points to BC3 phylotypes, which were further analyzed by SGC. Nanosalina (white) and Nanosalinarum phylotypes (blue squares) are enclosed within a grey line.
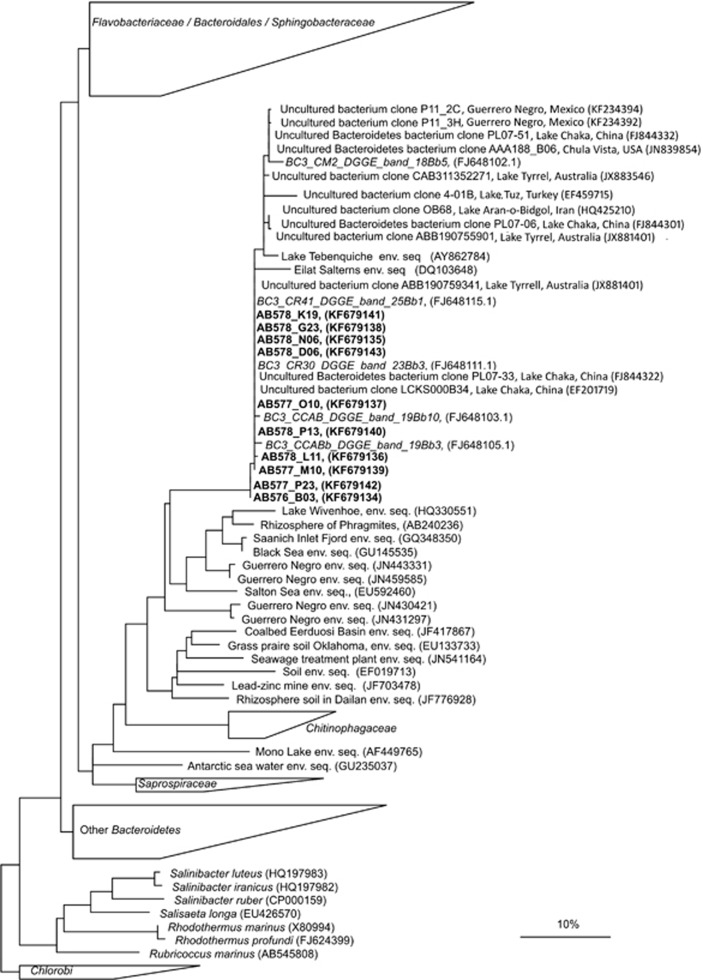
Phylotypes located in the center of the CCA biplot were found under a wide range of environmental conditions and could thus correspond to generalists. Among them, especially worth mentioning is the phylotype BC3 that was detected from 18% to 41% salinities in different samples taken throughout the whole year (see Figure 2). This phylotype (Figure 6) belongs to a clade merely formed by environmental clones of uncultured Bacteroidetes. The closest cultured bacterium, Cytophaga sp. Dex80-37, was only very distantly related to BC3 (87% sequence identity) and affiliates differently within the Flavobacteriaceae/Bacteriales/Sphingobacteriaceae branch (Figure 6). Database searches indicated that sequences very closely related (ore even identical) to BC3 were retrieved in the NGS study of the Santa Pola salterns mentioned above (Ghai et al., 2011), as well as in other studies from hypersaline environments worlwide, such as in Lake Chaka (China), Chula Vista salterns (California, USA), Lake Tyrrel (Australia) and Lake Aran-o-Bidgol (Iran) (Jiang et al., 2007; Wang et al., 2011; Podell et al., 2013). However, in none of these studies there is an explicit reference to this group nor is its ubiquity acknowledged. This lag between the first report of environmental sequences and the recognition of their ecological relevance is not uncommon, and it happened, for instance, with nanohaloarchaeal sequences, which were first reported in 1999 (Grant et al., 1999), but their abundance and worldwide distribution were not clearly stated until more than 10 years later (Narasingarao et al., 2012).
Figure 6.
Phylogenetic reconstruction based on a neighbor joining analysis of a subset of almost complete sequences belonging to the phylum Bacteroidetes. DGGE band sequences corresponding to phylotype BC3 are in italics and 16S rRNA corresponding to the SAGS obtained from a CR30 sample are in bold. Members of Chlorobi were included as outgroup. Partial sequences of DGGE and clones were added to the tree using the parsimony tool implemented in ARB (Ludwig et al., 2004). The bar indicates 1% sequence divergence.
Characterization of the phylotype BC3 by means of single-cell genomics
Despite its apparent abundance and wide distribution, the phylotype BC3 has not been cultured yet. For this reason, we decided to carry out a culture-independent approach to gain some insight into its metabolism by using high-throughput single-cell sequencing. For that purpose, we performed fluorescence-activated cell sorting in combination with whole-genome amplification to recover the genetic information for these prokaryotes in the crystallizer CR30. Results of the 16S rRNA gene PCR screening and sequencing for the generated SAGs showed that a total of 10 SAGs belonging to the Bacteroidetes BC3 phylotype were recovered (Figure 6), 5 of which were selected for further genome sequencing. The resulting genome assembly from the low-coverage Illumina sequencing for each SAGs was ≈0.4 Mpb (Supplementary Tables S1 and S2), recovering approximately 10% of the genome (considering that the mean of Bacteroidetes genomes is 4±1.7 Mpb; data obtained from IMG prokaryote database, dated 15 December 2012). The GC content was significantly lower (47%) than other sequenced extremely halophilic Bacteroidetes, such as S. ruber (66% GC). Comparison of average nucleotide identities (Supplementary Figure S3) among the sequenced SAGs indicated that all of them might belong to the same species as the average nucleotide identity threshold for species circumscription had been observed to be between 94% and 96% (Richter and Rosselló-Móra, 2009). This result must be taken cautiously as fragments accounting for at least 20% of the genomes of the strains under comparison are needed for stable average nucleotide identity calculations (Richter and Rosselló-Móra, 2009). However, except for L11, we are confident with regard to the results, given that the amount of aligned genome stretches for average nucleotide identity calculations were relatively high and ranged between 12% and 26% of the total sequenced DNA. The hypothesis that all cells belonged to the same species is reinforced by their almost identical 16S rRNA gene sequences.
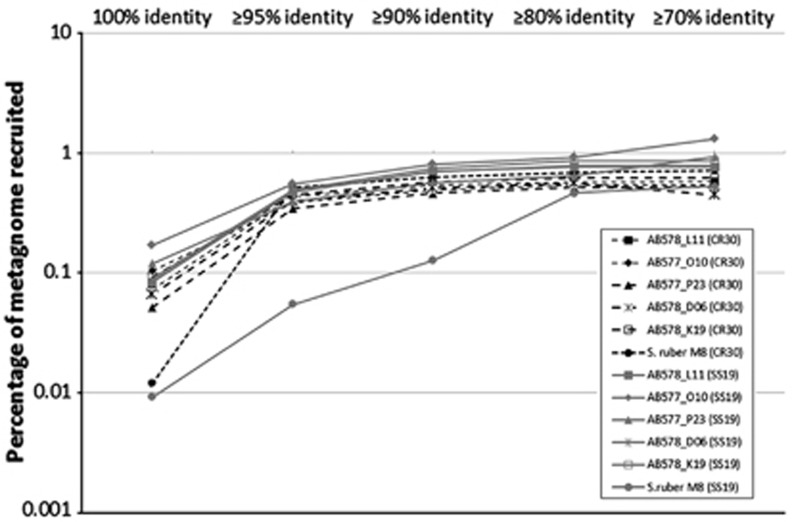
Metagenomic fragment recruitment data demonstrated that these sequenced SAGs were predominant in both the intermediate salinity pond of 19% (SS19) and the crystallizer CR30 (Ghai et al., 2011) as the recruitment ratio was as high as the very abundant S. ruber (Figure 7). Indeed, in the SS19 pond, the recruitment ratio of BC3 was higher than S. ruber, which points to a more generalist lifestyle of this new group in terms of salinity range.
Figure 7.
Metagenomic fragment recruitment of Bacteroidetes SAGs and the reference bacteroidetes species S. ruber. The fraction of recruited metagenome reads from the crystallizer pond CR30 and the medium-salinity pond SS19 was analyzed using a range of different DNA identity thresholds and normalized per Mbp genome size.
Genome annotation suggested that this hypersaline planktonic cluster contains unexpected metabolic capabilities. Arsenate respiration metabolic features were detected as a specific transporter to pump out the highly toxic form arsenite, and the key gene regulator arsR involved in arsenate respiration (Murphy and Saltikov, 2009) were found in this group. In addition, the ATP-dependent chaperone ClpB, which has been demonstrated to confer resistance to arsenic (Morgante and González-Pastor, personal communication), was not only detected in the phylotype BC3 but was also fairly abundant in the metagenome of CR30, indicating that this chaperone could be a common mechanism for resistance to arsenic in saline systems together with the use of the enzymes 3-isopropylmalate dehydrogenase, 5′phosphoribosyl anthranilate isomerase and aspartate semialdehyde dehydrogenase (Supplementary Figure S4).
Like other Bacteroidetes, this group also possesses a complete repertoire of gliding-motility proteins that have been related to degradation of complex polysaccharides (Gomez-Pereira et al., 2012). In addition, transporters for large biopolymers, glycosyl hydrolases and transferases were detected. For instance, SAGs AB578-K19 and AB578-D06 possessed a nucleotide-binding protein with the corresponding catalytic domain previously demonstrated to be involved in DNA uptake, which could be used either as a source of organic matter or for DNA transformation. Thus some traits of this group resembled those of other planktonic Bacteroidetes previously described to be involved in the remineralization of high-molecular-weight organic matter (Gomez-Pereira et al., 2012). The putative DNA-uptake systems found in the BC3 group could be related to its use as P source, as hypersaline systems are considered to be P-limited (in this study, N:P rations based on ammonia and phosphate concentrations ranged from 277 to 2770). In addition, a beta-lactamase and an ABC-antimicrobial peptide transporter were present at least in two of the single cells. These genes could be involved in interactions of BC3 with their accompanying microbiota. Indeed, a recent metabolomic characterization of S. ruber strains, with which BC3 shares its habitat, has shown that this bacterium produces a plethora of different antimicrobial-like compounds (Antón et al., 2013).
Furthermore, a nitroreductase enzyme involved in the utilization of complex aromatic and heterocyclic compounds with nitro- functional groups was detected. This enzyme has been reported to have potential applications in bioremediation, biocatalysis and biomedicine (Emptage et al., 2009). In addition, the cas genes 2 and 6 and the corresponding CRISPR arrays (Makarova et al., 2011), together with different components of the putative type 1 site-specific restriction modification systems (endonucleases and methyltransferases) involved in degrading foreign DNA, were found in some of the single cells, suggesting that this group might be under potential highly selective viral predatory pressure. This is not surprising given that hypersaline environments harbor the highest viral concentrations reported so far for aquatic systems (Santos et al., 2012). Finally, a gene coding for the light-driven proton pump bacteriorhodopsin was present at least in one of the sequenced SAGs.
Conclusions
Here, we have shown temporal changes in prokaryotic assemblages in a given pond as well as differences between ponds of very similar salinities, underscoring the need for caution when drawing general conclusions based on local data. In addition, we have found some traits that are considered to be characteristic of hypersaline systems, such as the abundance of Haloquadratum-related phylotypes and the predominance of Bacteroidetes representatives. The comparison of samples taken at different time points and different ponds has also enabled the detection of a new Bacteroidetes phylotype across a wide range of salinity and environmental conditions, which led us to propose its generalist nature. Phylotype BC3, now known to be widely distributed in salterns worldwide, had not been detected in Santa Pola saltern 10 years ago with techniques of the same resolution as the ones used here (Benlloch et al., 2002). Overall, our results show that solar saltern microbiota undergoes considerable changes throughout the year, and therefore the idea that hypersaline ponds are ‘steady-state' systems should be reconsidered.
Acknowledgments
This work was supported by the projects CGL2012-39627-C03-01 and 02 of the Spanish Ministry of Economy and Competitiveness, which were also co-financed with FEDER support from the European Union. TG group research is funded in part by a grant from the Spanish Ministry of Economy and Competitiveness (BIO2012-37161), a grant from the Qatar National Research Fund grant (NPRP 5-298-3-086) and a grant from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013)/ERC (grant agreement no. ERC-2012-StG-310325). We thank the staff of the Bras del Port salterns for their help throughout the sampling campaign. We thank Heinz Himmelbauer and the CRG's ultra-sequencing unit for their support and Esther Rubio-Portillo for her help with statistics sofware.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón J, Llobet-Brossa E, Rodríguez-Valera F, Amann R. Fluorescence in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ Microbiol. 1999;1:517–523. doi: 10.1046/j.1462-2920.1999.00065.x. [DOI] [PubMed] [Google Scholar]
- Antón J, Lucio M, Peña A, Cifuentes A, Brito-Echeverría J, Moritz F, et al. High metabolomic microdiversity within co-occurring isolates of the extremely halophilic bacterium Salinibacter ruber. PLoS One. 2013;8:5. doi: 10.1371/journal.pone.0064701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol. 2002;52:485–491. doi: 10.1099/00207713-52-2-485. [DOI] [PubMed] [Google Scholar]
- Antón J, Peña A, Santos F, Martínez-García M, Schmitt-Kopplin P, Rosselló-Mora R. Distribution, abundance and diversity of the extremely halophilic bacterium Salinibacter ruber. Saline Systems. 2008;28:4–15. doi: 10.1186/1746-1448-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón J, Rosselló-Mora R, Rodríguez-Valera F, Amann R. Extremely halophilic bacteria in crystallizer ponds from solar salterns. Appl Environ Microbiol. 2000;66:3052–3057. doi: 10.1128/aem.66.7.3052-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baati H, Guermazi S, Amdouni R, Gharsallah N, Sghir A, Ammar E. Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles. 2008;12:505–518. doi: 10.1007/s00792-008-0154-x. [DOI] [PubMed] [Google Scholar]
- Baati H, Guermazi S, Gharsallah N, Sghir A, Ammar E. Microbial community of salt crystals processed from Mediterranean seawater based on 16S rRNA analysis. Can J Microbiol. 2010;56:44. doi: 10.1139/w09-102. [DOI] [PubMed] [Google Scholar]
- Baati H, Jarboui R, Gharsallah N, Sghir A, Ammar E. Molecular community analysis of magnesium-rich bittern brine recovered from a Tunisian solar saltern. Can J Microbiol. 2011;57:975. doi: 10.1139/w11-088. [DOI] [PubMed] [Google Scholar]
- Benlloch S, López-López A, Casamayor E, Øvreås L, Goddard V, Daae FL, et al. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ Microbiol. 2002;4:349–360. doi: 10.1046/j.1462-2920.2002.00306.x. [DOI] [PubMed] [Google Scholar]
- Bolhuis H, Palm P, Wende A, Falb M, Rampp M, Rodriguez-Valera F, et al. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics. 2006;7:169. doi: 10.1186/1471-2164-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boujelben I, Gomariz M, Martínez-García M, Santos F, Peña A, López C, et al. Spatial and seasonal prokaryotic community dynamics in ponds of increasing salinity of Sfax solar saltern in Tunisia. Antonie Van Leeuwenhoek. 2012;101:845–857. doi: 10.1007/s10482-012-9701-7. [DOI] [PubMed] [Google Scholar]
- Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML. Cultivation of Walsby's square haloarchaeon. FEMS Microbiol Lett. 2004;238:469–473. doi: 10.1016/j.femsle.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Burns DG, Janssen PH, Itoh T, Kamekura M, Li Z, Jensen G, et al. Haloquadratum walsbyi gen. nov., sp. nov., the square haloarchaeon of Walsby, isolated from saltern crystallizers in Australia and Spain. Int J Syst Evol Microbiol. 2007;57:387–392. doi: 10.1099/ijs.0.64690-0. [DOI] [PubMed] [Google Scholar]
- Business Editors/High-Tech Writers SPSS enabling technologies partner program to provide embeddable analytic and predictive technologies to software and solutions providers. Business Wire. 2002;1 [Google Scholar]
- Casamayor EO, Schafer H, Baneras L, Pedros-Alio C, Muyzer G. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2000;66:499–508. doi: 10.1128/aem.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Elloumi J, Carrias JF, Ayadi H, Sime-Ngando T, Bouaïn A. Communities structure of the planktonic halophiles in the solar saltern of Sfax, Tunisia. Estuar Coast Shelf Sci. 2009;81:19–26. [Google Scholar]
- Emptage CD, Knox RJ, Danson MJ, Hough DW. Nitroreductase from Bacillus licheniformis: a stable enzyme for prodrug activation. Biochem Pharmacol. 2009;77:21–29. doi: 10.1016/j.bcp.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Estrada M, Henriksen P, Gasol JM, Casamayor EO, Pedrós-Alió C. Diversity of planktonic photoautotrophic microorganisms along a salinity gradient as depicted by microscopy, flow cytometry, pigment analysis and DNA-based methods. FEMS Microbiol Ecol. 2004;49:281–293. doi: 10.1016/j.femsec.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Gasol JM, Casamayor EO, Joint I, Garde K, Gustavson K, Benlloch S, et al. Control of heterotrophic prokaryotic abundance and growth rate in hypersaline planktonic environments. Aquat Microb Ecol. 2004;34:193–206. [Google Scholar]
- Ghai R, Pašić L, Fernández AB, Martin-Cuadrado AB, Mizuno CM, McMahon KD, et al. New abundant microbial groups in aquatic hypersaline environments. Sci Rep Nature. 2011;1:135. doi: 10.1038/srep00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pereira PR, Schueler M, Fuchs BM, Bennke C, Teeling H, Waldmann J, et al. Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environ Microbiol. 2012;14:52–66. doi: 10.1111/j.1462-2920.2011.02555.x. [DOI] [PubMed] [Google Scholar]
- Grant S, Grant WD, Jones BE, Kato C, Li L. Novel archaeal phylotypes from an East African alkaline saltern. Extremophiles. 1999;3:139–145. doi: 10.1007/s007920050109. [DOI] [PubMed] [Google Scholar]
- Guixa-Boixereu N, Calderón-Paz JI, Heldal M, Bratbak G, Pedrós-Alió C. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol. 1996;11:215–227. [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:1. [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse I, Bok J, Zwart G. A simple remedy against artificial double bands in denaturing gradient gel electrophoresis. J Microbiol Methods. 2004;57:279–281. doi: 10.1016/j.mimet.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Jiang H, Dong H, Yu B, Liu X, Li Y, Ji S, et al. Microbial response to salinity change in Lake Chaka, a hypersaline lake on Tibetan plateau. Environ Microbiol. 2007;9:2603–2621. doi: 10.1111/j.1462-2920.2007.01377.x. [DOI] [PubMed] [Google Scholar]
- Legault BA, Lopez-Lopez A, Alba-Casado JC, Doolittle WF, Bolhuis H, Rodriguez-Valera F, et al. Environmental genomics of "Haloquadratum walsbyi" in a saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species. BMC Genomics. 2006;4:7–171. doi: 10.1186/1471-2164-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García M, Brazel D, Swan B, Arnosti C, Chain P, Reitenga K, et al. Capturing single cell genomes of activepolysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS One. 2012;7:e35314. doi: 10.1371/journal.pone.0035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García M, Swan BK, Poulton NJ, Gomez ML, Masland D, Sieracki ME, et al. High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. ISME J. 2012;6:113–123. doi: 10.1038/ismej.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Murray AE, Preston CM, Delong EF. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maturrano L, Santos F, Rossello-Mora R, Anton J. Microbial diversity in Maras Salterns, a hypersaline environment in the Peruvian Andes. Appl Environ Microbiol. 2006;72:3887–3895. doi: 10.1128/AEM.02214-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JN, Saltikov CW. The ArsR repressor mediates arsenite-dependent regulation of arsenate respiration and detoxification operons of Shewanella sp. strain ANA-3. J Bacteriol. 2009;191:6722. doi: 10.1128/JB.00801-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu MB, Martínez-García M, Santos F, Peña A, Guven K, Antón J. Prokaryotic diversity in Tuz Lake, a hypersaline environment in Inland Turkey. FEMS Microbiol Ecol. 2008;65:474–483. doi: 10.1111/j.1574-6941.2008.00510.x. [DOI] [PubMed] [Google Scholar]
- Muyzer G, Hottenträger S, Teske A, Wawer C.1996Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA-a new molecular approach to analyse the genetic diversity of mixed microbial communitiesIn: Akkermans ADL, van Elsas JD, de Bruijn FJ, (eds).Molecular Microbial Ecology Manual Kluwer: Dordrecht, Netherland; 1–23. [Google Scholar]
- Narasingarao P, Podell S, Ugalde JA, Brochier-armanet C, Emerson JB, Brocks JJ, et al. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 2012;6:81–93. doi: 10.1038/ismej.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D, Porter K, Russ B, Burns D, Dyall-smith M. Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles. 2010;14:161–169. doi: 10.1007/s00792-009-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol Ecol. 2002;39:1–7. doi: 10.1111/j.1574-6941.2002.tb00900.x. [DOI] [PubMed] [Google Scholar]
- Pašić L, Bartual SG, Ulrih NP, Grabnar M, Velikonja BH. Diversity of halophilic archaea in the crystallizers of an Adriatic solar saltern. FEMS Microbiol Ecol. 2005;54:491–498. doi: 10.1016/j.femsec.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Pedrós-Alió C.2005Diversity of microbial communities: the case of solar salternsIn: Gunde-Cimerman N, Plemenitas A, Oren A, (eds).Adaptation to Life at High Salt Concentrations in Archaea, Bacteria, and Eukarya. Cellular Origins, Life in Extreme. Habitats and Astrobiology (COLE) Kluwer Academic Publishers: Netherlands; 71–90. [Google Scholar]
- Pedrós-Alió C. Marine microbial diversity: can it be determined. Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Pernthaler J, Amann R.2004Sensitive multicolor fluorescence in situ hybridization for the identification of environmental microorganismsIn: Kowalchuk GA, de Bruijn FJIM, Head IM, Akkermans AD, van Elsas JD, (eds).Molecular Microbial Ecology Manual2nd ednKluwer Academic Publishers: Dordrecht, The Netherlands; Boston, MA, USA; London, UK; 711–726. [Google Scholar]
- Podell S, Ugalde JA, Narasingarao P, Banfield JF, Heidelberg KB, Allen EE. Assembly-driven community genomics of a hypersaline microbial ecosystem. PLoS One. 2013;18:e61692. doi: 10.1371/journal.pone.0061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan A, Ferguson HR, Bornarth CJ, Song W, Driscoll M, Lasken RS. Genomic DNA amplification from a single bacterium. Appl Environ Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010;4:739–751. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Yarza P, Parro V, Meseguer I, Rosselló-Móra R, Antón J. Culture-independent approaches for studying viruses from hypersaline environments. Appl Environ Microbiol. 2012;78:1635. doi: 10.1128/AEM.07175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer HL, Bernard L, Courties C, Lebaron F, Servais P, et al. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol Ecol. 2001;34:243–253. doi: 10.1111/j.1574-6941.2001.tb00775.x. [DOI] [PubMed] [Google Scholar]
- Soerensen KB, Canfield DE, Teske AP, Oren A. Community composition of a hypersaline endoevaporitic microbial mat. Appl Environ Microbiol. 2005;71:7352–7365. doi: 10.1128/AEM.71.11.7352-7365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM. A place for DNA±DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- ter Braak CJF, Smilauer P.2002CANOCO Reference Manual & CanoDraw for Windows User's Guide: Software for Canonical Community Ordination (version 4.5) Microcomputer Power: Ithaca, New York, USA; p500 [Google Scholar]
- Thompson JR, Marcelino LA, Polz MF. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by 'reconditioning PCR'. Nucleic Acids Res. 2002;30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- Wang J, Yang D, Zhang Y, Shen J, van der Gast C, Hahn MW, et al. Do patterns of bacterial diversity along salinity gradients differ from those observed for macroorganisms. PLoS One. 2011;6:e27597. doi: 10.1371/journal.pone.0027597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O, Stepanauskas R, Mohan NR, Papke RT. Cell sorting analysis of geographically separated hypersaline environments. Extremophiles. 2013;17:265–275. doi: 10.1007/s00792-013-0514-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.