Abstract
The molecular immune response of the pulpal tissue during chronic carious infection is poorly characterized. Our objective was to examine the expression of potential molecular mediators of pulpal inflammation, correlate their levels with disease severity, and determine the cellular localization of key molecules. Results indicated that there was significantly increased transcriptional activity in carious compared to healthy pulp, and the increase correlated positively with disease severity. Semiquantitative reverse transcriptase PCR analysis in 10 carious and 10 healthy pulpal tissue samples of the S100 family members S100A8, S100A9, S100A10, S100A12, and S100A13; the cytokines tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), IL-8, IL-6, and epithelial cell-derived neutrophil attractant 78 (ENA-78); and the structural protein collagen-1α indicated that all genes tested, with the exception of S100A10, were more abundantly expressed in carious teeth. In addition, we found that the closer the carious lesion front was to the pulpal chamber the higher the expression was for all genes except S100A10. Multiple-regression analysis identified a significant positive correlation between the expression levels of S100A8 and IL-1β, ENA-78, and IL-6 and between collagen-1α and S100A8, TNF-α, IL-1β, IL-8, IL-6, and ENA-78. Immunohistochemical studies in carious pulpal tissue indicated that S100A8 and the S100A8/S100A9 complex were predominantly expressed by infiltrating neutrophils. Gene expression analyses in immune system cells supported these findings and indicated that bacterial activation of neutrophils caused upregulation of S100A8, S100A9, and S100A13. This study highlights the complex nature of the molecular immune response that occurs during carious infection.
While previous reports have shown that a variety of immune system cells are recruited to the dental pulp to respond to infection arising from caries (9, 13), the molecular mediators of this process are poorly characterized. The microbial populations associated with dental caries are highly complex and variable. The mutans group streptococci and lactobacilli are well documented as being key to caries initiation and development; however, during disease progression reports have associated other bacterial flora, such as anaerobic gram-positive cocci (e.g., peptostreptococci) and gram-negative rods (e.g., Fusobacterium, Prevotella, and Porphyromonas), with deeper lesions (19, 29).
Recently, molecules of the S100 family have attracted great interest due to their cellular and tissue expression profile, association with inflammatory disorders, utility in clinical diagnostics, and potential as novel therapeutic targets. The 18 S100 family members so far described are 10 to 14 kDa in size and contain two EF hand motifs capable of binding calcium. They function in a range of cellular activities, including signal transduction, cell differentiation, regulation of cell motility, transcription, and cell cycle progression (11). Two of the most well-characterized members are S100A8 and S100A9, which can form a heterodimeric complex with antimicrobial properties. In addition, this complex has an ability to stimulate neutrophil adhesion and chemotaxis and is also involved in arachidonic acid metabolism and transport (14, 15). Their elevated expression has previously been associated with immune and epithelial cells from a variety of inflammatory disorders (22).
S100A12 has homology with S100A8 and S100A9 but does not interact with either and plays a distinct role during inflammation (30). S100A12 interacts with the receptor for advanced glycation end products present on macrophages, lymphocytes, and endothelial cells. Subsequent receptor activation results in synthesis and secretion of proinflammatory mediators via the NF-κB pathway. High levels of expression of S100A12 have been associated with several inflammatory diseases (11, 26).
S100A10 and S100A13 are less well characterized. S100A10 forms a heterotetramer complex with annexin II, which localizes to the plasma membrane and the endosome and functions in a Ca2+-dependent manner to regulate membrane traffic and ion currents (33). In addition, S100A10 has been shown to be involved in the regulation of cytosolic phospholipase A2 synthesis of arcahidonic acid and tissue-type plasminogen activator-dependent plasminogen activation (7, 18). S100A13 is newly described and most probably represents a secreted protein involved in the release of fibroblast growth factor 1 and synaptotagmin 1 in response to temperature stress (16, 21).
Cytokines and chemokines are well studied in the host response to bacterial infection and are expressed by a variety of immune and structural cells. They are characterized by their pleiotropism and pluripotentiality and play vital roles in mediating the immune system response by exerting their biological effects on a range of target cells. Certain cytokines, such as tumor necrosis factor-alpha (TNF-α), interleukin-1β (IL-1β), IL-6, IL-8, and epithelial cell-derived neutrophil attractant 78 (ENA-78) are predominantly proinflammatory and mediate both local and systemic inflammatory responses, with increased levels being associated with a variety of diseases (4, 6, 17, 24, 31). Structural proteins, such as collagens, can also be increased due to tissue inflammation as a result of infection, and their overexpression can result in fibrotic tissue damage (23).
So far, only limited studies have been performed on the molecular nature of the dental tissue immune response. Our objective, therefore, was to better characterize the molecules involved by analyzing expression of members of the S100 and cytokine gene families, along with collagen-1α, in both healthy and carious teeth and to correlate these to disease severity. A better understanding of the molecular mediation of dental tissue inflammation will ultimately facilitate improved future diagnosis and treatment.
MATERIALS AND METHODS
Extraction of dental tissue from human teeth.
Carious and sound mature human premolar and molar teeth, extracted for orthodontic purposes from patients in the age range of 20 to 30 years, were obtained immediately postextraction from clinics at the Birmingham Dental Hospital following informed patient consent. Carious teeth exhibited disease ranging from enamel involvement only to deep dentinal lesions. Extracted teeth were either immediately frozen in liquid nitrogen or were submerged in the RNA stabilizing solution RNA Later (Sigma, London, United Kingdom). Teeth were subsequently longitudinally sliced with a segmented diamond-edged rotary saw (TAAB Laboratories, Aldermaston, United Kingdom) and cooled with phosphate-buffered saline (PBS), and the pulpal tissue was carefully removed intact with a sterile dental probe and forceps.
Histological analysis.
Both dentine and pulp samples were fixed for 24 h in 10% (wt/vol) neutral buffered formalin (Surgipath, Peterborough, United Kingdom). Following pulpal tissue extirpation, dentine samples were subsequently demineralized in 10% formic acid (Sigma) until they were fully decalcified (approximately 10 days) as assessed by radiography. Both pulp and dentine samples were routinely processed through a series of graded alcohols and xylene and were embedded in paraffin wax (Tissue Tek III). Serial sections 5 μm in thickness were cut, dried at 56°C overnight, and stained in Mayer's hematoxylin and eosin (H&E).
Measurement of disease severity.
After analysis of histologically prepared dentine serial sections derived from carious teeth, the shortest distance between the microbial front of the carious lesion and the pulp chamber was determined by using a Leitz Dialux 222 microscope fitted with a 10-μm ocular ruler at 50-fold magnification. Measurements were performed in triplicate, and the average distances were calculated.
Purification and activation of immune system cells.
Following informed patient consent, venous blood from medically healthy donors who had no history of inflammatory disease was collected under sterile conditions in vacutainer tubes containing either heparin sulfate (neutrophils) or sodium citrate (monocytes). Neutrophils were subsequently prepared from 7 ml of venous blood by using a previously described single-gradient (δ = 1.079) Percoll density centrifugation technique (1). In brief, blood was layered onto the 8-ml Percoll gradient (Amersham, London, United Kingdom) and was centrifuged for 8 min at 150 × g and then for a further 10 min at 1,200 × g to separate blood platelets and mononuclear cells. Neutrophils were subsequently harvested, and contaminating red blood cells were removed by addition of 30 ml of ice-cold lysis buffer (0.83% NH4Cl in solution). Cells were then subjected to two 2-ml washes, first in lysis buffer and then in PBS for 6 min at 360 × g, and finally were resuspended in 1 ml of PBS. Prior to stimulation, neutrophil cell counts and viability were determined by using the trypan blue (Sigma) exclusion method. A proportion of isolated neutrophil cells was stimulated by exposing 5 × 106 cells to 1 ml of GPBSS (PBS with the addition of 10 mM glucose, 1 mM CaCl2, 1.5 mM MgCl2) supplemented with 250 ng of Escherichia coli lipopolysaccharide (LPS) (Sigma) for 3 h at 37°C. As a control, 5 × 106 neutrophils were incubated for 3 h at 37°C in unsupplemented GPBSS. Following incubation cells were centrifuged at 1,200 × g for 2 min, the supernatant was removed, and the pellet was used for RNA isolation.
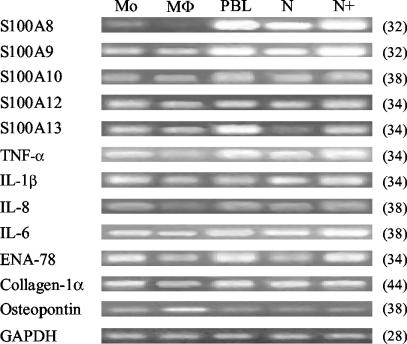
Monocytes were isolated from 200 ml of venous blood by using Ficoll Hypaque (Amersham-Pharmacia Biotech, London, United Kingdom) gradient centrifugation. Extracted blood was diluted at a ratio of 1:1 with Hanks' balanced salt solution (HBSS), pelleted by centrifugation, and rinsed twice more in HBSS. Cells were resuspended in RPMI 1640 medium prior to cell counting. To enrich for monocytes, cells were plated at a density of 4 × 106 cells/ml and were incubated at 37°C for 1 h. Plated cells were washed twice in HBSS to remove nonadherent lymphocytes. Monocyte RNA was subsequently harvested from adherent cells. Macrophages were matured by culturing monocytes for 5 days in RPMI 1640 plus 10% fetal calf serum with 50 ng of granulocyte-macrophage colony-stimulating factor (GM-CSF; R&D Systems)/ml. To confirm GM-CSF macrophage maturation, osteopontin gene expression levels were assayed in RNA derived from the monocytes and macrophages (see Fig. 6) (10).
FIG. 6.
Gene expression analysis of S100 family members, cytokines, collagen-1α, and osteopontin in monocytes (Mo), macrophages (Mφ), peripheral blood derived lymphocytes (PBL), neutrophils (N), and E. coli LPS-stimulated neutrophils (N+). Cycle number at which PCR samples were analyzed is shown in parenthesis. A representative image of duplicate analyses is shown.
RNA isolation and cDNA synthesis.
Total RNA was extracted by using the RNeasy mini kit (Qiagen, London, United Kingdom) and was eluted in a final volume of 30 μl of sterile water as recommended by the manufacturer. Prior to RNA extraction, pulpal tissue was homogenized by using an Ultra-Turrax T8 tissue disrupter (Fisher Scientific, Loughborough, United Kingdom). Subsequently, 0.5 to 1.8 μg of DNase-digested total RNA was used for oligo(dT) reverse transcription to generate single-stranded cDNA by using the Omniscript kit (Qiagen). Both RNA and cDNA concentrations were determined from absorbance values at a wavelength of 260 nm using a BioPhotometer (Eppendorf, Cambridge, United Kingdom). The total amount of RNA isolated per pulpal specimen was calculated by multiplying the concentration obtained by the final sample elution volume. All RNA samples used in this study had 260 nm:280 nm ratios greater than 1.8. RNA integrity was additionally verified by visual inspection of samples on 1% nondenaturing agarose gels stained with ethidium bromide.
Sq-RT-PCR analysis.
Semiquantitative reverse transcriptase PCR (Sq-RT-PCR) analysis was performed for the human genes encoding S100A8, S100A9, S100A10, S100A12, S100A13, TNF-α, IL-1β, IL-6, ENA-78, and collagen-1α with the housekeeping gene for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as a control. The primer sequences used are shown in Table 1. In summary, 50 ng of single-stranded cDNA was used to seed a 50-μl PCR, which was subjected to between 28 and 44 cycles. A typical amplification cycle of 95°C for 20 s, 51°C to 60°C for 20 s, and 72°C for 20 s was performed using a Mastercycler thermal cycler (Eppendorf). After the designated number of cycles, 8 μl of the PCR mix was removed and the product was separated and visualized on a 1.5% agarose gel containing 0.5 μg of ethidium bromide/ml. Scanned gel images were imported into AIDA image analysis software (Fuji, Sheffield, United Kingdom), and the volume density of amplified was products calculated and normalized against GAPDH housekeeping gene control values.
TABLE 1.
Primer sequences and cycling conditionsa
| Gene product | Primer sequence (5′→3′) | Tm (°C) | Product (bp) |
|---|---|---|---|
| S100A8 | (F)/ATTTCCATGCCGTCTACAGG | 60 | 247 |
| (R)/CAGCCTCTGGGCAGTAACTC | |||
| S100A9 | (F)/CAGCTGGAACGCAACATAGA | 60 | 229 |
| (R)/TCAGCATGATGAACTCCTCG | |||
| S100A10 | (F)/AAATTCGCTGGGGATAAAGG | 60 | 289 |
| (R)/TCTTATCAGGGAGGAGCGAA | |||
| S100A12 | (F)/CTTGAAGAGCATCTGGAGGG | 60 | 328 |
| (R)/CTCATTGAGGACATTGCTGG | |||
| S100A13 | (F)/AGCTAGCCCCTTGAGGACAT | 60 | 268 |
| (R)/TTGAGCTCCGAGTCCTGATT | |||
| TNF-α | (F)/AAGAATTCAAACTGGGGCCT | 61 | 432 |
| (R)/GGCTACATGGGAACAGCCTA | |||
| IL-1β | (F)/TCCAGGGACAGGATATGGAG | 61 | 308 |
| (R)/TTCTGCTTGAGAGGTGCTGA | |||
| IL-8 | (F)/TAGCAAAATTGAGGCCAAGG | 61 | 214 |
| (R)/GGACTTGTGGATCCTGGCTA | |||
| IL-6 | (F)/GAACTCCTTCTCCACAAGCG | 61 | 272 |
| (R)/TTTTCTGCCAGTGCCTCTTT | |||
| ENA-78 | (F)/TCTGCAAGTGTTCGCCATAG | 61 | 363 |
| (R)/GCTTAAGCGCCAAACATAGG | |||
| Collagen-1α | (F)/TGGGAGTGCAAGGATACTCTATATCG | 60 | 320 |
| (R)/CCCATCCCATCTTCGACGTAC | |||
| GAPDH | (F)/CCACCCATGGCAAATTCCATGGCA | 60 | 391 |
| (R)/TCTAGACGGCAGGTCAGGTCCACC |
Tm, annealing temperature; (F), forward primer; (R), reverse primer.
Immunohistochemical analysis.
Ten healthy and ten carious adult teeth were used for immunohistochemical analyses. Five-micron tissue sections were rehydrated through xylene and graded alcohols prior to pretreatment with 0.05% trypsin (1:250; Difco) in PBS for 15 min at room temperature and a 30-min microwave treatment in citrate buffer. Slides were washed with water followed by being washed with PBS, and endogenous peroxide activity was blocked with 3% hydrogen peroxidase in PBS (10 min) prior to sections being incubated with a 1:5 dilution of normal goat serum (30 min). Tissues were stained with affinity-purified mouse polyclonal antibodies to S100A8 (Bachem, St. Helens, United Kingdom), S100A8/S100A9 complex (Dako, Ely, United Kingdom), and neutrophil elastase (Binding Site, Birmingham, United Kingdom). Bound antibody was detected by using the streptavidin peroxidase kit method (StrAviGen; Biogenex) and 3,3′diaminobenzidine (0.05% for 15 min). Sections were counterstained with hematoxylin. Coronal areas of the pulp in healthy tissues and directly beneath carious lesions were examined by light microscopy. Negative controls included omission of primary antibody and substitution of the primary antibody with normal rabbit immunoglobulin. Positive control sections were immunostained human adult gingiva with chronic inflammation obtained from 6 patients exhibiting classic chronic periodontal disease diagnosed at the Birmingham Dental Hospital. Histological sections demonstrated the characteristic cell infiltrate seen during chronic inflammatory periodontal disease (28).
Statistical analysis.
Normalized expression levels for all genes studied were imported into the statistical analysis package SPSS (release 10.0.0) for multiple-regression analysis. t tests were performed to determine significance of difference between mean expression levels. P values are shown where appropriate.
RESULTS
Association between caries and quantity of RNA extracted.
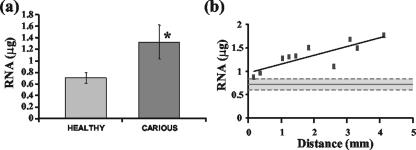
Analysis of the average isolated quantities of RNA indicated that significantly higher amounts were obtained from carious versus healthy dental tissue (Fig. 1a), although a wider distribution in the amount isolated from carious teeth (range, 0.87 to 1.76 μg) compared to that of healthy teeth (range, 0.57 to 0.84 μg) was observed. A statistically significant correlation was subsequently identified between carious RNA amounts and increasing disease severity, indicating that as the distance from the pulpal chamber to the microbial front decreased the amount of isolated RNA also decreased (Fig. 1b).
FIG. 1.
(a) Average amount of RNA isolated from 10 diseased and 10 healthy teeth. An asterisk indicates statistically significant difference. Error bars for one standard deviation from the means are shown. (b) Scatter plot displaying the relationship between the amount of RNA isolated and the distance from the microbial front for the 10 carious teeth analyzed (P < 0.003; equation of line was y = 0.1848x + 0.9689; R2 = 0.6896). The solid horizontal line represents the mean total RNA isolated from healthy dental tissue, and dashed lines indicate one standard deviation from the mean.
Gene expression analysis in carious and healthy teeth.
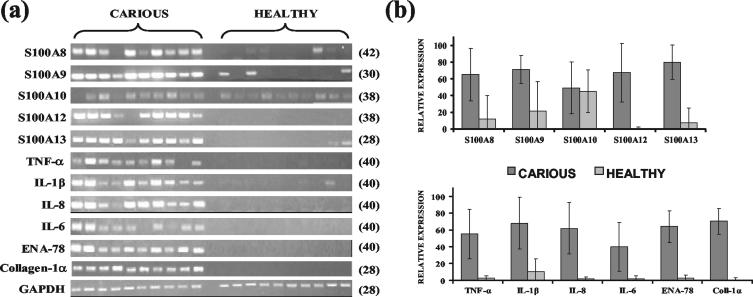
Sq-RT-PCR analysis was performed in the 10 carious and 10 healthy dental tissue-derived RNA samples with genes previously shown to be associated with the immune and host defense processes. The genes assayed included the S100 family members S100A8, S100A9, S100A10, S100A12, and S100A13, the genes encoding cytokines TNF-α, IL-1β, IL-8, IL-6, and ENA-78, and the gene for collagen-1α. Data indicated that transcript levels from S100 gene family members (except S100A10), the cytokines, and collagen-1α were statistically significantly higher (P value < 0.01) in carious than in healthy pulpal tissue samples (Fig. 2).
FIG. 2.
(a) Semiquantitative RT-PCR analysis showing differential gene expression analysis in carious and healthy dental tissue samples. The cycle number at which PCRs were analyzed is shown in parenthesis. (b) Densitometric analysis of gel images. Volume densities of amplified products were normalized against GAPDH control values, and average values for each group were calculated and expressed as a percentage of the highest normalized individual volume density obtained. Error bars for one standard deviation from the means are shown. All genes, except S100A10, showed statistically higher expression in carious teeth (P < 0.01).
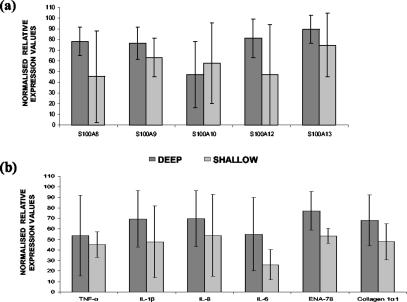
To determine whether expression levels could be used for classification of carious teeth into those exhibiting either shallow or deep carious lesions, the normalized expression levels for the genes assayed were correlated to distance measurements. The ratio of average normalized values at 0.5-mm intervals was subsequently calculated. The data indicated that a distance of less than 2 mm between the microbial front and the pulp chamber gave the highest average expression value for all genes except S100A10 (Fig. 3) and hence could be used to describe a deep carious lesion. Conversely, a distance of greater than 2 mm from the pulp chamber to the site of injury could be used to identify shallow carious lesions. Notably, when RNA quantities for carious teeth were also analyzed in a similar manner, a comparable result for the definition of deep and shallow carious lesions was obtained (data not shown).
FIG. 3.
Analysis of gene expression levels for S100 family members (a) and cytokines and collagen-1α (b) in teeth with deep and shallow carious lesions. Carious teeth with microbial fronts within 2 mm of the pulp chamber were classified as having deep lesions, and carious teeth with microbial fronts further than 2 mm from the pulp chamber were classified as having shallow lesions. Averaged normalized values for each classification group were calculated and plotted. Error bars for one standard deviation from the means are shown. ENA-78 exhibited a statistically significant difference in expression levels between deep and shallow carious lesions (P < 0.05).
Correlations between GAPDH-normalized gene expression levels.
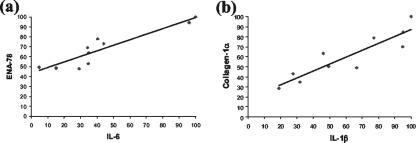
To identify correlations between GAPDH-normalized gene expression levels in the 10 carious disease samples, multiple regression analysis was performed using the SPSS statistical analysis software package. Eight significant positive correlations (P < 0.05) were subsequently identified between S100A8 and IL-1β, ENA-78, and IL-6 and between collagen-1α and S100A8, TNF-α, IL-1β, IL-8, IL-6, and ENA-78 (Table 2). R2 values and equation of lines, as determined from scatter plots (data not shown), are also provided for all significant correlations identified (Table 2). Representative scatter plots are shown for the correlations with the highest R2 values (Fig. 4).
TABLE 2.
Correlation matrix for cytokine, S100 family, and collagen-1α gene expression in human dental pulps
| Gene product | Gene producti
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S100A8 | S100A9 | S100A10 | S100A12 | S100A13 | TNF-α | IL-1β | IL-8 | IL-6 | ENA-78 | |
| S100A9 | 0.1416 | |||||||||
| S100A10 | 0.6429 | 0.6219 | ||||||||
| S100A12 | 0.8433 | 0.4643 | 0.8618 | |||||||
| S100A13 | 0.2462 | 0.4728 | 0.8081 | 0.1088 | ||||||
| TNF-α | 0.4721 | 0.9711 | 0.4412 | 0.8774 | 0.6514 | |||||
| IL-1β | 0.0196a | 0.5537 | 0.8909 | 0.8552 | 0.3441 | 0.0828 | ||||
| IL-8 | 0.1498 | 0.0754 | 0.7126 | 0.7513 | 0.8989 | 0.5760 | 0.0781 | |||
| IL-6 | 0.1722 | 0.3594 | 0.1687 | 0.7225 | 0.8758 | 0.3315 | 0.1345 | 0.4129 | ||
| ENA-78 | 0.0504 | 0.1796 | 0.1824 | 0.5999 | 0.9534 | 0.4242 | 0.0775 | 0.1020 | 0.0002b | |
| Collagen-1α | 0.0304c | 0.2722 | 0.7536 | 0.9013 | 0.8102 | 0.0362d | 0.0003e | 0.0248f | 0.0174g | 0.0052h |
y = 0.6824x + 16.351; R2 = 0.5141.
y = 0.5595x + 43.395; R2 = 0.849.
y = 0.4949x + 28.004; R2 = 0.4628.
y = 0.5181x + 34.073; R2 = 0.4412.
y = 0.6904x + 18.261; R2 = 0.8158.
y = 0.5195x + 27.287; R2 = 0.4871.
y = 0.5376x + 36.907; R2 = 0.5271.
y = 0.9786x − 5.9892; R2 = 0.644.
Statistically significant correlations (P < 0.05) are shown in italics.
FIG. 4.
Representative scatter plots of GAPDH-normalized gene expression in carious teeth for ENA-78 and IL-6 (R2 = 0.849) (a) as well as for collagen-1α and IL-1β (R2 = 0.816) (b).
Immunohistochemical analysis of S100A8 and S100A8/S100A9 complex.
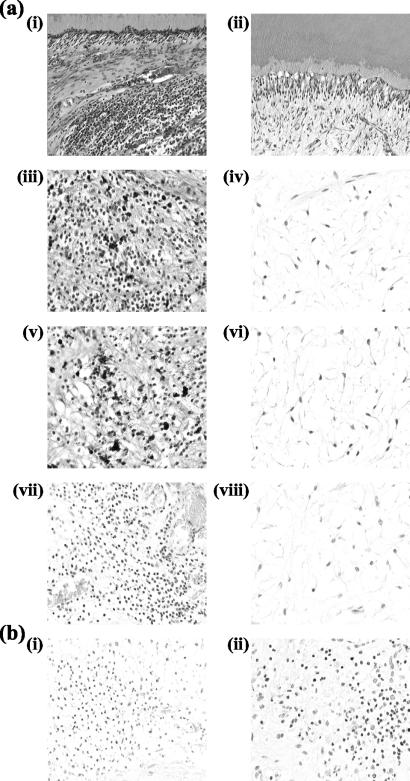
To determine the cellular expression of S100A8 and the S100A8/S100A9 complex, immunohistochemical analysis was performed. The infiltrating immune system cells examined in the coronal areas of the pulp directly beneath the carious lesion stained with antibodies to S100A8 and the S100A8/S100A9 protein complex more intensely than did pulpal fibroblasts and odontoblasts in carious dental tissues, while minimal staining was observed in similar regions of healthy pulps (Fig. 5). The nuclear morphology of adjacent H&E-stained sections indicated that these cells were neutrophils. To confirm this, sequential 5-μm histological sections from carious and healthy dental tissue and inflamed gingival samples were stained with an antibody specific to neutrophil elastase. The staining for both S100A8 and the S100A8/S100A9 protein complex and neutrophil elastase colocalized, confirming that the expression of S100A8 and the S100A8/S100A9 complex in carious samples was attributable to neutrophils (Fig. 5).
FIG. 5.
(a) Carious (i, iii, v, and vii) and healthy (ii, iv, vi, and viii) teeth stained with H&E (i and ii), anti-S100A8 (iii and iv), anti-S100A8/S100A9 (v and vi), and anti-neutrophil elastase (vii and viii). (b) Human inflamed gingiva stained with anti-S100A8. Shown are negative (omission of primary antibody and replacement with nonimmune rabbit immunoglobulin G) (i) and positive (ii) controls. Field widths: for i and ii of panel a, 479 μm; for iii to viii of panel a, 241 μm; for i and ii of panel b, 241 μm.
Gene expression analysis in immune system cells.
To determine the gene expression profile of the S100 family members, cytokines, and collagen-1α, Sq-RT-PCR analysis was performed using RNA derived from monocytes, macrophages, peripheral blood lymphocytes, neutrophils, and neutrophils activated by E. coli LPS. For S100 gene family members, data indicated that S100A8 and S100A9 were more abundantly expressed by neutrophils than by monocytes and macrophages and, as was also the case for S100A13, were upregulated in neutrophils following stimulation. S100A8, -A9, -A12, and -A13 appeared to be more abundantly expressed in monocytes than in GM-CSF-matured macrophages, although considerable differences in expression levels were not evident in all cases. Notably, S100A8, -A9, and -A13 also showed a relatively high expression in the lymphocyte preparation, indicating that they may be expressed by peripheral blood T and B cells.
While the gene expression levels of the cytokines also varied in the immune system cell types analyzed, the variations appeared less dramatic than those for S100 family members. Data indicated that for all cytokines analyzed, expression was lower in macrophages than in monocytes, although the degree of downregulation varied. In contrast, the cytokines in general appeared to be upregulated in neutrophils following stimulation with LPS. Collagen-1α levels were relatively equal in all RNA samples, although its expression appeared to be downregulated in the GM-CSF-induced macrophages.
DISCUSSION
Carious lesions have previously been shown to be dominated by a variety of infiltrating immune system cells (9, 13). The increase in cellularity accompanying caries therefore provides the simplest and most likely explanation with regards to the increase in the amount of RNA extracted from diseased compared to healthy pulps. In addition, however, it is also conceivable that an increase in transcription in cells of the dentine-pulp complex also accompanies caries as the pulp upregulates its activity in attempts to combat any invading bacteria and/or to initiate repair mechanisms. Notably, our data also indicated that there was an association between RNA amounts and the proximity of the pulp chamber to the microbial front and that the closer the carious lesion was the less the amount of RNA isolated. One possible explanation for this may be that lesions closer to the pulp chamber represent chronic carious infections, whereas shallow lesions may indicate the initial stages of the carious process. Pulps from teeth presenting chronic carious lesions may display inflammatory responses to the carious challenge that compromise localized cell vitality and thereby lead to decreased amounts of extractable RNA. Even prior to carious enamel cavitation, regressive changes can be observed in the pulp (2). Accumulation of inflammatory cells in the pulp may occur prior to caries invading dentine (3), although once caries has penetrated beyond enamel, pulpal inflammation becomes more prevalent (20). Such inflammatory events may locally compromise cell vitality.
As only a few limited studies have so far addressed the hypothesis that expression levels of inflammatory and immune system genes are elevated during caries (5, 12, 25, 27, 32), we analyzed transcript levels of 11 genes predicted to be associated with this process in 10 healthy and 10 carious pulpal specimens. Our data clearly indicated that the expression of all genes examined in this study, except S100A10, were more abundant in carious tissue, although heterogeneity in levels was present in carious samples. Initially, regression analysis was performed to determine if expression levels were directly related to the distance of the microbial front from the pulpal chamber, but significant positive correlation was observed only for S100A13 (data not shown). We therefore sought to determine whether the expression data could be used to define deep or shallow carious lesions and found that for carious teeth having less than 2 mm between the microbial front and the pulp chamber, higher average expression levels (except for S100A10) were observed. Teeth with less than 2 mm of remaining dentine coverage were therefore categorized as having deep carious lesions, and teeth with greater than 2 mm of remaining dentine coverage were categorized as having shallow carious lesions. Interestingly, Hahn et al. (8) previously used similar distances to classify teeth into deep- and shallow-caries categories; however, it appears that their classification system was purely arbitrary and not based on scientific evidence. In addition, they also found that mRNA levels of cytokines IL-4 and IL-10 were more abundant in deep carious lesions than they were in shallow carious lesions, although the opposite was true for gamma interferon.
Multiple regression analyses of gene expression levels also identified several significant positive correlations (Table 2). If gene expression levels had been assayed by using a quantitative approach, such as Northern blotting, real-time PCR, or RNase protection assays, more significant correlations may have been identified. For example, previous studies have shown that S100A8 and S100A9 proteins form a complex and therefore it is possible that their expression is coordinately regulated (15). If a quantitative assay had been applied followed by correlation analysis, such a relationship could have been more accurately ascertained. Further experiments in this area using quantitative gene expression analysis technology are, therefore, warranted. It is notable that the S100 proteins and the majority of cytokines studied here have been classified as markers of the acute phase of inflammation (6, 15), and several S100 family members, including S100A8, -A9, and -A12, have been implicated in the stimulation of proinflammatory cytokines via the NF-κB pathway (26). However, while it is possible that the observed significant positive correlations are a result of positive molecular feedback mechanisms, similar to the one highlighted above, it is equally possible that correlations are co-occurrences which result from independent events, such as bacterial stimulation, which coordinately activates gene expression. It is likely that both mechanisms occur during pulpal inflammation. The identified correlations could also represent changes in cell populations within the carious pulpal tissue indicative of different stages of the disease process and/or may even be due to differences in the composition of the microbial flora which occurs as caries progresses (19). Recently, Zehnder et al. (32) performed similar experiments analyzing the transcript levels of five cytokines in carious and healthy pulps obtained from 20 individuals. Their findings indicated that statistically significant higher levels of IL-6, IL-8, and IL-18 but not of IL-1α and IL-1β were present in carious pulps and that there was significant correlation between expression levels of IL-1α and IL-1β and between IL-6, IL-8, and IL-18 mRNA. In contrast, our data clearly showed that IL-1β levels are significantly higher in carious than in healthy pulp specimens (Fig. 2). Comparison of these two studies, therefore, suggests that analysis of a large sample population of carious tissue using a high-throughput gene expression analysis technique, such as microarrays, and taking into account other important disease factors, such as age of carious lesion, age of patient, disease severity, and infecting microbial populations, would provide a more comprehensive understanding of the multitude of genes important in carious disease. In addition, based on our findings, such analyses would facilitate determination of interrelationships between gene expression, potentially enabling elucidation of the biochemical and molecular networks and pathways involved.
Our immunohistochemical analyses showed that protein levels for S100A8 and the S100A8/S100A9 complex were elevated during carious disease, and serial sections stained with neutrophil elastase indicated that these molecules were predominantly expressed by neutrophils. These data are supported by other studies (15, 22) and by our gene expression analyses showing increased levels in neutrophils (Fig. 6). While the cells responsible for the expression of the other S100 family members have yet to be determined in carious disease, the data presented here suggests that activated neutrophils may also be a source for S100A13 (Fig. 6).
It is clear that the immune response due to dental tissue infection and the healing processes of the pulpal tissue are molecularly complex and highly heterogeneous. A more comprehensive knowledge of the pulp's own innate molecular and cellular responses will therefore provide the basis for the development of better diagnostic tools and more efficacious treatment modalities for dental caries.
Acknowledgments
J.L.M. is supported by a University of Birmingham Scientific Projects Committee grant (EBX 1193).
We thank Sally Kerr (Oral Surgery) for providing the teeth for this investigation, Robina Rayasat and Sue Finney for technical assistance, and Helen Wright for providing neutrophils.
Editor: A. D. O'Brien
REFERENCES
- 1.Bergstrom, K., and B. Asman. 1993. Luminol enhanced Fc-receptor dependant chemiluminescence from peripheral PMN cells. A methodological study. Scand. J. Clin. Lab. Investig. 53:171-177. [DOI] [PubMed] [Google Scholar]
- 2.Bjorndal, L., T. Darvann, and A. Thylstrup. 1998. A quantitative light microscopic study of the odontoblast and subodontoblastic reactions to active and arrested enamel caries without cavitation. Caries Res. 32:59-69. [DOI] [PubMed] [Google Scholar]
- 3.Brannstrom, M., and P. O. Lind. 1965. Pulpal response to early dental caries. J. Dent. Res. 44:1045-1150. [DOI] [PubMed] [Google Scholar]
- 4.Chung, K. F. 2001. Cytokines in chronic obstructive pulmonary disease. Eur. Respir. J. Suppl. 34:S50-S59. [PubMed] [Google Scholar]
- 5.D'Souza, R., L. R. Brown, J. R. Newland, B. M. Levy, and L. B. Lachman. 1989. Detection and characterization of interleukin-1 in human dental pulps. Arch. Oral Biol. 34:307-313. [DOI] [PubMed] [Google Scholar]
- 6.Feghali, C. A., and T. M. Wright. 1997. Cytokines in acute and chronic inflammation. Front. Biosci. 2:D12-D26. [DOI] [PubMed] [Google Scholar]
- 7.Gattaz, W. F., D. R. Lara, H. Elkis, L. V. Portela, C. A. Goncalves, A. B. Tort, J. Henna, and D. O. Souza. 2000. Decreased S100-beta protein in schizophrenia: preliminary evidence. Schizophr. Res. 43:91-95. [DOI] [PubMed] [Google Scholar]
- 8.Hahn, C. L., A. M. Best, and J. G. Tew. 2000. Cytokine induction by Streptococcus mutans and pulpal pathogenesis. Infect. Immun. 68:6785-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn, C. L., W. A. Falkler, Jr., and M. A. Siegel. 1989. A study of T and B cells in pulpal pathosis. J. Endod. 15:20-26. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto, S., T. Suzuki, H. Y. Dong, S. Nagai, N. Yamazaki, and K. Matsushima. 1999. Serial analysis of gene expression in human monocyte-derived dendritic cells. Blood 94:845-852. [PubMed] [Google Scholar]
- 11.Heizmann, C. W. 2002. The multifunctional S100 protein family. Methods Mol. Biol. 172:69-80. [DOI] [PubMed] [Google Scholar]
- 12.Huang, G. T., A. P. Potente, J. W. Kim, N. Chugal, and X. Zhang. 1999. Increased interleukin-8 expression in inflamed human dental pulps. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 88:214-220. [DOI] [PubMed] [Google Scholar]
- 13.Izumi, T., I. Kobayashi, K. Okamura, and H. Sakai. 1995. Immunohistochemical study on the immunocompetent cells of the pulp in human non-carious and carious teeth. Arch. Oral Biol. 40:609-614. [DOI] [PubMed] [Google Scholar]
- 14.Kerkhoff, C., M. Klempt, V. Kaever, and C. Sorg. 1999. The two calcium-binding proteins, S100A8 and S100A9, are involved in the metabolism of arachidonic acid in human neutrophils. J. Biol. Chem. 274:32672-36279. [DOI] [PubMed] [Google Scholar]
- 15.Kerkhoff, C., M. Klempt, and C. Sorg. 1998. Novel insights into structure and function of MRP8 (S100A8) and MRP14 (S100A9). Biochim. Biophys. Acta 1448:200-211. [DOI] [PubMed] [Google Scholar]
- 16.Landriscina, M., R. Soldi, C. Bagala, I. Micucci, S. Bellum, F. Tarantini, I. Prudovsky, and T. Maciag. 2001. S100A13 participates in the release of fibroblast growth factor 1 in response to heat shock in vitro. J. Biol. Chem. 276:22544-22552. [DOI] [PubMed] [Google Scholar]
- 17.MacDermott, R. P. 1999. Chemokines in the inflammatory bowel diseases. J. Clin. Immunol. 19:266-272. [DOI] [PubMed] [Google Scholar]
- 18.MacLeod, T. J., M. Kwon, N. R. Filipenko, and D. M. Waisman. 2003. Phospholipid-associated annexin A2-S100A10 heterotetramer and its subunits: characterization of the interaction with tissue plasminogen activator, plasminogen, and plasmin. J. Biol. Chem. 278:25577-25584. [DOI] [PubMed] [Google Scholar]
- 19.Martin, F. E., M. A. Nadkarni, N. A. Jacques, and N. Hunter. 2002. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 40:1698-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massler, M. 1967. Pulpal reactions to dental caries. Int. Dent. J. 17:441-460. [PubMed] [Google Scholar]
- 21.Mouta-Carreira, C., T. M. LaVallee, F. Tarantini, A. Jackson, J. T. Lathrop, B. Hampton, W. H. Burgess, and T. Maciag. 1998. S100A13 is involved in the regulation of fibroblast growth factor-1 and p40 synaptotagmin-1 release in vitro. J. Biol. Chem. 273:22224-22231. [DOI] [PubMed] [Google Scholar]
- 22.Nacken, W., J. Roth, C. Sorg, and C. Kerkhoff. 2003. S100A9/S100A8: myeloid representatives of the S100 protein family as prominent players in innate immunity. Microsc. Res. Tech. 60:569-580. [DOI] [PubMed] [Google Scholar]
- 23.Nathan, C. 2002. Points of control in inflammation. Nature 420:846-852. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen, O. H., B. Vainer, S. M. Madsen, J. B. Seidelin, and N. H. Heegaard. 2000. Established and emerging biological activity markers of inflammatory bowel disease. Am. J. Gastroenterol. 95:359-367. [DOI] [PubMed] [Google Scholar]
- 25.Pezelj-Ribaric, S., I. Anic, I. Brekalo, I. Miletic, M. Hasan, and M. Simunovic-Soskic. 2002. Detection of tumor necrosis factor alpha in normal and inflamed human dental pulps. Arch. Med. Res. 33:482-484. [DOI] [PubMed] [Google Scholar]
- 26.Roth, J., T. Vogl, C. Sorg, and C. Sunderkotter. 2003. Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends Immunol. 24:155-158. [DOI] [PubMed] [Google Scholar]
- 27.Sawa, Y., Y. Horie, Y. Yamaoka, N. Ebata, T. Kim, and S. Yoshida. 2003. Production of colony-stimulating factor in human dental pulp fibroblasts. J. Dent. Res. 82:96-100. [DOI] [PubMed] [Google Scholar]
- 28.Seymour, G. J., R. N. Powell, K. L. Cole, F. Aitken, D. Brookes, I. Beckman, H. Zola, J. Bradley, and G. F. Burns. 1983. Experimental gingivitis in humans: a histochemical and immunological characterisation of the lymphoid cell subpopulations. J. Perio. Res. 18:375-385. [DOI] [PubMed] [Google Scholar]
- 29.Tanzer, J. M., J. Livingston, and A. M. Thompson. 2001. The microbiology of primary dental caries in humans. J. Dent. Educ. 65:1028-1037. [PubMed] [Google Scholar]
- 30.Vogl, T., C. Propper, M. Hartmann, A. Strey, K. Strupat, C. van den Bos, C. Sorg, and J. Roth. 1999. S100A12 is expressed exclusively by granulocytes and acts independently from MRP8 and MRP14. J. Biol. Chem. 274:25291-25296. [DOI] [PubMed] [Google Scholar]
- 31.Walz, A., P. Schmutz, C. Mueller, and S. Schnyder-Candrian. 1997. Regulation and function of the CXC chemokine ENA-78 in monocytes and its role in disease. J. Leukoc. Biol. 62:604-611. [DOI] [PubMed] [Google Scholar]
- 32.Zehnder, M., N. Delaleu, Y. Du, and M. Bickel. 2003. Cytokine gene expression—part of host defence in pulpitis. Cytokine 22:84-88. [DOI] [PubMed] [Google Scholar]
- 33.Zobiack, N., U. Rescher, C. Ludwig, D. Zeuschner, and V. Gerke. 2003. The annexin 2/S100A10 complex controls the distribution of transferrin receptor-containing recycling endosomes. Mol. Biol. Cell. 14:4896-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]