Abstract
The staphylococcal superantigen-like proteins (SSLs) are a family of polymorphic paralogs encoded in the Staphylococcus aureus genome whose function is unknown. The crystal structure of SSL7 was determined and compared to that of SSL5 and that of a classical superantigen, streptococcal pyrogenic exotoxin. Although the overall architecture of the superantigen family is retained in both SSL7 and SSL5, there are significant differences in the structures which suggest that the characteristic major histocompatibility complex binding site of superantigens has been lost. To complement these data, the abilities of SSL7 and a closely related paralog, SSL9, to interact with cells of the immune system were investigated. In populations of human white blood cells, both SSLs interacted selectively with monocytes via specific saturable but separate binding sites, which led to rapid uptake of the SSLs. In addition, SSLs were rapidly taken up by dendritic cells, but not by macrophages, into the same endosomal compartment as dextran. The ability of these secreted proteins to target antigen-presenting cells may enhance a misplaced antibody response against the proteins, which may facilitate bacterial colonization rather than contribute to host protection. Like classical superantigens, therefore, SSLs may distract the host's immune system, but they may do so via entirely different molecular mechanisms.
Staphylococcus aureus is a major causative agent of community- and hospital-acquired infections worldwide (35). This organism is an important pathogen due to a combination of invasiveness, toxin production, and antibiotic resistance. S. aureus causes a wide variety of clinical syndromes, ranging from uncomplicated infections of the skin to life-threatening toxic shock syndrome (11, 32). This bacterium causes disease by producing large numbers of exoproteins or virulence factors. Among the many known virulence factors are two families of staphylococcal pyrogenic superantigens, staphylococcal enterotoxins and toxic shock syndrome toxin 1 (5, 16). Superantigens bind to major histocompatibility complex (MHC) class II on antigen-presenting cells (B cells, monocytes, and dendritic cells) outside the classical antigen-binding groove, and they activate T cells by binding with the variable region of the β chain of T-cell receptors (16, 29). This cross-linking triggers the nonspecific activation and proliferation of T cells, induces the production of high levels of a variety of cytokines (5, 20, 22), and causes toxic shock syndrome characterized by fever, rash, hypotension, and multiple-organ failure. Staphylococcal enterotoxins are responsible for many cases of food poisoning (intoxication) associated with ingestion of toxin-contaminated food (11, 35). To date, more than 13 staphylococcal enterotoxins have been described, and it is likely that additional toxins will continue to be discovered (34).
In addition to these classical superantigens, S. aureus also produces a family of proteins that have sequence homology to the superantigens, the staphylococcal exotoxin-like proteins (SETs). These proteins were first identified by identifying a genetic locus that encodes at least five SETs (SET1 to SET5), and they were reported to be able to stimulate the secretion of some proinflammatory cytokines by human peripheral blood mononuclear cells (PBMCs) (44). More recently, data resulting from sequencing of the genomes of several different S. aureus strains have revealed a large number of related (levels of homolgy, 36 to 67%) set genes clustered on a genomic island (3, 24). This putative pathogenicity island, which is present in all strains of S. aureus examined to date, contains between 7 and 11 set genes, which have various degrees of sequence homology. In addition, there appears to be extensive interstrain allelic polymorphism for each of the set genes (3, 14, 24; unpublished data). The International Nomenclature Committee for Staphylococcal Superantigen Nomenclature has recently recommended (27a) that the SETs should be renamed staphylococcal superantigen-like proteins (SSLs) and that the genes should be designated ssl1 to ssl11 in clockwise order from the replication origin of the chromosome based on homology to the full complement of genes found in strain MW2. This nomenclature is essentially that described by Fitzgerald et al. (14), except that the numbering of the genes is in the opposite direction. To differentiate between allelic variants, the ssl gene designation is preceded by the strain designation. This nomenclature is used throughout this paper. Arcus and colleagues (2) have determined the three-dimensional structure of one member of the family, SSL5 (previously SET3). The crystal structure of this protein shows that it has many of the characteristic structures of the superantigen superfamily, but there are also significant differences. In contrast to classical superantigens, SSLs do not have the main properties of superantigens, such as polyclonal T-cell activation, pyrogenicity, or enhancement of endotoxin shock (2, 14). The functions of SSLs, therefore, remain unclear, but their high degree of diversification and polymorphism suggest that they are subject to strong selective pressures and imply that they may play an important role in host-pathogen interactions.
In this study we determined the structure of SSL7 (previously SET1) (44) in order to gain better insight into the relationships among the different members of the SSL family. We also investigated the tropism of SSL7 and its close homologue SSL9 (previously SET5) (44) for different cells of the immune system. Our results demonstrate that both SSL7 and SSL9 interact with human antigen-presenting cells (monocytes and dendritic cells), although the two proteins interact with independent sites on the cell surface. Facilitated uptake of SSLs by antigen-presenting cells may play a role in the modulation of host immunity to S. aureus.
MATERIALS AND METHODS
Recombinant protein expression and purification.
Recombinant N-terminal histidine-tagged SSL7 and SSL9 from S. aureus strain NCTC6571 were produced in Escherichia coli by using the expression vector pQE30 containing the NCTC6571 ssl7 and NCTC6571 ssl9 genes, respectively, as previously described (44). Embp32 from Staphylococcus epidermidis was also expressed as a recombinant N-terminal histidine-tagged fusion protein in E. coli and was purified as previously described (43).
In order to produce recombinant SSL7 without the histidine tag, a cleavable affinity tag was introduced. The NCTC6571 ssl7 gene from pQE30 was subcloned into the glutathione S-transferase (GST) expression vector pGEX-4T-1 on a BamHI-SalI fragment and was transformed into E. coli JM109. For gene expression, an overnight culture was diluted 1:25 in fresh Luria-Bertani broth containing 100 μg of ampicillin per ml and incubated for 2 h at 37°C. Gene expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 37°C. Cells were harvested by centrifugation at 6,000 × g for 30 min, resuspended, and lysed for 20 min in B-PER protein extraction reagent (Pierce & Warriner Ltd.) containing protease inhibitors (Sigma). Lysates were clarified by centrifugation at 23,000 × g for 10 min. The lysate containing the GST-SSL7 fusion protein was passed through a GST-Sepharose column (Amersham-Pharmacia Biotech), and recombinant SSL7 without the GST tag was released from the column by digestion with thrombin essentially as described by the supplier of the column (Amersham-Pharmacia Biotech).
Crystallization.
Crystals of SSL7 were obtained by the hanging-drop vapor diffusion technique at room temperature. Crystals were obtained under two different conditions. For the first condition (form I), the well buffer contained 25 to 30% (wt/vol) polyethylene glycol monomethyl 2000, 0.2 M ammonium sulfate, and 0.1 M morpholineethansulfonic acid (MES) (pH 6.5). The drops consisted of 1 μl of recombinant SSL7 (10 mg/ml) and 1 μl of well buffer. The crystals had a flat plate morphology with dimensions up to 0.3 by 0.2 by 0.01 mm. For the second condition (form II) we used the same protein concentration and drop size, but the well buffer consisted of 28% (wt/vol) polyethylene glycol 2000 and 0.1 M Li2SO4 buffered with 0.1 M Tricine at pH 8.5. In this case the crystals were rod shaped, and the dimensions were approximately 0.3 by 0.05 by 0.05 mm.
X-ray data collection.
Data were collected from cryocooled crystals following immersion in mother liquor containing 30% (vol/vol) glycerol (as described by Garman and Schneider [15]).
(i) Form I.
The data were collected at a resolution of 2.75 Å on line BM14 at the European Synchrotron Radiation Facility (Grenoble, France) by using a Mar Research charge-coupled device detector. The data were indexed and integrated with Mosflm (27) and were scaled and merged by using Scala (13) from the CCP4 suite (8). The subsequent analysis was carried out by using programs from the CCP4 suite, unless indicated otherwise. Data collection statistics are shown in Table 1. Autoindexing indicated that the crystals belonged to point group P422 with cell dimensions a = b = 81.66 Å and c = 148.04 Å and a Matthews coefficient of 3.1 (corresponding to a solvent content of 60% [vol/vol]) for two molecules in the asymmetric unit. There was a significant peak in the native Patterson map at fractional coordinates (0.000, 0.372, and 0.500), and there were no peaks attributable to noncrystallographic symmetry in the self-rotation function, indicating that there were two molecules related by a twofold rotation parallel to the a axis in the asymmetric unit.
TABLE 1.
Data collection and refinement f
| Form | Resolution (Å) |
Nref
|
Rmerge (%)a
|
I/σI
|
Completeness (%)
|
Redundancy (%)
|
Protein
|
Water
|
R factor (%)c
|
rmsd
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Measured | Uni. | All | Highb | All | High | All | High | All | High | N | Mean B factor (Å2)c | N | Mean B factor (Å2) | Workingd | Freee | Bonds (Å) | Angles (°) | ||
| I (P43212) | 45.5-2.75 | 59,980 | 12,607 | 9.4 | 46.0 | 14.6 | 3.1 | 98.7 | 99.2 | 4.5 | 4.3 | 3,127 | 43.6 | 18 | 30.9 | 23.5 | 27.5 | 0.008 | 1.35 |
| II (P212121) | 30.0-2.7 | 99,169 | 11,049 | 14.0 | 48.0 | 4.6 | 1.5 | 99.9 | 99.9 | 4.5 | 3.9 | 42.6 | 24 | 42.5 | 23.4 | 29.9 | 0.011 | 1.3 | |
Rmerge = Σ|II − IM|/ΣIM, where II is the observed intensity of a reflection and IM is the mean intensity of all related reflections.
High, highest-resolution shell. Form 1, 2.9 to 2.75 Å; form II, 2.85 to 2.70 Å.
R factor = Σ|Fobs − Fcalc|/ΣFobs, where Fobs is the observed structure factor and Fcalc is the structure factor calculated from the atomic model.
R factor for the 95.1% of data included in the refinement.
R factor for the 4.9% of data randomly selected and excluded from refinement.
Nref, number of reflections; uni., unique; N, number of atoms.
(ii) Form II.
The data were collected at a resolution of 2.7 Å on line ID14-1 at the European Synchrotron Radiation Facility by using an ADSC Quantum 4R charge-coupled device detector. Data indexing, integration, scaling, and analysis were carried out as described above for form I. Autoindexing and analysis of systematic absences indicated space group P212121 with a = 51.65 Å, b = 71.59 Å, c = 103.47 Å, and a Matthews coefficient of 2.00, corresponding to a solvent content of 40%. There was a peak in the self-rotation function at κ = 180°, confirming the presence of two molecules in the asymmetric unit. Data collection statistics are shown in Table 1.
Molecular replacement.
The coordinates for one monomer of SSL5 (pdb-id: 1M4V) (2) were used for molecular replacement, which was carried out with Molrep (42).
(i) Form I.
There was a single clear peak in the rotation function (I/σI = 5.3; next peak I/σI = 3.58). The translation function was carried out for all nine possible space groups, and the best solution was for P43212, with a correlation coefficient of 32.6%. Side chains that were different in the SSL5 and SSL7 proteins were replaced by alanines in the correctly positioned model, and rigid-body refinement was carried out by using CNS, version 1.1 (6). At this point, the R factor was 52.2% and the Rfree factor was 54.6%. A round of simulated annealing reduced the R and Rfree factors to 44.6 and 48.9%, respectively.
(ii) Form II.
Molecular replacement for form II was performed as described above for form I. The best solution had a correlation coefficient of 37.4% for two molecules in the asymmetric unit. Following rigid-body and simulated annealing refinement in CNS, the R and Rfree factors were 34.16 and 42.06%, respectively.
Density modification.
For both crystal forms, cross-crystal averaging, noncrystallographic averaging, and phase improvement were carried out by using Dmmulti (9) in CCP4 (8) prior to calculation of maps for manual rebuilding of the model.
Model building and refinement.
In both cases, manual rebuilding was performed by using O (19), and in later refinement rounds XtalView (30) was used. Refinement was carried out with CNS. Alanine residues in the initial model were exchanged for the correct side chains where positive Fourier difference density could be seen. At a number of positions the sequence alignment was incorrect, and additional rebuilding of the chain was required. For the refinement, an overall anisotropic B-factor correction and bulk solvent scaling with k = 0.36 and B = 26.9 Å2 for form I and k = 0.59 and B = 101 Å2 for form II were applied. Noncrystallographic symmetry restraints were applied throughout refinement except in the later stages where there was clear evidence of a difference between the chains. After all protein residues had been included in refinement, a number of tightly bound waters were added, where there was a 3-root mean square (rms) peak in the difference Fourier and a 1-rms peak in the 2Fo-Fc map and there were appropriate protein-water hydrogen bonds. At the end of refinement, the R factor was 23.5% and the Rfree factor was 27.5% for form I and the R factor was 23.4% and the Rfree factor was 29.9% for form II. The final coordinates and structure factors have been submitted to the pdb database with identification codes 1V1O for form I and 1V1P for form II.
A homology model of SSL9 was created by using the program MODELLER (26) and both SSL7 and SSL5 as template structures.
FITC labeling of SSLs.
SSL7 and SSL9 were dialyzed against labeling buffer (0.2 M NaHCO3, pH 9.0) overnight at room temperature. Fifty microliters of a 1-mg/ml fluorescein isothiocyanate (FITC) (Sigma) solution in dimethyl sulfoxide was added to 1 ml of a 2-mg/ml protein solution. After 4 h of incubation at room temperature in the dark, unbound FITC was removed by size exclusion chromatography by using a PD-10 (Sephadex) column. The concentration of labeled protein and the FITC/protein ratio were determined by spectrophotometry. All preparations had FITC/protein ratios between 1:1 and 2:1.
Antibodies.
The following monoclonal antibodies (MAbs) were used: CD2 (mouse MAb MAS 593; immunoglobulin G2b [IgG2b]; Harlan), CD3 (supernatant mouse MAb UCHT1; IgG1; gift from P. C. L. Beverley, Edward Jenner Institute for Vaccine Research, Compton, United Kingdom), CD14 (supernatant mouse MAb HB246; IgG2b; gift from P. C. L. Beverley), CD19 (supernatant mouse MAb BU12; IgG1; gift from D. Hardie, Birmingham University, Birmingham, United Kingdom), and HLA-DR (supernatant mouse MAb L243; IgG2a; gift from P. C. L. Beverley).
Cell culture.
Human PBMC-derived dendritic cells were generated from fresh whole blood samples obtained from healthy volunteers (1, 33). Mononuclear cells separated with Lymphoprep (Nycomed Pharma) by centrifugation at 400 × g for 30 min were incubated in six-well tissue culture plates for 2 h at 37°C in the presence of 5% CO2 in complete medium (CM) (RPMI 1640 medium [Gibco] supplemented with 10% fetal calf serum [PAA Laboratories], 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM l-glutamine [Clare Hall Laboratories, Cancer Research UK]). The adherent cells were cultured in fresh CM supplemented with 100 ng of human recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF) per ml and 50 ng of interleukin-4 (IL-4) (Schering-Plough Research Institute) per ml. On day 4 of incubation, loosely adherent cells were collected, and contaminating T and B lymphocytes were removed by incubation with MAbs CD3, CD2, and CD19, followed by anti-mouse IgG-coated immunomagnetic Dynabeads (Dynal). The supernatant, containing highly purified dendritic cells, was cultured for another 3 days in fresh CM with GM-CSF and IL-4. Human PBMC-derived macrophages were obtained by using the same procedure for dendritic cell culture, except that 10% human serum was used and no cytokines were added (41).
Binding and uptake of FITC-labeled SSLs by human cells.
Binding assays were performed by incubating 106 cells/ml in CM with various concentrations of SSL-FITC (0.05 to 1.25 μM) for 1 h at 4 or 37°C. In some experiments, 8 μM unlabeled SSL was added to the cells together with the labeled protein. After incubation, cells were washed three times by centrifugation and examined by flow cytometry. In some experiments, cells were also stained for various surface markers after SSL uptake. Cells were incubated with the relevant MAb for 30 min at 4°C, washed, and then incubated in 1:25-diluted phycoerythrin-conjugated goat anti-mouse immunoglobulin (Jackson ImmunoResearch) for 30 min at 4°C. Cells were washed, fixed in 2% formaldehyde, and examined by using a FACScan flow cytometer (Becton Dickinson).
Confocal microscopy.
A total of 105 cells were seeded on 32-mm coverslips coated (for dendritic cells only) overnight at 4°C with 10 μg of fibronectin (Sigma) per ml in Hanks balanced salt solution (Gibco). After incubation for 2 h at 37°C in CM, cells were incubated with SSL-FITC (1.25 μM) and/or Texas Red-dextran (1 mg/ml; Molecular Probes) for 1 h at 37°C in CM. The coverslips were then washed three times in cold Hanks balanced salt solution and fixed in 2% paraformaldehyde. The slides were examined with a Bio-Rad confocal microscope. Images were acquired from 0.5-μm optical sections of individual cells.
RESULTS
Determination of SSL structure.
Recombinant SSL7, consisting of residues 36 to 231 of the sequence whose GenBank accession number is AF094826, was crystallized under two different conditions, each of which gave rise to a different crystal form.
(i) Form I.
The final, 2.75-Å-resolution model built into the electron density map contained two SSL7 molecules, which represented residues 18 to 213 of the recombinant SSL7. In addition, 18 water molecules were included at stereochemically sensible locations. Although the His tag and N-terminal tail were disordered and omitted from the density map, the majority of the residues had well-defined electron density. The final R and Rfree factors were 23.5 and 27.5%, respectively, and refinement statistics are shown in Table 1. The final model had good stereochemistry, with 98.6% of the residues in the most favored and additionally allowed regions of the Ramachandran plot and no residues in disallowed regions. The geometry was better than expected for the average 2.75-Å structure according to PROCHECK analyses (26).
(ii) Form II.
The form II model also contained two SSL7 molecules, and in this case residues 21 to 213 and residues 23 to 213 of the construct were in chains A and B, respectively. Twenty-four water molecules were added in this form. Once again, with the exception of the His tag and N-terminal tail, the majority of the molecule had well-defined electron density. The final R and Rfree factors were 23.4 and 29.9%, respectively. The final model had good stereochemistry, with 96.5% of the residues in the two most favored regions of the Ramachandran plot. The geometry was better than expected for the average 2.7-Å structure according to PROCHECK. Refinement statistics for both crystal forms are shown in Table 1.
(iii) SSL7 structure.
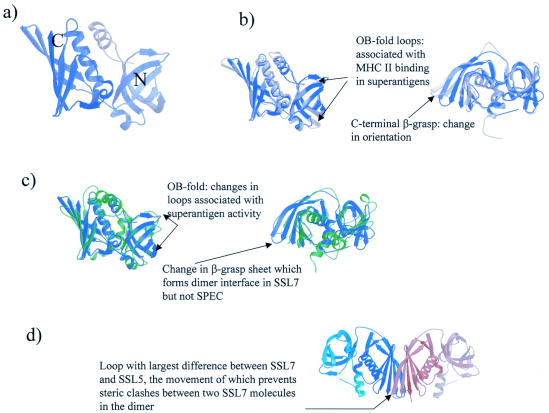
The structure of the SSL7 monomer is shown in Fig. 1a. As predicted from sequence comparisons, the fold is similar to that of the bacterial superantigens and consists of two domains. The N-terminal domain (residues 18 to 110) is an OB fold, a variety of a β-barrel associated with oligosaccharide and DNA binding, while the C-terminal domain (residues 111 to 213) forms a β-grasp domain (a series of β-strands wrapped around a helix).
FIG. 1.
(a) Structure of SSL7, with shading from the N terminus (gray) to the C terminus (lilac). This image and all other structural illustrations were drawn by using Bobscript (12), a modification of molscript (23), and were rendered with Raster3D (4, 31). (b) On the left the structure of SSL7 (lilac) is optimally superposed on the structure of SSL5 (gray). The arrows indicate the locations of the OB-fold loops that are equivalent to those associated with MHC class II binding in superantigens. On the right the image is rotated 90° to highlight the change in the C-terminal β-grasp domain orientation. The largest shift in loop position is indicated by an arrow. (c) On the left the structure of SSL7 (lilac) is optimally superposed on the structure of SPEC (green). The arrows indicate the locations of the OB-fold loops that are associated with superantigen-MHC class II interactions and are markedly different in SSL7. On the right the image is rotated 90° to highlight the change in β-grasp domain orientation. An arrow indicates the location of the largest shifts between SSL7 and SPEC. (d) SSL7 dimer found in both the form I and form II asymmetric units. The images is orientated to highlight the interaction of the two β-grasp domains to form a β-sandwich. One monomer is shaded from white to gray, the other monomer is shaded from dark blue to light blue, and an arrow indicates the loop whose position is most different in optimally superposed SSL5 and SSL7; this change in position prevents steric clashes in the SSL7 dimer.
Altogether, we obtained four copies of the SSL7 monomer (two from each crystal form), and they were all very similar, as shown by the Cα atom root mean square deviations (rmsds) (Table 2). The only differences between the structures arise from differences in the conformations of flexible loops and in the linker between the N- and C-terminal domains (residues 106 to 112). The relative orientations of the two domains with respect to one another and the orientations of the individual secondary structural elements were the same in the different copies of the monomer. For this reason, unless explicitly stated otherwise, we used one example monomer, chain A from form I, for the comparisons and discussions below.
TABLE 2.
rmsds between the different copies of the SSL7 monomer
| Form | Copy | rmsd (Å)
|
|||
|---|---|---|---|---|---|
| Form I
|
Form II
|
||||
| Copy A | Copy B | Copy A | Copy B | ||
| I | A | 0.228 | 0.701 | 0.703 | |
| B | 0.669 | 0.642 | |||
| II | A | 0.450 | |||
| B | |||||
(iv) SSL7 and other proteins.
When the SSL7 monomer was superposed on the structure of SSL5 (SET3) (2), the other member of the family whose three-dimensional structure is available, the two were found to have the same fold (Fig. 1b), as might be expected for proteins exhibiting 40% sequence identity. However, when the structures were optimally superposed, the rmsd, which was 1.33 Å over 157 spatially equivalent Cα atoms, was surprisingly high for two such highly related proteins (a value of about 1.1 Å over the whole structure would be anticipated based on the sequence identity [7]). This rmsd could be largely accounted for by changes in two regions of the structure. First, there was a change in the twist of the β-sheet in the C-terminal β-grasp domain (Fig. 1b, right structure). The change in the twist resulted in shifts in individual residue positions as large as 6.65 Å (for the Cα atom of G125 in the C-terminal domain). Second, there were changes in the conformations of the loops on the external face of the N-terminal OB fold (Fig. 1b); these loops were associated with a generic low affinity MHC class II binding site in superantigens (17, 21), and changes in them could suggest differences in the functions of the two proteins. These large movements accounted for the low contrast and large R factor for the initial molecular replacement solution prior to the simulated annealing which successfully realigned the sheet strands and some of the loop residues.
The SSL7 structure was also compared to a homology model of SSL9 based on both SSL7 and SSL5 as template structures (data not shown). The sequence of SSL9 had a much greater level of homology to SSL7 (sequence identity, 49%) than to SSL5 (sequence identity, 35%). This was reflected in the model of SSL9; when the structures were optimally superposed, SSL9 had a Cα atom rmsd of 0.6 Å over 188 spatially equivalent atoms from SSL7 and a Cα atom rmsd of 1.4 Å over 177 spatially equivalent atoms from SSL5.
When SSL7 and the superantigen with a known structure with which it exhibited the highest level of sequence identity (streptococcal pyrogenic exotoxin [SPEC]; level of sequence identity, 29%) (37) were optimally superposed (Fig. 1c), we found that once again the overall fold was conserved. SPEC had some extended loops, but the structures were otherwise very similar. Interestingly, despite the fact that the SPEC and SSL7 sequences were far more divergent than the SSL7 and SSL5 sequences, the two structures superposed nearly as well. The Cα atom rmsd for optimally superposed SSL7 and SPEC was 1.48 Å over 134 structurally equivalent atoms. The difference between the structures resulted from both a slight change in the orientation of the β-grasp domain and large differences in the orientations of the strands of the OB fold; the very large changes in conformation in this domain were not unexpected since the SSLs do not bind MHC class II proteins and this is the region primarily involved in this interaction in the superantigens.
(v) Dimerization.
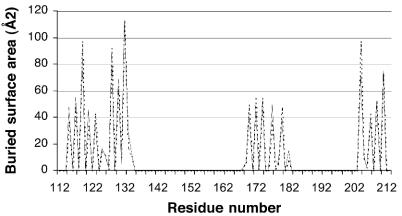
In both crystal form I and crystal form II there were two molecules in the crystallographic asymmetric unit; these two molecules were related by a proper 2 fold, which resulted in the formation of an intimate dimer (Fig. 1d). The dimer was virtually identical in the two crystal forms, as shown by a comparison of the residues buried in the dimer interface (Fig. 2) and by the fact that the form I and form II dimers could be superposed with an all-Cα atom rmsd of 0.881 or 0.902 Å depending on the orientation. The remainder of the crystal packing was entirely different for the two forms, and since the crystals grew under different conditions and at markedly different pH values, it is unlikely that the dimers formed solely as a result of crystal packing forces. As Fig. 1d shows, the dimer interface was the result of the two β-grasp domains interacting to create an intermolecular β-sandwich. In the process, 1,122 Å2 of the monomer surface in form I and 1,146 Å2 of the monomer surface in form II were buried; these values are in the range observed in biologically relevant dimers (18). Dimer formation resulted in burial of a number of hydrophobic residues (including F119, L128, and I132), and a number of neutral polar residues contributed to a hydrogen bonding network between the two monomers; however, no charge-charge interactions were created. It has been shown that protein-protein interaction surfaces differ little from the normal exterior surfaces of proteins but that they tend to contain additional neutral polar residues and fewer charged residues (28); the SSL7 dimer interface is entirely consistent with this.
FIG. 2.
Residues in the SSL7 dimer interface: plot of residue numbers versus the buried surface area (in square angstroms). Dotted line, form I; solid line, form II.
Cellular tropism of SSL7 and SSL9.
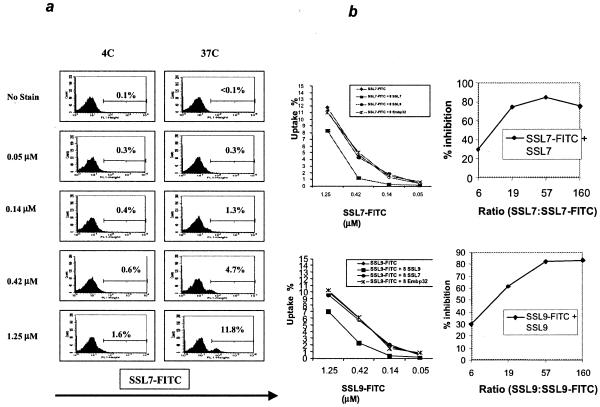
PBMCs were incubated with various concentrations of SSL7-FITC or SSL9-FITC for 1 h, and cell-associated fluorescence was measured by flow cytometry. Both SSL7 (Fig. 3a) and SSL9 (data not shown) stained a small proportion of PBMCs at 37°C but not at 4°C. The level of staining was dose dependent up to a maximum at a protein concentration of 1.25 μM and did not increase at higher concentrations. The mean percentages of cells stained with SSL7 (9.8% ± 1.8%; range, 7.1 to 12.2%; n = 7) and SSL9 (10.9% ± 1.1%; range, 9.4 to 12.6%; n = 5) were very similar. The mean fluorescence also increased with time between 5 and 120 min (data not shown), suggesting that there was progressive uptake of the SSLs by the cells.
FIG. 3.
(a) Dose and temperature dependence of SSL7 uptake by PBMCs. PBMCs were incubated for 60 min with various concentrations of SSL7-FITC at 37 or 4°C. The results are representative of the results of 10 separate experiments carried out with PBMCs from healthy donors. (b) Inhibition of binding of SSL-FITC to PBMCs by unlabeled SSL. Different concentrations of SSL7-FITC (upper graphs) or SSL9-FITC (lower graphs) were incubated with PBMCs with or without 8 μM unlabeled SSL7, SSL9, or Embp32 protein. The data are from one representative experiment of three experiments that were performed. The graphs on the right show the levels of inhibition observed at different ratios of unlabeled SSL7 to labeled SSL7 (top graph) and at different ratios of unlabeled SSL9 to labeled SSL9 (bottom graph).
In order to determine whether the interaction between SSLs and PBMCs was specific, competitive inhibition of SSL-FITC cell labeling by unlabeled SSL was investigated (Fig. 3b). Excess unlabeled SSL7 was able to block uptake of SSL7-FITC by up to 80% (Fig. 3b, graphs on the right). In contrast, neither SSL9 nor an unrelated bacterial protein also carrying a polyhistidine tag (Embp32) had any effect on the SSL7-FITC signal. Conversely, unlabeled SSL9, but not SSL7 or Embp32, blocked uptake of SSL9-FITC (Fig. 3b, lower graphs).
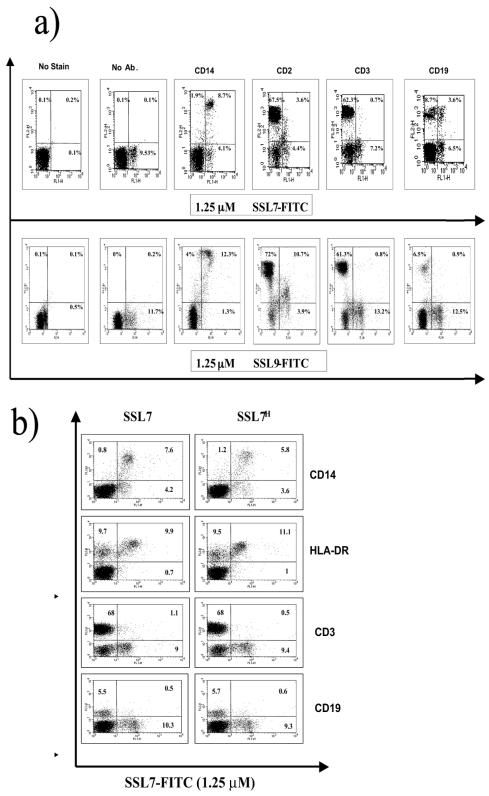
The PBMC subpopulations which were the targets for SSL7 and SSL9 were further characterized by immunophenotyping by using MAbs to the major surface markers CD2, CD3, CD14, and CD19 (Fig. 4). Both SSL7 and SSL9 were taken up by the majority of CD14-positive cells and by a population of CD2 low cells, a phenotype consistent with that of peripheral blood monocytes (10). Neither SSL7 nor SSL9 showed any interaction with CD3-positive T cells. Interestingly, SSL7-FITC, but not SSL9-FITC, stained a subpopulation of CD19 B cells in some individuals, providing further evidence that the receptor for these two SSLs is distinct. The interaction between SSL7 and B cells was not investigated further in this study.
FIG. 4.
Immunophenotype of PBMCs that interact with SSLs. PBMCs were stained for various cell surface markers after incubation with 1.25 μM SSL7-FITC or SSL9-FITC for 60 min at 37°C. The results are representative of the results of five separate experiments. (a) SSLs with histidine tag. Ab, antibody. (b) Comparison of staining observed with SSL7-FITC with the histidine tag (SSL7H) (graphs on the right) and without the histidine tag (graphs on the left).
In order to confirm that the binding of SSLs to PBMCs was not affected by the presence of the histidine tag, SSL7 was subcloned into pGEX-4T-1, expressed, and purified, and the GST tag was then removed by cleavage with thrombin (see Materials and Methods). FITC-labeled SSL7 lacking a tag showed binding characteristics very similar to those of the histidine-tagged version (Fig. 4b). In particular, both versions of SSL7 bound to only a subpopulation of HLA-DR-expressing cells (Fig. 4b), suggesting that the proteins did not bind directly to this receptor.
Uptake of SSLs by dendritic cells.
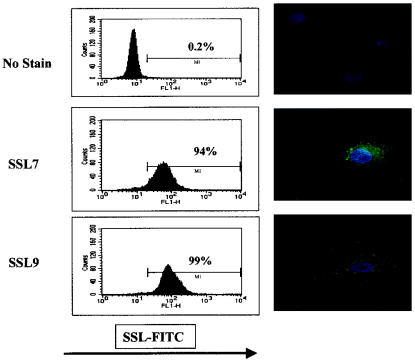
Peripheral blood monocytes were cultured in vitro in the presence of GM-CSF and IL-4 in order to drive differentiation into myeloid dendritic cells (39). After depletion of residual lymphocytes, the population obtained after 7 days of incubation consisted of >90% CD1a+ HLA-DRhigh CD14low dendritic cells (data not shown). These cells were incubated for 60 min at 37°C with either SSL7-FITC or SSL9-FITC and examined by flow cytometry and confocal microscopy (Fig. 5). Dendritic cells stained uniformly strongly positive for both SSL7 and SSL9. SSL7 lacking the histidine tag also bound strongly to dendritic cells (data not shown). Confocal microscopy confirmed that fluorescence was predominantly due to intracellular uptake of SSL rather than to surface staining. Both SSL7 and SSL9 were concentrated in small vesicular structures, which were localized particularly in the perinuclear region of the cell.
FIG. 5.
SSL binding to PBMC-derived dendritic cells. Dendritic cells derived from CD14+ cells were incubated with 1.25 μM SSL conjugated with FITC. Binding was analyzed by flow cytometry (graphs on the left) and confocal microscopy (images on the right). Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) and are blue. The results are representative of the results of a set of three separate experiments carried out with dendritic cells from healthy donors.
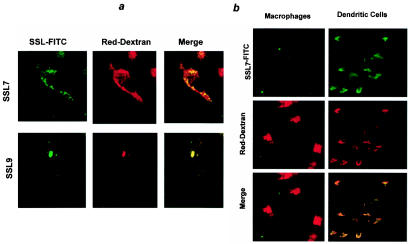
In order to examine the nature of these vesicles further, dendritic cells were cultured in the presence of SSL7 or SSL9-FITC and Texas Red dextran (Fig. 6a), which is avidly taken up by dendritic cells via mannose receptors (38). Texas Red dextran strongly labeled a large number of intracellular vesicles throughout the dendritic cell cytoplasm. The SSL distribution and the dextran distribution partially overlapped, and some intracellular vesicles clearly contained both markers. However, SSL-positive dextran-negative vesicles were also observed. In a small proportion of cells, vesicles containing SSL9 appeared to aggregate and to generate very large vesicles, which contained high concentrations of both SSL and dextran (Fig. 6a).
FIG. 6.
(a) Intracellular localization of SSLs in human PBMC-derived dendritic cells. Dendritic cells were cultured in the presence of 1.25 μM SSL7- or SSL9-FITC with 1 mg of Texas Red dextran per ml for 60 min at 37°C. Cells were analyzed by confocal microscopy. The results are representative of the results of six separate experiments. (b) Intracellular uptake of SSLs by PBMC-derived dendritic cells and PBMC-derived macrophages. Cells were incubated with 1.25 μM SSL7-FITC and 1 mg of Texas Red dextran per ml for 60 min at 37°C. The results are representative of the results of three separate experiments.
In order to determine whether uptake of SSLs was a generalized feature of endocytic cells, peripheral blood monocytes were differentiated into macrophages by culture in human serum without added cytokines. Under these culture conditions, the cells develop a completely different phenotype (CD1a−,HLA-DR−,CD14high) and morphology (lack of dendrite formation) (41). Macrophages, like dendritic cells, efficiently endocytosed Texas Red dextran but showed no uptake of either SSL7 (Fig. 6b) or SSL9 (data not shown).
DISCUSSION
One of the most exciting results of the structural studies of SSL7 was the identification of the same SSL7 homodimer in the asymmetric units of two otherwise very different crystal forms grown under very different solution conditions. This dimer has a number of characteristics that are seen in functionally relevant dimers, and we therefore suggest that it is not purely an artifact of crystallization. Interestingly, analysis of the structure of SSL5 did not reveal any such dimer formation (2), and the residues making up the interface are not conserved in the different SSLs. The change in the orientation of the β-grasp domain in SSL7 relative to the orientation in SSL5 is necessary to allow the dimer to form; if a similar dimer is created from SSL5 monomers, clashes occur between residues 110 to 114 in one monomer and residues 197 to 200 in the other monomer and more seriously between residues 118 to 125 and residues 161 to 165. These clashes are alleviated by the change in the orientation of the β-grasp domain β-strands in SSL7, further suggesting that SSL5 does not form a dimer in the same way.
Since the crystal structure of SSL9 is not available yet, a preliminary comparison with SSL7 was carried out by using a homology model. Like the SSL5 residues, the residues involved in the dimer interface in SSL7 are not conserved in SSL7 and SSL9. However, the sequence changes do not create steric or electrostatic clashes between the two monomers; rather, the changes are such that hydrogen bonding between the two molecules is maintained, and in some cases new hydrogen bonds are formed. It is therefore possible that SSL9 may form a dimer in the same manner as SSL7.
We have also commented on the differences between SSL7 and SSL5 in the region of the N-terminal domain that are implicated in a general low-affinity MHC class II binding site in superantigens. The homology model revealed that the residues in these loops are in general highly conserved in the SSL7 and SSL9 sequences. However, there are some important differences, including the change of P93 in SSL7 to a threonine residue in SSL9. This proline is part of a well-ordered β-turn in SSL7, while in SSL5 there is no ordered secondary structural element present at this position; in fact, the loop in SSL5 is rather disordered. The pattern of sequence conservation in the N-terminal loops suggests that the structures and functions of SSL7 and SSL9 are more related to one another than to the structures and functions of SSL5. However, the small number of nonconservative sequence differences between SSL7 and SSL9 are also entirely consistent with differences in the putative receptor binding, as discussed further below.
Studies of the superantigens have shown that their interactions with MHC class II and T-cell receptor molecules are very diverse, involving a number of different interaction surfaces and stoichiometries. This includes the formation of functionally important superantigen dimers for some superantigens (for example, the Zn2+-dependent dimers formed by staphylococcal exotoxin D [40]), which form via the C-terminal β-grasp domain, in a manner reminiscent of the homodimers which we have observed for SSL7. It also includes the formation of heterodimers with the same surface of the N-terminal OB fold but different surfaces of MHC molecules (as in the complexes of HLA-DR1 with staphylococcal enterotoxin B [17] and toxic shock syndrome toxin 1 [21]). In a similar manner, the functional and structural studies described in this paper showed that SSL7 and SSL5, which are among the most distantly related SSLs identified so far, may be functionally active in different quaternary states.
The significant differences among the structures of SSL7, SSL5, and other superantigens suggest that these molecules may interact with different binding partners, and this suggestion is supported by the cellular tropism data reported in this paper. The characteristic features of the interaction between SSL7 and SSL9 molecules and PBMCs are specificity, temperature dependence, and cell selectivity. Specificity, which indicates that the interaction is mediated by a cell surface receptor, was demonstrated by the fact that unlabeled SSL blocked uptake of SSL-FITC. This competition was observed for both SSL7 and SSL9. The lack of reciprocal inhibition between SSL7 and SSL9 suggests that these two molecules have different binding partners on the cell surface, although we cannot rule out the possibility that they bind to different sites on the same molecule. Since it was impossible to measure binding in the absence of uptake, true measurements of affinity could not be obtained. The concentrations required to obtain measurable uptake, however, were on the order of 0.1 μM, suggesting that the affinity of interaction with any putative receptor is relatively low. This is a characteristic of many classical superantigens (25).
The temperature dependence and time dependence of the SSL interaction suggest that there is receptor-mediated uptake rather than simple binding to the cell surface, and this was confirmed by the confocal microscopy studies discussed below. However, a small amount of surface binding was detected at 37°C but not at 4°C by using indirect labeling of intact cells with an antibody against the histidine tag (data not shown). The interaction of SSL with the receptor, as well as the subsequent uptake, was therefore temperature dependent. The colocalization of SSLs and FITC-dextran suggests that the proteins are taken up into the same compartment that is targeted by the mannose receptor, although the data do not suggest that SSLs themselves are taken up via this receptor. The very large vesicles observed upon SSL9 uptake (which were never seen in the presence of dextran alone) presumably resulted from fusion of many SSL-containing vesicles, and formation of these vesicles may have been driven by intramolecular interactions between SSL molecules (as observed during dimerization in crystal structures). Similar vesicle distortion was observed in the presence of excess invariant chain, again driven by multiple interactions between invariant chain molecules (36).
The third characteristic of SSLs observed in these cellular studies with PBMCs was the highly selective nature of the target population with which an interaction could be detected. In ex vivo PBMCs, the major target population is the monocyte population, which is characterized by high expression of CD14. Essentially all monocytes were found to interact with both SSL7 and SSL9. In contrast, neither SSL7 nor SSL9 interacted with T cells, as identified by expression of CD3 and high levels of CD2. Interestingly, SSL7, but not SSL9, also bound to a portion of the B cells (on the order of 30%, although the percentages for different individuals were significantly different), providing further evidence that the receptors for these two molecules are distinct. Experiments are in progress to further characterize the B-cell subpopulation which interacts with SSL7. Since a very significant proportion of T cells and all human B cells also express class II MHC proteins (e.g., HLA-DR), this result rules out direct binding of SSL7 or SSL9 to these molecules, thus clearly distinguishing them from classical superantigens.
Monocytes express both class I and class II MHC molecules and can act as antigen-presenting cells for the activation of CD4 or CD8 T cells. However, the prototype antigen-presenting cell, and the only cell type which can activate naïve T cells, is the dendritic cell. It was therefore interesting that both SSL7 and SSL9 were taken up efficiently by monocyte-derived dendritic cells. This cell type, which can be obtained by culturing peripheral blood monocytes in the presence of appropriate cytokines, provides a widely used model for myeloid dendritic cells. In contrast, neither SSL7 nor SSL9 showed any tropism for macrophages, a cell type which is also produced by in vitro culture of monocytes but which does not present antigen except when it is stimulated by appropriate cytokines, such as gamma interferon.
The functional consequence of SSL interaction with antigen-presenting cells remains to be determined. One possible model is that SSL7 and SSL9 induce alterations in the functional properties of these cells, preventing their normal function and thus moderating the response of the host to the bacterium. Studies in progress, however, suggest that SSLs are not toxic to antigen-presenting cells, and antigen-presenting cell activity is not adversely affected by the presence of SSLs. An alternative model, however, is that self-targeting to antigen-presenting cells may enhance the immunogenicity of these proteins. The uptake of SSL7 and SSL9 into an endosomal compartment which intersects with the dextran uptake pathway is certainly compatible with such a role, since uptake via the mannose receptor efficiently targets antigens to the class II MHC antigen-processing pathway (38). Although enhancing immunogenicity at first sight appears to be paradoxical, the generation of an antibody response to a secreted protein does not necessarily confer any advantage in bacterial clearance by the host. On the contrary, the interaction between secreted toxin and specific antibody in the microenvironment of the bacteria may activate complement and hence contribute to the breakdown of the physical barriers which restrict the invasiveness of the bacteria. Like classical superantigens, therefore, SSLs may provide S. aureus with an alternative molecular strategy with which to distract the protective adaptive immune response of the host and thus may contribute to bacterial pathogenicity.
Acknowledgments
Ali M. Al-Shangiti was supported by a studentship from the Ministry of Health, Kingdom of Saudi Arabia, Claire E. Naylor was supported by a Dorothy Hodgkin Research Fellowship from the Royal Society, and David C. Briggs was supported by a studentship from the BBSRC.
We thank Ajit K. Basak for help with data collection, Jane Skok for help with confocal microscopy, and David S. Moss for useful discussions.
Editor: J. B. Bliska
REFERENCES
- 1.Alderman, C. J., P. R. Bunyard, B. M. Chain, J. C. Foreman, D. S. Leake, and D. R. Katz. 2002. Effects of oxidised low density lipoprotein on dendritic cells: a possible immunoregulatory component of the atherogenic micro-environment? Cardiovasc. Res. 55:806-819. [DOI] [PubMed] [Google Scholar]
- 2.Arcus, V. L., R. Langley, T. Proft, J. D. Fraser, and E. N. Baker. 2002. The three-dimensional structure of a superantigen-like protein, SET3, from a pathogenicity island of the Staphylococcus aureus genome. J. Biol. Chem. 277:32274-32281. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 4.Bacon, D. J., and W. F. Anderson. 1988. A fast algorithm for rendering space-filling molecule pictures. J. Mol. Graph. 6:219-220. [Google Scholar]
- 5.Bohach, G. A., M. M. Dinges, D. Mitchell, D. Chlendere, and P. M. Schlievert. 1997. Exotoxins, p. 83-111. In K. Grossley (ed.),The staphylococci in human disease. Churchill Livingstone Inc., New York, N.Y.
- 6.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. Delano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 7.Chothia, C., and A. Lesk. 1986. The relation between the divergence of sequence and structure in proteins. EMBO J. 5:823-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collaborative Computational Project, no. 4. 1994. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 9.Cowtan, K. 1994. ‘dm’: an automated procedure for phase improvement by density modification. CCP4 Newsl. Protein Crystallogr. 1994.
- 10.Crawford, K., D. Gabuzda, V. Pantazopoulos, J. Xu, C. Clement, E. Reinherz, and C. A. Alper. 1999. Circulating CD2+ monocytes are dendritic cells. J. Immunol. 163:5920-5928. [PubMed] [Google Scholar]
- 11.Dinges, M. M., P. M. Orwin, and P. M. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13: 16-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esnouf, R. 1999. Further additions to Molscript version1.4, including reading and contouring of electron-density maps. Acta Crystallogr. Sect. D Biol. Crystallogr. 55:938-940. [DOI] [PubMed] [Google Scholar]
- 13.Evans, P. 1997. Scaling of MAD data, p. 97-102. In K. S. Wilson, G. Davies, and A. W. Ashton (ed.), Recent advances in phasing: Proceedings of the CCP4 Study Weekend. CCLRC, Daresbury Laboratory, Daresbury, United Kingdom.
- 14.Fitzgerald, J. R., S. D. Reid, E. Ruotsalainen, T. J. Tripp, M. Liu, R. Cole, P. Kuusela, P. M. Schlievert, A. Jarvinen, and J. M. Musser. 2003. Genome diversification in Staphylococcus aureus: molecular evolution of a highly variable chromosomal region encoding the staphylococcal exotoxin-like family of proteins. Infect. Immun. 71:2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garmen, E., and T. Schneider. 1997. Macromolecular cryocrystallography. J. Appl. Crystallogr. 30:211-237. [Google Scholar]
- 16.Herman, A., J. W. Kappler, P. Marrack, and A. M. Pullen. 1991. Superantigens: mechanism of T-cell stimulation and role in immune responses. Annu. Rev. Immunol. 9:745-772. [DOI] [PubMed] [Google Scholar]
- 17.Jardetzky, T., J. Brown, J. Gorga, L. Stern, R. Urban, Y. Chi, C. Stauffacher, J. Strominger, and D. Wiley. 1994. Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen. Nature 368:711-718. [DOI] [PubMed] [Google Scholar]
- 18.Jones, S., and J. M. Thornton. 1995. Protein-protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol. 63:31-65. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T., and M. Kjelgaard. 1997. Electron density map interpretation. Methods Enzymol. 277:173-208. [DOI] [PubMed] [Google Scholar]
- 20.Kawabe, Y., and A. Ochi. 1990. Selective anergy of V beta 8+,CD4+ T cells in Staphylococcus enterotoxin B-primed mice. J. Exp. Med. 172:1065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J., R. Urban, J. Strominger, and D. Wiley. 1994. Toxic shock syndrome toxin-1 complexed with a class II major histocompatibility molecule HLA-DR1. Science 266:1870-1874. [DOI] [PubMed] [Google Scholar]
- 22.Kotb, M. 1995. Bacterial pyrogenic exotoxins as superantigens. Clin. Microbiol. Rev. 8:411-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraulis, P. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 24.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 25.Labrecque, N., J. Thibodeau, and R. P. Sekaly. 1993. Interactions between staphylococcal superantigens and MHC class II molecules. Semin. Immunol. 5:23-32. [DOI] [PubMed] [Google Scholar]
- 26.Lawkowski, R., M. MacArthur, D. Moss, and J. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 27.Leslie, A. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 ESF-EAMCB Newsl. Protein Crystallogr. 1992.
- 27a.Lina, G., G. A. Bohach, S. P. Nair, K. Hiramatsa, E. Jouvin-Marche, and R. Mariurza. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis., in press. [DOI] [PubMed]
- 28.Lo Conte, L., C. Chothia, and J. Janin. 1999. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 285:2177-2198. [DOI] [PubMed] [Google Scholar]
- 29.Marrack, P., and J. Kappler. 1990. The staphylococcal enterotoxins and their relatives. Science 248:1066. [PubMed] [Google Scholar]
- 30.McRee, D. E. 1993. Practical protein crystallography. Academic Press, San Diego, Calif.
- 31.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 32.Nair, S. P., R. J. Williams, and B. Henderson. 2000. Advances in our understanding of the bone and joint pathology caused by Staphylococcus aureus infection. Rheumatology (Oxford) 39:821-834. [DOI] [PubMed] [Google Scholar]
- 33.Newton, P. J., I. V. Weller, D. R. Katz, and B. M. Chain. 2003. Autologous apoptotic T cells interact with dendritic cells, but do not affect their surface phenotype or their ability to induce recall immune responses. Clin. Exp. Immunol. 133:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orwin, P. M., J. R. Fitzgerald, D. Y. Leung, J. A. Gutierrez, G. A. Bohach, and P. M. Schlievert. 2003. Characterization of Staphylococcus aureus enterotoxin L. Infect. Immun. 71:2916-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orwin, P. M., D. Y. Leung, H. L. Donahue, R. P. Novick, and P. M. Schlievert. 2001. Biochemical and biological properties of staphylococcal enterotoxin K. Infect. Immun. 69:360-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romagnoli, P., C. Layet, J. Yewdell, O. Bakke, and R. N. Germain. 1993. Relationship between invariant chain expression and major histocompatibility complex class II transport into early and late endocytic compartments. J. Exp. Med. 177:583-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roussel, A., B. Anderson, H. Baker, J. Fraser, and E. Baker. 1997. Crystal structure of the streptococcal superantigen SPE-C: dimerization and zinc binding suggest a novel mode of interaction with MHC class II molecules. Nat. Struct. Biol. 4:635-643. [DOI] [PubMed] [Google Scholar]
- 38.Sallusto, F., M. Cella, C. Danieli, and A. Lanzavecchia. 1995. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J. Exp. Med. 182:389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sallusto, F., and A. Lanzavecchia. 1994. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 179:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundstrom, M., L. Abrahmsen, P. Antonsson, K. Mehindate, W. Mourad, and M. Dohlsten. 1996. The crystal structure of staphylococcal enterotoxin type D reveals Zn2+-mediated homodimerization. EMBO J. 15:6832-6840. [PMC free article] [PubMed] [Google Scholar]
- 41.Swetman, C. A., Y. Leverrier, R. Garg, C. H. Gan, A. J. Ridley, D. R. Katz, and B. M. Chain. 2002. Extension, retraction and contraction in the formation of a dendritic cell dendrite: distinct roles for Rho GTPases. Eur. J. Immunol. 32:2074-2083. [DOI] [PubMed] [Google Scholar]
- 42.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022-1025. [Google Scholar]
- 43.Williams, R. J., B. Henderson, L. J. Sharp, and S. P. Nair. 2002. Identification of a fibronectin-binding protein from Staphylococcus epidermidis. Infect. Immun. 70:6805-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams, R. J., J. M. Ward, B. Henderson, S. Poole, B. P. O'Hara, M. Wilson, and S. P. Nair. 2000. Identification of a novel gene cluster encoding staphylococcal exotoxin-like proteins: characterization of the prototypic gene and its protein product, SET1. Infect. Immun. 68:4407-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]