Abstract
Urinary extracellular vesicles provide a novel source for valuable biomarkers for kidney and urogenital diseases: Current isolation protocols include laborious, sequential centrifugation steps which hampers their widespread research and clinical use. Furthermore, large individual urine sample volumes or sizable target cohorts are to be processed (e.g. for biobanking), the storage capacity is an additional problem. Thus, alternative methods are necessary to overcome such limitations. We have developed a practical vesicle isolation technique to yield easily manageable sample volumes in an exceptionally cost efficient way to facilitate their full utilization in less privileged environments and maximize the benefit of biobanking. Urinary vesicles were isolated by hydrostatic dialysis with minimal interference of soluble proteins or vesicle loss. Large volumes of urine were concentrated up to 1/100 of original volume and the dialysis step allowed equalization of urine physico-chemical characteristics. Vesicle fractions were found suitable to any applications, including RNA analysis. In the yield, our hydrostatic filtration dialysis system outperforms the conventional ultracentrifugation-based methods and the labour intensive and potentially hazardous step of ultracentrifugations are eliminated. Likewise, the need for trained laboratory personnel and heavy initial investment is avoided. Thus, our method qualifies as a method for laboratories working with urinary vesicles and biobanking.
Most mammalian epithelial cell types actively secrete a surprising variety of vesicles such as exosomes, microvesicles, exosome-like vesicles, retrovirus-like particles and apoptotic blebs into their extracellular space1,2, accurately reflecting the exquisite intracellular processes. The various vesicle types have, accordingly, been identified and characterized in all bodily fluids, including urine2.
Extracellular vesicles (EVs) are proposed to act as a ubiquitous intercellular communication pathway, thus revealing an accurate fingerprint of processes and pathways3. This has led to an explosion of interest in EVs as potential source of biomarkers4. Many studies have identified fully functional specialized proteins as well as a variety of functional RNA species in EVs5.
In addition to the search for the biological relevance of EVs, isolation methods have been developed to yield distinct EV populations, as recently reviewed by Momen-Heravi et al7. Despite technical improvements, the isolation step remains one of the challenges8,9 especially for a diluted biofluid such as urine which is, however, the obvious source for kidney - urogenital derived biomarkers10,11,12. Consequently, very recent reports show the intriguing possibility of monitoring diabetic nephropathy by exosome profiling13,14.
Here we report our simplified new method to efficiently isolate urinary EVs for discovery research and clinical diagnostics practically without excessive interference from Tamm-Horsfall (THP) protein15.
Results
Vesicle Enrichment Methods
The workflow developed to isolate urine exosomal vesicles is summarised in figure 1. Urine samples were spun with 2,000 g to remove cells, bacteria, cellular casts and the bulk of Tamm-Horsfall protein (THP) macropolymers (Fig. 2A,asterisk).The supernatant (SN) 2000 g was used to isolate UEVs by our in-house system which consists of a separating funnel connected with a dialysis membrane with molecular weight cut-off (MWCO) of 1,000 kDa (Supplemental Fig. 1). Hydrostatic pressure of the urine in the funnel pushes the solvent through the mesh of dialysis membrane (filtration), together with all the analytes below the selected MWCO. This filtration-concentration-dialysis process is called “hydrostatic filtration dialysis” (HFD).
Figure 1. Urinary vesicles enrichment by hydrostatic dialysis and subsequent characterization.
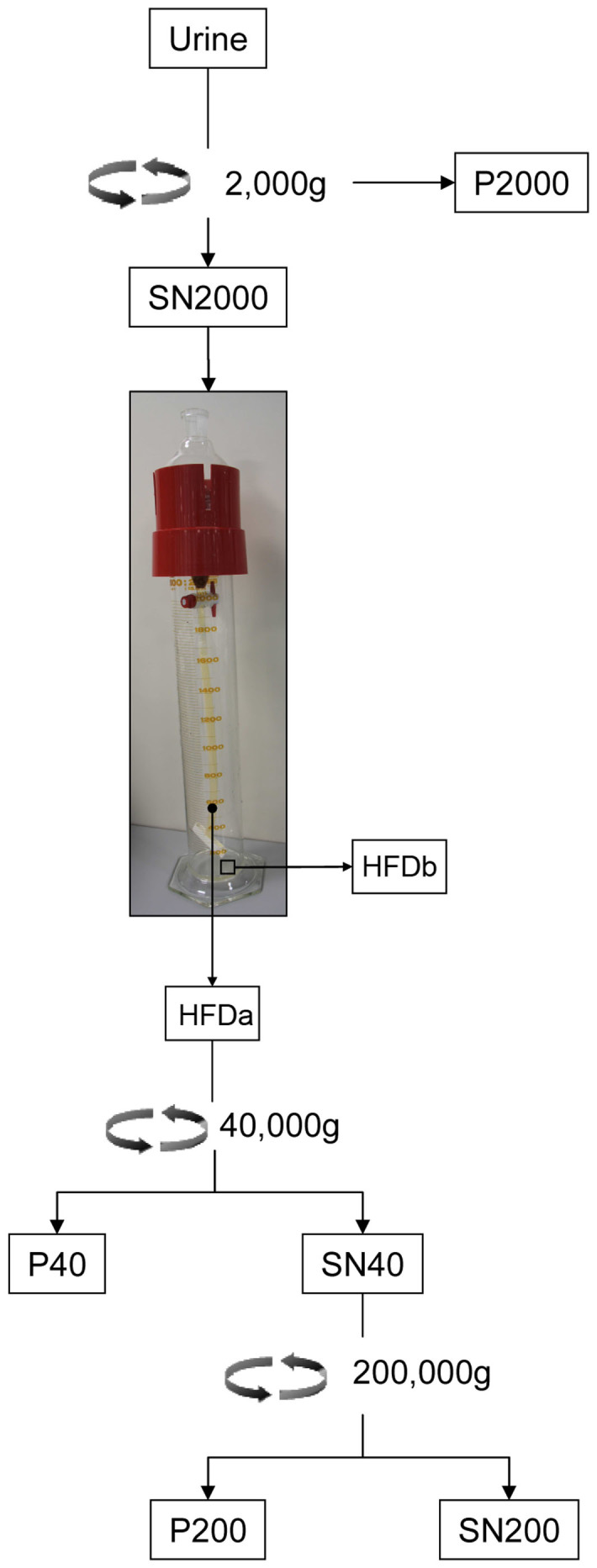
Workflow to isolate urinary vesicles starting from a pool of urine from healthy donors. HFDa hydrostatic dialysis retained fraction above 1,000 kDa (filled circle), HFDb hydrostatic dialysis below 1,000 kDa (open square). Assembly and principle of the system is detailed in Supplemental Figure 1.
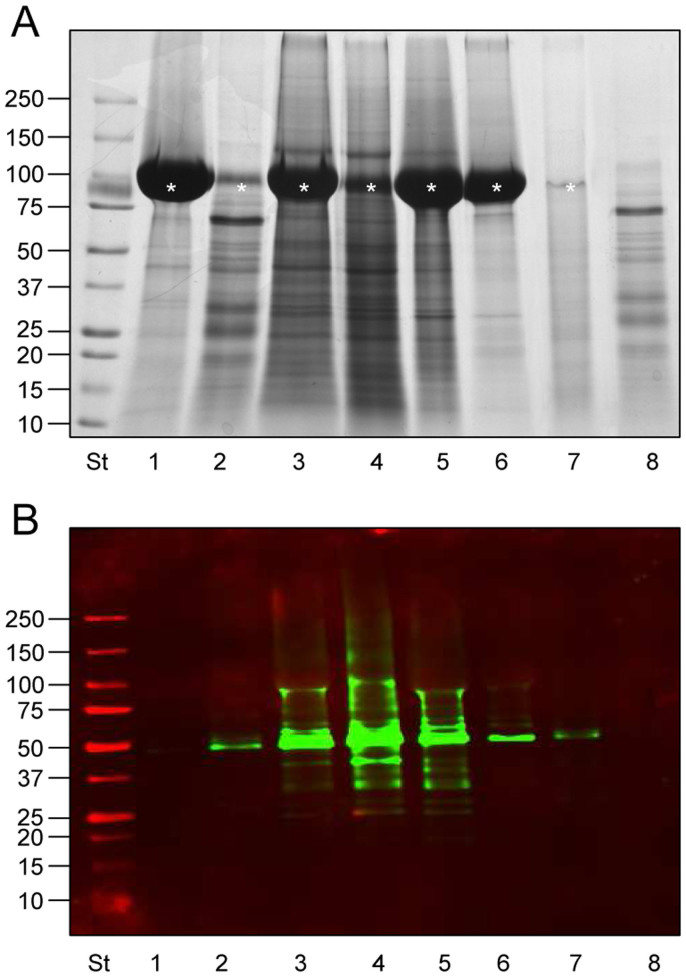
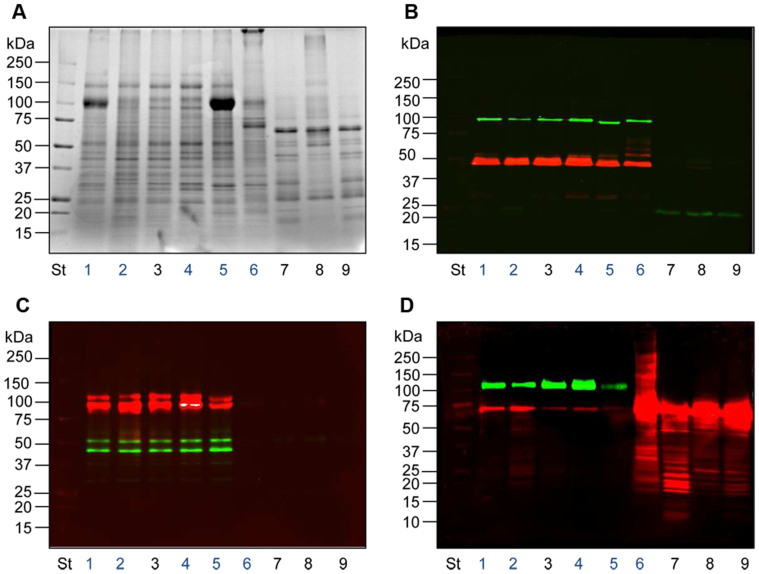
Figure 2. SDS-PAGE protein pattern and TSG101 detection of HFD fractions.
(A) Colloidal coomassie gel of all the fractions isolated following the workflow in figure 1. (B) Immunodetection of the exosome marker protein TSG101 from the same gel as A. Fifteen µg of protein (Bradford assay) were loaded per lane (A,B) in the same order: Lane 1, Pellet 2,000 g; Lane 2, Supernatant 2,000 g; Lane 3, Hydrostatic Filtration Dialysis retained above 1,000 kDa MWCO (HFDa); Lane 4, HFDa Pellet 40,000 g; Lane 5, HFDa Pellet 200,000 g; Lane 6, HFDa Supernatant 200,000 g; Lane 7, SDS elution of dialysis tube (15 µg of protein BCA assay); Lane 8, flow through solution below 1,000 kDa MWCO (HFDb). Asterisk in A at 100 kDa indicate the Tamm-Horsfall Protein. P indicates pellet; SN, supernatant. Molecular weights are expressed in kilo Dalton.
The retained solution above 1,000 kDa (HFDa) recovered from the dialysis tube was centrifuged at 40,000 g and 200,000 g, respectively, as standard application of differential centrifugation method for a further vesicle concentration.
Immunodetection of TSG101, an established exosomal marker involved in their biogenesis16 (Fig. 2B), revealed that essentially the entire signal was detected in the HFDa fractions (Fig. 2B, lanes3–6) with a minimal loss on the dialysis membrane (Fig. 2B, lane7). No TSG101 signal was detected in the fraction from below 1,000 kDa (HFDb, lane8). This reflects efficiency of the HFD method.
Notably, the ultracentrifugation step, as in the conventional serial centrifugations, showed to be ineffective in fully recovering TSG101 -positive exosomes, as substantial signal was left in the supernatant (Fig. 2B, lane6).
Transmission Electron Microscopy and Tunable Resistive Pulse Sensing Analysis
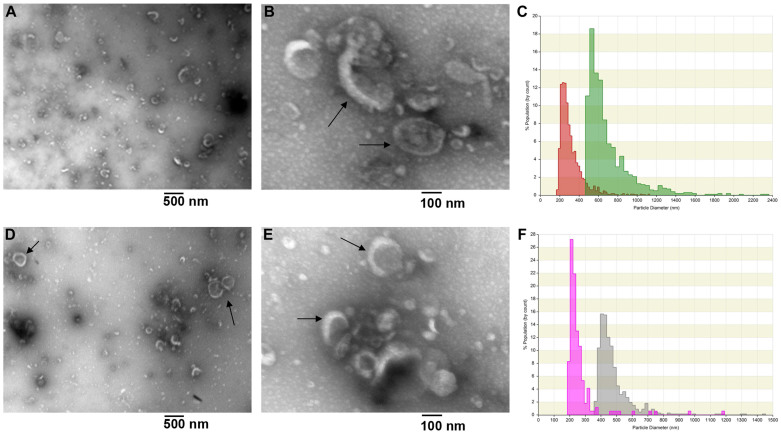
Transmission Electron Microscopy (TEM) analysis of the 40,000 g and 200,000 g pellets from the concentrated HFDa were used to visualise size and morphology of vesicles from HFDa (Fig. 3A,B,D,E). Notably, TEM pictures revealed a heterogeneous population of vesicles, including the ones 50–90 nm in diameter with cup shaped morphology, typical of exosome vesicles17 and similar structures with a wider diameter (arrows).
Figure 3. Transmission Electron micrographs and Tunable Resistive Pore Sensing size distribution of isolated urinary vesicles.
Enriched EVs adsorbed on formavar carbon-coated grids, fixed and negative stained by uranyl acetate. (A) Low magnification (x5000) of EVs recovered at 40,000 g pellet; scale bar 500 nm. (B) higher magnification (x25000) of 40,000 g pellet; scale bar 100 nm. (D) low magnification (x5000) of EVs recovered at 200,000 g; scale bar 500 nm. (E) high magnification (x25000) of 200,000 g pellet; scale bar 100 nm.Vesicle polydispersity of the two pellets was confirmed by TRPS analysis. (C,F) The measurements were performed on an Izon qNano. The polyurethane nano porous (NP) membrane sizes used for these experiments were rated for 100–400 nm (NP200) particles (green HFDa P40,000 g and red HFDa P200,000 g) and 200–800 nm (NP400) particles (gray HFDa P40,000 g and light purple HFDa P200,000 g). The measurements obtained were with a minimum of 1000 particles detected per measure. Wide range of particle sizes is obvious in both C and F.
We next used tunable resistive pulse sensing (TRPS) to measure the size distribution of vesicles18 using polystyrene nanobeads as a standard in the presence of 1% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)19 to favour the polydispersity, reduce vesicle aggregation and limit the blockage of the polyurethane membrane pores. The TRPS analysis confirmed the heterogeneity seen in TEM (Fig. 3C,F) and shows that our method efficiently catches all classes of vesicles beyond just exosomes.
Relative Quantification of TSG101 and THP Adsorption onto the Dialysis Membrane
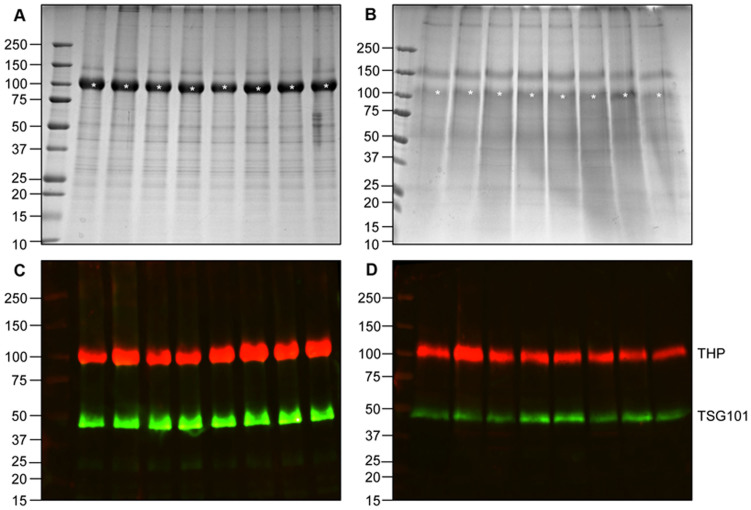
Adsorption of some soluble urinary proteins on top of exosomes, was established by the co-detection of markers Tamm-Horsfall Protein (THP) and TSG101 (Fig. 4B). Relative quantification of TSG101 and THP separately in HFDa, performed in 8 replicas, and in the dialysis membrane fraction (as extracted by 1% (w/v) SDS treatment), respectively, allowed us to estimate that THP and TSG101 integrated intensity (II) of SDS fractions (Fig. 4C) represents the 48% of THP and 18% of TSG101 signals detected for the respective HFDa fractions (Fig. 4C) loading 10 μg of total protein (BCA assay) per lane (Fig. 4 A,B).
Figure 4. Estimation of vesicle adsorption on the lumen of dialysis membranes.
(A, B) Colloidal Coomassie stained gel of HFDa and SDS elution of protein adsorbed on the surface of the membrane starting from 200 ml of urine, processed in 8 independent devices. Gel loading was based on the BCA assay with 10 µg of protein per lane. C and D, the respective gel as above immunostained for TSG101 and THP. Both membranes run from same experiment and at the same laser intensity set at a value to reach the limit of saturation point of the most abundant band. (TSG101: 153.69 ± 30.97 Kilo Integrated Intensity (KII), limits 190.06–138.14; CV 15.5%; THP 237.60 ± 80.09 KII, limits 332.57–127.28, CV 25.7%) and SDS dialysis membrane fraction (TSG101: 27.93 ± 11.22 KII, limits 46.78–16.05 CV 6.1%; THP 104.15 ± 62.90 KII, limits 267.05–62.56 CV 20.0%). Asterisk at 100 kDa indicates the Tamm-Horsfall Protein. Molecular weights are expressed in kilo Daltons.
Comparative performance of differential centrifugation protocol vs hydrostatic filtration dialysis
Two liters of first morning urine (pooled, several donors) was collected and split in two fractions of 1l each and further processed by HFD and differential centrifugation protocol15, respectively, with few modifications. Firstly, a centrifugation step at 2,000 g was introduced before the 17,000 g and both pellets were then treated with DTT and re-centrifuged at 17,000 g (Fig. 5A, B). Secondly, to test what was left in the SNs obtained after DTT treatment, SNs originated from the DTT treatment were not combined with the very first SN15. Finally, to check if vesicles were still present in the final DTT_SN200,000 g and SN200,000 g of the differential centrifugation approach, SNs were treated by HFD and, in parallel, the 1l HFDb underwent ultracentrifugation at 200,000 g to check potential vesicle loss.
Figure 5. Differential centrifugation protocol and HFD protein pattern and vesicle recovery.
Coomassie gels of all the fractions obtained from pellet 2,000 g (A), differential centrifugation protocol15 (C) and hydrostatic dialysis. (E). Panels B, D and F are the representative immunodetections of TSG101 as per A,C,E. Ten µg of protein (Bradford assay) were loaded per lane. Lane 1 crude urine, Lane 2, SN 2,000 g; Lane 3, Pellet 2,000 g; Lane 4, P2,000g_DTT_P17,000g; Lane 5, P2,000g_DTT_P200,000g; Lane 6, 4 P2,000g_DTT_SN 200,000 g; Lane 7, P17,000g; Lane 8, P17,000g_DTT_ P17,000g; Lane 9, P17,000g_DTT_ P200,000g; Lane 10, P17,000g_DTT_ SN 200,000g HFDa; Lane 11, P17,000g_DTT_ SN 200,000g HFDb; Lane 12, P200,00g; Lane 13, SN200,000g; Lane 14, SN200,000g HFDa; Lane 15, SN200,000g HFDb; Lane 16, HFDa; Lane 17, HFDa P17.000g; Lane 18, HFDa P200,000g; Lane 19, HFDa SN 200,000g; Lane 20, HFDb; Lane 21, HFDb P 200,000g; Lane22, HFDb SN 200,000g. (G) Plot of the protein amount recovered in each fraction (Bradford assay: Lane 1 653.0 ± 26.9 μg; Lane 7 413.3 ± 15.6 μg; Lane 8 1.79.5 ± 7.5 μg; Lane 9 71.0 ± 10.6 μg; Lane 10 93.1 ± 26.2 μg; Lane 12 1673.7 ± 116.6 μg; Lane 14 6.265.7 ± 84.1 μg; Lane 16 2194.2.20 ± 266.6 μg; Lane 17 447.6 ± 63.2 μg; Lane 18 884.7 ± 175.2 μg; Lane 19 355.2 ± 38.6 μg; Lane 21 391.3 ± 46.9 μg). (F) Fluorescence Integrated Intensity (II)of TSG101 signal ± standard deviation of 3 independent western blots (Supplemental Fig. 2) per each fraction (TSG101: Lane 1 0.91 ± 0.24 Kilo Integrated Intensity (KII), limits 1.14-0.6; CV 24.9%; Lane 2 2.45 ± 0.30 KII, limits 2.34-2.6, CV 12.1%; Lane 4 1.04 ± 0.26 KII, limits 0.68–1.3 CV 25.3%; Lane 7 8.15 ± 0.66 KII, limits 7.31–8.92 CV 8.1%; Lane 8 12.04 ± 1.44 KII, limits 10.23–13.76 CV 12.0%; Lane 9 26.82 ± 3.63 KII, limits 22.96–31.69 CV 13.6%; Lane 10 6.52 ± 0.71 KII, limits 5.86–7.50 CV 10.9%; Lane 12 16.13 ± 1.29 KII, limits 14.79–17.87 CV 8.0%; Lane 13 3.02 ± 0.526 KII, limits 2.42–3.68 CV 17.1%; Lane 14 18.61 ± 3.45 KII, limits 15.89–23.38 CV 18.6%; Lane 16 29.20 ± 2.31 KII, limits 26.44–32.09 CV 7.9%; Lane 17 31.41 ± 4.90 KII, limits 24.50–35.27 CV 15.6%; Lane 18 21.50 ± 1.97 KII, limits 19.43–24.14 CV 9.2%; Lane 19 0.96 ± 0.66 KII, limits 6.09–7.63 CV 9.5%). P pellet; SN supernatant. Molecular weights are in kilo Daltons.
Figure 5 resumes the results of this comparative analysis. Bradford protein assay (Fig. 5G) and immunodetection of TSG101 (Fig. 5H) were used to estimate the yield of vesicles (total protein amount) and exosomes (TSG101 II) respectively, in the two approaches. All in all, HFD permitted a superior recovery of vesicles (Fig. 5E and F lanes16,17,18,19) in a shorter time and inexpensive setup (see cost comparison in Table 1) than the conventional differential centrifugation protocol. For comparison, the conventional method has exosomes (TSG101 positive) in each fraction studied (Fig. 5D lanes7,8,9,10,12,13,14) to obscure its full efficiency in the recovery. The introduction of the centrifugation step at 2,000 g allowed elimination of the bulk of THP without losing exosomes. In fact, a minimal signal of TSG101 for exosomes was recovered in the 17,000 g pellet but not in the 200,000 g pellet after DTT treatment. Thus, the origin of TSG101 is most likely due to epithelial cells, cell debris or macrovesicles centrifuged in the first spin rather than exosomes entrapped in the THP filaments. Moreover, when lanes were loaded with the same amount of protein it was still possible to see and recover TSG101 positive vesicles in the pellet at 17,000 g after DTT treatment (Fig. 5D lane 8) showing a protein pattern very similar to the fraction recovered at 200,000 g after DTT (Fig. 5D lane 8). Finally, ultracentrifugation failed to fully recover all exosomes in both protocols with an important signal left in the final SN (Fig. 5 D,F lanes13,19) and despite the denaturation by DTT, THP is still recovered in both the ultracentrifugation pellet and HFDa (Fig. 5C lane9,10).
Table 1. Comparative performance of HFD vs differential centrifugation technique.
| Hydrostatic Filtration dialysis | Differential centrifugation | |
|---|---|---|
| Time | 9 hoursa,d | 40 hours (~5 working days)a,d |
| 27 nonstop hoursb,d | 30 hours (~4 working days)c,d | |
| Implementation costse | < €250 | ~ €130,000 |
| Estimated cost per samplef | €1.5 | €15 |
aTime to process 8 samples 200 ml each including differential centrifugation for HFD.
bTime to process 1l of urine in 1 l separating funnel device and 25 cm length membrane dialysis including differential centrifugation for HFD.
cTime to process 1l of urine by differential centrifugation.
dUtilising polycarbonate tubes (max volume 16.5 ml per tube) and Beckman 70 Ti rotor at 200,000 g, 2 hours per run.
eSet up investments.
fCost estimate based on Direct consumables without labour or investment costs.
Differential Centrifugation of HFDa and Pellet Recovery from HFDb
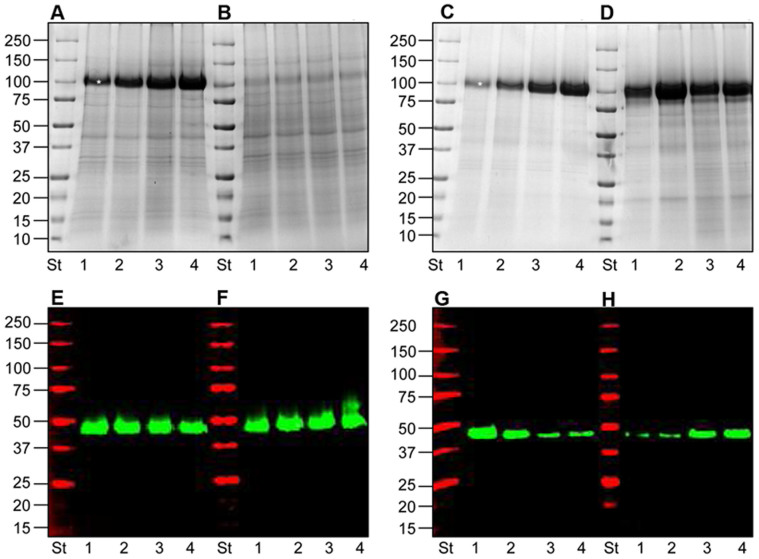
A systematic evaluation of centrifugation force effects of the conventional method vesicle recovery revealed that vesicles started to sediment already at 5,000 g and up to 40,000 g (Fig. 6A lane2,3,4). As THP was found deleterious for UEV recovery, we decided to set the speed at 40,000 g which allowed us to simply recover the bulk of vesicles including exosomes (positive for TSG101 and programmed cell death 6-interacting protein, ALIX,) and other vesicle markers: dipeptidyl peptidase 4 (DPP4), nephrilysin (NEP) and podocin (NPHS2) (Fig. 6A,B,C,D).
Figure 6. Differential centrifugation of HFDa and HFDb fractions.
(A) Coomassie gel of all the fractions isolated by centrifugation at 5,000 g, 20,000 g, 40,000 g and 200,00 g (B,C,D) Immunodetection of the exosome marker protein TSG101 (Panel B red), ALIX (Panel B green), DPP4 (panel D green), NEP (Panel C red), Podocin (Panel C green) and human serum albumin HSA (panel D red) from the same gel as A. Five µg of protein (Bradford assay) were loaded per lane: Lane 1, HFDa; Lane 2, HFDa P5,000g; Lane 3, HFDa P20,000g; Lane 4, HFDa P40,000g; Lane 5, HFDa P200,000g; Lane 6, HFDa SN200,000g; Lane 7, HFDb; Lane 8, HFDb P200,000g; Lane 9 HFDa SN200,000g. P indicates pellet; SN, supernatant. Molecular weights are expressed in kilo Dalton.
Exosomes and other UEVs are extensively recovered in the early stage while exosomes are left in late centrifugation steps
The final SN200,000 g (Fig. 6B lane6) showed still signal for TSG101 and ALIX but none for the other antigens, implying presence of a specific subpopulation of exosomes; usually discarded. This shows that, due to their physical characteristics, the exosomes are well retained in the HFDa and, as found above, that exosomes are extensively recovered in the earlier centrifugation steps while the wealth of the other vesicle markers are missing in the SN200,000. Thus, the HFDa fraction can be directly used also without further steps as a mixed UEV population.
HFD process does not lose exosomes in the flow-through
To check whether hydrostatic dialysis flow-through fraction (HFDb) leads to loss of exosome vesicles, a more deep analysis was done after steps up to 200,000 g centrifugation. Notably, none of the vesicle markers were detected (Fig. 6B,C,D lane8) while important soluble proteins like human serum albumin (HSA, Fig. 6D) in the 200,000 g pellet was seen. This shows that vesicles were not lost in the HFD process and the flow-through mostly contains common urinary proteins, like HSA, which precipitates at high speed (Fig. 6D lane8) and it becomes an important interfering element for the analysis of proteinuric samples.
The early HFDa fraction efficiently catches all UEV groups
Efficiency of the hydrostatic dialysis system to retain EV-associated proteins and to remove soluble proteins was further investigated by systematic immunodetection of some of the main vesicle proteins20, (Supplemental Figs. 3 and 4). As no substantial vesicles were found in the HFDb, profiling of the UEV associated proteins was performed only on HFDa fractions (Supplemental Fig. 3). This screening included tetraspanin, CD63, internal EV "cargo" proteins including ubiquitin-containing proteins, β-actin, glyceraldeyde-3-phosphate dehydrogenase (GAPDH) as well as proteins of the most proximal urogenital site, visceral epithelial cells (podocyte) of glomerular filtration barrier.
Most of interfering soluble proteins and THP are lost in the HFD process
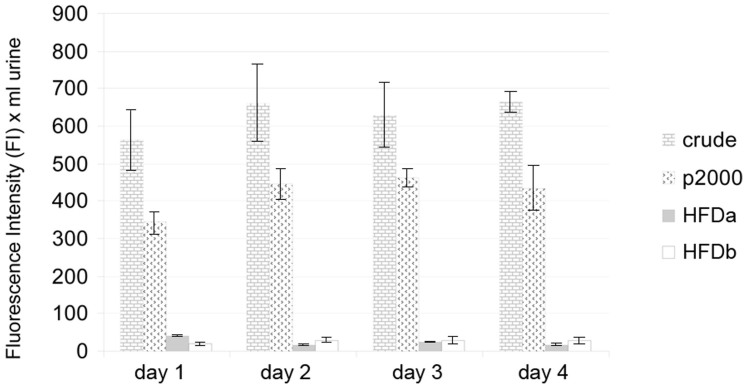
The selectivity of our method was established by detection of α1-glycoprotein acid (ORM) - which is not listed in the published EV proteomes6,20 and showed a full recovery in the HFDb (Supplemental Fig. 4, lane7). Despite the high membrane molecular weight cut-off in the process used, THP was still present in the HFDa. A relative quantitation was performed in the whole urine (crude), P2000g, HFDa and HFDb fractions by fluorescent intensity analysis of a Coomassie gel15 using the Odyssey laser scanner. Results were normalized for 1 ml of crude urine (Supplemental Fig. 5). From the fluorescent II we calculated that, on average 4.0% of THP is retained in the HFDa while an estimated 66.9% of THP sediments at 2,000 g with 4.2% left in HFDb (Fig. 7).
Figure 7. Relative quantification of THP in crude urine, P2000g, HFDa and HFDb fractions.
The average amounts of THP deduced from densitometry of Coomassie-stained gels (triplicate are represented in Supplemental Fig. 5). Fractions were loaded by volume (crude urine 50 μl, P2000 and HFDa equivalent to 100 μl and 1.5 ml of crude urine, respectively; HFDb 100 μl). Fluorescent integrated intensity (II) ± standard deviations were referred to the equivalent of 1 ml of urine (Day 1 crude 563.4 ± 82.03 Kilo integrated intensity (KII) limits 465.2–666.6 CV 14.6%; P2,000g 342.7 ± 31.5 KII limits 303.7–343.5 CV 9.2%; HFDa 49.1 ± 1.0 KII limits 40.1–42.4 CV 2.3%; HFDb 19.2 ± 3.1 KII limits 16.6–23.6 CV 16.3%; Day 2 crude 662.1 ± 103.0 KII limits 516.0–740.2 CV 14.6%; P2,000g 445.4 ± 40.3 KII limits 401.8–499.0 CV 9.1%; HFDa 17.8.5 ± 1.3 KII limits 16.7–19.7 CV 7.4%; HFDb 30.1 ± 5.6 KII limits 26.1–38.0 CV 18.6%; Day 3 crude 629.6 ± 86.9 KII limits 515.8–726.6 CV 13.8%; P2,000g 462.2.3 ± 24.8 KII limits 429.5–489.6 CV 5.4%; HFDa 24.25 ± 1.3 KII limits 23.1–25.9 CV 5.3%; HFDb 28.8 ± 10.2 KII limits 14.4–36.8 CV 35.4%; Day 4 crude 663.7.0 ± 27.9 KII limits 638.8–702.6 CV 4.2%; P2,000g 434.6 ± 59.0 KII limits 351.9–485.6 CV 13.6%; HFDa 17.3.5 ± 1.0 KII limits 16.9–18.6 CV 5.9%; HFDb 28.2 ± 7.2 KII limits 21.2–38.1 CV 25.6%).
Hydrostatic Filtration Dialysis is Superior to Differential Centrifugations Over a Wide Range of Sample Volumes
Having exhaustively characterised the method to enrich UEVs from a diluted solution we next tested its performance on different volumes of urine, an essential practical parameter, in eight replicas for each volume (15, 50, 100 and 200 ml). UEVs were first enriched from HFDa fraction after the initial 2000 g centrifugation and then HFDa was processed by differential centrifugation as a simple application of HFDa and to crosscheck further vesicles yield in supernatant and pellets. Vesicle recovery at each step was assessed by protein concentration measured by two methods, the Bradford and Bicinchoninic acid (BCA) assays. The BCA yielded as much as 2.5 ± 0.5 times more protein compared to Bradford (Supplemental Figs. 6A,7A,8A,9A). Presence of glycoproteins, like THP and urinary pigments are known to distort protein concentration analysis21,22.The coefficient of variation (CV) of the protein yield ranged between 30% (15 ml urine) to 12% (200 ml urine) with these two methods. This shows that starting with larger volume helps to decrease technical variability.
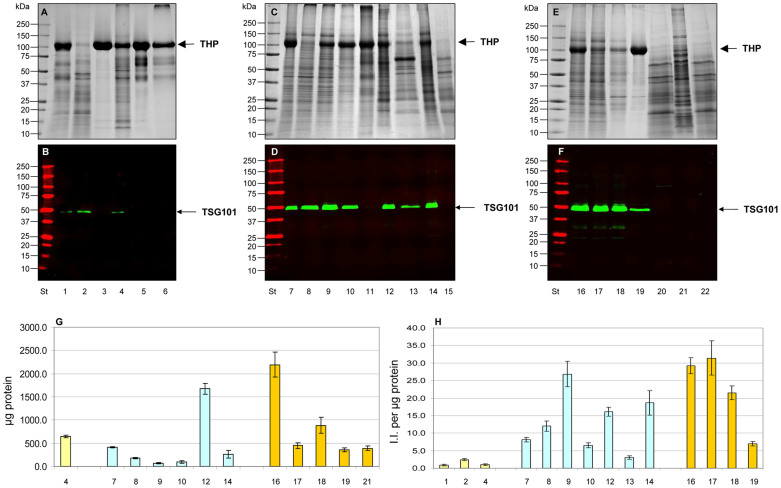
Figure 8 sums up the protein patterns obtained from each fraction (Fig. 8A,B,C,D) including TSG101 (Fig. 8E,F,G,H) detection as the marker of exosomes. Full set of results from the replicas can be seen in Supplemental Figs. 6,7,8,9.
Figure 8. Efficiency of hydrostatic filtration dialysis to enrich vesicle from different urine volumes.
Protein staining (Bradford, 5 µg per lane) of different urine volumes of vesicles obtained from HFDa (A), HFDa P40,000g (B); HFDa P200,000g (C), HFDa SN200,000g (D) and the respective immunostaining for exosomes (TSG101) in E,F,G,H. Lanes 1 to 4 contain 15, 50, 100 and 200 ml starting volume, respectively. Asterisk (*) in A and C at 100 kDa indicates the Tamm-Horsfall Protein. Molecular weights are expressed in kilo Dalton.
Concentrated HFDa pellets collected at 40,000 g (Fig. 8 and Supplemental Fig. 7B) proved once again to be an interesting fraction which enriches many types of UEVs with minimal interference from THP.
Ultracentrifugation leads to incomplete recovery yield of exosomes
When tested over a wide range of starting volumes (15–200 ml), HFDa solutions were, surprisingly, found to give a superior recovery of TSG101 positive UEVs in the 40,000 g (Fig. 8F, lane1–4). pellet over that of 200,000 g (Fig. 8G, lane1–4) independently from the starting volume. Our method was found to be independent of artefacts by THP which is mostly removed in the early steps of the HFD process (P2,000g). Independently from the starting volume our results also show that with the conventional centrifugations there is still a population of exosomes which do not sediment even at high centrifugation forces (Fig. 8H, lane 1–4)23 and are easily lost from subsequent analyses.
In summary, HFD showed to efficiently enrich vesicles in a more exhaustive way than differential centrifugation protocol in which an important fraction of exosome vesicles are lost in the final supernatant. Moreover, HFD resulted to a vastly superior cost-efficiency with a faster workflow than differential centrifugation as summarised in Table 1.
Hydrostatic Filtration Dialysis Yields Good Quality miRNA
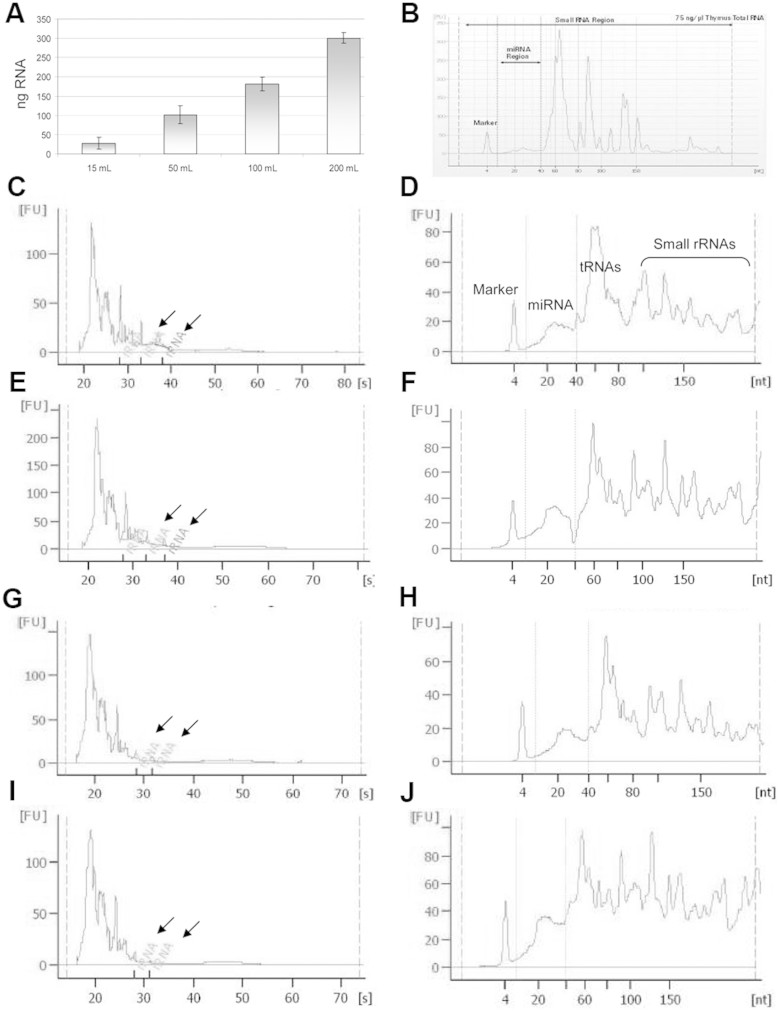
The shuttling of distinct RNA species, especially miRNA, within exosomes from the site of release to distant targets has been firmly established24. The exosomal RNA patterns shown thus far, however, differ significantly in respect to the method used for RNA extraction25. Here we adopted a column-based method specifically designed to enrich RNA from urinary exosomes. The RNA quality extracted from the HFDa fraction was investigated using the Agilent 2100 Bioanalyzer with 2 different chips dedicated to analyse small amounts of total RNA (Fig. 9C,E,G,I) and small RNA (<150 nt) (Fig. 9D,F,H,J). The analysis for total RNA showed enrichment below 1000 nt and absence of the ribosomal RNA (arrows in Fig. 9C,E,G,I). This can be considered as a sign of degradation. However, “degraded” RNA seems to be a physiological feature of the RNA contained in EVS26.
Figure 9. Isolated RNA yield and profile.
(A) Recovered RNA quantity was analyzed using the NanoDrop 1000 spectrophotometer (Thermo Scientific). RNA quality was assessed using the Agilent 2100 Bioanalyzer. (B) Illustrates an example small RNA chip profile (http://www.chem.agilent.com/Library/technicaloverviews/Public/5989-7002EN.pdf). Representative electropherograms of 10 ng of RNA extracted from HFDa obtained from 15, 50,100 and 200 ml of urine respectively were analysed using Pico RNA Chip and show an absence of rRNA peaks (arrows) (C,E,G,I). Electropherograms for small RNA species (D,F,H,J) extracted from HFDa obtained from 15, 50,100 and 200 ml of urine respectively and analysed using the small RNA Chip (Agilent) show the size distribution of small RNAs in interval of 10–150 nucleotides (nt) including microRNA in the size between 10 and 40 nt. The average concentration of the miRNA yield was, as based on the Bioanalyzer results, 4.58 ± 1.33 ng/µl (2.71–7.43 ng/µl, CV = 29%) per 10 ng/µl of total RNA loaded, corresponding to an average of 37.6% of miRNA in respect to small RNA amount.
A further analysis with the chip for small RNA revealed a typical electrophoretogram in repeated experiments (Fig. 9D,F,H,J). Accordingly, within the small RNA fraction (6–150 nt), miRNA with a size distribution of 10–40 nt was clearly detected.
Discussion
The explosion of interest into extracellular vesicles (EVs) continues, including the study of exosome patterns during diabetic nephropathy13,14. However, serious obstacles associated with the non-standardized EV isolation methods remain unresolved, thus limiting their full utilization. Here we provide a simple, inexpensive solution (Table 1) to replace the widely used method of serial ultracentrifugations. A better UEV recovery was achieved with our simple hydrostatic dialysis method. The results show its excellent performance and yield of both target indicator proteins and distinct RNA species, fully validating the usefulness of the method. With the key performance parameters provided, this hydrostatic dialysis method should become the golden standard for EV enrichment for analytics and/or starting point for discovery research.
The nomenclature of UEVs has not been firmly defined and, consequently, optimized standard isolation protocols are yet to evolve. This is mainly due to the overlapping biophysical and biochemical properties27 of UEVs, which can be divided into two main categories reflecting their release pathways28.
Herein we systematically and repeatedly outlined our optimized and versatile UEV isolation method to combat various procedural challenges including the non-availability of costly ultracentrifuges, expert trained personnel in the operation of highly specialized equipment such us an ultracentrifuge and concentration needs for sample storaging. Notably, we used here the conventional isolation protocol in parallel – the differential centrifugation- to allow performance comparisons.
A simple low-speed tabletop centrifugation (at 2,000 g) after urine collection was needed to remove cellular debris, bacteria and an estimated 67% of Tamm-Horsfall protein as urinary casts or large THP polymers. While THP may also entrap vesicles15 a low signal of the exosomal marker TSG101 (Fig. 5A lane 4) was seen in this fraction, due most likely to epithelial cells and/or debris therein while no TSG101 signal was detected in the 200,000 g pellet (Fig. 5B lane 5). Therefore we can assume vesicles are not lost at this step as entrapped within THP polymers. However, THP precipitated following the differential centrifugation protocol although subjected to denaturation by DTT (Fig. 5A lanes 4 and5; Fig. 5C lane 9).
The simple hydrostatic filtration/concentration/dialysis step (at MWCO of 1,000 kDa) showed the efficiency of the cellulose ester -membrane meshwork of the dialysis tube to yield a variety of UEVs. With the average of 75 ml per hour of urine filtered in multiple dialysis columns of 15 cm, a large number and volume (up to 1000 ml per device) of samples can be processed by this process by non-trained personnel in a single day (24 hours). However, with larger than 200 ml of sample volume the HFD system shows minor efficiency decrease mostly due to adsorption of soluble proteins while, to a lesser extent, exosomes in the dialysis tube (Fig. 4).
No signal for vesicle markers TSG101, ALIX, DPP4, NEP and podocin were recovered in HFDb (Fig. 6). Notably, even "enforced" ultracentrifugation at 200,000 g in which considerable amount of human serum albumin (HSA) was still recovered in the pellet (Fig. 6 lane 8). This reinforces the usefulness of hydrostatic dialysis to remove soluble proteins even before the set of differential centrifugations. As far as THP is concerned an estimated 4.0% of initial amount was recovered in the HFDa fraction while the 67% of THP precipitated with the first spin (Fig. 7) without loss of TSG101 positive exosome vesicles.
Concentration of urine is a recognized bottleneck for UEV isolation and use of e.g. nano-membrane concentrators (with polyethersulfone membrane)29,30,31 and microfiltration disc membranes (with hydrophilized polyvinylidene difluoride)32 have been proposed. However, these characteristically react with any soluble proteins and UEVs both clogging the respective nanomembranes. This interaction also occurs in a minor degree in our HFD system with large volumes of urine (Fig. 4); however, HFD is a much gentler way of concentrating samples and with the added benefits of simultaneous dialysis and a vastly superior cost-efficiency.
Here we routinely handled up to 1000 ml of single urine samples in a single device without considerable distortions in the protein patterns between different experiments. Furthermore, collection and analysis of the void first and second morning urines during four consecutive days showed a qualitative good intra-individual and inter-day repeatability (Supplemental Fig. 5). Moreover, the dialysis step allowed normalization in terms of levelling the physical-chemical parameters of HFDa fractions by eliminating all the analytes in the urine below the desired MWCO. This is an additional point of superiority of our HFD system over the conventional method in which the yield is greatly influenced by variations in viscosity, protein content and hydration status of the sample provider33,34,35.
The volume of urine needed for analytical purposes and sample storage is an important practical challenge. Accordingly, we tested here the performance of the HFD approach utilizing different urine volumes. No appreciable differences were seen in the protein pattern of the EVs recovered in the HFDa (Fig. 8 and Supplemental Fig. 6) or from pellets obtained after the conventional differential centrifugation protocol (Supplemental Figs. 7 and 8). Differential centrifugation was compared with HFDa keeping in mind all these aforementioned challenges39,40,41, with results overwhelmingly supporting the use of HFD. Furthermore, our method showed an excellent repeatability of protein patterns from all different fractions (HFDa, HFDa P40,000g, HFDa P200,000g and HFDa SN200,000g). The coefficient of variation in the protein amount ranged between 30% and 12% (Supplemental Figs. 6A,7A,8A,9A). In general, starting from larger volumes we obtained lower CVs. Moreover, results of small RNA extraction experiments confirm the excellent performance of the HFD also for transcriptomic studies.
In conclusion, the hydrostatic dialysis method introduced here is a highly efficient and dynamic method to enrich EVs from urine, with the additional benefit of efficient pre-processing and concentration of the samples, e.g. for biobanking purposes in one step. Moreover, the retained solution above 1,000 kDa of urinary samples appeared to be an excellent starting material of UEVs for any further applications like the conventional differential centrifugation protocols and preconcentration step for density ultracentifugation or size exclusion chromagraphy and for RNA extraction. Finally, the simplicity of the system and its function can be of great help to monitor potential biomarkers expressed in very low amount as a daily clinical routine. In the last decades the discovery phase aimed to dig up what was called the “rare proteome” and/or more in general, molecules expressed at very low level. UEVs provide an ideal platform for this. However, the lengthy process of vesicle isolation has prevented full utilization of UEVs for the day-to-day clinical monitoring of the patient. Thus, HFD, and its performance parameters appear as perfect fit for assessment of biomarkers in a short time and without the use of expensive detection systems. Potentially, following the simple instructions on how to operate HFD system patients themselves can easily be the end-users in the form of daily monitoring with dedicate point of care tests.
Methods
Urine samples
Urine samples were collected from healthy volunteers among the laboratory staff, aged 20–44 (N = 4), First morning void urine was processed within 3 h without adding protease inhibitors. Urine was anonymously labelled, tested with Combur 10 TestD Dipsticks (Roche Diagnostic, Basel, Switzerland) and pool together. Additionally, one volunteer assented to collect first and second morning urines during four consecutive days. Written informed consents were obtained from all participants. This study was approved by The Ethical Committee of Dublin City University. All experiments were performed in accordance with the declaration of Helsinki.
Vesicle purification
Hydrostatic Filtration Dialysis (HFD): A schematic representation of the methodology used for vesicle isolation is shown in figure 1. Pooled urine samples (50 ml per tube) were centrifuged at a Relative Centrifugal Force (RCF) of 2,000 g calculated at average radius of 100 mm in a swing bucket rotor Benchtop Universal 320 centrifuge (Hettich Zentrifugen, Tuttingen, Germany) for 30 min at room temperature (RT) (without braking). The supernatant (SN) ~ 0.5l was poured in a separating funnel connected with a dialysis membrane made of cellulose ester (CE) with molecular weight cut-off (MWCO) of 1,000 kDa (Spectra/Por Biotech MWCO 1,000,000 MWCO Catalogue number 131486; Spectrum Laboratories, Ca) (Supplemental Fig. 1).
The hydrostatic pressure of the urinary solution in the funnel pushes the solvent (water) through the mesh of dialysis membrane (filtration), together with all the analytes below the selected MWCO. After the first step resulting in sample concentration, the separating funnel was refilled with 200 ml of deionised filtrate (0.22 µm) water (R ≥ 18.2 MΩ·cm, mQ water) to rinse away remaining analytes below the MWCO until the volume of 5–8 ml of volume is reached. This filtration-concentration-dialysis process is called “hydrostatic filtration dialysis” (HFD).
HFD and differential centrifugation:The retained solution above the 1,000 kDa cut-off (HFDa) (5 ml) was then centrifuged at 5,000 g, 20,000 g and/or 40,000 g calculated at maximum radius 105 mm of a fixed angle JA-20 rotor (clearing factor or k factor = 770) (Beckman Coulter, Fullerton, Ca) for 1 h at RT. The retained 40,000 g supernatant (SN) fraction (5 ml) was then ultracentrifuged at 200,000 g calculated at maximum radius 91.9 mm of 70 Ti fixed-angle rotor (k factor = 44) (Beckman Coulter) for 2 h (RT) using a Beckman XL-80 Ultracentrifuge (Beckman Coulter). All the pellets were re-suspended in mQ water. For a pilot study, HFDa from the starting urinary volumes of 15, 50, 100 and 200 ml were concentrated to 3 ml. After determining the protein concentration, an equal amount of total protein was loaded in polycarbonate centrifugation tubes (3 ml). Ultracentrifugation was performed at 200,000 g calculated at the maximum radius 82.0 mm of 70.1Ti fixed-angle rotor (k factor = 36) (Beckman Coulter) for 2 h at RT.
Differential centrifugation and HFD: Comparative analysis was performed according to Fernández-Llama and colleagues15. Pellets from 2,000 g and 17,000 g were resuspended in 10 ml of 250 mM sucrose, 10 mM triethanolamine pH7.6 and 200 mg/ml of DTT for 10 minutes at 37°C vortexing every 2 minutes. Centrifugations at 17,000 g (42 ml per tube of urine and 10 ml of dithiothreitol (DTT) fraction) were performed in a fixed angle JA-20 rotor (clearing factor or k factor = 770) (Beckman Coulter) for 30 min at RT. RCF were calculated at average radius of 70 mm. Ultracentrifugations (16,5 ml urine per tube and 10 ml of DTT fraction) were performed at 200,000 g calculated at maximum radius 91.9 mm of 70 Ti fixed-angle rotor (k factor = 44) (Beckman Coulter) for 2 h (RT) using a Beckman XL-80 Ultracentrifuge (Beckman Coulter). All the final pellets were resuspended in 1 ml of purified water. The final SNs were poured in HFD system and processed as described above. Conversely, the urine solution below the 1,000 kDa cut-off (HFDb) was ultracentrifuged (16.5 ml per tube) at 200,000 g calculated at maximum radius 91.9 mm of 70 Ti fixed-angle rotor (k factor = 44) (Beckman Coulter) for 2 h (RT) using a Beckman XL-80 Ultracentrifuge (Beckman Coulter).
Protein assay, Gel electrophoresis and Western blot
Protein quantification was determined by Coomassie36 and bicinchoninic acid (BCA)37 microassays. Protein amounts were first dried by vacuum concentration and then resuspended in 7 M Urea, 2 M thiourea, 5% (w/v) SDS, 40 mM Tris-HCl, pH 6.8, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 20% (v/v) glycerol and 50 mM dithiothreitol (DTT) in a ratio of 0.25 μg of protein per μl of solution38. Protein denaturation was obtained after an overnight (ON) incubation at RT. Proteins were separated by SDS-PAGE39 and stained with colloidal Coomassie G-25040 or transferred to 0.45 μm nitrocellulose membrane (Whatman, Springfield, UK)41. For Western blotting, membranes were saturated with Odyssey blocking buffer (LI-COR Biosciences, Lincoln, MA) and incubated with specific antibody according to manufacture instructions: Anti-CD63 (Santa Cruz Biotechnologies, Santa Cruz, CA); anti-ALIX (Thermo Scientific, Waltham, Ma), anti-tumour suppressor gene (TSG101), anti-β-actin and anti-podocin (Sigma Aldrich; Dorset, UK); anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Abcam, Cambridge, UK); anti dipeptidyl dipeptidase 4 (DDP4 and anti-neprilysin (NEP) (R & D System Inc., Minneapolis, MN); anti-human IgA, polyclonal Rabbit anti-human alpha-1-antitrypsin, anti-human alpha-2-macroglobulin; anti-ubiquitin, anti-IgG, anti-IgM, polyclonal rabbit anti-human serum albumin (HSA) (Dako, Glostrup, Denmark), mouse anti-HSA (Medix Biochemica, Kauniainen, Finland) anti-non-catalytic region of tyrosine kinase adaptor protein 1 [Nck1] and rabbit anti-Tamm-Horsfall glycoprotein were obtained from University of Helsinki, Finland. After 6 washes in PBS-Tween (0.1%, v/v), membranes were incubated with infrared dye-coupled secondary antibody (LI-COR Biosciences); 1:5000 1 hour at RT. Acquisition of the fluorescent signal was performed by Odyssey infrared imaging system (LI-COR Biosciences). Determination of molecular weight of all bands of interest and quantification of the signal were performed by Odyssey Infrared Laser Scanner software (LI-COR Biosciences).
Transmission Electron Microscopy (TEM)
Vesicle preparations (50 μg) were fixed with 1% (v/v) glutaraldehyde in phosphate buffer solution (PBS). Fixed preparations were then spotted onto a Formvar/Carbon 300 mesh grid and dried at room temperature. The grids were washed twice with PBS and stained with 5% (w/v) uranyl acetate in water for 10 min. After staining, samples were imaged by JEM-2100 TEM (Jeol Ltd., Tokyo, Japan).
Tunable Resistive Pulse Sensing
qNano measurements were performed according to standard procedures18. Polyurethane tunable nanopores membrane NP400 (200–800 nm) and NP200 (100–400 nm) were placed on qNano (Izon Ltd., Christchurch, New Zealand) and stretched open 47 and 45 mm respectively. Electrolyte solution was made of 1% (w/v) 3-[(3-Cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)19 in PBS and placed in both sides of the fluid cell. Current pulse signals were collected using IZON proprietary software (Izon Ltd.). Blockade counts setting in this study was ranged at 1000 events. Calibration was made using standard polystyrene particles of 400 (CPC400) and 200 (CPC200) nm of diameter (Izon Ltd.).
RNA extraction and analysis
A Urine Exosome RNA isolation Kit (Norgen Biotek, Thorold, Canada) was used for RNA extraction according to the manufacturers' instructions. RNA quantity and quality was determined spectrophotometrically by Nanodrop ND-1000 and by capillary electrophoresis (Agilent 2100 Bioanalyzer, Agilent technologies, Foster City, Ca). Total RNA was analysed with the Agilent 6000 Pico kit and Small RNA kit (Agilent technologies) according to the manufacture's protocol, loading 10 ng of RNA per well.
Author Contributions
L.M. designed the study and developed the method. L.M., D.T., D.G., A.B.M., G.C., S.A. performed experiments and analysed data. L.M. and H.H. interpreted the results and wrote the manuscript. HH supervised the project.
Supplementary Material
Supplementary figures
Acknowledgments
This study was supported by The Irish Health Research Board HRB grant number HRA/09/62, European Union funded programs "Urosense" (IAAP-GA-2011-286386) and "KidneyConnect" (Grant number 602422). The authors thank Professor Martin Clynes from National Institute for Cellular Biotechnology (NICB) for the access to ultracentrifuge and Bioanalyzer.
References
- Raposo G. & Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akers J. C., Gonda D., Kim R., Carter B. S. & Chen C. C. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retovirus-like vesicles, and apoptotic bodies. J. Neuooncol. 113, 1–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turturici G., Tinnirello R., Sconzo G. & Geraci F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: advantages and disadvantages. Am. J. Physiol. Cell Physiol. 306, C621–633 (2014). [DOI] [PubMed] [Google Scholar]
- Shifrin D. A. Jr, Demory Beckler M., Coffey R. J. & Tyska M. J. Extracellular vesicles: communication, coercion, and conditioning. Mol. Biol. Cell 24, 1253–1259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller G. Microvesicles/exosomes as potential novel biomarkers of metabolic diseases. Diabetes Metab. Syndr. Obes. 5, 247–282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra H. et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 10, e1001450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen-Heravi F. et al. Current methods for the isolation of extracellular vesicles. Biol. Chem. 394, 1253–1262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan M. C. et al. Subfractionation, characterization, and in depth proteomic analysis of glomerular membrane vesicles in human urine. Kidney Int. 85, 1225–1237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L. L. et al. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int. J. Biol. 9, 1021–1031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear J. W., Street J. M. & Bailey M. A. Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics 13, 1572–1580 (2013). [DOI] [PubMed] [Google Scholar]
- Cravedi P., Ruggenenti P. & Remuzzi G. Proteinuria should be used as a surrogate in CKD. Nat Rev Nephrol. 8, 301–306 (2012). [DOI] [PubMed] [Google Scholar]
- Duijvesz D., Luider T., Bangma C. H. & Jenster G. Exosomes as biomarker treasure chests for prostate cancer. Eur. Urol. 59, 823–831 (2011). [DOI] [PubMed] [Google Scholar]
- Burger D. et al. Urinary Podocyte microparticles identify prealbuminuric diabetic glomerular injury. J. Am. Soc. Nephrol. 25, 1401–1407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubiri I. et al. Diabetic Nephropathy induces changes in the proteome of human urinary exosomes as revealed by label-free comparative analysis. J. Proteomics 96, 92–102 (2014). [DOI] [PubMed] [Google Scholar]
- Fernández-Llama P. et al. Tamm Horsfal protein and urinary exosomes isolation. Kidney Int. 77, 736–742 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falguieres T. et al. In Vitro Budding of Intralumenal Vesicles into Late Endosomes Is Regulated by Alix and Tsg101. Mol. Biol. Cell. 19, 4942–4955 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Février B. & Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell. Biol. 16, 415–421 (2004). [DOI] [PubMed] [Google Scholar]
- Vogel R. et al. Quantitative sizing of nano/microparticles with a tunable elastomeric pore sensor. Anal. Chem. 83, 3499–506 (2011). [DOI] [PubMed] [Google Scholar]
- Musante L. et al. Biochemical and physical characterisation of urinary nanovesicles following CHAPS treatment. PLoS One 7, 37279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales P. A. et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 20, 363–79 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountoulakis M., Juranville J. F. & Manneberg M. Comparison of the coomassie brilliant blue, bicinchoninic acid and lowry quantitation assays, using non glycosylated and glycosylated proteins. J. Biochem. Biophys. Meth. 24, 265–274 (1992). [DOI] [PubMed] [Google Scholar]
- Sampson D. L. et al. The highly abundant urinary metabolite urobilin interferes with the bicinchoninic acid assay. Anal. Biochem. 442, 110–117 (2013). [DOI] [PubMed] [Google Scholar]
- Musante L., Saraswat M., Ravidà A., Byrne B. & Holthofer H. Recovery of Urinary Nanovesicles from Ultracentrifugation Supernatants. Nephrol. Dial. Transplant. 28, 1425–1433 (2013). [DOI] [PubMed] [Google Scholar]
- Valadi H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- Eldh M., Lötvall J., Malmhäll C. & Ekström K. Importance of RNA isolation methods for analysis of exosomal RNA: evaluation of different methods. Molecular Immunology 50, 278–286 (2012). [DOI] [PubMed] [Google Scholar]
- Batagov A. O. & Kurochkin I. V. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3'-untranslated regions. Biol. Direct 8, 12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Ostrowski M. & Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 (2009). [DOI] [PubMed] [Google Scholar]
- EL Andaloussi S., Mäger I., Breakefield X. O. & Wood M. J. Extracellular vesicles:biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 347–357 (2013). [DOI] [PubMed] [Google Scholar]
- Cheruvanky A. et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. Renal Physiol. 292, 1657–1661 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood I. M. et al. (2010).Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 78, 810–816 (2010). [DOI] [PubMed] [Google Scholar]
- Alvarez M. L., Khosroheidari M., Kanchi Ravi R. & DiStefano J. K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 82, 1024–1032 (2012). [DOI] [PubMed] [Google Scholar]
- Merchant M. L. et al. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin. Appl. 4, 84–96 (2010). [DOI] [PubMed] [Google Scholar]
- Momen-Heravi F. et al. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front. Physiol. 3, 162–167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman B. A. et al. The impact of temperature and urinary constituents on urine viscosity and its relevance to bladder hyperthermia treatment. Int. J. Hyperthermia 29, 206–210 (2013). [DOI] [PubMed] [Google Scholar]
- Burton-Opitz R. & Dinegar R. The viscosity of urine. Am. J. Physiol. 47, 220–230 (1918). [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Smith P. K. et al. Measurement of protein using bicinchoninic acid. Analyt. Biochem. 150, 76–85 (1985). [DOI] [PubMed] [Google Scholar]
- Rabilloud T. Detergents and chaotropes for protein solubilization before two-dimensional electrophoresis. Methods Mol. Biol. 528, 259–67 (2009). [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227, 680–685 (1970). [DOI] [PubMed] [Google Scholar]
- Candiano G. et al. Blue Silver: A very Sensitive Colloidal Coomassie G-250 Staining for Proteome Analysis. Electrophoresis 25, 1327–1333 (2004). [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T. & Gordon J. Electrophoretic Transfer of Proteins from Polyacrylamide Gels to Nitrocellulose Sheets - Procedure and some Applications.". Proc. Natl. Acad. Sci. U S A. 76, 4350–4354 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures