Abstract
Eukaryotic release factor 3 (eRF3) is implicated in translation termination and also interacts with the poly(A)-binding protein (PABP, Pab1 in yeast), a major player in mRNA metabolism. Despite conservation of this interaction, its precise function remains elusive. First, we showed experimentally that yeast eRF3 does not contain any obvious consensus PAM2 (PABP-interacting motif 2). Thus, in yeast this association is different from the well described interaction between the metazoan factors. To gain insight into the exact function of this interaction, we then analyzed the phenotypes resulting from deleting the respective binding domains. Deletion of the Pab1 interaction domain on eRF3 did not affect general mRNA stability or nonsense-mediated mRNA decay (NMD) pathway and induced a decrease in translational readthrough. Furthermore, combined deletions of the respective interacting domains on eRF3 and on Pab1 were viable, did not affect Pab1 function in mRNA stability and harbored an antisuppression phenotype. Our results show that in Saccharomyces cerevisiae the role of the Pab1 C-terminal domain in mRNA stability is independent of eRF3 and the association of these two factors negatively regulates translation termination.
Keywords: Pab1, eRF3, mRNA decay, translation termination, NMD
INTRODUCTION
The relationship between translation and decay is a key to understanding the fate of mRNAs and the regulation of gene expression in a cell. In eukaryotes, the poly(A)-binding protein (PABP) binds to the poly(A) tail of mRNAs, and plays a major role in mRNA metabolism (Mangus et al. 2003). Among other partners, PABP interacts with eRF3 (eukaryotic Release Factor 3) (Hoshino et al. 1999; Cosson et al. 2002), which mediates translation termination with its partner eRF1 (Zhouravleva et al. 1995). eRF3–PABP association occurs in all eukaryotic organisms, and it has been proposed to mediate efficient termination (Cosson et al. 2002), couple translation termination with mRNA decay (Funakoshi et al. 2007), and protect mRNA from NMD (nonsense-mediated mRNA decay) degradation (Kervestin and Jacobson 2012). However, the exact function remains elusive despite more than a decade of investigation.
eRF3 (encoded by SUP35 in yeast) is a GTPase that stimulates eRF1 activity and recycling (Salas-Marco and Bedwell 2004; Eyler et al. 2013). For clarity, we will subsequently refer to Sup35 as eRF3. The GTPase motifs and the eRF1 binding site are located in the conserved and essential C-terminal region (Ter-Avanesyan et al. 1993; Stansfield et al. 1995). The N-terminal part which consists of two domains N and M has been proposed to regulate termination positively (Cosson et al. 2002) or negatively (Volkov et al. 2007), and interacts with PABP (Hoshino et al. 1999). Purification of the full-length human and yeast eRF3 proved challenging, thus limiting all the in vitro systems on translation termination to use a truncated version of eRF3 (eRF3-C), leaving the role of eRF3-NM domain in termination an open question.
PABPs (Pab1 in yeast) play a role in mRNA metabolism by organizing a scaffold of proteins on the poly(A) tail. Schematically, PABP contains four RNA binding domains (RRM) implicated in poly(A)-binding and translation regulation, followed by an unstructured proline-rich linker (L) and then a C-terminal domain (PABC) (Mangus et al. 2003). To avoid confusing with the gene name for mammalian cytoplasmic PAB1 (PABPC), this C-terminal part is also called the MLLE domain, referring to a conserved motif. Subsequently, we will use PABC/MLLE to address this domain (Kozlov et al. 2010). PABC/MLLE binds to a conserved PAM2 (PABP interaction motif) sequence (Kozlov et al. 2010) and utilizes the same interface for interacting with several factors, including the deadenylation complexes and eRF3. However, in yeast, sequence analysis did not identify a PAM2 motif in eRF3 (Kozlov et al. 2002).
PABP controls deadenylation, the first step of mRNA degradation (Garneau et al. 2007). It protects the mRNA from degradation by coating the poly(A) tail and also regulates the activity of the deadenylases complexes (Doidge et al. 2012; Ezzeddine et al. 2012). However, it remains unsolved if the regulation of poly(A) tail degradation by PABP involves a direct role of eRF3 (Funakoshi et al. 2007; Simon and Seraphin 2007; Yao et al. 2007). Finally the interaction between eRF3–PABP is also implicated in mRNA surveillance by the NMD pathway. The mRNP composition downstream from the termination event is the main criterion that distinguishes normal from premature stop codons. PABP plays a major role in organizing the normal mRNP downstream from the stop codon, possibly mediated by eRF3 binding (Amrani et al. 2004). However, we previously showed in vitro that Pab1–eRF3 interaction is not the main determinant inhibiting NMD activation at a normal stop codon in yeast (Kervestin et al. 2012).
To better understand the function of Pab1–eRF3 interaction in Saccharomyces cerevisiae, we first aimed at experimentally defining eRF3 PAM2 motif. Our data demonstrated that Pab1–eRF3 interaction implicated broader regions of these two factors than for the human complex. In yeast, Pab1 L and C domains, and eRF3 N and M domains are required. Moreover, despite identifying phenylalanine positions that, when mutated, affected Pab1 binding to eRF3 in the two-hybrid system, the two factors were still associated in vivo. Thus, eRF3 does not contain a consensus PAM2 motif and interacts with Pab1 through an alternative molecular interface. Deletion of the Pab1 binding domain on eRF3 did not affect mRNA stability of normal mRNA, in contrast to the reciprocal deletion of the Pab1-LC or C domain. Hence, the function of the Pab1-LC domain in deadenylation is not linked to eRF3. Further, none of eRF3 or Pab1 deletions affected the NMD pathway. Finally, deletions of the interacting domains decreased translational readthrough, reflecting more efficient termination. Our data indicate that eRF3–Pab1 interaction is not involved in mRNA decay or mRNA surveillance. eRF3-NM domain negatively regulates translation termination through Pab1 binding and maintains a basal level of translational readthrough.
RESULTS
Characterization of Pab1 and eRF3 interaction in the two-hybrid system
In order to generate mutations preventing eRF3 from interacting with Pab1, we set up a two-hybrid approach to identify the Pab1 binding site on eRF3.
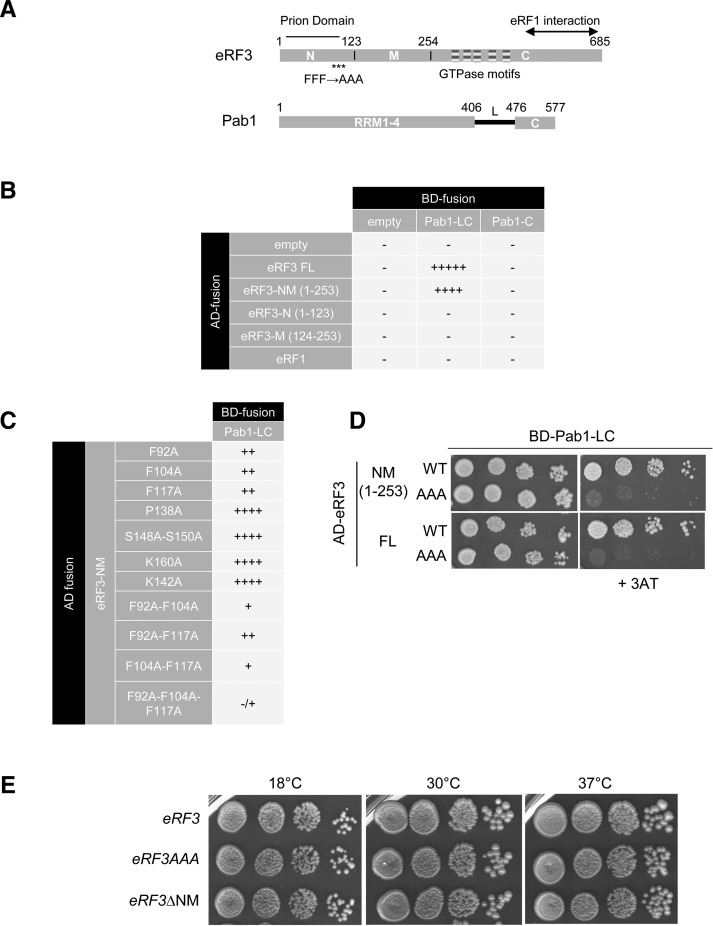
The interaction between eRF3 full-length (FL) or domains of eRF3 (1–253: NM domain, 1–123: N domain, 123–253: M domain) and Pab1 domains (405–577: LC domain, 476–577: C domain) was analyzed in the two-hybrid system (Fig. 1A). eRF3 exists in a prion state that causes the [PSI+] phenotype (Serio and Lindquist 2001). This phenotype can be induced by overexpressing the full-length or only the NM part of eRF3, results in a translation termination defect and can affect cell growth under certain conditions (Derkatch et al. 1998). Hence, the reporter strain (L40) was cured for the [PSI+] phenotype to prevent any artifact. The results of the two-hybrid analysis were analyzed by growth on selective medium without histidine and in the presence of the 3AT competitor (Fig. 1B). Binding of Pab1 on eRF3 required both N and M domains and could not be restricted to the N or the M part, making it more challenging to identify a minimal region containing a PAM2 sequence (Fig. 1B). Moreover, the Pab1-C domain alone did not interact with eRF3 and the L domain was required, although the KITGMLLE motif which binds PAM2 motif in human PABP is conserved (positions 529–536, KITGMILD) (Kozlov and Gehring 2010).
FIGURE 1.
Interaction between Pab1 and eRF3 in the two-hybrid reporter system. (A) Schematic representation of eRF3 and Pab1 functional domains. The eRF3 GTPase motifs are represented by striped boxes. Prion determinant and eRF1 interacting region are depicted. The asterisks indicate the location of the three mutated phenylalanine residues. (B) Identification of the eRF3 interacting domains with Pab1 in the two-hybrid reporter system. The fusion GAL4-AD-eRF1 was used as a negative control. The interaction was analyzed by scoring the growth on the appropriate medium after 6 d at 30°C; (−) no growth in absence of 3AT, (−/+) no growth on 1 mM 3AT, (+) no growth on 5 mM 3AT, (++) no growth on 10 mM 3AT, (++++) growth on 10 mM 3AT, (+++++) growth on 20 mM 3AT. (C) Mutations of eRF3-NM domain. The indicated mutations were introduced and the interaction with Pab1-LC domain was tested as described for B. (D) Mutation of phenylalanine residues in alanine at position 92, 140, and 117 affects eRF3 interaction with Pab1. Interaction of Pab1-LC domain with full length (FL) or the NM domain of eRF3 either wild-type (WT) or harboring the triple mutation (AAA) was analyzed by serially diluting and spotting the reporter strain expressing the different construction on selective medium containing 3AT (right panel). Growth was scored after 5 d of growth at 30°C. (E) Growth phenotype of eRF3 mutant strains. The indicated strains (SKY1085, SKY1081, and SKY1182) were grown overnight in YPDA at 30°, serially diluted and spotted onto YPDA plate. Growth was scored after 3 d of growth at 30°C and 37°C, and 4 d at 18°C.
Next, we generated alanine substitutions in eRF3 to define Pab1 interacting positions. Phenylalanine residues play a major role in PAM2–PABC/MLLE recognition (Siddiqui et al. 2007) so positions F92, F104, and F117 were targeted, and also other positions conserved in an alignment of eRF3 sequences from yeast species (data not shown). Only mutations of phenylalanine residues diminished eRF3 interaction with Pab1 (Fig. 1C) and the strongest decrease was observed for substitution of the three phenylalanine residues altogether in both the NM domain and the full-length factor (Fig. 1D). Finally, we generated mutant strains containing the triple mutation FFF 92-104-117 AAA (eRF3AAA) or lacking the entire NM domain (eRF3ΔNM) at the genomic locus. eRF3AAA and eRF3ΔNM strains were viable at 18°C, 30°C, and 37°C (Fig. 1E). In these strains, the level of Pab1 and eRF3 analyzed by Western blotting is not affected (data not shown).
Pab1 and eRF3 interacting domains in vivo
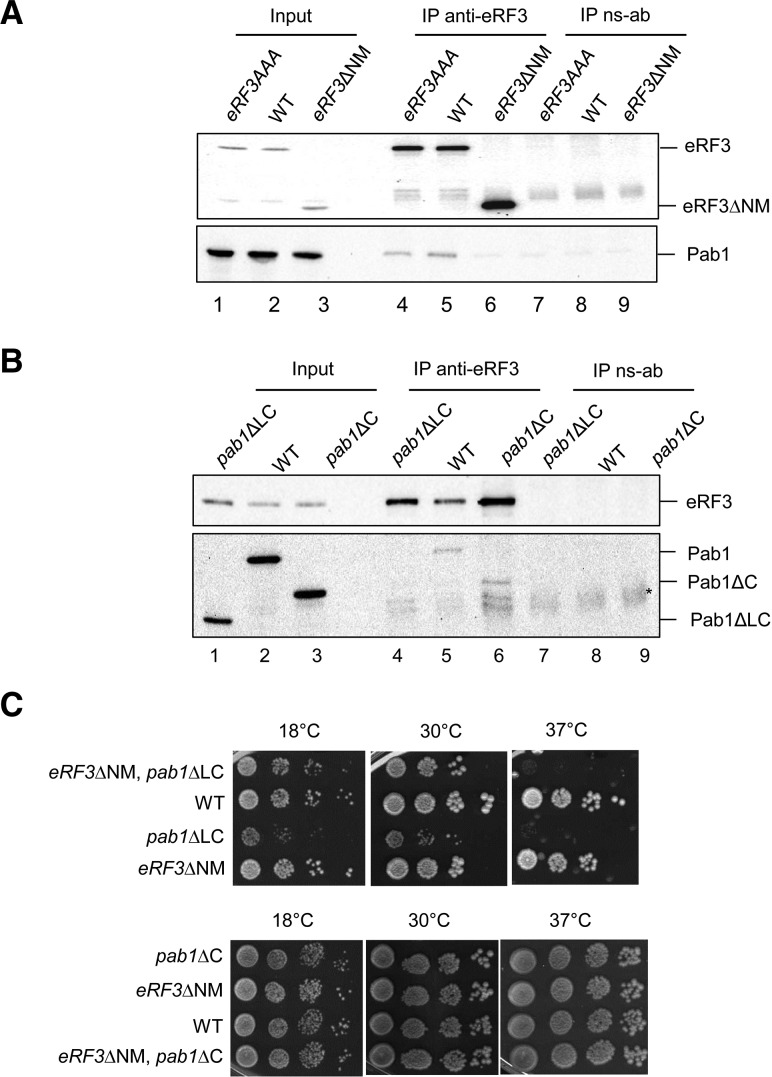
To explore the in vivo association of Pab1 and eRF3, yeast cells expressing wild-type eRF3, eRF3AAA, or eRF3ΔNM were lysed and immunoprecipitated using antiserum to eRF3 (anti-eRF3) or nonspecific antiserum (ns-ab) (Fig. 2A,B). In wild-type extracts, Pab1 coimmunoprecipitated with eRF3 (Fig. 2A, lane 5) and this association was RNAse A resistant (data not shown). For the triple mutant eRF3AAA, Pab1 was still present in the immunoprecipitation reaction (Fig. 2A, lane 4), whereas deletion of the entire NM domain abolished the Pab1 association (Fig. 2A, lane 6). The absence of the Pab1–eRF3 interaction in eRF3ΔNM extracts could not arise from destabilization of the release factor because the truncated protein was present in the extract (Fig. 2A, lane 3) and the strain was viable (SUP35 encoding eRF3 is an essential gene). Although mutation FFF 92-104-117 AAA affected Pab1–eRF3 interaction in the two-hybrid approach, the interaction was still maintained in vivo. Moreover, analysis of mRNA stability, NMD activation, polysomes profile and termination efficiency in eRF3AAA strain did not reveal any difference compared with the wild type (Supplemental Fig. S1). These results suggest that phenylalanine residues are implicated in mediating Pab1–eRF3 interaction but in contrast to human, they are not essential. These data were confirmed in vitro using purified factors (Supplemental Fig. S2). Association of eRF3 with Pab1 was also analyzed by immunoprecipitation in mutant strains lacking the Pab1-C domain (pab1ΔC) or both the linker and the C domain (pab1ΔLC) at the PAB1 locus. eRF3 was still associated with Pab1ΔC (Fig. 2B, lane 6) and only deletion of both the Pab1-L and -C domains abolished Pab1 binding to eRF3 (Fig. 2B, lane 4), confirming that the linker domain of Pab1 is essential for eRF3 binding.
FIGURE 2.
Interactions in strains deleted for various domains of Pab1 and eRF3. (A,B) Coimmunoprecipitation assays on yeast extracts from strains SKY1085, SKY1081, and SKY1182 expressing various versions of eRF3 as indicated in A; and BSY1552, BSY1537, and W303-1B expressing various versions of Pab1 as indicated in B using anti-eRF3 polyserum (IP anti-eRF3) or nonspecific anti-serum (IP ns-ab) as a negative control. Positions of the different forms of Pab1 and eRF3 are indicated. Aliquots of extract were analyzed (Input) and represent 1/80 of the quantity loaded in the IP lanes. The star indicates the position of residual signal from IgG. (C) Growth phenotype of the following strains: SKY1263, SKY1264, SKY1265, SKY1266 (upper panel); SKY1317, SKY1318, SKY1319, SKY1320 (lower panel) resulting from crossing strains eRF3ΔNM (SKY1182) with pab1ΔLC (BSY1552) or pab1ΔC (BSY1537), and isolated from representative tetrads. Serial dilutions of overnight liquid cultures were deposited on YPDA plates and their growth was scored after 3 d of growth for 30°C and 37°C, and 4 d for 18°C.
Next, we investigated the phenotypes resulting from combining deletions of Pab1 and eRF3 reciprocal binding domains, by crossing eRF3ΔNM with pab1ΔLC or pab1ΔC strains. Deletion of the eRF3-NM domain in mutant strains where Pab1-LC or -C domains are absent yielded viable cells (Fig. 2C). The double mutants did not show a defect in cell growth compared with the single mutants. As previously published (Simon and Seraphin 2007) pab1ΔLC cells grew slowly and were temperature sensitive (Fig. 2C, upper panel). The presence of the eRF3ΔNM allele in pab1ΔLC mutant slightly increased cell growth compared with the single pab1ΔLC mutant and yet, the strain was still temperature sensitive. This indicates that deletion of the eRF3-NM domain partially reverts the pab1ΔLC growth defect. This observation is puzzling because Pab1ΔLC alone has lost its ability to interact with eRF3. For the double mutant pab1ΔC with eRF3ΔNM, the cell growth was equivalent to the wild-type strain while the two factors did not interact (Fig. 2C, lower panel). These data demonstrate that the combination of both deletions is viable, and this association is not essential.
Disrupting the interaction of Pab1 with eRF3 did not affect mRNA stability or NMD
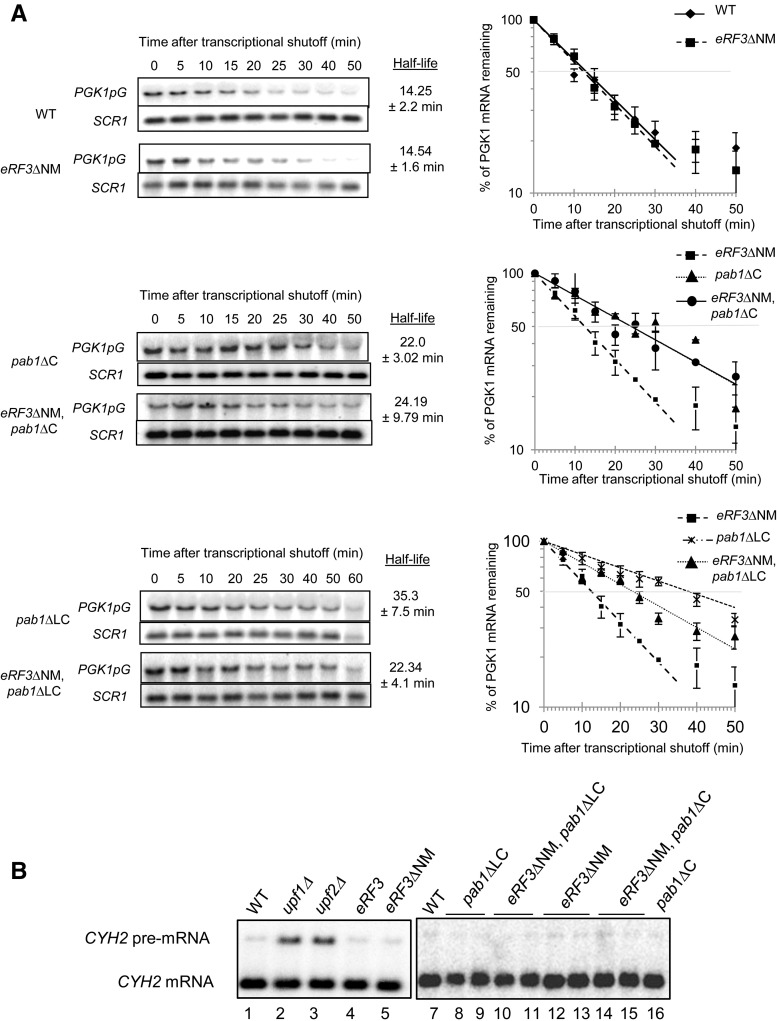
The interaction of Pab1 and eRF3 is proposed to couple the end of translation to mRNA decay and thus influence mRNA stability (Kobayashi et al. 2004; Funakoshi et al. 2007). Hence, we analyzed mRNA half-life in transcriptional chase experiments using a reporter PGK1pG mRNA under the control of a repressible promoter (tetO-driven promoter) (Simon and Seraphin 2007). In the mutant strain eRF3ΔNM, the PGK1pG half-life was roughly the same as for the wild-type cells, ∼14 min (Fig. 3A). To confirm this observation for endogenous mRNA, the drug thiolutin was used to inhibit transcription by RNA polymerase II. In wild-type cells, the half-life of TIF51a mRNA was measured to be ∼11 min (Supplemental Fig. S3), which was slightly shorter than the previously reported (Sinturel et al. 2012). The absence of the eRF3-NM domain did not affect the half-life of this mRNA (12.8 min). All these data indicate that the eRF3-NM domain does not influence mRNA stability.
FIGURE 3.
mRNA stability and NMD analysis. (A) Northern blot analysis of total RNA extracted from the following strains SKY1085, SKY1182 (upper panel), BSY1537, SKY1247 (central panel), BSY1552, SKY1263 (lower panel) at the indicated times after inhibition of transcription. The blots were probed to detect the reporter PGK1-pG mRNA and the noncoding SCR1 transcript that serves as a control. The same results were obtained for strains isolated from tetrad dissection. Experiments were reproduced at least three times independently. For quantification the RNA signal at time point 0 was set at 100% of PGK1pG mRNA and used to determine the percentage of mRNA remaining at the different time points and calculate the PGK1pG mRNA half-life. (B) Northern blot analysis of total RNA isolated from the following strains: W303-1B (lanes 1,7), HFY870, HFY1300, SKY1085, SKY 1182, SKY1265, BSY1552, SKY1259, SKY1263, SKY1182 (lanes 4,12), SKY1266, SKY1247, SKY1248, BSY1537. Signals corresponding to CYH2 mRNA and pre-mRNA are indicated.
Northern blot analysis of PGK1pG mRNA after transcriptional shut off in mutant strains pab1ΔLC or pab1ΔC showed mRNA stabilization reflected by an increase in PGK1pG mRNA half-life (Fig. 3; Supplemental Fig. S3). This confirms the role of Pab1-LC/C domain in regulating mRNA stability (Simon and Seraphin 2007). Combining the eRF3ΔNM deletion in pab1ΔC strain did not affect the mRNA stability observed for the single mutant (Fig. 3A), suggesting that this phenotype was independent of eRF3. As previously observed, the strain pab1ΔLC harbored a more severe increase of mRNA half-life (35.3 min for the mutant compared with 14.25 min for the WT) (Fig. 3A; Simon and Seraphin 2007). Combining pab1ΔLC with eRF3ΔNM led to an intermediate phenotype that could be linked to the cell growth phenotype of the double mutant (Fig. 3; Supplemental Fig. S3).
All these data indicate that deletion of eRF3-NM domain does not directly affect mRNA half-life and that the eRF3–Pab1 interaction is not implicated in mRNA stability. However, the role of the Pab1-LC/C domain in promoting mRNA decay is confirmed, and our data strongly suggest that this feature is independent of eRF3.
The distance between Pab1 and the termination event is a major determinant of NMD activation (Amrani et al. 2004; Kervestin and Jacobson 2012). Therefore, we analyzed mRNA decay by the NMD pathway by monitoring the levels of the CYH2 pre-mRNA, a well described NMD substrate (He et al. 1993). Deleting the eRF3-NM domain, the Pab1-LC or C domains or both did not affect CYH2 pre-mRNA levels (Fig. 3B). In contrast, deletion of the NMD genes UPF1 or UPF2 led to the stabilization of the pre-mRNA. This result indicates that the absence of interaction between eRF3 and Pab1 does not affect the NMD pathway.
The lack of interaction between eRF3 and Pab1 impacts translation termination
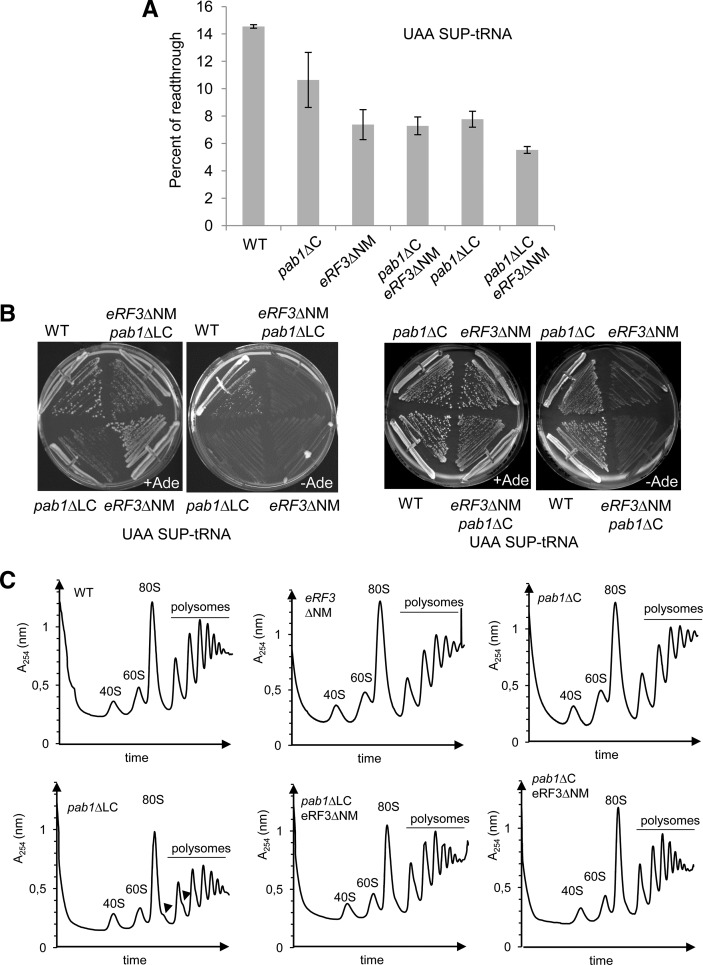
The main function of eRF3 is to mediate translation termination in association with eRF1. Although the eRF3-NM domain is not strictly required for translation termination, we analyzed translational readthrough using reporter plasmid encoding dual luciferases (renilla followed by firefly) separated by a stop codon, or a sense codon as a control (Keeling et al. 2004). Briefly, the activity of the firefly luciferase was quantified and normalized to the activity of the renilla luciferase and 100% of readthrough corresponded to the sense codon control. Translational readthrough was analyzed in the strains obtained by crossing eRF3ΔNM with pab1ΔLC or pab1ΔC, cured for the [PSI+] phenotype and in the presence of a mutant tRNA suppressor of UAA codon. This latter feature was required to increase the signal in wild-type cells and facilitate the detection of the readthrough levels in the mutants (Supplemental Fig. S4). Deletion of eRF3-NM as well as Pab1-LC domains led to almost a twofold decrease of readthrough and this effect is more pronounced on the mutant strain containing both deletion (Fig. 4A). Translational readthrough was not significantly affected for the pab1ΔC strain that still retains Pab1–eRF3 interaction, and the double mutant pab1ΔC, eRF3ΔNM harbored the same translational readthrough level as the eRF3ΔNM mutant alone (Fig. 4A). These results were confirmed in vivo by streaking the cells on medium without adenine to monitor suppression of the UAA nonsense codon in the ade2-1 gene present in the background of our strains. Introduction of the suppressor-tRNA in wild-type cells allowed growth on selective medium (−Ade). In contrast, the mutant strain deleted for eRF3-NM domain failed to grow indicating an antisuppressor phenotype (Fig. 4B). Overall, this set of experiments indicates that deletion of the eRF3-NM domain decreases translational readthrough. Reciprocal deletion of the Pab1-LC domain exhibited similar phenotypes (Fig. 4A,B) which are enhanced when the two mutations are combined. Growth on selective medium was observed for the mutant strain pab1ΔC, and not when combined to deletion of eRF3-NM domain, confirming that antisuppression phenotype is dependent on eRF3–Pab1 interaction (Fig. 4B). These data are in agreement with the previous report on the negative role of the eRF3-NM domain in translation termination (Volkov et al. 2007) and show that this effect is mediated through Pab1 interaction.
FIGURE 4.
Translational readthrough and polysome profiles of the mutant strains. (A) Translational readthrough levels determined with the dual luciferase reporter in strains SKY1405/1488, SKY1486, SKY1407, SKY1489, SKY1406, SKY1404 expressing the tRNA suppressor of UAA stop codon. Readthrough was quantified as described in the main text and in the Materials and Methods. SEM is indicated. The results were similar at 25°C or 30°C. (B) Strains used in A were streaked 3 d at 30°C on synthetic medium in the presence (+Ade) or the absence (−Ade) of adenine to assay the suppression of ade2–1 nonsense mutation. The result was identical at 25°C. (C) Extracts from strains SKY1264, SKY1266, BSY1537, SKY1265, SKY1263, and SKY1247 were separated on sucrose gradient by ultracentrifugation and absorbance at 254 nm was measured. The arrowheads indicate the position of stalled preinitiation complexes (halfmers). The same results were obtained with strains containing the same deleted or wild-type versions of the genes in parental strains or isolated from different tetrads.
To exclude that these results were linked to a more general role of eRF3–Pab1 in translation, we analyzed the polysome profiles of these strains by sucrose gradient ultracentrifugation. Profiles obtained for the strains eRF3ΔNM, pab1ΔC, and the double mutant eRF3ΔNM, pab1ΔC were identical to the wild-type (Fig. 4C). Deletion of the Pab1-LC domain led to a weak but reproducible decrease of the 80S level, decrease in the 60S large subunit level and the formation of stalled preinitiation complexes (halfmers) (Helser et al. 1981). These defects and more specifically the presence of the halfmers were partially suppressed by deletion of eRF3-NM domain. More specifically, the ratio 60S:40S is not back to the wild-type level indicating that translation is still affected, a defect likely linked to the deletion of Pab1-LC domain independently of eRF3 interaction. It is noteworthy that deletion of eRF3-NM domain in the pab1ΔLC strain partially reverted the defects on cell growth, mRNA stability and polysomes profiles but did not affect translation termination phenotype. Overall, these results indicate that the eRF3-NM domain, and thus the Pab1–eRF3 interaction, did not affect general translation and were specifically implicated in translation termination.
DISCUSSION
The poly(A)-binding protein Pab1 and the translation termination factor eRF3 were shown to interact more than a decade ago (Hoshino et al. 1999), but the role of this association remains under investigation. Herein, we have studied the biological functions of eRF3–Pab1 interaction in yeast where this association is prevented by deletion of their respective interaction domains. Our results demonstrate that this interaction is implicated in fine tuning of translation termination and not needed in vivo to stimulate mRNA decay.
The PABC/MLLE domain binds several regulatory proteins and translation factors by recognizing a conserved short sequence, PAM2. This motif is found in PABP partners in different organisms except in yeast. Of the three factors that interact with Pab1-C-terminal domain in yeast; eRF3, Pan3, and Pbp1, only Pan3 contains a PAM2 motif (Mangus et al. 1998; Siddiqui et al. 2007; Kozlov et al. 2010). Structures of PABC/MLLE have been obtained and the one from yeast is divergent (Kozlov et al. 2002). Our results confirm that the determinants of Pab1 recognition on eRF3 in yeast are different from the well described PABC/MLLE–PAM2 interactions in human (Kozlov and Gehring 2010). First, Pab1 binding domain on eRF3 could not be restricted to the N or the M domain and reciprocally, both domains L and C of Pab1 were required for eRF3 interaction. Thus, eRF3–Pab1 interaction requires broader binding domains than in human and involves mainly the linker (L) domain of Pab1 (Fig. 1; Mangus et al. 2004). Second, mutation of three conserved phenylalanine residues in the eRF3-NM domain affected Pab1 binding to eRF3 in yeast two-hybrid (Fig. 1) and in in vitro pull-down experiments (Supplemental Fig. S2), but failed to inhibit the association in vivo (Fig. 2). Thus, the phenylalanine residues are implicated in mediating Pab1–eRF3 interaction, but in contrast to human they are not essential. Interestingly, Pab1-L domain is also implicated in the interaction of Pab1 with Pbp1 and Pan3 (Mangus et al. 1998, 2004; Siddiqui et al. 2007). Mutation of conserved phenylalanine positions in the PAM2 motif identified in Pan3 weakened Pab1–Pan3 interaction without abolishing it (Siddiqui et al. 2007). Thus, the use in yeast of an alternative molecular interface from MLLE–PAM2 could be a general observation.
Hence, eRF3 does not contain a canonical PAM2 motif, which precludes the identification of specific residues that fully inhibit Pab1–eRF3 interaction when mutated. Therefore, to study eRF3–Pab1 interaction, we focused on the phenotypes of strains expressing a truncated version of eRF3 (eRF3ΔNM) or Pab1 (pab1ΔLC) that have lost their ability to interact with one another. Analysis of mRNA stability in strains where the Pab1 binding domain on eRF3 was deleted showed that mRNA stability was not affected (Fig. 3). This indicates that, in yeast, loss of the Pab1–eRF3 interaction does not affect the general stability of mRNAs. However, in pab1ΔLC and pab1ΔC strains, the mRNAs were stabilized. Thus, the Pab1-LC/C domain influences mRNA decay independently of eRF3. Our results contradict previous published work showing that eRF3 regulates mRNA stability through PABP binding in yeast and humans (Funakoshi et al. 2007). Our conclusions only apply to yeast and we do not exclude that the role of the Pab1–eRF3 interaction in mRNA decay is different in humans. In contrast to yeast, there is a physical interaction between PABP and the deadenylation complex CCR4/NOT through the TOB factor (Funakoshi et al. 2007). The deadenylation complex PAN2–PAN3 which binds PABC/MLLE is implicated in the first distributive steps of cytoplasmic deadenylation whereas in yeast its role in mRNA cytoplasmic decay is not clarified (Wolf and Passmore 2014). The original approach based on overexpression of the eRF3-NM domain to inhibit Pab1–eRF3 interaction could not be reproduced in our hands as overexpression strongly affected cell growth, most likely due to the induction of the [PSI+] phenotype. However, our results clearly show that deleting the eRF3-NM domain does not affect mRNA stability. Hence, our work supports the notion that in yeast the Pab1-LC domain is implicated in mRNA stability independently from eRF3, probably by modulating Pab1 packing on the poly(A) tail to control the accessibility of the deadenylases (Simon and Seraphin 2007; Yao et al. 2007).
At a premature stop codon, translation termination is aberrant and, in the presence of the Upf factors, activates mRNA decay by the NMD pathway (for review, see Kervestin and Jacobson 2012; Mühlemann and Jensen 2012). When tethered downstream from a premature termination codon, Pab1 is able to protect mRNA from NMD degradation (Amrani et al. 2004; Behm-Ansmant et al. 2007; Eberle et al. 2008; Singh et al. 2008). Previously we have shown that activation of NMD is not based on a simple competition between Pab1 and Upf1 for eRF3 binding (Kervestin et al. 2012). Here, we show that NMD is still active in all the mutant strains, thus eRF3–Pab1 is not required for this surveillance pathway (Fig. 3B). This result is in agreement with previous data showing that the absence of Pab1 does not inhibit NMD (Meaux et al. 2008). Further, preventing Pab1 binding to eRF3 did not destabilize normal mRNA nor decrease translation termination efficiency. Thus, the absence of Pab1–eRF3 interaction is not the determinant that activates NMD at a termination event. Moreover, deletion of the Pab1 and eRF3 interaction domains actually led to a more efficient termination in our reporter system (Fig. 4). This suggests that the efficiency of translation termination per se (stop codon recognition followed by peptidyl release) is not the trigger of the NMD pathway. Abnormal termination at a premature stop codon could reflect a poorly dissociable post-termination mRNP complex, a defect prevented by Pab1 at normal termination codons (Kervestin and Jacobson 2012).
The role of eRF3-NM domain and Pab1 binding in termination is still elusive because purification of full-length human and yeast eRF3 proved challenging, thus forcing use of a truncated version of eRF3 (eRF3-C) in all the in vitro systems on translation termination. Analysis of translation termination in mutant strains deleted for Pab1 and eRF3 interaction domains showed a decrease in translational readthrough (Fig. 4). These data are in agreement with a previous report on the negative role of the eRF3-NM domain in translation termination (Volkov et al. 2007). This phenotype was specific for Pab1–eRF3 interaction because this decrease was observed for eRF3ΔNM, pab1ΔLC, pab1ΔLC/eRF3ΔNM, and pab1ΔC/eRF3ΔNM mutant strains but not for the pab1ΔC strain that still retained Pab1–eRF3 interaction. These data confirm a role for the eRF3–Pab1 interaction in translation termination, and this association negatively affects the termination process. The double mutant eRF3ΔNM, pab1ΔLC showed a slight but significant higher decrease in translational readthrough suggesting that Pab1 and eRF3 could act in translation termination not solely through their association, possibly by titrating a common factor yet to identify. Interestingly, a role of Pab1 in efficient translation termination has been previously published, based on a decrease of translational readthrough when Pab1 is overexpressed (Cosson et al. 2002). We reproduced these initial data and confirmed that Pab1 overexpression also slowed cell growth (Supplemental Fig. S5). This indicates that Pab1ΔLC and eRF3ΔNM do not promote a better termination by relieving a titration of eRF3 by Pab1. Because Pab1 can oligomerize (Kuhn and Pieler 1996; Melo et al. 2003), overexpression of Pab1 must promote Pab1 polymerization and association on the poly(A) tail, a phenotype also shared by the pab1ΔLC strain, thus inhibiting interaction with eRF3 (Simon and Seraphin 2007; Yao et al. 2007). Overall, our data indicate that translation termination in wild-type cells is not at its maximal efficiency, possibly to tolerate recoding events implicating readthrough of a stop codon (Namy et al. 2004; Dunn et al. 2013). Pab1 interaction with eRF3 appears to play a role in maintaining this basal level of translational readthrough.
How does the Pab1–eRF3 interaction negatively regulate translation termination? First, Pab1 can affect eRF1–eRF3 interaction. However, in vitro binding data showed that eRF1 binding is more efficient to eRF3 full-length than eRF3ΔNM (Supplemental Fig. S2). Hence, our data do not support this hypothesis although a more formal conclusion requires a deeper quantitative analysis. Second, Pab1 could influence eRF3 GTPase activity, which is required for efficient polypeptide release (Frolova et al. 1996). The in vitro role of Pab1 in termination or eRF3 GTPase assays has never been analyzed due to the challenge of purifying full-length eRF3. Therefore, this remains an unexplored possibility. Finally, Pab1–eRF3 can influence post-termination events such as the recycling of the termination complex (Becker et al. 2012). The role of Pab1 in recycling has never been addressed. However, its implication in the NMD pathway and the link between NMD and ribosome recycling suggests that Pab1 can influence eRF3 association with the ribosome and thus the recycling as well as the termination process.
In conclusion, this study presents an analysis of the phenotypes induced by the lack of Pab1–eRF3 interaction in yeast. The data confirmed the role of the Pab1-C-terminal domain on mRNA stability, and demonstrated that this function is independent of eRF3. Moreover, this work revealed that the role of Pab1–eRF3 binding is not in regulation of deadenylation, but rather in fine tuning of translation termination. Future in vitro analyses will be required to understand how Pab1 influences the termination process.
MATERIALS AND METHODS
Yeast strains and growth conditions
Yeast cells were grown in standard YEPD medium supplemented with adenine 0.02 mg/mL (YPDA medium) or HC medium (Amberg et al. 2005). All the strains derived from W303-1B or BMA64 background and the genotypes are listed in Supplemental Table S1. Strains SKY1081, 1085, and 1182 were obtained by using the one-step gene-replacement method with PCR fragments resulting from amplifying the DNA fragment containing eRF3 sequences fused to HIS3MX6 from S. pombe with oligonucleotides SKO92 and SKO145 (sequences in Supplemental Table S2). DNA fragments containing (i) the eRF3 sequences were obtained by amplifying plasmids SKP239, SKP327, and SKP221 (plasmid description in Supplemental Table S3) using oligonucleotides SKO86 and SKO87, (ii) the HIS3MX6 sequence was obtained by PCR amplification using SKO144 and SKO145 oligonucleotides on the plasmid pYM15 from EUROSCARF. Strains SKY1259, 1263–1266 were obtained by crossing SKY1182 and BSY1552 following by tetrad dissection, strains SKY1247–1248 and SKY1317–1320 by crossing SKY1182 and BSY1537, and strains SKY1289–1290 by crossing SKY1081 and BSY1552. The strains SKY1247–1248, SKY1259/SKY1263, SKY1264/SKY1289/SKY1319 and SKY1266/SKY1318 have the same genotype and were isolated from different tetrads. When required, the [PSI+] phenotype was cured by streaking the strains three consecutive times on YPDA medium containing 5 mM guanidinium chloride GuHCl (Eaglestone et al. 2000). Mating, sporulation, and tetrad dissection were performed as described (Amberg et al. 2005).
Plasmid constructions
Standard cloning procedures were used. Unless indicated, genomic DNA extracted from the W303-1B strain was used as the template for PCR amplifications. The plasmids are described in Supplemental Table S3. The plasmids SKP239, SKP327, and SKP221 were derived from pRS313, and contain the eRF3 promoter, coding, and terminator sequences amplified by PCR and inserted at XbaI and XhoI cloning sites. For SKP239, the eRF3 genomic region was amplified using oligonucleotides SKO86–SKO87. SKP327 was derived from SKP239 after directed mutagenesis to introduce the F92A, F104A, and F117A mutations. For SKP221, the DNA fragment was obtained by fusion PCR using oligonucleotides SKO86 and SKO87 on a DNA matrix containing (i) the eRF3 promoter sequence amplified by PCR with oligonucleotides SKO86–SKO53, (ii) the eRF3-C coding sequence followed by the terminator sequence amplified by PCR with oligonucleotides SKO56–SKO87. The plasmids used for the two-hybrid assay were derived from pBTM116 (a gift from Benoit Palancade) and pACTII (a gift from P. Lesage) and contain the LexA-binding domain (BD) and the GAL4-activation domain (AD), respectively (Van Criekinge and Beyaert 1999). SKP190 and SKP196 contained fragments of PAB1 obtained by PCR amplification with oligonucleotides SKO73–SKO77 and SKO75–SKO77, respectively, and inserted at BamHI and PstI sites of pBTM116. The plasmids SKP86, SKP116, SKP275, SKP278 contained the eRF3 coding sequence for the full-length factor, the NM, the N, and the M domains, respectively, inserted in EcoRI–XhoI (SKP86, SKP116) or BamHI, XhoI (SKP275, SKP278) of pACTII. The DNA fragments were obtained by PCR amplification between oligonucleotides SKO1–SKO2, SKO1–SKO21, SKO9–SKO126, and SKO21–SKO112, respectively. LMP241 and LMP272 were derived from SKP116 and SKP86, respectively, after direct mutagenesis. SKP418 contained PGK1pG under the control of a tetO-regulated promoter. The coding sequences derived from pBS2813 (Simon and Seraphin 2007) after digestion by NotI, BamHI, and ApaI and insertion in the NotI–ApaI sites of pRS425. The plasmid containing the mutated tRNA suppressor of UAA stop codon with a LEU2 selection marker is a generous gift from I. Stansfield. Mutagenesis was performed with the Quick change mutagenesis protocol (Stratagen).
Two-hybrid analysis
The reporter strain (SKY1001) was transformed with plasmids containing the indicated regions of Pab1 and eRF3 (FL stands for full length) fused with the LexA-binding domain (BD) and GAL4-activation (AD) domain, respectively. Expression of all the different constructions was confirmed by Western blotting. Aliquots corresponding to A600 = 0.04 after overnight culture were spotted onto the appropriate medium lacking tryptophan and leucine for plasmids selection, lacking histidine and in the presence of increasing concentration of 3-amino-1,2,4-triazole (+3AT). Interaction was scored after 6 d at 30°C.
Immunoprecipitation
Log-phase cultures were harvested and the pellet was washed and resuspended in IP buffer (10 mM Tris–HCl pH 7.4, 30 mM MgCl2, 100 mM NaCl, 0.01% Triton, 0.1 mM DTT, 1 mM EDTA, Roche protease inhibitors). Lysates were cleared by centrifugation. Antibodies raised against Pab1 or eRF3 C-terminal domain (Agrobio) were conjugated to protein A-Sepharose resin from GE by incubation overnight at 4°C in buffer (1% NP40, 10 mM Tris–HCl pH 7.5, 100 mM NaCl) followed by covalent linkage using 1% glutaraldehyde in buffer (2 mM EDTA, 1 M NaCl) 1 h at 4°C. Extracts were diluted in IP buffer to reach 2 mg/mL of total protein. One milliliter of extract per reaction was incubated with the resin O/N at 4°C. Total protein and immunoprecipitation reactions were separated by 8% SDS-PAGE followed by Western blotting using standard procedures.
RNA analysis
All strains were grown to log phase in YPDA medium at 30°C. Inhibition of transcription of the reporter gene PGK1pG, RNA isolation, and Northern blotting procedures were performed as described previously (Simon and Seraphin 2007; He et al. 2008). For the mRNA stability analysis, the blots were probed with 32P-labeled oligonucleotides “pG” to detect PGK1pG mRNA and “SCR1” (sequences available in Supplemental Table S2). For steady-state analysis, the blot was probe for CYH2 transcript using randomly labeled DNA fragment as described in He et al. (2008). Signals were measured and quantified using a Typhoon Trio (GE Healthcare).
Dual-luciferase assays
Readthrough assays were performed using the dual-luciferase reporter assay system (Promega) and a XS3 LB960 Bertholds luminometer. Aliquots of log-phase cultures in the appropriate medium were lysed according to the manufacturer's protocol. Ten microliters of lysis extract was incubated with 50 µL of LARII reagent, followed by shaking for 2 sec and measurement of the firefly luciferase activity for 10 sec. Then 50 µL of STOP and GLO buffer were added, shake for 2 sec and Renilla luciferase activity was measured for 10 sec. Each experiment was performed using eight technical duplicates. Each individual culture was assayed several times, and the values were derived from at least three independent experiments.
Polysome profile analysis
Cycloheximide was added to a final concentration of 50 μg/mL to 150 mL of log-phase culture in rich medium (YPDA). Cells were harvested by centrifugation 5 min at 3500 rpm at 4°C, washed and resuspended to a final volume of 500 µL in lysis buffer (20 mM Tris–HCl pH7.4, 5 mM MgCl2, 20 mM KCl, 1 mM DTT) containing protease inhibitors without EDTA from Roche. Cells lysis was performed using glass beads and vortexing, and the lysate was cleared by two subsequent centrifugations at 16,000g, 5 min at 4°C. Absorbance was measured at 260 nm and the volume corresponding to 12 OD was fractionated by ultracentrifugation on a sucrose gradient (10%–50%) in a buffer containing 50 mM Tris–HCl pH 7.4, 12 mM MgCl2, 50 mM NH4Cl, and 1 mM dithiothreitol (DTT) for 2 h 45 min at 39,000 rpm and 4°C (SW41 rotor). The fractions were recovered with an ISCO fractionator, and the absorbance at 254 nm was measured.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Agence Nationale pour la Recherche (ANR-06-BLAN-0075-02); the Centre National pour la Recherche Scientifique; and the Human Frontier Science Program organism (RGP0018/2009-C). We thank Josette Banroques, Lionel Benard, Marc Graille, Emmeline Huvelle, Pascale Lesage, Benoit Palancade, Cosmin Saveanu, Bertrand Seraphin, Ian Stansfield, and Allan Jacobson as well as the members from his laboratory, past and present, for sharing reagents and advice. We thank Fabrice Lejeune, Domenico Libri, Tommaso Villa, and Kyle Tanner for critical reading of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.047282.114.
REFERENCES
- Amberg DC, Burke DJ, Strathern JN 2005. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118. [DOI] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. 2012. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482: 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E 2007. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26: 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G 2002. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI+] propagation. Mol Cell Biol 22: 3301–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Liebman SW 1998. Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc Natl Acad Sci 95: 2400–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doidge R, Mittal S, Aslam A, Winkler G 2012. The anti-proliferative activity of BTG/TOB proteins is mediated via the Caf1a (CNOT7) and Caf1b (CNOT8) deadenylase subunits of the Ccr4-not complex. PLoS One 7: e51331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS 2013. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife 2: e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Ruddock LW, Cox BS, Tuite MF 2000. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc Natl Acad Sci 97: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O 2008. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler DE, Wehner KA, Green R 2013. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J Biol Chem 288: 29530–29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Chen CY, Shyu AB 2012. Evidence providing new insights into TOB-promoted deadenylation and supporting a link between TOB's deadenylation-enhancing and antiproliferative activities. Mol Cell Biol 32: 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2: 334–341. [PMC free article] [PubMed] [Google Scholar]
- Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S 2007. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev 21: 3135–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ 2007. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126. [DOI] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc Natl Acad Sci 90: 7034–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Amrani N, Johansson MJ, Jacobson A 2008. Qualitative and quantitative assessment of the activity of the yeast nonsense-mediated mRNA decay pathway. Methods Enzymol 449: 127–147. [DOI] [PubMed] [Google Scholar]
- Helser TL, Baan RA, Dahlberg AE 1981. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide. Mol Cell Biol 1: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J Biol Chem 274: 16677–16680. [DOI] [PubMed] [Google Scholar]
- Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A 2012. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S, Li C, Buckingham R, Jacobson A 2012. Testing the faux-UTR model for NMD: analysis of Upf1p and Pab1p competition for binding to eRF3/Sup35p. Biochimie 94: 1560–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Funakoshi Y, Hoshino S, Katada T 2004. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J Biol Chem 279: 45693–45700. [DOI] [PubMed] [Google Scholar]
- Kozlov G, Gehring K 2010. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PLoS One 5: e10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Siddiqui N, Coillet-Matillon S, Trempe JF, Ekiel I, Sprules T, Gehring K 2002. Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein. J Biol Chem 277: 22822–22828. [DOI] [PubMed] [Google Scholar]
- Kozlov G, Ménade M, Rosenauer A, Nguyen L, Gehring K 2010. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J Mol Biol 397: 397–407. [DOI] [PubMed] [Google Scholar]
- Kuhn U, Pieler T 1996. Xenopus poly(A) binding protein: functional domains in RNA binding and protein–protein interaction. J Mol Biol 256: 20–30. [DOI] [PubMed] [Google Scholar]
- Mangus DA, Amrani N, Jacobson A 1998. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol Cell Biol 18: 7383–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Agrin NS, Smith M, Gongidi P, Jacobson A 2004. Positive and negative regulation of poly(A) nuclease. Mol Cell Biol 24: 5521–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaux S, van Hoof A, Baker KE 2008. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol Cell 29: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo EO, Dhalia R, Martins de Sa C, Standart N, de Melo Neto OP 2003. Identification of a C-terminal poly(A)-binding protein (PABP)–PABP interaction domain: role in cooperative binding to poly (A) and efficient cap distal translational repression. J Biol Chem 278: 46357–46368. [DOI] [PubMed] [Google Scholar]
- Mühlemann O, Jensen TH 2012. mRNP quality control goes regulatory. Trends Genet 28: 70–77. [DOI] [PubMed] [Google Scholar]
- Namy O, Rousset JP, Napthine S, Brierley I 2004. Reprogrammed genetic decoding in cellular gene expression. Mol Cell 13: 157–168. [DOI] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM 2004. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol 24: 7769–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Lindquist SL 2001. [PSI+], SUP35, and chaperones. Adv Protein Chem 57: 335–366. [DOI] [PubMed] [Google Scholar]
- Siddiqui N, Mangus D, Chang TC, Palermino JM, Shyu AB, Gehring K 2007. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J Biol Chem 282: 25067–25075. [DOI] [PubMed] [Google Scholar]
- Simon E, Seraphin B 2007. A specific role for the C-terminal region of the Poly(A)-binding protein in mRNA decay. Nucleic Acids Res 35: 6017–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinturel F, Bréchemier-Baey D, Kiledjian M, Condon C, Bénard L 2012. Activation of 5′-3′ exoribonuclease Xrn1 by cofactor Dcs1 is essential for mitochondrial function in yeast. Proc Natl Acad Sci 109: 8264–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J 14: 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN 1993. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol 7: 683–692. [DOI] [PubMed] [Google Scholar]
- Van Criekinge W, Beyaert R 1999. Yeast two-hybrid: state of the art. Biol Proced Online 2: 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov K, Osipov K, Valouev I, Inge-Vechtomov S, Mironova L 2007. N-terminal extension of Saccharomyces cerevisiae translation termination factor eRF3 influences the suppression efficiency of sup35 mutations. FEMS Yeast Res 7: 357–365. [DOI] [PubMed] [Google Scholar]
- Wolf J, Passmore LA 2014. mRNA deadenylation by Pan2–Pan3. Biochem Soc Trans 42: 184–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Chiang YC, Zhang C, Lee DJ, Laue TM, Denis CL 2007. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol Cell Biol 27: 6243–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 14: 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.