Abstract
Methanothermobacter marburgensis is a strictly anaerobic, thermophilic methanogenic archaeon that uses methanogenesis to convert H2 and CO2 to energy. M. marburgensis is one of the best-studied methanogens, and all genes required for methanogenic metabolism have been identified. Nonetheless, the present study describes a gene (Gene ID 9704440) coding for a putative NAD(P)H:quinone oxidoreductase that has not yet been identified as part of the metabolic machinery. The gene product, MmNQO, was successfully expressed, purified and characterized biochemically, as well as structurally. MmNQO was identified as a flavin-dependent NADH:quinone oxidoreductase with the capacity to oxidize NADH in the presence of a wide range of electron acceptors, whereas NADPH was oxidized with only three acceptors. The 1.50 Å crystal structure of MmNQO features a homodimeric enzyme where each monomer comprises 196 residues folding into flavodoxin-like α/β domains with non-covalently bound FMN (flavin mononucleotide). The closest structural homologue is the modulator of drug activity B from Streptococcus mutans with 1.6 Å root-mean-square deviation on 161 Cα atoms and 28% amino-acid sequence identity. The low similarity at sequence and structural level suggests that MmNQO is unique among NADH:quinone oxidoreductases characterized to date. Based on preliminary bioreactor experiments, MmNQO could provide a useful tool to prevent overflow metabolism in applications that require cells with high energy demand.
Keywords: crystal structure, cytoplasm, Methanothermobacter marburgensis, NADH regeneration, NADH:quinone oxidoreductase
Abbreviations: acetyl-CoA, acetyl coenzyme A; CER, CO2 evolution rate; CV, column volume; DCPIP, 2,6-dichloroindophenol sodium salt hydrate; FMN, flavin mononucleotide; IPTG, isopropyl-β-D-thiogalactopyranoside; MdaB, modulator of drug activity B; PMF, proton motive force; r.m.s.d., root-mean-square deviation; TCA, tricarboxylic acid
Short abstract
A novel NADH:quinone oxidoreductase, MmNQO, from Methanothermobacter marburgensis was identified. MmNQO oxidizes NADH with several electron acceptors and is structurally similar to bacterial MdaB. It is localized in the cytosol and may provide a useful tool to prevent overflow metabolism.
INTRODUCTION
Aerobic cellular respiration in eukaryotes involves glycolysis, TCA (tricarboxylic acid) cycle (also Krebs cycle or citric acid cycle) and respiratory chain. During glycolysis, glucose is converted to pyruvate, followed by oxidative decarboxylation to acetyl-CoA (acetyl coenzyme A). Acetyl-CoA enters the TCA cycle within the mitochondrial matrix where oxidation occurs concomitantly with reduction of NAD+ to NADH, and FAD to FADH2. The large quantities of NADH and FADH2 generated provide the high-potential reducing equivalents required for the ensuing electron-transfer events that build the PMF (proton motive force) necessary to power ATP synthesis through oxidative phosphorylation. This electron-transfer chain of inner mitochondrial-membrane protein complexes includes three proton pumps: NADH dehydrogenase (NADH:ubiqinone oxidoreductase; complex I [1]); cytochrome bc1 reductase (ubiquinol:cytochrom c oxidoreductase; complex III); and cytochrome c oxidase (complex IV). Succinate dehydrogenase (succinate:ubiquinone oxidoreductase; complex II) is part of the TCA cycle, and does not pump protons, but rather serves as a link between TCA cycle and respiratory chain. The electrochemical proton gradient formed across mitochondrial membrane couples the respiratory chain to production of ATP through oxidative phosphorylation catalysed by ATP synthase (complex V).
In prokaryotes, the respiratory chain is confined to the cytoplasmic membrane in the intermembrane space. For Escherichia coli, the proton gradient at the membrane is generated by two types of enzymes, many of which have been characterized [2] (Supplementary Table S1): (i) dehydrogenases, which oxidize organic substrates and reduce ubiquinone; and (ii) oxidases, which oxidize ubiquinol and reduce molecular oxygen to water. Two membrane-bound E. coli NADH:ubiquinone oxidoreductases, NDH-1 and NDH-2, have been described [3–6], where NDH-1 operates as proton pump and shows sequence homology to the eukaryotic mitochondrial complex I, whereas NDH-2 is a single-subunit enzyme that catalyses the same reaction but without generating a PMF. Although NDH-2 is not directly involved in energy formation in E. coli, it performs an essential role by oxidizing excessive NADH to NAD+. If the rate of glucose consumption is higher than the respiratory capacity, the cells use pathways that are normally activated under anaerobic conditions to regenerate NAD+ and produce lactate or acetate, a phenomenon called overflow metabolism [7,8]. Since these undesired metabolites inhibit cell growth, it is desirable to avoid NADH accumulation [9].
The delicate NAD+/NADH balance has been studied in detail for E. coli, where it was demonstrated that increased amounts of NADH induced a metabolic shift towards fermentation even in the presence of oxygen excess [10]. In another study, the nuo-operon, coding for the NADH:ubiquinone oxidoreductase complex, was overexpressed in E. coli and increased activity with NADH and ferricyanide as electron acceptor was observed [11]. Unfortunately, the authors did not comment on the viability of the cells, or changes in metabolic activity. This is relevant since overexpression of wild-type NADH:ubiquinone oxidoreductases usually correlates with negative, sometimes detrimental effects for the cells, which have been rationalized by cellular stress associated with overexpression of integral membrane proteins [12].
Methanogenic archaea (methanogens) derive their energy from methanogenesis, a type of anaerobic respiration, where CO2 is reduced to methane in the presence of hydrogen. Based on their 16S rRNA, methanogens can be grouped into different classes, one being Methanothermobacter [13]. One of the most studied representatives of this class is M. marburgensis (formerly M. thermoautotrophicum) [13] for which the methanogenic pathway has been analysed in detail. The M. marburgensis genome has been sequenced (GenBank CP001710; [14]), and all enzymes essential for methanogenesis have been identified [15].
Interestingly, despite M. marburgensis being strictly anaerobic, we identified a novel gene coding for a yet uncharacterized NADH-dehydrogenase-like enzyme, i.e. gene ID 9704440 (UniProt D9PVS9). This was unexpected since all enzymes involved in energy metabolism of M. marburgensis have supposedly been identified. In the present study, the gene product of gene ID 9704440, here referred to as MmNQO, was cloned, overexpressed in E. coli and characterized biochemically and structurally.
EXPERIMENTAL
Materials and reagents
Electron-acceptor substrates from Sigma Aldrich): coenzyme Q1, coenzyme Q10, DCPIP (2,6-dichloroindophenol sodium salt hydrate), potassium ferricyanide, 1,4-benzoquinone (1,4-BQ), ferrocenium hexafluorophosphate (Fc+), diethyl oxalacetate sodium salt, sodium fumarate dibasic, methyl red sodium (crystalline), 3,3′-methylene-bis(4-hydroxycoumarin). Electron acceptor substrates from Carl Roth (Germany): sodium thiosulphate anhydrous, DL-malic acid. Electron-donor substrates from Sigma Aldrich: β-nicotinamide adenine dinucleotide phosphate and reduced tetra (cyclohexylammonium) salt (NADPH).
Sequence analyses
Sequence analyses for the three cloned M. marburgensis genes were performed with BLAST (http://blast.ncbi.nlm.nih.gov) [16] against existing sequence databases. The TOPCONS server (http://topcons.cbr.su.se) [17] was used to analyse the presence of transmembrane helices, and analysis of sequence motifs and domains was performed using the SMART server (http://smart.embl-heidelberg.de) [18].
Preparation of genomic DNA and gene cloning
Genomic DNA from M. marburgensis was extracted with the peqGOLDTriFast kit (Peqlab). Frozen biomass of M. marburgensis was thawed and resuspended in 1.0 ml TriFast™ per 100 mg wet biomass. DNA concentration was measured using a Nanodrop device (ND1000 Spectrophotometer, Thermo Scientific). The gene 9704440 was amplified from genomic DNA using oligonucleotide primers listed in Supplementary Table S2 and cloned into a pET21a+ vector providing a C-terminal hexahistidine (His6) tag. PCRs were done in a total volume of 50 μl containing 2 μl dNTP mix (200 μM), 1 μl of each primer (50 μM), 10 μl 5× HF buffer, about 10 ng of genomic DNA and 0.5 μl Phusion Hot Start II Polymerase (2 units/μl, Finnzymes) in a S1000™ Thermal Cycler (Biorad) using the following program: 1 min at 98°C; 98°C 10 s—55°C 20 s–72°C 60 s (30 cycles); 72°C 10 min. The amplified PCR product was purified from the reaction mixture using a PCR purification kit (Qiagen), and cloned into the pET21a+ vector via the NheI and XhoI sites using standard cloning methods. The recombinant plasmid was transformed into E. coli BL21(DE3) by electroporation.
Expression in shake flask and bioreactor cultures
Shake flasks
Shake flasks were used for the production of protein for biochemical and structural studies. A 10-ml aliquot of an overnight culture was transferred into 100 ml TBamp medium in 1 litre baffled shake flasks and grown at 37°C with shaking (220 rpm). Induction at OD600 of about 0.6 was performed with 0.1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) at 25°C. After 24 h, biomass was harvested by centrifugation (4,500 g, 4°C, 20 min). E. coli BL21(DE3) harbouring the pET21a+ vector was included as negative control.
Bioreactor cultivations
For the pre-culture, fresh E. coli transformants were cultured in 100 ml DeLisa medium ([19]; Supplementary Table S3) in 1-litre baffled shake flasks at 37°C (220 rpm) for approximately 12 h. The grown pre-culture was transferred aseptically to the culture vessel. The inoculation volume was 10% of the final starting volume. Batch cultivations were carried out in a 3 l working volume bioreactor from Labfors. DeLisa medium was sterilized in the bioreactor and sterile glucose-monohydrate mixed with trace elements was added aseptically. The pH was adjusted to 7.4 using NH4OH solution (2.4–2.6 M), and 0.3 ml polypropylen glycol were added as anti-foam. The pO2 and pH levels were measured with sterile electrodes. Base consumption was determined gravimetrically. Cultivation temperature was set to 37°C and agitation was fixed to 1400 rpm. The culture was aerated with 2.0 vvm air and off-gas was measured. Recorded data were logged in a PIMS (process information management system; Lucullus, Securecell). After the complete consumption of the carbon source, as indicated by a drop in off-gas activity, the fed-batch was initiated.
For fed-batch cultivation, the feed with a glucose-monohydrate concentration of 440 g/l was aseptically added to the vessel. Using an in-house developed Kalman filter, the feed flow and specific growth rate (μ) was set to a value below the maximum specific growth rate (μmax) determined during the batch phase to prevent overflow metabolism. The two cultures of BL21(DE3) cells (one expressing 9704440 and a negative control) were induced with 0.1 mM IPTG after 7.5 h of fed-batch. Before IPTG was added, the temperature was decreased to 25°C to minimize the formation of inclusion bodies [20]. Samples were taken at the end of the batch, before induction, and at the end of cultivation. Dry cell weight was determined by centrifugation of 5 ml culture broth, washing the pellet with 5 ml deionized water und subsequent drying at 105°C to a constant weight in an oven. Substrate and metabolites in the supernatant were determined by HPLC analysis with an anion-exchange column (Supelcogel C-160H, Sigma Aldrich) equipped with a refractive index detector (Agilent Technologies). The flow rate of the mobile phase (0.1% H3PO4) was set to 0.5 ml/min and the system was run isocratically. Calibration for glucose, ethanol, acetate, citrate, malate, lactate, fumarate, oxaloacetate and pyruvate was done by measuring standards at concentrations of 0.1, 1.0 and 5.0 g/l, respectively.
Protein purification and biochemical characterization
After harvesting, cells were resuspended in homogenization buffer (50 mM Mops, 500 mM NaCl, pH 7.4) to a final concentration of 20 g wet biomass per litre and protease inhibitor was added (two tablets of Complete EDTA-free protease inhibitor from Roche per 100 ml). Cells were homogenized using an Emulsiflex C3 homogenizer (Avestin). The homogenized suspension was centrifuged (20000 g, 4°C, 15 min), and overall protein concentration and enzyme activity were measured. Soluble proteins and inclusion bodies were analysed by SDS–PAGE.
Purification of the 9704440 gene product was performed by IMAC (immobilized metal affinity chromatography). The protein solution was diafiltrated in binding buffer (20 mM Mops, 500 mM NaCl and 20 mM imidazole, pH 7.4) using a Centramate 500S system (PALL). A Ni2+-Sepharose 6 Fast Flow resin (GE Healthcare) resin was equilibrated with 5 CVs (column volumes) of binding buffer prior to sample loading. After loading, the column was washed with 5 CV of binding buffer, followed by elution of the target protein with a linear gradient (10 CV) of elution buffer (20 mM Mops, 500 mM NaCl, 500 mM imidazole, pH 7.4) and collection of 2.5-ml fractions. Sample binding and elution were performed at a flow rate of 60 cm/h, whereas equilibration and washing steps were done at 120 cm/h. Fractions containing the target protein were pooled, diafiltrated in 50 mM Mops (pH 7.4) and concentrated using Amicon Ultra-15 Centrifugal Filter Units with 10 kDa molecular weight cut-off (Merck-Millipore) to a final concentration of 3 mg/ml followed by purity analysis using SDS–PAGE and activity measurements. The protein content was measured spectrophotometrically at a wavelength of 595 nm according to the Bradford protocol using BSA as standard.
To identify the cofactor, the visible (VIS) spectrum was recorded in the range of 300–800 nm. Besides flavins, iron-sulphur clusters are found as prosthetic groups in NADH:quinone oxidoreductases with an absorption maximum at 455 nm [21]. The spectrum was recorded for 20 μl of purified 9704440 gene product using 50 mM MOPS buffer (pH 7.4) as blank. To analyse cofactor release, the enzyme was incubated with 5% trichloroacetic acid at 100°C for 2 h followed by centrifugation and repeating the measurement.
For measuring pH optimum and pH range, a 50 μl aliquot of enzyme solution was incubated with 50 μl of 100 mM buffer with the pH set to different pH values between 2.5 and 10.0 in steps of 0.5: citrate (2.5–5.5), carbonate (5.3–7.3), phosphate (6.2–8.2), Tris (7.5–9.0) and glycin (8.8–10.0). The enzyme was incubated at 30°C for 1 h and centrifuged at 20,000 g for 15 min to remove denaturated protein. Residual activity was measured in the supernatant using NADH and 1,4-BQ.
Enzyme kinetics
Enzymatic activity of the 9704440 gene product was measured using NADH as electron donor with either coenzyme Q1 (ubiquinone) or 1,4-BQ as electron acceptor. 1 ml reaction mixture contained 50 mM MOPS (pH 7.4), 1 mM of electron acceptor and 1 mM of electron donor. The oxidation of NADH was followed at 340 nm (ε340=6.22 mM/cm). One unit of activity was defined as the amount of enzyme catalysing the oxidation of 1 μmol NADH/min. All measurements were done in duplicates. Catalytic constants for both NADH and NADPH were determined using a range of electron acceptors (see the Experimental section for details). Approximately 1 ml reaction mixture contained 50 mM Mops (pH 7.4), different concentrations of electron donor (1.0–150 μM) and a saturating concentration of 1 mM electron acceptor. The reaction was initiated by adding 20 μl purified enzyme at a concentration of 3 mg/ml. Measurements were done at 340 nm at 30°C in a UV-1601 spectrophotometer (Shimadzu) and the absorbance was recorded for 240 s using the UVPC Optional Kinetics software (Shimadzu). All measurements were done in duplicates. Catalytic constants were calculated using the program SigmaPlot.
Crystallization and structure determination
Prior to crystallization, the buffer of the protein sample was exchanged by gel filtration to 20 mM Bis-Tris (pH 6.0), 150 mM NaCl and concentrated to 10 mg/ml. Crystallization screening was performed in Corning 3550 96-well sitting-drop plates using the sitting-drop vapour diffusion method, and 300 nl drops dispensed by a mosquito Crystal robotics (TTP Labtech) at protein-to-reservoir ratios of 1:1, 1:2 and 2:1. Well-formed crystals grew from a solution of 0.2 M NaCl, 0.1 M Bis-Tris (pH 5.5), and 25% PEG [poly(ethylene glycol)] 3350. Crystals were harvested from the mother liquor and vitrified in liquid nitrogen. X-ray intensity data to 1.50 Å resolution were collected at 100K at Diamond Light Source (UK) on macromolecular crystallography beamline I03, and the data processed (Supplementary Table S4) by the XDS package [22]. The resulting data indexed space group P21, with cell parameters a=56.60 Å, b=94.69 Å, c=72.13 Å and β=93.34°.
Phases were obtained by molecular replacement with the PHENIX package [23] using the NAD(P)H:quinone oxidoreductase MdaB (modulator of drug activity B) from Streptococcus mutans as search model (PDB code 3LCM; [24]). An initial model was generated with warpNtrace included in the ARP/wARP package [25]. The model was refined at 1.50 Å resolution using PHENIX against the maximum-likelihood target and adjusted manually with the graphics softwares COOT [26] and O [27]. Refinement in PHENIX included XYZ-coordinate refinement, real-space refinement, and refinement of individual anisotropic temperature factors. The final model comprises four protein chains, A, B, C and D, forming two dimers (A/B and C/D), and four FMN molecules (one per monomer), and 427 water molecules. The gene construct cloned in the pET21d+ vector adds the sequence −2MAS0 at the N-terminus and the sequence 197EHHHHHH203 corresponding to a non-cleavable hexahistidine tag at the C terminus. In the final model, each monomer is composed of residues 1-196 (UniProt D9PVS9). Additional residues from the cloning sequence modeled at the N-terminus include −1AS0 for monomers A-D, and 197EHHHHHH203 at the C-terminus in monomers A and C. Monomer B contains the sequence 197EHHHH201 of the C-terminal tag, whereas monomer D retains only 197E of the tag. Residues not modeled due to lack of electron density: monomer A, 113–118; monomer B, 114–117, monomer C, 113–118, monomer D, 115–116. Structure-based fold similarity analyses were performed using the DALI Lite server (http://ekhidna.biocenter.helsinki.fi/dali_server) [28] and PDBeFold (http://www.ebi.ac.uk/msd-srv/ssm) [29]. Coordinates and structure factors have been deposited in the Protein Data Bank with accession code 4R81.
RESULTS AND DISCUSSION
Sequence analyses
The sequence of M. marburgensis gene ID 9704440 (UniProt D9PVS9) was analysed using BLAST, SMART and TOPCONS. Based on the results from these analyses, gene ID 9704440 codes for a soluble protein of 196 amino acids. The sequence shows similarity to a large number of bacterial NAD(P)H:quinone reductases with a varying degree of sequence identity in the range of 30–40%, of which several hits were loosely annotated as belonging to the bacterial MdaB.
Expression in shake-flask and bioreactor cultures
Initially, the product for gene ID 9704440, hereafter referred to as MmNQO, was expressed in shake flask cultures. To monitor production, intracellular protein samples were analysed for enzymatic activity and by SDS–PAGE. To optimize the amount of expressed MmNQO, E. coli was cultivated in a bioreactor. Based on the data from the batch phase, the maximum specific growth rates (μmax) for both strains (one expressing 9704440 and a negative control) were calculated. The specific growth rate (μ) for each strain in the fed-batch phase was set below μmax/2 to avoid overflow metabolism [30]. Batch and fed-batch were performed at 37°C before temperature was decreased to 25°C upon induction. During induction, μ was set to only 10% of μmax (Table 1). Off-gas was analysed online, and the respective CER (CO2 evolution rate), depicting metabolic activity, was calculated for both cultures of BL21(DE3), one containing the recombinant pET21a+ vector with the MmNQO gene insert, and a negative control transformed with the empty pET21a+ (Supplementary Figure S1). Specific rates and yields were calculated from online and off-line data (Table 1). The batch phase for the two cultivations was similar, resulting in a similar amount of biomass. Nonetheless, the two cultures displayed different growth characteristics during the subsequent non-induced fed-batch resulting in different CER profiles and yields. For the negative control, more substrate was converted into CO2 than into biomass, whereas the opposite was observed for cells with MmNQO (Table 1). Except for cells containing the recombinant plasmid in the induction phase, the carbon balance (C-balance) did not close to 1.0. However, we identified several metabolites in the cell-free cultivation broths (Supplementary Figures S2 and S3), and when taking these metabolites into account, it was possible to close the C-balance for the batch and induction phases of both cultures. For the other cultivation phases, the C-balances did not close to 1.0. As indicated by increasing concentrations of extracellular DNA and protein, this may be due to cell lysis. The observation that the apparent μ of the cells was different to the set μ during the non-induced fed-batch phase for both cultures may also be attributed to cell lysis since the Kalman filter, regulating the feed according to intact cells, did not respond to cell lysis.
Table 1. Cultivation of E. coli BL21(DE3) carrying empty plasmid and plasmid with MmNQO insert.
| pET21d+ | pET21d+-MmNQO | |||||
|---|---|---|---|---|---|---|
| Batch | Fed-batch | Induction | Batch | Fed-batch | Induction | |
| μmax (h−1) | 0.52 | – | – | 0.53 | – | – |
| μset (h−1) | – | 0.2 | 0.05 | – | 0.15 | 0.05 |
| μreal (h−1) | – | 0.09 | 0.04 | – | 0.07 | 0.04 |
| biomass (g/l) | 5.1 | 20.9 | 44.8 | 5.2 | 16.2 | 35.6 |
| YX/S (c-mol/c-mol) | 0.33 | 0.34 | 0.24 | 0.33 | 0.47 | 0.44 |
| YCO2/S (c-mol/c-mol) | 0.44 | 0.40 | 0.34 | 0.44 | 0.35 | 0.53 |
| C-balance without metabolites | 0.77 | 0.74 | 0.58 | 0.77 | 0.82 | 0.97 |
| Metabolites (g/l) | ||||||
| Pyruvate | 2.9 | 2.8 | 0.37 | 0.38 | 0.67 | – |
| Lactate | – | 0.49 | 0.73 | 0.52 | 0.71 | 0.52 |
| Fumarate | – | – | 0.32 | – | – | – |
| Acetate | – | – | 0.23 | – | – | – |
| C-balance with metabolites | 0.94 | 0.79 | 0.60 | 1.03 | 0.85 | 1.05 |
The observed discrepancy between the two cultures is interesting, especially the difference in metabolite formation (Supplementary Figures S2 and S3). Although a specific growth rate below μmax/2 was applied, overflow metabolism during the non-induced fed-batch phase resulted in the production of metabolites. This may be interpreted as a very low respiratory capacity of cells in both cultures. It is also interesting to note that pyruvate was the principal metabolite, and not acetate, indicating that the cells responded to an increased energy demand (accumulation of pyruvate for increased ATP production). Similar results have been observed by others, where metabolic shifts from anabolic to catabolic pathways occur after induction to allocate glucose for recombinant protein expression at the expense of biomass production [30].
After induction of cells transformed with the recombinant plasmid, no more metabolites were generated, and the previously formed metabolites were consumed (Table 1, Supplementary Figure S3). Thus, recombinant expression of MmNQO appears to reduce overflow metabolism. This not only supports a function of MmNQO as a NADH:quinone oxidoreductase, but also shows the potential of the enzyme to relieve overflow metabolism in recombinant E. coli. To date, detrimental overflow metabolism in E. coli is either tackled by tailoring the bioprocess, which requires extensive metabolomic analyses and the development of reliable soft sensor tools [31,32] or by engineering the E. coli strain. Current strain engineering strategies focus on modifying the substrate uptake system [33,34] and not on overexpression of NDH enzyme complexes, since previous studies have shown cellular stress associated with overexpression of integral membrane proteins [12]. Although cells with a reduced substrate uptake capacity are not extensively affected by overflow metabolism, these cells are physiologically impaired and are thus not useful for industrial production processes. Consequently, E. coli cells, where excessive NADH is regenerated by a cytosolic enzyme, such as MmNQO, might describe an interesting host for energy demanding bioprocesses.
Protein purification and biochemical characterization
Recombinant His6-tagged MmNQO was purified to homogeneity, and the apparent molecular weight (Mw), as determined with about 24 000 Da by SDS–PAGE, agreed well with the theoretical Mw of 23 538 Da. Any activity detected in the flow through during IMAC purification was ascribed to endogeneous E. coli NADH:quinone oxidoreductases. Purified MmNQO was tested for NADH:quinone oxidoreductase activity using NADH or NADPH as electron donor and different electron acceptors (Supplementary Table S5). With NADH as electron donor, MmNQO showed NADH:quinone oxidoreductase activity with all electron acceptors tested, whereas NADPH was only oxidized in the presence of three electron acceptors. The activity with NADPH as substrate was typically 2- to 20-fold lower compared with NADH. MmNQO showed the highest specific activity with NADH and Ferrocenium hexafluorophosphate as electron acceptor.
FAD, FMN and sulphur–iron (Fe–S) clusters are common prosthetic groups in NADH:quinone oxidoreductases [2], where they typically show similar absorption maxima of 455 and 450 nm, respectively [21,35,36]. Purified MmNQO displayed yellow colour and spectral properties with an absorption maximum at 455 nm (Supplementary Table S1 and Supplementary Figure S4). Incubation with trichloroacetic acid resulted in the disappearance of the 455 nm signal, suggesting that the cofactor is tightly bound to the protein.
Enzyme kinetics
Michaelis–Mentens plots and kinetic constants were determined with either NADH (Table 2; Supplementary Figure S5) or NADPH (Table 3; Supplementary Figure S6) as electron donor using different electron acceptors. The apparent Km values for NADH and NADPH vary from about 17 to 258 μM depending on the electron-acceptor used, which is in the range previously observed for prokaryotic NADH:quinone oxidoreductases [4,37,38]. The turn-over rates (kcat) fall in the range 4.95–19.8 min−1, with NADH/DCPIP being the most efficient redox couple. DCPIP is known to be a good electron acceptor for prokaryotic NADH:quinone oxidoreductases [4,39,40], and the wild-type E. coli NADH:quinone oxidoreductase reduces coenzyme Q1, DCPIP and ferricyanide using NADH as redox partner [40]. Furthermore, as for MmNQO, E. coli NADH:quinone oxidoreductases display considerable variation in the kinetic constants depending on the electron acceptor used, as NDH-1 prefers ferricyanide as electron acceptor, whereas NDH-2, with higher affinity for NADH, prefers coenzyme Q1 [40]. With coenzyme Q1 as electron acceptor, MmNQO shows a Km value of 96.8 μM for NADH. This is more than 7-fold higher than the corresponding Km value for E. coli NADH:quinone oxidoreductase (14 μM; [40]), and almost 10-fold higher than that determined for Thermus thermophilus with the NADH/coenzyme Q1 substrate pair (10 μM; [41]). Based on the kinetic analyses, MmNQO shows a certain preference for NADH over NADPH. Moreover, NADH can be oxidized using all ten artificial electron acceptor substrates tested, whereas NADPH is only turned over in the presence of 1,4-BQ, coenzyme Q1 and potassium ferricyanide. These results are similar with what has been reported for wild-type E. coli NDH-1 [4], and to some extent also for Corynebacter glutamicum NADH:quinone oxidoreductase, which show higher affinity and turnover rates for NADH using coenzyme Q1 compared with NADPH [42].
Table 2. Kinetic constants of MmNQO with NADH and NADPH (0.1–150 μM) and different electron acceptors at saturating concentrations.
DCPIP, dichlorophenolindophenol; n.d., not determined (the affinity was too low and no reliable kinetic constants could be determined).
| NADH | Electron acceptor DCPIP | Km (μM) 17.5 | kcat (min−1) 4.95 | kcat/Km (μM−1·min−1) 0.283 |
|---|---|---|---|---|
| Coenzyme Q10 | 33.7 | 6.86 | 0.204 | |
| Potassium ferricyanide | 74.8 | 14.3 | 0.191 | |
| Fc+ | 64.8 | 11.2 | 0.173 | |
| 1,4-BQ | 56.5 | 9.74 | 0.172 | |
| Coenzyme Q1 | 96.8 | 15.6 | 0.161 | |
| Diethyl-oxalacetate sodium salt | 63.2 | 9.73 | 0.154 | |
| Sodium fumarate dibasic | 41.4 | 6.15 | 0.149 | |
| DL-malic acid | 102.5 | 14.2 | 0.139 | |
| Sodium thiosulphate | 257.9 | 19.8 | 0.077 | |
| NADPH | Coenzyme Q1 | 111.6 | 14.6 | 0.130 |
| 1,4-BQ | 48.1 | 6.0 | 0.125 | |
| Potassium ferricyanide | n.d. | n.d. | n.d. |
Stability of MmNQO activity was assessed at different pH values (2.5–10.0) using NADH and 1,4-BQ as electron donor and acceptor substrates, respectively. The enzyme was active in the pH range 4.5–8.5 with an optimum at 8.0 (Supplementary Figure S7).
Crystal structure of MmNQO
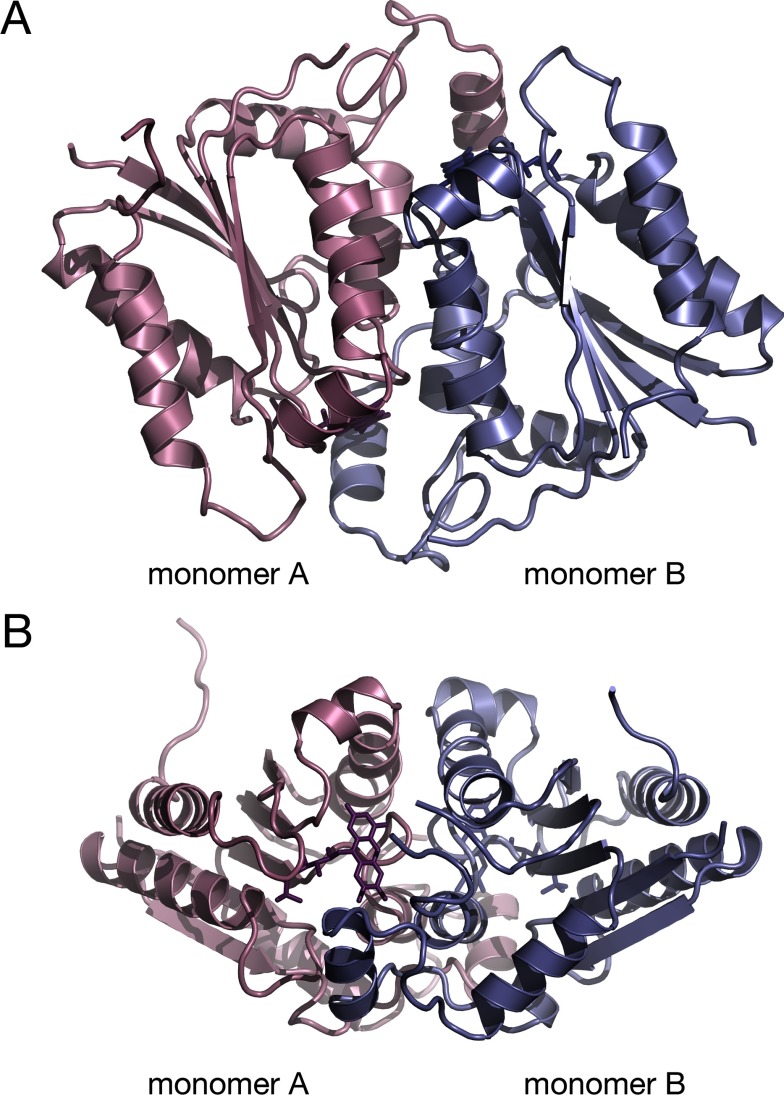
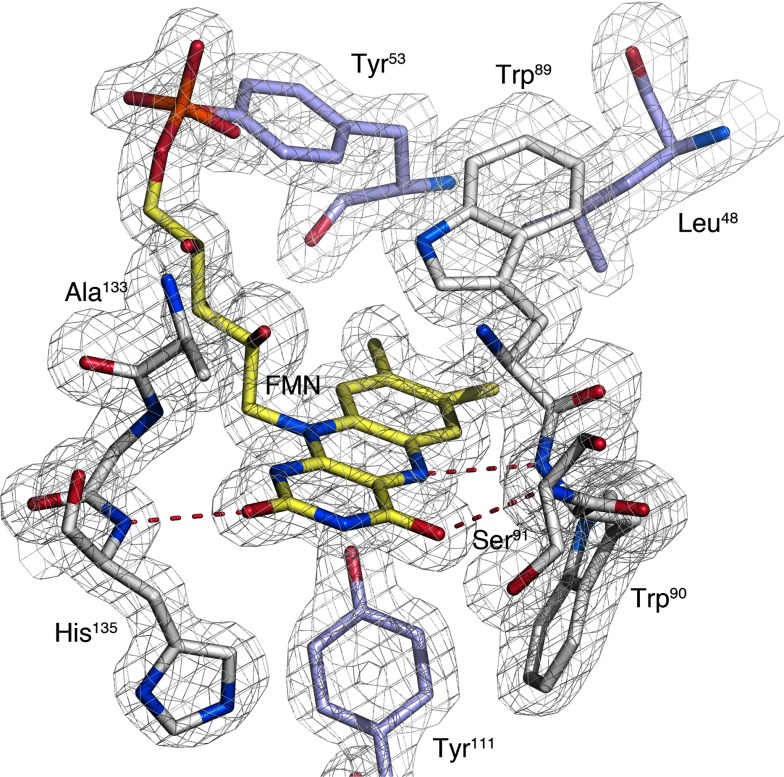
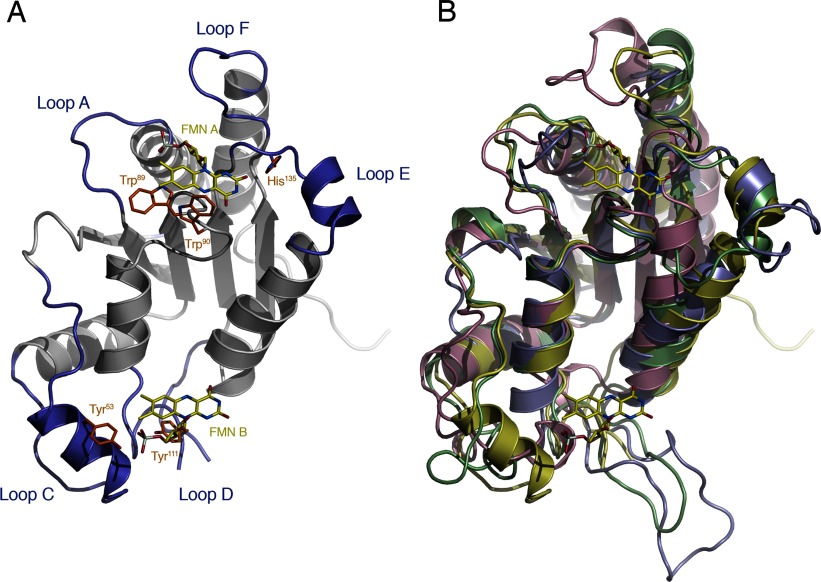
The crystal structure of MmNQO features a homodimeric enzyme where each monomer adopts the flavodoxin-like α/β-fold commonly observed for NAD(P)H:quinone oxidoreductases (Figure 1). The active sites of both monomers are situated at the dimer interface, each with a non-covalently bound FMN molecule bound approximately 8 Å below the molecule's surface. The flavin isoalloxazine ring is firmly bound by the protein (Figure 2) with its dimethylbenzene nucleus interacting with hydrophobic side chains (Trp89and Trp90 from molecule A; and Leu48 and Tyr53 from molecule B); the N5 atom of the central phenylenediamine ring forming a hydrogen bond to the backbone amide nitrogen of Trp90; and the O2 and O4 oxygen atoms in the pyrimidine of the isoalloxazine ring forming hydrogen bonds to the backbone amide groups of His135 and Ser91, respectively.
Figure 1. Overall fold of MmNQO.
(A) The flavodoxin-like fold of monomers A and B in the A/B dimer are coloured pink and blue, respectively. The FMN molecules are shown as stick models in darker colours. (B) Same as in panel (A) but rotated 180°.
Figure 2. FMN-binding pocket in the monomer of MmNQO.
The flavin-binding pocket in monomer A (white). The aromatic indole rings of Trp89 and Trp90 pack against the flavin ring (yellow). Two tyrosine and one leucine residue from the B monomer (blue) are shown.
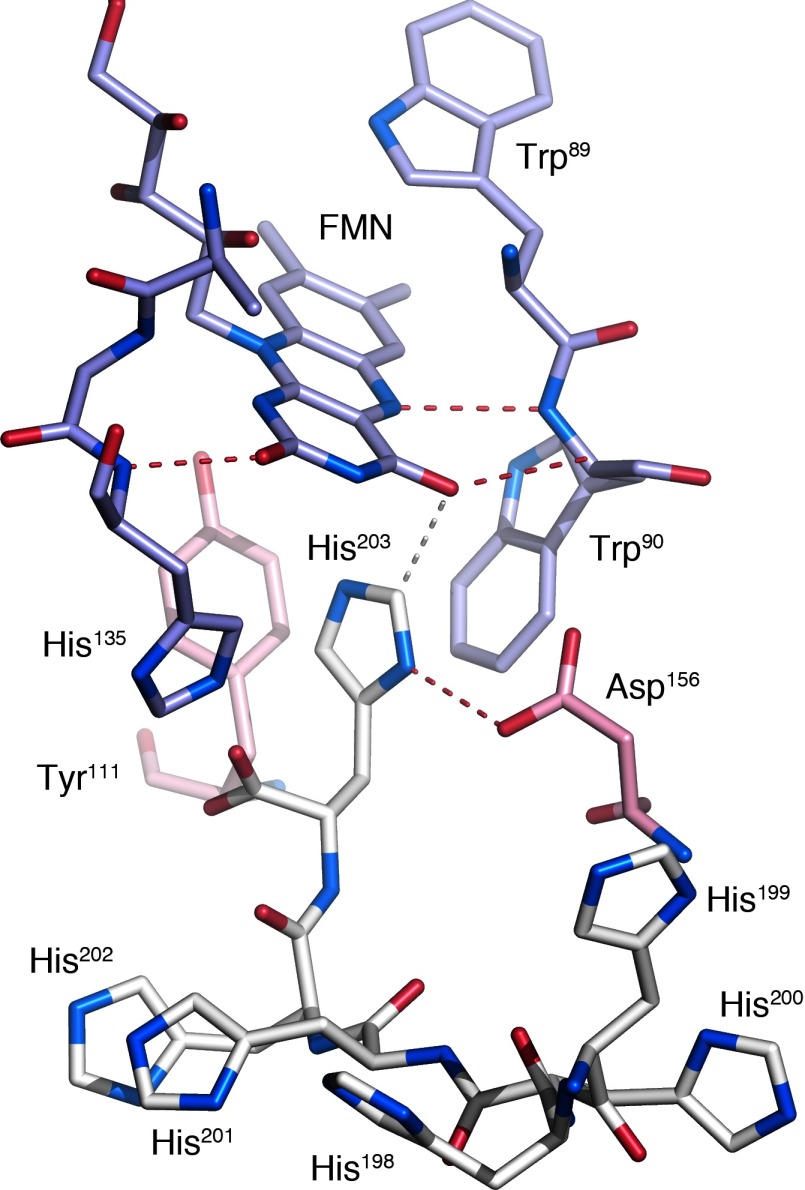
There are two non-crystallographically related homodimers (A/B and C/D) in the asymmetric unit of the crystal. In each dimer, one monomer (A in the A/B dimer and C in the C/D dimer) packs such that the ultimate histidine of the C-terminal hexahistidine tag is reaching into the flavin pocket to interact with the FMN ring in a crystallographically related molecule, i.e. His203 in molecule A interacts with the FMN in a crystallographically related D monomer and Asp156 in a crystallographically related C monomer (Figure 3). This interaction places the A-His203 CE1 atom 3.1 Å from the O4 atom of D-FMN. Similarly, His203 in molecule C interacts with crystallographically related C-Asp156 and B-FMN.
Figure 3. Interactions of the hexahistidine tag.
The hexahistidine tag (His198–His203) in molecule A (white) is interacting with the FMN-binding pocket of a crystallographically related C/D dimer (monomer C, pink; monomer D, blue). Red dashed lines represent hydrogen bonds and the white dashed line represents the separation between His203 CE1 and FMN O4.
Comparison of MmNQO with structurally related enzymes
Structure-based fold similarity analysis using the DALI Lite and PDBeFold servers returned three principal classes of NAD(P)H:quinone oxidoreductases: (1) modulator of drug activity B (MdaB); (2) bacterial FMN-dependent azoreductases (AzoR); and (3) mammalian FAD-dependent quinone reductases type 1 (QR1/NQR1) and 2 (QR2/NQR2). Additional protein structures occurred at lower scores, for instance, KefF of the KefC K+ efflux system, ChR (chromate reductase), EmoB of the EmoA/EmoB EDTA biodegradation system, and Trp-repressor-binding protein WrbA. The differences in sequence identity and r.m.s.d. (root-mean-square deviation) between the three principal groups are large according to the DALI analysis, typically 19–23% identity, and 2.3–2.8 Å r.m.s.d. on Cα positions. All structures display a structurally well-conserved homodimeric flavodoxin-like fold where the principal differences are noted for the precise length and conformation of mainly the dimer-interface loops delineating the flavin-binding pocket. It is thus difficult to speculate on the function of MmNQO based on global sequence comparison. The azoreductases, KefF, EmoB and ChR use FMN as cofactor, whereas NQRs and MdaBs use FAD.
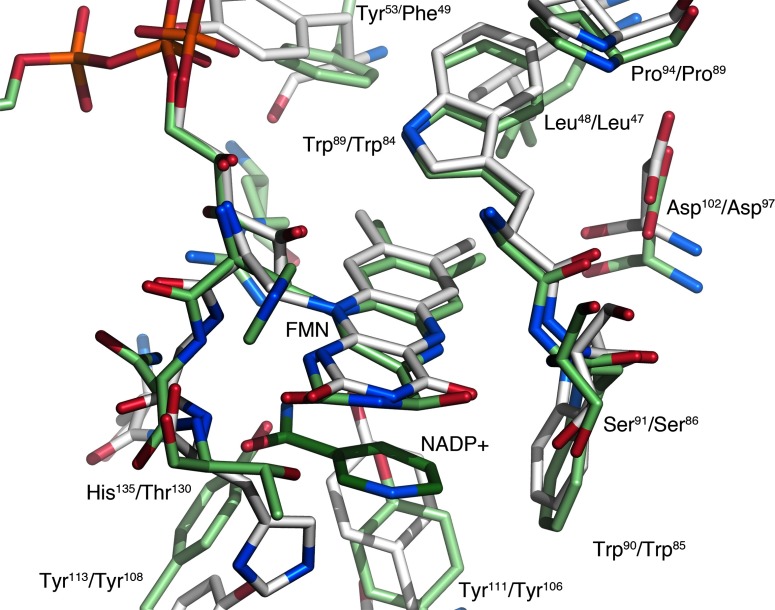
In the first group, the model of FAD-dependent MdaB from Streptococcus mutans scores as the most similar structure (PDB code 3LCM; [24]) and was thus used as search model in molecular replacement calculations. S. mutans MdaB is also the most similar structure at the local level in the immediate vicinity of the flavin. The side chains Trp89, Trp90, Tyr111, Tyr113, Asp102, Ser91 and Pro94 near the FMN group in MmNQO are all present in SmMdaB as Trp81, Trp82, Tyr106, Tyr108, Asp97, Ser86 and Pro89 (Figure 4). The higher local similarity in and near the flavin pocket suggests that MmNQO may play a similar role, or uses similar substrates. The function of MdaBs is not fully understood, but enzymes typically provide resistance towards certain drugs [43] and defense against oxidative stress [44]. SmMdaB has been confirmed to act as an FAD-dependent NADPH:quinone reductase [24]. MdaB can use menadione (vitamin K3) as electron acceptor, and binding of both NADP+ and menadione have been confirmed biochemically and crystallographically (PDB codes 3LCM and 4F8Y, respectively). Both ligands can also be accommodated in the active site of MmNQO, but with slight adjustments relative to the SmMdaB crystal structures.
Figure 4. Comparison of MmNQO with SmMdaB.
Overlay of the flavin pocket in MmNQO (white) and MdaB (light green) with part of the NADP+ molecule shown (dark green). Numbering of amino acids with MmNQO first.
Bacterial FMN-dependent azoreductases represent the second group. Although there are similarities to MmNQO in the active site, albeit lower than for MdaBs, closer inspection of existing azoreductase structures in complex with azo dyes and other ligands reveal important differences reducing the probability of MmNQO being an azoreductase. Structural comparisons with azoreductases in complex with cibacron blue (PDB code 3W78), methyl red (PDB code 2V9C and 3KEG), dicoumarol (PDB code 2Z9C), balsalazide (PDB code 3LT5), nitrofurazone (PDB code 3RW6) and anthraquinone-2-sulphonate (PDB code 4C0X), reveal that neither of these ligands can be easily accommodated in the MmNQO active site without steric hindrance. Cibacron blue is the most problematic ligand causing clashes with Trp90, His135 and Asp139 in monomer A, and Tyr111 and Tyr113 in monomer B of the dimer. For the other ligands, clashes are mainly with His135. Although local conformational changes may well change the precise details and relieve steric hindrance, His135 is very strategically positioned at one side of the FMN cofactor to restrict the substrate-binding region, a feature that does not exist in bacterial FMN azoreductases. To investigate this further, we tested two common azoreductase substrates, methyl red and dicoumarol, with MmNQO. Whereas no catalytic activity could be detected with dicoumarol, catalytic constants for methyl red were Km=683 μM and kcat=6.34 min−1 giving a very low specificity constant of 0.009 μM−1·min−1, which is 31-fold lower than that for NADH/DCPIP (Table 2). Thus, a role of MmNQO as an azoreductase is unlikely.
The third structurally related group is mammalian FAD-dependent quinone reductases. However, the active sites in mammalian NQRs have a lower local similarity to MmNQO than the MdaBs. A glycine in NQR1 and NQR2 is found at the position of His135 in MmNQO creating a more spacious active site at the pyrimidine side of the flavin ring. The only conserved side chain near the flavin ring is Trp90, which is occupied by Trp105 in NQR1 and NQR2.
As mentioned above, the quinone reductase most similar to MmNQO is the FAD-binding SmMdaB. Interestingly, the E. coli homologue of SmMdaB (EcMdaB; PDB code 2B3D [45]) is strikingly different than both MmNQO and SmMdaB. Based on pairwise structural alignment, the amino-acid sequence identity between MmNQO and SmMdaB is 23.5%, whereas the identity is only 16.5% between SmMdaB and EcMdaB (Figure 5), revealing a more distant relationship between the two MdaBs. In addition to bacterial MdaBs, MmNQO displays structural similarities to the FMN-dependent NAD(P)H:quinone oxidoreductase Saccharomyces cerevisiae Lot6p (former YLR011wp; PDB code 1T0I [46]). ScLot6p has been shown to act as an inducer of apoptosis [47] and is the only known quinone reductase in budding yeast [48]. The structure-based sequence identity between MmNQO and ScLot6p is 13.8%, which is slightly higher than the pairwise identity of 13.3% between MmNQO and EcMdaB.
Figure 5. Structure-based amino-acid sequence alignment.
Amino-acid sequence numbering is according to that of the coordinate files: S. mutans MdaB (SmMdaB; PDB code 3LCM), E. coli MdaB (EcMdaB; PDB code 2B3D), S. cerevisiae Lot6p (ScLot6p; PDB code 1T0I). Amino acids identical to MmNQO are gray-shaded. Pairwise identities based on structural alignment: MmNQO-SmMdaB, 23.5%; MmNQO-ScLot6p, 13.8%; MmNQO-EcMdaB, 12.8%; SmMdaB-ScLot6p, 13.3%; SmMdaB-EcMdaB, 16.5%; ScLot6p-EcMdaB, 7.3%. Amino acids highlighted by an asterisk: the restricting His135 in a subunit A at the flavin A Si-side; Trp89 and Trp90 in subunit A flanking the flavin A Re- and Si-side, respectively; Asp102 and Lys98 in subunit A forming a salt bridge at the flavin B Re-side; and subunit A Tyr111 at the flavin B Si-side. The five loop regions A–F discussed in the text are boxed.
When inspecting the structure-based sequence alignment for the four dimeric enzymes, six loop regions (loops A–F, Figures 5 and 6) reveal significant insertions or deletions. All loops except one (loop B) are involved in formation of the flavin- and substrate-binding region at the dimer interface. Loop A (residues 9–16 in MmNQO) binds the α-phosphate moiety in FAD or FMN and coincides with the flavodoxin loop. However, the flavodoxin fingerprint (T/S)XTGXT that is normally associated with this loop in flavodoxins is not present in any of the four enzymes. In EcMdaB, loop A is four residues longer compared with the other enzymes. The second loop, B, is present only in ScLot6p and is found at the face opposite to the dimer interface, and thus appears to have no role in flavin or substrate binding. Loop C (residues 44–67 in MmNQO) in one subunit closes off the dimethylbenze edge of the flavin pocket in the neighbouring subunit of the dimer. Loop C is absent altogether in EcMdaB, but its function appears to be partly compensated for by loop A (residues 23–28 in EcMdaB) that is absent in the other enzymes. Residues 104–124 (MmNQO) form an extensive D loop in MmNQO and MdaBs which is located at the Si-side of the flavin where it creates the ‘floor’ of the substrate-binding pocket. Loop D is absent in ScLot6p leaving the FMN fully exposed to the solvent. Loop E (134–145 in MmNQO) packs against the pyrimidine edge of the flavin in the same subunit. Also this loop is absent in ScLot6p. When the precise side chains that interact with the flavin ring are concerned, ScLot6p is clearly distinct from the MmNQO/MdaB group (Figure 5). Interestingly however, the restricting His135 in MmNQO is present also in ScLot6p (His127), but is displaced away from the flavin ring due to the absence of loop E. Loop F (173–182 in MmNQO) contributes to binding of the adenosine moiety in FAD. In the two FAD-binding MdaBs, this loop is folded away to accommodate the adenosine ring whereas in the FMN-binding MmNQO and ScLot6p loop F closes off around the phosphoribityl moiety of FMN.
Figure 6. Comparison of MmNQO with MdaBs and Lot6p.
(A) Ribbon drawing highlighting the six structurally variable loops (blue) in subunit A of MmNQO and side chains discussed in the text (orange) that interact with the FMN molecules in subunits A and B (yellow). The view is facing the dimer interface but for clarity subunit B has been omitted. Loop D in MmNQO is disordered without interpretable electron density and has not been modelled. (B) Structural superpositioning of MmNQO (yellow), SmMdaB (green; PDB code 3LCM), ScLot6p (pink; PDB code 1T0I) and EcMdaB (blue; PDB code 2B3D) with the FMN molecules in MmNQO shown.
It has been reported that during heterologous expression and purification of FAD-dependent human NQO2 in E. coli, a significant fraction of the enzyme binds either FMN or FAD, or a mixture thereof [49,50]. It is possible that the FMN bound to E. coli-expressed MmNQO may not represent the physiologically relevant co-factor. To evaluate the possible preference for FMN or FAD by MmNQO, a detailed comparison with the FAD-binding SmMdaB provides some information. At the sequence level, the region responsible for binding the adenosine part of FAD in SmMdaB is overall very similar in MmNQO, i.e. loop A (residues 9–15) and loop F (residues 173–182), including an important interaction offered by His9 to the FMN α-phosphate group. An additional interaction to the monophosphate is provided by Tyr53 in MmNQO, which is absent in SmMdaB. Although the overall sequence similarity of loops A and F is relatively high between the two enzymes, the conformation of loop F is fundamentally different. In the FMN-bound state of MmNQO, loop F is folded around the monophosphate moiety whereas in the FAD-bound state of SmMdaB, the F-loop is folded away to accommodate the adenosine ring. Whether this conformational difference reflects true differences in flavin co-factor specificity is not possible to deduce from the structure alone, but it should be emphasized that the density for the FMN is excellent and the existing interactions appear very favourable. ScLot6p also binds FMN, and as observed for MmNQO, loop F adopts a conformation that closes around the monophosphate group, which contrasts to the more open conformation observed for the FAD-binding enzymes SmMdaB and EcMdaB. Conceivably, loop F evolved an inherent ability to bind both FMN and FAD to allow either co-factor to be used by this group of enzymes.
Conclusions
The function of MmNQO inferred from structural similarity leans towards an MdaB-type activity, however it should be emphasized that the overall sequence identity is low, and r.m.s.d. values high. With respect to in vitro kinetics and structure, MmNQO shows similarities to several quinone reductase members such as MdaB, Lot6p, and NQO2. Structural comparisons are insufficient to conclusively answer whether MmNQO prefers FMN or FAD, and whether NADH or NADPH serves as coenzyme. Based on structural and biochemical data, we suggest that FMN and NADH are preferred, which justifies that MmNQO is annotated as an FMN-dependent NADH:quinone oxidoreductase. The biological function of MmNQO is yet to be elucidated, but from the data presented here it may constitute an archaeal counterpart of the modulator of drug activity B in bacteria. The NADH:quinone oxidoreductase activity of MmNQO and its localization to the cytoplasm suggest that it may be a useful tool for applications where E. coli cells require a high-energy metabolism and increased capacity of NADH regeneration, e.g. recombinant protein production processes. Hitherto, attempts to eliminate overflow metabolism in E. coli by overexpressing NADH:quinone oxidoreductases have been unsuccessful mainly due to the stress associated with overexpression of large integral membrane complexes. The comparably small size of soluble MmNQO makes it a potentially useful enzyme for regeneration of excessive NAD+ in bacterial hosts.
ACKNOWLEDGEMENTS
The authors want to thank Mr Simon Rittmann for providing M. marburgensis biomass and for his assistance in the first trial of extracting genomic DNA. The beamline staff scientists are acknowledged for support during data collection at beamline I03 at Diamond Light Source (U.K.). The work was facilitated by the Protein Science Facility at Karolinska Institutet/SciLifeLab (http://psf.ki.se).
AUTHOR CONTRIBUTION
Eva Ullmann and Thomas Gundinger performed cloning, expression and biochemical experiments. Christoph Herwig and Oliver Spadiut initiated, planned and supervised these experiments. Tien Chye Tan performed crystallization. Tien Chye Tan and Christina Divne processed the data and determined the crystal structure. Christina Divne refined and analysed the crystal. Oliver Spadiut was responsible for the experimental design of the biochemical assays and bioprocess experiments, and Christina Divne for the structural determination and analysis. All authors participated in writing the paper.
FUNDING
Funding by the Austrian Science Fund FWF [project number P24861-B19] and financial support to C.D. and T.C.T. from the Swedish Research Council FORMAS [grant number 2013-1741] and the Swedish Research Council VR [grant number 2011-5768] is acknowledged.
References
- 1.Hirst J. Mitochondrial complex I. Annu. Rev. Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 2.Anraku Y., Gennis R.B. The aerobic respiratory chain of Escherichia coli. Trends Biochem. Sci. 1987;12:262–266. doi: 10.1016/0968-0004(87)90131-9. [DOI] [Google Scholar]
- 3.Dancey G.F., Levine A.E., Shapiro B.M. The NADH dehydrogenase of the respiratory chain of Escherichia coli. I. Properties of the membrane-bound enzyme, its solubilization, and purification to near homogeneity. J. Biol. Chem. 1976;251:5911–5920. [PubMed] [Google Scholar]
- 4.Dancey G.F., Shapiro B.M. The NADH dehydrogenase of the respiratory chain of Escherichia coli. II. Kinetics of the purified enzyme and the effects of antibodies elicited against it on membrane-bound and free enzyme. J. Biol. Chem. 1976;251:5921–5928. [PubMed] [Google Scholar]
- 5.Friedrich T. The NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochim. Biophys. Acta. 1998;1364:134–146. doi: 10.1016/S0005-2728(98)00024-3. [DOI] [PubMed] [Google Scholar]
- 6.Melo A.M.P., Bandeiras T.M., Teixeira M. New insights into type II NAD(P)H:quinone oxidoreductases. Microbiol. Mol. Biol. Rev. 2004;68:603–616. doi: 10.1128/MMBR.68.4.603-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vemuri G.N., Eiteman M.A., Altman E. Increased recombinant protein production in Escherichia coli strains with overexpressed water-forming NADH oxidase and a deleted ArcA regulatory protein. Biotechnol. Bioeng. 2006;94:538–542. doi: 10.1002/bit.20853. [DOI] [PubMed] [Google Scholar]
- 8.Vemuri G.N., Eiteman M.A., McEwen J.E., Olsson L., Nielsen J. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2402–2407. doi: 10.1073/pnas.0607469104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akesson M., Karlsson E.N., Hagander P., Axelsson J.P., Tocaj A. On-line detection of acetate formation in Escherichia coli cultures using dissolved oxygen responses to feed transients. Biotechnol. Bioeng. 1999;64:590–598. doi: 10.1002/(SICI)1097-0290(19990905)64:5<590::AID-BIT9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 10.Berrios-Rivera S.J., Bennett G.N., San K.Y. Metabolic engineering of Escherichia coli: increase of NADH availability by overexpressing an NAD(+)-dependent formate dehydrogenase. Metab. Eng. 2002;4:217–229. doi: 10.1006/mben.2002.0227. [DOI] [PubMed] [Google Scholar]
- 11.Spehr V., Schlitt A., Scheide D., Guenebaut V., Friedrich T. Overexpression of the Escherichia coli nuo-operon and isolation of the overproduced NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 1999;38:16261–16267. doi: 10.1021/bi9919605. [DOI] [PubMed] [Google Scholar]
- 12.Wagner S., Klepsch M.M., Schlegel S., Appel A., Draheim R., Tarry M., Högbom M., van Wijk K.J., Slotboom D.J., Persson J.O., et al. Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wasserfallen A., Nölling J., Pfister P., Reeve J., Conway de Macario E. Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen. nov., and to reclassify several isolates in three species, Methanothermobacter thermautotrophicus comb. nov., Methanothermobacter wolfeii comb. nov., and Methanothermobacter marburgensis sp. nov. Int. J. Syst. Evol. Microbiol. 2000;50:43–53. doi: 10.1099/00207713-50-1-43. [DOI] [PubMed] [Google Scholar]
- 14.Liesegang H., Kaster A.K., Wiezer A., Goenrich M., Wollher A., Seedorf H., Gottschalk G., Thauer R.K. Complete genome sequence of Methanothermobacter marburgensis, a methanoarchaeon model organism. J. Bacteriol. 2010;192:5850–5851. doi: 10.1128/JB.00844-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaster A.K., Goenrich M., Seedorf H., Liesegang H., Wollher A., Gottschalk G., Thauer R.K. More than 200 genes required for methane formation from H(2) and CO(2) and energy conservation are present in Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus. Archaea. 2011;2011:973848. doi: 10.1155/2011/973848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Bernsel A., Viklund H., Hennerdal A., Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucl. Acids Res. 2009;37:W465–W468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schultz J., Milpetz F., Bork P., Ponting C.P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLisa M.P., Li J., Weigand W.A., Bentley W.E. Monitoring GFP-operon fusion protein expression during high cell density cultivation of Escherichia coli using an on-line optical sensor. Biotechnol. Bioeng. 1999;65:54–64. doi: 10.1002/(SICI)1097-0290(19991005)65:1<54::AID-BIT7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 20.Spadiut O., Posch G., Ludwig R., Haltrich D., Peterbauer C.K. Evaluation of different expression systems for the heterologous expression of pyranose 2-oxidase from Trametes multicolor in E. coli. Microb. Cell Fact. 2010;9:1–14. doi: 10.1186/1475-2859-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowal A.T., Morningstar J.E., Johnson M.K., Ramsay R.R., Singer T.P. Spectroscopic characterization of the number and type of iron-sulfur clusters in NADH:ubiquinone oxidoreductase. J. Biol. Chem. 1986;261:9239–9245. [PubMed] [Google Scholar]
- 22.Kabsch W. XDS. Acta Crystallogr. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Frosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z., Li L., Dong Y.H., Su X.D. Structural and biochemical characterization of MdaB from cariogenic Streptococcus mutans reveals an NADPH-specific quinone oxidoreductase. Acta Crystallogr. D. 2014;70:912–921. doi: 10.1107/S1399004713033749. [DOI] [PubMed] [Google Scholar]
- 25.Langer G., Cohen S.X., Lamzin V.S., Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/S0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Holm L., Rosenström P. Dali server: conservation mapping in 3D. Nucl. Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 30.Özkan P., Sariyar B., Ütkür Ö., Akman U., Hortacsu A. Metabolic flux analysis of recombinant protein overproduction in Escherichia coli. Biochem. Eng. J. 2005;22:167–195. doi: 10.1016/j.bej.2004.09.012. [DOI] [Google Scholar]
- 31.Insel G., Celikyilmaz G., Ucisik.Akkaya E., Yesiladali K., Cakar Z.P., Tamerler C., Orhon D. Respirometric evaluation and modeling of glucose utilization by Escherichia coli under aerobic and mesophilic cultivation conditions. Biotechnol. Bioeng. 2007;96:94–105. doi: 10.1002/bit.21163. [DOI] [PubMed] [Google Scholar]
- 32.Gustavsson R., Mandenius C.F. Soft sensor control of metabolic fluxes in a recombinant Escherichia coli fed-batch cultivation producing green fluorescence protein. Bioprocess. Biosyst. Eng. 2013;36:1375–1384. doi: 10.1007/s00449-012-0840-z. [DOI] [PubMed] [Google Scholar]
- 33.Knabben I., Regestein L., Marquering F., Steinbusch S., Lara A.R., Büchs J. High cell-density processes in batch mode of a genetically engineered Escherichia coli strain with minimized overflow metabolism using a pressurized bioreactor. J. Biotechnol. 2010;150:73–79. doi: 10.1016/j.jbiotec.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Lara A.R., Caspeta L., Gosset G., Bolivar F., Ramirez O.T. Utility of an Escherichia coli strain engineered in the substrate uptake system for improved culture performance at high glucose and cell concentrations: an alternative to fed-batch cultures. Biotechnol. Bioeng. 2008;99:893–901. doi: 10.1002/bit.21664. [DOI] [PubMed] [Google Scholar]
- 35.Pfenninger-Li X.D., Dimroth P. The Na(+)-translocating NADH:ubiquinone oxidoreductase from the marine bacterium Vibrio alginolyticus contains FAD but not FMN. FEBS Lett. 1995;369:173–176. doi: 10.1016/0014-5793(95)00745-U. [DOI] [PubMed] [Google Scholar]
- 36.Mains I., Power D.M., Thomas E.W., Buswell J.A. Purification of an NADH-(dichlorophenol-indophenol) oxidoreductase from Bacillus stearothermophilus. Biochem. J. 1980;191:457–465. doi: 10.1042/bj1910457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayashi M., Miyoshi T., Takashina S., Unemoto T. Purification of NADH-ferricyanide dehydrogenase and NADH-quinone reductase from Escherichia coli membranes and their roles in the respiratory chain. Biochim. Biophys. Acta. 1989;977:62–69. doi: 10.1016/S0005-2728(89)80009-X. [DOI] [PubMed] [Google Scholar]
- 38.Pohl T., Schneider D., Hielscher R., Stolpe S., Dömer K., Kohlstädt M., Böttcher B., Hellwig P., Friedrich T. Nucleotide-induced conformational changes in the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) Biochem. Soc. Trans. 2008;36:971–975. doi: 10.1042/BST0360971. [DOI] [PubMed] [Google Scholar]
- 39.Pruss B.M., Nelms J.M., Park C., Wolfe A.J. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsushita, Ohnishi K.T., Kaback H.R. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry. 1987;26:7732–7737. doi: 10.1021/bi00398a029. [DOI] [PubMed] [Google Scholar]
- 41.Yagi T. Bacterial NADH-quinone oxidoreductases. J. Bioenerg. Biomembr. 1991;23:211–225. doi: 10.1007/BF00762218. [DOI] [PubMed] [Google Scholar]
- 42.Nantapong N., Otofuji A., Migita C.T., Adachi O., Toyama H., Matsushita K. Electron transfer ability from NADH to menaquinone and from NADPH to oxygen of type II NADH dehydrogenase of Corynebacterium glutamicum. Biosci. Biotechnol. Biochem. 2005;69:149–159. doi: 10.1271/bbb.69.149. [DOI] [PubMed] [Google Scholar]
- 43.Chatterjee P.K., Sternberg N.L. A general genetic approach in Escherichia coli for determining the mechanism(s) of action of tumoricidal agents–application to Dmp-840, a tumoricidal agent. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8950–8954. doi: 10.1073/pnas.92.19.8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green L.K., La Flamme A.C., Ackerley D.F. Pseudomonas aeruginosa MdaB and WrbA are water-soluble two-electron quinone oxidoreductases with the potential to defend against oxidative stress. J. Microbiol. 2014;52:771–777. doi: 10.1007/s12275-014-4208-8. [DOI] [PubMed] [Google Scholar]
- 45.Adams M.A., Li Z. Modulator of drug activity B from Escherichia coli: crystal structure of a prokaryotic homologue of DT-diaphorase. J. Mol. Biol. 2006;359:455–465. doi: 10.1016/j.jmb.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 46.Liger D., Graille M., Zhou C.-Z., Leulliot N., Quevillon-Cheruel S., Blondeau K., Janin J., van Tilbeurgh H. Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)H-FMN and ferric iron reductase activities. J. Biol. Chem. 2004;279:34890–34897. doi: 10.1074/jbc.M405404200. [DOI] [PubMed] [Google Scholar]
- 47.Sollner S., Durchschlag M., Fröhlich K.-U., Macheroux P. The redox-sensing quinone reductase Lot6p acts as an inducer of yeast apoptosis. FEMS Yeast Res. 2009;9:885–891. doi: 10.1111/j.1567-1364.2009.00546.x. [DOI] [PubMed] [Google Scholar]
- 48.Megarity C.F., Looi H.K., Timson D.J. The Saccharomyces cerevisiae quinone oxidoreductase Lot6p: stability, inhibition and cooperativity. FEMS Yeast Res. 2014;14:797–807. doi: 10.1111/1567-1364.12167. [DOI] [PubMed] [Google Scholar]
- 49.Leung K.K.K., Litchfield D.W., Shilton B.H. Flavin adenine dinucleotide content of quinone reductase 2: analysis and optimization for structure-function studies. Anal. Biochem. 2012;420:84–89. doi: 10.1016/j.ab.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 50.Megarity C.F., Gill J.R.E., Caraher M.C., Stratford I.J., Nolan K.A., Timson D.J. The two common polymorphic forms of human NRH-quinone oxidoreductase 2 (NQO2) have different biochemical properties. FEBS Lett. 2014;588:1666–1672. doi: 10.1016/j.febslet.2014.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]