Abstract
Interferon-induced transmembrane (IFITM) proteins inhibit the infection of a wide range of viruses including human immunodeficiency virus type 1 (HIV-1). At present, little is known about how viruses overcome IFITM restriction. In this study, we have utilized HIV-1 as a model and selected IFITM1-resistant viruses after multiple passages of HIV-1 in IFITM1-expressing SupT1 cells. Sequencing the entire viral genome revealed several mutations in the vpu and envelope genes, among which mutations Vpu34 and EnvG367E together enable efficient HIV-1 replication in IFITM1-expressing cells. Vpu34 introduces a stop codon at amino acid position 35 of Vpu, whereas EnvG367E changes the G367 residue at the CD4-binding site of gp120. These two mutations do not appear to overcome the downregulation of viral p24 expression caused by IFITM1, but rather enhance HIV-1 replication by promoting cell-to-cell virus transmission. Altogether, our data demonstrate that HIV-1 can mutate to evade IFITM1 restriction by increasing cell-to-cell transmission.
Key words: HIV-1, IFITM1, Cell-to-cell transmission, Escape mutations
Highlights
-
•
IFITM1 inhibits HIV-1 replication in SupT1 cells.
-
•
HIV-1 evolves to escape IFITM1 inhibition by mutating Vpu and Env.
-
•
The Vpu and Env mutations overcome IFITM1 through promoting HIV-1 cell to cell transmission.
Introduction
Interferon (IFN) inhibits virus infection through inducing the expression of hundreds of cellular genes collectively known as interferon-stimulated genes (ISGs) (Sadler and Williams, 2008). The antiviral functions of some of these ISGs have been well characterized. Examples include the RNase L and OAS proteins, protein kinase R (PKR), myxovirus-resistance (Mx) proteins, ISG15, APOBEC3G (apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G), TRIM5α (Tripartite motif-containing protein 5α), tetherin, SAMHD1 (SAM domain and HD domain-containing protein 1) and Schlafen 11 (Hrecka et al., 2011, Laguette et al., 2011, Li et al., 2012, Neil et al., 2008, Sadler and Williams, 2008, Stremlau et al., 2004). A recent addition to this list is the interferon-induced transmembrane (IFITM) proteins which inhibit a number of highly pathogenic human viruses (Brass et al., 2009, Huang et al., 2011, Jiang et al., 2010, Lu et al., 2011, Schoggins et al., 2011, Weidner et al., 2010). IFITM proteins are distinct from all other ISGs in that they inhibit viral entry, particularly the step of membrane fusion (Brass et al., 2009, Feeley et al., 2011, Huang et al., 2011, Lu et al., 2011) (reviewed in (Diamond and Farzan, 2013; Perreira et al., 2013)).
Humans have IFITM1, 2, 3, 5 and 10, all of which are located on chromosome 11 (Hickford et al., 2012). IFITM1, 2, 3 and 5 are clustered in a 26.5 kb region, whereas IFITM10 is located 1.4 Mb apart. IFITM5 is also called bone-restricted IFITM-like (Bril) protein due to its strict expression in osteoblasts and its role in bone mineralization and maturation (Hanagata et al., 2010, Moffatt et al., 2008). The function of IFITM10 is unknown, but it is highly conserved in different species (Hickford et al., 2012). IFITM1, 2 and 3 have been shown to promote anti-proliferation, homotypic cell adhesion, and apoptosis (Brem et al., 2003, Deblandre et al., 1995, Evans et al., 1993, Evans et al., 1990). They are also overexpressed in some tumor cells, such as human colorectal tumors, suggesting a possible role in oncogenesis (Andreu et al., 2006, Siegrist et al., 2011). In addition to their ubiquitous expression in different tissues (Seo et al., 2010, Yamashita et al., 2000), IFITM1, 2 and 3 also respond to the stimulation by interferon (Lewin et al., 1991), supporting their roles in host antiviral defense. Indeed, IFITM1, 2 and 3 have been reported to inhibit the infections by influenza A virus (IAV), flavivirus (West Nile virus, Dengue virus, and Yellow Fever virus), filovirus (Ebola virus and Marburg virus), SARS coronavirus, vesicular stomatitis virus (VSV), hepatitis C virus (HCV), reovirus, Rift Valley fever virus, as well as human immunodeficiency virus type 1 (HIV-1) (Anafu et al., 2013, Brass et al., 2009, Feeley et al., 2011, Jiang et al., 2010, Lu et al., 2011, Mudhasani et al., 2013, Raychoudhuri et al., 2011, Schoggins et al., 2011, Weidner et al., 2010). In addition to this broad antiviral activity, knockout of ifitm3 in mice or IFITM3 deficiency in humans renders the hosts highly vulnerable to IAV infection (Bailey et al., 2012, Everitt et al., 2012, Wakim et al., 2013), highlighting the importance of IFITM proteins in host antiviral defense in vivo.
Human IFITM1, 2 and 3 are of 125, 132 and 133 amino acids in length, respectively. They are predicted to have two transmembrane domains (Siegrist et al., 2011). Results of cell-surface immunostaining and flow cytometry experiments suggest that their amino- and carboxy-termini project toward the extracellular space or luminal compartments (Brass et al., 2009, Weidner et al., 2010). However, recent evidence also supports the cytoplasmic localization of the N-terminus (Bailey et al., 2013, Yount et al., 2012). In addition to the plasma membrane, IFITM proteins are also observed in the endoplasmic reticulum (ER) and endosomes (Alber and Staeheli, 1996, Brass et al., 2009, Feeley et al., 2011, Jia et al., 2012, Lu et al., 2011, Yang et al., 2007, Yount et al., 2010, Zucchi et al., 2004). The localization of IFITM3 in late endosomes is important for inhibiting IAV infection, because ectopic expression of IFITM3, or its induced expression by interferon, causes expansion of late endosomes and lysosomes and results in the sequestration of endocytosed IAV particles in these acidic membrane compartments (Feeley et al., 2011, Huang et al., 2011). By taking advantage of lipid analogs and fluorescence labeling, we recently showed that oleic acid (OA), but not chlorpromazine (CPZ), rescues the inhibitory effect of IFITMs on cell-to-cell fusion induced by Jaagsiekte sheep retrovirus (JSRV) Env and IAV hemagglutinin (HA), indicating that IFITM proteins interfere with the hemifusion stage of virus entry, possibly by changing membrane fluidity and curvature (Li et al., 2013). This conclusion is further strengthened by the fact that IFITM proteins increase lipid order of membranes (Li et al., 2013). This latter property of IFITM proteins is at least partially attributed to their interaction with VAPA (vesicle-membrane-protein-associated protein A) and consequent disturbance of cholesterol homeostasis (Amini-Bavil-Olyaee et al., 2013).
Viruses often evolve mechanisms to evade or antagonize host restrictions (Malim and Bieniasz, 2012), and this strategy should also be operative for the IFITM proteins. Indeed, HCV infection increases the expression of miR-130a that targets the 3′ untranslated region of IFITM1 mRNA and thus diminishes IFITM1 expression (Bhanja Chowdhury et al., 2012). Additionally, arenaviruses, which require low pH for entry, are refractory to IFITM restriction (Brass et al., 2009), although the underlying mechanism still remains unclear. In order to better understand the viral evasion of IFITM restriction, we investigated whether HIV-1 can develop resistance to IFITM1 in CD4+ SupT1 cells. The results showed that long-term culture led to the emergence of IFITM1-resistant HIV-1 mutants, and the escape mutations were mapped to the viral Vpu and Env proteins.
Results
HIV-1 mutates to escape from the inhibition by IFITM1 in SupT1 cells
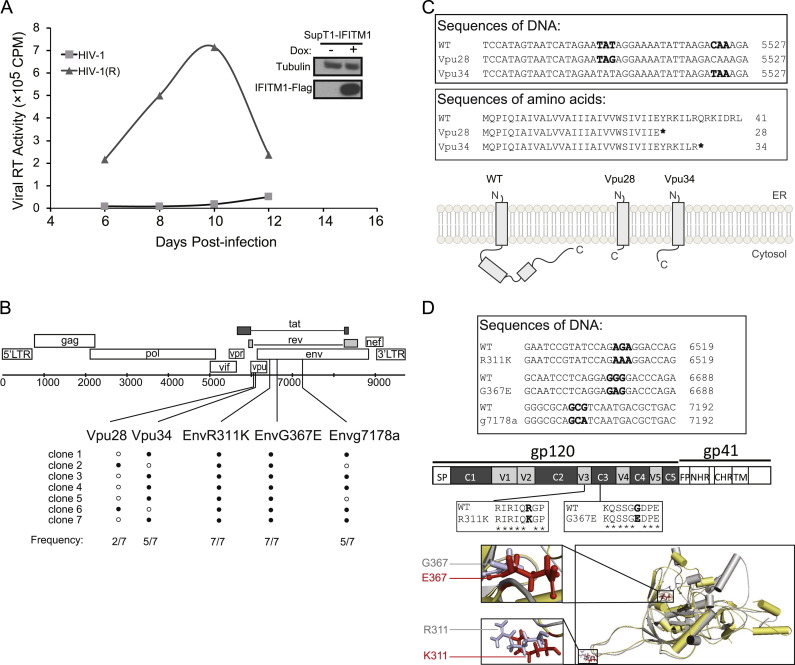
We previously reported that IFITM1, 2 and 3 suppressed HIV-1 replication in SupT1 cells, with IFITM1 exhibiting the greatest inhibition (Lu et al., 2011). In order to investigate whether HIV-1 is able to develop resistance to IFITM restriction, we grew HIV-1 in IFITM1-expressing SupT1 cells and observed that the virus gradually became refractory to IFITM1 inhibition and replicated to high levels ( Fig. 1A). As a control, we also grew HIV-1 in SupT1 cells without ectopic expression of IFITM1 for the same time interval. We then sequenced the entire genomes of these two virus populations. Five mutations were identified only in IFITM1-resistant viruses, not in those that had replicated in the control SupT1 cells (Fig. 1B). Two mutations are located in Vpu, namely Vpu28 and Vpu34, respectively. Vpu28 was seen in two out of the seven sequenced viral DNA clones, Vpu34 in five clones, indicating that the virus either carried the Vpu28 or the Vpu34 mutation. Both mutations created a stop-codon, resulting in the premature termination of Vpu translation (Fig. 1C). The other three mutations were found in the Env coding region (Fig. 1B). EnvR311K resides in the variable loop 3 (V3), which is critical for co-receptor-binding (Fig. 1D). EnvG367E is in the CD4-binding site in constant region 3 (C3) (Fig. 1D). The g7178a mutation does not cause any amino acid change in envelope, nor in the Rev or Tat proteins. This mutation was found in five out of the seven sequenced DNA clones. These data suggest that HIV-1 is able to escape from IFITM1 inhibition by mutating its Vpu and Env proteins.
Fig. 1.
Identification of escape mutations. (A) Replication of wild type HIV-1 and the escape viruses named HIV-1(R) in IFITM1-expressing SupT1 cells. The same amounts of viruses were used to infect SupT1 cells expressing IFITM1 with induction by doxycycline. Amounts of viruses in the supernatants were determined by measuring the viral reverse transcriptase activity. The expression of IFITM1-Flag in SupT1 cells was examined by Western blotting with doxycycline (Dox) induction. (B) Illustration of the escape mutations in HIV-1 genome. Seven viral DNA clones were sequenced. The frequency of each mutation in seven clones is shown. (C) Vpu28 and Vpu34 mutations cause premature termination of Vpu translation. Part of the DNA sequences and amino acid sequences of the wild type and mutated Vpu are shown. The Vpu fragments of the Vpu28 and Vpu34 mutants are illustrated. (D) Mutations EnvR311K and EnvG367E are located in the V3 and C3 regions of envelope protein, respectively. The g7178a mutation does not cause amino acid change. Part of the DNA sequences and amino acid sequences of the EnvR311K and EnvG367E mutations are presented. Locations of these two mutations in the gp120 structural model are shown. The structural models were generated using the I-TASSER server for protein 3D structure prediction (Zhang, 2008).
The Vpu34 and EnvG367E mutations rescue HIV-1 replication in IFITM1-expressing SupT1 cells
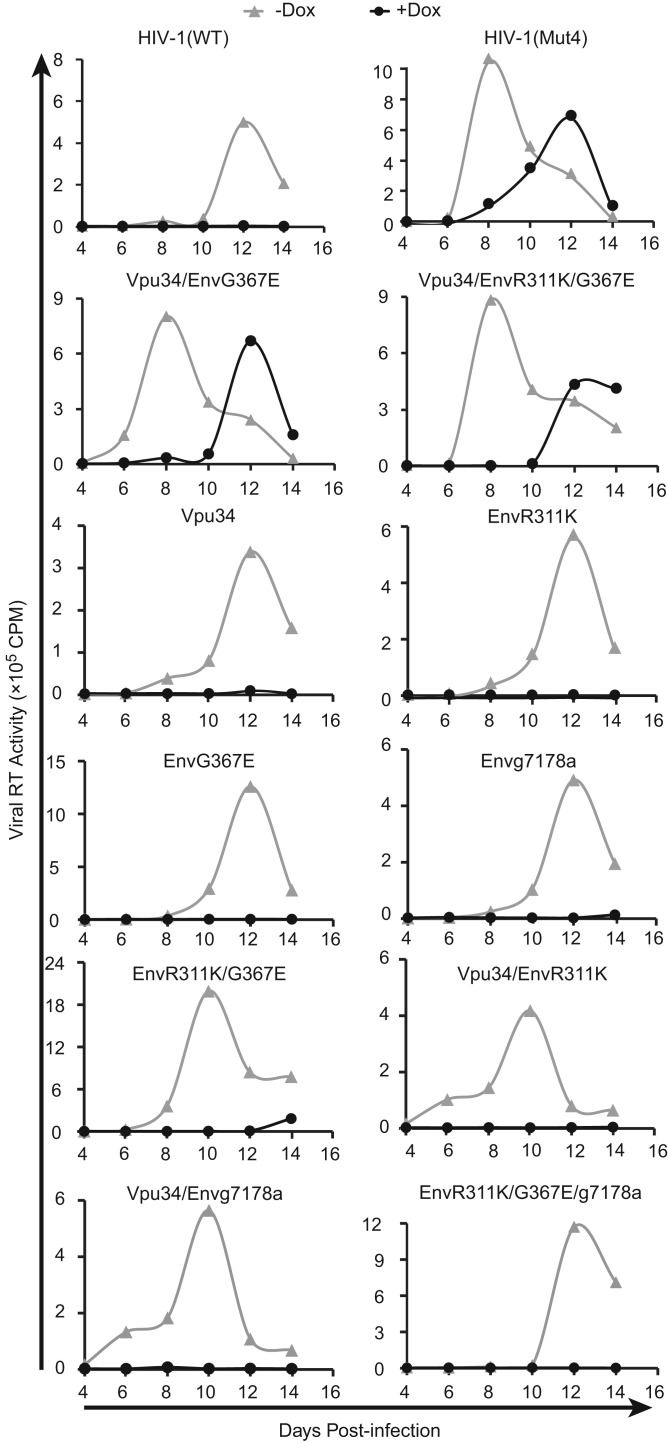
We next asked which of these five mutations are sufficient to rescue the IFITM1 inhibition of HIV-1. Since Vpu28 and Vpu34 mutations both resulted in truncation of Vpu, and Vpu34 was seen in five out of seven viral DNA clones, we chose to further characterize Vpu34 by performing the following experiments. We inserted Vpu34, EnvR311K, EnvG367E and g7178a into the HIV-1 proviral DNA either individually or in different combinations, and generated 11 mutated HIV-1 DNA clones ( Table 1). We then transfected these viral DNA clones into HEK293T cells to produce viruses, and used viruses of the same viral p24 amounts to infect SupT1 cells that were induced with doxycycline to express IFITM1. Infection experiments were also performed without doxycycline as the control for the IFITM1-negative condition. In the absence of IFITM1 expression, HIV-1(Mut4) (containing Vpu34, EnvR311K, EnvG367E and g7178a mutations), Vpu34/EnvG367E, and Vpu34/EnvR311K/G367E grew slightly faster than the wild type virus and other viruses ( Fig. 2). With IFITM1 expression, viruses HIV-1(Mut4), Vpu34/EnvR311K/G367E, and Vpu34/EnvG367E exhibited much robust replication, in strong comparison to the wild type and other mutants which were inhibited by IFITM1 (Fig. 2). We thus conclude that the Vpu34 and EnvG367E mutations together are sufficient to overcome IFITM1 inhibition, and that EnvR311K and g7178a do not play an important role.
Table 1.
Mutations in each HIV-1 mutant.
| HIV-1 mutants |
Mutations |
|||
|---|---|---|---|---|
| Vpu34 | R311K | G367E | g7178a | |
| HIV-1(Mut4) | √ | √ | √ | √ |
| Vpu34 | √ | |||
| EnvR311K | √ | |||
| EnvG367E | √ | |||
| Envg7178a | √ | |||
| EnvR311KG367E | √ | √ | ||
| Vpu34/EnvR311K | √ | √ | ||
| Vpu34/EnvG367E | √ | √ | ||
| Vpu34/Envg7178a | √ | √ | ||
| EnvR311K/G367E/g7178a | √ | √ | √ | |
| Vpu34/EnvR311K/G367E | √ | √ | √ | |
Fig. 2.
Mutations of Vpu34 and EnvG367E rescue HIV-1 replication in IFITM1-expressing SupT1 cells. The mutations Vpu34, EnvR311K, EnvG367E and g7178a were inserted into HIV-1 DNA either individually or in different combinations. The HIV-1(Mut4) virus contains all four mutations. These viruses were used to infect SupT1 cells with or without IFITM1 induction by doxycycline. Viral replication was determined by measuring levels of viral reverse transcriptase activity in culture supernatants. Results shown represent three independent infection experiments.
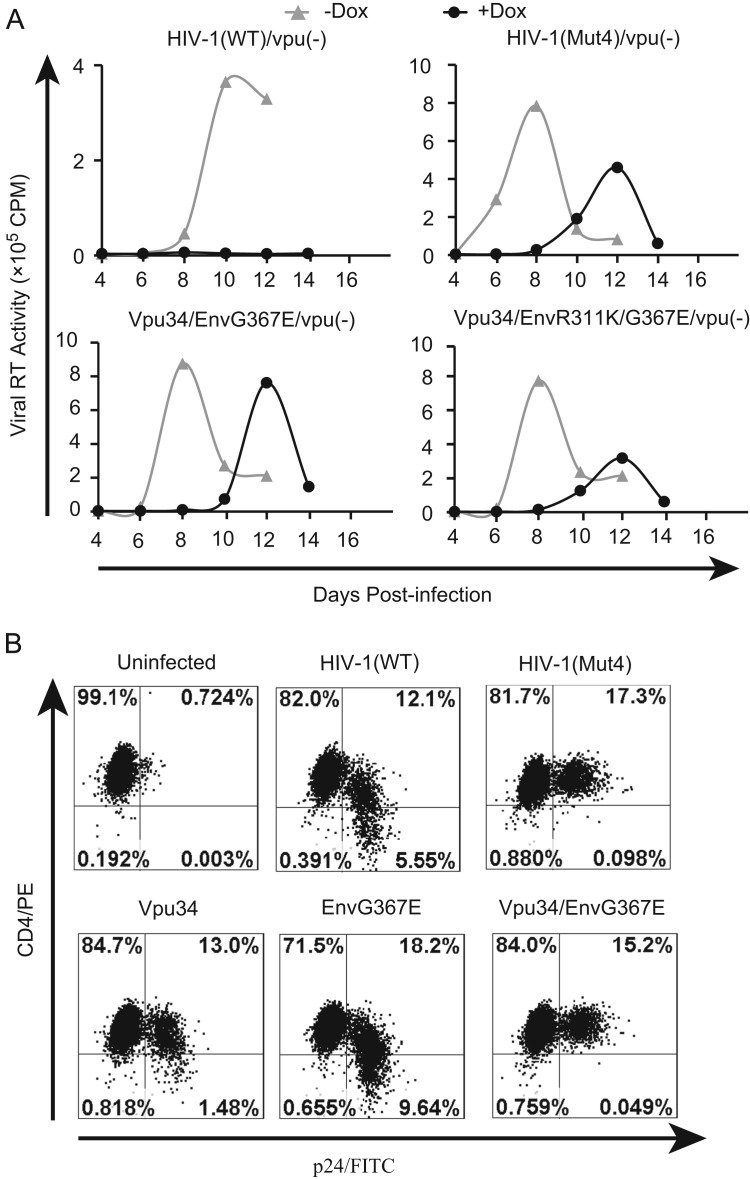
The Vpu34 mutation allows the expression of the first 34 amino acids of Vpu (Fig. 1C). In order to test whether this truncated form of Vpu contributes to the rescue of HIV-1, we mutated the translation start codon AUG of Vpu to ACG in order to completely eliminate the Vpu expression. We inserted this Vpu(−) mutation into the wild type and the mutated HIV-1 DNA, and generated mutants HIV-1/Vpu(−), HIV-1(Mut4)/Vpu(−), Vpu34/EnvG367E/Vpu(−), and Vpu34/EnvR311K/G367E/Vpu(−). We observed that, in contrast to the complete inhibition of HIV-1/Vpu(−) by IFITM1, the other three viruses replicated efficiently ( Fig. 3A). We further showed that the Vpu34 mutation was sufficient to block the downregulation of cell surface CD4 by HIV-1 (Fig. 3B). Therefore, the loss of Vpu expression, but not the expression of the first 34 amino acids of Vpu, enables the virus to escape IFITM1 inhibition.
Fig. 3.
Deleting Vpu facilitates HIV-1 to escape IFITM1 inhibition. (A) Abolishing Vpu expression does not affect the ability of Vpu34 mutation to rescue HIV-1 replication in IFITM1-expressing SupT1 cells. The first ATG of Vpu was changed to ACG, which is named Vpu(−). This Vpu(−) mutation was inserted into viral DNA clones HIV-1(Mut4), Vpu34/EnvG367E, and Vpu34/R311K/EnvG367E such that the expression of the first 34 amino acids of Vpu in these viruses are eliminated. Virus replication in SupT1 cells, with or without IFITM1 induction, was monitored by measuring levels of viral reverse transcriptase activity in culture supernatants. One representative result of three independent infections is shown. (B) The Vpu34 mutation abolishes the ability of HIV-1 to downregulate cell surface CD4. The wild type HIV-1 and its mutants were used to infect SupT1 cells. Forty hours after infection, cells were stained with PE-conjugated anti-CD4 antibody and FITC-conjugated anti-p24 antibody followed by flow cytometry analysis. Results shown represent three independent experiments.
The Vpu34 and EnvG367E mutations do not rescue the defect in HIV-1 p24 expression caused by IFITM1
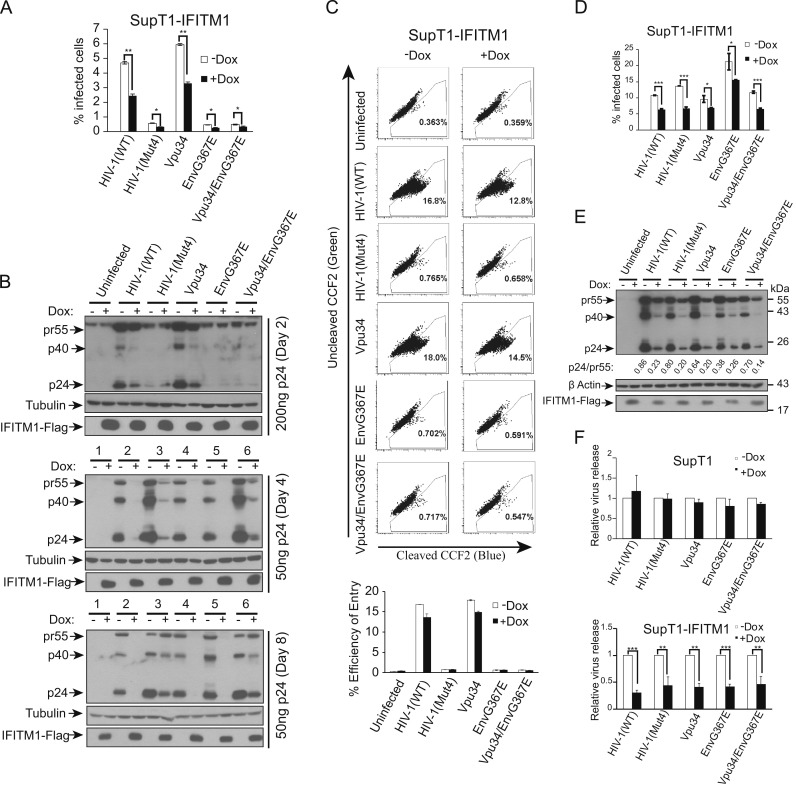
We next performed short-term infection to investigate the rescue mechanism conferred by Vpu34 and EnvG367E. We have previously shown that IFITM1 decreases the expression of HIV-1 p24 (Lu et al., 2011), an observation that is further supported by the study of Chutiwitoonchai et al. (2013). To test whether the Vpu34 and EnvG367E mutations can overcome this defect, SupT1 cells were infected with the same amounts of wild type and mutated HIV-1 particles, and viral Gag/p24 expression was measured by staining cells with FITC-conjugated anti-p24 antibody. In the absence of IFITM1 expression, viruses HIV-1(Mut4), Vpu34/EnvG367E and EnvG367E generated 10-fold fewer p24-positive SupT1 cells than the wild type virus and the Vpu34 mutant ( Fig. 4A). This observation was confirmed by the results of Western blots measuring cell-associated HIV-1 Gag and p24 (Fig. 4B, 200 ng p24 virus infection). This deficiency was likely a result of the impaired virus entry, since results of HIV-1 virion fusion experiments showed that the HIV-1(Mut4), Vpu34/EnvG367E and EnvG367E mutants were more than 10 fold less efficient in entering cells than the wild type and the Vp34 mutant (Fig. 4C). Given that HIV-1(Mut4), Vpu34/EnvG367E and EnvG367E all bear the EnvG367E mutation, we suspected that this envelope protein mutation likely have dramatically diminished HIV-1 infectivity. Supporting this speculation, we found that this defect in infection was fully corrected when the wild type and mutated viruses carried the VSV G protein that is not affected by IFITM1 (Weidner et al., 2010) (Fig. 4D). Interestingly, when IFITM1 was induced to express, both the wild type and the mutated viruses either bearing HIV-1 envelope or VSV G protein, still gave 50% fewer p24-positive SupT1 cells (Fig. 4A and D). This decrease is more pronounced when levels of cell-associated HIV-1 p24 were measured by Western blotting, and this decrease persisted until later time points of infection such as day 4 and day 8 (Fig. 4B). In addition, the results of Western blots showed decreased ratio of p24 to pr55 in IFITM1-expressing cells (Fig. 4E), which indicates a defect in Gag processing. These defects were further reflected by a similar reduction in virus particles that were released into the supernatants (Fig. 4F). Therefore, the Vpu34 and EnvG367E mutations are unable to repair the defect in viral p24 expression caused by IFITM1, suggesting that they must have rescued HIV-1 replication in IFITM1-expressing SupT1 cells by a new mechanism.
Fig. 4.
The Vpu34 and EnvG367E mutations do not overcome Gag downregulation caused by IFITM1 in the short-term infection of HIV-1. (A) The wild type and mutant viruses were used to infect SupT1 cells with or without IFITM1 induction. Forty hours after infection, the infected cells were stained with FITC-conjugated anti-p24 antibody and scored by flow cytometry. Results of three independent infections are summarized in the bar graph. The p values were calculated and the significance is indicated by ⁎ (<0.05) and ⁎⁎ (<0.01). (B) The infected cells were collected 2, 4 and 8 days after infection. Levels of HIV-1 Gag/p24 were examined in Western blots. (C) The cell entry efficiency of wild type and mutant viruses were examined by Blam-Vpr virion fusion assay. The cleavage of CCF2/AM by Blam-Vpr was measured by flow cytometry. Results of three independent infections are summarized in the bar graph. (D) The wild type and mutant HIV-1 were pseudotyped with VSV G protein and used to infect SupT1 cells with or without IFITM1 induction. Forty hours after infection, the infected cells were stained with FITC-conjugated anti-p24 antibody and scored by flow cytometry. Results of three independent infections are summarized in the bar graph. The p values were calculated and the significance is indicated by ⁎ (<0.05) and ⁎⁎⁎ (<0.001). (E) Levels of viral Gag/p24 expression in the infected cells were determined by Western blotting. The intensities of pr55 and p24 protein bands were determined with the Image J software (NIH). The ratios of p24 to pr55 were calculated and shown below the Western blot. (F) Amounts of viruses in the culture supernatants were determined by measuring viral reverse transcriptase activity. Virus amount that was produced by the wild type HIV-1 in the absence of doxycycline induction is arbitrarily set as 1. Results shown are the averages of three independent infections. The p values were calculated and the significance is indicated by ⁎⁎ (<0.01) and ⁎⁎⁎ (≤0.001).
The EnvG367E mutation impairs the usage of CD4 receptor
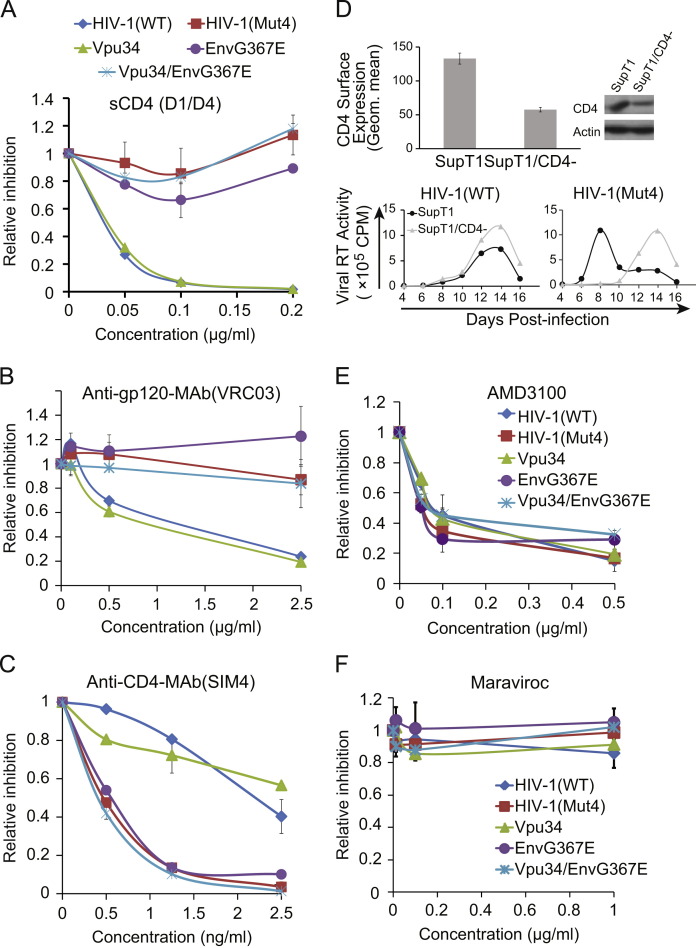
It is not surprising that the EnvG367E mutant is poorly infectious because the EnvG367E mutation alters the conserved G367 amino acid at the CD4-binding site (Fig. 1D). Such a mutation would be expected to diminish the affinity of envelope for CD4. Indeed, we observed that as little as 0.1 µg/ml of soluble CD4 (sCD4(D1/D4)) was able to reduce the infection of wild type HIV-1 and the Vpu34 virus by 10-fold, as opposed to less than 30% decrease for the EnvG367E, Vpu34/EnvG367E and HIV-1(Mut4) viruses ( Fig. 5A). We further tested the usage of CD4 receptor using an antibody named VRC03 that recognizes the CD4-binding site on gp120 (Wu et al., 2010, Zhou et al., 2010). Again, viruses HIV-1(Mut4), EnvG367E and Vpu34/EnvG367E, which all carry the EnvG367E mutation, exhibited greater resistance to VRC03 inhibition than the wild type virus (Fig. 5B).
Fig. 5.
The EnvG367E mutation diminishes the usage of CD4 receptor. (A) The same amounts of wild type or mutated HIV-1 were used to infect the CEM-Rev-Luc indicator cells in the presence of increasing amounts of soluble CD4 (sCD4). Virus infection was determined by measuring levels of luciferase activity in the infected CEM cells. Infection by each virus without sCD4 is arbitrarily set as 1. Results are the averages of three independent infections. (B) Sensitivity of the wild type and HIV-1 mutants to the inhibition by the VRC03 antibody that recognizes the CD4-binding site on gp120. (C) Inhibition of the wild type and mutated viruses by the anti-CD4 antibody SIM4. (D) Knockdown of CD4 delays the replication of HIV-1(Mut4). The shRNA targeting CD4 mRNA was used to create a stable SupT1 cell line. The cell surface level of CD4 was determined by staining with anti-CD4 antibody followed by flow cytometry, the result is presented in the bar graph. The total amount of CD4 was assessed by Western blotting. Replication of the wild type and the HIV-1(Mut4) viruses was examined in the CD4-knockdown SupT1 cells and the control SupT1 cells by measuring levels of viral reverse transcriptase. (E) Inhibition of the wild type and mutated viruses by the CXCR4 antagonist AMD3100. (F) Sensitivity of the wild type and mutated viruses to the CCR5 antagonist maraviroc.
We speculated that the diminished usage of CD4 by the EnvG367E mutant may render the virus more sensitive to the cell surface CD4 level. To test this possibility, we first used the CD4 antibody SIM4 to block cell surface CD4 and then measured its effect on HIV-1 infection. The results showed a much greater inhibition of the Vpu34/EnvG367E and EnvG367E viruses as compared to the wild-type HIV-1 and the Vpu34 mutant (Fig. 5C). Next, we used shRNA to knock down CD4 expression in SupT1 cells. Out of the four shRNA clones tested, one clone diminished CD4 level by approximately 40% (Fig. 5D), the other three did not significantly affect CD4 expression (data not shown). This CD4-knockdown SupT1 cell line was then infected by either the wild type HIV-1 or the HIV-1(Mut4) virus. Compared to their replication in the control SupT1 cells, the growth of HIV-1(Mut4), but not the wild type HIV-1, was delayed in the CD4-knockdown cells (Fig. 5D). Results of Fig. 5E and F further showed that the Vpu34 and EnvG367E mutations did not affect the usage of CXCR4 as the co-receptor, since all mutated viruses were as sensitive as the wild type virus to the inhibition by the CXCR4 antagonist AMD3100 (Fig. 5E), and none was inhibited by the CCR5 antagonist maraviroc (Fig. 5F). These results suggest a deficient engagement of CD4 by the mutated viral envelope EnvG367E.
The Vpu34 and EnvG367E mutations enhance HIV-1 cell-to-cell transmission
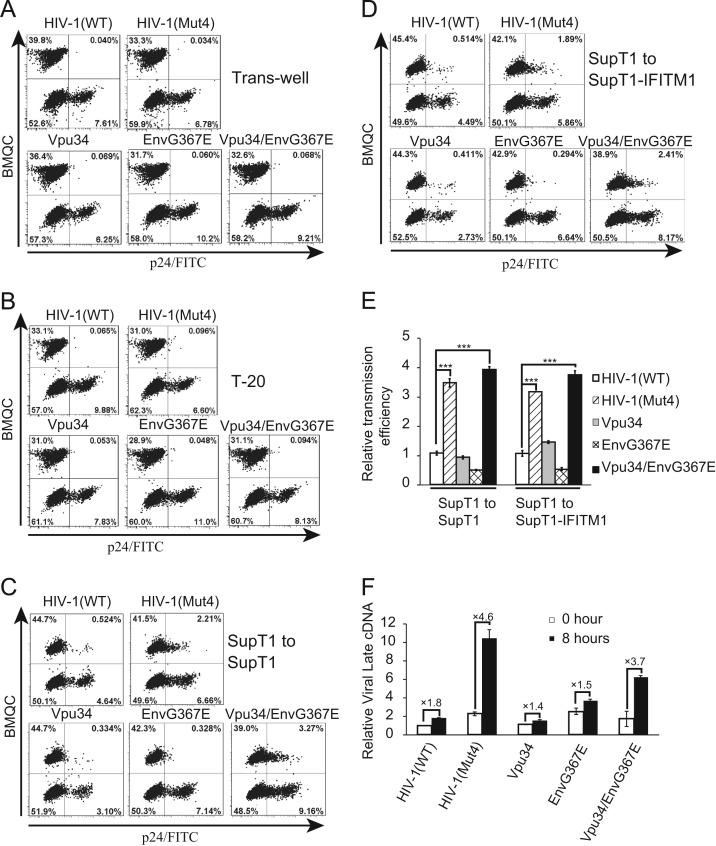
It is intriguing that the Vpu34/EnvG367E virus particles were 10-fold less infectious than the wild type HIV-1 (Fig. 4), yet this mutant replicates as efficiently as does the wild type virus in SupT1 cells (Fig. 2). We also noted that, although infection of the mutated viruses was initially impaired as shown by the results of Western blotting at day 2 after infection, the HIV-1(Mut4) and Vpu34/EnvG367E mutants produced similar levels of viral p24 as compared to wild type HIV-1 at day 4 and day 8 after infection (Fig. 4B). Given that cell-to-cell transmission of HIV-1 is known to dominate virus spread over free-virus infection (Sattentau, 2008), it is possible that the Vpu34/EnvG367E mutant compensates for its reduced infectivity by enhancing cell-to-cell transmission. To test this, we first infected the donor SupT1 cells with the wild type or mutated HIV-1 particles bearing the VSV-G protein, ensuring that both wild type and mutated HIV-1 would generate similar numbers of infected donor cells. Forty hours after infection, donor cells were mixed with target SupT1 cells that were labeled with a cell tracker BMQC, and viral p24 was examined with FITC-conjugated anti-p24 antibody. In order to distinguish cell-to-cell transmission from free virus particles, we used trans-wells to separate donor cells from target cells. As would be expected, few p24-positive cells were detected in the trans-well infection ( Fig. 6A), indicating that free HIV-1 particles did not lead to significant infection of target cells. We then added HIV-1 fusion inhibitor T-20 into the mixed cell population and did not observe significant infection of target cells (Fig. 6B), confirming that the cell-to-cell virus transmission shown in Fig. 6C is viral envelope-dependent. When the control SupT1 cells (without IFITM1 expression) were used as the target cells in the transmission assay, the HIV-1(Mut4) and Vpu34/EnvG367E mutants were 3–4 fold more efficient at cell-to-cell transmission than the wild type virus or the Vpu34 and EnvG367E mutants (Fig. 6C). We next used IFITM1-expressing SupT1 cells as target cells, and observed that IFITM1 did not markedly affect HIV-1 cell-to-cell transmission (Fig. 6D). Again, Vpu34 and EnvG367E mutations together enhanced the transmission of viruses from donor cells to IFITM1-expressing target cells (Fig. 6D). In order to validate these data, we measured the amount of HIV-1 late cDNA in the mixed of donor and target SupT1 cells at time 0 and 8 h after mixing. The increases of HIV-1 late cDNA over a 8-h period of co-culture are 4.6 and 3.7 folds for HIV-1(Mut4) and Vpu34/EnvG367E, but are less than 2-fold for wild type HIV-1, Vpu34 and Env367E (Fig. 6F), which suggests a much higher cell-to-cell transmission efficiency for HIV-1(Mut4) and Vpu34/EnvG367E. Therefore, we conclude that the Vpu34 and EnvG367E mutations rescue HIV-1 replication in IFITM1-expressing SupT1 cells through promoting cell-to-cell virus transmission.
Fig. 6.
The Vpu34 and EnvG367E mutations enhance HIV-1 transmission between SupT1 cells. SupT1 cells were first infected with HIV-1 or its mutants (equivalent to 50 ng of viral p24) for 40 h before being washed with complete RPMI1640 media and used in the following experiments. (A) The HIV-1 infected donor cells and the BMQC-labeled target cells were separated by trans-wells that allow free virus infection. The infected cells were harvested 8 h after and stained with FITC-conjugated anti-p24 antibody and scored by flow cytometry. (B) HIV-1 fusion inhibitor T-20 was added to the mixture of donor and target cells to block HIV-1 cell-to-cell transmission. Cells were harvested 8 h after incubation and stained for viral p24. (C) SupT1 cells were first infected with the wild type or mutated viruses before mixing with the BMQC-labeled target SupT1 cells. After 8 h, the infected target cells were stained with FITC-conjugated anti-p24 antibody and scored by flow cytometry. (D) The infected SupT1 cells were mixed with the BMQC-labeled IFITM1-expressing SupT1 cells to assess the effect of IFITM1 on HIV-1 cell-to-cell transmission. (E) The relative transmission efficiency was calculated for each experiment with the value of wild type HIV-1 transmission arbitrarily set as 1. The results of three independent transmission experiments are averaged and shown in the bar graph. The p values were calculated and the significance is indicated by ⁎⁎⁎ (<0.001). (F) Cell to cell transmission of the wild type and mutant viruses was determined by measuring viral late cDNA amounts. The donor and target cells were collected before mixing (time 0) and 8 h after mixing. Total DNA was extracted, HIV-1 late cDNA was measured by real-time PCR. The GAPDH DNA was also quantified by real-time PCR and the results serve as the internal control for each sample. The amount of late DNA of the wild type HIV-1 at time 0 is arbitrarily set as 1. The results shown are the average of three independent infections.
The effect of Vpu34 and EnvG367E on cell-to-cell transmission is cell type-specific
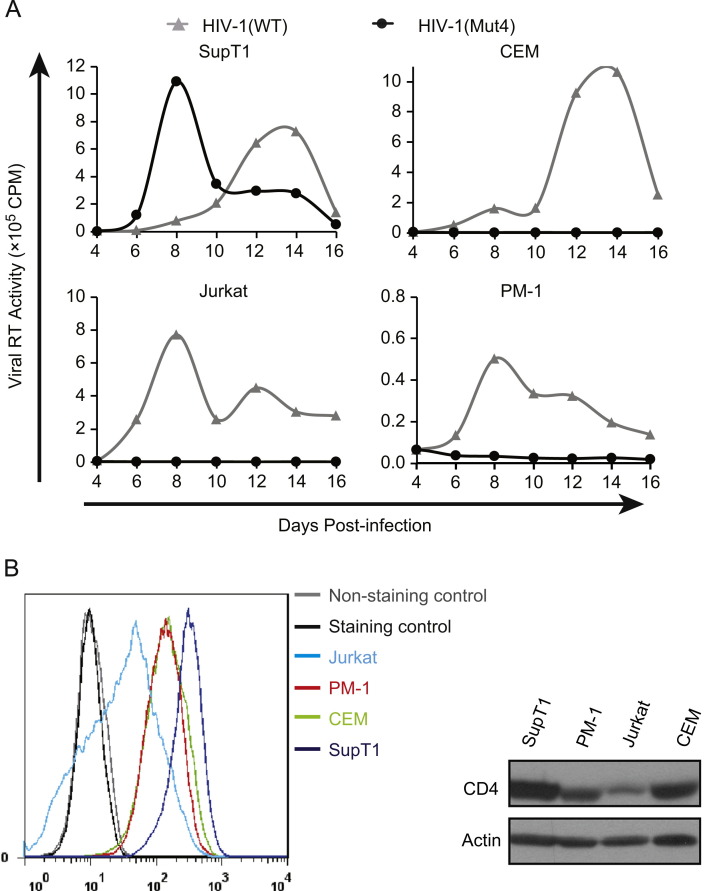
Since the IFITM1-resistant mutations arose in SupT1 cells by selection, we wished to examine the replication capacity of the escape virus in other CD4+ T cell lines. Surprisingly, in contrast to its higher-than wild type level replication in SupT1 cells, the HIV-1(Mut4) mutant, which carried all escape mutations, was unable to grow in CEM, Jurkat, and PM-1 cells ( Fig. 7A), suggesting that this escape mutant has adapted to SupT1 cells for efficient replication. We noticed that SupT1 cells express a relatively higher level of CD4 than CEM, Jurkat and PM-1 cells (Fig. 7B), which may partially underlie the much more efficient replication of EnvG367E-containing HIV-1 in SupT1 cells.
Fig. 7.
Replication of the wild type HIV-1 and the HIV-1(Mut4) mutant in SupT1, CEM, Jurkat and PM-1 cells. (A) Levels of virus production were monitored by measuring viral reverse transcriptase activity in the supernatants at various time intervals. Results shown represent three independent infections. (B) Cell surface CD4 in SupT1, CEM, Jurkat and PM-1 cells was stained with anti-CD4 antibody and its level was determined by flow cytometry. The total levels of cellular CD4 were measured by Western blotting.
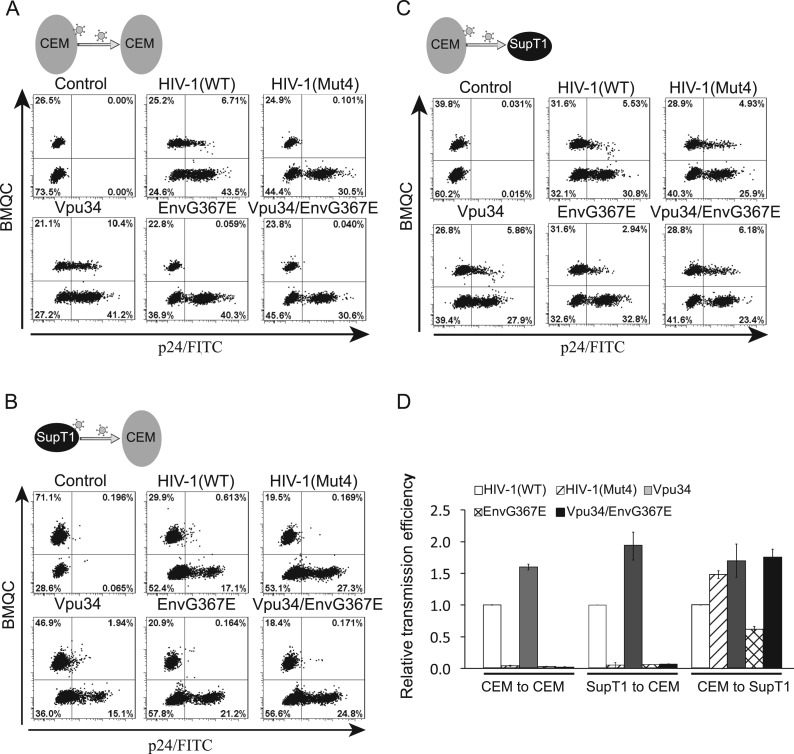
We next examined the cell-to-cell transmission efficiency of HIV-1(Mut4) between CEM cells, and found that HIV-1(Mut4) was unable to spread, consistent with its inability to grow in CEM cells ( Fig. 8A). This transmission deficiency was likely caused by the EnvG367E mutation, because both EnvG367E and Vpu34/EnvG367E were incompetent in cell-to-cell transmission, yet the Vpu34 mutant spread more efficiently than the wild type virus (Fig. 8A). To dissect whether this transmission deficiency is of donor or target cell-effect, we performed additional cell-to-cell transmission assays using SupT1 as donor and CEM cells as target cells, or vice versa. The results showed that the HIV-1(Mut4), EnvG367E, and Vpu34/EnvG367E mutants transmitted efficiently only when SupT1 served as target cells (Fig. 8B–D), suggesting that the target cell type determines the cell-to-cell transmission efficiency of the EnvG367E-containing HIV-1 mutants. Altogether, these data suggest that the Vpu34 and EnvG367E mutations were selected to circumvent IFITM1 restriction in a cell type-specific manner.
Fig. 8.
Cell-to-cell transmission of the HIV-1 wild type and mutants between different pairs of donor and target cells. (A) Transmission of the wild type and mutated HIV-1 between CEM cells. (B) HIV-1 transmission from SupT1 to CEM cells. (C) HIV-1 transmission from CEM to SupT1 cells. (D) Results of three independent transmission experiments are summarized in the bar graph. The transmission efficiency of wild type HIV-1 is arbitrarily set as 1.
Discussion
In this study, we have identified two mutations Vpu34 and EnvG367E that act together to rescue HIV-1 from IFITM1 restriction. The Vpu34 mutation creates a translation stop codon at the amino acid position 35 (Fig. 1C), resulting in the expression of the first 34 amino acids of Vpu that form the transmembrane domain. We ruled out the involvement of this 34-amino-acid Vpu fragment in antagonizing IFITM1, because eliminating the full-length Vpu expression by mutating the translation start codon AUG also enabled the EnvG367E mutation to rescue HIV-1 replication (Fig. 3). Together, our results indicate that the loss of Vpu and a mutation in Env (EnvG367E) are both critical for HIV-1 to evade IFITM1 restriction. We also detected the R311K mutation in the V3 loop of Env, but this mutation does not appear to contribute significantly to circumventing IFITM1. Nonetheless, mutating the V3 loop may represent one way for the virus to counter different inhibitory pressures. This scenario is supported by a recent study showing that feline immunodeficiency virus is able to escape from inhibition by a N-terminal fragment of TSG101 by acquiring a K410N mutation in the V3 loop of its Env protein (Luttge et al., 2013).
Vpu plays two major roles in HIV-1 replication; one is to down-regulate the cell surface CD4, and another is to promote the virus release by counteracting tetherin (Dube et al., 2010). Because the Vpu34 mutant replicated almost as efficiently as the wild type HIV-1 in SupT1 cells (Fig. 2), despite of its inability to downregulate the cell surface CD4 (Fig. 3B), Vpu is dispensable for HIV-1 to replicate in SupT1 cells. This could result from the lack of endogenous tetherin expression in SupT1 (Rong et al., 2009), which alleviates the need for Vpu to antagonize tetherin. It is possible that, in the face of both tetherin and IFITM1, both of which can be induced by IFN, HIV-1 would have to overcome these two restrictions by evolving pathways other than mutating Vpu alone.
Our data show that Vpu34 mutation alone is insufficient to antagonize IFITM1, it needs to act together with the EnvG367E mutation. The G367 amino acid in gp120 is highly conserved among different HIV-1 strains (HIV DATABASES, http://www.hiv.lanl.gov/content/index), as it serves as a key residue for CD4-binding (Fig. 1D). Indeed, we found that the G367E mutation alone causes a 10-fold decrease in the infectivity of HIV-1 particles. Interestingly, this defect does not apparently affect the replication of the EnvG367E mutant in SupT1 cells, and Vpu34 and EnvG367E together enhance the viral growth in SupT1 cells as compared to the wild type virus. How can HIV-1 tolerate such a deleterious mutation as EnvG367E? One possibility is that SupT1 cells express relatively high level of CD4 (Fig. 7B), which could support the EnvG367E mutant to efficiently transmit from cell to cell (Fig. 6). In addition, given that EnvG367E is unable to replicate in T cell lines other than SupT1, SupT1 cells may express a factor(s) that allow the replication of EnvG367E mutant.
We found that the Vpu34 and EnvG367E mutations were unable to overcome the defect in viral p24 expression caused by IFITM1 (Fig. 4). Instead, they enhanced the HIV-1 transmission between SupT1 cells by ~4-fold, indicating that the enhanced replication of these mutants in SupT1 cells are due to cell-to-cell transmission. It is possible that such a gain in cell-to-cell transmission allows HIV-1 to evade different types of restrictions. This Vpu34/EnvG367E-dependent pattern requires SupT1 as the target cells (Fig. 8), which suggests that certain features of the infected target cells promote the formation of virological synapse. These features may include high CD4 level at cell surface as a result of losing Vpu and high level of the EnvG367E envelope that has a weak affinity for CD4 and therefore is less likely trapped at the ER as a result of CD4 binding. Alternatively, the Vpu34/EnvG367E mutant may be efficient in entry via cell-associated routes. It is currently unclear which of these possibilities promotes HIV-1 cell-to-cell transmission. It is interesting to note that a previous study by Gummuluru and colleagues has reported a Vpu mutation called Rap5, which is very similar to Vpu34 (Gummuluru et al., 2000). Rap5 was selected in a rapid turnover assay performed in Jurkat cells, resulting in a frameshift of Vpu and leading to the expression of its first 32 amino acids, which is remarkably similar to the Vpu34 mutation reported here. Importantly, Rap5 also rescued HIV-1 replication by promoting cell-to-cell spread of virus, which is related to accumulation of Rap5 viruses on the cell surface, despite that Rap5 had no apparent effect on the expression of Env. This mechanism was further supported by a later study showing that, in the absence of Vpu, tetherin promotes HIV-1 cell to cell transmission (Jolly et al., 2010). Although this virus particle retention mechanism may not operate for Vpu34 in SupT1 cells that do not express tetherin, the virus manages to evolve a second mutation G367E in envelope protein that, together with Vpu34, achieves higher transmission efficiency to escape IFITM1 inhibition. Apart from enhanced cell-to-cell transmission, we also observed that the EnvG367E virus caused a delayed cytopathogenecity during infection of SupT1 cells as compared to the wild type virus (data not shown), suggesting that the attenuated binding of EnvG367E to CD4 likely also diminishes cell fusion/killing and thereby allows the infected cells to produce more viruses.
IFITM1 has been shown to inhibit entry of a number of enveloped viruses, including IAV, Yellow Fever virus, SARS coronavirus, Ebola virus, etc. (Brass et al., 2009, Feeley et al., 2011, Huang et al., 2011, Li et al., 2013). Interestingly, it does not appear to restrict HIV-1 by impeding entry (Lu et al., 2011). Instead, IFITM1 reduces HIV-1 Gag/p24 expression in virus producer cells, which results in diminished virus production. Interestingly, we found no evidence that the escape mutations would overcome this latter defect. While it is possible that the effect of IFITM1 on HIV-1 is cell type specific and virus-strain dependent (Brass et al., 2009), the identification of the escape mutations that promote HIV-1 cell-to-cell transmission, as reported here, demonstrates that HIV-1 can evolve a mechanism to evade IFITM1 restriction. We envision that similar mechanisms are also operative for other viruses that are restricted by IFITMs.
Conclusions
Our results demonstrate that HIV-1 is able to circumvent the inhibition by IFITM1 in tissue culture by acquiring mutations in viral Vpu and Env proteins. These mutations, namely Vpu34 and G367E, do not seem to overcome the Gag downregulation caused by IFITM1, but rather enable the virus to regain efficient replication by promoting virus spread between cells.
Materials and methods
Plasmids and antibodies
The tetracycline-inducible IFITM1 SupT1 cell line was generated as previously described (Lu et al., 2011). The HIV-1 BH10 proviral DNA clone was obtained from the NIH AIDS Reference and Reagent Program. The mutations Vpu34, EnvR311K, EnvG367E and g7178a were engineered using the site-directed mutagenesis kit (Stratagene). The anti-Flag and anti-β actin antibodies were purchased from Sigma, anti-tubulin antibody from Santa Cruz biotechnology, anti-HIV-1 p24 antibody from ID Lab Inc., phycoerythrin (PE)-conjugated anti-human CD4 antibody from BD Biosciences, Dylight-649-conjugated anti-Flag antibody from Rockland, FITC-conjugated anti-HIV-p24 antibody from Beckman. G418 was purchased from Invitrogen, puromycin and doxycycline from Sigma.
Virus infection
HIV-1 stocks were produced by transfection of the human embryonic kidney cell line (HEK293T) with HIV-1 proviral DNA. The culture supernatants were clarified by passing through the 0.2 µm filter (VWR) to remove the cell debris. Amounts of viruses were determined by measuring viral p24 (CA) levels using the HIV-1 p24 Antigen Capture Assay kit (Cat. 5447, ABL Inc.).
Virus infection was measured by four assays. First, infection of the TZM-bl indicator cells that express CD4/CXCR4/CCR5 and contain the HIV-1 LTR-Luc reporter (Wei et al., 2002). TZM-bl cells were first seeded into 24-well plates (4×104 cells/well) one day before virus infection. Forty hours after infection, cells were lysed with 1× passive lysis buffer (Promega) and the levels of luciferase activity were measured using the luciferase assay kit (Promega). Second, short-term infection of the SupT1 cells. SupT1 cells were first exposed to viruses equivalent to 200 ng viral p24 antigen. Forty hours after infection, the infected cells were washed with cold 1× phosphate-buffered saline and fixed with 1% paraformaldehyde. The infected cells were stained with anti-HIV-1 p24 antibody and scored by flow cytometry. Third, long-term infection of SupT1 cells. SupT1 cells were infected with virus equivalent to 10 ng viral p24. Viral replication was monitored by measuring the levels of viral reverse transcriptase activity in supernatants over various time intervals. Lastly, we used the CEM-Rev-Luc cells to examine the sensitivity of HIV-1 to inhibition by different agents. The CEM-Rev-Luc cells express luciferase in a qHIV-1 Rev-dependent manner (Wu et al., 2007). These cells are a gift of Dr. Yuntao Wu (George Mason University). Cells were first pretreated for 1 h with different doses of each of the five agents, including soluble CD4 (sCD4), broadly neutralizing antibody VRC03, anti-CD4 antibody (SIM4), CXCR4 antagonist (AMD3100) and CCR5 antagonist maraviroc (all agents were obtained from the NIH AIDS Reference and Reagent Program). The cells were then infected with the same amounts of different HIV-1 stocks. Forty hours after infection, cells were lysed and luciferase activity was measured to determine virus infection.
Identification and cloning of the escape viruses
We started by infecting IFITM1-expressing SupT1 cells with HIV-1 to monitor the development of resistant viruses. When marked cytopathogenic effect and high levels of viral RT activity in the supernatant were detected 3 weeks after infection, we used these newly produced viruses to infect fresh IFITM1-expressing SupT1 cells. After five such passages, HIV-1 was able to reach peak level of its replication at 6–8 days instead of 20 days as observed in the initial round of infection. We harvested SupT1 cells that were infected by the highly replicable HIV-1 and extracted the total cellular DNA using the DNeasy Blood & Tissue kit (Qiagen). Viral genomic DNA was amplified with three primer pairs to cover the entire genome. The PCR products were cloned into the PCR-Blunt II-TOPO vector with Zero Blunt TOPO PCR Cloning Kit (Invitrogen). An average of seven positive DNA clones for each PCR reaction was sent to McGill University and Quebec Innovation Center for sequencing. We also grew HIV-1 in the control SupT1 cells that do not express IFITM1 for the same period of time, and viral genomes were similarly amplified by PCR, cloned, and sequenced.
shRNA knockdown of CD4 in SupT1 cells
We purchased four shRNA clones that target human CD4 mRNA (Sigma-Aldrich, catalog no. SHCLNG-NM_000616). The sequences of these four shRNA are:
shRNA1-CCGGCCAGATAAAGATTCTGGGAAACTCGAGTTTCCCAGAATCTTTA TCTGGTTTTTG,
shRNA2-CCGGCCTTCTTAACTAAAGGTCCATCTCGAGATG GACCTTTAGTTAA GAAGGTTTTTG,
shRNA3-CCGGCCTGATCATCAAGAATCTTA ACTCGAGTTAAGATTCTTGATGA TCAGGTTTTTG, and
shRNA4-CCGGAGAGCGGATG TCTCAGATCAACTCGAGTTGATCTGAGACATC CGCTCTTTTTTG.
These shRNA clones were transfected into HEK293T cells together with plasmid DNA pLP1 (encoding HIV-1 Gag and Gag-Pol), pLP2 (encoding HIV-1 rev) and pVSV-G (encoding VSV G protein) to produce VSV-G pseudotyped lentiviral particles that package the shRNA. SupT1 cells were then infected with these virus particles and subject to selection with 2 µg/ml puromycin for stably transduced cells. The empty shRNA vector was used to generate the control cell line. The total amounts of CD4 in these SupT1 cell lines were determined by Western blotting. To determine the cell surface level of CD4, cells were first incubated with the PE-conjugated anti-human CD4 antibody on ice for 30 min, then fixed with 1% paraformaldehyde. The positively stained cells were scored by flow cytometry. Only shRNA3 significantly decreased CD4 level.
HIV-1 virion fusion assay
The experiment was performed as described previously (Cavrois et al., 2002, Lu et al., 2011). Briefly, 3 μg of HIV-1 DNA was co-transfected with 1 μg pCMV-BlaM-Vpr DNA into 293T cells. Supernatants containing virions were filtered with 0.22 μm filter and then concentrated by ultracentrifugation at 100,000g for 1 h at 4 °C. Pelleted viruses were suspended with DMEM, quantified for viral RT activity, aliquoted and stored at −80 °C. For virion fusion assay, SupT1 cells were infected with the same amounts of wild type HIV-1 and mutant viruses by spinoculation for 45 min at room temperature, followed by incubation for 2 h at 37 °C. Cells were then washed with CO2-independent medium (Invitrogen), mixed with 100 μl loading solution (CCF2/AM substrate, Invitrogen) for 1 h at room temperature in the dark. After washing off the loading solution, cells were incubated in 200 μl of development medium in dark for 16 h at room temperature. Cells were washed and fixed with 3.7% formaldehyde. The cleavage of CCF2/AM was measured by flow cytometry.
Western blotting
Cell lysates were separated in 1% sodium dodecyl sulfate-12% polyacrylamide gels (SDS-PAGE) by electrophoresis and then transferred onto polyvinylidene difluoride (PVDF) membranes (Roche). The membranes were blocked with 4% skim milk (in 1× phosphate-buffered saline) and further probed with anti-p24 (1:5000), anti-β actin (1:5000) or anti-tubulin (1:5000) antibodies. After a further incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (GE Healthcare), the protein bands were visualized by exposure to X-ray films following a brief treatment of the membranes with the ECL (enhanced chemiluminescence) reagents.
Cell-to-cell transmission assay
SupT1 cells were first infected with HIV-1 particles bearing VSV-G protein. VSV-G protein was used to effectively increase the infection rate of donor cells; however, the subsequent virus transmission from donor cells to target cells was mediated by HIV-1 Env. Forty hours after, the infected cells (designated as donor cells) were washed with complete medium prior to mixing with un-infected SupT1 cells (designated as target cells) that had been labeled with cell tracker BMQC (Invitrogen). After 8 h, the cell mixtures were fixed with 1% paraformaldehyde and permealized with 0.1% Triton X-100. After staining with FITC-conjugated anti-HIV-1 p24 antibody, the p24-positive donor and target cells were scored by flow cytometry. In order to control for the infection by free HIV-1 particles, infections were also set up with trans-wells that separated the donor from target cells but allowed the free movement of HIV-1 particles. In addition, we performed control experiments by adding the HIV-1 fusion inhibitor T-20 (2 mM, obtained from the NIH AIDS Reference and Reagent Program) to the mixed donor and target cells in order to block HIV-1 envelope-dependent cell-to-cell fusion.
HIV-1 transmission between cells was also determined by measuring the production of viral late cDNA as described by Jolly et al., (2010). Briefly, the donor and target SupT1 cells were collected at time 0 (before mixing) and 8 h after mixing. Total cellular DNA was prepared using the DNeasy Blood & Tissue kit (Qiagen). The same amounts of DNA from different infection samples were subjected to real-time PCR to quantify HIV-1 late DNA and cellular GAPDH DNA as described in (Lu et al., 2011). The amounts of GAPDH DNA serve as internal controls.
Acknowledgments
We are grateful to Dr. Yuntao Wu for providing the CEM-Rev-Luc cell line. This study was supported by funding from Canadian Institutes for Health Research (CIHR) (HVI-98828 and MOP-77649) and the US National Institutes of Health (NIH) (R21AI105584 and R01AI112381).
Contributor Information
Shan-Lu Liu, Email: liushan@missouri.edu.
Chen Liang, Email: chen.liang@mcgill.ca.
References
- Alber D., Staeheli P. Partial inhibition of vesicular stomatitis virus by the interferon-induced human 9-27 protein. J. Interferon Cytokine Res. 1996;16(5):375–380. doi: 10.1089/jir.1996.16.375. [DOI] [PubMed] [Google Scholar]
- Amini-Bavil-Olyaee S., Choi Y.J., Lee J.H., Shi M., Huang I.C., Farzan M., Jung J.U. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe. 2013;13(4):452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anafu A.A., Bowen C.H., Chin C.R., Brass A.L., Holm G.H. Interferon inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J. Biol. Chem. 2013;288(24):17261–17271. doi: 10.1074/jbc.M112.438515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu P., Colnot S., Godard C., Laurent-Puig P., Lamarque D., Kahn A., Perret C., Romagnolo B. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 2006;66(4):1949–1955. doi: 10.1158/0008-5472.CAN-05-2731. [DOI] [PubMed] [Google Scholar]
- Bailey C.C., Huang I.C., Kam C., Farzan M. IFITM3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8(9):e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.C., Kondur H.R., Huang I.C., Farzan M. Interferon-induced transmembrane protein 3 is a Type II transmembrane protein. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.514356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanja Chowdhury J., Shrivastava S., Steele R., Di Bisceglie A.M., Ray R., Ray R.B. Hepatitis C virus infection modulates expression of interferon stimulatory gene IFITM1 by upregulating miR-130A. J. Virol. 2012;86(18):10221–10225. doi: 10.1128/JVI.00882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E., Adams D.J., Xavier R.J., Farzan M., Elledge S.J. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R., Oraszlan-Szovik K., Foser S., Bohrmann B., Certa U. Inhibition of proliferation by 1-8U in interferon-alpha-responsive and non-responsive cell lines. Cell. Mol. Life Sci. 2003;60(6):1235–1248. doi: 10.1007/s00018-003-3016-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavrois M., De Noronha C., Greene W.C. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 2002;20(11):1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- Chutiwitoonchai N., Hiyoshi M., Hiyoshi-Yoshidomi Y., Hashimoto M., Tokunaga K., Suzu S. Characteristics of IFITM, the newly identified IFN-inducible anti-HIV-1 family proteins. Microbes Infect. 2013;15(4):280–290. doi: 10.1016/j.micinf.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblandre G.A., Marinx O.P., Evans S.S., Majjaj S., Leo O., Caput D., Huez G.A., Wathelet M.G. Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. J. Biol. Chem. 1995;270(40):23860–23866. doi: 10.1074/jbc.270.40.23860. [DOI] [PubMed] [Google Scholar]
- Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13(1):46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M., Bego M.G., Paquay C., Cohen E.A. Modulation of HIV-1-host interaction: role of the Vpu accessory protein. Retrovirology. 2010;7:114. doi: 10.1186/1742-4690-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.S., Collea R.P., Leasure J.A., Lee D.B. IFN-alpha induces homotypic adhesion and Leu-13 expression in human B lymphoid cells. J. Immunol. 1993;150(3):736–747. [PubMed] [Google Scholar]
- Evans S.S., Lee D.B., Han T., Tomasi T.B., Evans R.L. Monoclonal antibody to the interferon-inducible protein Leu-13 triggers aggregation and inhibits proliferation of leukemic B cells. Blood. 1990;76(12):2583–2593. [PubMed] [Google Scholar]
- Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J., Wise H.M., Kane L., Goulding D., Digard P., Anttila V., Baillie J.K., Walsh T.S., Hume D.A., Palotie A., Xue Y., Colonna V., Tyler-Smith C., Dunning J., Gordon S.B., Smyth R.L., Openshaw P.J., Dougan G., Brass A.L., Kellam P. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484(7395):519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley E.M., Sims J.S., John S.P., Chin C.R., Pertel T., Chen L.M., Gaiha G.D., Ryan B.J., Donis R.O., Elledge S.J., Brass A.L. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7(10):e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummuluru S., Kinsey C.M., Emerman M. An in vitro rapid-turnover assay for human immunodeficiency virus type 1 replication selects for cell-to-cell spread of virus. J. Virol. 2000;74(23):10882–10891. doi: 10.1128/jvi.74.23.10882-10891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanagata N., Li X., Morita H., Takemura T., Li J., Minowa T. Characterization of the osteoblast-specific transmembrane protein IFITM5 and analysis of IFITM5-deficient mice. J. Bone Miner. Metab. 2010;29(3):279–290. doi: 10.1007/s00774-010-0221-0. [DOI] [PubMed] [Google Scholar]
- Hickford D.E., Frankenberg S.R., Shaw G., Renfree M.B. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genomics. 2012;13(1):155. doi: 10.1186/1471-2164-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K., Hao C., Gierszewska M., Swanson S.K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M.P., Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., Longobardi L.E., Boltz D., Kuhn J.H., Elledge S.J., Bavari S., Denison M.R., Choe H., Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R., Pan Q., Ding S., Rong L., Liu S.L., Geng Y., Qiao W., Liang C. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 2012;86(24):13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Weidner J.M., Qing M., Pan X.B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P.Y., Block T.M., Guo J. Identification of five interferon-induced cellular proteins that inhibit west Nile virus and dengue virus infections. J. Virol. 2010;84(16):8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Booth N.J., Neil S.J. Cell-cell spread of human immunodeficiency virus type 1 overcomes tetherin/BST-2-mediated restriction in T cells. J. Virol. 2010;84(23):12185–12199. doi: 10.1128/JVI.01447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Segeral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin A.R., Reid L.E., McMahon M., Stark G.R., Kerr I.M. Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 1991;199(2):417–423. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- Li K., Markosyan R.M., Zheng Y.M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J.C., Gratton E., Cohen F.S., Liu S.L. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9(1):e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Kao E., Gao X., Sandig H., Limmer K., Pavon-Eternod M., Jones T.E., Landry S., Pan T., Weitzman M.D., David M. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491(7422):125–128. doi: 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Pan Q., Rong L., He W., Liu S.L., Liang C. The IFITM proteins inhibit HIV-1 infection. J. Virol. 2011;85(5):2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttge B.G., Panchal P., Puri V., Checkley M.A., Freed E.O. Mutations in the feline immunodeficiency virus envelope glycoprotein confer resistance to a dominant-negative fragment of Tsg101 by enhancing infectivity and cell-to-cell virus transmission. Biochim. Biophys. Acta. 2013 doi: 10.1016/j.bbamem.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M.H., Bieniasz P.D. HIV restriction factors and mechanisms of evasion. Cold Spring Harb. Perspect. Med. 2012;2(5):a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt P., Gaumond M.H., Salois P., Sellin K., Bessette M.C., Godin E., de Oliveira P.T., Atkins G.J., Nanci A., Thomas G. Bril: a novel bone-specific modulator of mineralization. J. Bone Miner. Res. 2008;23(9):1497–1508. doi: 10.1359/jbmr.080412. [DOI] [PubMed] [Google Scholar]
- Mudhasani R., Tran J.P., Retterer C., Radoshitzky S.R., Kota K.P., Altamura L.A., Smith J.M., Packard B.Z., Kuhn J.H., Costantino J., Garrison A.R., Schmaljohn C.S., Huang I.C., Farzan M., Bavari S. IFITM-2 and IFITM-3 but Not IFITM-1 restrict Rift Valley Fever Virus. J. Virol. 2013;87(15):8451–8464. doi: 10.1128/JVI.03382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S.J., Zang T., Bieniasz P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Perreira, J.M., Chin, C.R., Feeley, E.M., and Brass, A.L. (2013). IFITMs restrict the replication of multiple pathogenic viruses. J. Mol. Biol. [DOI] [PMC free article] [PubMed]
- Raychoudhuri A., Shrivastava S., Steele R., Kim H., Ray R., Ray R.B. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J. Virol. 2011;85(24):12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L., Zhang J., Lu J., Pan Q., Lorgeoux R.P., Aloysius C., Guo F., Liu S.L., Wainberg M.A., Liang C. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. J. Virol. 2009;83(15):7536–7546. doi: 10.1128/JVI.00620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 2008;6(11):815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo G.S., Lee J.K., Yu J.I., Yun K.J., Chae S.C., Choi S.C. Identification of the polymorphisms in IFITM3 gene and their association in a Korean population with ulcerative colitis. Exp. Mol. Med. 2010;42(2):99–104. doi: 10.3858/emm.2010.42.2.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist F., Ebeling M., Certa U. The small interferon-induced transmembrane genes and proteins. J. Interferon Cytokine Res. 2011;31(1):183–197. doi: 10.1089/jir.2010.0112. [DOI] [PubMed] [Google Scholar]
- Stremlau M., Owens C.M., Perron M.J., Kiessling M., Autissier P., Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Wakim L.M., Gupta N., Mintern J.D., Villadangos J.A. Enhanced survival of lung tissue-resident memory CD8(+) T cells during infection with influenza virus due to selective expression of IFITM3. Nat. Immunol. 2013;14(3):238–245. doi: 10.1038/ni.2525. [DOI] [PubMed] [Google Scholar]
- Wei X., Decker J.M., Liu H., Zhang Z., Arani R.B., Kilby J.M., Saag M.S., Wu X., Shaw G.M., Kappes J.C. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002;46(6):1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner J.M., Jiang D., Pan X.B., Chang J., Block T.M., Guo J.T. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 2010;84(24):12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N.S., Louder M., McKee K., O׳Dell S., Perfetto S., Schmidt S.D., Shi W., Wu L., Yang Y., Yang Z.Y., Yang Z., Zhang Z., Bonsignori M., Crump J.A., Kapiga S.H., Sam N.E., Haynes B.F., Simek M., Burton D.R., Koff W.C., Doria-Rose N.A., Connors M., Mullikin J.C., Nabel G.J., Roederer M., Shapiro L., Kwong P.D., Mascola J.R. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2010;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Beddall M.H., Marsh J.W. Rev-dependent indicator T cell line. Curr. HIV Res. 2007;5(4):394–402. doi: 10.2174/157016207781024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T., Hashimoto S., Kaneko S., Nagai S., Toyoda N., Suzuki T., Kobayashi K., Matsushima K. Comprehensive gene expression profile of a normal human liver. Biochem Biophys Res Commun. 2000;269(1):110–116. doi: 10.1006/bbrc.2000.2272. [DOI] [PubMed] [Google Scholar]
- Yang G., Xu Y., Chen X., Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007;26(4):594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]
- Yount J.S., Karssemeijer R.A., Hang H.C. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 2012;287(23):19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount J.S., Moltedo B., Yang Y.Y., Charron G., Moran T.M., Lopez C.B., Hang H.C. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 2010;6(8):610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinforma. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Georgiev I., Wu X., Yang Z.Y., Dai K., Finzi A., Kwon Y.D., Scheid J.F., Shi W., Xu L., Yang Y., Zhu J., Nussenzweig M.C., Sodroski J., Shapiro L., Nabel G.J., Mascola J.R., Kwong P.D. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329(5993):811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchi I., Prinetti A., Scotti M., Valsecchi V., Valaperta R., Mento E., Reinbold R., Vezzoni P., Sonnino S., Albertini A., Dulbecco R. Association of rat8 with Fyn protein kinase via lipid rafts is required for rat mammary cell differentiation in vitro. Proc. Natl. Acad. Sci. USA. 2004;101(7):1880–1885. doi: 10.1073/pnas.0307292101. [DOI] [PMC free article] [PubMed] [Google Scholar]