Abstract
NadA is a novel vaccine candidate recently identified in Neisseria meningitidis and involved in adhesion to host tissues. The nadA gene has been found in approximately 50% of the strains isolated from patients and in three of the four hypervirulent lineages of non-serogroup A strains. Here we investigated the presence of the nadA gene in 154 meningococcal strains isolated from healthy people (carrier strains). Only 25 (16.2%) of the 154 carrier isolates harbored the nadA gene. The commensal species Neisseria lactamica was also found not to have the nadA gene. Eighteen of the carrier strains belonged to the ET-5 and ET-37 hypervirulent clusters, indicating that only the 5.1% of the genuine carrier population actually harbored nadA (7 of 136 strains). Five of the seven strains harbored a novel allele of the nadA gene that was designated nadA4. The NadA4 protein was present on the bacterial surface as heat-stable high-molecular-weight oligomers. Antibodies against the recombinant NadA4 protein were bactericidal against homologous strains, whereas the activity against other NadA alleles was weak. In conclusion, the nadA gene segregates differently in the population of strains isolated from healthy individuals and in the population of strains isolated from patients. The presence of NadA can therefore be used as a tool to study the dynamics of meningococcal infections and understand why this bacterium, which is mostly a commensal, can become a severe pathogen.
Neisseria meningitidis, a gram-negative diplococcus that is able to colonize the upper respiratory tract of humans, is a major cause of meningitis and septicemia. In most industrialized countries, even if about 10% of the human population is colonized by the bacterium in endemic periods, only 1 to 3 individuals per 100,000 people develop disease (13). Therefore, N. meningitidis should be considered a commensal which on rare occasions becomes a very dangerous pathogen. The reasons why meningococcus can be a commensal and a pathogen at the same time are poorly understood (23).
Meningococci are classified in serogroups based on the chemical composition of the polysaccharide capsule. Serogroups A, B, C, Y, and W-135 are associated with disease. Most of the meningococcal strains isolated from invasive disease have been classified by multilocus enzyme electrophoresis (4) into hypervirulent lineages (electrophoretic types ET-37 and ET-5, cluster A4, lineage III, and subgroups I, III, and IV-1) or by multilocus sequence typing (MLST) (17) into sequence type complexes (ST-11, ST-32, ST-8, ST-41/44, ST-1, ST-5, and ST-4). Meningococcal carrier populations are much less defined, and they include some proportion of strains which belong to hypervirulent clusters that apparently are identical to the strains isolated from patients. However, most strains normally isolated from healthy individuals are rarely able to cause the disease (14).
Meningococcal carriage is believed to be the natural reservoir of strains responsible for outbreaks. The appearance of strains belonging to an hypervirulent cluster correlates with increased disease frequency, and meningococcal carriage may increase during outbreaks (up to 50%) compared to the carriage during endemic periods (average, 10%) (2, 19). Meningococcal carriage has been shown to be an immunizing event both in children and in adults (3, 10) and is able to induce a bactericidal response. However, the efficacy of immunization for preventing disease is controversial since it has also been shown that carriage cannot induce protection against colonization and invasion (1). Carrier strains are genetically quite diverse compared to disease-associated strains (5, 14). The polysaccharide capsule, which is the most important pathogenicity factor of meningococci, is frequently missing in carrier strains, and this may partially account for the difference in pathogenicity (5, 8). While capsule switching could temporarily occur as a consequence of phase variation (20), the absence of the capsule is often due to the absence of the whole capsule operon (6). Along with the hypervariability of serotype and subserotype antigens, this is the main cause of the inadequacy of the conventional serological markers for tracing the fate of meningococcal isolates, particularly carrier strains (1). In this work we studied the presence and molecular features of NadA, a new adhesin, potential virulence factor, and vaccine candidate recently identified in N. meningitidis, in a large panel of carrier strains obtained from various sources and in the commensal species Neisseria lactamica. The nadA gene is known to be present in approximately 50% of meningococcal isolates and is more frequently associated with strains which belong to hypervirulent clusters. It is always present in members of three of the four major non-serogroup A meningococcal hypervirulent clusters, namely, the ET-5 complex, the ET-37 complex, and the cluster A4, whereas it is never present in lineage III strains. NadA is an adhesin which exhibits homology with a family of proteins involved in invasion and pathogenesis. Its sequence is unusually well conserved, and only three alleles (alleles 1, 2, and 3) have been identified. Allele 1 is harbored by all of the ET-5 strains tested so far, whereas alleles 2 and 3 are present mainly in strains belonging to the ET-37 complex and A4 cluster and also in strains not belonging to any hypervirulent cluster. Our analysis showed that the nadA gene is underrepresented in carrier strains and that in a subset of these strains there is a new allele that we designated allele 4 (nadA4).
MATERIALS AND METHODS
Bacterial strains, culture conditions, and chromosomal DNA isolation.
A total of 154 carrier strains of N. meningitidis and 18 strains of N. lactamica were selected for analysis. The meningococcal strains came from six different countries (Norway, People's Republic of China, United States, Chile, Iceland, and Oman), whereas the N. lactamica strains came from England and Oman. The years of isolation ranged from 1972 to 2000. Strain 2996, an example of a clinical isolate harboring allele 3 of nadA, and NGH38, a strain that did not contain the nadA gene, were used in functional assays as positive and negative controls, respectively (7). In order to extract chromosomal DNA, bacteria were grown overnight at 37°C in the presence of 5% CO2 in gonococcus agar supplemented with Kellog's solution (0.22 M d-glucose, 0.03 M l-glutamine, 0.001 M ferric nitrate, and 0.02 M cocarboxylase) (Sigma Aldrich). A starting culture of each strain at an optical density at 600 nm of 0.05 was grown to the stationary phase in 5 ml of gonococcus medium. Then 1.5 ml was pelleted by centrifugation at 16,060 × g for 15 min, and chromosomal DNA was prepared by using a Macherey-Nagel NucleoSpin tissue kit according to the manufacturer's protocol. Escherichia coli DH5α and BL21(DE3) were used as a cloning strain and an expression host, respectively, and were used as indicated by the supplier (Invitrogen).
PCR and nucleotide sequencing.
PCR amplification of the nadA gene was performed with about 10 ng of chromosomal DNA by using primers located upstream and downstream of the nadA gene coding region. The forward primer was A1 (GTGGACGTACTCGACTACGAAGG), and the reverse primer was B2 (CGAGGCGATTGTCAAACCGTTC). The Invitrogen Accuprime Taq DNA polymerase system was used. The PCR conditions were as follows: 30 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 68°C for 1 min. The amplified fragments were purified with a QIAquik PCR purification kit (QIAGEN) and were sequenced with an ABI 377 automatic sequencer (Applied Biosystems). A sequence analysis was performed by using the A1 and B2 primers used for gene amplification and primers 1 (TATGTAAACAAACTTGGTGGGG), 2 (GAAATAGAAAAGTTAACAACCAAGTT), 3 (GACATCAAAGCTGATATCGCTAC), 4 (TTTCGAGGTGGCGCGTTCGGG), 5 (GTAGCGATATCAGCTTTGATGTC), and 6 (CTTGGTTGTTAACTTTTCTATTTC). Sequences were analyzed by using the EditView program (Perkin-Elmer), a GeneJockey II sequence processor (Biosoft), MacBoxshade, and the GCG Wisconsin package.
Western blot analysis.
Total cell extracts and outer membrane vesicles (OMVs) were prepared as described previously (7, 22). The protein concentration in each sample was evaluated by Bradford analysis. Equal amounts of total proteins and OMVs were boiled for 10 or 30 min, either alone, in the presence of the zwitterionic detergent Triton X-100 at a concentration of 2%, or in the presence of the anionic detergent sodium dodecyl sulfate (SDS) at a concentration of 2%. Samples were subjected to SDS-polyacrylamide gel electrophoresis on 5 to 15% Bio-Rad polyacrylamide gels, transferred onto nitrocellulose transfer membranes (Schleicher & Schuell), and hybridized with a 1:200 dilution of an anti-NadA4 mouse polyclonal antiserum. The blots were incubated with a 1:2,000 dilution of the DAKO anti-mouse peroxidase conjugate secondary antibody, and the signal was developed with a Bio-Rad Opti-4CN substrate kit.
nadA4 gene cloning and NadA4 protein expression and purification.
The nadA4 gene (encoding amino acids 28 to 268) was amplified by PCR by using chromosomal DNA of strain NGE28, forward primer 961-13F (CGCGGATCCCATATGCCACCGCTGACGAAATTG), and reverse primer 961-C REV (CCCGCTCGAGACCCACGTTGTAAGGTTG) (an NdeI restriction site in the forward primer and an XhoI restriction site in the reverse primer are underlined). The PCR conditions were as follows: five cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and elongation at 68°C for 1 min and 25 cycles of denaturation at 94°C for 30 s, annealing at 65°C for 30 s, and elongation at 68°C for 1 min. The PCR was performed by using 10 ng of chromosomal DNA and High Fidelity Taq DNA polymerase (Invitrogen). The amplified DNA, after purification and NdeI/XhoI digestion, was cloned into the pET21b+ vector (Novagen). The plasmids were transformed into E. coli BL21(DE3) to express the proteins as COOH-terminal histidine fusions. Recombinant proteins were expressed and purified as previously described (18).
Dot blotting.
Aliquots (10 μl) of total extracts of each strain were boiled for 10 min and spotted onto nylon membranes (Boehringer Mannheim). The DNA on the membranes were cross-linked by 2 min of exposure to UV light, and the membranes were stored at room temperature. For probe preparation, the nadA gene was amplified from strain 2996 by using inner primers 961-F (GCCACAAGCGACGACG) and 961-R (CTCGTAATTGACGCC) and Platinum High Fidelity Taq DNA polymerase (Invitrogen). The PCR conditions were as follows: 30 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and elongation at 68°C for 1 min. After purification with a QIAquik column (QIAGEN), the DNA (∼350 ng) was labeled with digoxigenin by using a DIG High Prime DNA labeling and detection starter kit II (Roche). Prehybridization, hybridization, and detection of digoxigenin-labeled nucleic acids were performed as recommended by the DIG system user's guide (Boehringer Mannheim).
Binding assay.
Chang epithelial cells (Wong-Kilbourne derivative, clone 1-5c-4, human conjunctiva) were maintained in Dulbecco modified Eagle medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 15 mM l-glutamine, and antibiotics. Cells were nonenzymatically detached from the support by using cell dissociation solution (Sigma), harvested, and resuspended in RPMI medium supplemented with 1% FBS. Then 1 × 105 cells were mixed with either medium alone or three concentrations (175, 43, and 10 μg/ml) of histidine-tagged NadA3, NadA4, or GNA2132 proteins and incubated for 30 to 40 min at 4°C. After two washes with 1% FBS in phosphate-buffered saline, cells were incubated with mouse polyclonal antiserum against each protein for 1 h on ice. After two washes, the preparations were incubated on ice for 30 min with R-phycoerythrin-conjugated goat F(ab)2 antibody to mouse immunoglobulin, and the cells were finally analyzed with a FACS-Scan flow cytometer. The mean fluorescence intensity for each population was calculated.
Serum bactericidal assay.
Twenty micrograms of purified recombinant protein was used to immunize 6-week-old CD1 female mice (Charles River Laboratories) intraperitoneally (four to six mice per group). Immunization was performed on days 0, 21, and 35 by using Freund's adjuvant or a solution containing 3 mg of aluminum hydroxide per ml. Blood samples for analysis were taken on day 49. Serum bactericidal activity was evaluated as described previously (7) by using baby rabbit serum (CedarLane) as the complement source. The serum bactericidal titer was defined as the serum dilution that resulted in a 50% decrease in the number of CFU per milliliter after 60 min of incubation compared with the number of CFU per milliliter at time zero.
RESULTS
Distribution of NadA among N. meningitidis carrier strains and N. lactamica.
We used a panel of 154 carrier strains isolated from healthy individuals; 74 strains were nongroupable, 52 strains belonged to serogroup B, 6 strains belonged to serogroup C, 1 strain belonged to serogroup K, 1 strain belonged to serogroup I, 4 strains belonged to serogroup W-135, 6 strains belonged to serogroup Y, 3 strains belonged to serogroup X, and 7 strains belonged to serogroup Z. By using PCR and dot blotting, 25 (16.2%) of the 154 strains were found to be positive for the nadA gene. This frequency is much lower than the frequency observed for strains isolated from patients (approximately 50%). The presence of NadA in the different strains is shown in Table 1. The genes in all positive strains were sequenced, and the sequences were compared to the sequences of the three previously described nadA alleles. The strains harboring NadA are described in Table 2. Allele 1 was found in all strains belonging to the ET-5 hypervirulent cluster as previously described for ET-5 strains, isolated from patients (7). Allele 2 was found in all strains belonging to the ET-37 hypervirulent cluster and in the 16747 strain (ST-189). Strain 2022 carried a nadA2 gene interrupted by the insertion sequence IS1301 (11), as sometimes occurs in clinical isolates. The remaining five positive strains (3.2% of the total), which did not belong to any hypervirulent cluster, had a new nadA allele that we designated nadA4 (associated with carrier strains). The nadA4 allele is located in the meningococcal chromosome like the other nadA alleles, and there are direct repeats before and after the gene insertion, putative promoter and terminator sequences, and a variable number of tetranucleotide repeats at the promoter level (7). All the nadA4-harboring strains were isolated in Norway during three different surveys spanning 8 years. These strains have different serotypes and serosubtypes, and from a genetic point of view, four of them are closely related. They belong to ST-26 and ST-1301, which differ by one allele only in the MLST scheme, whereas one strain (16260) belongs to a very different genetic environment (ST-37), which differs from ST-26 and ST-1301 in all seven MLST genes. NadA is not present in the 18 strains of the nonpathogenic human commensal N. lactamica examined. By analyzing the PCR patterns obtained with primers external to the nadA gene insertion site (7) of eight N. lactamica strains, we found that four of them gave the results typical of meningococcal strains lacking the nadA gene (that is, a 400-bp PCR product). Interestingly, the other four N. lactamica strains had in addition a 542-nucleotide insertion, a result different from those obtained for the other neisserial strains lacking the nadA gene. The insertion was sequenced and analyzed. There were no homologous sequences in the database. The insertion is flanked by 17-nucleotide direct repeats and is located 10 nucleotides upstream of the locus of insertion of the other nadA alleles (Fig. 1). The presence of this sequence in a subset of strains could be considered a relic of the early differentiation of two N. lactamica subgroups. This sequence is found only in this species.
TABLE 1.
Presence and distribution of nadA alleles
| Organism | ET cluster | No. of strains with the following NadA alleles:
|
|||
|---|---|---|---|---|---|
| Allele 1 | Allele 2 | Allele 4 | None | ||
| Neisseria meningitidis | ET-5 | 13 | |||
| ET-37 | 5 | ||||
| Others | 2 | 5 | 129 | ||
| Neisseria lactamica | 18 | ||||
TABLE 2.
Description of the 25 NadA-positive carrier strains
| Allele or insertion | Strain (cluster)a | Classification | Country |
|---|---|---|---|
| Allele 1 | 2034 | B:15:P1.16 | Norway |
| 727/6 | B:15:P1.3 | Chile | |
| 569/6 | B:15:P1.3 | Chile | |
| 64/69 | NG:15:P1.7,16 | Norway | |
| 220173I | NG:4:P1.15 | Iceland | |
| 16148 | B:15:P1.5,2 | Norway | |
| 16471 | NG:15:P1.7,16 | Norway | |
| 16194 | B:15:P1.7,16 | Norway | |
| 16516 | NG:15:- | Norway | |
| 16467 | NG:15:P1.17,16 | Norway | |
| 30973 | NG:NT:NST | Iceland | |
| 40475 | NG:4:P1.15 | Iceland | |
| 40973 | NG:4:P1.15 | Iceland | |
| Allele 2 | E013-15 (ET-37) | Y | United States |
| 16381 (ET-37) | C:2a:P1.5,2 | Norway | |
| 16438 (ET-37) | C:2a:- | Norway | |
| 38VI (ET-37) | B | Norway | |
| 50775 (ET-37) | C:2a:- | Iceland | |
| 16747 (ST-189) | C:NT:P1.10 | Norway | |
| Allele 4 | NGE28 (ST-26) | B:4:- | Norway |
| 65/96 (ST-26) | B:4:P1.14 | Norway | |
| 149/96 (ST-1301) | B:1,19:P1.5,2 | Norway | |
| 16260 (ST-37) | B:11:P1.1,7 | Norway | |
| 16282 (ST-1301) | NG:1:P1.5,2 | Norway | |
| IS1301 | 2022 | NG:4:P1.10 | Norway |
All strains in allele 1 belong to cluster ET-5.
FIG. 1.
Nucleotide sequence of the nadA locus of the four N. lactamica strains, showing the 542-bp insertion. The insertion sequence is indicated by red type. The sequences located upstream and downstream of the insertion site of nadA gene and found in all meningococcal strains are indicated by black type. Blue type indicates the 16 bp present in all N. lactamica strains and in the N. meningitidis strains lacking nadA. When the nadA gene is present in a meningococcus, it always replaces these 16 bp. The direct repeats of the N. lactamica insertion and the 6-nucleotide direct repeats found in all meningococcal strains are underlined.
nadA4 and its encoded peptide.
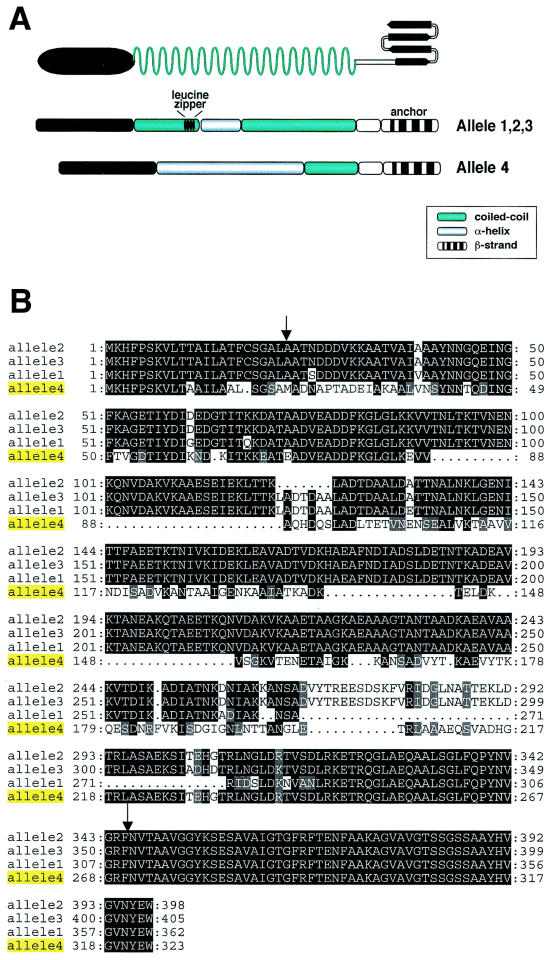
The nadA4 gene codes for a protein (NadA4) consisting of 323 amino acids, which is shorter than the proteins encoded by alleles 1, 2, and 3 (NadA1, NadA2, and NadA3), which are composed of 362, 398, and 405 amino acids, respectively. An alignment of the four NadA alleles and the overall similar secondary structures of NadA4 and the other alleles are shown in Fig. 2. Secondary-structure analysis of NadA1, NadA2, and NadA3 revealed that each protein can be divided into three domains: an N-terminal globular head, an internal region with high coiled-coil propensity, and a C-terminal membrane anchoring region. Secondary-structure analysis of NadA4 revealed that the amino-terminal region (amino acids 24 to 90) exhibits 63% homology with the proteins encoded by the other alleles and has an undefined secondary structure. The internal region exhibits only 25% identity with other NadA forms, but surprisingly it maintains the α-helix propensity (60%). The carboxy-terminal region (amino acids 244 to 323), which spans the four amphipatic β-strands and is predicted to form the membrane anchor of the protein, is perfectly conserved (100%). Compared to other forms, NadA4 has a disrupted form of the leucine zipper motif.
FIG. 2.
(A) Predicted topology of NadA and comparison of the secondary structures of NadA4 and other NadA forms. (B) Alignment of the four nadA-encoded peptide sequences. The arrows indicate the end of the leader peptide and the first amino acid of the membrane anchoring domain.
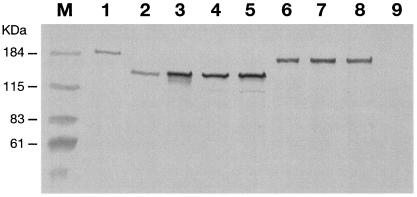
To verify that the carrier strains were able to express NadA, total cell extracts and/or OMVs were loaded on an SDS gel and incubated with an anti-NadA4 mouse polyclonal antibody. All carrier strains carrying NadA1 or NadA2 were able to express detectable levels of the protein as high-molecular-mass oligomers, like the strains isolated from patients (data not shown). The results of the Western blot analysis of strain 65/96, bearing NadA4, are shown in Fig. 3. NadA4 was present in the total extract of strain 65/96 (Fig. 3, lane 2) and migrated at a molecular mass lower than that of NadA3 (Fig. 3, lane 1). The protein migrated at an apparent molecular mass of approximately 150 kDa and probably represented high-molecular-mass oligomers, most probably trimers (Fig. 3). As expected, NadA was not present in the NGH38 strain used as a negative control (Fig. 3, lane 9). The presence of NadA4 in the OMV fractions (strain 65/96) suggests that the protein is successfully exported to the outer membrane. Interestingly, NadA4 was recognized in Western blots by antisera raised against each of the three alleles, indicating that the differences in the sequences do not impair immune recognition (data not shown). To confirm that NadA4 forms stable high-molecular-weight oligomers, like the other forms, the OMV fractions were subjected to particularly severe denaturing conditions (Fig. 3, lanes 3 to 8). The remarkable stability in the presence of heat and reducing agents even in the absence of the leucine zipper motif (Fig. 2A) suggests that the coiled-coil region alone is able to promote stable oligomerization of protein monomers. The lack of cysteine residues in NadA4, like the lack of such residues in the other NadA forms, indicates that disulfide bonds are not involved in this phenomenon either. This result is consistent with results obtained for YadA (an adhesin/invasin of Yersinia species), which is homologous to NadA (12) and which is able to form stable high-molecular-mass aggregates, even if cysteine residues and a leucine zipper motif are not present.
FIG. 3.
Evaluation of the presence and stability of the NadA4 protein. Unless indicated otherwise, all samples were loaded on the gel after addition of sample buffer and boiling for 10 min. Lane 1, strain 2996 (NadA3) total protein; lane 2, strain 65/96 (NadA4) total protein;lanes 3, 4, and 5, strain 65/96 (NadA4) OMVs boiled for 30 min, resuspended in 2% Triton X-100, and resuspended in 2% SDS, respectively; lanes 6, 7, and 8, strain 2996 (NadA3) OMVs boiled for 30 min, resuspended in 2% Triton X-100, and resuspended in 2% SDS, respectively; lane 9, strain NGH38 OMVs (negative control).
NadA4 is able to induce antibodies with bactericidal activity and adhere to epithelial cells.
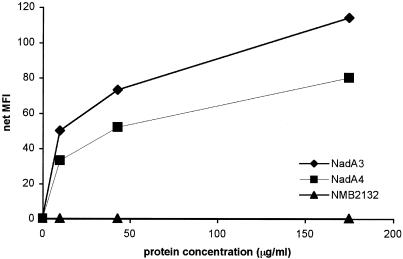
To check whether NadA4 is able to induce antibodies with bactericidal activity, the protein, devoid of the leader peptide and of the anchoring domain (Fig. 2A), was expressed in E. coli as a His fusion protein, purified, and used to immunize mice in the presence of either Freund's adjuvant or alum as an adjuvant. Immune sera were then tested with a bactericidal assay by using strains 65/96 and 2996, which harbor NadA4 and NadA3, respectively. The results (Table 3) showed that NadA4 was able to induce antibodies which were bactericidal against the homologous strain when they were used in combination with either Freund's adjuvant or alum. However, the bactericidal activity against a NadA3-harboring strain was very low or absent. These results indicate that NadA4 is not able to mount a cross-bactericidal response, unlike NadA1, NadA2, and NadA3, which have been proven to induce bactericidal antibodies against each other, regardless of the sequence variations. Antibodies against NadA3 were highly bactericidal against the homologous strain (2996), whereas the bactericidal titer against strain 65/96, expressing NadA4, was low and present only when immunization was performed in the presence of aluminum hydroxide as the adjuvant. These results suggest that there are some protective epitopes which are common to NadA3 and NadA4, whose conformation could be affected by the adjuvant. NadA is able to bind epithelial cells. To verify that NadA4 has the binding activity of the other forms, recombinant NadA4 was used in a binding assay. Chang epithelial cells were incubated with increasing concentrations of recombinant NadA4 and with either recombinant NadA3 as a positive control or an unrelated protein, recombinant protein NMB2132 (18), as a negative control. Binding was detected by fluorescence-activated cell sorting. As shown in Fig. 4, NadA4 binding was dose dependent, and the mean fluorescence index was approximately 30% reduced compared to that of NadA3, probably indicating reduced binding affinity.
TABLE 3.
Comparison of the serum bactericidal activity titers induced by NadA3 and NadA4 against homologous and heterologous strains
| Antigen | Adjuvant | Serum bactericidal activity titer
|
|
|---|---|---|---|
| Strain 2996 (allele 3) | Strain 65/96 (allele 4) | ||
| NadA3 | Freund's | 32,768 | <16 |
| Alum | 16,384 | 256 | |
| NadA4 | Freund's | 32 | 4,096 |
| Alum | 128 | 2,048 | |
FIG. 4.
Fluorescence-activated cell sorting analysis to compare the binding ability of NadA4 with the binding ability of NadA3. Chang epithelial cells were incubated with medium alone or with three concentrations of the recombinant proteins. MFI, mean fluorescence intensity.
DISCUSSION
A major unanswered question about N. meningitidis is the degree of similarity between the strains isolated from healthy individuals (carrier strains) and the strains isolated from people with disease. Apart from the capsule (6), which can account for the overall higher rate of nongroupable strains in carriage strains than in pathogens, no other pathogenicity factors could consistently explain the difference between strains able to evoke disease and commensals. In this work, we found that the nadA gene is an additional genetic marker that segregates differently in the two populations; while it is present in 50% of the strains isolated from patients and in members of three of the four hypervirulent clusters, it is largely absent in most of the strains isolated from healthy people (only 16.2% of such strains carry the gene). Remarkably, in this population, most of the nadA-positive strains (18 of 25 strains) belonged to the ET-5 and ET-37 hypervirulent lineages. Strains belonging to hypervirulent clusters are seldom isolated from healthy individuals (less than 10%) (5) and are apparently indistinguishable from homologous strains isolated from patients. The ET-5 and ET-37 strains used in this study represent the overlap between the two populations. The differences between hypervirulent cluster strains isolated from patients and hypervirulent cluster strains isolated from healthy individuals should be analyzed further in order to understand whether the different behaviors of genetically similar, if not identical, strains could be due to different abilities of the isolates to cause disease or due to different susceptibilities to disease of the different hosts. When we examined the carrier strains used in the present work that did not belong to any hypervirulent cluster, we found that the nadA gene was present in only 5.1% of them. Interestingly, all but two of these strains harbored a new nadA allele which we found only in carrier strains and which we designated nadA4. The NadA4 protein is expressed and found in the bacterial surface, and it is able to induce antibodies with bactericidal activity and to bind to epithelial cells like the other three forms of NadA. Questions to be answered in the future are whether we identified, in the carrier strain population, a subgroup of isolates which are potentially more virulent than the isolates lacking NadA and, if NadA4 does not confer any additional virulence properties, what the function of this protein is. In conclusion, we believe that a partial answer to the question of why in most countries only a few individuals colonized by N. meningitidis develop disease is beginning to emerge. Most of the people are colonized by strains which lack factors that are essential for virulence, such as the capsule, or that are potentially important for virulence, like NadA. These strains are unlikely to be able to cause disease and should be considered commensals, like N. lactamica, which also lacks the capsule and which, as shown in this work, also lacks the nadA gene. However, the commensal strains are likely to play an important role in providing exchange of genetic material and in priming immunity against the virulent strains (9, 15, 16, 21).
Acknowledgments
We thank C. Mallia for editing the manuscript, G. Corsi for providing artwork, R. Rossi for providing the OMV preparation, and R. Moxon for providing eight N. lactamica strains. In this work we used the MLST website (http://www.mlst.net) developed by Man-Suen Chan and Keith Jolley; the development of this site is funded by the Wellcome Trust.
Editor: J. N. Weiser
REFERENCES
- 1.Ala'Aldeen, D. A., K. R. Neal, K. Ait-Tahar, J. S. Nguyen-Van-Tam, A. English, T. J. Falla, P. M. Hawkey, and R. C. Slack. 2000. Dynamics of meningococcal long-term carriage among students and their implications for mass vaccination. J. Clin. Microbiol. 38:2311-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cartwright, K. A. (ed.). 1995. Meningococcal carriage and disease, p. 114-146. John Wiley and Sons, Chichester, United Kingdom.
- 3.Caugant, D. A., E. A. Høiby, E. Rosenqvist, L. O. Froholm, and R. K. Selander. 1992. Transmission of Neisseria meningitidis among asymptomatic military recruits and antibody analysis. Epidemiol. Infect. 109:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caugant, D. A., K. Bøvre, P. Gaustad, K. Bryn, E. Holten, E. A. Hoiby, and L. O. Froholm. 1986. Multilocus genotypes determined by enzyme electrophoresis of Neisseria meningitidis isolated from patients with systemic disease and from healthy carriers. J. Gen. Microbiol. 132:641-652. [DOI] [PubMed] [Google Scholar]
- 5.Caugant, D. A., E. A. Hoiby, P. Magnus, O. Scheel, T. Hoel, G. Bjune, E. Wedege, J. Eng, and L. O. Froholm. 1994. Asymptomatic carriage of Neisseria meningitidis in a randomly sampled population. J. Clin. Microbiol. 32:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claus, H., M. C. Maiden, R. Maag, M. Frosch, and U. Vogel. 2002. Many carried meningococci lack the genes required for capsule synthesis and transport. Microbiology 148:1813-1819. [DOI] [PubMed] [Google Scholar]
- 7.Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Aricò, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 11:1445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frasch, C. E. 1979. Non-capsular surface antigens of Neisseria meningitidis. Semin. Infect. Dis. 2:304-337. [Google Scholar]
- 9.Gold, R., I. Goldschneider, M. L. Lepow, T. F. Draper, and M. Randolph. 1978. Carriage of Neisseria meningitidis and Neisseria lactamica in infants and children. J. Infect. Dis. 137:12-121. [DOI] [PubMed] [Google Scholar]
- 10.Goldschneider, I., E. C. Gotschlich, and M. S. Artenstein. 1969. Human immunity to the meningococcus. II. Development of human immunity. J. Exp. Med. 129:1327-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilse, R., S. Hammerschmidt, W. Bautsch, and M. Frosch. 1996. Site-specific insertion of IS1301 and distribution in Neisseria meningitidis strains. J. Bacteriol. 178:2527-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoiczyk, E., A. Roggencamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jodar L., I. M. Feavers, D. Salisbury, and D. M. Granoff. 2002. Development of vaccines against meningococcal disease. Lancet 359:1499-1508. [DOI] [PubMed] [Google Scholar]
- 14.Jolley, K. A., J. Kalmusova, E. J. Feil, S. Gupta, M. Musilek, P. Kriz, and M. C. Maiden. 2000. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 38:4492-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremastinou, J., G. Tzanakaki, A. Pagalis, M. Theodondou, D. M. Weir, and C. C. Blackwell. 1999. Detection of IgG and IgM to meningococcal outer membrane proteins in relation to carriage of Neisseria meningitidis or Neisseria lactamica. FEMS Immunol. Med. Microbiol. 24:73-78. [DOI] [PubMed] [Google Scholar]
- 16.Linz, B., M. Schenker, P. Zhu, and M. Achtman. 2000. Frequent interspecific genetic exchange between commensal neisseriae and Neisseria meningitidis. Mol. Microbiol. 36:1049-1058. [DOI] [PubMed] [Google Scholar]
- 17.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, J. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Aricò, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 19.Simmons, G., D. Martin, J. Stewart, N. Jones, L. Calder, and D. Bremmer. 2001. Carriage of Neisseria meningitidis among household contacts of patients with meningococcal disease in New Zealand. Eur. J. Clin. Microbiol. Infect. Dis. 20:237-242. [DOI] [PubMed] [Google Scholar]
- 20.Swartley, J. S., A. A. Marfin, S. Edupuganti, L.-J. Liu, P. Cieslak, B. Perkins, J. D. Wenger, and D. S. Stephens. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 94:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troncoso, G., S. Sanchez, M. Moreda, M. T. Criado, and C. M. Ferreiros. 2000. Antigenic cross-reactivity between outer membrane proteins of Neisseria meningitidis and commensal Neisseria species. FEMS Immunol. Med. Microbiol. 27:103-109. [DOI] [PubMed] [Google Scholar]
- 22.van der Ley, P., J. E. Heckels, M. Virji, P. Hoogerhout, and J. T. Poolman. 1991. Topology of outer membrane porins in pathogenic spp. Infect. Immun. 59:2963-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Deuren, M., P. Brandtzaeg, and J. W. M. van der Meer. 2000. Update on meningococcal disease with emphasis on pathogenesis and clinical management. Clin. Microbiol. Rev. 13:144-166. [DOI] [PMC free article] [PubMed] [Google Scholar]