Abstract
Fragile X syndrome (FXS) is the most common inherited form of intellectual disability caused by the CGG trinucleotide expansion in the 3′-untranslated region of the FMR1 gene on the X chromosome, that silences the expression of the Fragile X mental retardation protein (FMRP). FMRP has been shown to bind to a G-rich region within the PSD-95 mRNA which encodes for the postsynaptic density protein 95 (PSD-95), and together with the microRNA miR-125a, to play an important role in the reversible inhibition of the PSD-95 mRNA translation in neurons. The loss of FMRP in Fmr1 KO mice disables this translation control in the production of the PSD-95 protein. Interestingly, the miR-125a binding site on PSD-95 mRNA is embedded in the G-rich region bound by FMRP and postulated to adopt one or more G quadruplex structures. In this study, we have used different biophysical techniques to validate and characterize the formation of parallel G quadruplex structures and binding of miR-125a to its complementary sequence located within the 3′ UTR of PSD-95 mRNA. Our results indicate that the PSD-95 mRNA G-rich region folds into alternate G quadruplex conformations that coexist in equilibrium. miR-125a forms a stable complex with PSD-95 mRNA, as evident by characteristic Watson–Crick base-pairing that coexists with one of the G quadruplex forms, suggesting a novel mechanism for G quadruplex structures to regulate the access of miR-125a to its binding site.
Keywords: Fragile X, PSD-95 mRNA, miR-125a
INTRODUCTION
Fragile X syndrome (FXS) represents the most common form of inherited intellectual disability, being caused by the silencing of the fragile X mental retardation (FMR1) gene, which is located on the X chromosome and encodes for the fragile X mental retardation protein (FMRP). Due to the abnormal expansion of a trinucleotide CGG sequence within its 5′-untranslated region, the FMR1 gene becomes silenced, resulting in the loss of FMRP, an mRNA-binding protein whose role in messenger RNA (mRNA) translation control is essential for normal brain function (Bassell and Warren 2008). It is evaluated that nearly one in 4000 males and one in 6000 females are affected by FXS (Coffee et al. 2009). FMRP is part of a small family of RNA-binding proteins, being most abundantly expressed in the brain and testes and characteristic in both fetal and adult tissues (Devys et al. 1993). FMRP is mainly located in the cytoplasm, however, due to a nuclear localization signal (NLS) and a nuclear export signal (NES) recognized within its sequence, it can be transported between the cytoplasm and nucleus (Feng et al. 1997). Once in the nucleus, FMRP associates with specific mRNAs and other proteins as part of larger ribonucleoprotein complexes that can be transported to dendrites where they control the expression of certain proteins in response to synaptic stimulation signal (Dictenberg et al. 2008). RNA-binding motifs of special interest for FMRP–RNA binding, are its two ribonucleoprotein K homology domains (KH1 and KH2) and a specific group of arginine-glycine-glycine repeats, referred to as RGG box (Siomi et al. 1993). It was identified that the FMRP RGG box has high affinity for specific G quadruplex structures of neuronal mRNA targets (Darnell et al. 2001; Schaeffer et al. 2001; Menon and Mihailescu 2007), whereas the KH domains recognize another specific RNA structure called a kissing complex (Darnell et al. 2005). G quadruplex structures are formed when four guanine nucleotides connected through Hoogsteen hydrogen bonding assemble into a square planar arrangement that is stabilized by potassium ions (Sen and Gilbert 1990; Hud et al. 1996).
FMRP normally undergoes phosphorylation on its highly conserved serine 500 residue, an important chemical post-translational modification believed to have a prime role in regulatory function of FMRP (Ceman et al. 2003; Narayanan et al. 2007). The exact details of the mechanisms by which FMRP controls the translation of its mRNA targets are not known. However, at least for a subset of these mRNAs, it has been proposed that their translation regulation in neurons is mediated through the interactions of FMRP with the microRNA pathway (Edbauer et al. 2010; Muddashetty et al. 2011). Among these mRNAs targets whose translation is regulated by FMRP is PSD-95 mRNA (Todd et al. 2003; Muddashetty et al. 2007, 2011), which encodes for the postsynaptic density protein 95, a member of the membrane-associated guanylate kinase family (MAGUK) of the postsynaptic scaffold proteins essential for glutamate receptor localization and synapse function (Keith and El-Husseini 2008). Active control of PSD-95 expression in response to neurotransmitter signals is required to allow proper dendritic spine development as this protein plays an important role in synaptic plasticity necessary for learning and memory (Colledge et al. 2003; De Roo et al 2008; Xu et al. 2008). FMRP and microRNAs have affinity to target localized mRNAs and regulate protein synthesis. It has been demonstrated by Muddashetty et al. (2011) that the 3′ UTR of PSD-95 mRNA contains specific binding sites for both FMRP and the microRNA miR-125a, that appear to cooperate to regulate PSD-95 mRNA translation which is dysregulated in neurons obtained from FMRP knockout mice. Previous work has shown that FMRP binds a G-rich region on the 3′ UTR of PSD-95 mRNA (Zalfa et al. 2007) that overlaps with miR-125 binding site (Muddashetty et al. 2011). However, it remains unclear whether this G-rich region forms G quadruplex structures that could play a role to mediate miR-125a interactions. In an effort to elucidate the mechanism by which miR-125a regulates PSD-95 mRNA translation, we designed in vitro experiments that analyze the interactions between miR-125a and PSD-95 mRNA. This study focuses on the analysis of the interactions between miR-125a and its proposed complementary sequence within the 3′ UTR of PSD-95 mRNA. Our results indicate that the PSD-95 mRNA G-rich region folds into alternate G quadruplex conformations which coexist in equilibrium with the miR-125a binding site being accessible only in one of the conformations. These data suggest a potential mechanism that involves interplay between FMRP, G quadruplex and microRNA to regulate translation.
RESULTS AND DISCUSSION
The guanine rich region within PSD-95 mRNA 3′ UTR is predicted to form multiple G quadruplex structures
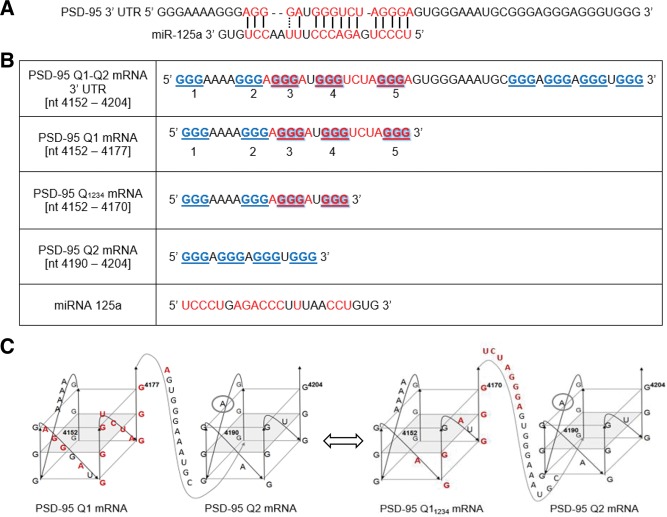
The translation of PSD-95 mRNA is suggested to be controlled by the cooperative action of FMRP and miR-125a which interact with the PSD-95 mRNA 3′-UTR region (Muddashetty et al. 2011). miR-125a is proposed to use its seed sequence to recognize the specific “AGGGA” target sequence within PSD-95 mRNA 3′ UTR (Fig. 1A), that is embedded in a G-rich region (Fig. 1B). Given that G-rich sequences containing at least four repeats of consecutive Gs have the potential to fold into G quadruplex structures, we and others (Todd et al. 2003; Zalfa et al. 2007; Subramanian et al. 2011) predicted the existence of G quadruplex structures within PSD-95 mRNA 3′ UTR. We have used the QGRS Mapper software (http://bioinformatics.ramapo.edu/QGRS/analyze.php) to predict G quadruplex formation in the 3′ UTR of PSD-95 mRNA in a specific sequence surrounding the miR-125a binding site [nucleotides 4152–4204]. Two G quadruplex structures were predicted to form, named Q1 and Q2, with the complementary target site for miR-125a being folded within the Q1 G quadruplex (Fig. 1B,C). Muddashetty et al. (2011) have indirectly shown in vivo that miR-125a binds to its proposed complementary seed sequence within PSD-95 mRNA 3′ UTR (Fig. 1A,B, in which the putative miR-125a binding site is highlighted in red), as mutations within this region abolish the effect miR-125a has upon the regulation of the PSD-95 mRNA translation. According to the QGRS Mapper prediction software, however, the miR-125a complementary sequence on PSD-95 mRNA is fully embedded into the predicted Q1 G quadruplex structure, raising the question of how is this microRNA able to recognize and bind its mRNA target.
FIGURE 1.
(A) G-rich segment of the PSD-95 mRNA with the putative miR-125a binding site highlighted in red (Muddashetty et al. 2011). Base-pairing between mRNA and microRNA is highlighted in red. (B) Four PSD-95 mRNA sequences were used in various experiments to characterize the existence of predicted G quadruplexes and assess formation of a microRNA complex. Five stretches of GGG repeats are involved in G quadruplex conformations; those base-paired with the miR-125a (red) or external (blue) are highlighted. The long sequence spanning from [nucleotides 4152–4204] is truncated into shorter sequences that involve predicted Q1 and Q2 G quadruplexes. (C) Arrangement of the predicted Q1 and Q2 G quadruplex structures in the PSD-95 mRNA showing the putative miR-125a binding site in Q1 sequence (highlighted in red). (Left) Four conformations are predicted to have the miR-125a binding site inaccessible within the G quadruplex (one shown for Q1 conformation, the other three are shown in Supplemental Fig. 1). (Right) One conformation (Q11234) is predicted to have an exposed miR-125a binding site. QGRS Mapper software was used for the prediction (http://bioinformatics.ramapo.edu/QGRS/analyze.php).
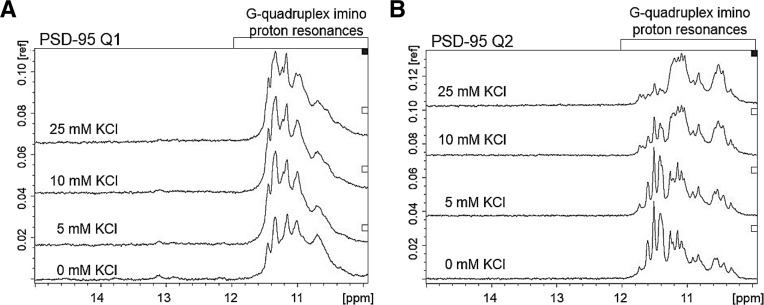
Thus, we used several biophysical methods to determine if indeed the PSD-95 G-rich region surrounding the miR-125a binding site folds into the predicted Q1 and Q2 G quadruplex structures. Two PSD-95 mRNA fragments encompassing the predicted G quadruplex structures Q1 [nucleotides 4152–4177] and Q2 [nucleotides 4190–4204] (Fig. 1B) were produced by T7 RNA polymerase driven in vitro transcription reactions. First, we used 1D 1H NMR spectroscopy to analyze their imino proton resonance region, and as seen in Figure 2, resonances are present in the region 10–12 ppm, for both PSD-95 Q1 (Fig. 2A) and PSD-95 Q2 (Fig. 2B), even in the absence of KCl (bottom spectra). Imino proton resonances in the region 10–12 ppm have been assigned to guanine imino protons engaged in Hoogsteen base pairs within individual G quartets and are considered signatures of G quadruplex structure formation (Fürtig et al. 2003; Menon et al. 2008; Nambiar et al. 2011). While DNA G quadruplexes require the presence of potassium ions for folding, RNA G quadruplexes of identical sequence can fold even in the absence of these ions, but have low stability (Joachimi et al. 2009). In the case of PSD-95 Q1 RNA, low-intensity resonances in the range 12–14 ppm, corresponding to guanine and uracil imino protons involved in Watson–Crick base pairs, are also present (Fig. 2A, bottom spectrum), indicating the formation of an alternate Q1 conformation. As KCl was titrated in the samples in the range 0–25 mM, the intensity of the Q1 G quadruplex imino proton resonances increased with the concomitant disappearance of the Watson–Crick imino proton resonances (Fig. 2A), indicating that in the presence of K+ ions this alternate structure is converted to a G quadruplex. No Watson–Crick imino proton resonances are present for Q2 RNA, however, for both the PSD-95 Q1 and Q2 sequences, the relative intensities and broadness of the G quadruplex resonances changed as the KCl concentration was increased above 10 mM, indicating either a dynamic exchange between differently folded G quadruplex conformations, or an exchange between conformations that involve monomeric G quadruplex and complexes formed through the stacking of such monomeric G quadruplex structures. Because the PSD-95 Q2 sequence contains GGG repeats separated by single nucleotide loops, it is not prone to the formation of alternate G quadruplex structures, and is predicted to form a very stable G quadruplex structure, even at low KCl concentrations (Fig. 1B,C). Thus, in the case of PSD-95 Q2 we attribute the broadness of the imino proton resonances to the formation of dynamic stacked G quadruplex structures stabilized by high KCl concentrations, which are favored by the high RNA concentration used in the NMR experiments.
FIGURE 2.
1H NMR spectra of (A) PSD-95 Q1 mRNA (350 µM) and (B) PSD-95 Q2 mRNA (350 µM) at various concentrations of K+ ions in 10 mM cacodylic acid buffer, pH 6.5, at 25°C.
Next, we analyzed the PSD-95 Q1 and Q2 G quadruplexes by 20% nondenaturing polyacrylamide gel in 0.5× TBE buffer (Fig. 3A,B), varying the KCl concentration in the range 0–100 mM. Two bands were observed for PSD-95 Q1 RNA at all KCl concentrations investigated (Fig. 3A, black arrows), their relative intensities changing as the KCl concentration was increased, suggesting that this sequence could adopt multiple conformations, which is consistent with the NMR spectra (Fig. 2). In contrast, the PSD-95 Q2 G quadruplex migrated as a single band at all KCl concentrations investigated (Fig. 3B), suggesting the formation of a single G quadruplex conformation.
FIGURE 3.
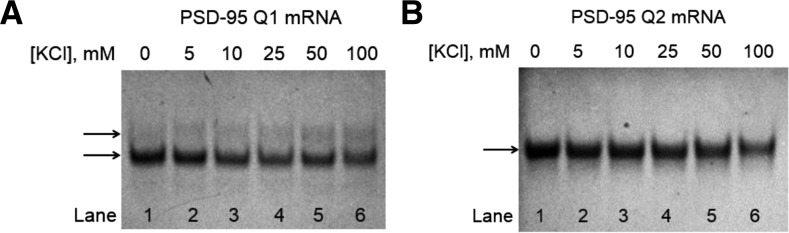
Twenty percent nondenaturing polyacrylamide gel of (A) PSD-95 Q1 mRNA and (B) PSD-95 Q2 mRNA in 0.5× TBE buffer in increasing salt concentrations. Lane 1 (A): 20 μM PSD-95 Q1 mRNA in the absence of KCl; Lane 1 (B): 20 μM PSD-95 Q2 mRNA in the absence of KCl; lanes 2–6 (A,B): 5–100 mM KCl concentration. The RNA samples were annealed by boiling for 5 min in the presence of salt and then incubated at room temperature for 10 min. The gels were visualized by UV shadowing at 254 nm.
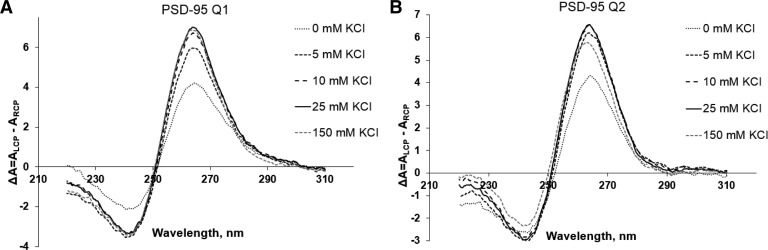
To obtain additional information about the PSD-95 Q1 and Q2 G quadruplex folds we acquired their CD spectra in the presence of increasing KCl concentrations in the range 5–150 mM. For parallel G quadruplex structure formation, a positive band at ∼265 nm and a negative band at ∼240 nm have been observed, whereas the signatures of an antiparallel G quadruplex structure are a negative band at ∼260 nm and a positive band at ∼295 nm (Miyoshi et al. 2003; Paramasivan et al. 2007; Kypr et al. 2009; Vorlíčková et al. 2012; Randazzo et al. 2013). Both PSD-95 Q1 and Q2 exhibit the signature of a parallel G quadruplex structure, even in the absence of K+ ions (Fig. 4A,B). As KCl levels increased, the intensities of these bands increased, indicating the K+ ions driven G quadruplex stability. However, minimal changes were observed in the spectra upon the increase in salt concentration from 10 to 150 mM implying that these sequences require a low ionic strength to achieve a fully stable G quadruplex structure.
FIGURE 4.
CD spectra of (A) PSD-95 Q1 mRNA and (B) PSD-95 Q2 mRNA in increasing KCl concentrations at 25°C. 10 µM RNA sample was suspended in 10 mM cacodylic acid buffer, pH 6.5 and KCl was titrated in the ratio of 1–150 mM. The spectra were corrected by subtracting the buffer contributions.
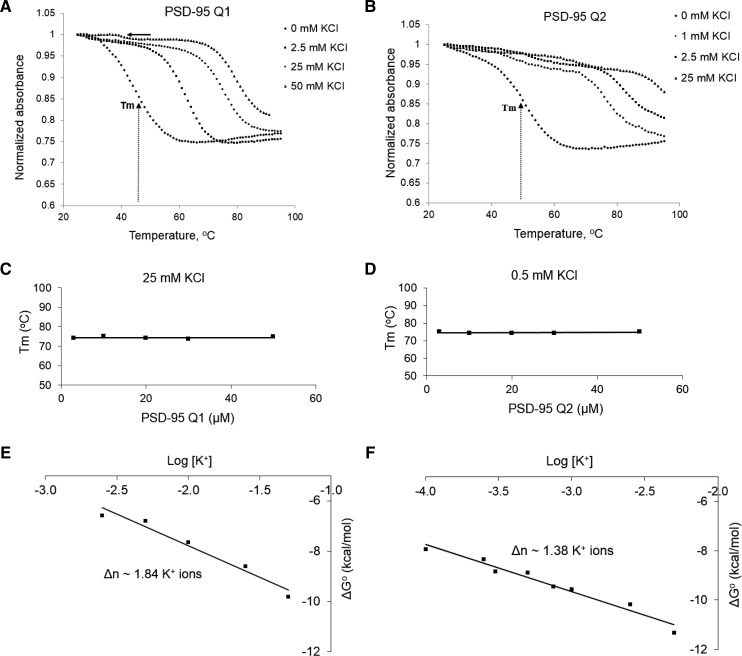
To obtain information about the overall stability of the PSD-95 Q1 and Q2 G quadruplex structures, we performed UV thermal denaturation experiments. The changes in absorbance as the temperature was raised from 25°C to 95°C were monitored at 295 nm, wavelength sensitive to G quadruplex denaturation (Mergny et al. 1998). For both sequences a typical hypochromic transition due to G quadruplex unfolding at elevated temperatures was observed at KCl concentrations in the range 0–25 mM KCl (Fig. 5A,B; Hud et al. 1996). A second hypochromic transition became apparent for the PSD-95 Q1 sequence in the presence of 50 mM KCl (Fig. 5A, black arrow). This result is consistent with the native gel electrophoresis results, which showed a second upper band corresponding to one or more alternate conformations of PSD-95 Q1 that became more intense at 50 and 100 mM KCl (Fig. 3A, lanes 5 and 6). A second minor hypochromic transition, not as clearly defined, seems to be present for the PSD-95 Q2 at high KCl concentrations, which could be due to the dissociation of stacked Q2 G quadruplex monomers.
FIGURE 5.
UV spectroscopy thermal denaturation data of (A) 10 µM PSD-95 Q1 mRNA and (B) 10 µM PSD-95 Q2 mRNA in increasing KCl concentration in 10 mM cacodylic acid buffer, pH 6.5. Tm represents the melting temperature at which half of the RNA has dissociated. G quadruplex melting temperatures of (C) PSD-95 Q1 mRNA at 25 mM KCl and (D) PSD-95 Q2 mRNA at 0.5 mM KCl as a function of RNA concentration, indicating “intramolecular” structure formation in both RNAs. Plots of (E) PSD-95 Q1 mRNA and (F) PSD-95 Q2 mRNA ΔG°s as a function of logarithm of K+ ion concentration, where Δn represents the number of K+ ions released upon G quadruplex unfolding.
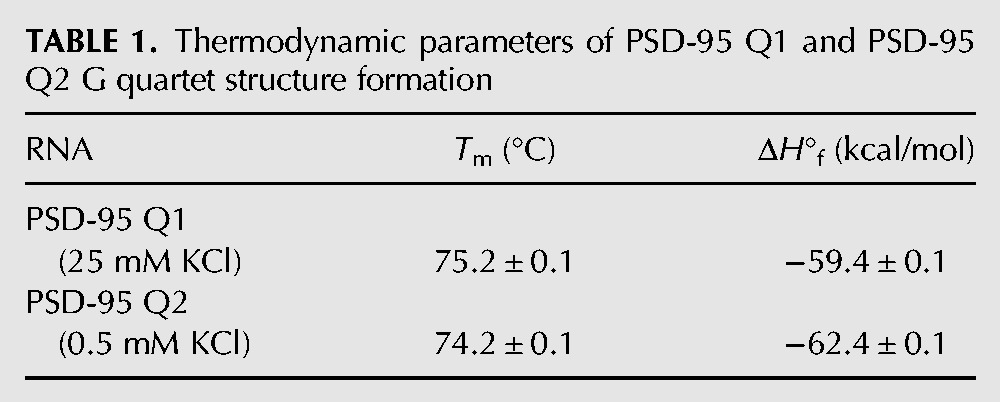
To determine if the main G quadruplex conformations in PSD-95 Q1 and Q2 RNAs are intramolecular we performed UV thermal denaturation experiments at a fixed KCl concentration and variable RNA concentration in the range 3–50 µM. This analysis was performed in the presence of 25 mM KCl for PSD-95 Q1 RNA and only in 0.5 mM KCl for PSD-95 Q2 RNA as this G quadruplex is very stable and in the presence of 25 mM KCl is only partially melted at 95°C (Fig. 5B). For intramolecular species the melting temperature is independent of the RNA concentration (Materials and Methods, Equations 1 and 2) and this was observed for both PSD-95 Q1 (Tm = 75°C in the presence of 25 mM KCl) and Q2 sequences (Tm = 74°C in the presence of 0.5 mM KCl) (Fig. 5C,D).
The thermodynamic parameters of G quadruplex formation were obtained by fitting the 50°C–90°C region and 60°C–95°C region of the PSD-95 Q1 and Q2 thermal denaturation curves, respectively, with Equation 3 (Materials and Methods) that assumes a two-state model (Table 1; Menon et al. 2008). The enthalpy of formation of a single G-quartet plane has been reported to range between −18 and −25 kcal/mol (Hardin et al. 2000). Thus, the values obtained for the enthalpy of G quadruplex formation in PSD-95 Q1 (–59.4 ± 0.1 kcal/mol) and PSD-95 Q2 (–62.4 ± 0.1), respectively, are consistent with the presence of three G quartet planes in each of these structures, as predicted by the QGRS mapper software (Fig. 1B).
TABLE 1.
Thermodynamic parameters of PSD-95 Q1 and PSD-95 Q2 G quartet structure formation
This thermodynamics analysis was expanded to obtain information about the number of K+ ions coordinated within the main G quadruplex conformations of PSD-95 Q1 and Q2 sequences (Materials and Methods, Equation 4). On average ∼1.9 K+ ions coordinate the main G quadruplex structure within the PSD-95 Q1 G quadruplex, whereas ∼1.4 K+ ions coordinate the PSD-95 Q2 G quadruplex (Fig. 5E,F).
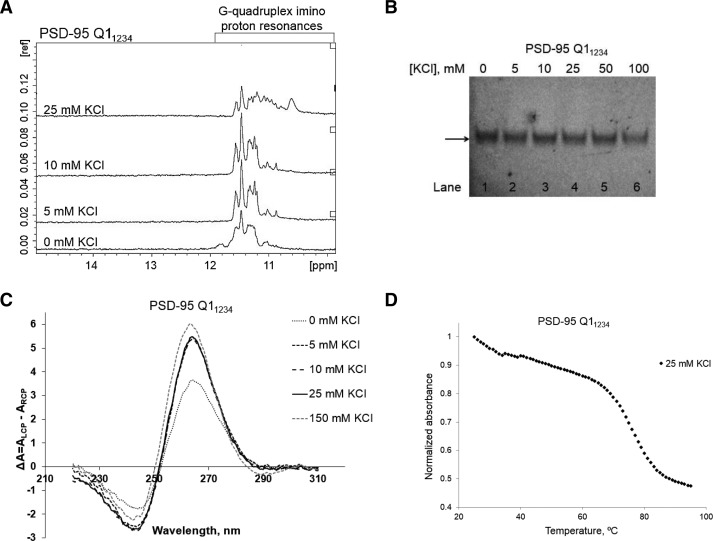
Taken together these biophysical experiments provide direct evidence that the PSD-95 sequence surrounding the miR-125 binding site adopts several stable G quadruplex structures. A closer analysis of the PSD-95 Q1 RNA revealed that there are five stretches of GGG repeats in its sequence (labeled 1–5 in Fig. 1A), allowing for the formation of several potential G quadruplex conformations: Q11245, which uses the GGG repeats 1, 2, 4, and 5 in the formation of the G quartet planes (Fig. 1B, left) (predicted by the QGRS software mapper), Q11234, which uses the GGG repeats 1–4 (Fig. 1B, right), Q11235, Q11345, and Q12345 (Supplemental Fig. 1). It is interesting to note that among these possible structures there is only one, Q11234, that exposes the miR-125 seed sequence binding site (Fig. 1C, right). To determine if the Q11234 quadruplex could actually form as predicted, we transcribed its sequence (nucleotides 4152–4170 in Fig. 1C, right), and analyzed it by 1D 1H NMR spectroscopy. As seen in Figure 6A, even in the absence of KCl, there are resonances in the imino proton region centered ∼11.5 ppm which correspond to G imino protons involved in G quartet formation. As the KCl concentration is increased these resonances increase in intensity becoming more defined, indicating that the G quadruplex structure formed by Q11234 is further stabilized by the presence of K+ ions.
FIGURE 6.
(A) 1D 1H NMR spectra of PSD-95 Q11234 mRNA at various concentrations of K+ ions in 10 mM cacodylic acid buffer, pH 6.5, at 25°C. (B) Twenty percent of nondenaturing polyacrylamide gel in 0.5× TBE buffer in increasing salt concentrations. Lane 1: PSD-95 Q11234 mRNA in the absence of KCl; lanes 2–6: 0–100 mM KCl concentration. The gel was visualized by UV shadowing at 254 nm. (C) CD spectra of PSD-95 Q11234 mRNA in increasing KCl concentration at 25°C. 10 µM RNA sample was suspended in 10 mM cacodylic acid buffer, pH 6.5 and KCl was titrated in the ratio of 1–150 mM. The spectra were corrected by subtracting the buffer contributions. (D) UV denaturation of 10 µM PSD-95 Q11234 mRNA at 25 mM KCl in 10 mM cacodylic acid buffer, pH 6.5.
As expected, PSD-95 Q11234 migrates as a single band in a 20% nondenaturing polyacrylamide gel in 0.5× TBE buffer (Fig. 6B) at all KCl concentrations investigated, as no alternate G quadruplex conformations are predicted to form in this truncated sequence. Similarly to PSD-95 Q1 and Q2, Q11234 exhibits the CD spectrum signature of a parallel G quadruplex structure (Fig. 6C) that is stabilized by K+ ions. The UV thermal denaturation profile of PSD-95 Q11234 reveals a single hypochromic transition with a melting temperature of 76°C, which is in very good agreement with the Tm of 75°C determined for the main hypochromic transition of PSD-95 Q1 RNA (Fig. 6D). These experiments indicate that the conformation Q11234, which exposes the miR-125a seed sequence binding site is accessible to the PSD-95 Q1 quadruplex.
miR-125a interactions with the PSD-95 mRNA Q1 region
As described above, PSD-95 Q1 RNA, spanning nucleotides 4152–4177, is dynamic in forming several alternate G quadruplex structures which are differently populated and only one of which (Q11234) we predict exposes the “AGGGA” sequence complementary to the miR-125a seed sequence.
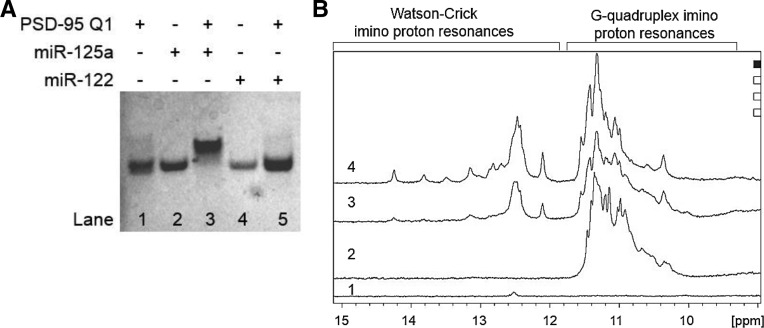
To test this prediction, we performed electromobility shift assays in 20% nondenaturing polyacrylamide gels analyzing the PSD-95 Q1 binding with miR-125a. As a negative control we used the liver-specific microRNA miR-122 that has no complementarity with PSD-95 Q1. As seen in Figure 7A, in the presence of a 1:1 ratio of PSD-95 Q1 and miR-125a there is a new band corresponding to the stable PSD-95 Q1- miR-125a complex (lane 3) that migrates slower than the free PSD-95 Q1 (lane 1) or miR-125a (lane 2). In contrast, in the presence of a 1:1 ratio of PSD-95 Q1 and miR-122 only the overlapped bands corresponding to the free PSD-95 Q1 RNA and miR-122 are observed (Fig. 7A, compare lanes 1, 4, and 5). In control experiments we have shown that the short PSD-95 Q1234 G quadruplex that lacks the target mRNA sequence complementary to the miR-125a seed sequence is not able to bind to miR-125a (Supplemental Fig. 2A). Similarly, as expected, miR-125a does not bind to the PSD-95 Q2 quadruplex that does not contain a region complementary to its seed (Supplemental Fig. 2B).
FIGURE 7.
(A) EMSA of PSD-95 Q1 mRNA and miR-125a/miR-122 in a 20% nondenaturing polyacrylamide gel in 0.5× TBE buffer and 5 mM KCl. miR-122 was used as a negative control. Lane 1: PSD-95 Q1 mRNA (15 μM); lane 2: miR-125a (15 μM); lane 3: PSD-95 Q1 and miR-125a RNAs in a 1:1 ratio (15 μM); lane 4: miR-122 (15 μM); lane 5: PSD-95 Q1 and miR-122 RNAs in a 1:1 ratio (15 μM). PSD-95 Q1 samples were annealed by boiling in 5 mM KCl and cooled at room temperature for 5 min. miR-125a/miR-122 was then added and incubated at room temperature for 30 min. The gel was visualized by UV shadowing at 254 nm. (B) 1D 1H NMR spectra of (1) 350 μM miR-125a, (2) 350 μM PSD-95 Q1 mRNA, (3) PSD-95 Q1 mRNA and miR-125a incubated at room temperature in a 1:1 ratio (350 μM), (4) PSD-95 Q1 mRNA and miR-125a in a 1:1 ratio (350 μM) boiled together to promote annealing. All RNA samples were prepared in 25 mM KCl in 10 mM cacodylic acid buffer, pH 6.5. 1H NMR experiments were run at 25°C.
These results suggest that the dominant G quadruplex conformation in PSD-95 Q1 RNA is Q11234, which exposes the “AGGGA” sequence recognized by the miR-125a seed sequence. An alternate possibility is that in the presence of miR-125a the G quadruplex structures adopted by PSD-95 Q1 sequence are unwound, allowing for the formation of the PSD-95 Q1- miR-125a complex. To distinguish between these possibilities we analyzed the binding of miR-125a to PSD-95 Q1 RNA by 1D 1H NMR spectroscopy, a method that reports on modifications of the G quadruplex structure through its G imino proton resonance changes. As seen in Figure 7B bottom spectrum, a single low-intensity resonance was observed at ∼12.6 ppm in the imino proton resonance region of the free miR-125a indicating that some secondary structure involving the formation of a G–C Watson–Crick base pair is formed in the free miR-125a RNA. The second spectrum in Figure 7B shows the G imino proton resonances involved in G quadruplex formation in PSD-95 Q1 centered ∼11 ppm. When PSD-95 Q1 and miR-125a are mixed in a 1:1 ratio at room temperature, a new set of well-defined imino proton resonances appears at 12.1, 12.4, 12.5, and 12.6 ppm in the region corresponding to Watson–Crick G–C base pairs (Fig. 7B, third spectrum; Fürtig et al. 2003). Broader resonances are also visible at 13.2 and 14.3 ppm, corresponding to U imino protons involved in Watson–Crick A–U base pairs. These resonances report on the formation of G–C and A–U base pairs between miR-125a RNA and complementary bases in PSD-95 Q1 RNA. Interestingly, the G quadruplex imino proton resonances did not change significantly, indicating that the structure is not unwound upon miR-125a binding. When miR-125a and PSD-95 Q1 were boiled together in a 1:1 ratio to promote their annealing, the PSD-95 Q1 G quadruplex structure still forms as indicated by the resonances centered ∼11 ppm (Fig. 7B, fourth spectrum). The Watson–Crick imino proton resonances described above are well defined, and several new resonances become visible at 12.7, 12.8, 12.9, 13.5 and 13.8 ppm, indicating the formation of additional base pairs between miR-125a and PSD-95 Q1 RNA. Taken together, these results indicate that the dominant G quadruplex conformation in PSD-95 Q1 RNA is Q11234, which exposes the binding site for miR-125a and that this structure is not unwound upon miR-125a binding.
miR-125a interactions with the PSD-95 mRNA Q1–Q2 region
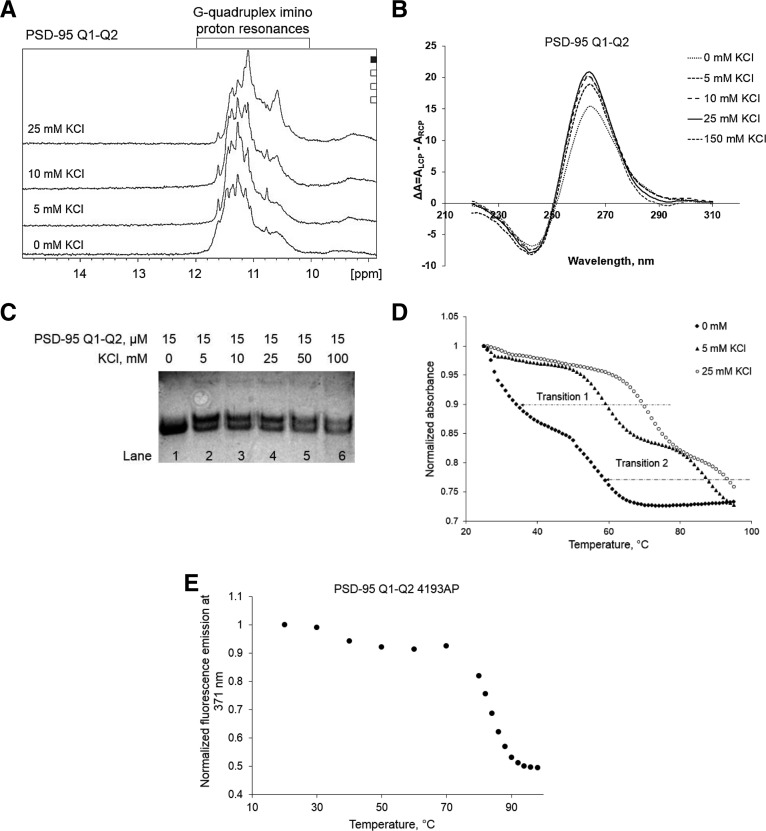
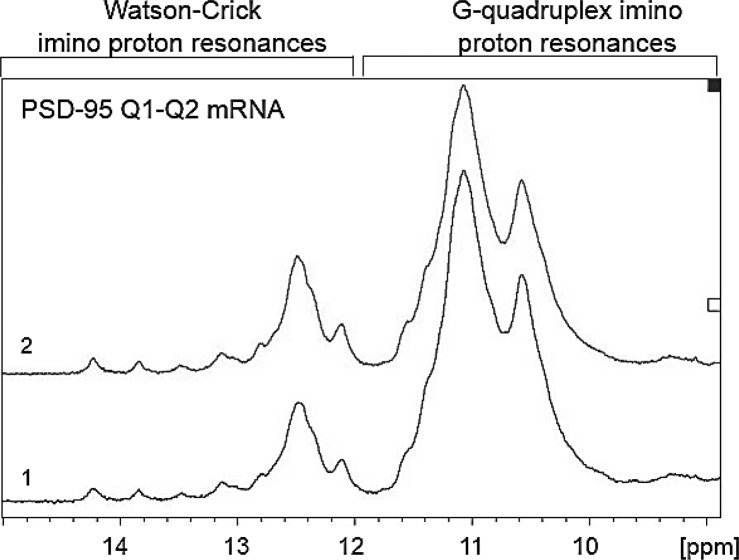
We have confirmed that the isolated PSD-95 Q1 and Q2 sequences form stable G quadruplex structures and that miR-125a interacts with its complementary sequence located within the Q1 segment. Subsequently, we investigated if miR-125a can still bind its complementary seed sequence within the Q1 region in the context of an extended region spanning nucleotides 4152–4204 which comprises both Q1 and Q2 sequences. This sequence, named PSD-95 Q1–Q2 mRNA [nucleotides 4152–4204] (Fig. 1B), was transcribed in vitro by T7 RNA polymerase driven transcription reaction. First, we performed 1D 1H NMR spectroscopy monitoring the imino proton resonance region changes upon KCl titration to observe G quadruplex formation. As seen in Figure 8A, imino proton resonances are observed between 10 and 12 ppm even in the absence of K+ ions, indicating G quadruplex formation, consistent with the 1H NMR spectra of the isolated PSD-95 Q1 and Q2 G quadruplexes (Fig. 2A,B). Once the concentration of KCl was increased the intensities of these resonances increased indicating the G quadruplex structure(s) stabilization. To analyze the fold of these G quadruplex structure(s), we performed CD spectroscopy experiments in the presence of increasing KCl concentrations in the range 5–150 mM. A positive band at ∼265 nm and a negative band at ∼240 nm were observed, indicating a parallel G quadruplex fold (Fig. 8B). The CD signal of the expanded PSD-95 Q1–Q2 mRNA was significantly higher than the signals of the isolated Q1 and Q2 sequences (Fig. 4), consistent with the expectation that more than one G quadruplex structure is formed in PSD-95 Q1–Q2 mRNA.
FIGURE 8.
(A) 1D 1H NMR spectra of PSD-95 Q1–Q2 mRNA at various concentrations of K+ ions in 10-mM cacodylic acid buffer, pH 6.5, at 25°C. (B) CD spectra of PSD-95 Q1–Q2 mRNA in increasing salt concentration at 25°C. 10 µM RNA sample was suspended in 10 mM cacodylic acid buffer, pH 6.5 and KCl was titrated in the ratio of 1–150 mM. The spectra were corrected by subtracting the buffer contributions. (C) Twenty percent of nondenaturing polyacrylamide gel in 0.5× TBE buffer in increasing KCl concentrations. Lane 1: PSD-95 Q1–Q2 mRNA in the absence of KCl; lanes 2–6: 0–100 mM KCl concentration. The gel was visualized by UV shadowing at 254 nm. (D) UV denaturation of 10 µM PSD-95 Q1–Q2 mRNA at 0, 5, and 25 mM KCl in 10 mM cacodylic acid buffer, pH 6.5. (E) Fluorescence spectroscopy thermal denaturation of PSD-95 Q1–Q2 4193AP mRNA in the presence of 5 mM KCl.
PSD-95 Q1–Q2 mRNA was also analyzed by 20% native polyacrylamide gel in 0.5× TBE buffer, varying the KCl concentration in the range 0–100 mM. As seen in Figure 8C, one band was observed in the absence of KCl (lane 1), whereas upon the titration of KCl, the intensity of this band decreased, with the concomitant apparition of a second main band (lanes 2–6), indicating the presence of two distinct conformations in PSD-95 Q1–Q2 mRNA. A faint retarded upper band becomes also visible upon the titration of KCl, which we attribute to the formation of a dimeric species.
To explore the origin of these two distinct bands observed in PSD-95 Q1–Q2 mRNA native gel in the presence of K+ ions, we performed thermal UV denaturation experiments in the presence of different KCl concentrations (Fig. 8D). At 5 mM KCl, two distinct hypochromic transitions are observed (labeled transition 1 and 2 in Fig. 8D, black triangles), which we attributed to the melting of the Q1 and Q2 G quadruplexes. The melting point of transition 1 was determined to be Tm Q1–Q2 transition 1 = 59°C, whereas that of transition 2 was Tm Q1–Q2 transition 2 = 87°C. Based on the melting temperatures of the isolated Q1 and Q2 G quadruplexes in 5 mM KCl (Tm Q1 = 64°C and Tm Q2 = 86°C), we tentatively assigned transition 1 to correspond to the Q1 G quadruplex unfolding and transition 2 to the Q2 G quadruplex unfolding. To confirm these assignments we have designed two fluorescently labeled RNAs: the isolated PSD-95 Q2 and the extended PSD-95 Q1–Q2 mRNA sequences, in which the adenine at position 4193 of Q2 (Q2 4193AP mRNA) and the same corresponding adenine in Q1–Q2 mRNA (Q1–Q2 4193AP mRNA) were replaced with 2-aminopurine (2AP) (Fig. 1C, adenine 4193 is circled). 2AP is a highly fluorescent analog of adenine whose steady-state fluorescence is sensitive to changes in its microenvironment (Serrano-Andrés et al. 2006; Bharill et al. 2008). We performed fluorescence spectroscopy thermal denaturation experiments of PSD-95 Q1–Q2 mRNA in the presence of 5 mM KCl, monitoring the changes of the 2AP reporter steady-state fluorescence. In these experiments only the 2AP reporter located in the Q2 G quadruplex contributes to the signal, thus, although the unfolding of the Q1 quadruplex also takes place, just the transition corresponding to the unfolding of the Q2 quadruplex is expected to be observed. This is in contrast to the UV thermal denaturation experiment where all nucleotides contribute to the absorbance changes as the Q1 and Q2 G quadruplexes unfold and two transitions are observed (Fig. 8D). A single transition was observed in the fluorescence spectroscopy thermal denaturation profile of PSD-95 Q1–Q2 4193AP mRNA, with a Tm of 84°C (Fig 8E). This Tm is in close agreement with the melting temperature of the transition 2, Tm Q1–Q2 transition 2 = 87°C, observed in the UV thermal denaturation profile of PSD-95 Q1–Q2 (Fig. 8D, black triangles), confirming our assignment of this transition to the unfolding of the Q2 G quadruplex. As a positive control, we also performed fluorescence spectroscopy thermal denaturation of the isolated Q2 4193AP mRNA in 5 mM KCl and observed a single transition with the Tm of 86°C (Supplemental Fig. 3).
Next we performed UV thermal denaturation experiments of PSD-95 Q1–Q2 mRNA in the absence of KCl, conditions in which a single band is observed in its native gel (Fig. 8C, lane 1). As seen in Figure 8D (black diamonds) transition 2 corresponding to the unfolding of the Q2 G quadruplex has a Tm Q1–Q2 = 56°C, in good agreement with the Tm Q2 = 51°C measured for the isolated Q2 sequence in the absence of KCl (Fig. 5B). Transition 1 is not clearly defined in the absence of KCl, indicating that in the context of the extended Q1–Q2 sequence, the Q1 sequence does not fold into a stable G quadruplex, in contrast to the isolated Q1 sequence which had a Tm Q1 = 44°C in the absence of KCl (Fig. 5A). Only upon the addition of K+ ions is the Q1 sequence folding into a stable G quadruplex (transition 1: black diamonds, 5 mM KCl; open circles, 25 mM KCl). This result suggests that in the context of PSD-95 Q1–Q2, the Q2 G quadruplex is stably folded independent of the presence of KCl, whereas the Q1 sequence is dynamic coexisting in equilibrium between stably folded and unfolded G quadruplex conformations.
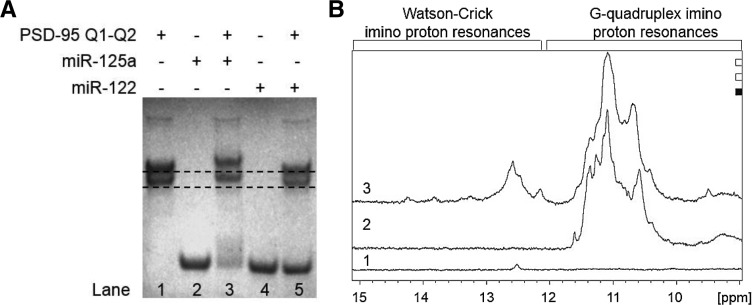
The PSD-95 Q1–Q2 mRNA interactions with miR-125a were analyzed by electromobility shift assays in 20% nondenaturing polyacrylamide. As seen in Figure 9A, in the presence of a 1:1 ratio of PSD-95 Q1–Q2 and miR-125a in 25 mM KCl both bands present in the free PSD-95 Q1–Q2 (lane 1) are shifted (lane 3), with the concomitant disappearance of the band corresponding to the free miR-125a (lane 2), indicating that miR-125a binds to both conformations adopted by the Q1 sequence. When Q1 is unfolded it is expected that miR-125a will bind to PSD-95 Q1–Q2 as its seed sequence target on PSD-95 mRNA will be accessible (Fig. 9A, compare lanes 1 and 3, lower band). However, even when Q1 is folded into a stable G quadruplex, miR-125a is still able to bind (Fig. 9A, compare lanes 1 and 3, upper band) indicating that the Q1 sequence adopts the Q11234 G quadruplex conformation in which the miR-125a seed sequence is exposed in the loop region between the Q1 and Q2 G quadruplexes (Fig. 1C, right). This result is in agreement with our previous findings that the main G quadruplex conformation in the isolated Q1 sequence was Q11234. As a negative control we used the liver-specific microRNA miR-122 and determined that in the presence of a 1:1 ratio of PSD-95 Q1–Q2 and miR-122 in 25 mM KCl no binding is observed (Fig. 9A, compare lanes 1, 4, and 5).
FIGURE 9.
(A) EMSA of PSD-95 Q1–Q2 mRNA and miR-125a/miR-122 in a 20% nondenaturing polyacrylamide gel in 0.5× TBE buffer and 25 mM KCl. miR-122 was used as a negative control. Lane 1: PSD-95 Q1–Q2 mRNA (15 μM); lane 2: miR-125a (15 μM); lane 3: PSD-95 Q1–Q2 and miR-125a RNAs in a 1:1 ratio (15 μM); lane 4: miR-122 (15 μM); lane 5: PSD-95 Q1–Q2 and miR-122 RNAs in a 1:1 ratio (15 μM). PSD-95 Q1–Q2 sample was annealed by boiling in 25 mM KCl and cooled at room temperature for 5 min. miR-125a/miR-122 was then added and incubated at room temperature for 30 min. The gel was visualized by UV shadowing at 254 nm. (B) 1D 1H NMR spectra of (1) 350 μM miR-125a, (2) 350 μM PSD-95 Q1–Q2 mRNA, (3) PSD-95 Q1–Q2 mRNA and miR-125a incubated at room temperature in a 1:1 ratio (350 μM). All RNA samples were prepared in 25 mM KCl in 10 mM cacodylic acid buffer, pH 6.5. 1H NMR experiments were run at 25°C.
Finally, we confirmed the binding between miR-125a and PSD-95 Q1–Q2 mRNA by 1D 1H NMR spectroscopy. As seen in Figure 9B, Watson–Crick base-pairing was observed when miR-125a was mixed at room temperature in a 1:1 ratio with PSD-95 Q1–Q2 mRNA in the presence of 25 mM KCl, while the G quadruplex structures remained folded (compare spectra 2 and 3), similarly to the results obtained for the isolated Q1 sequence (Fig. 7B).
Our study shows that the guanine rich area in the 3′ UTR of PSD-95 mRNA in close proximity to the miR-125a binding site adopts two intramolecular G quadruplex structures with parallel-topology: PSD-95 Q2 that is very stable independent of the presence of K+ ions, and PSD-95 Q1 that is very dynamic, existing in an equilibrium between folded and stably folded G quadruplex structure only in the presence of K+ ions. The PSD-95 Q1–Q2 sequence contains an additional GGG stretch in the linker region connecting the Q1 and Q2 quadruplexes, which theoretically could participate in the formation of alternate Q1 or Q2 quadruplex structures. We rule out the participation of the linker GGG in the folding of an alternate Q2 quadruplex, as such a structure would have the first loop formed from 6 nt, hence being less stable that the Q2 quadruplex pictured in Figure 1C. The results of our UV and fluorescence spectroscopy melting experiments indicate that the melting temperatures of the isolated Q2 quadruplex and of the Q2 quadruplex within the Q1–Q2 sequence are within 1°C from each other. Similarly, we rule out the participation of the linker GGG stretch in the formation of an alternate Q1 G quadruplex structure, as such a structure would completely embed the miR-125a target binding site, validated by in vivo experiments (Muddashetty et al. 2011), preventing the binding of miR-125a, and this contradicts our findings that both Q1 conformations are bound by miR125a (Fig. 9A, lane 3). While many G quadruplex conformations are accessible to the Q1 sequence, in vitro, in the absence of any other cellular factors, the Q1234 conformation in which the miR-125a seed is exposed is preferred. As far as we know, this is the first identified case in which the G quadruplex structure is implicated in the micro RNA pathway, expanding the repertoire of biological functions for this structure.
It was shown that FMRP works in conjunction with the microRNA pathway to regulate the PSD-95 translation (Muddashetty et al. 2011). It is possible that the equilibrium between the different structures adopted by the Q1 sequence will be shifted in the presence of FMRP, which could control the accessibility of miR-125a to its target sequence on PSD-95 mRNA. We used 1H NMR spectroscopy to determine if the FMRP RGG box, its domain shown to bind G quadruplex RNA with high affinity and specificity, has an effect upon miR-125a binding to its target sequence on PSD-95 Q1–Q2 mRNA. PSD-95 Q1–Q2 mRNA was first incubated with the FMRP RGG box in a 1:1 ratio, followed by the addition of miR-125a in a 1:1 ratio (Fig. 10, spectrum 1), or PSD-95 Q1–Q2 mRNA was first incubated with miR-125a in a 1:1 ratio, followed by the addition of the FMRP RGG box in a 1:1 ratio (Fig. 10, spectrum 2). Regardless of the addition order of the FMRP RGG box, the same Watson–Crick imino proton resonances are present, indicating that the presence of the FMRP RGG box does not disrupt the formation of a complex between miR-125a and PSD-95 Q1–Q2 mRNA (Fig. 10, spectra 1 and 2).
FIGURE 10.
1D 1H NMR spectra of 1:1:1 ratio of PSD-95 Q1–Q2, miR-125a, and FMRP RGG (300 μM). (1) PSD-95 Q1–Q2 was preincubated with the FMRP RGG box for 30 min at room temperature, followed by the additon of miR-125a and incubation for an additional 30 min at room temperature. (2) PSD-95 Q1–Q2 was preincubated with miR-125a for 30 min at room temperature, followed by the addition of the FMRP RGG box and incubation for an additional 30 min at room temperature. RNA samples were prepared in 10 mM cacodylic acid buffer, pH 6.5, in the presence of 150 mM KCl. 1H NMR experiments were run at 25°C.
These results were also confirmed by EMSA experiments in 15% nondenaturing polyacrylamide gel, in which PSD-95 Q1–Q2 mRNA was incubated with miR-125a fluorescently labeled by DY547 at the 3′ end, in the presence of the FMRP RGG box (Fig. 11). The native gel was visualized both, by UV shadowing detecting all RNA bands, as well as by exciting and monitoring the DY547 fluorescence emission, detecting only the bands corresponding to complexes containing miR-125a. As expected, the free PSD-95 Q1–Q2 mRNA forms two conformations (lane 1), visible by UV shadowing (bottom gel), but absent when the gel was visualized by DY547 fluorescence emission (top spectrum). Both conformations are bound by miR-125a (lane 2), with the unbound and complexed miR-125a being clearly detected by the DY547 fluorescence emission (top spectrum). When the FMRP RGG box is bound to PSD-95 Q1–Q2 mRNA (lane 3, bottom gel), the top band corresponding to the stable folded Q1–Q2 quadruplexes disappears, indicating the formation of a complex with the FMRP RGG box (likely in a 1:2 ratio), which due to its overall positive charge does not migrate in the gel. The second conformation (lower band) is also bound by the FMRP RGG box, possibly in a 1:1 ratio, as in this conformation the Q1 G quadruplex is not stably folded. PSD-95 Q1–Q2 mRNA was then first incubated with miR-125a, followed by the addition of the FMRP RGG box (lane 4), or first incubated with the FMRP RGG box, followed by the addition of miR-125a (lane 5), or incubated at the same time with both the FMRP RGG box and miR-125a (lane 6). In all cases, miR-125a is able to form a complex with PSD-95 Q1–Q2 mRNA, regardless of the presence of the FMRP RGG box. The formation of a complex containing two FMRP RGG boxes and miR-125a cannot be excluded, as such a complex would have an overall positive charge, preventing its migration through the gel.
FIGURE 11.
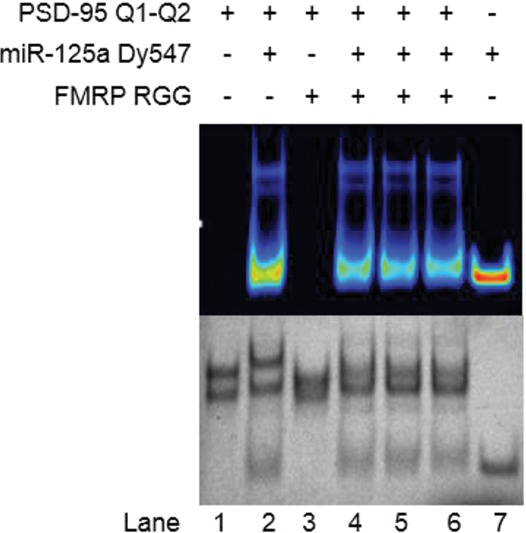
EMSA of PSD-95 Q1–Q2 mRNA, miR-125a, and FMRP RGG in a 15% nondenaturing polyacrylamide gel in 0.5× TBE buffer and 25 mM KCl. miR-125a was fluorescently labeled by Dy547 at its 3′ end. Lane 1: PSD-95 Q1–Q2 mRNA (15 μM); lane 2: PSD-95 Q1–Q2 and miR-125a RNAs in a 1:1 ratio (15 μM); lane 3: PSD-95 Q1–Q2 and FMRP RGG in a 1:2 ratio (15:30 μM); lane 4: PSD-95 Q1–Q2, miR-125a and FMRP RGG in a 1:1:2 ratio (15:15:30 μM). PSD-95 Q1–Q2 and miR-125a were preincubated first for 30 min at room temperature, and FMRP RGG was added and incubated for 30 min at room temperature. Lane 5: PSD-95 Q1–Q2, miR-125a, and FMRP RGG in a 1:1:2 ratio (15:15:30 μM). PSD-95 Q1–Q2 and FMRP RGG were preincubated first for 30 min at room temperature, and miR-125a was added and incubated for 30 min at room temperature. Lane 6: PSD-95 Q1–Q2, miR-125a, and FMRP RGG in a 1:1:2 ratio (15:15:30 μM). All three were added at the same time and incubated at room temperature for 30 min. Lane 7: miR-125a (15 μM). PSD-95 Q1–Q2 sample was annealed by boiling in 25-mM KCl and cooled at room temperature for 5 min. miR-125a/FMRP RGG was then added and incubated at room temperature. The gel was first visualized by UV shadowing at 254 nm (bottom gel picture), and then exposed to the excitation/emission spectra of 488/590 nm, to observe the fluorescence of the Dy547 label within miR-125a (top gel picture).
The FMRP regulation of PSD-95 mRNA translation is complex, being mediated by the FMRP phosphorylation. FMRP in its phosphorylated state and miR-125a containing RISC synergistically inhibit the PSD-95 mRNA translation, whereas upon FMRP dephosphorylation triggered by synaptic input, the RISC complex is no longer bound to PSD-95 mRNA, allowing for its translation to occur. While the requirement for FMRP in the regulation of PSD-95 mRNA translation is clearly established, the exact mechanism by which this is achieved is not known. Future work is needed to assess the role of FMRP phosphorylation in these models and experiments are in progress in our laboratories. It is anticipated that this work will motivate future studies to examine mechanisms of microRNA/RISC targeting by interplay between mRNA-binding proteins and G quadruplex structures.
MATERIALS AND METHODS
RNA samples
PSD-95 Q1 [nucleotides 4152–4177] (5′-GGGAAAAGGGAGGGAUGGGUCUAGGG-3′, miR-125a target site underlined), PSD-95 Q1234 [nucleotides 4152–4170] (5′-GGGAAAAGGGAGGGAUGGG-3′, miR-125a target site underlined, but lacking UCUAGGG seed sequence), PSD-95 Q2 [nucleotides 4190–4204] (5′-GGGAGGGAGGGUGGG-3′, no miR-125a target site), PSD-95 Q1–Q2 [nucleotides 4152–4204] (5′-GGGAAAAGGGAGGGAUGGGUCUAGGGAGUGGGAAAUGCGGGAGGGAGGGUGGG-3′, miR-125a target site underlined) were transcribed using the synthetic DNA templates purchased from TriLink BioTechnologies, Inc. and expressed by T7 RNA polymerase driven in vitro transcription reactions. The RNAs were purified by 20% polyacrylamide, 8-M urea gel electrophoresis and electroelution and subsequently dialyzed against 10-mM cacodylic acid, pH 6.5. miR-125a (5′-UCCCUGAGACCCUUUAACCUGUG-3′) and Dy547 labeled miR-125a (5′-UCCCUGAGACCCUUUAACCUGUG Dy547-3′) were chemically synthesized by Dharmacon, Inc.
Native gel electrophoresis
Prior to their use in the native gels, the RNA samples (typically 10–20 µM) were annealed by boiling for 5 min followed by incubation at room temperature for 10 min. KCl was added to the RNA samples at the desired concentration, prior to their annealing. To examine the effect of microRNA binding, miR-125a was added to the RNA after cooling in equal molar ratios and incubated for 30 min at room temperature. The native gels in 0.5× Tris/Borate/EDTA buffer were run at 4°C at 85 V and visualized by UV shadowing at 254 nm using an AlphaImager (AlphaInnotech, Inc.). In addition, miR-125a was fluorescently labeled with Dy457 synthetic dye at its 3′ end and binding with the PSD-95 Q1–Q2 mRNA was performed in the same fashion as for the unlabeled miR-125a. The position of the labeled miR-125a was detected by observing the fluorescence of Dy547 upon excitation at 488 nm.
UV spectroscopy thermal denaturation
UV thermal denaturation experiments were performed on a Cary 3E UV-VIS Spectrophotometer (Varian, Inc.) equipped with a peltier cell. Two hundred microliter samples containing 10 µM RNA in 10 mM cacodylic acid buffer, pH 6.5 and in the presence of 0–25 mM KCl were annealed as described above and thermally denatured by varying the temperature in the range 20°C–95°C, at a rate of 0.2°C/min and monitoring the absorbance changes at 295 nm, wavelength sensitive to G quadruplex denaturation (Mergny et al. 1998). In order to prevent sample evaporation, a layer of mineral oil was added to the cuvettes.
To study if an intermolecular or intramolecular quadruplex is formed within PSD-95 mRNA G-rich region, the UV thermal denaturation experiments were performed at variable RNA concentrations ranging from 3 to 30 μM and a fixed KCl concentration in a 10 mM cacodylic acid buffer, pH 6.5. In the case of G quadruplex structure formation between n number of RNA strands, the melting temperature (Tm) depends on the total RNA concentration (Equation 1) whereas the melting temperature of the G quadruplex structure formed within a strand (n = 1) is independent of the RNA concentration (Equation 2) (Hardin et al. 2000):
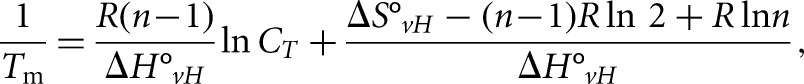 |
(1) |
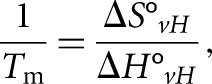 |
(2) |
where R is the gas constant and ΔH°vH and ΔS°vH are the Van't Hoff thermodynamic parameters.
The thermodynamic parameters of G quadruplex structures were obtained by fitting their UV thermal denaturation curves to Equation 3, which assumes a two-state model (Menon et al. 2008):
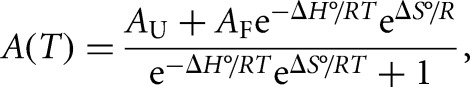 |
(3) |
where AU and AF represent the absorbance of the unfolded and native G quadruplex RNA structure, respectively, and R is the universal gas constant.
To calculate the number of K+ ions bound specifically by the G quadruplex structure, a simple model for folded to unfolded G quadruplex was assumed in which Δn K+ ions are lost upon heat treatment and G quadruplex unfolding. Δn is equal to the slope of the plot of ΔG° as a function of logarithm of K+ concentration:
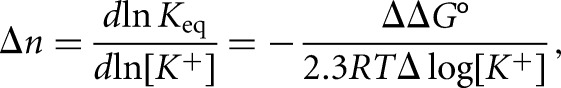 |
(4) |
where lnKeq=(ΔG°/RT) and ΔΔG°/Δlog[K+] is the slope of the plot of ΔG° as a function of the logarithm of K+ ion concentration (Hardin et al. 2000).
CD spectroscopy
All experiments were performed on a Jasco J-810 spectropolarimeter at 25°C, using a 1-mm path-length quartz cuvette (Starna Cells). Two hundred microliter volumes of 10 µM samples of annealed RNA were prepared in 10 mM cacodylic acid buffer, pH 6.5. The G quadruplex formation was monitored between 200 and 350 nm by titrating KCl in the range 5–150 mM, and averaging a series of seven scans with a 1 sec response time and a 2 nm bandwidth. The spectra were corrected by subtracting the cacodylic acid buffer contributions.
1H NMR spectroscopy
The G quadruplex formation was monitored by acquiring 1D 1H NMR spectra of RNA sequences at 25°C on a 500-MHz Bruker AVANCE spectrometer. Three hundred fifty micromolar RNA samples were prepared in 10-mM cacodylic acid buffer, pH 6.5 in a 90%H2O/10%D2O ratio and KCl was titrated in the range 5–25 mM. The water suppression was accomplished using the jump-and-return pulse sequence (Plateau and Gueron 1982) with the maximum of excitation set at 11 ppm or by using the Watergate pulse sequence (Piotto et al. 1992). To investigate the binding between PSD-95 mRNA (350 µM) and miR-125a (350 µM) in the presence or in the absence of FMRP RGG at a fixed KCl concentration of 25 mM or 150 mM, miR-125a/FMRP RGG was added to the annealed PSD-95 mRNA, followed by incubation at room temperature for 30 min prior to the acquisition of the NMR experiments.
Fluorescence spectroscopy
Steady-state fluorescence spectroscopy experiments of PSD-95 Q2 4 2AP mRNA [nucleotides 4190–4204] (5′-GGG(2AP)GGGAGGGUGGG-3′) and PSD-95 Q1–Q2 42 2AP mRNA [nucleotides 4190–4204] (5′-GGGAAAAGGGAGGGAUGGGUCUAGGGAGUGGGAAAUGCGGG(2AP)GGGAGGGUGGG-3′) were performed on a Horiba Jobin Yvon Fluoromax-3 and accompanying software fitted with a 150-W ozone-free xenon arclamp. Experiments were performed in a 150-μL sample volume, 3-mm path-length quartz cuvette (Starna Cells). Excitation wavelength was set to 310 nm, the emission spectrum was recorded in the range of 330–450 nm, and the bandpass for excitation and emission monochromators were both set to 3 nm. Both RNAs were melted in 5-mM KCl in the temperature range 20°C–90°C and at each temperature point the steady-state fluorescence was corrected to account for the known dependence of the free 2AP emission at 371 nm.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) grants 9R15HD078017-03A1 to M.R.M. and R21DA033478 to G.J.B.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.046722.114.
REFERENCES
- Bassell GJ, Warren ST 2008. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron 60: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharill S, Sarkar P, Ballin JD, Gryczynski I, Wilson GM, Gryczynski Z 2008. Fluorescence intensity decays of 2-aminopurine solutions: lifetime distribution approach. Anal Biochem 377: 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST 2003. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet 12: 3295–3305. [DOI] [PubMed] [Google Scholar]
- Coffee B, Keith K, Albizua I, Malone T, Mowrey J, Sherman SL, Warren ST 2009. Incidence of Fragile X syndrome by newborn screening for methylated FMR1 DNA. Am J Hum Genet 85: 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD 2003. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 40: 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB 2001. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107: 489–499. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB 2005. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev 19: 903–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roo M, Klauser P, Mendez P, Poglia L, Muller D 2008. Activity-dependent PSD formation and stabilization of newly formed spines in hippocampal slice cultures. Cereb Cortex 18: 151–161. [DOI] [PubMed] [Google Scholar]
- Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL 1993. The FMR–1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet 4: 335–340. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ 2008. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to Fragile X syndrome. Dev Cell 14: 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M 2010. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM 1997. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci 17: 1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürtig B, Richter C, Wöhnert J, Schwalbe H 2003. NMR spectroscopy of RNA. ChemBioChem 4: 936–962. [DOI] [PubMed] [Google Scholar]
- Hardin CC, Perry AG, White K 2000. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers 56: 147–194. [DOI] [PubMed] [Google Scholar]
- Hud NV, Smith FW, Anet FA, Feigon J 1996. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry 35: 15383–15390. [DOI] [PubMed] [Google Scholar]
- Joachimi A, Benz A, Hartig JS 2009. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg Med Chem 17: 6811–6815. [DOI] [PubMed] [Google Scholar]
- Keith D, El-Husseini A 2008. Excitation control: balancing PSD-95 function at the synapse. Front Mol Neurosci 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypr J, Kejnovska I, Renciuk D, Vorlickova M 2009. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res 37: 1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon L, Mihailescu MR 2007. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res 35: 5379–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon L, Mader SA, Mihailescu MR 2008. Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA 14: 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergny JL, Phan AT, Lacroix L 1998. Following G-quartet formation by UV-spectroscopy. FEBS Lett 435: 74–78. [DOI] [PubMed] [Google Scholar]
- Miyoshi D, Nakao A, Sugimoto N 2003. Structural transition from antiparallel to parallel G-quadruplex of d(G4T4G4) induced by Ca2+. Nucleic Acids Res 31: 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Kelić S, Gross C, Xu M, Bassell GJ 2007. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci 27: 5338–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ 2011. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell 42: 673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar M, Goldsmith G, Moorthy BT, Lieber MR, Joshi MV, Choudhary B, Hosur RV, Raghavan SC 2011. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res 39: 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST 2007. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci 27: 14349–14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramasivan S, Rujan I, Bolton PH 2007. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods 43: 324–331. [DOI] [PubMed] [Google Scholar]
- Piotto M, Saudek V, Sklenár V 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR 2: 661–665. [DOI] [PubMed] [Google Scholar]
- Plateau P, Gueron M 1982. Exchangeable protons without base line distortion using a new strong pulse sequence. J Am Chem Soc 104: 7310–7311. [Google Scholar]
- Randazzo A, Spada GP, da Silva MW 2013. Circular dichroism of quadruplex structures. Top Curr Chem 330: 67–86. [DOI] [PubMed] [Google Scholar]
- Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H 2001. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J 20: 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D, Gilbert W 1990. A sodium–potassium switch in the formation of four-stranded G4-DNA. Nature 344: 410–414. [DOI] [PubMed] [Google Scholar]
- Serrano-Andrés L, Merchán M, Borin AC 2006. Adenine and 2-aminopurine: paradigms of modern theoretical photochemistry. Proc Natl Acad Sci 103: 8691–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G 1993. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell 74: 291–298. [DOI] [PubMed] [Google Scholar]
- Subramanian M, Rage F, Tabet R, Flatter E, Mandel JL, Moine H 2011. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep 12: 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, Malter JS 2003. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci 100: 14374–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlíčková M, Kejnovská I, Sagi J, Renčiuk D, Bednářová K, Motlová J, Kypr J 2012. Circular dichroism and guanine quadruplexes. Methods 57: 64–75. [DOI] [PubMed] [Google Scholar]
- Xu W, Schlüter OM, Steiner P, Czervionke BL, Sabatini B, Malenka RC 2008. Molecular dissociation of the role of PSD-95 in regulating synaptic strength and LTD. Neuron 57: 248–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, et al. 2007. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci 10: 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.