Abstract
Delineating differences in gut microbiomes of human and animal hosts contributes towards understanding human health and enables new strategies for detecting reservoirs of waterborne human pathogens. We focused upon Blautia, a single microbial genus that is important for nutrient assimilation as preliminary work suggested host-related patterns within members of this genus. In our dataset of 57 M sequence reads of the V6 region of the 16S ribosomal RNA gene in samples collected from seven host species, we identified 200 high-resolution taxonomic units within Blautia using oligotyping. Our analysis revealed 13 host-specific oligotypes that occurred exclusively in fecal samples of humans (three oligotypes), swine (six oligotypes), cows (one oligotype), deer (one oligotype), or chickens (two oligotypes). We identified an additional 171 oligotypes that exhibited differential abundance patterns among all the host species. Blautia oligotypes in the human population obtained from sewage and fecal samples displayed remarkable continuity. Oligotypes from only 10 Brazilian human fecal samples collected from individuals in a rural village encompassed 97% of all Blautia oligotypes found in a Brazilian sewage sample from a city of three million people. Further, 75% of the oligotypes in Brazilian human fecal samples matched those in US sewage samples, implying that a universal set of Blautia strains may be shared among culturally and geographically distinct human populations. Such strains can serve as universal markers to assess human fecal contamination in environmental samples. Our results indicate that host-specificity and host-preference patterns of organisms within this genus are driven by host physiology more than dietary habits.
Introduction
Microbial mutualisms, commensalisms and pathogenic relationships influence host development (McFall-Ngai, 2002; Nyholm and Graf, 2012; McFall-Ngai et al., 2013), defense (May and Nelson, 2014), nutrient assimilation (Walter and Ley, 2011; Webster and Taylor, 2011; El Kaoutari et al., 2013) and disease in humans and other animals (Turnbaugh et al., 2006; Dethlefsen et al., 2007; Walter and Ley, 2011; Eloe-Fadrosh & Rasko, 2013). Microbial community datasets from fecal samples have described patterns of host-association and external factors that shape the gut microbiome (Ley et al., 2008; Muegge et al., 2011; Lozupone et al., 2012; Yatsunenko et al., 2012). Within a mammalian species, host diet may influence microbial composition and diversity more than genetics, geography or other factors (Ochman et al., 2010; Yatsunenko et al., 2012). However, across different host species, adaptation to host physiology as well as long term dietary patterns likely play important roles. Unraveling the basis for host-associated patterns in the microbiome would provide insight into a broad range of research initiatives in human and public health. Beyond describing healthy human microbiomes and potential therapeutic interventions, high-resolution descriptions of gut microbiota in humans and other organisms would provide a basis for identifying host sources of fecal pollution, estimating disease risk, and formulating mitigation strategies.
Only a few studies have explored differential distribution of closely related organisms (that is, within genera). These previous studies reveal that core genes may define shared traits for a given genus whereas accessory genes account for specialization within a certain environment (Frese et al., 2011; Oh et al., 2010, 2012). Multilocus sequence typing of Escherichia coli from animals and the environment revealed clusters distinct from human isolates, where these strains lacked certain stress and adherence genes (Oh et al., 2012). In Lactobaccillus reuteri, strains characterized by multilocus sequence typing mapped to different hosts (Oh et al., 2010) and specific genomic traits occurred in murine hosts, while human-associated strains comprised a larger pan genome (Frese et al., 2011). Identifying ecologically relevant subpopulations within closely related microorganisms represents a first step toward delineating the genomic basis for host-associated differences.
To explore relationships between specific microorganisms and different human and animal hosts, we previously used next generation sequencing to compare rapidly evolving ribosomal RNA (rRNA) gene regions for PCR amplicons from fecal microbial communities (McLellan et al., 2013; Shanks et al., 2013). Fecal samples from humans, cattle and chickens yielded distinct V6 rRNA gene sequences for each host that resolved to Blautia (McLellan et al., 2013), a genus in the bacterial family Lachnospiraceae that phylogenetic analysis places within the Clostridium coccoides group, also referred to as the Clostridium Cluster XIVa (Hayashi et al., 2006). Lachnospiraceae constitutes one of the major taxonomic groups of the human gut microbiota where they degrade complex polysaccharides to short chain fatty acids including acetate, butyrate, and propionate that can be used for energy by the host (Biddle et al., 2013). Other animals commonly harbor Lachnospiraceae, with herbivores having a higher abundance than carnivores (Furet et al., 2009). The wide range of functions carried out by Lachnospiraceae may influence their relative abundance in gut communities of different hosts.
The extension of rRNA amplicon pyrosequencing to sewage samples provided an opportunity to investigate the relative abundance patterns of different Blautia in samples that represent large human populations (McLellan et al., 2013). The most common Blautia V6 sequences recovered repeatedly in untreated sewage from a single major city perfectly matched those found in sewage samples in other US cities (Shanks et al., 2013). Next generation sequencing analyses of rRNA amplicons have identified differences in the microbiomes of various host species, enabling the tracking of subpopulations. However, the use of low-resolution genus-level taxon assignments that rely upon sequence similarities to a reference database and 97% sequence similarity thresholds to identify operational taxonomic units de-novo may be too coarse of measures to detect subtle variations in the 16S rRNA genes that represent different ecotypes (Eren et al., 2013a; Ward, 1998).
To investigate closely related Blautia populations in animal and human fecal samples, we employed oligotyping, a supervised computational method that allows the identification of very closely related but distinct organisms represented in next generation sequencing datasets (Eren et al., 2013a). An ‘oligotype' corresponds to a collection of high-entropy nucleotide positions within a defined region of microbial rRNA genes. By focusing on highly variable nucleotide positions, oligotyping can distinguish between organisms that display more than 99% identity in the sequenced region of the 16S rRNA gene (Eren et al., 2011, 2013a). The classification algorithm random forests (Breiman, 2001) tested the host classification performance of Blautia oligotypes; the biomarker discovery package LEfSe (Segata et al., 2011) identified oligotypes that were differentially abundant among animal groups; and the observation matrix from oligotyping revealed host-specificity patterns.
Beyond providing important information about the drivers of adaptation between bacterial organisms and their hosts, these patterns also guide the development of novel tools for environmental monitoring (Dubinsky et al., 2013; Newton et al., 2013). Human fecal pollution provides a reservoir for hundreds of waterborne disease agents and contributes worldwide to significant morbidity and mortality (Fewtrell et al., 2005). Elucidating bacterial taxa that discriminate human and animal sources of fecal pollution can identify new indicators for waterborne disease and contribute to health risk management.
Materials and methods
Sample collection and DNA extraction
Fecal samples from USA dogs, cats, chickens, cows, mule deer, and swine were collected into sterile tubes, kept on ice during transit from the field to the lab, and stored at −80 °C until DNA extraction. In addition, we collected ∼1 ml of freshly produced swine fecal samples from Brazil with a plastic scoop and stored at −20 °C until further processing. DNA was obtained from archived human fecal samples from a Schistosomiasis survey performed in Brazil in 2009 (Blanton et al., 2011). Composite (24-hr) sewage samples from Jones Island Waste Water Treatment Plant (Milwaukee, WI, USA) were provided by the Milwaukee Metropolitan Sewerage District (MMSD) and 10 ml were filtered onto 0.22 μm 47 mm filters (S-Pak, Millipore, Billerica, MA, USA). Influent sewage from the Empresa Baiana de Águas e Saneamento (EMBASA)—The Bahian Water and Sanitation Company in Salvador, Brazil was collected after primary settling and 10 ml was filtered and stored for DNA extraction. Filters were kept frozen (−80 °C) until processed for DNA extraction. Supplementary Table S1 provides details on sample sources. The GeneRite DNA-EZ kit (#DNA-EZ RW02, North Brunswick, NJ, USA) was used for US animal fecal extractions according to the manufacturer's instructions by researchers at the USEPA in Cincinnati, OH, USA. A 200-μg sample was taken from each tube of Brazil animal feces and processed with the Fast DNA Spin kit for Feces (MP Biomedicals, Solon, OH, USA) as per manufacturer's recommendations. For sewage samples, filters were crushed, and DNA was extracted with a FastSpin Soil DNA kit (MP Biomedicals). Human fecal DNA was extracted previously (Blanton et al., 2011); briefly, frozen fecal pellets were treated with lysis buffer, phenol and chloroform. DNA was stored at −20 °C until used. DNA purity was assessed using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA).
Library preparation and sequencing
Supplementary Table S1 documents the source and the previous use of the 69 samples in this analysis. The DNA extracted from each sample was sequenced as described previously (Eren et al., 2013b). Briefly, we amplified the V6 hypervariable region of the 16 S rRNA gene using custom fusion primers (Eren et al., 2013b). Primers consisted of the oligonucleotides (Integrated DNA Technologies, Coralville, Iowa) with, 8 different inline barcodes (forward primer) or 12 dedicated indices (reverse primer), and conserved sequences flanking the V6 region. Unique barcode-index combinations allowed multiplexing 96 samples per lane. We amplified libraries from three independent PCRs for each sample to minimize the impact of potential early-round PCR errors. Cycling conditions were: an initial 94 °C, 3 min denaturation step; 30 cycles of 94 °C for 30 s, 60 °C for 45 s and 72 °C for 60 s; and a final 2 min extension at 72 °C. Triplicate reactions were pooled, cleaned, size-selected and quantitated (Eren et al., 2013b). Samples were sequenced on one lane of an Illumina HiSeq (Illumina, Inc., San Diego, CA, USA) 100 cycle paired-end run along with a high-complexity shotgun metagenomic sample (40:60 ratio) to promote accurate cluster identification and phasing. We used CASAVA 1.8.2 (Illumina, Inc.) to call bases and demultiplex by index and custom scripts to resolve reads from each index bin into samples by barcode. Sequences are stored in VAMPS (http://vamps.mbl.edu) (Huse et al., 2014) under the project name SLM_NIH2_Bv6. Actual sample names corresponding to the aliases used in figures, and number of reads per sample are listed in Supplementary Table S1.
Quality control
Our amplicon design allowed both the first and second reads to span the entire V6 region resulting in complete overlaps plus 15–20 nucleotide extension into the proximal and the distal PCR primer sites. We required 100% consensus between the overlapping regions of the forward and reverse reads to minimize the impact of sequencing errors (Eren et al., 2013b). The URL https://github.com/meren/illumina-utils provides access for the open-source implementation of the V6 complete overlap analysis program.
Taxonomical classification and oligotyping
We used GAST (Huse et al., 2008) to assign taxonomy for each quality filtered read. We identified reads that classified to the genus Blautia for oligotyping analysis. Because homopolymer region-associated insertion and deletion errors occur rarely on the Illumina HiSeq platform (Loman et al., 2012), we padded shorter reads with end gaps to mend the length variation among V6 reads. We performed oligotyping (Eren et al., 2013a) using the oligotyping pipeline version 0.96 (available from http://oligotyping.org). As a result of the initial entropy analysis and supervision, 24 high-entropy positions were identified for oligotyping analysis. Supplementary Figure S1 shows the distribution of entropy along the V6 Blautia reads and positions used for oligotyping. To minimize the impact of noise, we set the minimum substantive abundance parameter to 100 (-M 100), which instructs the oligotyping pipeline to eliminate any oligotype whose most abundant unique sequence has a frequency that is smaller than 100 (Eren et al., 2013a). Any oligotype that appeared in less than three samples was also removed from the analysis (-s 3). Noise filtering based on –s and –M parameters removed 2.66% of reads from the dataset. We provide the observation matrix for oligotypes recovered and their representative V6 tags in Supplementary Table S1.
Downstream bioinformatics analyses and visualization
To investigate the efficacy of Blautia oligotypes for identifying the host source of fecal samples, we created a classification model using random forests (Breiman, 2001), a robust machine-learning algorithm for classification and regression that is suitable for microbial population data (Statnikov et al., 2013). A random forest consists of many decision trees that are grown during training using random sampling of both samples, and units (in our case oligotypes) in the training dataset. Random forest analysis generates an unbiased, ‘out-of-bag' (OOB) estimate of error during the training phase (Breiman, 2001) to assess how well the model generalizes unseen data without requiring some portion of the training data to be left out for testing the classifier. High ‘out-of-bag' errors represent low accuracy of the classifier. We performed random forest analysis using random Forest module 4.6-7 (Liaw and Wiener, 2002) implemented for R (R Development Core Team, 2011) by growing 2,000 trees with default parameters. We used LEfSe version 1.0 (Segata et al., 2011) with default parameters (minimum linear discriminant analysis score of 2.0, 30 bootstrap iterations for linear discriminant analysis) to identify oligotypes that show differential abundance patterns among different animal groups. For network analysis we used Gephi, an open-source software for exploring and manipulating networks (Bastian et al., 2009). For clustering analyses we used R functions with distance matrices implemented in the vegan package (Dixon 2003). We visualized the dendrogram for oligotypes using iTOL (Letunic and Bork, 2007) and used the ggplot2 (Ginestet, 2011) for all other visualizations. We edited all figures using Inkscape, an open-source graphics editor that is available from http://inkscape.org/.
Sequence data
Sequences have been deposited in NCBI under the accession number SRA # SRP041262.
Results
Oligotyping profiles
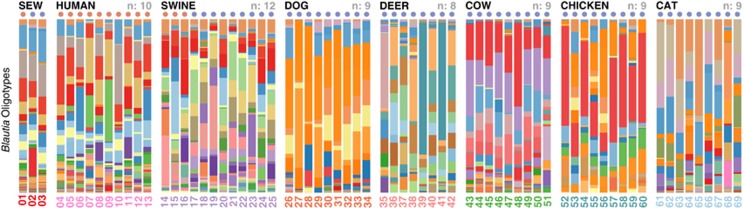
To explore differences between Blautia populations in fecal samples from geographically separate human populations, and to realize the full potential of Blautia rRNA gene sequences to distinguish host organisms, we collected 57 716 475 V6 rRNA amplicon sequences from 66 fecal samples (10 human samples from rural Jenipapo, Brazil, 56 animals, and 3 untreated sewage influent samples from Milwaukee, WI and Salvador, Brazil). The taxon assignment algorithm GAST (Huse et al., 2008) resolved 925 061 sequences from this dataset to the genus Blautia. The human, dog and cat fecal samples had a significantly higher proportion of Blautia (1.2%, 1.3% and 5.8% respectively) compared with a tenfold lower relative abundance in chickens, pigs, cattle and deer (0.16%, 0.27%, 0.29% and 0.30% respectively; Student's t-test, P<0.05). For the combined dataset we identified 200 distinct Blautia oligotypes. Figure 1 displays the relative abundance of different Blautia oligotypes for sewage, human and animal samples, Supplementary Figure S2 displays the number of V6 reads recovered from each sample, and the percentage of V6 reads that classified to genus Blautia. Supplementary Figure S3 shows a heat map of community similarity based upon oligotype analysis, and Supplementary Table S1 reports the observation matrix for Blautia oligotypes and representative V6 tag sequences for each oligotype.
Figure 1.
Oligotype distribution among sample groups. Red dots below group names indicate samples collected from Brazil, and blue dots identify samples collected from the United States.
Human Blautia oligotypes at the individual and population level
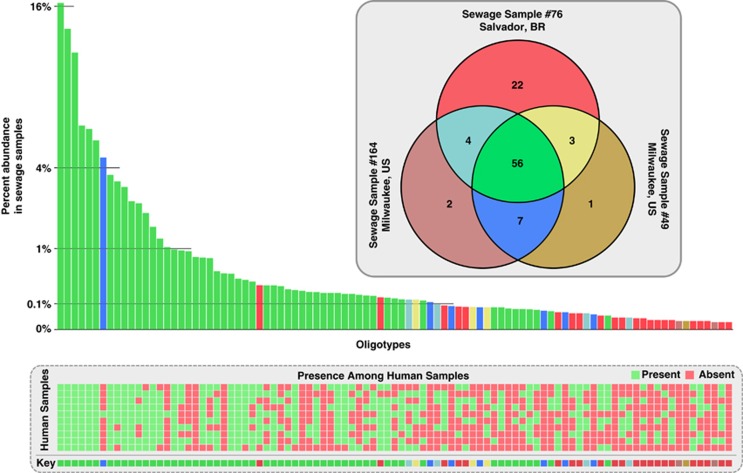
The untreated sewage from the Jones Island treatment plant (Milwaukee, WI, USA) represents a random sampling of human fecal inputs from >500 000 individuals, while the EMBASA sewage treatment plant (Salvador, Brazil) represents a random sampling of ∼three million individuals. On average, Blautia represented 0.9% of the sequences from Milwaukee sewage and 2.0% for Salvador samples. The two Milwaukee sewage samples shared 63 Blautia oligotypes that ranged in relative abundance from 0.03% to as high as ∼18.7% (Figure 2). The Salvador and combined Milwaukee sewage samples shared 56 oligotypes. Oligotypes not shared by two or more samples occurred at very low frequencies (relative abundance from 0.029% to ∼0.16%).
Figure 2.
Shared and distinct oligotypes in sewage and Brazilian human fecal samples. The Venn diagram shows the number of shared oligotypes between the three sewage samples. The bar plot shows the abundance of each oligotype found in sewage samples. The color of each bar corresponds to the regions of the same color in the Venn diagram. The lower panel is a heat map that uses presence (green) and absence (red) to show the distribution of each oligotype among 10 samples collected from Brazilian humans. Colors listed in the key below the heat map follow the colors given to corresponding bars and Venn diagram.
The Salvador sewage contained 79 of the 81 Blautia oligotypes detected in Brazilian human fecal samples whereas the combined Milwaukee sewage contained only 61 (75.0%) (Figure 2). The occurrence of the most common Blautia oligotypes in each of the human fecal samples supports the idea that sewage samples reflect microbial populations in humans. A combined analysis of all sewage samples identified 17 additional oligotypes that we did not detect in the Brazil humans, 11 of which occured in Milwaukee sewage. These data reveal similar but non-identical oligotype compositions for the Milwaukee and Salvador sewage samples, which implies significant overlap of Blautia in human microbiomes in Milwaukee and Jenipapo/Salvador. Two moderately abundant oligotypes (>0.1% of the Blautia reads) in the Salvador sewage occur in 9 of the 10 Brazilian human samples, but we did not detect those sequences in either of the Milwaukee sewage samples. Differences were mainly seen in the presence/absence of rare oligotypes, which suggest that very low-abundance taxa might define differences between the microbiomes of geographically separated human populations.
Host classification by Blautia oligotypes
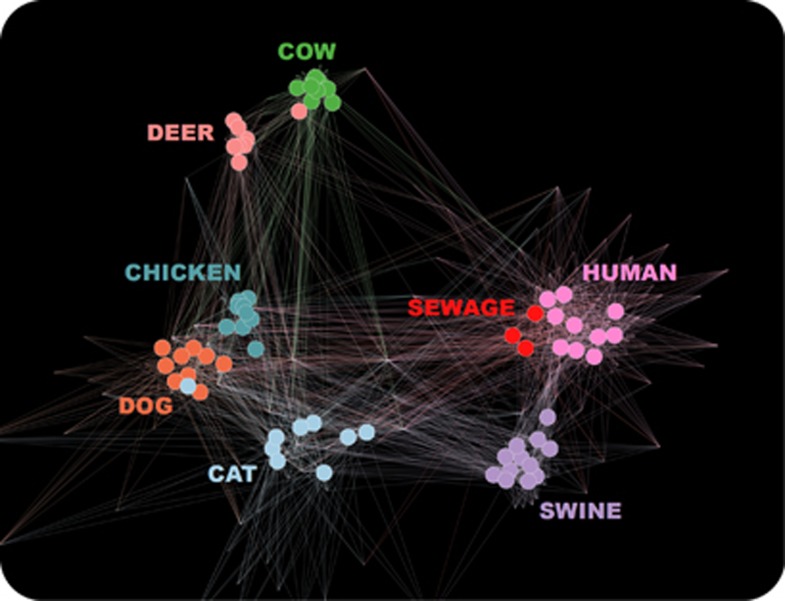
Network analysis showed that Blautia oligotypes for each animal species including humans formed distinct clusters (Figure 3) that were tightest for cow, deer and chicken. Only a single deer sample interrupted the Blautia oligotype clusters for cow and a single cat sample interrupted the Blautia oligotype cluster for dog. The network analysis also showed a close relationship between sewage and the human fecal samples. The Multi-Dimensional Scaling ordination using a Morisita-Horn similarity index exhibited a similar distribution of samples (Supplementary Figure S4).
Figure 3.
Network analysis of samples with respect to Blautia oligotypes. Edges connect samples to oligotypes and are colored based on the sample. A force-directed algorithm is used to analyze the occurrence of oligotypes in samples to reach equilibrium. Force-directed algorithms mimic basic physical properties such as repulsion and gravity, through which samples that share similar oligotypes are drawn together, while samples that have different oligotype profiles are pushed away from each other.
Identification of host sources using oligotypes
To investigate the efficacy of Blautia oligotypes for identifying the host source of fecal samples, we created a classification model using random forests (Breiman, 2001), a robust machine-learning algorithm for classification and regression (see Materials and methods). All fecal samples classified into cognate groups during the random forest analysis, with the exception of single fecal samples from deer (confused with cow) and from cat (confused with dog). Supplementary Figure S5 shows the confusion matrix based on out-of-bag generalized error. Very low ‘out-of-bag' error rates generated during the training phase confirm that Blautia oligotypes describe robust host associations with the potential for accurately classifying samples of unknown origin to specific source hosts.
Differential abundance of Blautia oligotypes in hosts
We used LEfSe (Segata et al., 2011) to identify Blautia oligotypes that show statistically significant differential abundances among groups of human and sewage samples versus animals. LEfSe identifies units that are highly associated with one or more previously defined classes in a dataset by utilizing non-parametric statistical tests, and estimates a size effect score for each differentially abundant feature using linear discriminant analysis. LEfSe identified 171 oligotypes with an linear discriminant analysis score that exceeded a threshold of 2.0, whereas 154 oligotypes distributed among human (44), cat (26), swine (23), cow (18), dog (16), deer (15) and chicken (12), and an additional 17 oligotypes associated with sewage. A total of 65% of all oligotypes matched at least one entry found in NCBI's non-redundant database with 100% identity and coverage. While only 13% of oligotypes associated with deer samples by LEfSe matched a sequence identically in NCBI's non-redundant database, 89% of oligotypes that occurred mostly in human and sewage samples had identical hits. The percentage of remaining oligotypes with identical hits was 83% for chicken, 70% for cat, 67% for dog, 61% for cow and 61% for swine groups (Supplementary Table S1). Supplementary Table S1 also reports the LEfSe results and lists the oligotypes that can contribute to the identification of host source of fecal samples. We found no correlation (r=0.28, P=0.53) between the number of group-associated oligotypes recovered by LEfSe and the average number of Blautia reads in these groups; the abundance of Blautia reads do not explain the number of distinct host-associated oligotypes.
Host-specific, host-associated and host-preferred oligotypes
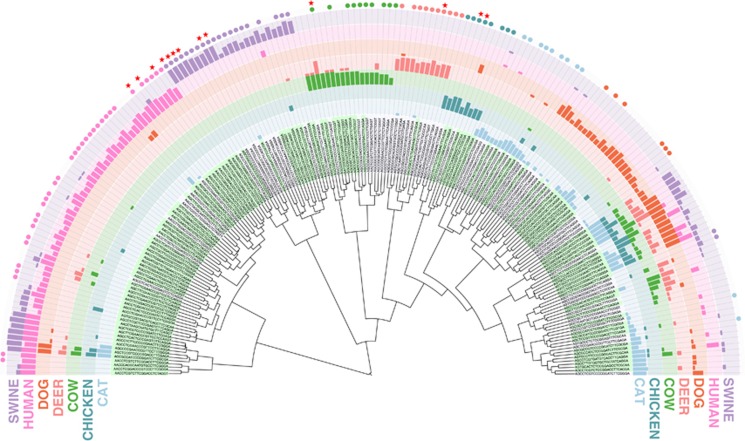
Based upon the distribution patterns of 200 distinct oligotypes in different hosts we have adopted three terms with different properties to describe the differential distribution of an oligotype among samples from different host groups: ‘host-specific', ‘host-associated' and ‘host-preferred'. A ‘host-specific' oligotype must occur in all samples for a single animal species and must be absent in fecal samples from all other groups. A ‘host-associated' oligotype only occurs in fecal samples from one animal species but not necessarily in every fecal sample from that species. A ‘host-preferred' oligotype occurs at a statistically significant higher abundance in fecal samples from a particular animal but can also occur at low-abundance in fecal samples from other hosts. Figure 4 exemplifies hypothetical distribution patterns of cosmopolitan and ‘host-specific', ‘host-associated' and ‘host-preferred' oligotypes. Our analysis identified 13 host-specific Blautia oligotypes in fecal samples from humans (three oligotypes), swine (six oligotypes), cow (one oligotype), deer (one oligotype), and chicken (two oligotypes). Table 1 lists 13 host-specific oligotypes and their representative V6 tag sequences. Figure 5 shows the distribution profile of these oligotypes among human and animal samples. Fecal samples from dogs and cats did not contain oligotypes that met the definition criteria for host-specific oligotypes. Most oligotypes that occurred in every dog and cat fecal sample also occurred in other groups of animals, however, some oligotypes classified to host-preferred because of an elevated relative abundance in dogs or cats compared with their fractional representation in other animal groups. There were also 11 host-associated oligotypes that occurred in cat and 6 in dog fecal samples (Figure 5). Both the host-associated and host-specific Blautia oligotypes potentially offer molecular markers for discriminating between fecal pollution sources.
Figure 4.
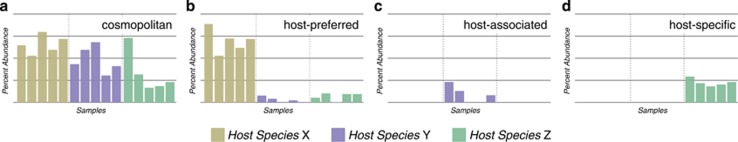
Example distribution patterns for (a) ‘cosmopolitan', (b) ‘host-preferred', (c) ‘host-associated' and (d) ‘host-specific' oligotypes. Bars show relative abundances of hypothetical oligotypes and are colored based on host species— X, Y and Z.
Table 1. Host-specific oligotypes and their V6 tag sequences.
| Source | Oligotype | Representative V6 Tag | Average % abundance | Total no. of reads | Average no. of reads per sample |
|---|---|---|---|---|---|
| Human | AACCTCGGACCCGTCCCCTTAGGA | AAATCTTGACATCCCTCTGACCGGGACTTAACCGTCCCCTTTCTTCGGAACAGAGGAGAC | 0.0859 | 9,874 | 987 |
| Human | AGCCTCGTCTTCGGACCTCTAGGT | AAGTCTTGACATCCCTCTGACCGGTCTTTAATCGGACCTTCTCTTCGGAGCAGAGGTGAC | 0.0038 | 446 | 45 |
| Human | AGCCTGACTCCCGAGTCTTCAGGA | AAGTCTTGACATCCCTCTGACGGACTCTTAACCGAGTCTTTCCTTCGGGACAGAGGAGAC | 0.0088 | 1,019 | 47 |
| Swine | AGCGGCGGACCCGTCCCTCCCCGA | AAGTCTTGACATCCGGATGACCGGGACTTAACCGTCCCTTCCCTTCGGGGCATCCGAGAC | 0.0130 | 2,022 | 168 |
| Swine | AGCGGATCCTCCGGGACTCCCCGA | AAGTCTTGACATCCGGATGACAGTCCTTTAACCGGGACTTCCCTTCGGGGCATCCGAGAC | 0.0127 | 1,980 | 165 |
| Swine | AGCCTCGGGCCCGCCCCTTCAGGA | AAGTCTTGACATCCCTCTGACCGGGGCTTAACCGCCCCTTTCCTTCGGGACAGAGGAGAC | 0.0092 | 1,433 | 119 |
| Swine | AGCCTCGGGCCCGTCCCTTCAGGA | AAGTCTTGACATCCCTCTGACCGGGGCTTAACCGTCCCTTTCCTTCGGGACAGAGGAGAC | 0.0048 | 758 | 63 |
| Swine | AGCGGCGCCGCGCGGCCTCCCCGA | AAGTCTTGACATCCGGATGACCGGCCGGTAACGCGGCCTTCCCTTCGGGGCATCCGAGAC | 0.0029 | 452 | 38 |
| Swine | AGCCTTATCCCGGGATCTTCAGGA | AAGTCTTGACATCCCTCTGACTGATCCGTAACGGGATCTTTCCTTCGGGACAGAGGAGAC | 0.0015 | 239 | 20 |
| Cow | GGCCTCTTCCCCGGAACTTCAGGT | AGGACTTGACATCCCTCTGACCGTTCCTTAACCGGAACTTTCCTTCGGGACAGAGGTGAC | 0.0545 | 3,794 | 421 |
| Deer | AGCCGCGAACCGGTTCCTCTCGGA | AAGTCTTGACATCCCGATGACCGGAACGTAACGGTTCCTTCTCTTCGGAGCATCGGAGAC | 0.0232 | 1,492 | 186 |
| Chicken | AGCCTCTTCTCCGGAACTTCAGGA | AAGTCTTGACATCCCTCTGACCGTTCTTTAACCGGAACTTTCCTTCGGGACAGAGGAGAC | 0.0133 | 610 | 68 |
| Chicken | AGCCTCCGCCCCGGCGCTTCAGGA | AAGTCTTGACATCCCTCTGACCGCGCCTTAACCGGCGCTTTCCTTCGGGACAGAGGAGAC | 0.0031 | 142 | 16 |
Average percent abundance column lists the average relative abundance of each oligotype among the source host. Total number of reads column shows the size of the oligotype.
Figure 5.
Dendrogram of Blautia oligotypes and their host occurrence patterns. Bars show the proportion of animal or human samples in which the given oligotype is present. Circles indicate host-associated oligotypes. Host-specific oligotypes are denoted with stars. Background colors for oligotypes alternate between green and gray, where green indicates that a perfect hit with 100% coverage is found for a given oligotype in NCBI's non redundant database.
Discussion
Blautia as a marker for sewage and human fecal contamination
The Clostridium coccoides group (Clostridium Cluster XIVa), which encompasses the genus Blautia, can represent up to 30% of the human gut microbiome (Franks et al., 1998). Oligonucleotide primers that target the C. coccoides group have detected fecal pollution in quantitative PCR assays (Bonkosky et al., 2009), and pyrosequencing suggests that Blautia might serve as an indicator of human fecal pollution (Newton et al., 2011; McLellan et al., 2013). Both the depth of sequencing (number of reads) and the breadth of different host species in an analysis will influence the development of models that can differentiate between host sources of fecal pollution. This study included a larger number of host species relative to our initial use of next generation sequencing to identify candidate human fecal indicators, and it took advantage of a larger number of sequence reads with improved accuracy afforded by the use of enhanced quality filtering methods (Eren et al., 2013b).
Although there is growing evidence for host-specificity of microbial strains through whole genome sequencing, microarray analyses, or multilocus sequence typing (Oh et al., 2010, 2012; Parsons et al., 2010; Frese et al., 2011), the basis for host-specificity is largely unknown. Overall community analyses through the 16S rRNA gene data mostly provide indirect evidence for patterns of specificity between animals and their microbial communities implied by differential abundances of taxa that occur in multiple hosts. The combination of deep sequencing, stringent quality filtering and high-resolution taxonomic units can utilize the 16S rRNA gene data to be used as an exploratory approach to identify marker sequences for host-specific or host-associated strains. Besides the immediate environmental applications, these findings can guide targeted approaches to reveal host-microbe interactions and can shed light on the genetic disposition of these stable associations.
Rather than attempting to identify whole community differences, in this study we focused on the finer architecture of Blautia populations and their distribution in the host groups. Blautia oligotypes accurately identified different hosts, and the analysis of sewage captured population-level human microbiota across large geographical distances. The 200 Blautia oligotypes differentially distributed among fecal samples from humans and animals (Figure 1, Supplementary Figure S3) and included oligotypes that are specific to human, swine, cow, deer and chicken hosts (Table 1). The occurrence of human-specific and human-associated oligotypes provided unambiguous identification of human sources in all three sewage samples. A random forest classifier trained with Blautia oligotypes from each of the host species identified the origin of 67 of our 69 samples, including all human and sewage samples. A LEfSe analysis that explored differential abundance and linkage to different hosts identified 44 oligotypes in sewage that link with humans. Together these analyses reinforce the working hypothesis that Blautia oligotypes can differentiate between sewage-derived human and animal fecal contamination. Future work to expand the number of animals and host types will validate the host-related patterns identified in these studies.
The three classes ‘host-specific', ‘host-associated' and ‘host-preferred' (Figure 4) set different expectations about outcomes for molecular detection schemes. Overall, we identified a low number of cosmopolitan oligotypes (n=23) relative to those that showed differential abundance patterns (n=177). A host-specific Blautia oligotype provides a positive signal from all fecal samples for an animal species, whereas host-associated oligotypes are not found in all samples from an animal species. Deeper sequencing may reveal that a certain oligotype are indeed in all animals within a species, making host-associated an operational definition. The host-specific Blautia oligotypes will provide more reliable determinants of fecal pollution from a given host than would host-associated oligotypes.
Host-specific oligotypes that are rare everywhere
All host-specific oligotypes represent low-abundance organisms in the sampled microbial community. For example, only 0.17% of the V6 reads from chicken fecal samples resolved to Blautia (Supplementary Figure S2, Supplementary Table S1), but we recovered two chicken-specific Blautia oligotypes. The abundance of the chicken-specific oligotype AGCCTCCGCCCCGGCGCTTCAGGA had an average of 16 reads per chicken sample, which represents a very low relative abundance for this oligotype (∼0.003%) in the nine chicken samples. BLAST search of the representative sequence of this rare oligotype against NCBI's non-redundant database returns only one identical hit, with the annotation ‘caeca content of chicken gut' (GenBank accession GQ175463.1, accessed on 21 April 2014). The rarity of host-specific oligotypes was not limited to chickens. The fourth column of Table 1 reports the average abundance of each host-specific oligotype, which ranges from 0.001% to only 0.09%.
Despite use of stringent quality filtering that eliminates the vast majority of sequencing errors (Eren et al., 2013b), technical artifacts could have generated rare and seemingly host-specific oligotypes. In large amplicon datasets, random sequencing errors for an abundant parent template might produce multiple identical erroneous reads that may form a false operational taxonomic unit. However, a search of all potential parent sequences within a one-nucleotide similarity neighborhood exhibited no correlation between abundant reads in our dataset and very rare sequences that define host-specific oligotypes (see Supplementary Text S1). Sequencing error does not appear to account for the low frequency of host-specific rare oligotypes (Supplementary Table S1). Instead we interpret this pattern as evidence of taxa that are rare wherever they appear.
Geographical and ecological distribution of Blautia populations
Oligotype analysis of this dataset revealed other ecologically important patterns in host Blautia population structures. Despite a small sample size (n=10) for human fecal samples, Brazil sewage was primarily comprised of these human oligotypes, with only five additional ones found in the EMBASA wastewater treatment plant. This observation supports the hypothesis that microbial diversity from a limited number of fecal samples from the human population of a small village can account for most of the Blautia diversity in a sewage system servicing millions of individuals, which is consistent with previous reports using sewage as a representative sample of microbiomes of the human population (McLellan et al., 2013). In addition, the geographically distant Milwaukee sewage system only contained an additional 12 Blautia oligotypes beyond what was recovered from Brazilian human fecal samples. The sharing of oligotypes between the Milwaukee and the Salvador, Brazil samples suggests the widespread distribution of most Blautia taxa over continental scales. There were some notable differences in oligotypes found only in Brazil or only in US samples (Figure 2). For example, two moderately abundant oligotypes (>0.1% of the sewage-associated Blautia reads) in the EMBASA sewage occur in 9 of the 10 Brazilian human samples, but we did not detect those sequences in the Milwaukee sewage samples. We cannot exclude the possibility that more extensive sequencing efforts might detect unobserved oligotypes if they occur in vanishingly low numbers, but their distribution patterns reinforce the idea that differential abundance of Blautia oligotypes characterize geographically separated human populations. Significant differences in the presence/absence of rare oligotypes suggest that very low-abundance taxa might define differences between the microbiomes of geographically separated populations of humans. Overall, the strong human signature in the three sewage samples reflects the anticipated influence of human fecal input on these systems.
Blautia within the family Lachnospiraceae, provide energy to their host from polysaccharides that other gut microorganisms cannot degrade (Flint et al., 2008; Biddle et al., 2013). Different Blautia strains have specialized functions such as alpha-(1,6)-galactosidase activity in Ruminococcus gnavus, which is proposed to fall within the Blautia cluster but has not been renamed (Ludwig et al., 2008; Walker et al., 2011; Aguilera et al., 2012; Cervera-Tison et al., 2012), or H2 consumption by B. hydrogenotrophicus during acetogenesis (Bernalier et al., 1996). The colonization patterns we observed could relate to the selection of different Blautia organisms by diet, which can account for differences in animal groups and changes in the community over time (Ley et al., 2008; Muegge et al., 2011; Walker et al., 2011; Maga et al., 2012; Martínez et al., 2013). However, our results demonstrate very similar Blautia distribution patterns between the US sewage samples and samples collected from Brazilian humans. This stable diversity pattern extends to rare, host-specific Blautia oligotypes across large geographical distances for all three sewage systems, human fecal samples, as well as fecal samples from swine that feed upon typical agriculture feed in the US and food scraps in rural Brazilian villages. Because the urban and rural human populations on two different continents have different dietary habits (Yatsunenko et al., 2012) and the swine populations in Brazil and the US consume different feedstuff, the occurrence of very similar collections of Blautia oligotypes in a particular group of animals in both communities demonstrates that there are certain niches of microbial members affected by host physiology more so than diet. With respect to stability of rare but host species-specific oligotypes, we hypothesize that these organisms have adapted to the subtle physiological characteristics of their hosts and they fill a functional niche that might include fulfilling a keystone metabolic requirement.
Underrepresented Blautia diversity
We found an array of Blautia oligotypes that were largely unique for a host and common across all the samples in the host group (Figure S1). A BLAST search against 91 cultured Blautia isolates obtained from the RDP database (Cole et al., 2009) demonstrated that only 15 of 2055 unique V6 sequences generated in this study perfectly matched the sequences from cultivars (n=20). Some V6 sequences matched two or three cultured species, illustrating possible redundancy or high relatedness of cultured representatives. In a previous study, none of the near full-length environmental 16S sequences from sewage that resolved to Lachnospiraceae identically matched reference sequences from cultured species (McLellan et al., 2013). Both investigations imply greater, yet to be discovered, diversity for Blautia. Interpreting the significance of metabolic diversity—revealed through shotgun metagenomic investigations—and understanding specialized roles or host adaptation strategies of Blautia strains in fecal samples from different animals will require genome analyses from novel Blautia cultivars from different host environments.
In summary, we found remarkable population structure in a single genus of bacteria for all seven of the host species. The profiles of Blautia oligotypes likely represent strains that comprise a pool of metabolic capacity optimized for a host, as well as genomic traits that are linked to adaptation to a host environment. Our findings also suggest the existence of rare taxa that may be specific to certain host species universally, and the host physiology to be the major determinant of the community membership in certain niches of the gut microbiomes. Melding genomic studies with surveys of natural populations occurring in human and animal reservoirs could reveal the important role of Blautia in metabolism and health in different host ecological niches while at the same time suggesting additional determinants to identify sources of fecal pollution.
Acknowledgments
We thank Orin Shanks and Hyatt Green of the USEPA for providing US animal fecal samples. We also thank Ron Blanton from Case Western Reserve University for assisting with obtaining human fecal samples. This study was funded by the NIH grant R01AI091829-01A1 to SLM.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aguilera M, Rakotoarivonina H, Brutus A, Giardina T, Simon G, Fons M. Aga1, the first alpha-Galactosidase from the human bacteria Ruminococcusgnavus E1, efficiently transcribed in gut conditions. Res Microbiol. 2012;163:14–21. doi: 10.1016/j.resmic.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. ICWSM. 2009;2:361–362. [Google Scholar]
- Bernalier A, Willems A, Leclerc M, Rochet V, Collins MD. Ruminococcushydrogenotrophicus sp. nov., a new H2/CO2-utilizing acetogenic bacterium isolated from human feces. Arch Microbiol. 1996;166:176–183. doi: 10.1007/s002030050373. [DOI] [PubMed] [Google Scholar]
- Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. 2013;5:627–640. [Google Scholar]
- Blanton RE, Blank WA, Costa JM, Carmo TM, Reis EA, Silva LK, et al. Schistosoma mansoni population structure and persistence after praziquantel treatment in two villages of Bahia, Brazil. Int J Parasitol. 2011;41:1093–1099. doi: 10.1016/j.ijpara.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkosky M, Hernández-Delgado EA, Sandoz B, Robledo IE, Norat-Ramírez J, Mattei H. Detection of spatial fluctuations of non-point source fecal pollution in coral reef surrounding waters in southwestern Puerto Rico using PCR-based assays. Mar Pollut Bull. 2009;58:45–54. doi: 10.1016/j.marpolbul.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- Cervera-Tison M, Tailford LE, Fuell C, Bruel L, Sulzenbacher G, Henrissat B, et al. Functional analysis of family GH36 α-galactosidases from Ruminococcusgnavus E1: insights into the metabolism of a plant oligosaccharide by a human gut symbiont. Appl Environ Microbiol. 2012;78:7720–7732. doi: 10.1128/AEM.01350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–930. [Google Scholar]
- Dubinsky EA, Conrad ME, Chakraborty R, Bill M, Borglin SE, Hollibaugh JT, et al. Succession of hydrocarbon-degrading bacteria in the aftermath of the deepwater horizon oil spill in the gulf of Mexico. Environ Sci Technol. 2013;47:10860–10867. doi: 10.1021/es401676y. [DOI] [PubMed] [Google Scholar]
- Eloe-Fadrosh EA, Rasko DA. The human microbiome: from symbiosis to pathogenesis. Annu Rev Med. 2013;64:145–163. doi: 10.1146/annurev-med-010312-133513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, et al. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 2013;4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Vineis JH, Morrison HG, Sogin ML. A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS One. 2013;8:e66643. doi: 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ. Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS One. 2011;6:e26732. doi: 10.1371/journal.pone.0026732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewtrell L, Kaufmann RB, Kay D, Enanoria W, Haller L, Colford JM. Water, sanitation, and hygiene interventions to reduce diarrhoea in less developed countries: a systematic review and meta-analysis. Lancet Infect Dis. 2005;5:42–52. doi: 10.1016/S1473-3099(04)01253-8. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Franks AH, Harmsen HJ, Raangs GC, Jansen GJ, Schut F, Welling GW. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, et al. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 2011;7:e1001314. doi: 10.1371/journal.pgen.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furet J-P, Firmesse O, Gourmelon M, Bridonneau C, Tap J, Mondot S, et al. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol Ecol. 2009;68:351–362. doi: 10.1111/j.1574-6941.2009.00671.x. [DOI] [PubMed] [Google Scholar]
- Ginestet C. ggplot2: Elegant graphics for data analysis. J R Stat Soc Ser. 2011;174:245–246. [Google Scholar]
- Hayashi H, Sakamoto M, Kitahara M, Benno Y. Diversity of the Clostridium coccoides group in human fecal microbiota as determined by 16S rRNA gene library. FEMS Microbiol Lett. 2006;257:202–207. doi: 10.1111/j.1574-6968.2006.00171.x. [DOI] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Mark Welch D, Welch DM, Relman DA, et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Mark Welch DB, Voorhis A, Shipunova A, Morrison HG, Eren AM, et al. VAMPS: a website for visualization and analysis of microbial population structures. BMC Bioinformatics. 2014;15:41. doi: 10.1186/1471-2105-15-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Schleifer K-H, Whitman WB.Revised road map to the phylum Firmicutes Bergey's Manual of Systematic Bacteriology2nd edn,Vol. 3Springer: New York, NY, USA; 2008 [Google Scholar]
- Maga EA, Desai PT, Weimer BC, Dao N, Kültz D, Murray JD. Consumption of lysozyme-rich milk can alter microbial fecal populations. Appl Environ Microbiol. 2012;78:6153–6160. doi: 10.1128/AEM.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013;7:269–280. doi: 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May G, Nelson P. Defensive mutualisms: do microbial interactions within hosts drive the evolution of defensive traits. Funct Ecol. 2014;28:356–363. [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ. Unseen forces: the influence of bacteria on animal development. Dev Biol. 2002;242:1–14. doi: 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- McLellan SL, Newton RJ, Vandewalle JL, Shanks OC, Huse SM, Eren AM, et al. Sewage reflects the distribution of human faecal Lachnospiraceae. Environ Microbiol. 2013;15:2213–2227. doi: 10.1111/1462-2920.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, Bootsma MJ, Morrison HG, Sogin ML, McLellan SL. A microbial signature approach to identify fecal pollution in the waters off an urbanized coast of Lake Michigan. Microb Ecol. 2013;65:1011–1023. doi: 10.1007/s00248-013-0200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, Vandewalle JL, Borchardt MA, Gorelick MH, McLellan SL. Lachnospiraceae and Bacteroidales alternative fecal indicators reveal chronic human sewage contamination in an urban harbor. Appl Environ Microbiol. 2011;77:6972–6981. doi: 10.1128/AEM.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Graf J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol. 2012;10:815–827. doi: 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, et al. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 2010;4:377–387. doi: 10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- Oh S, Buddenborg S, Yoder-Himes DR, Tiedje JM, Konstantinidis KT. Genomic diversity of Escherichia isolates from diverse habitats. PLoS One. 2012;7:e47005. doi: 10.1371/journal.pone.0047005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Worobey M, Kuo C-H, Ndjango J-BN, Peeters M, Hahn BH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons SK, Bull CM, Gordon DM. Low prevalence of Salmonella enterica in Australian wildlife. Environ Microbiol Rep. 2010;2:657–659. doi: 10.1111/j.1758-2229.2010.00152.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria; 2011;vol. 1:409. [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, Newton RJ, Kelty CA, Huse SM, Sogin ML, McLellan SL. Comparison of the microbial community structures of untreated wastewaters from different geographic locales. Appl Environ Microbiol. 2013;79:2906–2913. doi: 10.1128/AEM.03448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statnikov A, Henaff M, Narendra V, Konganti K, Li Z, Yang L, et al. A comprehensive evaluation of multicategory classification methods for microbiomic data. Microbiome. 2013;1:11. doi: 10.1186/2049-2618-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- Ward DM. A natural species concept for prokaryotes. Curr Opin Microbiol. 1998;1:271–277. doi: 10.1016/s1369-5274(98)80029-5. [DOI] [PubMed] [Google Scholar]
- Webster NS, Taylor MW. Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol. 2011;14:335–346. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.