Abstract
Genes involved in the process of memory formation have been studied intensively in model organisms; however, little is known about the mechanisms that are responsible for natural variation in memory dynamics. There is substantial variation in memory retention among closely related species in the parasitic wasp genus Nasonia. After a single olfactory conditioning trial, N. vitripennis consolidates long-term memory that lasts at least 6 days. Memory of the closely related species N. giraulti is present at 24 h but is lost within 2 days after a single trial. The genetic basis of this interspecific difference in memory retention was studied in a backcrossing experiment in which the phenotype of N. giraulti was selected for in the background of N. vitripennis for up to five generations. A genotyping microarray revealed five regions that were retained in wasps with decreased memory retention. Independent introgressions of individual candidate regions were created using linked molecular markers and tested for memory retention. One region on chromosome 1 (spanning ∼5.8 cM) and another on chromosome 5 (spanning ∼25.6 cM) resulted in decreased memory after 72 h, without affecting 24-h-memory retention. This phenotype was observed in both heterozygous and homozygous individuals. Transcription factor CCAAT/enhancer-binding protein and a dopamine receptor, both with a known function in memory formation, are within these genomic regions and are candidates for the regulation of memory retention. Concluding, this study demonstrates a powerful approach to study variation in memory retention and provides a basis for future research on its genetic basis.
Introduction
The ability to learn and form memory is vital for animal species. Learning allows individuals to adjust to variation in their environment. A learning experience can result in the formation of different types of memory, which differ in their characteristics and the cellular pathways involved. Immediately after a learning experience, short-term memory is formed, which is a labile type of memory that typically lasts for minutes up to hours at most (Menzel, 1999; Margulies et al., 2005; van den Berg et al., 2011). This type of memory can be disrupted by anaesthesia, such as a cold shock in insects and is, therefore, also classified as anaesthesia-sensitive memory. More durable and less labile types of memory include anaesthesia-resistant memory (ARM), which can last from hours to days, and long-term memory (LTM), which is dependent on protein synthesis and can last up to the entire life time of an animal (Margulies et al., 2005; Eisenhardt, 2006; Smid et al., 2007).
Both the process of memory formation and the cellular mechanisms involved are highly conserved among distant animal phyla (Dubnau, 2003). There is, however, interspecific and intraspecific natural variation in memory dynamics (Hoedjes et al., 2011). In most cases, a single conditioning trial results in the formation of short-term memory and/or ARM, which is lost after hours to days. Many animal species will only form LTM after multiple conditioning trials, which are spaced in time (Margulies et al., 2005; Eisenhardt, 2006; Smid et al., 2007). However, there is variation in the number of trials required to form LTM. Some species, including a number of parasitic wasps, even consolidate LTM after a single conditioning trial (Krashes and Waddell, 2008; Hoedjes et al., 2011). Parasitic wasps are instrumental for studying natural variation in memory retention and the number of conditioning trials required to form LTM, as there is substantial variation between closely related parasitic wasp species (Hoedjes et al., 2011).
The mechanisms that are responsible for natural variation in memory dynamics between individuals or species are largely unknown (Hoedjes et al., 2011). One gene that is known to be involved in natural variation in both short-term memory and LTM formation in Drosophila melanogaster is the cGMP-dependent protein kinase foraging (Mery et al., 2007). Other studies have investigated natural variation in memory dynamics in parasitic wasps and in fruit flies using an experimental evolution procedure but did not study the genetic basis of this trait (Mery and Kawecki, 2002; van den Berg et al., 2011). In contrast, studies using laboratory-generated mutants of D. melanogaster have been highly successful in identifying single loci with large effects on memory formation, including genes that are specifically involved in LTM formation (for example, Margulies et al., 2005; Keene and Waddell, 2007; Song et al., 2009; Copf et al., 2011). For example, the cAMP-responsive transcription factor CREB is known to regulate the number of trials required to form LTM in D. melanogaster; induced expression of a specific splice variant resulted in LTM formation after a single conditioning trial, whereas 10 spaced conditioning trials were required normally (Tubon et al., 2013). Overexpression of tyrosine phosphatase SHP2 (corkscrew), also in D. melanogaster, induces LTM formation after 10 massed conditioning trials, that is, trials with a short intertrial interval (Pagani et al., 2009). It remains to be investigated whether these and other known LTM genes are also involved in natural variation in memory retention, which is the focus of this study.
Parasitic wasp species of the genus Nasonia provide unique opportunities to study natural variation in memory retention and LTM formation. Nasonia vitripennis will form LTM, which will last over 6 days, after a single Pavlovian conditioning trial in which an odour is associated with the reward of finding a host. In contrast, a single conditioning trial results in the formation of ARM in the closely related species N. giraulti and this memory will be lost within 1–2 days after conditioning. Only multiple conditioning trials will result in the formation of long-lasting memory in N. giraulti (Hoedjes et al., 2012; Hoedjes and Smid, 2014). The genus Nasonia has emerged as a model system with powerful genetic tools to study interspecific variation, which include sequenced genomes and a high-density genetic map (Werren and Loehlin, 2009; Werren et al., 2010; Desjardins et al., 2013). The four described species of the genus can interbreed when cured of a Wolbachia infection that prevents hybridization in nature, which provides excellent opportunities for quantitative trait locus (QTL) mapping and cloning. Recent research has successfully backcrossed traits of N. giraulti into a N. vitripennis background in order to study the genetic basis of interspecies differences in wing size and morphology, female host preference behaviour, hybrid incompatibilities and pheromone composition (Niehuis et al., 2008; Desjardins et al., 2010; Loehlin and Werren, 2012; Niehuis et al., 2013). We hypothesized that memory retention can also be introgressed from one Nasonia species to another, which facilitates studies on the genetic basis of this trait. Introgression of memory-related phenotypes was successful in a number of studies on mice and rats (Wehner et al., 1997; Jarome et al., 2010). Compared with these mammalian species, Nasonia has a much shorter generation time and is easier to rear and handle. Furthermore, these Hymenopteran species have a haplodiploid mating system, that is, females are diploids, whereas males are haploids that develop from unfertilized eggs, which make them particularly suitable for introgression and genetic dissection (Werren and Loehlin, 2009). This study is the first to investigate QTLs that underlie a naturally occurring difference in memory retention and LTM formation between closely related species, which enables us to address the number of loci involved in this phenotype and their dominance.
We studied memory retention 24 h after conditioning (likely a form of ARM), which we refer to as short-lasting memory in this study, and after 48 h or more following conditioning (which can include ARM and LTM), referred to as long-lasting memory. The aim of this study is to specifically identify genes involved in reduced long-lasting memory (N. giraulti phenotype, that is, short-memory retention), without affecting short-lasting memory (observed in both species). The genetic introgression process was carried out for four generations while selecting for giraulti-like memory retention. Then, selected introgression lines were genotyped (Desjardins et al., 2013). Individual candidate memory retention QTLs were then independently introgressed and their effects on memory retention determined, resulting in confirmation of two chromosomal regions containing memory retention QTLs.
Materials and methods
Nasonia strains and maintenance
Experiments were performed with N. vitripennis (AsymCx) and N. giraulti (RV2x(U)) strains, which are completely homozygous and have sequenced genomes (Werren et al., 2010), or on hybrids and introgression lines derived from a cross between these two strains. Wasps were reared on Calliphora vomitoria fly pupae as described in Hoedjes et al. (2012). Female wasps were collected on the day of emergence, and were then mated, provided honey and water and kept in a climate cabinet at a temperature of 25 °C and a photoperiod of 16L:8D for 1–3 days until conditioning.
Olfactory conditioning and memory retention test
Female parasitic wasps were conditioned using a Pavlovian conditioning assay in which an odour (chocolate or vanilla odour, the conditioned stimulus (CS+)) was associated with the reward of access to a host (the unconditioned stimulus (US)), a C. vomitoria pupa. The protocol for individual conditioning as described in Hoedjes et al. (2012) was adapted to allow conditioning of groups of wasps in order to obtain large numbers of conditioned wasps that were required for the experiments (see Supplementary Information for more details).
Memory retention was tested in a T-maze olfactometer as described in Hoedjes et al. (2012). Briefly, the olfactometer consists of two tubular arms, which are connected to a middle tube, with a continuous airflow of 100 ml min−1 through each arm. Chocolate odour was offered on one side and vanilla odour was offered on the other side. Groups of 10–12 wasps were released in the middle of the T-maze, the numbers of wasps in the two arms were recorded after 10 min and the percentage that had chosen each odour was calculated. Immediately afterwards, a reciprocal group of wasps was tested. The difference in odour preference between the two reciprocal groups, that is, one group conditioned with vanilla as CS+ and chocolate as CS−, and a second group conditioned with chocolate as CS+ and vanilla as CS− (respectively, group 1 and group 2), is traditionally used as a measure for memory retention and is represented by the Performance Index (PI; Tully et al., 1994). PI was calculated as follows: group 1 (% CS+)−group 2 (% CS−). A t-test is used to test for significant memory retention, that is, whether a PI is significantly different from 0. Univariate analysis of variance (ANOVA) was used to test for effects of selection procedure or genotype on memory retention. If applicable, the effect of time point of the memory retention test (a covariate) and the interaction between genotype and time point was tested as well. A Tukey-HSD (honestly significant difference) post hoc test was used when appropriate (SPSS version 19; IBM, Armonk, NY, USA).
Initial introgression of memory retention genes from N. giraulti into N. vitripennis background
F1 hybrids were generated by mating N. vitripennis females to N. giraulti males. This cross was chosen because nuclear–mitochondrial incompatibilities can complicate introgressions in the reciprocal cross (Breeuwer and Werren, 1995). Memory retention of the hybrids was compared with both parental strains to assess which phenotype was dominant. Memory retention was tested 24 (±1) and 48 (±1) h after conditioning. For the introgression experiment, the aim was to select wasps that demonstrate memory retention after 24 h, but not after 48 h or later (similar to N. giraulti), and to backcross this short-memory retention into the background of N. vitripennis. Virgin female offspring were mated to N. vitripennis males. An entire group of sisters was subsequently conditioned on either vanilla or chocolate odour (CS+) as described above; approximately similar numbers of wasps were conditioned on each of the two odours. Wasps were tested 20–24 h after conditioning and wasps that failed to walk towards the learned odour were removed from the experiment to avoid selecting wasps with general defects in the learning and memory pathways. The remaining wasps were tested twice between 60 and 72 h after conditioning, which was a more convenient time frame than 48 h after conditioning. Wasps that walked toward vanilla and chocolate were collected separately after the first memory test and then tested as two separate groups for the second time in order to select wasps that chose the ‘correct' and ‘wrong' odour twice. Wasps that chose the ‘wrong' odour (CS−) twice were considered to have lost their memory, similar to N. giraulti, and these wasps were selected to continue introgression of this memory phenotype. Wasps that chose the learned odour (CS+) twice were considered to be wasps with long-lasting memory and were selected to create long (N. vitripennis-like) memory retention lines during introgression. These lines were used as controls with which short-memory retention lines (N. giraulti-like) were compared. Additional experiments that test effects of multiple memory retention tests on the observed memory behaviour of N. vitripennis and N. giraulti are described in the Supplementary Information. Selected females were individually provided three hosts in a glass tube closed with a cotton wool plug to generate offspring.
Initially, a total of 20 F1 hybrid females were mated to N. vitripennis males and their female offspring were conditioned and tested for memory retention. Offspring were further backcrossed with N. vitripennis and conditioned and tested as described above up to the 4th backcross generation (Figure 1a). Short-memory retention lines and control lines were created in the first backcross generation and were maintained separately from each other. Every generation, a total of 15–25 female offspring from wasps of the short-memory retention lines were selected to continue independent introgression lines. Similarly, 15–25 female offspring from wasps with long-memory retention (that is, N. vitripennis-like) were selected to continues independent control lines. Offspring from females that were conditioned on vanilla were conditioned on chocolate the next generation to avoid selection for a specific odour preference.
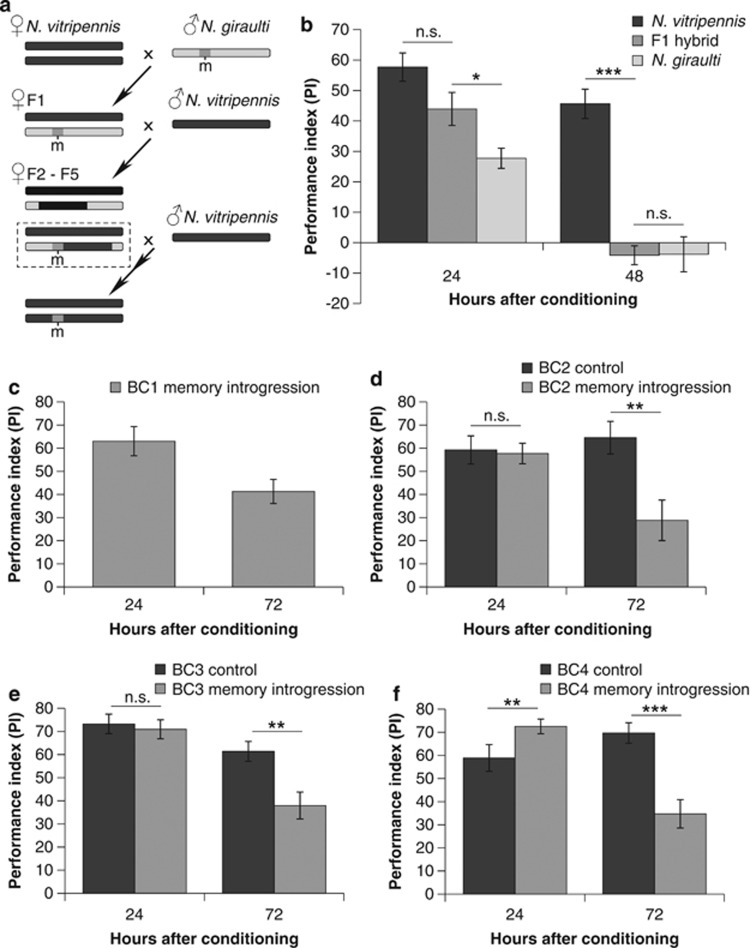
Figure 1.
Initial introgression of memory retention genes. (a) Genes that control long-lasting memory (‘m') were backcrossed from N. giraulti (<48 h memory retention) into the background of N. vitripennis (>144 h memory retention). (b) Memory retention of F1 hybrids was compared with both parentals. (c) Selection for decreased long-lasting memory while backcrossing was started in the BC1 generation and resulted in decreased memory scores in the memory introgression lines when tested after 72 h, but not after 24 h in the (d) BC2, (e) BC3 and (f) BC4 generations. Level of significance: ***P<0.001, **<P<0.01, *0.01<P<0.05.
As entire groups of sisters were conditioned on a single odour, an adjusted PI was calculated for this experiment. Approximately 50% of unconditioned wasps will choose the ‘learned odour' (as shown in Hoedjes et al. (2012)) and ‘half' of a PI can be calculated by subtracting 50% from the percentage of wasps that chose the learned odour. The PI is obtained by multiplying this number by two: (% CS+ −50%) × 2.
A back-up was created during the third generation of backcrossing to ensure continuation of the project during transition from the United States of America to the Netherlands. Experiments on these back-up lines are described in the Supplementary Information.
Genotyping of memory introgression lines
A total of eight samples were genotyped, each of which consisted of the pooled DNA of up to 10 wasps from a sibship. Four pairs of samples were taken from four independent short-memory retention lines. One sample of each pair consisted of up to 10 sisters that had chosen the ‘wrong' odour twice (short-memory retention), whereas the other sample consisted of up to 10 sisters that had chosen the learned odour twice (N. vitripennis-like memory retention, which was used as a control). Samples were genotyped using the high-density comparative genomic hybridization (CGH) genotyping microarray for Nasonia, which contains more than 20 000 markers, and were analysed using a bulk-segregant analysis (Desjardins et al., 2013). Two pairs of samples were taken from the third backcross generation and two from the fifth backcross generation. The four pairs of samples were composed of groups of sisters that each had been derived from a different female selected in the first backcross generation. The genotype of individual wasps was confirmed using indel markers within observed introgressed regions in a polymerase chain reaction (PCR). The genotyping analyses have been described more in detail in the Supplementary Information.
Confirmation of memory retention QTLs by independent introgressions
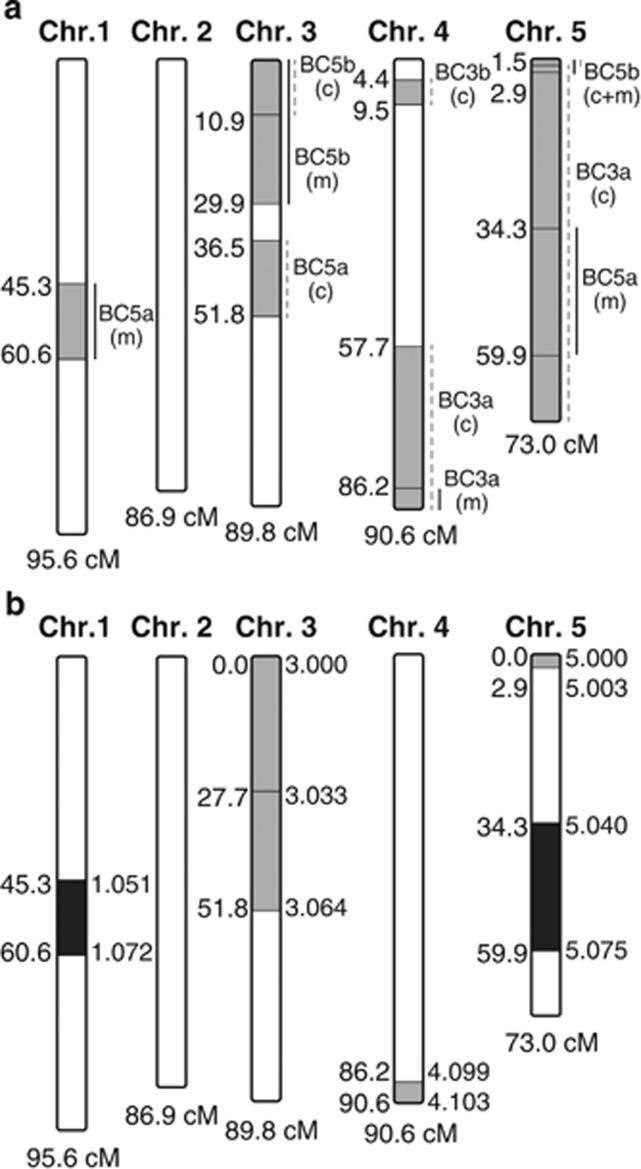
A total of five introgressed regions were observed in wasps with decreased long-lasting memory retention in the microarray experiment. To independently and individually test these candidate regions, each was then backcrossed from N. giraulti into the background of N. vitripennis for a confirmation experiment, using linked molecular markers to track each region during introgression by PCR genotyping (see Supplementary Information for more details). The experiment is independent from the initial introgression experiment and selection depended on genotype alone and not on phenotype. F1 hybrids were generated as described above and backcrossed to N. vitripennis up to the third to fifth backcross generation, before the effects of the introgressed regions on memory retention were tested. For each of the five regions of interest, multiple primers were used that are located close to the border of the region and/or within the region. Each of these introgression lines had a single region of interest that was backcrossed. The locations of the regions on the linkage map of Desjardins et al. (2013) are given. A single region on chromosome 1 (location: 45.3–60.6 cM), a small region on chromosome 4 (location: 86.2–90.6 cM) and two regions on chromosome 5 (locations: 0.0–2.9 and 34.3–59.9 cM) were introgressed and tested. Two lines with different parts of the region of interest on chromosome 3 were created (locations: 0.0–27.7 and 27.7–51.8 cM).
The introgressed regions were maintained heterozygously throughout this experiment. Hybrid females were mated to N. vitripennis, which, therefore, results in a mix of female offspring with and without the introgressed region. To test the effect of an introgressed region on memory retention, sibling females were individually provided five hosts in a glass vial as described above. Their female offspring were conditioned and tested for memory retention after 72 (±2) h; this time point was chosen instead of 48 h to match the time point of selection in the initial introgression experiment. Initially, memory retention was tested after three (chromosomes 1 and 3), four (chromosomes 3 and 5) or five (chromosome 4) generations of backcrossing. Offspring of wasps that were heterozygous for the region of interest were compared with offspring of sister wasps that were homozygous N. vitripennis for the region of interest (control group). This approach controls for unlinked regions segregating in the offspring that could affect memory retention.
Isogenic sibships were then produced, which allows for more reliable mapping of phenotype to genotype (Velthuis et al., 2005). Regions that were found to have an effect on PI were investigated further by testing memory retention of offspring of males with the introgressed region, which were mated to N. vitripennis females to produce isogenic female sibships containing the heterozygous region of interest. Wasps were conditioned and memory retention was tested after 24 (±2) and 72 (±2) h as described earlier. Offspring of males that carried the region of interest (that is, offspring were heterozygous for the markers linked to memory retention) were compared with offspring of brothers without the introgressed region (that is, homozygous N. vitripennis for the markers; control group). The design controls for unlinked segregating genetic variation because sibships are compared from the same introgressed lineage with and without the target region.
Experiments were carried out to generate homozygous strains for regions of interest that affected long-lasting memory, in combination with further partitioning of the genomic regions by recombination. Using recombination, we succeeded in generating a strain that was homozygous for a part of the introgressed region on chromosome 1 (location: 54.8–60.6 cM). This strain was named ‘SIL_LTM1A_gV' and the strain was tested for memory retention after 24 (±2), 72 (±2) and 120 (±2) h after a single conditioning. Memory retention of this segmental introgression line was compared with N. vitripennis and N. giraulti. The size of the introgressed region was determined using PCR as described above.
Results
Initial introgression of memory retention genes from N. giraulti to N. vitripennis
Memory retention of N. vitripennis, N. giraulti and F1 hybrids (N. vitripennis females × N. giraulti males) was tested (Figure 1b). There was an effect of genotype on the PI both after 24 and 48 h (24 h: F2,27=10.93, P<0.001; 48 h: F2,27=37.65, P<0.001; n=10 PIs for each genotype and time point). After 24 h, memory of N. vitripennis (NV) and N. giraulti (NG) differ significantly from each other (Tukey–HSD: N. vitripennis vs N. giraulti <0.001) and the PIs of the F1 hybrids were almost intermediate to the parentals (Tukey–HSD: N. vitripennis vs F1=0.099, N. giraulti vs F1=0.045). The finding that F1 hybrids are intermediate suggests the presence of one or more loci affecting short-lasting memory. This result suggests at least some co-dominance, although a combination of fully dominant and fully recessive loci is an option as well. By 48 h, memory of N. giraulti has been lost, whereas N. vitripennis still had significant memory retention. Memory retention of the F1 hybrids is similar to N. giraulti (Tukey–HSD: N. vitripennis vs F1<0.001, N. giraulti vs F1=1.000).
The dominant N. giraulti phenotype was backcrossed into N. vitripennis genetic background (Figure 1a). The BC1 generation (Figure 1c) is a mixture of different genotypes and memory retention phenotypes, from which introgression lines (N. giraulti phenotype) and control lines were selected. In the BC2 generation (Figure 1d), both control and introgression lines have similar PIs at 24 h after conditioning (F1,40=0.44, P=0.835, introgression: n=22 PIs, control: n=20 PIs). After 72 h the PIs of the introgression lines have decreased in comparison with the control lines (F1,30=9.73, P=0.004, introgression: n=17 PIs, control: n=15 PIs). A similar pattern of memory retention is visible in the BC3 generation (Figure 1e; 24 h: F1,50=0.15, P=0.699, introgression: n=27 PIs, control: n=25 PIs; 72 h: F1,42=10.25, P=0.003, introgression: n=23 PIs, control: n=21 PIs). In the BC4 generation (Figure 1f), the introgression line has higher PIs compared with the control at 24 h after conditioning (F1,57=7.55, P=0.008, introgression: n=35 PIs, control: n=24 PIs) but again a decreased memory retention after 72 h (F1,46=17.36, P<0.001, introgression: n=29 PIs, control: n=19 PIs).
Genotyping of memory retention introgression lines
A total of four selected introgression lines were genotyped using the microarray (Desjardins et al., 2013). Two pairs of samples (decreased long-lasting memory and control) were taken from the third backcross generation and two pairs were taken from the fifth backcross generation. The number and characteristics of regions that were retained during the introgression process are shown in Figure 2a and Table 1. A total of five regions was found in wasps with decreased long-lasting memory. Two of these regions (on chromosome 1 and part of the region on chromosome 3) were only detected in wasps with decreased long-lasting memory, whereas the other regions were also detected in control samples (which consist of sisters that demonstrate long-memory retention). None of these five regions was detected in multiple samples. Two regions were detected only in control samples.
Figure 2.
Potential memory retention QTLs. (a) Multiple heterozygous regions were detected by genotyping microarray in the four pairs of samples (BC3a, BC3b, BC5a and BC5b). The grey sections show the location of these heterozygous regions on each of the five chromosomes of Nasonia, which is also given in cM on the left. The lines on the right of each section show the size of the introgressed region in each of the samples (m—decreased long-lasting memory, black lines; c—control, dashed grey lines). (b) A total of six introgression lines were generated to test the effects of individual QTLs on memory retention. The grey sections indicate the location of these regions on the chromosomes of Nasonia, the location is given in cM on the left of each region and the marker cluster on the linkage map of Desjardins et al. (2013) is given on the right. The two dark grey regions affect memory retention, whereas the light grey regions did not.
Table 1. Potential memory retention QTLs.
| Sample | QTL location | QTL size | Marker clusters | Number with QTL | |
|---|---|---|---|---|---|
| BC3a-memory | Chr. 4, 86.2–90.6 cM | ∼4.4 cM | ∼2.1 Mb | 4.099–4.103 | 2 of 3 (66.7%) |
| BC3a-control | Chr. 4, 57.7–90.6 cM | ∼32.9 cM | ∼6.3 Mb | 4.071–4.103 | 5 of 10 (50.0%) |
| Chr. 5, 1.5–73.0 cM | ∼71.5 cM | ∼32.4 Mb | 5.001–5.091 | 8 of 10 (80.0%) | |
| BC3b-memory | None observed | n.a. | n.a. | n.a. | 0 of 4 (0%) |
| BC3b-control | Chr. 4, 4.4–9.5 cM | ∼5.1 cM | ∼1.0 Mb | 4.005–4.011 | 7 of 10 (70.0%) |
| BC5a-memory | Chr. 1, 45.3–60.6 cM | ∼15.3 cM | ∼44.0 Mb | 1.051–1.072 | 1 of 2 (50.0%) |
| Chr. 5, 34.3–59.9 cM | ∼25.6 cM | ∼19.4 Mb | 5.040–5.075 | 1 of 2 (50.0%) | |
| BC5a-control | Chr. 3, 36.5–51.8 cM | ∼15.3 cM | ∼23.9 Mb | 3.043–3.064 | 1 of 3 (33.3%) |
| BC5b-memory | Chr. 3, 0.0–29.2 cM | ∼29.2 cM | ∼8.0 Mb | 3.000–3.035 | 7 of 10 (70.0%) |
| Chr. 5, 0.0–2.9 cM | ∼2.9 cM | ∼1.8 Mb | 5.000–5.003 | 3 of 10 (30.0%) | |
| BC5b-control | Chr. 3, 0.0–10.9 cM | ∼10.9 cM | ∼4.0 Mb | 3.000–3.014 | 6 of 10 (60.0%) |
| Chr. 5, 0.0–1.5 cM | ∼1.5 cM | ∼0.8 Mb | 5.000–5.001 | 2 of 10 (20.0%) | |
This table shows the regions that were detected in each of the samples by genotyping microarray analyses and their characteristics; the location of the QTL on the linkage map (cM), the size in cM and the location of the QTL within the marker clusters on the linkage map of Nasonia (Desjardins et al., 2013). The QTL size is also estimated in Mb, by adding up scaffold sizes that were mapped to the genetic map by Desjardins et al. (2013). The QTL size can be larger than estimated because of gaps between scaffolds and a number of scaffolds that could not be mapped to the genetic map. Genotyping PCR was used to confirm the presence of heterozygous regions (N. vitripennis/N. giraulti) at the location of the QTL; the number of wasps with such a heterozygous region is given.
Confirmation of memory retention QTLs by independent introgressions
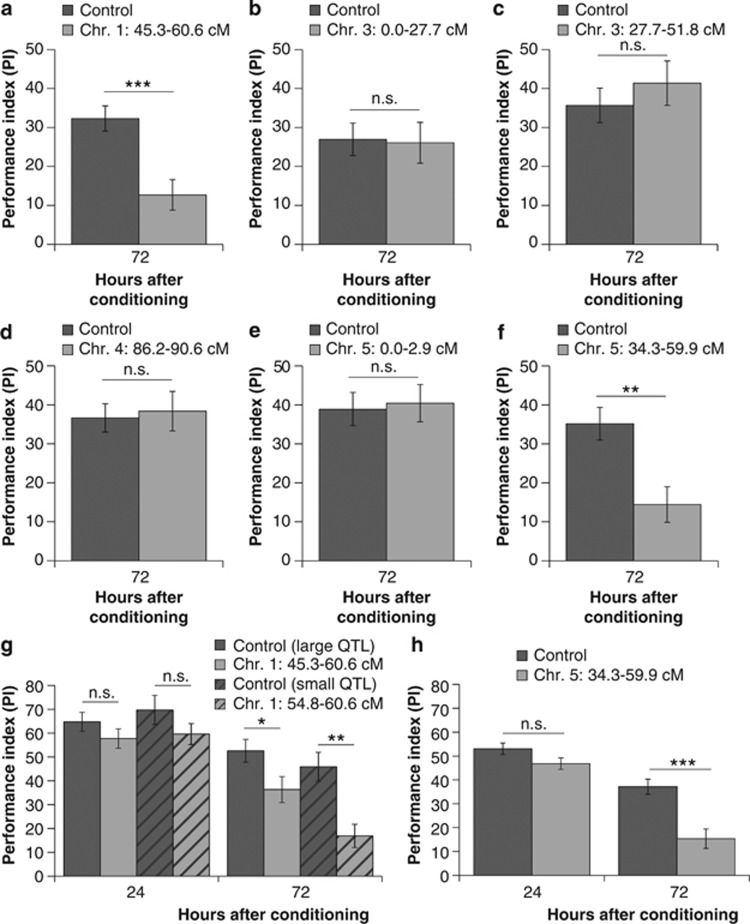
Following identification of candidate regions for memory retention, six different regions were separately introgressed (Figure 2b) and tested for memory retention. This time molecular markers were used to track the introgressions created for three to five generations, rather than selecting for memory retention during the introgression as before. Offspring of females that were heterozygous for these regions were tested for memory retention 72 h after conditioning (Figures 3a–f and Table 2). Two of the six QTLs resulted in a decreased 72-h memory compared with control wasps (offspring from the same family without the QTL region), although this memory had not completely disappeared, as is the case in N. giraulti. These two QTLs are located on chromosome 1 (location: 45.3–60.6 cM; Figure 3a) and chromosome 5 (location: 34.4–59.9 cM; Figure 3f). No decrease in memory retention was observed for the two tested QTLs on chromosome 3 (location: 0.0–27.7 and, 27.7–51.8 cM; Figures 3b and c), the QTL on chromosome 4 (location: 86.2–90.6 cM; Figure 3d), and the second tested QTL on chromosome 5 (location: 0.0–2.9 cM; Figure 3e).
Figure 3.
Confirming memory retention QTLs. A total of six regions were introgressed and tested for effects on 72 h memory (a–f). Additional experiments on the QTLs on (g) chromosome 1 and (h) chromosome 5 confirmed that these affected the 72-h memory, but not 24-h memory. Level of significance: ***P<0.001, **<P<0.01, *0.01<P<0.05.
Table 2. Statistical results are presented of univariate ANOVA tests on memory retention of the 6 introgression lines with potential memory retention QTLs (Figure 3).
| Figure | Genomic region | Time | F | P-value | n (PI's) |
|---|---|---|---|---|---|
| 3a | Chr. 1 (45.3–60.6 cM) | 72 h | F1,43=15.22 | <0.001 | control: 25, QTL: 20 |
| 3b | Chr. 3 (0.0–27.7 cM) | 72 h | F1,28=0.02 | 0.895 | control: 17, QTL: 13 |
| 3c | Chr. 3 (27.7–51.8 cM) | 72 h | F1,32=0.65 | 0.426 | control: 19, QTL: 15 |
| 3d | Chr. 4 (86.2–90.6 cM) | 72 h | F1,31=0.08 | 0.786 | control: 15, QTL: 18 |
| 3e | Chr. 5 (0.0–2.9 cM) | 72 h | F1,26=0.06 | 0.810 | control: 14, QTL: 14 |
| 3f | Chr. 5 (34.4–59.9 cM) | 72 h | F1,25=10.73 | 0.003 | control: 16, QTL: 11 |
| 3g | Chr. 1 (45.3–60.6 cM) | 24 h | F1,26=1,38 | 0.251 | control: 17, QTL: 11 |
| (i.e. large QTL) | 72 h | F1,27=4,75 | 0.038 | control: 18, QTL: 11 | |
| 3g | Chr. 1 (54.8–60.6 cM) | 24 h | F1,19=1,61 | 0.220 | control: 6, QTL: 15 |
| (i.e. small QTL) | 72 h | F1,21=10,37 | 0.004 | control: 6, QTL: 17 | |
| 3h | Chr. 5 (34.4–59.9 cM) | 24 h | F1,51=3.31 | 0.075 | control: 24, QTL: 29 |
| 72 h | F1,51=16.69 | <0.001 | control: 24, QTL: 29 |
Additional confirmation experiments were carried out for the region on chromosome 5 (location: 34.4–59.9 cM), and for the region on chromosome 1 (location: 45.3–60.6 cM; ‘large QTL') and a smaller subset of this region (location: 54.8–60.6 cM; ‘small QTL') resulting from a recombination event within the introgressed region on chromosome 1 (Figures 3g and h and Table 2). In these experiments, isogenic offspring of the inbred N. vitripennis line, mated to males with or without the introgressed region, were tested at 24 and 72 h after conditioning.
The QTL on chromosome 5 affects the 72-h memory retention, but not the 24-h memory retention when compared with controls (offspring from the same family without the QTL region). There was a significant interaction between the factors time point and genotype (F1,102=6,04, P=0.016; Figure 3h and Table 2).
Similarly, a comparison of the PI's of each genotype per time point showed that the QTLs on chromosome 1 also did not affect the 24-h memory, compared with the controls, but only the 72-h memory retention. This was observed for both the large and the small regions (Figure 3g and Table 2). This result suggests that the QTL that regulates memory retention is located within the smaller region on chromosome 1.
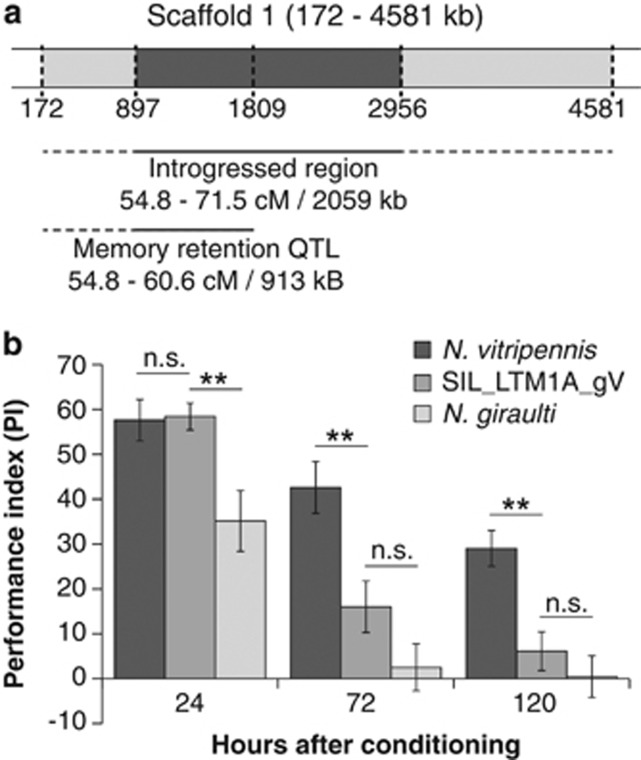
In addition, a homozygous introgression line, named ‘SIL_LTM1A_gV', was created for the QTL of interest on the smaller introgressed region on chromosome 1 (location: 54.8–60.6 cM; scaffold 1; 913 kb according to N. vitripennis genome assembly v1.0). The size of the introgressed region in SIL_LTM1A_gV was characterized (Figure 4a). The region results from one recombination event at ∼54.8 cM and a second recombination event between 71.5 and 83.2 cM. Consequently, the introgressed region has a size between 2059 and 4409 kb.
Figure 4.
Memory retention of the homozygous introgression line ‘SIL_LTM1A_gV'. A homozygous segmental introgression line was generated (a) that includes the memory retention QTL on chromosome 1, scaffold 1. The five markers used for genotyping this region are shown (vertical dashed lines). Recombination had occurred between 172–897 and 2956–4581 kb (within the light grey areas and dashed line below figure), which implicates that the introgressed region can have a size of up to 4409 kb. A homozygous N. giraulti introgressed region of at least 2059 kb (shown in dark grey and solid line below figure) was confirmed. The second line below the figure indicates the location of the QTL that was observed in the genotyping microarray experiment. (b) SIL_LTM1A_gV was compared with N. vitripennis and N. giraulti at 24, 72 and 120 h after conditioning. Level of significance: **0.001<P<0.01.
Memory retention of SIL_LTM1a_gV (SIL) was compared with memory retention of N. vitripennis (Figure 4b and Table 3). Comparison of the PIs of SIL_LTM1a_gV with N. vitripennis reveals that the PIs were not significantly different at 24 h after conditioning, indicating that the introgressed QTL does not affect the 24-h memory retention. At 72 h, memory retention of SIL_LTM1a_gV has decreased significantly compared with N. vitripennis (that is, the PI has decreased 62.4%), there is, however, still significant memory retention in SIL_LTM1a_gV (t9=2.79, P=0.021). After 120 h, the PI of SIL_LTM1a_gV is also significantly lower compared with N. vitripennis (that is, the PI has decreased 78.9%) and it is no longer significantly different from 0 (t9=1.42, P=0.189), that is, the memory trace appears to be completely lost in SIL_LTM1a_gV when measured after 120 h. The interaction of genotype and time point significantly affects PI (F2,56=5,81, P=0.019). Comparing SIL_LTM1a_gV with N. giraulti revealed that the PIs of the introgression line were higher at 24 h after conditioning, but are not significantly different from N. giraulti at both 72 and 120 h after conditioning (Figure 4b and Table 3).
Table 3. Statistical results are presented of univariate ANOVA tests on memory retention of the homozygous introgression line ‘SIL_LTM1A_gV', N. vitripennis and N. giraulti (Figure 4).
| Time | F | P-value | n (PI's) | Tukey-HSD post-hoc test |
|---|---|---|---|---|
| 24 h | F2,27=6.89 | 0.004 | 10 for each genotype | NV vs SIL=0.993; NG vs SIL=0.008 |
| 72 h | F2,27=13.34 | <0.001 | 10 for each genotype | NV vs SIL=0.006; NG vs SIL=0.220 |
| 120 h | F2,27=12.24 | <0.001 | 10 for each genotype | NV vs SIL=0.002; NG vs SIL=0.628 |
Discussion
Introgression of memory retention
The memory retention phenotype of N. giraulti was introgressed into the background of N. vitripennis. This procedure was chosen because the phenotype of N. giraulti was dominant in F1 hybrids and could be tracked in individuals heterozygous for memory retention QTL during introgression. A concern with selection for decreased memory retention is that mechanisms of trait loss can be numerous and potentially unrelated to the process of memory formation. Negative epistatic interactions between nuclear genes of the two species occur in hybrids of N. vitripennis and N. giraulti, which cause reduced viability (Breeuwer and Werren, 1995; Gadau et al., 1999) and ‘behaviour sterility' in some genotypes of hybrid males (Beukeboom and van den Assem, 2001; Clark et al., 2010; Koevoets et al., 2012). Hybrid breakdown in learning behaviour could also occur in hybrids, for example, as a result of decreased perception or ability to discriminate odours, disturbed perception or behaviour towards other wasps or hosts, or general defects of cognitive skills. These factors could result in a decreased performance during memory retention tests, which can incorrectly be interpreted as decreased memory retention (Tully et al., 1994; Mery and Kawecki, 2002). However, our tested memory retention QTLs did not affect memory retention when tested at 24 h (Figures 3g, h and 4b). In addition, no reduced response levels were observed. Therefore, we can conclude that these QTLs are not causing general disruption of learning ability. Throughout most of this study, the introgressed regions were maintained heterozygously in females, thereby further reducing expression of potential hybrid incompatibilities, which have been observed to be mostly recessive (Breeuwer and Werren, 1995). Hybrid breakdown as a result of interspecific introgression appears not to affect the ability to learn and form memories, at least for the regions under study. The initial introgression experiment specifically targeted a decreased long-lasting memory while retaining shorter-lasting memory, which allowed further studies on the genetic basis of this trait.
Memory retention QTLs
A QTL on chromosome 1 (∼5.8 cM in size) and a QTL on chromosome 5 (∼25,5 cM in size) both reduced memory retention after 72 h. Long-lasting memory was, however, not completely lost, as is the case for N. giraulti. This phenotype was observed both when the QTL was maintained heterozygously (both QTLs) and homozygously (only tested for the QTL on chromosome 1). This indicates that the effect of both QTLs on memory retention is dominant but that neither region alone results in the complete N. giraulti memory retention phenotype. Considering the complexity of this behavioural phenotype, it was expected that multiple loci with potential epistatic interactions among them control memory retention. Long-lasting memory (>48 h) of N. vitripennis is known to consist of multiple memory types that may occur in parallel. These include a form of ARM, which is expressed between approximately 72 h up to 96 h after a single conditioning trial (Schurmann et al., 2009), and LTM, which is observed at 96 h (Schurmann et al., 2012; Hoedjes and Smid, 2014).
The two QTLs identified in this study may affect a single memory type, which could explain a reduction, but not a complete loss of 72 h memory. A complete loss of memory after 120 h was observed for the QTL on chromosome 1 (SIL_LTM1a_gV), which suggests that this QTL affects LTM formation. Testing the effect of multiple conditioning trials on LTM formation in SIL_LTM1A_gV can reveal whether the LTM phenotype is similar to N. giraulti, that is, whether LTM is formed after two conditioning trials (Hoedjes and Smid, 2014). The two QTLs may have an additive effect, especially if they affect different memory types: combining both QTLs into a single introgression line may result in N. giraulti phenotype. In addition, the QTLs may interact with the other observed introgressed regions, which did not affect memory retention on their own when tested after 72 h (Carlborg and Haley, 2004). Further research is required to elucidate possible epistatic interactions.
Genetic factors that regulate memory retention
The goal of this study was to determine genetic factors that underlie natural variation in (long-lasting) memory retention in the genus Nasonia. The observed QTLs provide a basis for further research to determine the exact mechanisms involved. Transcription factor CREB, tyrosine phosphatase corkscrew and cGMP-dependent protein kinase foraging are not located within the two identified genomic regions; however, several other genes with a known function in the memory formation process are. These include the transcription factor CCAAT/enhancer-binding protein, which can interact with CREB, and a dopamine receptor (Alberini, 2005; El-Ghundi et al., 2007). It is, however, preliminary to select such potential candidates for further research, as the introgressed regions with memory retention QTLs contain hundreds of genes. Excellent opportunities for further partitioning of the genomic regions by recombination, combined with mapping and molecular tools available in Nasonia, will allow fine-scaling of memory retention loci (Werren et al., 2010; Desjardins et al., 2013).
Conclusion
Learning and memory formation are universal animal traits; however, there is variation in memory retention. We have introgressed the short-memory retention of N. giraulti into the genetic background of N. vitripennis and have identified two QTLs, which result in decreased long-lasting memory. Variation in learning and memory performance, both between and within species, may have large implications for host-finding behaviour in parasitic wasps, and likely represents an important evolutionary adaptation to changing environmental conditions (Hoedjes et al., 2011). Our study with Nasonia provides novel insight in the genetic mechanisms responsible for natural variation in memory retention. Further studies are required to fine-scale the identified QTLs and to investigate epistatic interaction among QTLs, in order to identify genetic factors that regulate memory retention. The Nasonia model system provides excellent possibilities to pursue these experiments. Knowledge of the genetic basis of natural variation in memory retention is important for our understanding of the evolution of this variation, not only in Nasonia, but also in other animal species.
Data archiving
Data of memory retention tests and the genotyping array available from the Dryad Digital Repository: 10.5061/dryad.18v85.
Acknowledgments
We thank J Lopez and C Desjardins for their assistance with the genotyping microarray analysis, R Edwards for assistance with rearing wasps, D Wheeler for assistance with genome information and W van Tol for assistance with genotyping PCRs. This study was funded by NWO/ALW Open Programme Grant 819.01.011 to HMS, a US NSF EAGER award (ID 1250790) to JHW and a grant from the Dr JL Dobberke Foundation to KMH.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Alberini CM. Mechanisms of memory stabilization: are consolidation and reconsolidation similar or distinct processes. Trends Neurosci. 2005;28:51–56. doi: 10.1016/j.tins.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Beukeboom LW, van den Assem J. Courtship and mating behaviour of interspecific Nasonia hybrids (Hymenoptera, Pteromalidae): a grandfather effect. Behav Genet. 2001;31:167–177. doi: 10.1023/a:1010201427204. [DOI] [PubMed] [Google Scholar]
- Breeuwer JAJ, Werren JH. Hybrid breakdown between two haplodiploid species—the role of nuclear and cytoplasmic genes. Evolution. 1995;49:705–717. doi: 10.1111/j.1558-5646.1995.tb02307.x. [DOI] [PubMed] [Google Scholar]
- Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies. Nat Rev Genet. 2004;5:618–U614. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Clark ME, O'Hara FP, Chawla A, Werren JH. Behavioral and spermatogenic hybrid male breakdown in Nasonia. Heredity. 2010;104:289–301. doi: 10.1038/hdy.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copf T, Goguel V, Lampin-Saint-Amaux A, Scaplehorn N, Preat T. Cytokine signaling through the JAK/STAT pathway is required for long-term memory in Drosophila. Proc Natl Acad Sci USA. 2011;108:8059–8064. doi: 10.1073/pnas.1012919108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Gadau J, Lopez JA, Niehuis O, Avery AR, Loehlin DW, et al. Fine-scale mapping of the Nasonia genome to chromosomes using a high-density genotyping microarray. G3. 2013;3:205–215. doi: 10.1534/g3.112.004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Perfectti F, Bartos JD, Enders LS, Werren JH. The genetic basis of interspecies host preference differences in the model parasitoid Nasonia. Heredity. 2010;104:270–277. doi: 10.1038/hdy.2009.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J. Neurogenetic dissection of conditioned behavior: evolution by analogy or homology. J Neurogenet. 2003;17:295–326. doi: 10.1080/01677060390441859. [DOI] [PubMed] [Google Scholar]
- Eisenhardt D. Learning and memory formation in the honeybee (Apis mellifera) and its dependency on the cAMP-protein kinase A pathway. Anim Biol. 2006;56:259–278. [Google Scholar]
- El-Ghundi M, O'Dowd BF, George SR. Insights into the role of dopamine receptor systems in learning and memory. Rev Neurosci. 2007;18:37–66. doi: 10.1515/revneuro.2007.18.1.37. [DOI] [PubMed] [Google Scholar]
- Gadau J, Page RE, Werren JH. Mapping of hybrid incompatibility loci in Nasonia. Genetics. 1999;153:1731–1741. doi: 10.1093/genetics/153.4.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoedjes KM, Kruidhof HM, Huigens ME, Dicke M, Vet LEM, Smid HM. Natural variation in learning rate and memory dynamics in parasitoid wasps: opportunities for converging ecology and neuroscience. Proc R Soc B. 2011;278:889–897. doi: 10.1098/rspb.2010.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoedjes KM, Steidle JLM, Werren JH, Vet LEM, Smid HM. High-throughput olfactory conditioning and memory retention test show variation in Nasonia parasitic wasps. Genes Brain Behav. 2012;11:879–887. doi: 10.1111/j.1601-183X.2012.00823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoedjes KM, Smid HM. Natural variation in long-term memory formation among Nasonia parasitic wasp species. Behav Process. 2014;105:40–45. doi: 10.1016/j.beproc.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Jarome TJ, Kwapis JL, Nye SH, Helmstetter FJ. Introgression of Brown Norway chromosome 1 onto the fawn hooded hypertensive background rescues long-term fear memory deficits. Behav Genet. 2010;40:85–92. doi: 10.1007/s10519-009-9297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Waddell S. Drosophila olfactory memory: single genes to complex neural circuits. Nat Rev Neurosci. 2007;8:341–354. doi: 10.1038/nrn2098. [DOI] [PubMed] [Google Scholar]
- Koevoets T, Niehuis O, van de Zande L, Beukeboom LW. Hybrid incompatibilities in the parasitic wasp genus Nasonia: negative effects of hemizygosity and the identification of transmission ratio distortion loci. Heredity. 2012;108:302–311. doi: 10.1038/hdy.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, Waddell S. Rapid consolidation to a radish and protein synthesis-dependent long-term memory after single-session appetitive olfactory conditioning in Drosophila. J Neurosci. 2008;28:3103–3113. doi: 10.1523/JNEUROSCI.5333-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehlin DW, Werren JH. Evolution of shape by multiple regulatory changes to a growth gene. Science. 2012;335:943–947. doi: 10.1126/science.1215193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies C, Tully T, Dubnau J. Deconstructing memory in Drosophila. Curr Biol. 2005;15:R700–R713. doi: 10.1016/j.cub.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R. Memory dynamics in the honeybee. J Comp Physiol A. 1999;185:323–340. [Google Scholar]
- Mery F, Belay AT, So AKC, Sokolowski MB, Kawecki TJ. Natural polymorphism affecting learning and memory in Drosophila. Proc Natl Acad Sci USA. 2007;104:13051–13055. doi: 10.1073/pnas.0702923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mery F, Kawecki TJ. Experimental evolution of learning ability in fruit flies. Proc Nat Acad Sci USA. 2002;99:14274–14279. doi: 10.1073/pnas.222371199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis O, Buellesbach J, Gibson JD, Pothmann D, Hanner C, Mutti NS, et al. Behavioural and genetic analyses of Nasonia shed light on the evolution of sex pheromones. Nature. 2013;494:345–348. doi: 10.1038/nature11838. [DOI] [PubMed] [Google Scholar]
- Niehuis O, Judson AK, Gadau J. Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and N. vitripennis. Genetics. 2008;178:413–426. doi: 10.1534/genetics.107.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani MR, Oishi K, Gelb BD, Zhong Y. The phosphatase SHP2 regulates the spacing effect for long-term memory induction. Cell. 2009;139:186–198. doi: 10.1016/j.cell.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmann D, Collatz J, Hagenbucher S, Ruther J, Steidle JLM. Olfactory host finding, intermediate memory and its potential ecological adaptation in Nasonia vitripennis. Naturwissenschaften. 2009;96:383–391. doi: 10.1007/s00114-008-0490-9. [DOI] [PubMed] [Google Scholar]
- Schurmann D, Sommer C, Schinko APB, Greschista M, Smid H, Steidle JLM. Demonstration of long-term memory in the parasitic wasp Nasonia vitripennis. Entomol Exp Appl. 2012;143:199–206. [Google Scholar]
- Smid HM, Wang GH, Bukovinszky T, Steidle JLM, Bleeker MAK, van Loon JJA, et al. Species-specific acquisition and consolidation of long-term memory in parasitic wasps. Proc R Soc B. 2007;274:1539–1546. doi: 10.1098/rspb.2007.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Sun K, Shuai Y, Lin R, You WJ, Wang LZ, et al. Suppressor of hairless is required for long-term memory formation in Drosophila. J Neurogenet. 2009;23:405–411. doi: 10.3109/01677060903096133. [DOI] [PubMed] [Google Scholar]
- Tubon TC, Zhang J, Friedman EL, Jin H, Gonzales ED, Zhou H, et al. dCREB2-mediated enhancement of memory formation. J Neurosci. 2013;33:7475–7487. doi: 10.1523/JNEUROSCI.4387-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- van den Berg M, Duivenvoorde L, Wang GH, Tribuhl S, Bukovinszky T, Vet LEM, et al. Natural variation in learning and memory dynamics studied by artificial selection on learning rate in parasitic wasps. Anim Behav. 2011;81:325–333. [Google Scholar]
- Velthuis BJ, Yang WC, van Opijnen T, Werren JH. Genetics of female mate discrimination of heterospecific males in Nasonia (Hymenoptera: Pteromalidae) Anim Behav. 2005;69:1107–1120. [Google Scholar]
- Wehner JM, Radcliffe RA, Rosmann ST, Christensen SC, Rasmussen DL, Fulker DW, et al. Quantitative trait locus analysis of contextual fear conditioning in mice. Nat Genet. 1997;17:331–334. doi: 10.1038/ng1197-331. [DOI] [PubMed] [Google Scholar]
- Werren JH, Loehlin DW. The parasitoid wasp Nasonia: an emerging model system with haploid male genetics. Cold Spring Harb Protoc. 2009;2009:pdb.emo134. doi: 10.1101/pdb.emo134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.