Abstract
SR proteins are a well-conserved class of RNA-binding proteins that are essential for regulation of splice-site selection, and have also been implicated as key regulators during other stages of RNA metabolism. For many SR proteins, the complexity of the RNA targets and specificity of RNA-binding location are poorly understood. It is also unclear if general rules governing SR protein alternative pre-mRNA splicing (AS) regulation uncovered for individual SR proteins on few model genes, apply to the activity of all SR proteins on endogenous targets. Using RNA-seq, we characterize the global AS regulation of the eight Drosophila SR protein family members. We find that a majority of AS events are regulated by multiple SR proteins, and that all SR proteins can promote exon inclusion, but also exon skipping. Most coregulated targets exhibit cooperative regulation, but some AS events are antagonistically regulated. Additionally, we found that SR protein levels can affect alternative promoter choices and polyadenylation site selection, as well as overall transcript levels. Cross-linking and immunoprecipitation coupled with high-throughput sequencing (iCLIP-seq), reveals that SR proteins bind a distinct and functionally diverse class of RNAs, which includes several classes of noncoding RNAs, uncovering possible novel functions of the SR protein family. Finally, we find that SR proteins exhibit positional RNA binding around regulated AS events. Therefore, regulation of AS by the SR proteins is the result of combinatorial regulation by multiple SR protein family members on most endogenous targets, and SR proteins have a broader role in integrating multiple layers of gene expression regulation.
Keywords: alternative pre-mRNA splicing, SR proteins, RNA processing, regulation of gene expression
INTRODUCTION
Gene expression in higher eukaryotes requires proper formation of mRNA from immature, pre-mRNA. This process entails removal of noncoding (intron) sequences by pre-mRNA splicing, so that coding (exon) sequences can be spliced together. Alternative splicing (AS) is the process of generating multiple mRNA isoforms from a single pre-mRNA by joining the exons together in different combinations. AS significantly expands the diversity of the proteome, with recent estimates suggesting that 90%–95% of human multiexon transcripts undergo AS (Wang et al. 2008). Proteome diversity is further increased by the fact that many mRNA variants are expressed differentially in different cell types and during different developmental stages (Pan et al. 2008). Given that AS is both pervasive and essential for tissue development and homeostasis, it is critical that we understand how it is regulated. Indeed, misregulation of specific AS events is the cause of several genetic diseases (e.g., spinal muscular atrophy) (Cartegni and Krainer 2002; Kashima and Manley 2003). Similarly, mutations that affect AS of tumor suppressors, leading to their inactivation, are responsible for some inherited and sporadic cancers (Venables 2004; Karni et al. 2007; Misquitta-Ali et al. 2011).
The splicing reaction is carried out by the spliceosome—a complex of five small nuclear ribonucleoprotein particles (snRNPs) and hundreds of accessory proteins. The spliceosome promotes precise excision of introns, by recognizing pre-mRNA sequence elements that define intron boundaries. The sequence elements are highly degenerate, and occur frequently within the genome (Kramer 1996; Will and Lührmann 2001).
Proper splicing outcome is ensured by multiple factors. First, accurate splice-site recognition by the spliceosome is facilitated by auxiliary proteins that bind cis-acting elements in pre-mRNA, and recruit and stabilize the spliceosome. Splice-site recognition is also determined by the specific combinations of cis-acting elements, their proximity to splice sites, and specific splicing factors’ expression levels. Lastly, splicing at a particular site can be activated or repressed when these auxiliary proteins bind nearby enhancer or silencer regions—to recruit or inhibit assembly/stability of spliceosomal components, respectively (Black 2003; Pagani and Baralle 2004; House and Lynch 2006; Chen and Manley 2009).
An essential class of splicing regulators is the serine/arginine (SR) family of proteins. SR proteins are a highly conserved family of splicing regulators—with one or two N-terminal RNA-recognition motifs (RRM), and a C-terminal (RS) domain enriched in arginine and serine dipeptides. SR proteins have primarily been shown to be regulators of splice-site selection, but have also been implicated in all crucial aspects of mRNA metabolism—including export, localization, translation, and nonsense-mediated decay (NMD) (Krainer et al. 1990; Long and Caceres 2009; Shepard and Hertel 2009). In AS, SR proteins appear to function as general activators—by RRM-domain-dependent binding of exonic-splicing enhancers (ESEs) in pre-mRNA and by recruiting the splicing machinery, via RS-domain–protein interactions, to the splice site (e.g., U1 snRNP to the 5′ splice site or U2AF to the 3′ site) (Graveley 2000; Lam and Hertel 2002; Matlin et al. 2005). Binding sites on pre-mRNA for SR proteins and other splicing regulators (e.g., hnRNP family) are typically in close proximity, thus suggesting that interplay between activation and repression modulates the frequency of exon inclusion (Zhu et al. 2001; Zahler et al. 2004).
While most SR protein family members have been implicated in AS regulation, many of the studies were performed in vitro, or based on single-gene analyses. A more comprehensive analysis is now possible due to recent advances in the ability to rapidly characterize AS. For example, RNA sequencing can identify genome-wide AS events (Brooks et al. 2011; Huelga et al. 2012), and immunoprecipitation approaches can identify precise binding sites for RNA-binding proteins across the transcriptome (Konig et al. 2010; Änko et al. 2012; Huelga et al. 2012). A study mapping SRSF3 and SRSF4 binding sites revealed that these two SR proteins bind a small, but distinct, subset of genes with very little overlap of endogenous targets (Änko et al. 2012). This implies that individual SR proteins bind distinct endogenous transcripts—with little cooperativity between family members. In contrast, another recent study mapped the in vivo, endogenous binding targets of SRSF1 and SRSF2, and found extensive overlap (Pandit et al. 2013). Furthermore, the study found SRSF1 and SRSF2 coordinated with and compensated for one another to regulate exon inclusion and skipping. Thus, while these studies suggest that SR proteins control AS through position-dependent pre-mRNA binding, it is unclear to what extent SR proteins cooperate with each other to regulate AS in a combinatorial fashion.
In this study, we identified AS events regulated by individual SR proteins, by utilizing RNAi of each of the eight identified SR proteins in Drosophila (SC35, SF2, SRp54, XL6, Rbp1, B52, Rsf1, and Rbp1-like), followed by RNA-seq analysis. This global approach revealed that a majority of SR-regulated AS events are controlled in a combinatorial manner by multiple SR proteins. In addition, all SR proteins can promote exon inclusion, and exon skipping. Utilizing SR protein-domain mutants, we show that in vivo AS regulation by the SR proteins requires an RS-domain, and that target specificity is dictated by the RRM-domain. Indicating that the binding location on the target RNA is responsible for the SR protein's positive or negative effect on AS. Using iCLIP-seq, we identified direct RNA-binding sites that contribute to regulation of AS, and produced RNA-splicing maps for the SR protein family. We find that multiple SR proteins associate with most transcripts, but can bind to specific locations. This suggests that AS is generally regulated by the collective contribution of multiple SR proteins, and presumably other splicing regulators, on most endogenous targets. Finally, beyond AS, we characterized the roles of the eight SR proteins in RNA metabolism—including transcript expression, and alternative promoter and polyadenylation selection—suggesting a role for SR proteins in the integration of multiple levels of gene expression regulation.
RESULTS
Identification of genome-wide SR-dependent AS events
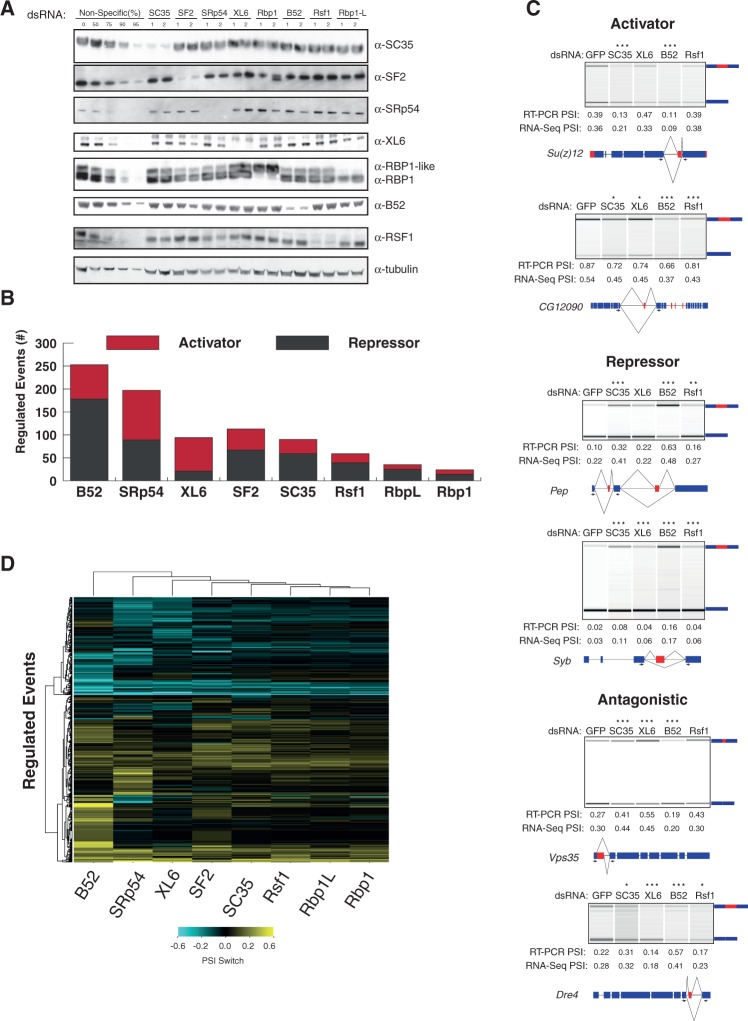
To identify the SR protein network regulating AS, we first reduced levels of each of the eight known SR proteins in Drosophila S2 cells, and then performed RNA-seq on RNA isolated from control (GFP)—and SR-RNAi-treated cells—to detect altered AS events (Fig. 1). Each protein was knocked down to <10% by RNAi. Individual SR proteins were reduced specifically with no appreciable changes in other SR protein family members (Fig. 1A); the only exception was up-regulation of Rbp1-like when Rbp1 was reduced, which has been previously noted (Kumar and Lopez 2005).
FIGURE 1.
Genome-wide analysis of SR-dependent AS events. Drosophila melanogaster S2 cells were treated with SR-RNAi. (A) Western blot analysis of S2 cells treated with two nonoverlapping SR-specific dsRNAs versus S2 cells treated with nonspecific dsRNA (GFP); loading control, α-tubulin. (B) RNA-seq analysis showing the total number of simple AS events regulated by each SR protein; each bar shows the proportion of events that are less included (SR-activated, red) or more included (SR-repressed, dark gray). (C) RT-PCR validation of putative SR-regulated AS events identified by RNA-seq analysis (significantly changed events [*] P ≤ 0.05, [**] P ≤ 0.01, [***] P ≤ 0.001 Fisher's exact test); events separated into activator, repressor, or antagonistic. PSI values calculated from the RT-PCR and RNA-seq experiments are shown for comparison below each lane. Gene model is displayed below each gel with arrows indicating primer locations for PCR of the alternative region (red). (D) Heatmap reveals hierarchical clustering of PSI-switch scores for SR-regulated AS events (y-axis) for each SR protein (x-axis).
To facilitate the RNA-seq analysis, we focused on the five simplest types of AS: cassette exons, competing donor or acceptor sites, mutually exclusive exons, and intron retention. In the eight SR protein-depleted S2 cells, we identified a total of 561 altered simple AS events, representing 405 genes (Supplemental Fig. S1A). This represents >10% of the 5472 reliably detected simple AS events in S2 cells, indicating that the SR protein family is important for extensive AS regulation. Many of the changes observed may be due to indirect or secondary effects of RNAi, since SR proteins alter the AS of many transcription and splicing factors. However, these changes are part of a large regulatory network where SR proteins are key regulators. Each SR protein alters different numbers of AS events (Fig. 1B; Supplemental Fig. S1A)—ranging from 253 for B52 to 24 for Rbp1. Previous studies of SR proteins focused only on their regulation of cassette exons or competing 5′ and 3′ splice sites. To determine whether SR proteins are important for only one versus a few types of simple AS events, we subdivided the affected events into the five types of simple AS, and found that SR proteins affect all types in similar proportions (Supplemental Fig. S1A,B).
In Drosophila, SR proteins are essential for tissue development and sex determination (Lynch and Maniatis 1996; Hoffman and Lis 2000; Gabut et al. 2007). Gene ontology analysis of SR-dependent targets revealed overrepresented terms corresponding to cell growth and proliferation, cell signaling, tissue and organ development, and neuronal processes (generation and differentiation of neurons and behavior) (Supplemental Table S2). Thus, SR proteins affect targets important in many physiological processes, but especially developmental.
SR proteins both activate and repress splicing events
SR proteins function as general activators of exon definition by binding cis-elements in pre-mRNA, known as exonic-splicing enhancer elements (ESEs), and thereby facilitate recruitment and/or stabilization of components of the core spliceosome. To determine whether SR proteins also repress AS events, we analyzed the proportion of events showing increased inclusion versus exclusion, when individual SR proteins are depleted. We found that all eight SR proteins function as both activators and repressors of AS events (Fig. 1B). Surprisingly, six of the eight SR proteins repressed more than half of their regulated events (ranging from 58.3% for Rbp1 to 71.4% for Rbp1-like); on the other hand, SRp54 and XL6 activated 54.8% and 77.7% of events, respectively.
To determine if SR proteins preferentially activate or repress a specific AS type, we separated the included or excluded events by AS type (Supplemental Fig. S2A–E). Seven of the eight SR proteins activated more cassette exons than other types of AS; the exception was B52, which repressed 53.1% of cassette exon events. Additionally, SR proteins were important for repression of most intron-retention splicing events—except for XL6, which activated 84.4% of its regulated intron-retention events. Since SR proteins repress a majority of intron-retention events, and intron-retention events are the largest class of SR-regulated AS, these events contribute significantly to the overall repression of AS observed. These observations of cassette exon activation and intron-retention repression are consistent with reports that SR proteins activate exon recognition (Lam and Hertel 2002). However, there are many examples where SR proteins silence inclusion of AS regions, and to different extents.
Multiple SR proteins can affect the same AS event
Semiquantitative RT-PCR of predicted targets demonstrated that multiple SR proteins can activate (Su(z)12 and CG12090) and repress (Pep and Syb) the same splicing event (Fig. 1C). Surprisingly, we also identified events where some SR proteins activate inclusion, while others repress inclusion: for example, enhanced skipping of a cassette exon in the Dre4 transcript when XL6 or Rsf1 are depleted, versus increased inclusion when SC35 or B52 are depleted. Such antagonism suggests that SR protein regulation is not always redundant, but different SR proteins can affect an AS region differently.
SR proteins collaborate to regulate a majority of AS events
Many AS events are regulated by multiple RNA-splicing factors in a combinatorial manner (Blanchette et al. 2005). For example, in Drosophila, SR proteins Rbp1 and SF2 cooperate with sex-specific splicing regulators Tra and Tra-2 to regulate AS in Doublesex (Lynch and Maniatis 1996). In addition, in Drosophila and mammals, AS is cooperatively regulated genome wide by another well-characterized splicing regulator family, hnRNPs (Blanchette et al. 2009; Huelga et al. 2012). Comparing affected AS events across all eight SR proteins revealed clustering of SR protein-dependent AS events (Fig. 1D). Similarly, when we compared the number of unique and shared AS events of each SR protein, we found that while individual SR proteins affect unique AS events, the majority of AS events were affected by more than one of the eight SR proteins (Fig. 2A). The one exception was B52, which regulated only 36% of its targets with other SR protein family members (Fig. 2A). To determine the extent of SR protein cooperative or antagonistic coregulation, we noted the direction of change (increased inclusion or increased exclusion) for each coregulated AS event, and clustered them accordingly. The vast majority of these events were regulated similarly by multiple SR proteins (Fig. 2B)—greater than 87.7% of coregulated events demonstrated cooperative regulation for each SR protein. Also, while individual events can be regulated by specific sets of SR proteins, there were no distinct pair-wise relationships between individual SR protein family members (Fig. 2B).
FIGURE 2.
Combinatorial regulation of AS by SR proteins. Drosophila melanogaster S2 cells were treated with RNAi against one or more of the eight SR proteins and RNA-seq was performed to detect the changes in AS. (A) Proportion of AS events regulated by one (dark gray) or more than one (red) SR protein. (B) Heatmap of coregulated AS events identified by RNA-seq subdivided by those with increased inclusion or exclusion. (C) Western blot analysis of lysates from S2 cells treated with dsRNA against one or two SR proteins and dsRNA against GFP as a control; α-tubulin used as a loading control. (D–F) S2 cells were treated with RNAi against one SR protein (SC35, B52 or XL6) or two (SC35 and B52 or XL6 and B52) and RNA isolated. RNA-seq was used to create a heatmap showing hierarchical clustering of PSI-switch scores for the resulting changes in AS events (D), graph displaying the total number of significantly changed events in each SR protein-depleted RNA-seq experiment (E), and number of coregulated events that displayed a greater (enhanced; red), equal or less (cooperative; blue), or opposite (compensatory; green) PSI change in the simultaneous knockdown compared with individual SR knockdown (F). (G) RT-PCR of coregulated AS events. PSI values and mean ΔPSI values from control displayed below each lane. Gene model is displayed below each gel with arrows indicating primer locations for PCR of the alternative region (red).
One of the more surprising findings was for Rsf1, which coregulated 81.4% of its AS events with other SR proteins (Fig. 2A), and 97.9% of those events were regulated in the same direction. Therefore, Rsf1 acts, almost exclusively, to cooperatively coregulate AS events with the other SR proteins. This finding is in contrast to a previous study, suggesting that Rsf1 acts as an antagonist of other SR proteins for splice-site recognition through protein interactions via its GRS domain (Labourier et al. 1999).
The extensive overlap of affected AS transcripts suggests that SR proteins control specific AS events either redundantly or combinatorially. To test these hypotheses, we reduced the expression of two pairs of SR proteins, simultaneously, in S2 cells and then performed RNA-seq to monitor changes in AS. To first demonstrate that individual and pair-wise SR protein expression is specifically reduced by RNAi treatment, we used Western blot analysis of the three SR proteins in S2 cell lysates (Fig. 2C). As expected, individual and pair-wise SR protein expression were reduced specifically. When we examined changes in AS, we found that in addition to the affected AS events identified when individual SR proteins were reduced, there were new events significantly changed when the SR pairs SC35:B52 and XL6:B52 protein levels are reduced together, 205 and 195 new events, respectively (Fig. 2D,E). Also, the overall magnitude of AS changes seemed to be greater for some AS events when two SR protein levels were reduced (Fig. 2D), supporting the combinatorial hypothesis. To determine if there is, indeed, an increase in the magnitude of AS change when two SR proteins are knocked down, we compared the PSI (# Inclusive reads/[# Inclusive reads + # Exclusive reads]) switch scores (PSI SR RNAi—PSI control) of the 23 and 26 coregulated AS events between SC35 and B52 and XL6 and B52, respectively. In 14 of the 23 events coregulated by SC35 and B52, and 12 of the 26 events coregulated by XL6 and B52, the PSI-switch score was increased (enhanced) when both SR proteins were knocked down simultaneously (Fig. 2F). One event coregulated by SC35 and B52 displayed a greater splicing change in the opposite direction (compensatory), and all of the other coregulated events for SC35, XL6, and B52 had PSI-switch scores remain the same or decrease in the simultaneous knockdown demonstrating cooperative regulation (Fig. 2F).
As a final test of combinatorial regulation, we performed semiquantitative RT-PCR on coregulated AS events. The AS cassette exon in Su(Z)12 exhibits less inclusion when either SC35 or B52 proteins are reduced, and a further decrease in inclusion when both proteins are reduced (Fig. 2G). Similarly, when XL6 and B52 are acting as repressors of the cassette exon in CG31638, an increase in magnitude of repression is observed when both proteins are knocked down (Fig. 2G). Surprisingly, this effect is even observed when the two SR proteins have antagonistic functions. XL6 acts as an activator, and B52 as a repressor of the cassette exon in the Dre4 transcript. When both XL6 and B52 are knocked down together the result is a splicing change that is near wild-type levels (Fig. 2G), suggesting that these two SR opposing proteins cancel each other out, and providing further support for combinatorial coregulation by SR proteins.
SR proteins regulate promoter selection and 3′ end processing of target transcripts
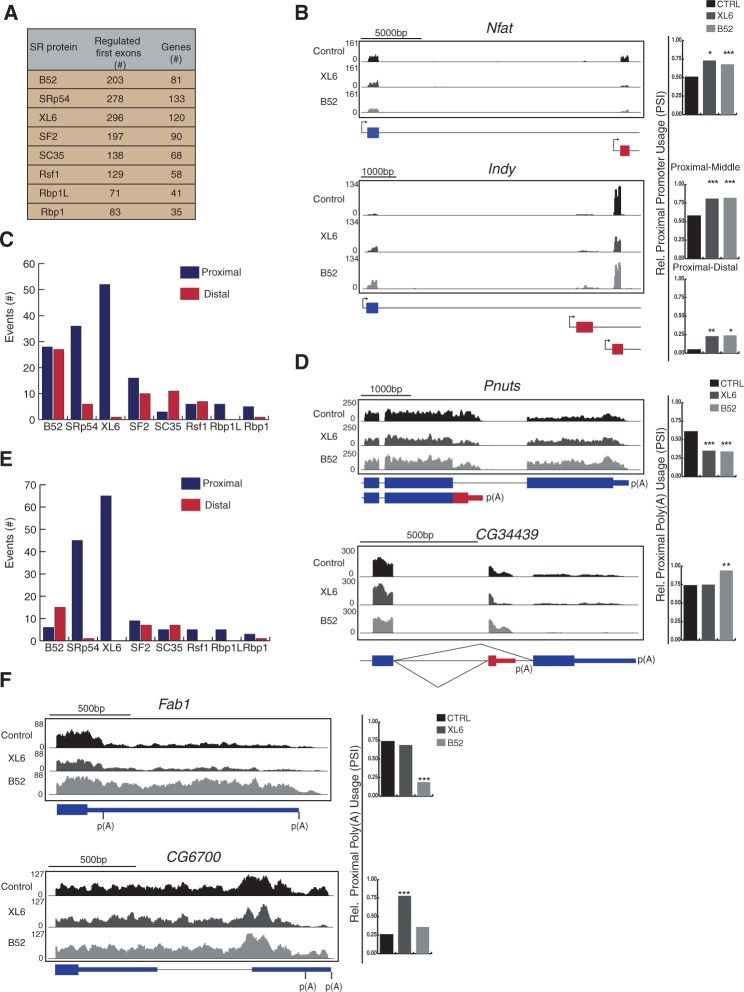
Many eukaryotic genes contain multiple promoters that determine a different start site/first exon of a transcript, thereby modulating expression at the transcriptional level. To investigate if SR proteins could affect promoter selection, we analyzed transcripts that were altered when an individual SR protein's expression was reduced that had annotated alternative start sites by RNA-seq (Fig. 1). Surprisingly, SR proteins affected the frequency of inclusion of many alternative first exons (Fig. 3A). We found that, of the eight SR proteins, Rbp1L affected the fewest first exons (71 in 41 genes) and SRp54 affected the most (278 in 133 genes). Two striking examples of differential promoter selection when XL6 or B52 was reduced were the usage of the distal promoter of the Nfat gene was decreased and usage of the two proximal promoters of the Indy gene was increased (Fig. 3B).
FIGURE 3.
SR proteins regulate promoter choice and 3′ end processing. Transcripts with alternative promoter and polyadenylation sites were analyzed for changes when individual SR proteins are depleted (Fig. 1). (A) Total number of transcripts with differential promoter selection when individual SR proteins are knocked down. (B) RNA-seq tracks for representative transcripts with altered promoters. Shaded gray regions above the gene model correspond to the density of mapped RNA-seq reads from each sample. Charts on the right indicate the relative frequency of proximal promoter usage (PSI; significantly changed events [*] P ≤ 0.05, [**] P ≤ 0.01, [***] P ≤ 0.001 Fisher's exact test). (C,E) Number of altered events within transcripts with CR-APA sites (C) and UTR-APA sites (E), subdivided into shifts to proximal or distal polyadenylation sites. (D,F) RNA-seq tracks for representative transcripts with CR-APA (D) and UTR-APA (F) (blue boxes, constitutive regions; red boxes, alternative regions and poly(A) sites; thin lines, 3′ UTR). Shaded gray regions above the gene model correspond to the density of mapped RNA-seq reads from each sample. Charts on the right indicate the relative frequency of proximal APA usage (PSI; significantly changed events [***] P ≤ 0.001 Fisher's exact test).
In addition to regulating the location of transcript initiation, transcripts can have alternative termination sites. Such alternative 3′ processing results from a dynamic interplay between cleavage/polyadenylation machineries and splicing regulators (Kyburz et al. 2006; Blechingberg et al. 2007). In some cases, alternative polyadenylation (APA) sites in protein-coding regions (CR-APA), yield different protein isoforms. To test whether SR proteins could affect transcript termination, we analyzed transcripts with annotated alternative termination from the RNA-seq results (Fig. 1). When SR proteins were depleted, APA sites were significantly changed. XL6 affected the largest number of CR-APA events (53), and Rbp1 the smallest number (6) (Fig. 3C). These large numbers indicate that regulation of CR-APA sites is an important function of SR proteins.
An example of CR-APA regulation was the Pnuts gene with two APA sites; use of the proximal site results in a transcript that is truncated by 2000 nt, and a protein that is half as large as when the distal site is used (Fig. 3D). When XL6 or B52 were reduced, use of the proximal APA site was reduced. The CG34439 gene, which also has two APA sites, provided a second example of CR-APA regulation; use of the proximal site yields a protein that is 74 amino acids, instead of 123. When B52 was reduced, use of the distal APA site was reduced (Fig. 3D), indicating that B52 is required for ensuring production of the longer protein isoform.
In addition to APA sites in the coding region, there are APA sites in the 3′ untranslated region (3′ UTR). These 3′-UTR alternative polyadenylation (UTR-APA) do not alter protein composition, but can affect protein expression. For example, using a more distal UTR-APA can introduce microRNA (miRNA)-binding sites, or other regulatory sequences such as AU-rich elements (AREs) and RNA localization signals (Sandberg et al. 2008). To determine whether SR proteins affect the use of UTR-APA sites, we analyzed transcripts in our RNA-seq results (Fig. 1) that had annotated APA sites within the 3′ UTR that were altered when individual SR proteins were knocked down. Surprisingly, SR proteins affected many UTR-APA events (Fig. 3C). Similar to CR-APA regulated events, in the majority of cases, we observed a shift toward usage of the proximal UTR-APA site (Fig. 3C,E). The two exceptions are B52 and SC35, for which many more events result in use of the distal site. Rbp1 affects the smallest number (4 events), and XL6 controls the largest (65 events). An example of SR protein regulation of UTR-APA is provided by Fab1 (Fig. 3F). Reduction in B52 increased use of the distal site, suggesting that B52 is required to produce a shorter 3′ UTR that excludes 1000 nt of potential regulatory sequences. A second example of SR protein regulation of UTR-APA is provided by CG6700. When XL6 is reduced there is a switch to use of the proximal poly(A) site (Fig. 3F). Therefore, XL6 is responsible for maintaining the long 3′ UTR of this transcript. Both SR proteins bind near regulated poly(A) sites, indicating a potential direct role in poly(A) selection (Supplemental Fig. S9). These data suggest that SR proteins affect UTR-APA selection—in the vast majority of cases, ensuring that longer transcripts are produced. The differences in proximal or distal poly(A) site regulation by the SR proteins could be a result of cell or tissue specific expression regulation. One striking difference between CR-APA and UTR-APA is the mechanism that may be used by SR proteins to regulate APA site choice. CR-APA could be the result of differential selection of splice sites, by the SR proteins, that include or remove poly(A) selection regulatory sequences from the transcript, and ultimately change where the transcript is polyadenylated. However, UTR-APA involves differential selection of poly(A) sites independent of splice-site selection. Many RNA-binding proteins have been implicated in UTR-APA control by enhancing or antagonizing the action of the formation of the 3′ processing complex, but SR proteins have only been implicated in CR-APA (Lou et al. 1998). These results support the notion that SR proteins might play a role in UTR-APA regulation, and also may suggest SR protein interaction with the 3′ end processing machineries.
SR proteins are required for maintaining proper transcript expression levels
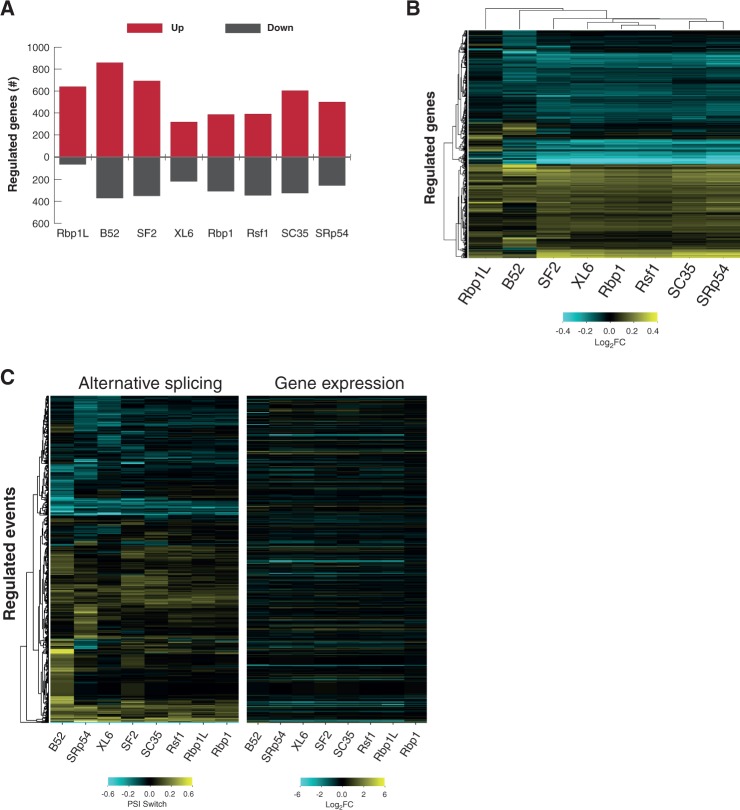
In addition to regulating transcript length and sequence composition, SR proteins modulate gene expression levels at other stages of RNA metabolism—such as mRNA export, stability, and translation (Long and Caceres 2009; Zhong et al. 2009). They also affect transcriptional elongation and associate with sites of active chromatin (Champlin et al. 1991; Lin et al. 2008); for example, SRSF2 activates transcription by mediating both release of P-TEFb from the 7SK complex, and then recruitment of P-TEFb and other transcriptional factors to the gene promoter (Ji et al. 2013). To examine genome-wide effects of SR proteins on gene expression, we examined the RNA-seq data for changes in overall transcript abundance in the absence of individual SR proteins (Fig. 1). We found that expression of a large number of genes underwent a greater than twofold change (Fig. 4A)—ranging from 539 genes for XL6 to 1230 for B52 (i.e., 6.1% and 13.8%, respectively, of the 8891 reliably detected genes). Expression of 2283 genes was either up-regulated (1661) or down-regulated (722). Thus, SR proteins can affect expression of 25.7% of expressed genes in S2 cells. Hierarchal clustering revealed that transcripts whose expression increased in the absence of SR proteins play key roles in cell growth and signaling, and transcripts whose expression decreased are important for reacting to external stimuli and tissue development (Fig. 4B; Supplemental Table S3). SR proteins did not affect the expression levels of many common housekeeping genes (HSP90, Act5C, RPL23; data not shown).
FIGURE 4.
SR proteins regulate RNA transcript levels. Transcript abundance was calculated and normalized for control and SR-depleted RNA-seq samples (Fig. 1). (A) Number of genes with differential expression (red, increased; dark gray, decreased). (B) Heatmap showing hierarchical clustering of calculated fold-change (log2) for genes with significantly altered transcript levels. (C) Heatmaps comparing identified AS-regulated events (Fig. 1D) (left) and their corresponding gene expression changes (fold-change [log2]) (right).
The number of genes whose expression was affected by SR protein knockdown (2283) exceeds the number that exhibited changes in simple AS events (405). This suggests that these two functions of SR proteins are independent. To test this hypothesis, we analyzed regulated AS events (Fig. 1D) to determine if there were significant changes in expression levels of those genes. The vast majority of genes (>62.5% for Rbp1 and >80% for all other SR proteins) affected at the level of AS exhibited no significant change in transcript abundance (Fig. 4C; Supplemental Fig. S3). This result suggests that SR protein-regulation of transcript expression is downstream from direct AS targets (e.g., regulation of transcription factor AS), or is independently regulated by a mechanism other than AS.
SR protein AS specificity is dictated by RRM domain, and requires an RS domain
We have shown that SR proteins regulate AS events in a combinatorial manner, in vivo. However, there are differences in the magnitude of splicing change observed depending on which SR protein is reduced. Additionally, there are examples of AS events where SR proteins can act antagonistically. RS domains from different SR proteins differ in sequence composition and length, but were found to functionally substitute for one another, in vitro. However, there are differences in potency of RS domains when tethered to a similar ESE (Graveley et al. 1998). We next wished to determine which SR protein domains are required for regulation of AS targets in vivo. To determine whether the RS domain is required, and whether different RS domains can functionally substitute for antagonistic and specific XL6 AS targets, we engineered stably transfected S2 cells that can be induced by a metallothionein promoter to express one of three constructs: Flag-HA-tagged XL6 that is full length (XL6-FL), lacks the RS domain (XL6▵RS), or has the RS domain of B52 (XL6-B52RS) (Fig. 5A). To eliminate effects of endogenous XL6, we reduced its expression with dsRNA that specifically targets the 3′ UTR of the endogenous copy (XL6) (Fig. 5B, lanes 1–12), without affecting expression of XL6-FL (Fig. 5B, lanes 1–4).
FIGURE 5.
SR protein regulation of AS depends on RRM-domain and requires RS-domain. (A) Schematic showing relevant domains of three copper-inducible proteins—all tagged with 2x Flag and 2x HA: RRM, RNA-binding; Zn, zinc knuckle; RS, C-terminal arginine/serine. (B) Western blot of endogenous and recombinant XL6 in Drosophila S2 cells assayed in the presence or absence of dsRNA against endogenous XL6 and induction or recombinant XL6 isoforms; loading control, histone H3. (C) Semiquantitative RT-PCR of XL6-regulated targets (Dre4, Vps35, DppIII) and B52-regulated controls (CG12065 and Syb) in transfected S2 cells expressing wild-type or mutant XL6 (Expressed) and cultured in the presence or absence of dsRNA against endogenous XL6 (XL6 RNAi). Transcript models showing exon and intron structure (exon, thick box; intron, thin line; alternative region, red). PSI values are displayed below each lane. (D) Semiquantitative RT-PCR of AS targets in untransfected S2 cells cultured in the presence of dsRNA to either nonspecific sequence (control) or B52. PSI values are displayed below each lane.
We next performed RT-PCR on RNA from the transfected S2 cells—examining 2 AS events, Dre4 and Vps35, which are antagonistically regulated by XL6 and B52; XL6 activates the Dre4 cassette exon and represses Vps35 AS events; B52 has the opposite effects (Fig. 5C, cf. lanes 1,2 and Fig. 5D). Overexpression of XL6-FL enhanced activation and repression of Dre4 and Vps35 AS events, respectively, even when endogenous XL6 protein was reduced (Fig. 5C, cf. lanes 3,4 and 1,2). However, when XL6▵RS was overexpressed this enhancement was not seen and AS regulation was still sensitive to endogenous XL6 protein (Fig. 5C, cf. lanes 7,8 and lanes 5,6). Surprisingly, overexpression of XL6-B52RS yielded RT-PCR AS data that was indistinguishable from XL6-FL (Fig. 5C, cf. lanes 1–4 and 9–12), even though endogenous B52 affects Dre4 and Vps35 AS events antagonistically of XL6. Finally, the cassette exon in DppIII is regulated by XL6 but is insensitive to the B52 protein level (Fig. 5C, cf. lanes 1,2 and Fig. 5D). Overexpression of XL6-FL or XL6-B52RS, but not XL6▵RS, enhanced DppIII AS events. Demonstrating that the specificity for SR-regulated AS targets is determined by the RRM domain, since XL6 with a B52 RS domain can still regulate DppIII AS, which in insensitive to B52. Furthermore, overexpression of XL6-FL, XL6ΔRS, and XL6-B52RS have smaller effects on AS targets that are not sensitive to XL6 protein levels, but are regulated by B52 (Fig. 5C,D; CG12065 and Syb).
Thus, XL6 activation and repression of Dre4 and Vps35 AS events, respectively, requires (1) an RS domain, (2) that RS domains from at least some different SR proteins be functionally interchangeable, (3) that SR proteins are not redundant splicing regulators, but can independently contribute to regulation of shared and independent AS events, and (4) that SR protein specificity is dictated by the RRM domain, not the RS domain.
iCLIP seq reveals distribution of genome-wide SR protein–RNA binding
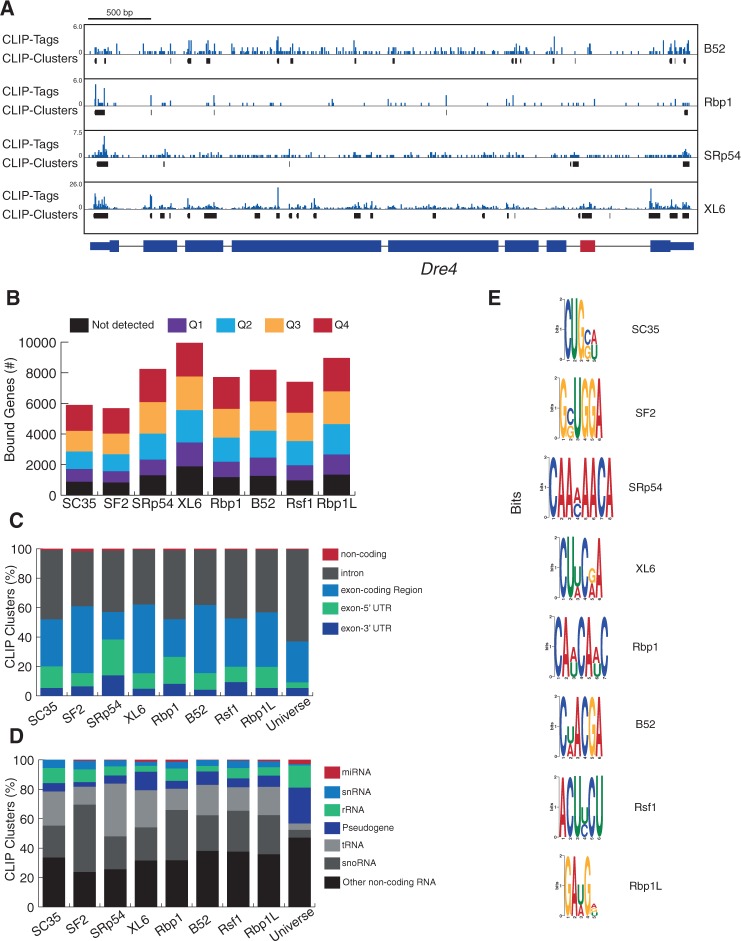
SR proteins have been shown to bind near splice sites in order to regulate AS. To determine if the observed combinatorial regulation of AS by SR proteins requires direct binding of SR proteins to a given transcript, we identified the precise genome-wide binding sites for all SR proteins, using iCLIP-seq. SR proteins were encoded on stable transgenes, transfected in S2 cells, FLAG-tag immunopurified, and the bound RNA purified and subjected to RNA-seq (Supplemental Fig. S4). The resulting reads (CLIP tags) were aligned to the Drosophila genome and generated 38,695–5,900,000 unique CLIP tags for each SR-protein replicate (Supplemental Table S4). From clusters of CLIP tags, we identified 16,004–102,040 significant CLIP clusters (i.e., SR-protein-binding sites) for each SR protein (Supplemental Table S4), using a cluster-finding algorithm; our conservative threshold (P ≤ 0.01) limited our analysis to the most significantly bound regions, but may have removed some true binding sites. Examination of the SR-protein CLIP clusters revealed SR proteins binding to multiple locations, and cross-linking by different SR proteins on the same transcript (Supplemental Fig. S5). For example, the SR-regulated transcript Dre4 displays CLIP clusters for four SR proteins (B52, Rbp1, SRp54, and XL6) (Fig. 6A); while some of these CLIP clusters overlap, many map to distinct regions. In addition, B52 and XL6 have many more clusters than Rbp1 and SRp54 (Fig. 6A). These data demonstrate that SR proteins bind overlapping as well as distinct sites on target RNAs, and do so with different specificities.
FIGURE 6.
Global landscape of RNA transcripts bound by SR proteins. (A) Dre4 gene (alternatively spliced exon is red) displaying density of CLIP tags (blue) and CLIP clusters (black) for four representative SR proteins across the transcript. (B) Number of genes that contain at least one SR protein CLIP cluster, grouped by gene expression level—ranging from “not detected” (below RNA-seq detection levels) to Q4, the highest level of expression. (C,D) Proportion of CLIP clusters that mapped to five different regions on SR-protein-bound transcripts (C) and common types of noncoding RNA for each SR protein (D); Universe, reference showing the proportion of genomic space for each feature. (E) Graphical representation of consensus-binding motifs derived for each SR protein. Height indicates the information content at each position of the binding motif in bits (log-odds in base 2).
Global landscape of RNA transcripts bound by SR proteins
To further study SR protein binding genome-wide, we profiled binding on all annotated transcripts for each SR protein. CLIP clusters were detected on 5679 (SF2) to 9952 (XL6) genes (Fig. 6B)—representing >90% of genes that are AS regulated by SR proteins (data not shown). To test whether iCLIP captured binding on only highly expressed genes, we compared bound genes with our S2-cell RNA-seq/expression data. SR protein CLIP clusters were identified in genes at all four quartiles of expression, in addition to genes expressed below RNA-seq detection levels (Fig. 6B).
Mapping analysis revealed that >98% of CLIP clusters bind protein-coding genes, and >53% bind exonic sequences (Fig. 6C); this represents a significant enrichment (P ≤ 0.001 χ2 test), given that 62.5% of the genic region is intronic, and is consistent with the fact that SR proteins bind ESE elements in pre-mRNA. Within exonic sequences, SR proteins mostly bind exons in the coding region, with a slight enrichment for the 5′-UTR region (Fig. 6C; P ≤ 0.001 χ2 test). SR protein CLIP clusters were also found in all major classes of noncoding RNAs (Fig. 6C,D); the most abundant were snoRNAs and tRNAs.
Consensus in vivo binding motifs for the SR proteins
SR protein-binding motifs identified using different experimental methods (mostly SELEX-based techniques) are degenerate purine-rich sequences that resemble identified ESEs or 5′ splice sites. However, most SR protein motifs are derived in vitro, using artificial constructs, and can differ in specificity depending on the experimental method used. We set out to identify the in vivo binding motifs for the eight Drosophila SR proteins using the CLIP clusters (Fig. 6A). To do this, we used the MEME-CHIP suite, which allows for identification of short binding motifs from large data sets, to identify enriched binding motifs from SR protein CLIP clusters. This will determine if individual SR proteins bind to specific or shared binding motifs.
XL6, Rbp1, and SRp54 bind CU-rich motifs that lack Gs (Fig. 6E), similar to the binding motif of their mammalian homolog, SRSF3 (Änko et al. 2012). Interestingly, Rbp1 has a different binding motif than Rbp1L, even though their RRM domains are 99% similar (differ by only two amino acids) (Fig. 6E). Rbp1L's motif includes Gs, but is very similar to the ESE motif (GAAGAA) that enhances splicing in vivo (Fairbrother et al. 2002).
Mammalian SF2/ASF (SRSF1) binds a purine-rich motif in vivo (Sanford et al. 2008), and Drosophila SF2 does as well (Fig. 6E). Binding motifs for the mammalian homologs of Drosophila SC35, B52, and Rsf1 have not been determined in vivo. However, SC35 and B52 bind a similar motif that was determined using SELEX (Shi et al. 1997; Cavaloc et al. 1999). The binding motif for Rsf1 is very similar to XL6's, and resembles an ESE (Fig. 6E).
We next examined iCLIP-identified SR-protein binding motifs in the subset of genes that exhibit differential AS when individual SR proteins are depleted. Most genes had at least one occurrence of the SR protein-binding motif (Supplemental Fig. S6, blue line). We compared the distribution of this fraction with the distribution of the fraction of genes that had an enriched motif score in the genes that are not AS regulated by the given SR protein, and found that as the motif score increased there was a smaller fraction of genes that contained the motif in the unregulated genes (Supplemental Fig. S6, red line). This indicates that there is a slight enrichment of identified SR-protein motifs in AS-regulated genes, compared with genes that are not AS regulated.
This analysis reveals that SR proteins bind to elements that resemble ESE motifs and are similar to previously identified binding motifs for their mammalian homologs. In addition, most SR proteins have a unique binding motif, but have a sequence motif that is closely related to other SR proteins.
Position-dependent regulation of AS by the SR proteins
In order to regulate pre-mRNA splicing, previous studies suggest that SR proteins bind to pre-mRNAs, primarily within exons. Indeed, we observe a higher percentage of CLIP clusters in exonic regions of annotated transcripts (Fig. 6C). To determine SR protein-binding positions in pre-mRNAs, looking for a possible exon bias (Fig. 6C), we mapped the relative density of SR protein CLIP tags within 300 nt of all 3′ and 5′ splice sites. SR protein CLIP tags showed a very strong preference for exonic regions, as expected—with the highest density of tags within 75 nt of 3′ and 5′ splice sites (Fig. 7A).
FIGURE 7.
SR proteins bind in a position-specific manner to regulate splicing. Mapping of SR protein CLIP-Tags near splice sites and alternatively spliced regions. (A) Relative density of CLIP-Tags 150 nt upstream of and downstream from the splicing acceptor (3′ splice site) and splicing donor (5′ splice site) for each SR protein iCLIP experiment on all bound transcripts. Wide gray bar, exon sequence; black line, intron sequence; colored line, individual SR protein binding. (B) RNA-splicing maps of four representative SR proteins on regulated cassette exon AS events. Bootstrapping analysis was used to test for robustness. Shaded regions correspond to the 95% confidence interval and solid lines are the estimated value of relative density of CLIP-Tags on cassette exons that have increased exclusion (red), increased inclusion (blue), or no change (dashed line) upon SR protein knockdown. The alternative exon is red and flanking constitutive regions are blue. n, total number of regulated AS events or unaffected AS events (middle).
To determine whether there is a relationship between SR protein binding pattern and AS regulation, we mapped the relative density of the iCLIP tags in a 400-nt region around the splice sites for the AS events that were activated, repressed, or unaffected and tested the robustness of the pattern using bootstrapping analysis (Fig. 7B). We focused our attention on the four SR proteins with the largest number of bound and regulated cassette exon events. Although the differences in binding patterns from the affected AS were not robustly different from the pattern of the unaffected events, this analysis suggests features of binding that inform SR protein function. Binding density was higher on activated cassette exons compared with repressed (Fig. 7B). When SR proteins repressed the alternative exon, there was a higher density of binding to introns and flanking constitutive exons compared with when the SR protein activated the inclusion of the alternative exon (Fig. 7B). Similar to our previous analysis of the binding density around 3′ and 5′ splice sites (Fig. 7A), SR protein binding was clustered near the splice sites. In addition to these common patterns, SR protein binding exhibited unique features. For example, while SRp54 binding on the alternative exon was diffuse—B52, XL6, and SF2 exhibited more focal binding patterns (Fig. 7B). When we examined SR protein binding patterns on competing donor AS events, our findings were similar (Supplemental Fig. S7). Specifically, SRp54, XL6, and SF2 binding densities were higher near the distal 5′ss when the SR proteins activate its usage, and higher near the proximal 5′ss when the three SR proteins activate its usage (Supplemental Fig. S7). Density of CLIP tags for AS events were mapped individually to demonstrate that the observed binding pattern was not due to the over-representation of one or few AS events (Supplemental Fig. S8). Thus, SR proteins bound to specific locations to activate or repress the inclusion of alternatively spliced regions.
DISCUSSION
In this study, we examined the cooperative nature of SR protein splicing regulation, by profiling the endogenously affected AS targets for all eight SR protein family members in Drosophila. We found that this SR protein family affects 561 endogenous simple AS events—the largest number of AS-regulated events profiled for SR proteins. Although our analysis cannot distinguish if SR proteins directly or indirectly regulate each target, it demonstrates that SR proteins are key regulators of a large AS regulatory network. We show that single SR proteins can regulate specific AS events by acting as both enhancer and repressor. Moreover, this analysis identified clear examples of AS events antagonistically regulated by multiple SR proteins. This result confirms and extends previous findings showing that two human SR proteins, SRSF1 and SRSF2, both activate and repress exon inclusion (Pandit et al. 2013); it also challenges the model that exon inclusion is solely dependent on a balance between antagonizing SR and hnRNP proteins.
The observation that multiple SR proteins could substitute for one another in the splicing of in vitro substrates had previously been interpreted as a redundant function role of SR proteins on regulating alternative splicing (Fu et al. 1992). However, the fact that SR-protein knockouts are embryonic lethal suggests that specific SR proteins are required for regulating AS of specific pre-mRNAs. In this study, we show that while some AS events are regulated by a single SR protein, most are coregulated by more than one. This suggests that, for the most part, SR proteins function in a combinatorial fashion to regulate AS events. Coregulation of AS events has been observed previously in Drosophila and mammals (Lynch and Maniatis 1996; Blanchette et al. 2005; Huelga et al. 2012; Pandit et al. 2013)—for example, by SR and hnRNP proteins (Blanchette et al. 2009; Huelga et al. 2012; Pandit et al. 2013). However, two recent RNA-mapping studies of mammalian SR proteins showed that while SRSF3 and SRSF4 exhibit unique binding patterns (i.e., share few, if any, RNA targets) (Änko et al. 2012), SRSF1 and SRSF2 show extensive overlap in their RNA targets (Pandit et al. 2013). This raises the possibility that SR proteins may preferentially share RNA targets.
In our analysis, we found that 87.7% of all coregulated AS events are regulated in the same direction, but there were also examples of antagonistic regulation. By reducing expression of two SR proteins simultaneously, we demonstrate that AS outcome of coregulated targets (both cooperative and antagonistic) is dictated by the contribution of the individual SR protein regulation (Fig. 2). Thus, even though compensatory binding of SR proteins to regulatory regions has been noted in mammals (Pandit et al. 2013), our data suggest that a large fraction of SR-regulated AS events are additive and noncompensatory. In future studies, it will be interesting to examine the extent of combinatorial regulation between splicing factors, and the role that this combinatorial regulation plays in tissue or developmentally regulated AS events.
It has been suggested that SR protein recognition of 5′ or 3′ splice sites involves SR protein binding of spliceosomal components (e.g., U1 snRNP and U2AF)—most likely via the RS domains (Wu and Maniatis 1993). However, splicing of several pre-mRNAs in vitro does not require an RS domain, and inclusion directly correlates with the strength of the 3′ splice site (Zhu and Krainer 2000). We demonstrate that AS regulation by the SR protein XL6 requires an RS domain, and the specificity and directionality of AS modulation is dependent on the RRM domain (i.e., RNA-binding location). We show that RS domains from different SR proteins can functionally substitute for one another in vivo—in spite of the fact that they differ in sequence composition, length and in their potency when tethered to a similar ESE (Graveley et al. 1998). The RS domain from B52 could functionally substitute for the XL6 RS domain, in vivo. This adds to the notion that RS domains are functionally interchangeable, on endogenous AS targets.
We mapped SR protein binding with high precision, and found that SR proteins bind to most transcripts, like mammalian SRSF1 and SRSF2 (Pandit et al. 2013). This is consistent with other ubiquitously expressed splicing regulators, such as the hnRNP family (Huelga et al. 2012), and is likely due to the SR protein family's broad role in RNA metabolism. Despite binding to many of the same transcripts, SR protein binding sites were distinct and a single SR protein often bound multiple sites on a single transcript. Indeed, most SR proteins exhibited a specific RNA-binding motif. The identified, in vivo, RNA-binding motifs closely resemble motifs previously identified in vitro, and mammalian SR binding motifs identified in vitro and in vivo. This suggests a deep evolutionary conservation of SR RNA-binding motifs, an observation that was recently extended to many RNA-binding proteins (Ray et al. 2013). In addition, SR proteins exhibited position-dependent regulation of AS, and were primarily associated with exonic sequences near the splice sites that they regulated. These RNA-binding characteristics are similar to those of mammalian SR proteins. These observations are also consistent with the notion that SR proteins activate splicing by recruiting nearby spliceosomal components (Wu and Maniatis 1993).
Initially identified as splicing regulators, SR proteins bind a diverse set of RNAs, including ncRNAs. Previous studies have described SR protein binding to long ncRNAs for transport and stability, and regulation of miRNA biogenesis (Tripathi et al. 2010; Wu et al. 2010). SR proteins were previously shown to associate with snoRNAS, but have never been implicated in the biogenesis of this class of ncRNA. These data suggest a novel role for SR proteins in regulation of ncRNA function, which could ultimately affect gene regulation.
SR proteins interact with components of the transcriptional machinery, including Pol II (Misteli and Spector 1999). SRSF2, for example, is recruited to the promoter region of genes through interaction with the 7SK complex, and is important for transcriptional activation (Ji et al. 2013). We show that all eight SR proteins regulate promoter selection for a number of targets. GO analysis of AS regulated targets revealed genes involved in cell signaling and development. Indeed, SR proteins regulate AS of key transcription factors (Su(z)12,Iswif,Hsf,Skd for examples). Thus, by altering the splicing of a transcriptional or signal transduction pathway regulator, an SR protein could contribute to widespread changes in downstream gene expression. Alternatively, SR proteins may regulate gene expression of targets by directly affecting their transcription and/or stability. By any or all of these mechanisms, SR proteins could precisely and rapidly regulate an organism's gene expression profile.
In addition to regulating promoter selection, we found that SR proteins regulate poly(A) site selection. Interaction between splicing regulators and the polyadenylation machinery has been demonstrated previously, but never for SR proteins (Lou et al. 1998; Licatalosi et al. 2008; Di Giammartino et al. 2011; Berg et al. 2012). We also noted differences in proximal versus distal poly(A) site regulation by SR proteins, which shorten or lengthen the transcript's 3′ UTR, respectively. This could result in changes in protein expression, which could be regulated cell- or tissue-specifically.
Understanding how SR proteins regulate AS is crucial to understanding the role AS plays in tissue development and homeostasis. Much progress has been made studying SR proteins in vitro or their regulation on a small number of genes. Recent studies have highlighted the cooperative nature of SR protein regulation. Our findings have confirmed and extended the combinatorial regulation of AS to the entire family of SR proteins, on a large number of endogenous targets. We show that this regulation requires an RS domain, and the specificity relies on the RNA-binding location. Furthermore, we demonstrated that SR proteins can affect promoter and poly(A) site choice, transcript expression, and can bind to diverse classes of RNAs. These findings highlight the complex and dynamic nature of SR protein transcript regulation. Alternative isoform selection is a product of coordinated promoter selection, AS regulation, and poly(A) site selection—all regulated by SR proteins and other transcriptional and RNA-processing machineries. Understanding SR protein regulation beyond AS will allow greater understanding of the integration of gene expression regulation between multiple regulatory networks.
MATERIALS AND METHODS
Cell culture
Drosophila S2 cells were maintained at 25°C in Drosophila Schneider's media (GIBCO # 21720) plus 10% fetal bovine serum (PAA #A15-701).
SR protein RNAi and mRNA-seq
RNAi knockdowns were performed, in duplicate, by seeding 2 × 106 cells per well in a six-well plate and adding 10-µg nonoverlapping dsRNA. After 48 h of incubation, a second round of 10-µg dsRNA was added, and incubated for an additional 48 h. The cells were harvested and the RNA purified using the RNeasy kit following the manufacturer's protocol (Qiagen #74106). For experiments where two SR proteins were knocked down simultaneously, 10 µg of targeting dsRNA for each SR protein was added. Specificity and efficiency of knockdown were monitored by Western blot using antibodies specifically generated to each SR protein.
We performed RNA-seq experiments on two replicate samples from each individual SR protein knockdown, two replicates of simultaneous SR protein knockdown (XL6:B52 & SC35:B52), as well as from control S2 cells. Library preparation was performed according to the mRNA sequencing preparation kit (Illumina). The samples were loaded onto a flow-cell for cluster generation, and sequenced using an Illumina Genome Analyzer II or Illumina HiSeq 2000 using single-read protocols (Illumina).
Alternative splicing event analysis (ASEA) and analysis of transcript abundance
Bowtie (Langmead et al. 2009) was used to perform alignment of sequence reads, from SR-depleted and control samples, to the Drosophila melanogaster (v5.22) genome, and a database of splice junctions (36 nt on each side of the splice junction) allowing for up to two mismatches and single alignment. Next, for each annotated alternative splicing event (ASE), the number of reads were tabulated that support the inclusive, exclusive, or both (constitutive) forms. Two Fisher tests are used to compare the tabulated read counts occurring in the SR-depleted and control results for each ASE. One test for significant changes in inclusive counts and another for changes in exclusive counts. To determine the magnitude and direction of splicing change PSI (percent spliced in) values are calculated. This is the length-normalized read count for the inclusive region over the sum of the length-normalized read count of the inclusive and constitutive region [(inclusive/inclusive length)/[(inclusive/inclusive length) + (constitutive/constitutive length)]]. Inclusive Simple ASEs (cassette exon, alt. acceptor, alt. donor, retained-intron, and mutually exclusive exon) that were changed significantly (P ≤ 0.05), and changed by at least 10% (PSI-switch greater than or equal to ±0.1) were included for downstream analysis. In addition, the same filter was used for ASEs that differed in the 5′ and 3′ ends.
To identify changes in overall transcript abundance, the mRNA RPKM (reads per kilobase of million mapped reads) was computed for each gene to measure relative expression. To detect differentially expressed genes in SR-depleted samples, the bioconductor edgeR package was used. We limited our analysis to significantly changed genes that varied twofold or more.
RT-PCR validation of affected targets
cDNA was generated by reverse transcribing ∼5 µg of the total RNA extracted from the control and SR-depleted cells using a mixture of oligo(dT) and random hexamer (N6) primers, and superscript III reverse transcriptase (Invitrogen) according to the manufacturer's protocol. PCR amplification of predicted targets was performed for 25–30 cycles with primers falling in exons flanking the alternative spliced region. PCR products were analyzed on an Agilent Technologies 2100 Bioanalyzer, using a DNA chip (Agilent DNA-1000 kit) according to the manufacturer's instructions.
Construction of 2x Flag-HA SR WT and mutant stable cell lines
Full-length SR proteins, XL6▵RS, and XL6-B52RS were PCR amplified and cloned into pDONR221 by BP reaction (Gateway cloning; Invitrogen). XL6-B52RS was amplified using a two-step PCR. First, the RRM-domain of XL6 and RS-domain of B52 were amplified, separately. Next, the purified PCR products were mixed together and amplified with outside primers to generate the fusion. Next, the constructs were inserted into a destination vector that contained the metallothionein promoter and a C-terminal 2x Flag and 2x HA tag (pMT-C2FL2HA) by LR reaction (Gateway cloning; Invitrogen).
Four hundred nanograms of pMT-C2FL2HA plasmid DNA containing full-length SR proteins, XL6▵RS, or XL6-B52RS and 40 ng of a plasmid carrying the gene for blasticidin resistance (pCO-BLAST) were transfected into 2.0 × 106 Drosophila S2 cells, seeded in a six-well dish, using Effectene transfection reagent (Qiagen) according to the manufacturer's protocol. After 48 h, 25 µg/mL of blasticidin (Invivogen) was added to the cells. Resistant cells were expanded and maintained in Schneider media with 10% FBS and 25 µg/mL blasticidin.
iCLIP-Seq of individual SR proteins
All iCLIP experiments were performed as previously described (Konig et al. 2011). Of note, 6.0 × 106 SR protein stably transfected S2 cells were seeded in a 10-cm dish, and protein expression induced by adding 100 µM CuSO4 for 48 h. Immunoprecipitation was performed using anti-FLAG M2 magnetic beads (Sigma, M882). High-throughput sequencing of iCLIP cDNA libraries from two replicate experiments for each SR protein were sequenced multiple times on an Illumina HiSeq 2000 with a 50-nt run length.
Identification of iCLIP-Seq clusters
The sequence analysis proceeded on the premise that each sequence read is comprised of a 9-nt segment of barcode, with the remaining sequence coming from a putative crosslink site and the sequence leading up to it with, potentially, extra sequences from the downstream PCR adapter. The nine bases of barcode consist of a 4-nt segment of known/designed experimental barcode (eb) at position 4 to 7 inserted in the middle of 3 and 2 nt of random sequences constituting the molecular barcode (mb). The eb allows for multiplexing experiments within a single lane on the sequencer. The mb provides for identifying PCR duplicates as described below.
The 51-bp fastq formatted sequence reads were first reformatted into fasta format (using: fastq_to_fasta -Q33 -n). Their read names assigned by Illumina CASAVA pipeline were replaced with sequence numbers (using: fastx_renamer -n COUNT). They were filtered to eliminate those lacking the expected sequencing vector, which was clipped from the unfiltered reads (using: fastx_clipper -n -a “TGAGATCGGAAGAGCGGTTCAGC” -M 2 -l 24 -v; hannonlab.cshl.edu/fastx_toolkit/).
The sequence of these reformatted, renamed, and adapter-clipped reads retained both the experimental and molecular (random) barcode (eb and mb).
Each set of perfectly identical sequences was then collapsed into a single (unique) representative sequence, and the sequence name was modified to hold a “cm” tag (collapsed multiplicity), being the count of collapsed sequences (using: fastx_collapser).
Each sequence was further tagged with its “eb,” being nucleotides 4 to 7, after adapter trimming, and its “mb,” being the bases at position 1 to 3 and 8-9 (using: custom Perl scripts).
By this point, each trimmed and collapsed sequence has been assigned three tags: cm, eb, and mb. This modified fastq was then aligned in a splice-aware fashion to the fly genome, allowing up to two mismatches. The splice-awareness was implemented by a two stage process: First reads were aligned to the Drosophila melanogaster genome (v5.22) with bowtie version 1.0.0; reads that failed to align in this way were aligned to an artificial “splice-junction” database and the coordinates of successful junction alignments were projected back into genomic coordinates (using: custom Perl scripts). Both stages allowed up to two mismatches, and used bowtie's “-trim5 9” option to cause the leading 9 bp (harboring the combined eb and mb) to be skipped during the alignment. The results of both stages were concatenated into a single SAM file, which was stream-edited (using: custom Perl scripts) to transfer the three tags (cm, eb, mb) out of the QNAME column and append them as new TAGs on the SAM file, allowing them to be used in downstream analysis.
The SAM formatted alignments were analyzed to identify coalignment sets (alignments to the same chromosome at the same locus). Within each coalignment set, sequencing errors in the eb and mb barcode segments were identified and corrected using a greedy approach in which less frequent values were replaced with the most frequent value within a hamming radius of 1. The error-corrected eb and mb values were assigned to new tags RG (read group) and mg (molecular group), respectively.
The error-corrected coalignment sets are demultiplexed on the RG, assigning reads to experiments. Within this, coalignment sets are further collapsed on the mg, thereby avoiding double counting PCR duplicates, and thus providing a read-out of the relative count of coaligned putatively unique (cDNA) molecules, which are finally aggregated based on commonality of the first position of the alignment, being the putative cross-link site, producing xlSites records, each of which is tagged with the count of distinct mg from which it was collapsed.
The significance of putative cross-link sites was evaluated by contrasting the distribution of their local density with what might be expected were cross-linking distributed by a Poisson process (Polymenidou et al. 2011); all sites whose local density was sufficiently unlikely to occur randomly were deemed “significant” and used in downstream analyses (i.e., the construction of RNA-binding maps). This analysis was conducted separately on the intronic and exonic regions within each gene, since their affinity is known to be different between these regions. The local density of all observed putative sites was determined in a “window” of 31 bases centered on each single nucleotide site (15 bp on either side). An empirical cumulative density function (eCDF) was constructed from these observed local density values. Then, for each region (either exonic or intronic), a Poisson distribution was calculated with its λ parameter taken as the number of putative sites in the entire region divided by the ratio of the window width (31) to the overall number of bases in the region. Finally, the lowest value of the observed eCDF whose probability of occurring was less than a threshold of 0.01 according to the region's Poisson distribution was taken as a threshold: All sites whose local density was at least this threshold were deemed “significant.”
Identified CLIP Clusters from each replicate experiment were merged into experiment CLIP Clusters. CLIP Cluster genomic annotations were assigned based on gene annotations give in FlyBase (v5.22).
Motif analysis
De novo motif finding was implemented using 1000 random SR protein CLIP-Clusters and the MEME-CHIP suite that includes DREME (Bailey 2011). The background was the 1000 input sequences shuffled preserving nucleotide composition.
MotifBS (http://compbio.berkeley.edu/people/ed/motifBS.html) was used to search the SR protein regulated AS genes with the identified motif, and return scores for the presence of the motif in the gene as compared with unaffected genes. The cumulative distributions of scores for the regulated and background genes were plotted.
RNA-splicing maps
iCLIP CLIP-Tags were mapped to the affected cassette exons AS events detected by RNA-seq at each nucleotide position in four regions (400 nt centered at each splice site). The relative density was calculated at each nucleotide in the region, by dividing the number of CLIP-Tags at a given nucleotide position by the total number of CLIP-Tags in all regions. Reliability was tested by performing bootstrapping analysis of the basis of 10,000 replicates to obtain the estimate value and 95% confidence interval. For each SR protein, cassette exons were separated into three groups, repressed, activated, and unchanged. Using R, we plotted at each nucleotide position the relative density of CLIP-Tags of activated, repressed, and unchanged cassette exons. The same was performed on competing acceptor (5′ss) splicing events.
DATA DEPOSITION
RNA-seq and iCLIP-Seq data files are available at the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63261) under accession number # GSE63261.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Baumann for support and critical comments throughout the development of this story and Donald Rio for critically reading the manuscript and helpful comments. We are also grateful for the technical expertise and help provided by Jerne Ule and Jeremy Sanford. This work has been supported by the Stowers Institute for Medical Research.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.043893.113.
Freely available online through the RNA Open Access option.
REFERENCES
- Änkö ML, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM 2012. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. 2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Green RE, Brenner SE, Rio DC 2005. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev 19: 1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Green RE, MacArthur S, Brooks AN, Brenner SE, Eisen MB, Rio DC 2009. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Mol Cell 33: 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechingberg J, Lykke-Andersen S, Jensen TH, Jorgensen AL, Nielsen AL 2007. Regulatory mechanisms for 3′-end alternative splicing and polyadenylation of the Glial Fibrillary Acidic Protein, GFAP, transcript. Nucleic Acids Res 35: 7636–7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Yang L, Duff MO, Hansen KD, Park JW, Dudoit S, Brenner SE, Graveley BR 2011. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res 21: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30: 377–384. [DOI] [PubMed] [Google Scholar]
- Cavaloc Y, Bourgeois CF, Kister L, Stévenin J 1999. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5: 468–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champlin DT, Frasch M, Saumweber H, Lis JT 1991. Characterization of a Drosophila protein associated with boundaries of transcriptionally active chromatin. Genes Dev 5: 1611–1621. [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Fu XD, Mayeda A, Maniatis T, Krainer AR 1992. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci 89: 11224–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Dejardin J, Tazi J, Soret J 2007. The SR family proteins B52 and dASF/SF2 modulate development of the Drosophila visual system by regulating specific RNA targets. Mol Cell Biol 27: 3087–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Hertel KJ, Maniatis T 1998. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J 17: 6747–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BE, Lis JT 2000. Pre-mRNA splicing by the essential Drosophila protein B52: tissue and target specificity. Mol Cell Biol 20: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House AE, Lynch KW 2006. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat Struct Mol Biol 13: 937–944. [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD 2013. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153: 855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol 14: 185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T, Manley JL 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 34: 460–463. [DOI] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J 2011. iCLIP—transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J Vis Exp 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D 1990. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell 62: 35–42. [DOI] [PubMed] [Google Scholar]
- Kramer A 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem 65: 367–409. [DOI] [PubMed] [Google Scholar]
- Kumar S, Lopez AJ 2005. Negative feedback regulation among SR splicing factors encoded by Rbp1 and Rbp1-like in Drosophila. EMBO J 24: 2646–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W 2006. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 23: 195–205. [DOI] [PubMed] [Google Scholar]
- Labourier E, Bourbon HM, Gallouzi IE, Fostier M, Allemand E, Tazi J 1999. Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev 13: 740–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam BJ, Hertel KJ 2002. A general role for splicing enhancers in exon definition. RNA 8: 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 15: 819–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417: 15–27. [DOI] [PubMed] [Google Scholar]
- Lou H, Neugebauer KM, Gagel RF, Berget SM 1998. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol 18: 4977–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW, Maniatis T 1996. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev 10: 2089–2101. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW 2005. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6: 386–398. [DOI] [PubMed] [Google Scholar]
- Misquitta-Ali CM, Cheng E, O'Hanlon D, Liu N, McGlade CJ, Tsao MS, Blencowe BJ 2011. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol Cell Biol 31: 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell 3: 697–705. [DOI] [PubMed] [Google Scholar]
- Pagani F, Baralle FE 2004. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet 5: 389–396. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M Jr, Fu XD 2013. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell 50: 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. 2011. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14: 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, Caceres JF 2008. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One 3: e3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ 2009. The SR protein family. Genome Biol 10: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hoffman BE, Lis JT 1997. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol Cell Biol 17: 2649–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP 2004. Aberrant and alternative splicing in cancer. Cancer Res 64: 7647–7654. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75: 1061–1070. [DOI] [PubMed] [Google Scholar]
- Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J 2010. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell 38: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Damgaard CK, Kjems J, Caputi M 2004. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem 279: 10077–10084. [DOI] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 35: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Krainer AR 2000. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev 14: 3166–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mayeda A, Krainer AR 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell 8: 1351–1361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.