Abstract
PCR was employed to determine the presence of all known superantigen genes (sea, seq, and tst) and of the exotoxin-like gene cluster (set) in 40 Staphylococcus aureus isolates from blood cultures and throat swabs; 28 isolates harbored superantigen genes, five on average, and this strictly correlated with their ability to stimulate T-cell proliferation. In contrast, the set gene cluster was detected in every S. aureus strain, suggesting a nonredundant function for these genes which is different from T-cell activation. No more than 10% of normal human serum samples inhibited the T-cell stimulation elicited by egc-encoded enterotoxins (staphylococcal enterotoxins G, I, M, N, and O), whereas between 32 and 86% neutralized the classical superantigens. Similarly, intravenous human immunoglobulin G preparations inhibited egc-encoded superantigens with 10- to 100-fold-reduced potency compared with the classical enterotoxins. Thus, there are surprisingly large gaps in the capacity of human serum samples to neutralize S. aureus superantigens.
Staphylococcus aureus persists as a commensal microorganism in 10 to 30% of the population, but the organism is also a common cause of food poisoning and infections of different severity such as skin abscesses and wound infections, osteomyelitis, endocarditis, pneumonia, toxic shock syndrome, and staphylococcal scarlet fever (21). S. aureus is one of the most frequent causes of hospital-acquired infections, and the emergence and spread of multiresistant strains give rise to concern. The pathogenicity of S. aureus is multifactorial, and the versatility of this organism is underscored by recent clinical studies (4, 14, 16, 31, 37, 38).
Superantigens activate large subpopulations of T lymphocytes by directly cross-linking certain T-cell receptor Vβ domains with conserved structures on major histocompatibility complex class II molecules (32). They belong to the most potent T-cell mitogens known and can induce massive systemic cytokine release, leading to the symptoms of toxic shock syndrome (22). Among the virulence factors of S. aureus are the staphylococcal enterotoxins, the causative agents of food poisoning. They also act as superantigens. Whole-genome sequencing of several S. aureus clinical isolates has revealed that all 17 known staphylococcal enterotoxins (staphylococcal enterotoxins A to E and G to Q and toxic shock syndrome toxin 1) are encoded on mobile genetic elements together with other virulence factors (3, 18, 40). For example, the recently described enterotoxin gene cluster egc, which contains the five superantigen genes seg, sei, sem, sen, and seo, as well as two pseudogenes is located on the genomic island SaPI3 (18).
egc is special in that it functions as an operon and its genes are transcribed into a single polycistronic mRNA (13). In addition, a large cluster of up to 11 genes with sequence homology to superantigens has been discovered on the genomic island SaPI2; they have been termed staphylococcal exotoxin-like genes, or set (3, 10, 18, 39). For an overview of the organization and nomenclature of the set gene cluster, see supplemental Fig. S1 at http://www.medizin.uni-greifswald.de/immun/gk840/holtfreters1.pdf.
It has been known for some time that superantigens and lipopolysaccharides of gram-negative bacteria act synergistically. In mice, lipopolysaccharide and staphylococcal enterotoxin A are effective at 100-fold-reduced doses if both agents are applied simultaneously (7). In general, superantigens sensitize rodents to the lethal effects of lipopolysaccharide (9). Such findings form the basis of the two-hit model of sepsis, which suggests that superantigens, besides being the causative agents of gram-positive toxic shock syndrome, may also contribute to septic shock induced by gram-negative or polymicrobial sepsis (5). Evidence for the two-hit model has been hard to find in humans for a number of reasons. First, most studies have concentrated on a few superantigens, the effects of which may have been masked by the presence of others. Second, host factors such as the HLA haplotype modulate the superantigen effects (17, 27, 28). Third, the serum of many healthy individuals contains antibodies, which can neutralize the T-cell-stimulatory effects of superantigens; 85 of 100 human serum samples fully inhibited the T-cell proliferation induced by all 11 isoforms of the streptococcal superantigen SMEZ, and the remaining 15 serum samples at least partially neutralized a subset of the variants (33). In mice, such antibodies have been shown to protect the animals from the toxic effects of superantigens as well as from the lethal consequences of S. aureus infection (1, 20, 26, 35). In humans, lack of detectable antibodies to toxic shock syndrome-associated superantigens in the serum was predictive of susceptibility to toxic shock syndrome (34, 36), and there is evidence that intravenous immunoglobulin preparations improve the survival of patients with streptococcal toxic shock syndrome (8, 15, 29).
The present study addresses whether it is possible, on the basis of our current knowledge about superantigens and exotoxin-like genes, to reliably predict the T-cell-stimulating properties of a given S. aureus clinical isolate. In addition, in this study, the prevalence of serum factors which can inhibit the T-cell stimulation induced by staphylococcal secretion products has been determined.
MATERIALS AND METHODS
Bacterial strains and secretion products.
Sequence information was available from the following S. aureus reference strains: N315, Mu50, and Col (http://www.tigr.org/), MW2 (http://www.cib.nig.ac.jp/), and NCTC6571, FRI326, and NCTC8325-4 (http://ncbi.nlm.nih.gov/). The strains FRI722, FRI955, FRI918, and FRI169 were from M. Bentley, University of Wisconsin, Madison (6). Twenty S. aureus clinical isolates from throat swabs of asymptomatic individuals (aSA1 to -20) and 20 clinical isolates from blood cultures (pSA1 to -20) were collected by the Friedrich-Loeffler-Institut für Medizinische Mikrobiologie from hospitals in northeast Germany in 2000. They were identified as S. aureus by their ability to produce clumping factor and/or protein A (Murex Staphaurex test; Murex Biotech Ltd., Dartford, United Kingdom). All 40 isolates were methicillin sensitive in an antibiogram. The isolates were cultured in Luria broth (LB) medium (10 g of peptone per liter, 0.5 g of yeast extract per liter, 10 g of NaCl per liter, and 1 mM NaOH) at 37°C up to an optical density at 540 nm of 3, corresponding to the postexponential growth phase. After centrifugation, the remaining cell debris was removed from the culture supernatants by filtration through a 0.02-μm filter, and the cell-free supernatants were stored at −70°C.
Serum samples, immunoglobulin preparations, blood cells, and proliferation assays.
For the initial experiment, serum samples were obtained from 100 consecutive blood donors from the Department of Transfusion Medicine. These had an average age of 30.1 years; 49% were female and 51% were male. For all further experiments, serum samples from 23 healthy volunteers (18 female and 5 male), average age 21.8 ± 1.1 years, were used.
The design of this study was approved by the Ethics Committee of the Medical Association of the Land Mecklenburg-Vorpommern (Ethikkommission der Ärztekammer Mecklenburg-Vorpommern bei der Ernst-Moritz-Arndt-Universität Greifswald), and informed consent was obtained from all blood donors.
Three different preparations of human immunoglobulin for intravenous application were purchased from Novartis Pharma (Nürnberg, Germany; Sandoglobin), Baxter Immuno (Vienna, Austria; Gammagard), and Octapharma Pharmaceuticals (Vienna, Austria; Octagam). Peripheral blood mononuclear cells (PBMC) from healthy blood donors were isolated by density centrifugation over Ficoll. Cells were cultured in 96-well flat-bottomed plates at a density of 105/well in RPMI with l-glutamine and penicillin-streptomycin. Heat-inactivated fetal bovine serum was added to a final concentration of 10%. To test for neutralizing serum factors, fetal bovine serum was replaced by individual heat-inactivated human serum where indicated. In the initial experiment, the fetal bovine serum was completely replaced by 10% heat-inactivated human serum. Since direct comparisons showed very similar results with only 2% human serum and 8% fetal bovine serum, these conditions were used in all further experiments to save material and thus enable extensive titration experiments with bacterial supernatants and recombinant superantigens.
The cells were stimulated for proliferation by incubation with bacterial supernatants, which were titrated over a wide range. The mitogen phytohemagglutinin (Abbot, Wiesbaden, Germany) was used at a final concentration of 0.5 μg/ml. After 72 h of culture in a humidified incubator in the presence of 5% CO2 at 37°C, [3H]thymidine (Amersham, Freiburg, Germany) was added at 0.5 μCi/well for 16 h. The cells were then harvested, and the incorporated radioactivity was determined. All measurements were performed in triplicate, and the standard errors of the mean were below 20% except for values of <1,000 cpm.
Sequence comparisons.
The deduced amino acid sequences of the set genes were compared with the Blast and Blast2 programs (http://ncbi.nlm.nih.gov/; http://www.tigr.org/ http://ncbi.nlm.nih.gov/; http://www.tigr.org/).
Detection of enterotoxin and set genes.
DNA was extracted from staphylococcal cultures and used as the template in PCRs for the detection of sequences corresponding to sea, seq, and tst with the primers shown in Table 1 (13, 24). For the analysis of the set cluster seven primer pairs were designed to amplify groups of set sequences covering the 11 loci as well as their known allelic variants (see supplemental online material, Fig. S1) as shown in Table 1. The nomenclature for the set genes in this paper follows that used by Kuroda et al. for the fully sequenced S. aureus reference strain N315 (18). The amplifications were performed with Taq polymerase in a Biometra thermocycler with the following conditions: initial denaturation at 95°C for 5 min, followed by 30 stringent cycles (1 min of denaturation at 95°C, 1 min of annealing at the temperature indicated in Table 1, and 1 min of extension at 72°C), and a final extension step at 72°C for 5 min. The quality of the DNA extracts and the absence of PCR inhibitors were confirmed by amplification of glyceraldehhyde-3-phosphate dehydrogenase or of 16S rRNA. The PCR products were then analyzed by electrophoresis through a 1% agarose gel. At least two independent experiments were performed for each determination.
TABLE 1.
Nucleotide sequences of enterotoxin and exotoxin-like gene-specific primers used in this study, annealing temperatures, and anticipated PCR products
| Gene | Oligonucleotide sequencea | Tanneal (°C) | Fragment length (bases) | Reference |
|---|---|---|---|---|
| tst | 5′ GCT TGC GAC AAC TGC TAC AG | 56.2 | 559 | 24 |
| 3′ TGG ATC CGT CAT TCA TTG TTA A | ||||
| sea | 5′ GCA GGG AAC AGC TTT AGG C | 63.0 | 520 | 24 |
| 3′ GTT CTG TAG AAG TAT GAA ACA CG | ||||
| seb-sec | 5′ ATG TAA TTT TGA TAT TCG CAG TG | 64.0 | 683 | 24 |
| 3′ TGC AGG CAT CAT ATC ATA CCA | ||||
| sec3 | 5′ CTT GTA TGT ATG GAG GAA TAA CAA | 59.2 | 283 | 24 |
| 3′ TGC AGG CAT CAT ATC ATA CCA | ||||
| sed | 5′ GTG GTG AAA TAG ATA GGA CTG C | 61.6 | 384 | 24 |
| 3′ ATA TGA AGG TGC TCT GTG G | ||||
| see | 5′ TAC CAA TTA ACT TGT GGA TAG AC | 61.6 | 170 | 24 |
| 3′ CTC TTT GCA CCT TAC CGC | ||||
| seg | 5′ CGT CTC CAC CTG TTG AAG G | 66.0 | 327 | 24 |
| 3′ CCA AGT GAT TGT CTA TTG TCG | ||||
| seh | 5′ CAA CTG CTG ATT TAG CTC AG | 58.0 | 360 | 24 |
| 3′ GTC GAA TGA GTA ATC TCT AGG | ||||
| sei | 5′ CAA CTC GAA TTT TCA ACA GGT AC | 67.2 | 465 | 24 |
| 3′ CAG GCA GTC CAT CTC CTG | ||||
| sej | 5′ CAT CAG AAC TGT TGT TCC GCT AG | 61.6 | 142 | 24 |
| 3′ CTG AAT TTT ACC ATC AAA GGT AC | ||||
| sek | 5′ ATG GCG GAG TCA CAG CTA CT | 62.0 | 197 | |
| 3′ TGC CGT TAT GTC CAT AAA TGT T | ||||
| sel | 5′ CAC CAG AAT CAC ACC GCT TA | 63.1 | 410 | |
| 3′ TCC CCT TAT CAA AAC CGC TAT | ||||
| sem | 5′ CTA TTA ATC TTT GGG TTA ATG GAG AAC | 62.2 | 325 | 13 |
| 3′ TTC AGT TTC GAC AGT TTT GTT GTC AT | ||||
| sen | 5′ ACG TGG CAA TTA GAC GAG TC | 61.0 | 475 | 13 |
| 3′ GAT TGA TCT TGA TGA TTA TGA G | ||||
| seo | 5′ AGT TTG TGT AAG AAG TCA AGT GTA GA ATC TTT AAA TTC AGC AGA TAT TCC ATC TAA C | 62.2 | 179 | 13 |
| sep | 5′ CTG AAT TGC AGG GAA CTG CT | 64.0 | 187 | |
| 3′ ATT GGC GGT GTC TTT TGA AC | ||||
| seq | 5′ GAA CCT GAA AAG CTT CAA GGA | 64.0 | 209 | |
| 3′ ATT CGC CAA CGT AAT TCC AC | ||||
| set2, set 2b, set2c, set8, set9 | 5′ AAG AGC GTA TTA TAC GAA ACC | 46.2 | 398 | |
| 3′ TTT CAA TAA GTT GTT TTC TCA A | ||||
| set5, set13 | 5′ CTG GTC ACG CGA AAG TAG AA | 61.0 | 293 | |
| 3′ CTT TGT TAT ACC GCC AAC GC | ||||
| set7, set2a | 5′ AGC AAC AGG TGT AAA CAC TAC AA | 52.1 | 308 | |
| 3′ TAG AGT ACT TTG CAC CTT CAA ATC | ||||
| set 3, set3a, set6, set10 | 5′ GAA AGC AAG TTT AGC ATT AGG | 54.0 | 239 | |
| 3′ TCT GTA CTC TTG TGA ATT TTC TA | ||||
| set5, set12 | 5′ AGC TAA AGC GAT ATT TGT ATT AGG | 59.0 | 396 | |
| 3′ TTC GGC GTT CTT AGA GAC TCA | ||||
| set 1a, set1b, set1c, set4, set11, set14 | 5′ AAA GCA ACA TTA GCA TTA GG | 56.0 | 344 | |
| 3′ TTC TTT GTT ACA CCA CCA AC | ||||
| set3b, set15 | 5′ GCT AAA GCA AGT TTA GCA CTA GG | 48.6 | 543 | |
| 3′ TTT ACT GTC TTT AGG TTC TGT CTT A | ||||
| 16S rRNA | 5′ GTA GGT GGC AAG CGT TAT CC | 58.0 | 228 | 24 |
| 3′ CGC ACA TCA GCG TCA G | ||||
| G3PDH | 5′ ACC ACA GTC CAT GCC ATC AC | 452 | ||
| 3′ TCC ACC ACC CTG TTG CTG TA |
Nucleotide sequences for the amplification of sek, sel, sep, and seq as well as the set genes and anticipated sizes of the PCR products were derived from published sequences of reference strains (see Materials and Methods). The nomenclature of the set genes corresponds to that in the supplemental online material, Fig. S1.
Production and purification of recombinant enterotoxins.
Recombinant enterotoxins were produced as previously described (13). Briefly, primers were designed for the amplification of full-length sea (5′CAGAATTCAGCGAGAAAAGCGAAGAAATAAATG and 3′GCCTGCAGTTAACTTGTATATAAATATATATCAATATGAATGTTTTCAG) and sei (5′CAGAATTCCAAGGTGATATTGGTGTAGGTAACTTAA and 3′GCCTGCAGTTAGTTACTATCTACATATGATATTTCGACATCAAG; restriction sites are italic). The 5′ primers were chosen within the coding sequence of the genes, omitting the region predicted to encode the signal peptide as determined by hydrophobicity analysis according to Kyte and Doolittle (19), and they contained restriction sites for EcoRI and PstI. After digestion with these enzymes, the PCR products were ligated into the pMAL-c2 expression vector from New England Biolabs (Ozyme), which was restricted with the same enzymes. The resulting plasmids were transfected into Escherichia coli TG1. The integrity of the open reading frames was confirmed by DNA sequencing of the junction between pMAL-c2 and the inserts. The fusion proteins were purified from cell lysates of transformed E. coli by affinity chromatography on an amylose column according to the manufacturer′s instructions (New England Biolabs). pMAL-c2 without an insert was also transfected into E. coli TG1, and maltose-binding protein (MBP) without enterotoxin was purified in a similar way and used as a control.
RESULTS
Occurrence of superantigen genes in clinical isolates of S. aureus.
In this study, 40 clinical S. aureus strains were analyzed for their ability to secrete T-cell-stimulating factors. Supernatants from high-density bacterial cultures were incubated with PBMC from three different donors in the presence of fetal bovine serum. Thymidine incorporation was used as a readout for T-cell activation. Both the degree of T-cell stimulation and the titers of the supernatants, which caused maximal proliferation, were highly reproducible between the three experiments (data not shown); 16 of 20 S. aureus isolates from blood cultures and 12 of 20 commensal S. aureus isolates from throat swabs were able to stimulate T cells.
To address the question of whether superantigens and/or staphylococcal enterotoxins could account for the T-cell activation, the 40 clinical isolates were analyzed by PCR for the presence of all 17 superantigen genes. For the characterization of the set cluster, a PCR system which allowed the amplification of seven groups of set genes covering the whole cluster was developed (see Materials and Methods). The results of this analysis were compared with the maximal T-cell proliferation which could be induced by the secretion products of individual bacterial strains (Table 2).
TABLE 2.
Induction of T cell proliferation by the secretion products of S. aureus clinical isolates correlates with the presence of enterotoxin genes but not with that of staphylococcal exotoxin-like genes (set)
| Sample | S. aureus isolate | Max T-cell activation (cpm)a | Staphylococcal enterotoxinsb | egc-encoded enterotoxinsb | setc |
|---|---|---|---|---|---|
| Blood culture | pSA4 | 152,914 | B, D, J, K, Q | + | |
| pSA2 | 79,281 | M, N, O | + | ||
| pSA16 | 65,158 | G, I, M, N, O | + | ||
| pSA12 | 54,439 | G, I, M, N, O | + | ||
| pSA10 | 52,038 | A, D, J, L | G, I, M, N, O | + | |
| pSA19 | 48,624 | C, L | G, I, M, N, O | + | |
| pSA17 | 45,499 | B, P | + | ||
| pSA18 | 44,338 | C, L | G, I, M, N, O | + | |
| pSA11 | 36,239 | C, L | G, I, M, N, O | + | |
| pSA5 | 33,833 | A, D | N, O | + | |
| pSA13 | 24,941 | B | + | ||
| pSA6 | 23,866 | M, N, O | + | ||
| pSA20 | 18,426 | C, L, P | + | ||
| pSA15 | 18,156 | C, L | G, I, M, N, O | + | |
| pSA8 | 14,375 | N, O | + | ||
| pSA7 | 5,751 | N, O | + | ||
| pSA1 | 1,609 | + | |||
| pSA3 | 1,495 | + | |||
| pSA14 | 871 | + | |||
| pSA9 | 274 | + | |||
| Throat swab | aSA4 | 72,879 | M, N, O | + | |
| aSA2 | 68,290 | G, I, M, N, O | + | ||
| aSA12 | 66,989 | C, L | G, I, M, N, O | + | |
| aSA1 | 56,741 | A, TSST | G, I, N, O | + | |
| aSA3 | 48,447 | A, TSST | G, I, N, O | + | |
| aSA19 | 44,926 | C, L, TSST | G, I, M, N, O | + | |
| aSA14 | 42,492 | C, L | G, I, M, N, O | + | |
| aSA16 | 42,241 | G, I, M, N, O | + | ||
| aSA18 | 41,699 | D, J, P | G, I, M, N, O | + | |
| aSA6 | 34,671 | A, TSST | G, I, N, O | + | |
| aSA11 | 20,062 | D, J | + | ||
| aSA5 | 15,994 | D, J | + | ||
| aSA7 | 1,530 | + | |||
| aSA17 | 774 | + | |||
| aSA10 | 752 | + | |||
| aSA15 | 717 | + | |||
| aSA13 | 686 | + | |||
| aSA20 | 658 | + | |||
| aSA9 | 467 | + | |||
| aSA8 | 397 | + |
Human PBMC were cultured for 72 h in the presence of 10% FBS and their proliferation was determined by thymidine incorporation. The cells were stimulated with serial dilutions (1:40 to 1:40,000,000) of sterile culture supernatants from 40 S. aureus clinical isolates. The titration curves peaked reproducibly at different dilutions; the maximal thymidine incorporation is indicated. Background values in the absence of bacterial supernatant ranged between 175 and 1450 cpm in different experiments. Values greater than three times the background were considered to be significant T-cell activation.
The presence of enterotoxin genes was determined by PCR.
Seven different PCRs were performed to cover all the loci of the set gene cluster. There was substancial heterogeneity in the composition of the set cluster, but this did not correlate with T-cell proliferation and is not shown.
Every strain harbored members of the set cluster regardless of whether it was able to stimulate T cells or not. Thus, the proteins encoded by the cluster of set genes do not appear to be responsible for the T-cell stimulation induced by S. aureus. In contrast, the 40 strains were very heterogeneous with regard to the 17 enterotoxin loci. There was an absolute correlation between the presence of members of the enterotoxin gene family in an S. aureus isolate and its secretion of T-cell-activating substances (Table 2), supporting the notion that enterotoxin gene products were the T-cell-stimulating agents in the bacterial supernatants.
Neutralizing antibodies in normal human serum.
Serum antibodies which can inhibit the T-cell activation by individual superantigens are frequently present in healthy individuals. However, since the majority of clinical samples of S. aureus have multiple superantigen genes (Table 2), we wished to determine whether the complex mixture of superantigens secreted by clinical S. aureus isolates can also be neutralized with similar frequency and efficiency. For an initial screen, T-cell-activating supernatants from 11 S. aureus isolates differing in their superantigen gene spectrum were selected. The S. aureus reference strain FRI 918, which expresses only a single superantigen (staphylococcal enterotoxin E), was also included. Serum samples from 100 healthy adult blood donors were then tested for their ability to inhibit the T-cell activation induced by the secretion products of these 12 S. aureus strains. The neutralizing response was determined as the percent inhibition of thymidine incorporation in the presence of 10% human serum versus 10% fetal bovine serum. To exclude nonspecific inhibition, the mitogen phytohemagglutinin was used as a control. Table 3 shows the neutralizing activity of 10 representative human serum samples.
TABLE 3.
Inhibition of T-cell proliferation elicited by culture supernatants of different S. aureus strains by 10 individual human sera
| S. aureus isolate | Enterotoxins | egc-encoded enterotoxins | % Inhibition with human serum no.:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 55 | 49 | 53 | 39 | 40 | 54 | 65 | 95 | 100 | 88 | |||
| PHA | −101 | −118 | −109 | −74 | −57 | −136 | −80 | −83 | −75 | −100 | ||
| FRI722 | A, B, O, K | 99 | 83 | 93 | 80 | 99 | −10 | −234 | −230 | 86 | 19 | |
| pSA5 | A, D | N, O | 79 | 25 | 50 | 32 | 42 | −73 | −59 | −7 | −9 | −16 |
| pSA17 | B, P | 68 | 56 | 84 | 20 | 72 | 1 | −20 | −3 | 12 | −52 | |
| pSA20 | C, L, P | 99 | 98 | 99 | 98 | 99 | −46 | −22 | 92 | −64 | 98 | |
| FRI955 | D, J | 99 | 53 | 98 | 99 | 98 | −51 | −19 | 36 | −83 | 93 | |
| FRI918 | E | 99 | 26 | 91 | 98 | 97 | −135 | −48 | −85 | −55 | 100 | |
| aSA2 | G, I, M, N, O | −81 | 28 | 29 | 3 | 22 | −15 | −85 | −54 | 2 | −48 | |
| pSA12 | G, I, M, N, O | −67 | 1 | 16 | −39 | 3 | −71 | −195 | −61 | −19 | −79 | |
| FRI169 | O, K, TSST | 68 | −12 | 97 | 89 | 87 | −312 | 81 | −231 | 93 | 123 | |
| pSA2 | M, N, O | −34 | 29 | 34 | −37 | −44 | −39 | −160 | −19 | −8 | −35 | |
| pSA16 | G, I, M, N, O | −187 | −50 | −49 | −119 | −101 | −98 | −246 | −45 | −99 | −68 | |
| aSA4 | M, N, O | 8 | 100 | 6 | −84 | −63 | −142 | −278 | −116 | −32 | −227 | |
Proliferation in the presence of 10% human serum was compared with that in the presence of 10% FBS (100%), and inhibition is expressed as a percentage of this value. Negative values indicate that there was more vigorous proliferation in the presence of human serum. The bacterial culture supernatants were used at a concentration which, in the presence of FBS, induced a level of proliferation which was just below the plateau.
None of the 100 human serum samples inhibited the phytohemagglutinin-induced proliferation; on the contrary, the T cells proliferated much more vigorously in the presence of human serum. It is a common observation that human serum under most conditions supports the proliferation of human T cells much better than fetal bovine serum. This was also frequently observed after stimulation with bacterial supernatants, most impressively after stimulation with secretion products of S. aureus isolate pSA16 (Table 3). Therefore, we considered a reduction in proliferation in the presence of human serum of more than 25% to be significant.
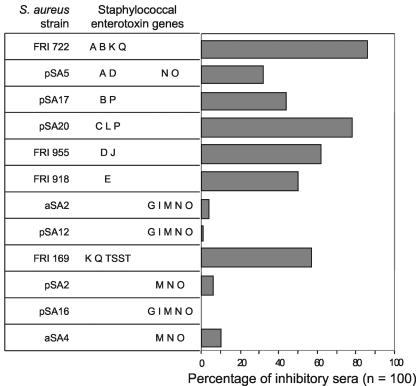
Table 3 shows 10 serum samples with individual patterns of neutralizing capacity: serum 54 did not neutralize any of the S. aureus-derived mitogenic factors, whereas others inhibited most T-cell-stimulating supernatants. Figure 1 summarizes the data for all 100 serum samples. There was a correlation between the enterotoxin genes of an isolate and the presence of inhibiting serum factors; the effects of secretion products from S. aureus isolates which harbored only members of the egc cluster of enterotoxins were inhibited only very rarely.
FIG. 1.
Inhibition of T-cell proliferation induced by S. aureus secretion products. Twelve S. aureus isolates were grown to high density, and their supernatants were titrated and used to stimulate proliferation of human PBMC in the presence of 10% fetal bovine serum. A concentration just off the plateau was then chosen for assessment of the proliferation elicited in the presence of 10% fetal bovine serum compared to 10% human serum, and 100 serum samples from healthy individuals were screened. A reduction of thymidine incorporation of at least 25% was taken as significant inhibition, because cells which were stimulated with the mitogen phytohemagglutinin as a control uniformly proliferated more vigorously in the presence of human serum.
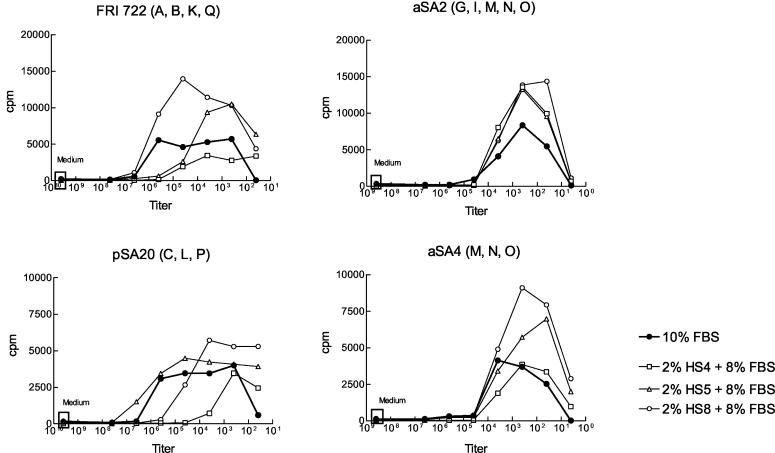
A more detailed analysis comparing two egc-containing S. aureus isolates (aSA2 and aSA4) with strains FRI 722 (sea, seb, seq, and sek) and pSA20 (sec, sel, and sep) with 23 new human serum samples confirmed the results obtained in the survey. In this experiment, the bacterial supernatants were titrated over a wide range of concentrations and inhibition was defined as a reduction of the mitogenic potency by a factor of at least 10 in the presence of 2% human serum. For example, with this criterion, the proliferation induced by S. aureus FRI722 was inhibited by human serum samples HS4 and HS5, and that induced by S. aureus pSA20 was inhibited by HS4 and HS8 (Fig. 2). As in the initial experiment, however, the addition of human serum frequently enhanced the T-cell proliferation. The patterns of inhibition and/or stimulation of T-cell activation by individual human serum samples were reproducible in repeat experiments. The results obtained with 23 different serum samples are summarized in Table 4. They suggest that the gene products of the egc very rarely elicit a strong neutralizing antibody response despite the fact that members of the egc cluster were the most frequent superantigen genes in our S. aureus isolates.
FIG. 2.
Neutralization capacity of human serum samples. Four S. aureus strains were grown to high density in culture, and the supernatants were titrated over a large range and used to stimulate proliferation of human PBMC in the presence of 10% fetal bovine serum (FBS). Proliferation was assessed by thymidine incorporation after 72 h of culture; 23 serum samples from healthy individuals (HS) were tested for their ability to inhibit this proliferation when used at a final concentration of 2% (in the presence of 8% fetal bovine serum). A right shift of the titration curve by at least a factor of 10 in the presence of human serum was considered to indicate significant inhibition. The effect of three typical human serum samples is shown as an example.
TABLE 4.
Sera from 23 healthy volunteers were tested for their abilitiy to inhibit the T-cell proliferation elicited by the secretion products of four different S. aureus strainsa
| Inhibition factor | No. of sera with neutralizing activity against:
|
|||
|---|---|---|---|---|
| FRI722 | pSA20 | aSA2 | aSA4 | |
| 0 | 14 | 8 | 22 | 21 |
| 10 | 6 | 2 | 1 | 2 |
| 100 | 2 | 5 | 0 | 0 |
| ≥1000 | 1 | 8 | 0 | 0 |
Experiments were conducted as shown in Fig. 3. PBMC were stimulated with bacterial supernatants over a large range of concentrations in the presence of 10% fetal bovine serum. The inhibition factor was determined from the right shift of the response curve when 2% of the fetal bovine serum was replaced with human serum. Proliferation induced by the mitogen phytohemagglutinin was never inhibited by human serum factors.
Neutralization of recombinant superantigens.
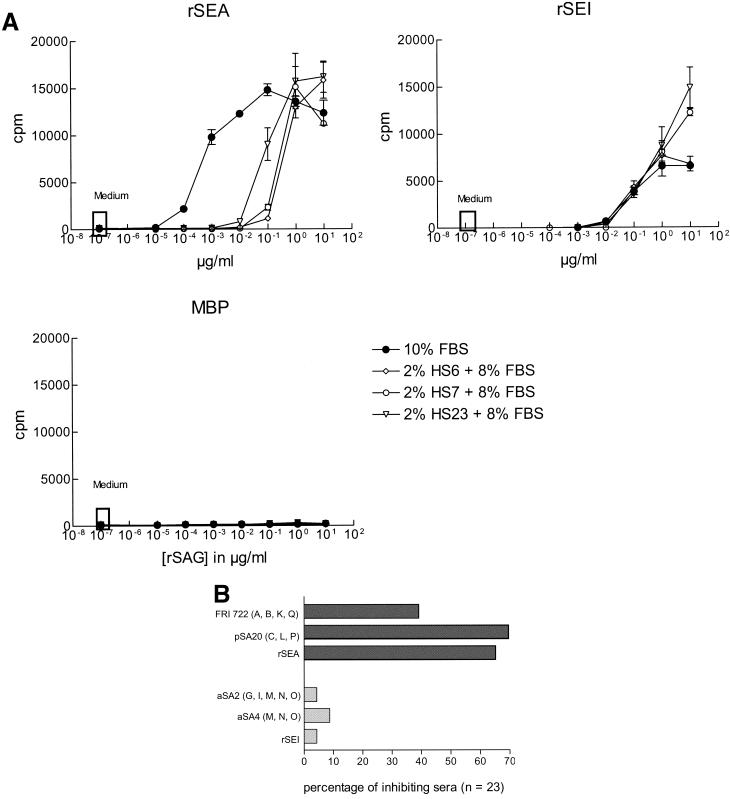
To find out whether these differences are an inherent property of the individual superantigens or whether they are caused by other bacterial secretion products contained in the culture supernatants, the experiment was repeated with recombinant superantigens; recombinant staphylococcal enterotoxin I (rSEI), which is encoded by egc, was compared with rSEA. The recombinant superantigens were expressed as fusion products with MBP, which did not induce proliferation by itself (Fig. 3A). Only 1 of 23 human serum samples reduced the potency of rSEI by a factor of 10, whereas by the same criteria rSEA was inhibited by 16 serum samples. This is in good agreement with the results obtained with the bacterial supernatants (Fig. 3B).
FIG. 3.
(A) Inhibition of T-cell proliferation induced by the recombinant superantigens rSEA and rSEI. Fusion proteins of MBP and staphylococcal enterotoxin A or I, respectively, were used to stimulate proliferation of human PBMC in the presence of 10% fetal bovine serum (FBS). PBMC which proliferated in response to MBP alone were excluded from the analysis; 23 serum samples from healthy individuals were then tested for their ability to inhibit this proliferation when used at a final concentration of 2% (in the presence of 8% fetal bovine serum). A right shift of the titration curve by at least a factor of 10 in the presence of human serum was considered to indicate significant inhibition. The effect of three typical human serum samples is shown as an example. (B) Frequency of human serum samples able to inhibit the proliferative effects of S. aureus secretion products or of recombinant superantigens. This figure summarizes the data obtained with 23 serum samples which were tested as shown in Fig. 2 and panel A.
Neutralization by pooled human immunoglobulin.
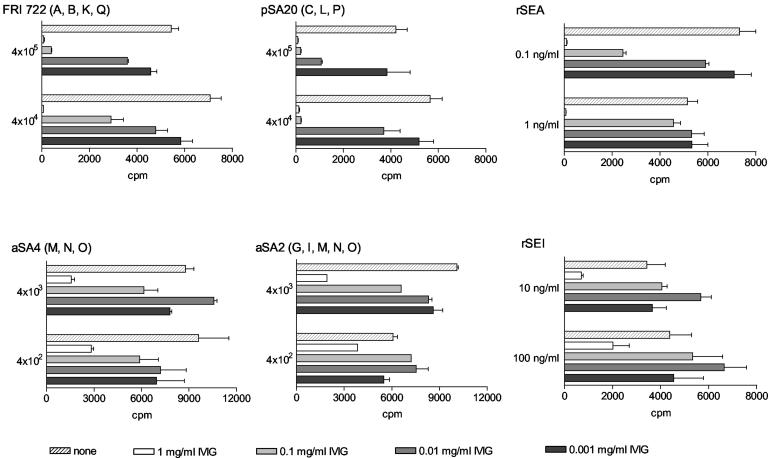
A similar difference in neutralizing capacity was also observed in three different intravenous immunoglobulin preparations from pooled human serum samples. Inhibition of T-cell proliferation induced by FRI722, pSA20, or rSEA was around 100-fold more efficient than that induced by aSA2, aSA4, or rSEI (Fig. 4). In contrast to human serum, the addition of purified immunoglobulin G did not increase the T-cell proliferation, suggesting that this enhancement of proliferation was likely due to human growth factors or hormones rather than to specific interactions of antibodies with superantigens.
FIG. 4.
Therapeutic preparations of human immunoglobulin G (IVIG) inhibit egc-derived enterotoxins less efficiently than other superantigens. Human PBMC were stimulated with bacterial supernatants or rSEA and rSEI at two different titers or concentrations as indicated on the left of each panel. They were cultured in the presence of 10% fetal bovine serum, and increasing concentrations of intravenous immunoglobulin preparations were added where indicated. In this figure, the effect of a preparation of Sandoglobin (Novartis Pharma) is shown; two other intravenous immunoglobulin preparations (see Materials and Methods) gave very similar results.
DISCUSSION
This survey of all known superantigen genes confirms earlier findings that most S. aureus clinical isolates harbor subsets of enterotoxin genes. There was extensive variation between individual strains, and generally the isolates harbored multiple enterotoxin genes, usually five. In this investigation, we observed a strict correlation between the presence of enterotoxin genes in an S. aureus isolate and its ability to elicit T-cell proliferation. Therefore, a genetic analysis of the superantigen gene loci could be useful for the prediction of the functional properties of unknown clinical isolates. There are two limitations: First, the presence of a superantigen gene does not necessarily mean that the protein is expressed at mitogenic levels. Second, while it appears unlikely that many more superantigen gene loci still await discovery, allelic variation at the known gene loci could be extensive. Point mutations, however, can abolish primer binding to a functional superantigen gene variant (C. Feig, unpublished observation).
In contrast to the enterotoxins, the presence of exotoxin-like genes, which are encoded by the set cluster on genomic island SaPI2, did not correlate with T-cell stimulation. This is in agreement with the observation that recombinant staphylococcal exotoxin-like proteins 1, 3, 10 and 15 had no superantigenic properties (2, 10). The data reported in this study as well as the published S. aureus sequences show that the set gene cluster is present in every S. aureus strain and that its composition is highly variable (10). This suggests a nonredundant function of its gene products in S. aureus, and the location of the set cluster on a genomic island indicates that this function may be relevant for host-pathogen interactions. What that function may be remains to be defined.
Testing of 123 serum samples from healthy blood donors for their ability to inhibit T-cell activation by the secretion products of 12 different S. aureus strains revealed remarkable heterogeneity and surprisingly large holes in neutralizing capacity. None of the tested serum samples neutralized all the S. aureus culture supernatants in this investigation, and the secretion products from isolates harboring egc but no other enterotoxin genes were neutralized only rarely. This was confirmed by experiments with recombinant staphylococcal enterotoxins A and I, which excludes that other factors contained in the bacterial supernatants are responsible (Table 4). As expected, this difference in neutralizing capacity was also mirrored by three different intravenous immunoglobulin preparations, which are prepared from very large pools of human serum (Fig. 4). The inhibition of egc gene products was about 100-fold less efficient than that of other superantigens. These differences in neutralizing capacity may result in variations in their therapeutic efficacy in patients infected with different S. aureus strains.
In agreement with our results, Banks and colleagues detected significantly lower serum antibody binding to recombinant staphylococcal enterotoxins G and I compared with A, B, or C2 in an enzyme-linked immunosorbent assay (4). Therefore, low concentrations of antibodies specific for egc-encoded proteins could at least partially explain the failure of many serum samples to neutralize these superantigens. This is unexpected because egc was the most frequent superantigen locus in S. aureus in the present as well as in earlier studies (12, 13). A survey of around 200 cases of toxic shock syndrome and staphylococcal scarlet fever demonstrated that egc-encoded su-perantigens are able to cause symptomatic staphylococcal toxemias but that these are probably rare (11, 23). Therefore, in spite of the low level of antibody responses against egc-encoded superantigens in the population, these do not appear to be a major threat to health.
What might be the molecular reasons for the low efficiency of neutralization of egc-encoded superantigens by serum factors? On the basis of sequence comparisons, the known superantigens of S. aureus and Streptococcus pyogenes have been grouped into three clusters. Each cluster contains at least one egc-encoded superantigen, which means that the egc genes differ strongly from each other (13). This makes it unlikely that they share unique structural features which interfere with a strong immune response against them. In addition, our data show that in vitro-generated supernatants of egc-harboring S. aureus isolates as well as recombinant staphylococcal enterotoxin I can efficiently stimulate T cells, further arguing against an inherent lack of immunogenicity. On the other hand, there are indications that egc-encoded superantigen expression may be regulated differently from classical superantigens. First, with an enzyme-linked immunosorbent assay method, Omoe and colleagues have shown that only a minority of S. aureus strains which harbored seg and sei genes secreted the staphylococcal enterotoxin G and I proteins in detectable amounts in vitro, which was in contrast to the classical staphylococcal enterotoxin H (30). Furthermore seg mRNA accumulated in the logarithmic growth phase, in contrast to the other staphylococcal enterotoxins and TSST-1, which are primarily transcribed during postexponential bacterial growth (25; G. Lina, unpublished data). Superantigen protein expression appears to correspond to the mRNA levels (S. Holtfreter, unpublished data). Unfortunately, little information is available about the regulatory mechanisms which are effective at different stages of S. aureus interaction with its host in vivo. It is conceivable that not only the amounts but also the spectrum of secreted superantigens differ between S. aureus carriage and S. aureus infection.
Finally, our data underscore the specificity of the superantigen-neutralizing antibodies. The lack of cross-inhibition has to be taken into account when designing superantigen vaccines.
Acknowledgments
We are grateful to Jan-Michael Heinrich for many helpful technical hints and to Bernhard Fleischer, Robert S. Jack, and Christine Schütt for critical comments on the manuscript.
The work was financially supported by the DFG (GK840, project A1) and by the Forschungsverbund Molekulare Medizin of the University of Greifswald.
We have no potentially conflicting commercial interests.
Editor: J. T. Barbieri
REFERENCES
- 1.Arad, G., R. Levy, D. Hillman, and R. Kaempfer. 2000. Superantigen antagonist protects against lethal shock and defines a new domain for T-cell activation. Nat. Med. 6:414-421. [DOI] [PubMed] [Google Scholar]
- 2.Arcus, V. L., R. Langley, T. Proft, J. D. Fraser, and E. N. Baker. 2002. The three-dimensional structure of a superantigen-like protein, SET3, from a pathogenicity island of the Staphylococcus aureus genome. J. Biol. Chem. 277:32274-32281. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 4.Banks, M. C., N. S. Kamel, J. B. Zabriskie, D. H. Larone, D. Ursea, and D. N. Posnett. 2003. Staphylococcus aureus express unique superantigens depending on the tissue source. J. Infect. Dis. 187:77-86. [DOI] [PubMed] [Google Scholar]
- 5.Bannan, J., K. Visvanathan, and J. B. Zabriskie. 1999. Structure and function of streptococcal superantigens in septic shock. Bacterial Sepsis Septic Shock 13:387-396. [DOI] [PubMed] [Google Scholar]
- 6.Bentley, M., D. Borst, and L. Regassa. 1992. Staphylococcal enterotoxins, toxic shock syndrome toxin and streptococcal pyrogenic exotoxins: a compararive study of their molecular biology. Chem. Immunol. 55:1-35. [PubMed] [Google Scholar]
- 7.Blank, C., A. Luz, S. Bendigs, A. Erdmann, H. Wagner, and K. Heeg. 1997. Superantigen and endotoxin synergize in the induction of lethal shock. Eur. J. Immunol. 27:825-833. [DOI] [PubMed] [Google Scholar]
- 8.Darenberg, J., N. Ihendyane, J. Sjolin, E. Aufwerber, S. Haidl, P. Follin, J. Andersson, and A. Norrby-Teglund. 2003. Intravenous immunoglobulin G therapy in streptococcal toxic shock syndrome: a European randomized, double-blind, placebo-controlled trial. Clin. Infect. Dis. 37:333-340. [DOI] [PubMed] [Google Scholar]
- 9.Dinges, M. M., and P. Schlievert. 2001. Role of T cells and gamma interferon during induction of hypersensitivity to lipopolysaccharide by toxic shock syndrome toxin 1 in mice. Infect. Immun. 69:1256-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald, J., S. Reid, E. Ruotsalainen, T. Tripp, M. Liu, R. Cole, P. Kuusela, P. Schlievert, A. Järvinen, and J. Musser. 2003. Genome diversification in Staphylococcus aureus: Molecular evolution of a highly variable chromosomal region encoding the staphylococcal exotoxin-like family of proteins. Infect. Immun. 71:2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jarraud, S., G. Cozon, F. Vandenesch, M. Bes, J. Etienne, and G. Lina. 1999. Involvement of enterotoxin G and I in staphylococcal toxic shock syndrome and styphylococcal scarlet fever. J. Clin. Microbiol. 37:2446-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarraud, S., M. A. Peyrat, A. Lim, A. Tristan, M. Bes, C. Mougel, J. Etienne, F. Vandenesch, M. Bonneville, and G. Lina. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166:669-677. [DOI] [PubMed] [Google Scholar]
- 14.Kalmeijer, M., H. Coertjens, P. von Nieuwland-Bollen, D. Bogaers-Hofman, G. de Baere, A. Stuurman, A. van Belkum, and J. Kluytmans. 2002. Surgical site infections in orthopedic surgery: the effect of mupirocin nasal ointment in a double blind, randomized placebo-controlled study. Clin. Infect. Dis. 35:353-358. [DOI] [PubMed] [Google Scholar]
- 15.Kaul, R., A. McGeer, A. Norrby-Teglund, M. Kotb, B. Schwartz, K. O'Rourke, J. Talbot, and D. E. Low. 1999. Intravenous immunoglobulin therapy for streptococcal toxic shock syndrome—a comparative observational study. The Canadian Streptococcal Study Group. Clin. Infect. Dis. 28:800-807. [DOI] [PubMed] [Google Scholar]
- 16.Kluytmans, J., J. Mouton, E. Ijzerman, Vandenbroucke-Grauls, A. Maat, J. Wagenvoort, and H. Verbrugh. 1995. Nasal carriage of Staphylococcus aureus as a major risk factor for wound infections after cardiac surgery. J. Infect. Dis. 171:216-219. [DOI] [PubMed] [Google Scholar]
- 17.Kotb, M., A. Norrby-Teglund, A. McGeer, H. El-Sherbini, M. T. Dorak, A. Khurshid, K. Green, J. Peeples, J. Wade, G. Thomson, B. Schwartz, and D. E. Low. 2002. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat. Med. 8:1398-1404. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 19.Kyte, J., and F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105. [DOI] [PubMed] [Google Scholar]
- 20.LeClaire, R. D., and S. Bavari. 2001. Human antibodies to bacterial superantigens and their ability to inhibit T-cell activation and lethality. Antimicrob. Agents Chemother. 45:460-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowy, F. 1988. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-529. [DOI] [PubMed] [Google Scholar]
- 22.McCormick, J. K., J. M. Yarwood, and P. M. Schlievert. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 55:77-104. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, R., S. Monday, G. Bohach, and S. Schlievert. 2001. Prolonged course of toxic shock syndrome associated with methicillin-resistant Staphylococcus aureus enterotoxins G and I. Int. J. Infect. Dis. 5:163-166. [DOI] [PubMed] [Google Scholar]
- 24.Monday, S., and G. Bohach. 1999. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 37:3411-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munson, S. H., M. T. Tremaine, M. J. Betley, and R. A. Welch. 1998. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect. Immun. 66:3337-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson, I.-M., M. Vedrengh, R. Ulrich, S. Bavari, and A. Tarkowski. 1999. Protection against Staphylococccus aureus Sepsis by vaccination with recombinant staphylococcal enterotoxin A devoid of superantigenicity. J. Infect. Dis. 180:1370-1373. [DOI] [PubMed] [Google Scholar]
- 27.Norrby-Teglund, A., S. Chatellier, D. E. Low, A. McGeer, K. Green, and M. Kotb. 2000. Host variation in cytokine responses to superantigens determine the severity of invasive group A streptococcal infection. Eur. J. Immunol. 30:3247-3255. [DOI] [PubMed] [Google Scholar]
- 28.Norrby-Teglund, A., G. T. Nepom, and M. Kotb. 2002. Differential presentation of group A streptococcal superantigens by HLA class II DQ and DR alleles. Eur. J. Immunol. 32:2570-2577. [DOI] [PubMed] [Google Scholar]
- 29.Norrby-Teglund, A., S. R. Norrby, and D. E. Low. 2003. The treatment of severe group a streptococcal infections. Curr. Infect. Dis. Rep. 5:28-37. [DOI] [PubMed] [Google Scholar]
- 30.Omoe, K., M. Ishikawa, Y. Shimoda, D.-L. Hu, S. Ueda, and K. Shinagawa. 2002. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates harboring seg, seh, or sei genes. J. Clin. Microbiol. 40:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perl, T., J. Cullen, R. Wenzel, M. Zimmerman, M. Pfaller, D. Sheppard, J. Twombley, P. French, and L. Herwaldt. 2002. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N. Engl. J. Med. 346:1871-1877. [DOI] [PubMed] [Google Scholar]
- 32.Proft, T., and J. Fraser. 1998. Superantigens: just like peptides, only different. J. Exp. Med. 187:819-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proft, T., S. L. Moffatt, K. D. Weller, A. Paterson, D. Martin, and J. D. Fraser. 2000. The streptococcal superantigen SMEZ exhibits wide allelic variation, mosaic structure, and significant antigenic variation. J. Exp. Med. 191:1765-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stolz, S., J. Davis, J. Vergeront, B. Crass, P. Chesney, P. Wand, and M. Bergdoll. 1885. Development of serum antibody to toxic shock toxin among individuals with toxic shock syndrome in Wisconsin. J. Infect. Dis. 151:883-889. [DOI] [PubMed] [Google Scholar]
- 35.Ulrich, R. G., M. Olson, and S. Bavari. 1998. Development of engineered vaccines effective against structurally related bacterial superantigens. Vaccine 16:1857-1864. [DOI] [PubMed] [Google Scholar]
- 36.Vergeront, J., S. Stolz, B. Crass, D. Nelson, J. Davis, and M. Bergdoll. 1983. Prevalence of serum antibody to staphylococcal enterotoxin F among Wisconsin residents: implication for toxic-shock syndrome. J. Infect. Dis. 4:692-698. [DOI] [PubMed] [Google Scholar]
- 37.von Eiff, C., K. Becker, K. Machka, H. Stammer, and G. Peters. 2001. Nasal Carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11-16. [DOI] [PubMed] [Google Scholar]
- 38.Wenzel, R., and T. Perl. 1995. The significance of nasal carriage of Staphylococcus aureus in the incidence ot postoperative wound infection. J. Hosp. Infect. 1:13-24. [DOI] [PubMed] [Google Scholar]
- 39.Williams, R. J., J. M. Ward, B. Henderson, S. Poole, B. P. O'Hara, M. Wilson, and S. P. Nair. 2000. Identification of a novel gene cluster encoding staphylococcal exotoxin-like proteins: characterization of the prototypic gene and its protein product, SET1. Infect. Immun. 68:4407-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarwood, J. M., J. K. McCormick, M. L. Paustian, P. M. Orwin, V. Kapur, and P. M. Schlievert. 2002. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. J. Biol. Chem. 277:13138-13147. [DOI] [PubMed] [Google Scholar]