Abstract
This unit describes the experimental procedures for the steady-state kinetic analysis of DNA synthesis across DNA nucleotides (native or modified) by DNA polymerases. In vitro primer extension experiments with a single nucleoside triphosphate species followed by denaturing polyacrylamide gel electrophoresis of the extended products is described. Data analysis procedures and fitting to steady-state kinetic models is presented to highlight the kinetic differences involved in the bypass of damaged versus undamaged DNA. Moreover, explanations concerning problems encountered in these experiments are addressed. This approach provides useful quantitative parameters for the processing of damaged DNA by DNA polymerases.
Keywords: DNA polymerase, translesion synthesis, steady-state kinetics
INTRODUCTION
The protocol describes the steady-state primer incorporation of a single nucleoside triphosphate across, or past, a DNA lesion by DNA polymerases, specifically translesion DNA polymerases. As a model system the damaged DNA contained an O4-methylthymidine (O4MeT Figure 1) insert and human DNA polymerase η (hpol η) was the Y-Family translesion polymerase of choice. The complete procedure for the single nucleotide primer extension assay across the DNA lesion, followed by primer extension product and substrate resolution by polyacrylamide gel electrophoresis (PAGE) are described here. Lastly, data fitting to the Michaelis-Menten kinetic model is presented to enable a quantitative comparison of kinetic parameters for the damaged and wild-type DNA.
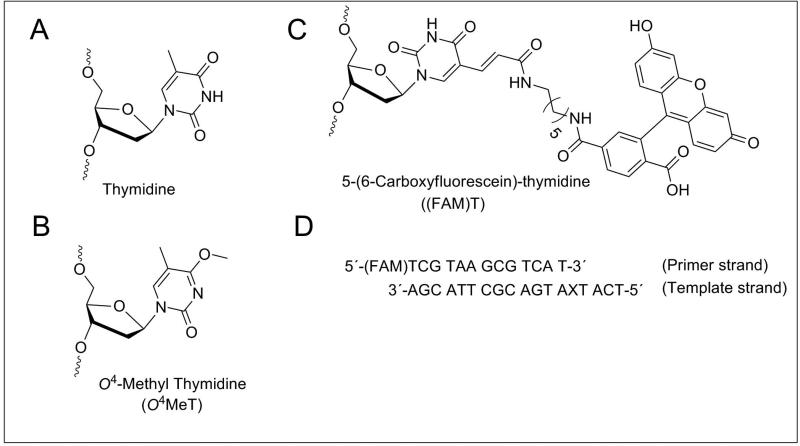
Figure 1.
Chemical structures of (A) thymidine (T), (B) O4-methylthymidine (O4MeT) inserts, (C) 5’-(6-carboxyfluorescein)-thymidine ((FAM)T) and (D) sequences of DNA template and primer indicating positions of the modification (X) and (FAM)T.
BASIC PROTOCOL 1
SINGLE NUCLEOTIDE INCORPORATION REACTION OF 2’-DEOXYNUCLEOSIDE TRIPHOSPHATES OPPOSITE A TEMPLATE CONTAINING AN O4-METHYLTHYMIDINE INSERT
Strategic Planning
Lesion-containing DNA template
Modified oligomers template can be acquired via commercial sources or can be synthesized using automated solid phase synthesis employing a phosphoramidite strategy. The lability of the particular modification is paramount and should be taken into consideration when planning these experiments.
Design of the DNA primer
The primer should have complementarity with the template sequence. Unlike extension product analysis by liquid-chromatography mass spectrometry (LCMS, UNIT 7.16; (Chowdhury and Guengerich, 2011)), no 2’-deoxyuridine inserts are required. Monitoring of the DNA extension products can normally be accomplished one of two ways, by 32P-phosphorimaging or by fluorescence (Guengerich, 2006; Patra et al., 2014). In the former case, an unmodified DNA primer can simply be radiolabeled with [γ-32P]-ATP and T4 polynucleotide kinase. Hazard precautions must be taken when working with radioactive materials, often necessitating safety training for personnel and designated radiation-approved laboratory space. Alternatively, the primer can be functionalized with a fluorescent tag to circumvent the latter hurdles associated with using radioactivity as a detection method. Limits of detection of fluorescently-labeled primers remain well-above the requirements for the experiments described in this unit. Engineering a primer with a fluorescent tag near the 5’-end of the primer does not interfere with the fidelity or kinetics of the polymerases, despite the relatively large sizes of some fluorescent moieties such as the 5’ 6-carboxyfluorescein-label (FAM-label, used herein). Akin to the DNA templates, DNA primers may be purchased commercially or prepared via automated solid phase synthesis employing a phosphoramidite strategy. The primer and template sequences were as follows, where (FAM)T and X indicate the 5’-6-carboxyfluorescein-thymidine and O4MeT inserts, respectively: 5’-(FAM)TCG TAA GCG TCA T-3’ and 3’-AGC ATT CGC AGT AXT ACT-5’.
The resulting DNA duplex can be of variable size, provided that the duplex stability remains above physiologically relevant temperature (Tm generally greater than 40 °C). This can be verified by UV thermal denaturation experiments prior to the enzymatic assays. However, the use of very long primers can limit the resolution of the substrate and product. Generally, primers should not exceed approximately 25-30 nucleotides in length and should have a minimum of approximately 10 base pairs.
DNA polymerases
DNA polymerase preparation and purification can be accomplished using standard protocols. In this case, the catalytic core construct of human DNA polymerase η (hpol η), which includes amino acids 1–432, was prepared according to reported procedures (Biertümpfel et al., 2010). Certain DNA polymerases require auxiliary factors for proper processivity of DNA extension. One example is mammalian DNA polymerase δ, which requires the proliferating cell nuclear antigen (PCNA) accessory protein, even with short primer-template complexes (Prelich and Stillman, 1988; Waga and Stillman, 1994). Experiments should be planned accordingly if auxiliary proteins are required.
Primer extension assay
Generally, the primer extension assay conditions necessitate optimization given bypass by translesion DNA polymerases varies greatly depending on the nature of the DNA modification, incoming dNTP, and/or DNA polymerase. Previous literature on a particular modification may serve as a basis for initial conditions. The initial screen entails investigating a broad range of dNTP concentrations, DNA polymerase concentration and reaction time in order to divulge a set of conditions to remain in the confines of the steady-state kinetic model (e.g. remaining below 20% of product formation). All analyses are monitored by PAGE and visualized/quantified by fluorescence (or 32P-phosphorimaging). In our hands, screening three values for each parameter normally supplied sufficient information to establish an appropriate set of conditions for the primer extension assay.
Materials
40% Acrylamide / bis-acrylamide solution (19:1, w/w (5% crosslinker), electrophoresis purity reagent) (Bio-Rad Laboratories)
Ammonium persulfate (Bio-Rad Laboratories)
Aqueous ethanol (70% v/v, reagent grade)
Bovine serum albumin (BSA), standard solution (2 mg/mL) (Pierce Protein Biology Products) Bromophenol blue (SigmaAldrich)
2’-Deoxyoribonucleoside triphosphate solutions (dNTP, 100 mM; New England Biolabs)
D,L-Dithiothreitol (DTT) (Research Products International)
DNA template (see Strategic Planning)
Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA, SigmaAldrich)
Formamide (Roche)
Glycerol (SigmaAldrich)
Human DNA polymerase η (hpol η; see Strategic Planning)
Magnesium chloride solution (25 mM) (Applied Biosystems)
N,N,Ń,Ń-Tetramethylethylenediamine (TEMED; Bio-Rad Laboratories)
Potassium chloride (SigmaAldrich)
Primer (see Strategic Planning)
Siliconizing reagent for glass (e.g. Sigmacote®; SigmaAldrich)
Tris-HCl buffer (1 M), pH 7.5 (Trizma® hydrochloride buffer solution, SigmaAldrich)
10 X Tris-Borate-EDTA (TBE) buffer (0.89 M Tris·Cl, 0.89 M boric acid, 20 mM EDTA) (individual solid components, SigmaAldrich)
Urea (electrophoresis grade, SigmaAldrich)
Xylene cyanol FF (SigmaAldrich)
Centrifuge
Centrifuge tube holder (UNITED Laboratory Plastics)
Cling plastic wrap
Digital timer (model # 62344-641, VWR International)
Disposable needle (18 G)
Disposable syringe (25 mL)
Dry block heater (e.g. VWR International, model # 946310) fitted with a modular heating block (e.g. VWR International, 30 wells to hold 0.65 mL centrifuge tubes) and thermometer (e.g. VWR International).
Erlenmeyer flask (125 mL)
Flat end sequencing tips (0.37 mm, 1-200 μL; Phenix Research Products)
Graduated cylinder (5 L)
GraphPad Prism® software (GraphPad Software)
Image J software (National Institutes of Health)
Lint-free tissue (e.g. Kimwipes, Kimberly-Clark)
Magnetic stir plate and stir bar
Microcentrifuge tubes (0.65 mL, VWR International)
Microsoft ExcelTM (Microsoft Corp.)
Nucleic acid Electrophoresis standard vertical apparatus (e.g. Sequi-Gen® GT DNA electrophoresis cell, 38 × 50 cm (Bio-Rad Laboratories))
Single edge razor blade or scalpel
Standard set of micropipettors (1000 μL, 200 μL, 20 μL, 10 μL, and 2 μL) with disposable tips Typhoon Trio Variable Mode Imager (GE Healthcare Life Sciences)
Vortex mixing device
Anneal the primer-template complex
If the single strand DNA stocks are in powder form, resuspend the individual oligomers in 18 mΩ water to reach a concentration of 1 mM.
Mix equimolar amounts of the DNA template containing the adduct and the primer containing (FAM)T; dilute with 18 mΩ water to reach a final concentration of 200 μM DNA duplex. Typically, a 50 - 75 μL DNA duplex stock is sufficient for the steady-state kinetics involving the four natural dNTPs.
Heat the DNA solution at 90 °C for 5 min in a dry heating block and then allow to slowly cool to room temperature for proper hybridization of the DNA duplex. The annealed DNA duplex stock should normally be stored at 4 °C. It should be noted that other storage conditions may have to be used depending on the stability of modification in the DNA template strand.
Single-nucleotide extension of the primer by DNA polymerase
Stock aqueous solutions of DTT (100 mM), glycerol (50% v/v), Tris (pH 7.5, 500 mM), KCl (2.5 M) and quench solution (95% v/v formamide, 20 mM EDTA pH 8.0, 0.01% (w/v) bromophenol blue, 0.01% (w/v) xylene cyanol FF) should be prepared prior to beginning the experiments. Aliquoting these solutions into small volumes (300-500 μL) is recommended, with storage according to the supplier's specifications. Note that stored solutions should be mixed prior to use. Each single-nucleotide extension experiment should be performed in duplicate.
Set dry block heater to 37 °C and allow to stabilize for about 10 min.
Preheat empty microcentrifuge in dry block heater for 5 min.
- Prepare dNTP stocks with varying concentrations. Label appropriately.Initial screen dNTP concentrations (5X) can be 50 μM, 500 μM, and 5000 μM. dNTP stocks of 50 μL are normally sufficient.
Place labeled centrifuge tubes in a centrifuge tube holder containing quenching solution (9 μL)
- Prepare premixes according to Table 1 at 0 °C (in an ice bucket). DNA polymerases are normally added last and should be thawed right before their addition to the premix. Label appropriately and ensure proper mixing by pipetting the content up and down a few times (do not mix solutions containing proteins vigorously using a vortex device).Nucleoside triphosphate incorporation reactions are initiated by the addition of a given dNTP (added last). Note that the concentrations of reagents are corrected for dilution in the final reactions from the addition of dNTP aqueous solutions. For each DNA template, protein polymerase, and protein concentration tested, a new premix must be prepared. For instance, in the case of the initial screen where one DNA template-primer, three protein concentrations, three dNTP concentration, and three time points are tested, a total of three premixes are prepared.
- Using the timer, add aliquots (8 μL) of premix in corresponding preheated tubes at 15 s intervals. Allow to equilibrate at 37 °C for five minutes.If the timer is turned on at t = 0 s, then the first addition will be at t = 15 s, the second at t = 30 s, etc. For the initial screen, nine preheated tubes will be required (three different protein concentrations, and three different dNTP concentrations). Note that using a time interval of 5 minutes limits the experiment to 20 reaction vessels (four additions per minute for 5 minutes).
Add dNTP aliquots (2 μL) at the 15 s intervals after equilibration step to initiate the DNA extension reaction. Ensure proper mixing by pipetting up and down a few times during the additions.
- At the desired time points, remove 1 μL from each reaction mixture and place in pre-prepared tubes containing quench buffer (9 μL). Ensure proper mixing by pipetting up and down a few times.Reaction volumes are 10 μL, allowing for multiple reaction time sampling. Make sure to note if a time point is taken at a later or an earlier time than planned. These values can be factored in the data analysis section.
Repeat step 8 for other time points.
Quenched reaction samples may be stored at −20 °C for subsequent analysis
Table 1.
Stock concentrations, final reaction concentrations and volumes to add for the preparation of individual premixes sufficient for 10 reaction points. Concentrations are corrected for the dilution with the final addition of dNTP
| Component | [Stock] | [Reaction] | Volume (μL) to add |
|---|---|---|---|
| DTT (mM) | 100 | 10 | 11 |
| BSA (mg/mL) | 2 | 0.1 | 5.5 |
| Glycerol (%) | 50 | 5 | 11 |
| MgCl2 (mM) | 25 | 5 | 22 |
| KCl (mM) | 2500 | 100 | 4.4 |
| Tris-Cl, pH 7.5 (mM) | 500 | 40 | 8.8 |
| DNA duplex (μM) | 200 | 5 | 2.75 |
| DNA polymerase (nM) | 20 | 2.5 | 13.75 |
| H2O | — | — | 8.8 |
| Total (μL) | 88 | ||
Analyze DNA extension products by separation using denaturing PAGE and detection
Bring DNA extension samples to room temperature if necessary.
- Prepare the 18% (v/v) 7.5 M urea denaturing gel solution by combining and mixing the urea (45 g), 40% (v/v) acrylamide /bis-acrylamide solution (19:1, w/w; 45 mL), and 10 X TBE (10 mL) and diluting the contents to 100 mL with 18 mΩ water in an Erlenmeyer flask (125 mL).This solution can be degassed under vacuum for 5 min.
Clean the glass plates of with 70% aqueous ethanol (v/v) using a lint-free tissue and allow to air dry.
Apply a thin coat of siliconizing reagent on one of the gel plates to facilitate the dislodging of the plate assembly when the experiment is complete.
Assemble glass plates in the standard vertical electrophoresis unit according to manufacturer's specifications.
Prepare and add fresh 17% (w/v) ammonium persulfate (170 mg diluted to 1 mL with 18 mΩ water) to a gently stirring gel solution
- Add TEMED (35 μL) to the gel solutionOnce both TEMED and APS have been added, polymerization of the acrylamide and bisacrylamide will be initiated. Introducing the gel into the apparatus and placing the comb must be carried out relatively quickly (in less than 150 s). Introduce the gel solution slowly (40 - 45 s) between the plates to prevent bubble formation.
Place comb (50 teeth) after the introduction of gel solution.
- Allow polymerization reaction to proceed for 1 h.Gel polymerization may be monitored using the unused stock solution left in the flask.
- Remove comb carefully so as not to rip the well walls.This step may be accomplished by evenly and slowly removing the comb.
- Prewarm the gel for 30 min – 60 min at 98 W (approximately 2500 V and 40 mA)Wells should be filled with running buffer (1X TBE) and free of bubbles.
- Wash individual wells using TBE (1X) using a 25 mL disposable syringe fitted with a disposable needle (18G)Step 12 is crucial for proper sample resolution by PAGE sharpness of the individual bands.
- Load quenched DNA extension products on gel (2 μL) with a micropipettor fitted with flat end tips, changing the tip for each sample.Introduction of samples at the base of each well must be done gently. Dispensing the sample out of the tip vigorously may result in dispersion/loss of the sample. Bubble formation should be avoided when dispensing samples.
- Carry out electrophoresis at 98 W for 2 h. The bromophenol blue and xylene cyanol FF dyes should be approximately 15 cm apart for the particular oligomer length tested herein.The electrophoresis length will vary depending on the size of the oligomers and acrylamide/bis-acrylamide composition. Generally, longer primers will require longer electrophoresis periods. DNA fragments of interest should migrate approximately half of the gel. Refer to dye migration patterns in order to plan accordingly.
Turn off the power supply and dismantle the gel apparatus.
- Set the plate assembly horizontal and carefully separate plates so as not to rip the gel.Make sure to note which plate the gel resides on. Flip the plates if the gel is adhered to the top plate.
- Cover the gel with plastic cling wrap paying attention not to trap air bubbles or generate ripples in the plastic.Air bubbles trapped between the cling wrap and the gel may be removed by removing the plastic cling wrap and reapplying it slowly. Alternatively, a piece of lint-free tissue may be used to gently push the bubbles out.
Clean both sides of a smaller glass plate (40 cm x 16 cm) with 70% aqueous ethanol and allow to air dry.
Place the small plate over the gel region containing the DNA extension products, as estimated from the migration of the two gel dyes.
Excise the gel region along the border of the small glass plate using a razor blade.
Remove the small plate without disturbing the cling wrap.
Transfer the excised gel of interest onto the small glass plate.
Remove bubbles by gently pushing them to the extremities of the plate using a lint-free tissue.
- Detect and visualize DNA substrate and extension products using a Typhoon® Trio Variable Mode Imager in the fluorescence mode with the green (532 nm) laser in conjunction with the 526-nm short-pass filter. Typically, the instrument is set to normal sensitivity, PTM = 600, pixels = 200 microns, and a flatten focal.Alternatively, detection and visualization may be accomplished using autoradiography (in this case 32P radiography). This methodology involves first exposing the gel, containing 32P-radiolabelled species, to an erased storage phosphor screen (set in an autoradiography cassette, e.g. Amersham Hypercassette Autoradiography Cassette 35 × 43 cm (GE Healthcare Life Sciences)). The exposure time will depend on the concentration of the species and the half-life of the radiolabel (about 14.3 days for 32P). Typically, exposure times of 5-15 min are common for freshly labelled DNA primers. The imprint of the samples on the screen is then analyzed using a Typhoon® Trio Variable Mode Imager in the storage phosphor mode.
Data processing and analysis by Image J® and Graphpad Prism®
Open data file in Image J and correct for contrast.
Using the ROI manager function, measure the intensity density of each band and its respective background value.
Transfer the intensity density values to Microsoft Excel™.
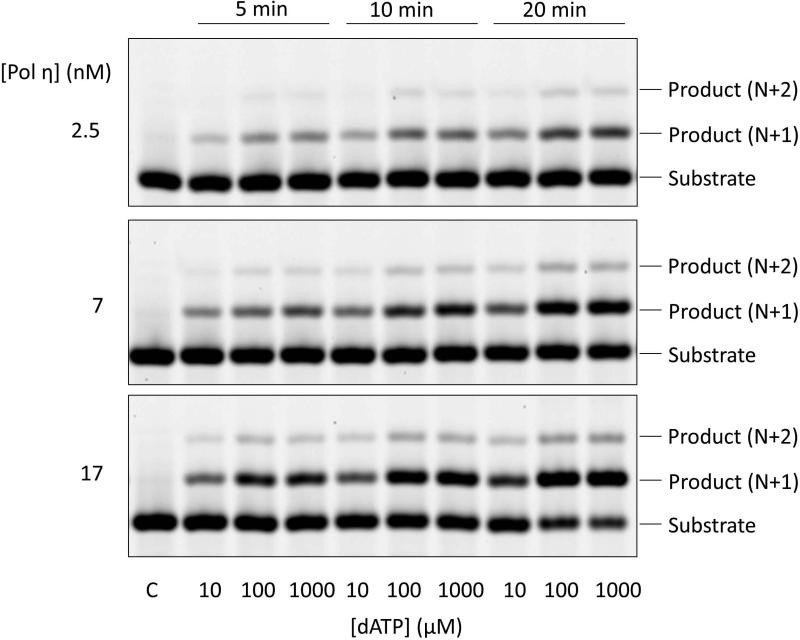
- Subtract the background values from the band readings and calculate the ratio (Rp) of product formation with respect to sum of the product (P) and substrate (S) (as shown in Table 2 and Table 3).The initial screen will supply information for a new set of conditions in order to sample [dNTP] concentrations that generate DNA extension product yields between 0 and 20% (of primer extension). Figure 2 displays the initial screen of extension across an O4MedT insert. To establish the optimal conditions, select an appropriate [E] and time value. Note that the N+2 product (where N is the oligomer length of the substrate; 13-mer in this case) must be minimized. In this case, [E] and t of 2.5 nM and 5 min (respectively) are most suitable given the low formation of N+1 at the highest [dATP] of 1000 μM and the virtual lack of the N+2 species. Also the product ratio is below 20% for all three [dATP] screened. The last parameter to select is the [dNTP] range. Since similar Rp are observed for [dATP] of 100 μM and 1000 μM, the 100 μM [dATP] should reside on the upper bound of the range (i.e. well above the KM value). For this study, we have chosen the following [dATP] concentrations (μM); 0.5, 1, 2.5, 5, 8, 15, 30, 60, 100, 150.
Table 2.
Product formation (N+1) ratio for primer extension screening across O4MeT using hpol η (2.5 nM) with varying [dATP] and time parameters (top panel of Figure 1 only).
| Time (min) | [dATP] (μM) | Substrate (S) integrated density | Product (P) integrated density | Rp (P/(P+S)) |
|---|---|---|---|---|
| 10 | 2544 | 195 | 0.071 | |
| 5 | 100 | 2989 | 415 | 0.12 |
| 1000 | 3106 | 463 | 0.13 | |
| 10 | 2568 | 294 | 0.10 | |
| 10 | 100 | 2993 | 619 | 0.17 |
| 1000 | 2982 | 645 | 0.18 | |
| 10 | 2915 | 516 | 0.15 | |
| 20 | 100 | 2808 | 902 | 0.24 |
| 1000 | 2573 | 887 | 0.26 |
Table 3.
Product formation (N+1) ratio for primer extension reaction (5 min), with insertion across from O4MeT using hpol η (2.5 nM) with varying [dATP].
| Trial 1 | Trial 2 | |||
|---|---|---|---|---|
| [dATP] (μM) | Rp (P/(P+S)) | v (min−1) | Rp (P/(P+S)) | v (min−1) |
| 0.5 | 0.0321 | 13 | 0.0219 | 8.7 |
| 1 | 0.0355 | 14 | 0.0298 | 12 |
| 2.5 | 0.0420 | 17 | 0.0423 | 17 |
| 5 | 0.0559 | 22 | 0.0533 | 21 |
| 8 | 0.0623 | 25 | 0.0655 | 26 |
| 15 | 0.0692 | 28 | 0.0812 | 33 |
| 30 | 0.0828 | 33 | 0.0931 | 37 |
| 60 | 0.0947 | 38 | 0.0957 | 38 |
| 100 | 0.110 | 44 | 0.101 | 40 |
| 150 | 0.114 | 46 | 0.105 | 42 |
Figure 2.
18% (19:1, w/w) Denaturing PAGE analysis of the extension reaction screen by hpol η inserting dATP across from O4MeT. hpol η concentrations are reported on the left, reaction time at the top, dNTP concentration at the bottom, and band identity at the right.
The following steps (6-10) are not for initial screening but rather for the actual experiments to carefully measure kinetic parameters (as shown in Figure 3).
-
5Calculate the turnover value (v in min-1) according to equation 1, where Rp is unitless, Di is the initial DNA substrate concentration (nM), E is the protein concentration (nM), and t is time (min) (as shown in Table 3).
-
6
Open Graphpad Prism® software, select multiple entries.
-
7Import the various tested [dNTP] as the independent (x) variable and the set of corresponding turnover values v (min-1) as the dependent (y) variables.Choose standard deviation (SD) instead of standard error of the mean (SEM) to display DNA polymerase extension variation.
-
8Choose the Analysis function and select the Michaelis-Menten entry (plot shown in Figure 4).The software will provide a best-fit of the data to the Michaelis-Menten model. A correlation coefficient (R2) will give an indication of the data fit to the model. Typically, values greater than 0.90 are acceptable. Parameters and graphical representation may be found in the left-hand column. That is, the maximum turnover value (kcat) and Michaelis constant (KM) can be found with their respective standard deviation.
-
9
Convert the kcat value to s−1 units and calculate the extension efficiency (kcat/KM,μM−1s−1) with the respective standard error.
-
10An f coefficient is calculated according to the equation below and expresses the comparison of the less efficiently incorporated dNTPs to the most-efficiently incorporated.
Figure 3.
18% (19:1, w/w) Denaturing PAGE analysis of the extension reaction (5 min) by hpol η (2.5 nM) across O4MeT with varying dATP. dNTP concentrations are reported at the bottom and band identity at the right.
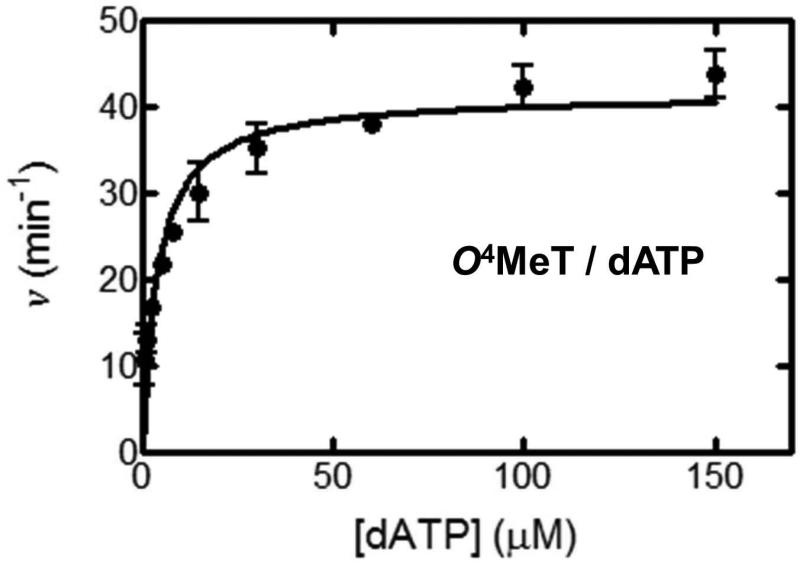
Figure 4.
Steady-state kinetic analysis of dATP incorporation by hpol η (v (min−1) versus [dATP] (plotted using Graphpad® Prism software).
COMMENTARY
Background Information
DNA is routinely assaulted by various endogenous and or exogenous agents, which may cause DNA damage. Taking into account that DNA serves as the organismal storage of genetic information, proper maintenance and propagation of DNA is of the utmost importance. Introduction of chemical modifications in DNA interferes with vital processes such as replication and transcription. Fortunately, organisms have developed cellular repair machineries to reverse the DNA damage back to its wild type. Despite the presence of these sophisticated repair mechanisms, certain DNA damages remain elusive or irreparable and as a result trigger critical cellular events such as steric blockage of polymerases, chromosomal instability, mutations in daughter strands, and activation of the apoptotic pathway (Shrivastav et al., 2010; Magaňa-Schwencke et al., 1982; Dronkert and Kanaar, 2001; Lawley and Phillips, 1996).
O4-Methylthymidine (O4MeT) was investigated in this protocol as the form of DNA damage in the template strand. This specific alkylation can arise from DNA exposure to tobacco derived N-nitrosamines and certain chemotherapeutic agents (Hecht, 1998; Singer, 1986; Lawley et al., 1973). It is highly mutagenic presumably because of the interference of the alkyl group with the Watson-Crick face causing the formation of altered hydrogen bonding patterns (Singer, 1986; Kalnik et al., 1988). Replication across O4MeT lowers the fidelity of polymerases leading to an O4MeT:dG pair, which will subsequently cause a dT to dC transition mutation (Singer et al., 1986). Moreover, the O4MeT:dG mismatch is a poor substrate for mismatch repair and direct repair (Samson et al., 1997; Duckett et al., 1996). Notably, O4MeT adducts have been detected in liver tissues of healthy volunteers without particular exposure to exogenous alkylating agents, epitomizing its importance as a biologically relevant adduct (Kang et al., 1995).
Illuminating a DNA lesion's potential to cause mutations from the DNA polymerase processivity is an important biological consequence. Misincorporation of nucleotides across, or past a specific lesion, is normally accomplished by DNA polymerase-catalyzed primer extension assays performed in vitro (Boosalis et al., 1987). Traditionally, DNA primers are labeled at the 5’ end with a 32P-containing phosphate with a combination of T4 polynucleotide kinase and [γ-32P]-ATP. More-recently, however, DNA primers functionalized with the fluorescent tag fluorescein have been employed instead of using radioactivity (e.g., Patra et al., 2014). DNA primers are extended by DNA polymerases across a single nucleotide in the template containing the lesion of interest in the presence of a single dNTP. Moreover, conditions are optimized to limit the formation of the extended product to less than 20% to prevent product inhibition and depletion of substrate, and limit the reaction to a single nucleotide extension (Guengerich, 2006). Reactions are carried out with constant DNA substrate concentration, DNA polymerase concentration, and time parameters while monitoring the formation of N+1 products at varying dNTP concentrations by denaturing PAGE analysis (Ellington and Pollard, 1998). By examining the incorporation of different dNTPs, Michaelis-Menten kinetic analyses provide quantitative information regarding the DNA polymerase's incorporation preference.
The Michaelis-Menten model (Berg et al., 2012; Michaelis and Menten, 1913) is described by the following relationship:
Where the reaction rate can be extrapolated to:
This model is governed by a number of assumptions. First, the components of system must be in a free diffusion state. Substrate concentrations are in large excess over the enzyme concentration, to push the equilibrium towards the DNA-enzyme complex [ES] (Berg et al., 2012). As stated above, product formation must be kept <20%, hence maintaining [S] >> [P] throughout the experiment.
Alternatively, the full-length extension assay analyzed by LC-MS analysis (UNIT 7.16; (Chowdhury and Guengerich, 2011)) furnishes insights on the ability of a DNA polymerase to extend across and beyond the modified nucleotide. Unlike the analysis by PAGE, this approach has an added facet of detecting and quantifying frameshift adducts, blunt-end additions, and incomplete primer extension occurrences (Zang et al., 2005; Christov et al., 2009). Despite these limitations, the single nucleotide extension reactions monitored by denaturing PAGE experiments are relatively straightforward to carry out while supplying central insights concerning the selectivity of the DNA polymerases for one nucleotide insertion over another.
Single nucleotide extension of DNA primers by DNA polymerase and analysis by denaturing PAGE
The general methodology of these studies begins with an initial experimental condition screen for the DNA primers extension with DNA polymerases. The screen will identify optimal polymerase concentration ([E]), reaction time (t), and dNTP concentration ([dNTP]) ranges to use. An example is presented in Figure 2 for the insertion of dATP directly across the O4MeT lesion. Note that the following base adjacent to the lesion in the template is a thymidine. In this respect, care must be taken to minimize the second round of DNA polymerization (generating the N+2 product). It is clear from the denaturing PAGE analysis and product quantification that appropriate experimental conditions would be as follows: [E] = 2.5nM, t = 5 min, and [dNTP] ranges = 0.5, 1, 2.5, 5, 8, 15, 30, 60, 100, 150 μM. The only variable that may be ambiguous from the selection is the [dNTP] range, because all other conditions generate a significant amount of N+2 product. The difference in extended product formation between 100 μM and 1000 μM [dATP] is not very large, which would suggest that these points lie on the plateau part of the kinetic curve (as observed in Figure 4). Typically, accurate data fitting to the Michaelis-Menten model should involve sampling the curve at [S] values below and above the KM value (2-3 points on either side of the KM value). In this case, it is safe to assume that the KM value will be much lower than 100 μM given that this point is projected to be on the upper limit of the curve. Measurements of kcat and KM can normally be achieved using as few as six to eight data points, if properly spaced. However, 10 data points were used in the experiment given the number of lanes found in the denaturing gel (40, e.g. four experiments of 10 samplings). An example of duplicate experiments is shown in Figure 3.
Critical Parameters and Troubleshooting
The success of this experiment primarily depends on the yield of the extension products, which precisely explains why the initial screen is very important. If the tested nucleoside triphosphate is the favored one for the first extension as well as the second, conditions must be optimized to generate a minimal amount of N+2 product while generating a sufficient amount of N+1 product. Achieving this may require additional screening of conditions. DNA polymerase concentration and time are normally identified first, followed by the dNTP concentrations. The latter parameter should sample points on the curve (of v vs. [S]) at substrate concentrations both below and above the KM.
The presence of bubbles in a gel is detrimental to the separation of the DNA fragments during PAGE. To circumvent the formation of bubbles in the gel, carefully follow the manufacturer's specifications for setting up gels. Generally, introducing the polymerizing gel solution between the plates slowly will prevent the formation of bubbles. Lanes in which bubbles have formed should be avoided. Wells should be rinsed with running buffer (1X TBE) very well right before loading the samples. Failing to do so will lower the resolution of the PAGE experiment.
Anticipated Results
Generally, the steady-state kinetic analysis of DNA primer extension by DNA polymerases supplies steady-state kinetic parameters, i.e. the Michealis constant (KM) and the maximal reaction velocity (kcat). These values can be used to characterize the impact of a given DNA modification on the processing by different DNA polymerases, relative to that of an undamaged DNA control. In the case described in this unit, hpol η inserted dATP across from an unmodified T in the template with > 85% fidelity. In contrast, hpol η misinserted dGTP with the most efficiency across from O4MeT, with slightly more efficiency compared to the dATP insertion (kcat/KM 0.19 versus 0.18 μM−1 s−1, Table 4). Insights are thus gained on the preferences of dNTP incorporation by a DNA polymerase.
Table 4.
Steady-state kinetics of incorporation of dATP, dGTP, dCTP, and dTTP opposite T and O4MeT by hpol η.
| Template base | dNTP | kcat (s−1) | KM (μM) | kcat/KM (μM−1s−1) | fa |
|---|---|---|---|---|---|
| T | dATP | 1.60 ± 0.03 | 1.4 ± 0.1 | 1.2 ± 0.1 | |
| T | dGTP | 0.71 ± 0.03 | 4.6 ± 0.7 | 0.15 ± 0.02 | 0.13 |
| T | dCTP | 0.16 ± 0.01 | 29 ± 7 | 0.006 ± 0.001 | 0.005 |
| T | dTTP | 0.37 ± 0.01 | 24 ± 3 | 0.015 ± 0.002 | 0.013 |
| O4MeT | dATP | 0.69 ± 0.03 | 3.9 ± 0.6 | 0.18 ± 0.03 | 0.94 |
| O4MeT | dGTP | 1.18 ± 0.02 | 6.2 ± 0.4 | 0.19 ± 0.01 | |
| O4MeT | dCTP | 0.11 ± 0.01 | 15 ± 3 | 0.007 ± 0.002 | 0.039 |
| O4MeT | dTTP | 0.25 ± 0.02 | 17 ± 4 | 0.015 ± 0.004 | 0.077 |
f = (kcat/KM)dNTP / (kcat/KM)max
It should be emphasized, as in all enzymology, that the determination of steady-state parameters is only a beginning in terms of understanding the kinetics of complex enzymes, such as DNA polymerases. With DNA polymerases, the rate-limiting step usually occurs following product formation, and kcat and KM are rather obscure collections of forward and reverse rate constants for six or more individuals steps in these reactions (e.g., Johnson, 2003; Guengerich, 2006). More detailed analysis requires the use of transient (“pre-steady-state”) kinetics, which use gel-based assays similar to those described here to analyze the products of reactions done at sub-second reaction times (in a rapid-quench apparatus).
Time Considerations
The primer extension assays have a varied timeframe depending on the screening step. Typically, condition screening experiments for each dNTP can be achieved in half of a day. Once conditions are optimized, a single PAGE experiment can analyze two DNA primer extension experiments with individual dNTPs (e.g. independent extension profiles with two different dNTPs, in duplicates). Thus, each DNA duplex investigated can be screened for the four standard dNTPs by a given DNA polymerase in approximately one to two weeks. Multiple PAGE experiments may carried out simultaneously to increase productivity.
Acknowledgments
DKO was a recipient of a postgraduate fellowship (CGS-D) from Natural Sciences and Engineering Research Council of Canada (NSERC) and funded by the Bionano Exchange Program from the Create Training Program in Bionanomachines (CTPB). We are grateful to Dr. Christopher J. Wilds for supplying the DNA template containing an O4MeT insert and Dr. Martin Egli for helpful discussions. This work was supported by grants from the National Institutes of Health (R01 ES010375 to F. P. Guengerich and M. Egli).
Literature Cited
- Berg JM, Tymoczko JL, Stryer L. Enzymes: Basic concepts and kinetics. In: Samols L, Hadler GL, Zimmerman P, Brooks N, editors. Biochemistry. Seventh Ed. W. H. Freeman; New York: 2012. pp. 229–237. [Google Scholar]
- Biertümpfel C, Zhao Y, Kondo Y, Ramón-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–8. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity: gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–96. [PubMed] [Google Scholar]
- Chowdhury G, Guengerich FP. Liquid chromatography-mass spectrometry analysis of DNA polymerase reaction products. Curr. Protoc. Nucleic Acid Chem. 2011;7:7.16.1–11. doi: 10.1002/0471142700.nc0716s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christov PP, Angel KC, Guengerich FP, Rizzo CJ. Replication past the N5-methyl-formamidopyrimidine lesion of deoxyguanosine by DNA polymerases and an improved procedure for sequence analysis of in vitro bypass products by mass spectrometry. Chem. Res. Toxicol. 2009;22:1086–95. doi: 10.1021/tx900047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–47. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- Duckett DR, Drummond JT, Murchiet AIH, Reardont JT, Sancart A, Lilleyt DMJ, Modrich P. Human MutScv recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymidine, or the cisplatin-d(GpG) adduct. Proc. Natl. Acad. Sci. USA. 1996;93:6443–47. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A, Pollard JD. Purification of oligonucleotides using denaturing polyacrylamide gel electrophoresis. Curr. Protoc. Mol. Biol. 1998;42:2.12.1–2.12.7. doi: 10.1002/0471142727.mb0212s42. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem. Rev. 2006;106:420–52. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- Johnson KA. Introduction to kinetic analysis of enzyme systems. In: Johnson KA, editor. Kinetic Analysis of Macromolecules. A Practical Approach. Oxford University Press; New York: 2003. pp. 1–18. [Google Scholar]
- Kalnik MW, Kouchakdjian M, Li BFL, Swann PF, Patel DJ. Base pair mismatches and carcinogen-modified bases in DNA: An NMR study of A•C and A•O4meT pairing in dodecanucleotide duplexes. Biochemistry. 1988;27:100–8. doi: 10.1021/bi00401a017. [DOI] [PubMed] [Google Scholar]
- Kang H, Konishi C, Kuroki T, Huh N. Detection of O6-methylguanine, O4-methylthymine and O4-ethylthymine in human liver and peripheral blood leukocyte DNA. Carcinogenesis. 1995;16:1277–80. doi: 10.1093/carcin/16.6.1277. [DOI] [PubMed] [Google Scholar]
- Lawley PD, Orr DJ, Shah SA, Farmer PB, Jarman M. Reaction products from N-methyl-N-nitrosourea and deoxyribonucleic acid containing thymidine residues. Biochem. J. 1973;135:193–201. doi: 10.1042/bj1350193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley PD, Phillips DH. DNA adducts from chemotherapeutic. Mutat. Res. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- Magaňa-Schwencke N, Henriques J-AP, Chanet R, Moustacchi E. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: Comparison of wild-type and repair-deficient strains. Proc. Natl. Acad. Sci. USA. 1982;79:1722–26. doi: 10.1073/pnas.79.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis L, Menten ML. Kinetik der Invertinwirkung. Biochem. Z. 1913;49:333–69. [Google Scholar]
- Patra A, Nagy LD, Zhang Q, Su Y, Müller L, Guengerich FP, Egli M. Kinetics, structure, and mechanism of 8-Oxo-7,8-dihydro-2′-deoxyguanosine bypass by human DNA polymerase η. J Biol Chem. 2014;289:16867–82. doi: 10.1074/jbc.M114.551820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G, Stillman B. Coordinated leading and lagging during SV40 DNA replication in vitro requires PCNA strand synthesis. Cell. 1988;53:117–126. doi: 10.1016/0092-8674(88)90493-x. [DOI] [PubMed] [Google Scholar]
- Samson L, Han S, Marquis JC. Mammalian DNA repair methyltransferases shield O4MeT from nucleotide excision repair. Carcinogenesis. 1997;18:919–24. doi: 10.1093/carcin/18.5.919. [DOI] [PubMed] [Google Scholar]
- Shrivastav N, Li D, Essigmann JM. Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. O-Alkyl pyrimidines in mutagenesis and carcinogenesis: Occurrence and significance. Cancer Res. 1986;46:4879–85. [PubMed] [Google Scholar]
- Singer B, Spengler SJ, Fraenkel-Conrat H, Kuśmierek JT. O4-Methyl, -ethyl, or -isopropyl substituents on thymidine in poly(dA-dT) all lead to transitions upon replication. Proc. Natl. Acad. Sci. USA. 1986;83:28–32. doi: 10.1073/pnas.83.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–12. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- Zang H, Goodenough AK, Choi J-Y, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J. Biol.Chem. 2005;280:29750–64. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]