Abstract
Porphyromonas gingivalis, one of the causative agents of adult periodontitis, can invade and survive within host epithelial cells. The molecular mechanisms by which P. gingivalis induces uptake and adapts to an intracellular environment are not fully understood. In this study, we have investigated the genetic responses of P. gingivalis internalized within human gingival epithelial cells (GECs) in order to identify factors involved in invasion and survival. We compared the differential display of arbitrarily PCR-amplified gene transcripts in P. gingivalis recovered from GECs with the display of transcripts in P. gingivalis control cultures. Over 20 potential differentially expressed transcripts were identified. Among these, pepO, encoding an endopeptidase, and genes encoding an ATP-binding cassette (ABC) transporter and a cation-transporting ATPase were upregulated in GECs. To investigate the functionality of these gene products, mutants were generated by insertional inactivation. Compared to the parental strain, mutants of each gene showed a significant reduction in their invasion capabilities. In addition, GEC cytoskeletal responses to the mutants were distinct from those induced by the parent. In contrast, adhesion of the mutant strains to GECs was not affected by lack of expression of the gene products. These results suggest that PepO, a cation-transporting ATPase, and an ABC transporter are required for the intracellular lifestyle of P. gingivalis.
Porphyromonas gingivalis, a gram-negative anaerobe, is a major pathogen in severe adult periodontitis. Similar to many other pathogens, P. gingivalis has the ability to invade various host cells, including gingival epithelial cells (GECs) (20), multilayered pocket epithelial cells (30), transformed epithelial cells (14), and endothelial cells (12, 13). The invasion of P. gingivalis into GECs is complete within 20 min, and intracellularly the organism accumulates in the perinuclear region (7). P. gingivalis remains viable and capable of replication inside of GECs. Infected GECs do not undergo apoptosis and maintain physiological integrity for extended periods (7, 19, 25). Thus, both bacteria and host cells adapt to their association. The major fimbriae of P. gingivalis are involved both in adhesion to epithelial cells and in the consequent signal transduction associated with invasion (27, 36). Fimbriae mediate adhesion to GECs, and stimulate subsequent invasion, through engagement of an integrin receptor (24, 37). Downstream signaling events include recruitment and activation of paxillin and focal adhesion kinase (38), transient elevation of Ca2+ ion levels (16), phosphorylation of c-jun N-terminal kinase, and down-regulation of extracellular signal-regulated kinase (35). This results in remodeling of the actin cytoskeleton (38) and ultimately impacts transcriptional activity, including genes encoding interleukin-8 and Bcl-2 (11, 25).
Expression of many bacterial genes, including genes for virulence determinants and invasion effectors, can be regulated in response to environmental conditions. Therefore, in order to identify genes that may be involved in the P. gingivalis invasion process, we sought to detect genes that are differentially regulated within epithelial cells. A number of molecular techniques have been developed that can identify differentially expressed genes in intracellular bacteria, including subtractive hybridization (29), in vivo expression technology (34), and differential display reverse transcription-PCR (DDRT-PCR) (1, 2, 8). DDRT-PCR has proven successful in the identification of a locus in Legionella pneumophila that is transiently induced during the first few hours of the intracellular infection (1, 2) and in the detection of Vibrio cholerae genes differentially expressed in an animal host (8). In this study, we adapted DDRT-PCR to display P. gingivalis genes differentially expressed during prolonged incubation with GECs. Such genes can be theorized to be important in the overall invasive process that spans adhesion, internalization, and intracellular survival. By disrupting these genes in P. gingivalis, we were able to reveal novel factors required for P. gingivalis internalization within GECs, including an endopeptidase, a cation-transporting ATPase, and an ATP-binding cassette (ABC) transporter.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis 33277 and derivatives were grown anaerobically at 37°C in Trypticase soy broth (TSB) supplemented per liter with 1 g of yeast extract, 5 mg of hemin, and 1 mg of menadione. When necessary, gentamicin and erythromycin were added to the medium at final concentrations of 200 and 10 μg/ml, respectively. Solid medium was prepared by supplementation with 5% sheep blood and 1.5% agar. Escherichia coli strains were grown in Luria-Bertani broth containing the necessary antibiotics (100 μg of ampicillin/ml or 200 μg of trimethoprim/ml).
Epithelial cell culture.
Primary low-passage-number cultures of GECs were prepared from gingival explants and cultured as monolayers in serum-free keratinocyte growth medium (Cambrex Bio Science) as described previously (19). Cells were used for experimentation during passage 4 when at 70 to 80% confluence.
RNA isolation from P. gingivalis cultivated in vitro or recovered from GECs.
Total RNA was isolated from P. gingivalis 33277 cultivated in TSB or recovered from GECs using a Totally RNA kit (Ambion) as described by the manufacturer. For intracellular P. gingivalis RNA, bacteria were reacted with GECs for 18 h, and the GECs were washed four times to remove external organisms. Fluorescence microscopy (7) has demonstrated that the vast majority of P. gingivalis bacteria are internalized under these conditions. Infected GECs were lysed with ice-cold deionized water, and P. gingivalis was harvested and treated with DNase I and RNase A in phosphate-buffered saline (PBS) supplemented with 1 mM MgCl2 to remove epithelial cell DNA and RNA prior to total RNA isolation.
DDRT-PCR.
DD-RTPCR was performed as described previously (10). Briefly, cDNA was synthesized from RNA isolated from P. gingivalis cells in culture medium or in GECs with Moloney murine leukemia virus reverse transcriptase (RETROscript; Ambion) using random hexamer primers. PCR was performed under conditions of 1 min of denaturation at 94°C, low-stringency annealing at 30°C for 2 min followed by 1 min of extension for 40 to 50 cycles, and a final extension at 72°C for 10 min. Primers 10 to 20 nucleotides in length were randomly selected from those used in our laboratory for other purposes, as the genome sequence of P. gingivalis was not available at the start of the study.
DDRT-PCR products were separated by agarose gel electrophoresis, and products specific to cDNA from intracellular P. gingivalis were eluted from the gel (QIAquick gel extraction kit; QIAGEN, Inc.) and cloned into pCRII-TOPO (Invitrogen). DNA sequences of the cloned fragments were determined on an ABI 377 DNA analyzer and were analyzed by BLAST searching the TIGR database for P. gingivalis (TIGR Comprehensive Microbial Resource; http://tigrblast.tigr.org/cmr-BLAST/).
RT-PCR.
Differential expression of a subset of the cloned genes was confirmed by RT-PCR using primers designed based on the DNA sequences of the genes in the TIGR database. The primers were the following: for 16S rRNA, 5′-TGGGTTTAAAGGGTGCGTAG-3′ and 5′-ACAACCATGCAGCACCTACA-3′; for luxS, 5′-CAGCACTGTGCTTCTCCAA-3′ and 5′-GAGGAGCAGGACTTTGTTCG-3′; for PG0147, 5′-ATAGAACCCTCGGGCGATAC-3′ and 5′-ACCTGCTTCACCAAGTCGTC-3′; for PG0159, 5′-GCTACCCGTATTGCCAAGAA-3′ and 5′-TTGTCGTCATGCATCCATTT-3′; for PG0165, 5′-GAGGTAAGGATAGACCGTTGGA-3′ and 5′-TGCGTTGCATTTCGAGTATC-3′; for PG1642, 5′-TCCGTAGAAGGCATGATGTG-3′ and 5′-CCGAAGCATCGAAGTAGAGG-3′; for PG2206, 5′-TTCGGCAAGCGAATACTTTT-3′ and 5′-GCTGGATTCGTACCAGAAGC-3′. cDNA was synthesized from the same amount of total RNA isolated under two different conditions as described above. PCR was performed under conditions of 1 min of denaturation at 94°C and annealing at 55°C for 1 min, followed by 1 min of extension at 72°C for 30 cycles.
Construction of mutants.
Insertional mutants for the cloned genes were constructed as described previously (10). Briefly, internal fragments of each gene were amplified by PCR with primers designed based on the DNA sequences in the TIGR database and cloned into pCRII-TOPO. The cloned fragments were obtained by digestion with BamHI and XbaI and elution from an agarose gel after electrophoresis. Each BamHI-XbaI fragment was cloned into suicide plasmid pVA3000 (21), carrying the erythromycin resistance gene cassette ermAM/ermF. These recombinant plasmids were introduced into E. coli S17-1 for mating with P. gingivalis. Conjugation between E. coli and P. gingivalis was performed as described previously (28). Transconjugants were selected on TSB-blood agar plates supplemented with erythromycin (20 μg/ml) and gentamicin (100 μg/ml).
Immunofluorescence microscopy.
Immunofluorescence labeling and microscopy were performed as described by Yilmaz et al. (38). Briefly, GECs cultivated on four-well chambered coverglass slides (Nalge-Nunc International Corp.) were infected with P. gingivalis strains at a multiplicity of infection (MOI) of 100 at 37°C for 30 min. The slides were washed four times with PBS containing 0.1% Tween 20 (PBS-T) to remove the nonadherent bacteria. Cells were fixed in 10% neutral buffered formalin, rinsed in PBS, and permeabilized for 15 min with 0.1% Triton X-100 in PBS at room temperature. Samples were incubated for 30 min in blocking solution (PBS-T supplemented with 5% goat serum) to mask nonspecific binding sites prior to the fluorescence labeling.
Visualization of intracellular bacteria was performed by indirect immunofluorescence microscopy using rabbit polyclonal antiserum to P. gingivalis 33277 and fluorescein isothiocyanate-conjugated Affini-pure F(ab′)2 fragment goat anti-rabbit immunoglobulin G (H+L; Jackson ImmunoResearch Lab.). F-actin was also labeled with phalloidin-tetramethylrhodamine B-isothiocyanate (Sigma). The labeled samples were examined using an epifluorescence microscope (Zeiss Axioskope). Single exposure images were captured sequentially using a cooled charge-coupled device camera (Qimaging) and saved by Qcapture software version 1394. Collected image layers were superimposed into a single image using Adobe Photoshop 6.0 software. At least 10 separate fields containing an average of 25 GECs were studied for each strain in triplicate assays.
Invasion assay.
Quantitation of viable P. gingivalis cells recovered intracellularly from GEC was determined by a conventional antibiotic protection assay, as modified for P. gingivalis (19). P. gingivalis cells were incubated with GECs at an MOI of 100 for 2 h.
Adherence assay.
Adherence of P. gingivalis strains to GECs was detected with an enzyme-linked immunosorbent assay (ELISA). GECs cultivated on 96-well plates were first fixed with 5% buffered formalin to inhibit P. gingivalis invasion. Viable P. gingivalis cells at an MOI of 100 were reacted with the fixed GECs for 30 min at 37°C and then washed five times with PBS to removed nonadherent bacteria. Samples were immunolabeled using the same protocol as described above for immunofluorescence labeling, except horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (ICN Biochemicals) was used as the secondary antibody. Adherence of each strain was determined by a colorimetric reaction using 3,3′,5,5′-tetramethylbenzidine liquid substrate system for ELISA (Sigma). Wells with uninfected GECs were used as a negative control. Preliminary experiments showed that the mutant and parent strains exhibited an equivalent degree of reactivity with the 33277 antibodies.
RESULTS
DDRT-PCR.
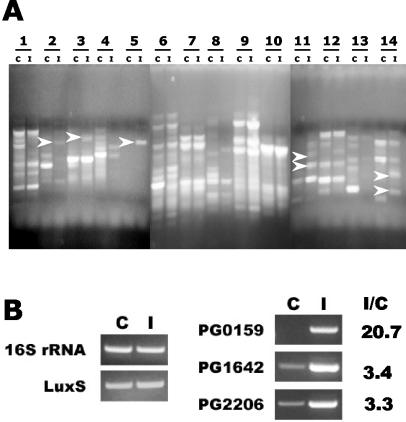
Figure 1A shows DDRT-PCR products from in vitro-cultivated and intracellular P. gingivalis (lanes C and I, respectively). The number of displayed bands was dependent on the primers used. However, independent of the number of bands, patterns from control and intracellular organisms were comparable, indicating that there was no major contamination with GEC RNA during intracellular bacterial RNA preparation. Distinct differences, however, could be observed between control and intracellular bacteria with several primers used (Fig. 1A). Over 20 bands representing potential genes upregulated in intracellular P. gingivalis were cloned and sequenced. The identifications of the genes based on BLAST analysis of the P. gingivalis genome database are listed in Table 1. PG0147 (heat shock protein gene, hslR) and PG2206 (ABC transporter gene) were isolated on two separate occasions with different arbitrary primers. In addition, in two clones the cloned fragments contained the overlapping region of two different coding regions (PG0147 and PG0148, and PG0155 and PG0156), as shown in Table 1. One contained the region encoding the 5′ end of PG0147 and 3′ end of PG0148, suggesting cotranscription of two genes. The other contained the overlapping 5′ ends of PG0155 and PG0156. Based on potential relevant functionality, PG0159, PG0165, PG1642, and PG2206 were selected for further studies. PG0147, encoding a hypothetical protein, was also selected, since it was identified from two different clones.
FIG. 1.
(A) Examples of DDRT-PCR products of in vitro-cultivated (C lanes) and intracellular (I lanes) P. gingivalis. Numbers indicate the pairs of different arbitrary primers used for PCR. Arrows indicate examples of differentially displayed bands eluted from the gel for cloning. (B) Confirmation by RT-PCR of differential expression of the genes for which mutations were generated. C lanes are RT-PCR products with RNA from in vitro-cultivated P. gingivalis, I lanes are the products with RNA from intracellular P. gingivalis. 16S rRNA and luxS are controls for total mRNA levels. The ratio of intracellular induction (I/C) was calculated from the relative intensity of amplification products normalized to levels of luxS mRNA, using NIH Image software.
TABLE 1.
List of open reading frames of P. gingivalis which were identified by DDRT-PCR as upregulated within GECs
| Locusa | Putative identification | Gene | Cellular role(s) |
|---|---|---|---|
| PG0130 | Phosphoglycerate mutase | pgm | Energy metabolism, glycolysis-gluconeogenesis |
| PG0147b,c | Hypothetical protein | ||
| PG0148c | Sigma 54-dependent transcriptional regulator | Regulatory functions, DNA interactions | |
| PG0155d | Riboflavin biosynthesis protein RibD | ribD | Biosynthesis of cofactors, prosthetic groups, and carriers: riboflavin, FMN, and FAD |
| PG0156d | Modification methylase, HemK family | Unknown function, general | |
| PG0159 | Endopeptidase PepO | pepO | Protein fate, degradation of proteins, peptides, and glycopeptides |
| PG0165 | Heat shock protein 15 | hslR | Protein synthesis, other |
| PG0212 | Precorrin-6y c5, 15-methyltransferase, putative | Biosynthesis of cofactors, prosthetic groups, and carriers: heme, porphyrin, and cobalamin | |
| PG0326 | Hypothetical protein | ||
| PG0338 | Hypothetical protein | ||
| PG0481 | 8-amino-7-oxononanoate synthase | bioF-1 | Biosynthesis of cofactors, prosthetic groups, and carriers: biotin |
| PG0525 | CTP synthase | pyrG | Purines, pyrimidines, nucleosides, and nucleotides: pyrimidine ribonucleotide biosynthesis |
| PG0534 | Hypothetical protein | ||
| PG1592 | HDIG domain protein | Energy metabolism: electron transport; hypothetical protein: domain | |
| PG1638 | Thioredoxin family protein | Energy metabolism, electron transport | |
| PG1642 | Cation-transporting ATPase, EI-E2 family, authentic frameshift | Transport and binding proteins: cations | |
| PG1683c | Conserved hypothetical protein | ||
| PG1684c | Hypothetical protein | ||
| PG1750 | Alpha-1,3/4-fucosidase, putative | Cell envelope, biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | |
| PG1780 | 8-amino-7-oxononanoate synthase | bioF-3 | Biosynthesis of cofactors, prosthetic groups, and carriers: biotin |
| PG2080 | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase | bioA | Biosynthesis of cofactors, prosthetic groups, and carriers: biotin |
| PG2165 | Glycyl-tRNA synthetase | glyS | Protein synthesis, tRNA aminoacylation |
| PG2205 | 2-Dehydropantoate 2-reductase, putative | Biosynthesis of cofactors, prosthetic groups, and carriers: pantothenate and coenzyme A | |
| PG2206b | ABC transporter, ATP-binding protein | Transport and binding proteins, unknown substrate | |
| PG2227 | Hypothetical protein |
TIGR locus designation.
Identified from more than one clone.
Two loci with the consecutive numbers were identified from the same clone. The cloned fragments contained the 5′ end of one and 3′ end of the other.
Two loci were identified from one clone. The cloned fragment contained 5′ ends of the two loci.
Verification of differentially expressed genes.
Differential expression of the selected genes after 18 h of association with GECs was confirmed by RT-PCR using primers for each gene based on DNA sequences of the genes from the TIGR CMR database (Fig. 1B) While no reliable housekeeping gene has been identified in P. gingivalis to use as a control for mRNA loading, no difference was detected in the level of expression of luxS, which is not involved in invasion (10), or 16S rRNA. Quantitation by densitometry and NIH Image software showed that expression of pg0159 was induced over 20-fold by GEC association, while pg1642 and pg2206 were induced over 3-fold. Equivalent results were obtained when RT-PCR was performed after 2 h of association with the GECs (data not shown).
Mutant construction.
In order to investigate the role of the selected subset of the differentially regulated genes in the invasive process of P. gingivalis in GECs, we attempted to construct mutant strains. Insertional mutagenesis for PG0159, PG1642, and PG2206 was confirmed by Southern hybridization with the chromosomal DNA of the transconjugants by using the ermAM/ermF cassette and the original cloned inserts as probes. All the transconjugants tested showed that the plasmids integrated into the proper location and disrupted the genes of interest (data not shown). Strains with a mutation in PG0159, PG1642, and PG2206 were designated as P. gingivalis YPEP, YATP, and YABC, respectively. However, mutants for PG0147 and PG0165 (hslR) could not be constructed despite several attempts, indicating that these gene products may play an essential role for survival of P. gingivalis.
Invasion and adherence characteristics of the YPEP, YATP, and YABC mutants.
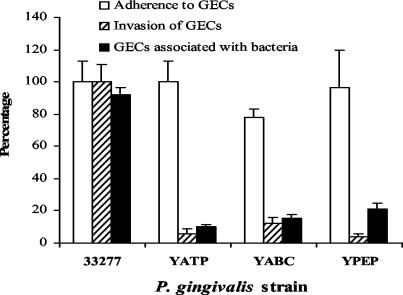
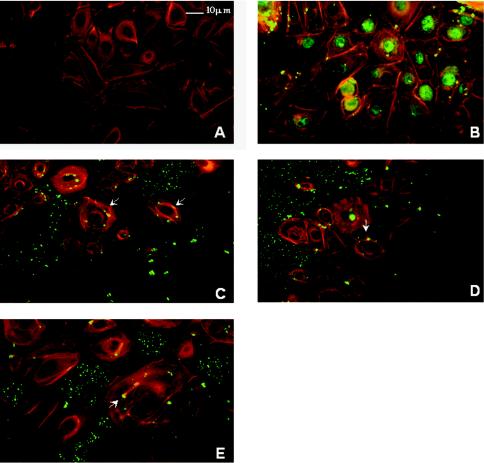
Genes differentially regulated within GECs after 18 h of incubation could potentially be involved in any stage of the invasive process, including adhesion, internalization, and intracellular survival and replication. To begin to sort through these possibilities, we examined the invasion characteristics of the mutant strains (Fig. 2). In a conventional antibiotic protection assay, which measures recovery of viable intracellular bacteria, the three mutants YABC, YATP, and YPEP demonstrated a reduction in invasion compared to the parent strain. Adhesion of the mutants to GECs was then analyzed and compared with the parental strain by ELISA. No significant difference between the parent strain and the mutants could be detected in adherence to GECs. Thus, disruption of the target genes did not affect binding to GECs, indicating that the lower numbers of intracellular bacteria are the result of a defect either in the internalization process or in intracellular survival. Immunofluorescence microscopy was used to examine the ability of the mutants to internalize within the GECs. As shown in Fig. 2, after 30 min of exposure less than 20% of GECs contained mutant P. gingivalis cells compared to over 90% of GECs with intracellular parental 33277 cells. Immunofluorescence microscopy also demonstrated that the parental strain 33277 internalized within GECs in high numbers and accumulated in the perinuclear area (Fig. 3B), consistent with our group's previous observations (38). In contrast, mutants YABC (Fig. 3C), YATP (Fig. 3D), and YPEP (Fig. 3E) showed a dramatic reduction in numbers of internalized bacteria. Furthermore, Fig. 3 also shows that the mutants failed to induce formation of actin stress fibers to the same degree as the parent strain. However, the mutant organisms triggered distinct cortical actin filament accumulations and actin cup formations in the GEC cytoskeleton (Fig. 3C, D, and E). There was also some degree of indistinct microvillus formation of the actin cytoskeleton in all mutant treated cells. Thus, disruption of the differentially regulated genes impairs the ability of P. gingivalis to initially localize within GECs, and this defect is associated with altered cytoskeletal responses to the organism.
FIG. 2.
Adherence and invasion properties of P. gingivalis mutant strains YABC, YATP, and YPEP relative to properties of strain 33277. Adherence of P. gingivalis to formalin-fixed GECs was determined by using an ELISA. Invasion and recovery of P. gingivalis cells after infection of GECs for 2 h were determined in an antibiotic protection assay. The percentage of GECs containing P. gingivalis after infection of GECs for 30 min was determined by fluorescence microscopy. At least 10 separate fields containing an average of 25 GECs were studied for each strain in triplicate assays.
FIG. 3.
Invasion of P. gingivalis mutants into GECs and reorganization of GEC actin filaments by P. gingivalis mutants. GECs were double labeled with phalloidin (shown in red) and P. gingivalis polyclonal antibodies (shown in green). Stained cells were viewed by indirect fluorescence microscopy. (A) Uninfected GECs with phalloidin and P. gingivalis antibodies were used as a control. (B to E) GECs were infected for 30 min with P. gingivalis 33277 (B), YABC (C), YATP (D), or YPEP (E). Arrows indicate mutant-specific actin rearrangements.
DISCUSSION
P. gingivalis, a major periodontal pathogen, is able to invade epithelial cells derived from the human gingiva in a short period of time and in high numbers. Internalized P. gingivalis bacteria inhibit GEC apoptosis and multiply and persist with the host cells (19, 22, 25). However, information on invasion- and survival-related factors of P. gingivalis and its molecular responses to the intracellular environment is very limited. To date, only the major fimbriae (FimA) and cysteine proteinases (gingipains) have been demonstrated to be involved in attachment to, and invasion of, oral epithelial cells (9, 27, 33, 36, 37). In this study, we adapted DDRT-PCR to identify a subset of genes that are upregulated within GECs and can thus be theorized to be important in the overall invasion process of P. gingivalis. DDRT-PCR has several advantages for such an approach, including the use of small amounts of RNA and the lack of a requirement for specifically designed genetic tools, which are very limited for P. gingivalis. In addition, the recent completion of the genome sequence of P. gingivalis (26) has made possible the rapid identification of nucleotide sequences by BLAST searching. Disadvantages to DDRT-PCR include a high false-positive rate and uncertainty regarding the degree of genome coverage. In the future, microarrays based on the genome sequence are likely to be the method of choice for similar gene expression studies.
A total of 25 different coding regions of P. gingivalis were identified by DDRT-PCR (Table 1). These include the genes whose products are involved in energy metabolism, biosynthesis of cofactors, prosthetic groups, and carriers, biosynthesis of cell envelope, and transport, as well as seven genes encoding hypothetical proteins. The high proportion of regulated genes that are involved in metabolism indicates that the physiological adaptation of P. gingivalis to an intracellular environment involves broadly based regulation of metabolic pathways at the transcriptional level.
On the basis of potential functionality, five different genes, PG0147, PG0159, PG0165, PG1643, and PG2206, were selected for mutational analysis. Among these five, we were able to construct mutants for PG0159 (endopeptidase PepO), PG1642 (cation-transporting ATPase), and PG2206 (ABC transporter). To assess the involvement of the identified genes in the invasion process, the invasion capability of the mutants was tested by an antibiotic protection-based intracellular survival assay. All of the mutants, YABC, YATP, and YPEP, demonstrated a significant reduction in recovery of intracellular organisms. This defect did not appear to be the result of diminished binding to the GECs, as the adherence properties of the mutants were similar to those of the parent. In contrast, immunofluorescence microscopy revealed that the mutants were significantly impaired in their ability to internalize within GECs both in terms of numbers of internal bacteria and numbers of GECs containing bacteria. Thus, genes encoding PepO, a cation-transporting ATPase, and an ABC transporter are required for optimal entry of P. gingivalis into GECs. The aberrant GEC actin rearrangements observed with the mutants also supports the concept that the gene products are involved in the mechanism of P. gingivalis internalization.
P. gingivalis PepO, a metalloendopeptidase, has significant homology with human endothelin-converting enzyme 1, which converts big endothelin-1 to endothelin-1 (4). Since elevation of endothelin-1 levels has been implicated in the pathogenesis of atherosclerosis and heart failure, the presence of PepO in P. gingivalis could have relevance for the pathogenesis of atherosclerosis should P. gingivalis be involved with this disease, as has been suggested (5, 6). The present study also implicates a role for PepO in invasion of P. gingivalis into host epithelial cells. These data corroborate and extend the observations of Ansai et al. (3), who reported that a PepO mutant of P. gingivalis is deficient in invasion of HeLa cells. The cation-transporting ATPase is homologous to proteins involved in transport of cations across bacterial membranes. In addition, in Proteus mirabilis this protein can control expression of the flagellin filament gene fliC, the flagellar master operon flhDC, and a leucine-responsive regulatory protein gene, lrp (18). The involvement of the cation-transporting ATPase in invasion may, therefore, be dependent on indirect effects on expression of other genes. Work in our laboratory to investigate the genes regulated by the cation-transporting ATPase is under way. ABC transporters are associated with many physiological processes, including the uptake of nutrients, the nonclassical secretion of signaling molecules and toxins, multidrug resistance, and the development of human disease. A role for ABC transporters in cell-to-surface and/or cell-to-cell interactions and biofilm development has been proposed in different organisms (15, 17, 23, 31). ABC transporters constitute one of the largest and most highly conserved superfamilies and are found in large numbers in all organisms (32). A total of 39 different ABC transporter genes have been identified in the genome of P. gingivalis W83. In this study, the ABC transporter gene PG2206 was identified as being up-regulated inside of GECs when different arbitrary primers were used for DDRT-PCR, suggesting the importance of the intracellular expression of the gene. The function of the gene product, as well as that of the substrate, remains to be determined.
In conclusion, DDRT-PCR identified several P. gingivalis genes specifically expressed in GECs. The products of three genes, encoding endopeptidase O, a cation-dependent ATPase, and an ABC transporter, have been demonstrated to play important roles in the invasion of P. gingivalis.
Acknowledgments
This work was supported by grants DE11111 and DE14168 from the National Institute of Dental and Craniofacial Research.
Editor: V. J. DiRita
REFERENCES
- 1.Abu Kwaik, Y. 1998. Induced expression of the Legionella pneumophila gene encoding a 20-kilodalton protein during intracellular infection. Infect. Immun. 66:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y., and L. L. Pederson. 1996. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol. Microbiol. 21:543-556. [DOI] [PubMed] [Google Scholar]
- 3.Ansai, T., W. Yu, S. Urnowey, S. Barik, and T. Taehara. 2003. Construction of a pepO gene-deficient mutant of Porphyromonas gingivalis: potential role of endopeptidase O in the invasion of host cells. Oral Microbiol. Immunol. 18:398-400. [DOI] [PubMed] [Google Scholar]
- 4.Awano, S., T. Ansai, H. Mochizuki, W. Yu, K. Tanzawa, A. J. Turner, and T. Takehara. 1999. Sequencing, expression and biochemical characterization of the Porphyromonas gingivalis pepO gene encoding a protein homologous to human endothelin-converting enzyme. FEBS Lett. 460:139-144. [DOI] [PubMed] [Google Scholar]
- 5.Beck, J., R. Garcia, G. Heiss, P. S. Vokonas, and S. Offenbacher. 1996. Periodontal disease and cardiovascular disease. J. Periodontol. 67:1123-1137. [DOI] [PubMed] [Google Scholar]
- 6.Beck, J. D., and S. Offenbacher. 1998. Oral health and systemic disease: periodontitis and cardiovascular disease. J. Dent. Edu. 62:859-870. [PubMed] [Google Scholar]
- 7.Belton, C. M., K. T. Izutsu, P. C. Goodwin, Y. Park, and R. J. Lamont. 1999. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell. Microbiol. 1:215-223. [DOI] [PubMed] [Google Scholar]
- 8.Chakrabortty, A., S. Das, S. Majumadar, K. Mukhopadhyay, S. Roychoudhury, and K. Chaudhuri. 2000. Use of RNA arbitrarily primed-PCR fingerprinting to identify Vibrio cholerae genes differentially expressed in the host following infection. Infect. Immun. 68:3878-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, T., K. Nakayama, L. Belliveau, and M. J. Duncan. 2001. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect. Immun. 69:3048-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, R. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darveau, R. P., C. M. Belton, R. A. Reife, and R. J. Lamont. 1998. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 66:1660-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desphpande, R. G., M. B. Khan, and C. A. Genco. 1998. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect. Immun. 66:5337-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorn, B. R., W. A. Dunn, Jr., and A. Progulske-Fox. 1999. Invasion of human coronary artery cells by periodontal pathogens. Infect. Immun. 67:5792-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan, M. J., S. Nakao, Z. Skobe, and H. Xie. 1993. Interactions of Porphyromonas gingivalis with epithelial cells. Infect. Immun. 61:2260-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905-918. [DOI] [PubMed] [Google Scholar]
- 16.Izutsu, K. T., C. M. Belton, A. Chan, S. Fatherazi, J. P. Kanter, Y. Park, and R. J. Lamont. 1996. Involvement of calcium in interactions between gingival epithelial cells and Porphyromonas gingivalis. FEMS Microbiol. Lett. 144:145-150. [DOI] [PubMed] [Google Scholar]
- 17.Kolenbrander, P. E., R. N. Andersen, and N. Ganeshkumar. 1994. Nucleotide sequence of the Streptococcus gordonii PK488 coaggregation adhesin gene, scaA, and ATP-binding cassette. Infect. Immun. 62:4469-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, H. C., D. Gygi, G. M. Fraser, and C. Hughes. 1998. A swarming-defective mutant of Proteus mirabilis lacking a putative cation-transporting membrane P-type ATPase. Microbiology 144:1957-1961. [DOI] [PubMed] [Google Scholar]
- 19.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamont, R. J., D. Oda, R. E. Persson, and G. R. Persson. 1992. Interaction of Porphyromonas gingivalis with gingival epithelial cells maintained in culture. Oral Microbiol. Immunol. 7:364-367. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S.-W., J. D. Hillman, and A. Progulske-Fox. 1996. The hemagglutinin genes hagB and hagC of Porphyromonas gingivalis are transcribed in vivo as shown by use of a new expression vector. Infect. Immun. 64:4802-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madianos, P. N., P. N. Papapanou, U. Nannmark, G. Dahlen, and J. Sandros. 1996. Porphyromonas gingivalis FDC381 multiplies and persists within human oral epithelial cells in vitro. Infect. Immun. 64:660-664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthysse, A. G., H. A. Yarnall, and N. Young. 1996. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J. Bacteriol. 178:5302-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakagawa, I., A. Amano, M. Kuboniwa, T. Nakamura, S. Kawabata, and S. Hamada. 2002. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect. Immun. 70:277-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakhjiri, S. F., Y. Park, O. Yilmaz, W. O. Chung, K. Watanabe, A. El-Sabaeny, K. Park, and R. J. Lamont. 2001. Inhibition of epithelial cell apoptosis by Porphyromonas gingivalis. FEMS Microbiol. Lett. 200:145-149. [DOI] [PubMed] [Google Scholar]
- 26.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Njoroge, T., R. J. Genco, H. T. Sojar, and C. A. Genco. 1997. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect. Immun. 65:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, Y., and B. C. McBride. 1993. Characterization of the tpr gene product and isolation of a specific protease-deficient mutant of Porphyromonas gingivalis W83. Infect. Immun. 61:4139-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plum, G., and J. E. Clark-Curtiss. 1994. Induction of Mycobcterium avium gene expression following phagocytosis by human macrophages. Infect. Immun. 62:476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandros, J., P. N. Papapanou, U. Nannmark, and G. Dahlen. 1994. Porphyromonas gingivalis invades human pocket epithelium in vitro. J. Periodont. Res. 29:62-69. [DOI] [PubMed] [Google Scholar]
- 31.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt, L., and R. Tampae. 2002. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 12:754-760. [DOI] [PubMed] [Google Scholar]
- 33.Tokuda, M., M. Duncan, M. I. Cho, and H. K. Kuramitsu. 1996. Role of Porphyromonas gingivalis protease activity in colonization of oral surfaces. Infect. Immun. 64:4067-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vriesema, A. J. M., H. Beekhuizen, M. Hamdi, A. Soufan, A. Lammers, B. Willekens, O. Bakker, A. G. A. Welten, M. H. A. M. Veltrop, J. S. van de Gevel, J. Dankert, and S. A. J. Zaat. 2000. Altered gene expression in Staphylococcus aureus upon interaction with human endothelial cells. Infect. Immun. 68:1765-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe, K., O. Yilmaz, S. F. Nakhjiri, C. M. Belton, and R. J. Lamont. 2001. Association of mitogen-activated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect. Immun. 69:6731-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yilmaz, O., K. Watanabe, and R. J. Lamont. 2002. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell. Microbiol. 4:305-314. [DOI] [PubMed] [Google Scholar]
- 38.Yilmaz, O., P. A. Young, R. J. Lamont, and G. E. Kenny. 2003. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology 149:2417-2426. [DOI] [PubMed] [Google Scholar]