Abstract
Candida albicans and Staphylococcus aureus are often co-isolated in cases of biofilm-associated infections. C. albicans can cause systemic disease through morphological switch from the rounded yeast to the invasive hyphal form. Alternatively, systemic S. aureus infections arise from seeding through breaks in host epithelial layers although many patients have no documented portal of entry. We describe a novel strategy by which S. aureus is able to invade host tissue and disseminate via adherence to the invasive hyphal elements of Candida albicans. In vitro and ex vivo findings demonstrate a specific binding of the staphylococci to the candida hyphal elements. The C. albicans cell wall adhesin Als3p binds to multiple staphylococcal adhesins. Furthermore, Als3p is required for C. albicans to transport S. aureus into the tissue and cause a disseminated infection in an oral co-colonization model. These findings suggest that C. albicans can facilitate the invasion of S. aureus across mucosal barriers, leading to systemic infection in co-colonized patients.
Introduction
Polymicrobial infections tend to be complex and result in aggressive forms of diseases that often exhibit increased resistance to antimicrobials and impact therapeutic measures (Brogden, 2003; Harriott & Noverr, 2009; Jenkinson & Douglas, 2002; Klotz et al., 2007; Lynch & Robertson, 2008). Yet, despite the gravity of such infections, studies of mechanisms underlying polymicrobial infections are in their infancy.
Among the vast number of human pathogens, the bacterial species Staphylococcus aureus and the fungal species Candida albicans are currently the second and third most commonly isolated bloodstream pathogens (Goetghebeur et al., 2007; Klevens et al., 2007; Perlroth et al., 2007). In particular, S. aureus has gained considerable attention from the medical community due to its involvement in the increasing number of nosocomial and community acquired infections resulting in nearly half a million hospitalizations and 50 000 deaths each year in the USA alone (Goetghebeur et al., 2007; Gordon & Lowy, 2008). This bacterial species is armed with an array of virulence factors including toxins and immunoavoidance strategies for invading and destroying host tissue during infection (Bien et al., 2011; Ferry et al., 2005). Despite its pathogenic potential, S. aureus is typically a non-invasive commensal and has been historically identified as a common nasopharyngeal resident, but is also found localized associated with moist skin areas of the axillae and groin. However, this microbial species has more recently been found to commonly exist in the oral cavity. S. aureus typically requires a breach in mucosal barriers to gain entry into the epithelium (Acton et al., 2009; Ohara-Nemoto et al., 2008; Smith et al., 2003; Zimmerli et al., 2009). Yet, it has been reported that a significant number of patients with staphylococcal bloodstream infections have no documented portal of entry (del Rio et al., 2009).
Similarly, C. albicans is the most frequently encountered pathogenic human fungal species and commonly colonizes host mucosal and moist skin surfaces (Calderone & Clancy, 2012; Cannon & Chaffin, 2001). However, under conditions of immune dysfunction, this opportunistic microbe can rapidly transition from commensal to pathogen, causing an array of infections ranging from localized mucosal to severe systemic infections with high morbidity and mortality rates (Calderone & Clancy, 2012; de Repentigny et al., 2004; Perlroth et al., 2007). Oral candidiasis or thrush is the most common opportunistic infection in HIV-infected population with 80–90 % of these individuals developing oropharyngeal candidiasis during the course of their illness (de Repentigny et al., 2004; Fidel, 2006). In addition, recent longitudinal studies have shown that in the ageing population, C. albicans is even more frequently encountered in the oral cavity, especially in edentulous elderly populations (Budtz-Jłrgensen et al., 1996; Kulak-Ozkan et al., 2002; Zaremba et al., 2006). The success of this species as an opportunistic pathogen is the result of its repertoire of virulence factors, including the ability to switch between a yeast and hyphal morphology, a property crucial to its pathogenicity (Calderone & Clancy, 2012).
Several studies have reported the co-isolation of S. aureus and C. albicans from numerous biofilm-associated diseases such as periodontitis, denture stomatitis, cystic fibrosis, keratitis, ventilator-associated pneumonia and urinary tract and burn wound infections (Adam et al., 2002; Baena-Monroy et al., 2005; Costerton et al., 1985; Cuesta et al., 2010; Gupta et al., 2005; Pate et al., 2006; Tawara et al., 1996; Timsit et al., 2001; Valenza et al., 2008). Although these studies only reported associations and did not prove causation, the frequency with which S. aureus and C. albicans are co-isolated merits further study. More directly relevant to bacteraemia, a study by Klotz et al. (2007) investigating the incidence of candidal bloodstream infections in hospitalized patients reported that C. albicans was co-isolated with S. aureus in 20 % of the cases. Further, animal studies by Carlson et al. (1983, 1985) demonstrated a significant increase in mortality in mice co-infected intraperitoneally with sublethal levels of C. albicans and S. aureus. This lethal synergism was also found in a more recent study by Peters & Noverr (2013) where co-infection led to a 40 % mortality rate and increased microbial burden in the spleen and kidney. The interaction between S. aureus and C. albicans does not appear to be strain specific, and it is significantly higher than the interaction between C. albicans and other microbial species (Staphylococcus epidermidis, Streptococcus pyogenes, Pseudomonas aeruginosa, Escherichia coli and Bacillus subtilis) (Peters et al., 2010, 2012).
Collectively, these observations seem to indicate a potential synergy in virulence when these species co-exist in a host. These speculations were recently validated by our in vitro studies, where microscopic images revealed a complex physical interaction with S. aureus demonstrating high affinity to the hyphal elements of C. albicans (Peters et al., 2010, 2012). Further analysis revealed that S. aureus bound specifically to the hyphae form of C. albicans and not to the yeast form (Peters et al., 2010). The main C. albicans target for S. aureus binding was shown to be the agglutinin-like sequence 3 adhesin (Als3p). Purified recombinant Als3p has been shown to bind S. aureus in in vitro assays, and binding of S. aureus to the C. albicans hyphae is significantly reduced in a mutant of Als3p compared to WT or other adhesin-deficient mutant strains (Peters et al., 2012). However, previous studies did not confirm this specific interaction by restoration of the WT binding phenotype through complementation of the C. albicans Als3p gene.
Given the propensity of C. albicans to adhere to and penetrate tissue via its invasive hyphae, combined with the high in vitro affinity of S. aureus to the hyphae, it was feasible to speculate that co-colonization with C. albicans may provide S. aureus with the means to gain entry into the vascular system (Cannon & Chaffin, 2001; Sudbery et al., 2004). Since S. aureus has more recently been identified in the oral cavity along with C. albicans, we sought to test the effects of their co-colonization in the oral cavity using a standard murine model of candidiasis. Tongue histopathological analysis was performed and subepithelial co-penetration of the tissue by invasive C. albicans hyphae with adherent S. aureus was shown. This interaction was absent in mutants of the C. albicans Als3p hyphal protein but was restored when Als3p was restored through complementation. We developed a novel murine model of oral co-colonization to monitor the development and potential progression of mucosal co-colonization to systemic disease in an immunocompromised host. In this model, mice suffered disseminated staphylococcal disease with high morbidity and mortality only when co-colonized by S. aureus and C. albicans. While this ‘microbial hitchhiking’ phenomenon has been identified in other in vitro systems (Edwards et al., 2006; Saito et al., 2012), this is the first report, to our knowledge, of systemic disease associated with multi-Kingdom interactions. The clinical implications are far-reaching, especially if this microbial infectious synergy occurs in other areas where co-colonization can commonly occur such as the intestinal tract, genital tract, infected and devitalized skin or axillary or groin areas of the skin. This study also provides crucial insight into one of the complex mechanisms behind polymicrobial interactions.
Methods
Strains.
The genotypes of the strains used can be found in the corresponding references: C. albicans strains SC5314 WT (SC5314) (Gillum et al., 1984), ALS3 mutant in SC5314 (als3Δ/Δ) (Phan et al., 2007), ALS3 mutant in SC5314 complemented (als3Δ/Δ ALS3) (Phan et al., 2007), the methicillin-resistant S. aureus WT strains M2 (Brady et al., 2006) and USA300 JE2 (Kennedy et al., 2010). In order to determine potential adhesins on S. aureus to which C. albicans was binding, 25 different S. aureus mutant strains were selected based on a review of microbial surface components recognizing adhesive matrix molecules in S. aureus (Clarke & Foster, 2006). The parent strain USA300 JE2 and the 25 S. aureus mutant strains that were used in the experiment were obtained from The Nebraska Transposon Mutant Library from Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) and were constructed by the Center for Staphylococcal Research (CSR) at the University of Nebraska Medical Center (http://app1.unmc.edu/fgx/). A description of all strains used can be found in Table 1.
Table 1. Strains used in this study.
| Strain | Designation | Genotype | Reference |
| S. aureus M2 | WT (SA) | ST30, spa type T019 and agrIII | Ali et al. (1997); Brady et al. (2006); Brady et al. (2007); Brady et al. (2011); Jabra-Rizk et al. (2006); Leid et al. (2002); Peters et al. (2010); Peters et al. (2012); Prabhakara et al. (2011a, b); Shirtliff et al. (2002) |
| S. aureus JE2 | JE2 | USA300 JE2 – a plasmid cured strain from WT USA300-LAC | Kennedy et al. (2008); Kennedy et al. (2010); http://appl.unmc.edu/fgx/ |
| NE1 | ΔSAUSA300_1327 – Cell Wall Protein (protein G-related albumin-binding (GA) module related to streptococcal emb adhesin) | ||
| NE26 | ΔSAUSA300_0224 – coa (staphylocoagulase precursor) | ||
| NE33 | ΔSAUSA300_2589 – sasA (Staphylococcus aureus surface protein A and LPXTG-motif cell wall surface anchor family protein) | ||
| NE98 | ΔSAUSA300_0548 – sdrE (SdrE protein binds host Factor H) | ||
| NE186 | ΔSAUSA300_2441 – fnbA (fibronectin binding protein A) | ||
| NE286 | ΔSAUSA300_0113 – spa (immunoglobulin G binding protein A) | ||
| NE295 | ΔSAUSA300_0025 – hypothetical 5-nucleotidase family protein | ||
| NE332 | ΔSAUSA300_1029 – sasE (iron transport associated domain protein) | ||
| NE391 | ΔSAUSA300_2565 – clfB (clumping factor B) | ||
| NE432 | ΔSAUSA300_0546 – sdrC (SdrC protein) | ||
| NE460 | ΔSAUSA300_0955 – atl (autolysin) | ||
| NE472 | ΔSAUSA300_2109 – fmtB (truncated FmtB protein) | ||
| NE510 | ΔSAUSA300_2110 – fmtB (truncated FmtB protein) | ||
| NE543 | ΔSAUSA300_0772 – clfA (clumping factor A) | ||
| NE557 | ΔSAUSA300_1030 – sirD (iron transport associated domain protein) | ||
| NE728 | ΔSAUSA300_2440 – fnbB (fibronectin binding protein B) | ||
| NE800 | ΔSAUSA300_2581 – sasF (putative surface anchored protein) | ||
| NE825 | ΔSAUSA300_2436 – hypothetical putative cell wall surface anchor family protein | ||
| NE1032 | ΔSAUSA300_0136 – sasD (cell wall surface anchor family protein) | ||
| NE1075 | ΔSAUSA300_1677 – sasI (cell wall surface anchor family protein) | ||
| NE1102 | ΔSAUSA300_1028 – sirH (iron transport associated domain-containing protein) | ||
| NE1190 | ΔSAUSA300_2256 – Atl (Bifunctional autolysin Atl [N-acetylmuramoyl-l-alanine amidase/Endo-beta-N-acetylglucosaminidase | ||
| NE1289 | ΔSAUSA300_0547 – sdrD (SdrD protein) | ||
| NE1558 | ΔSAUSA300_0774 – empbp (secretory extracellular matrix and plasma binding protein | ||
| NE1561 | ΔSAUSA300_1370 – ebpS (cell surface elastin binding protein) | ||
| C. albicans SC5314 | SC5314 | URA3 ALS3 ARG4 HIS1 URA3 ALS3 ARG4 HIS1 | Gillum et al. (1984) |
| C. albicans CAYF178U | als3Δ/Δ | ura3Δ : : λimm434 : : URA3-IRO1 als3 : : ARG4 arg4 : : hisG his1 : : hisG ura3Δ : : λimm434 als3 : : HIS1 arg4 : : hisG his1 : : hisG | Phan et al. (2007) |
| C. albicans CAQTP178U | als3Δ/Δ ALS3 | ura3Δ : : λimm434 : : URA3-IRO1 als3 : : ARG4 : : ALS3 arg4 : : hisG his1 : : hisG ura3Δ : : λimm434 als3 : : HIS1 arg4 : : hisG his1 : : hisG | Phan et al. (2007) |
Hyphal–bacterial attachment assay.
For all attachment assays carried out in order to identify the S. aureus adhesin responsible for C. albicans hyphal binding, an aliquot of a glycerol stock of C. albicans strain was grown and maintained on Sabouraud dextrose agar (BBL). Cultures were grown overnight in yeast–peptone–glucose (BBL) in an orbital shaker (120 r.p.m.) at 30 °C under aerobic conditions. Yeast cells were harvested and washed twice in sterile PBS. Hyphae formation was induced by first growing C. albicans as noted above on plastic Permanox slides (Electron Microscopy Sciences) in polystyrene six well plates (Corning) in 3 ml RPMI 1640 for 3 h. Nonadherent hyphae were removed by gently washing the slides in PBS, followed by the addition of 3 ml of fresh RPMI 1640.
Starter cultures of the WT S. aureus strain JE2 and 25 different S. aureus mutants strains were grown in trypticase soy broth (TSB; Remel) and incubated overnight at 37 °C. Erythromycin (10 µg ml−1) was added into the TSB media for the S. aureus mutant strains. Fresh exponential-phase S. aureus starter cultures were grown by diluting the overnight culture 1 : 100 in fresh TSB for 3 h. Exponential-phase S. aureus cell suspensions were washed in PBS, diluted to an OD600 of 0.1, and added to SC5314 C. albicans biofilms (WT and als3Δ/Δ). Dual-species biofilms were grown in RPMI 1640 buffered with HEPES and supplemented with l-glutamine (Invitrogen). Plates were placed on a rotary shaker to distribute the bacteria evenly and incubated for 1 h at 37 °C. Following incubation, non-adherent cells were removed by gently washing the slides in PBS and then examined using phase-contrast microscopy under a 100× oil-immersion objective. The total number of bacterial cells per field and attached bacteria per hyphae were counted. Per cent attachment was calculated by dividing the number of attached bacteria by the total number of bacteria. A total of ten random fields per coverslip were analysed. Each assay was done in triplicate.
Real-time adhesion analysis.
Real-time adhesion was visualized using the Bioflux 200 system (Fluxion) combined with an EVOS fl digital fluorescence microscope (AMG). To analyse adhesion of S. aureus to hyphae of SC5314 C. albicans in real-time, a GFP labelled S. aureus (S. aureusGFP) strain was used (Li et al., 2011). C. albicans suspended at 107 cells ml−1 in PBS was allowed to adhere to glass under static conditions at 37 °C. Following 2 h incubation, non-adherent cells were removed by flow with 0.2 Pa and yeast nitrogen base pH 7 containing 0.5 % d-glucose (YNB) was flowed through the channel. The flow was stopped and the channel was incubated at 37 °C for 3 h to allow hyphae to form (Jarosz et al., 2009), then YNB was removed by flow (0.2 Pa). Pilot experiments using different shear rates showed that hyphae remained attached to the glass surface up to a maximum of 2 Pa (not shown). Overnight cultures of S. aureusGFP, grown at 37 °C while shaking at 150 r.p.m., were harvested and resuspended in PBS. This suspension was flowed over the adherent hyphae of C. albicans at 0.2 Pa and images were obtained using an EVOS fl digital fluorescence microscope using a 10× objective and the appropriate filter set. Images were taken every 10 s for a total of 5 min and a movie was created at 2 frames s−1.
Microscopic analysis.
Peptide-nucleic acid fluorescent in situ hybridization (PNA-FISH) staining and confocal scanning laser microscopy was utilized to visualize the architecture of mixed biofilms in vitro as described previously (Peters et al., 2010). PNA-FISH employs fluorescent-labelled PNA probes to target the species-specific rRNA sequences in a specific FISH assay that enables whole cell visualization. Hybridization was performed following the manufacturer’s protocol (Advandx) using a cocktail of Cy2-labelled S. aureus and FITC-labelled C. albicans PNA probe mixtures. Briefly, dual-species biofilms were grown on glass coverslips in polystyrene six well plates (Corning). S. aureus and C. albicans were diluted in PBS to an OD600 of 0.1 and 1.0, respectively. This represented approximately 1×108 c.f.u. ml−1 for S. aureus and 2×107 c.f.u. ml−1 for C. albicans. Fifty microlitres of each cell suspension (representing a total of 5×106 c.f.u. of S. aureus and 1×106 c.f.u. of C. albicans) was added to wells containing 5 ml of RPMI 1640 buffered with HEPES and supplemented with l-glutamine (Invitrogen) and 5 % heat-inactivated FBS (Hyclone) and plates were incubated for 24 h at 37 °C. Following incubation, coverslips were gently rinsed with PBS to remove non-adherent cells. Imaging was performed by confocal scanning laser microscopy with a Zeiss LSM 510 confocal microscope (Carl Zeiss) using FITC filter sets.
In addition, microscopy was also performed on excised mouse tongues using a previously described ex vivo model (Kamai et al., 2002; Peters et al., 2012). Briefly, tongues were excised from euthanized CD1 mice and placed into polystyrene well plates containing 5 ml of RPMI 1640 per well. S. aureus and C. albicans were added in an identical manner as in the in vitro attachment assay: 50 µl of cell suspensions of each microbe for a total of 5×106 c.f.u. of S. aureus and 1×106 c.f.u. of C. albicans. Tongues were then washed several times in sterile PBS, transferred to fresh RPMI 1640 and incubated overnight at 37 °C with 5 % CO2. Tongues were then fixed, embedded in paraffin and tissue sections stained with PNA-FISH probes and examined by confocal microscopy. Fluorescent microscopy could not be performed on tissue recovered from animals infected in vivo due to interference in the hybridization process by host erythrocytes and inflammatory cells.
Murine in vivo model of co-infection.
Strains were grown on appropriate growth media and mid exponential-phase cultures were used in experiments. Animal studies were approved by the University of Maryland Animal Care and Use Committee. The animal model for oral co-infection was designed based on an established protocol for oral candidiasis (Dwivedi et al., 2011). Briefly, oral bacterial flora was suppressed by the addition of 300 µg ml−1 ampicillin into the drinking water and mice (C57BL/6J) were immunosuppressed by three subcutaneous injections of 225 mg kg−1 cortisone acetate, one given prior to inoculation and the other two given following inoculation 2 days apart. Anaesthetized animals were orally administered with standardized cell suspensions of C. albicans (100 µl of a 6×108 c.f.u. ml−1 suspension administered on a small cotton pad), S. aureus (50 µl of a 1×108 c.f.u. ml−1 suspension), or in combination (four mice per group in triplicate) on days 1 and 3, and were continually exposed to C. albicans (6×106 c.f.u. ml−1) through drinking water. During the course of the experiments, animals were weighed every 3 days, and on day 6 post-infection, were euthanized and tongues and kidneys were harvested. Tissue homogenates were assessed for microbial burden (cells g−1 tissue) by culturing on selective microbial media. Mice exhibiting greater than 20 % weight loss were euthanized and considered deceased, as established as a humane end point by the University of Maryland Institutional Animal Care and Use Committee.
Statistical analysis.
Experiments were performed in triplicate. Data analysis was performed using the two-tailed Student’s t-test, the Log Rank Test, or the Mann–Whitney U Test (two tailed), where appropriate. Values of P<0.05 were considered statistically significant.
Ethics statement.
The experiments in this study were approved under Protocol 1111010 of the Dental School IACUC committee and adhered to Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals (Animal Welfare Assurance A3200-01) and United States Department of Agriculture Animal Welfare Act & Animal Welfare Regulations.
Results
Hyphal–bacterial attachment assay: in vitro analysis of polymicrobial biofilm development using confocal and time lapse microscopy
As colonization and biofilm formation is a prerequisite for the development of the pathogenic process, in vitro studies were performed in order to visualize the interaction between S. aureus and C. albicans strains as they co-existed in a polymicrobial biofilm. Mixed-species biofilms of S. aureus and C. albicans mutant and complemented strains were grown in vitro and visualized by PNA-FISH staining. Microscopic images of the mixed biofilms paralleled those from our previous studies (Peters et al., 2010, 2012) where a significant decrease in S. aureus adherence to the hyphae of the C. albicans strain lacking the Als3p (als3Δ/Δ) was observed compared to the strains expressing the receptor. Additionally, there was no appreciable difference between the WT C. albicans or the als3Δ/Δ ALS3 complemented strain (data not shown).
Importantly, the initial adherence of S. aureus to the hyphae leading to the development of a mature biofilm was monitored and captured in real-time by fluorescent microscopy in a biofilm flow system. A movie was created of compiled images taken every 10 s over a 5 min time frame (Video S1, available in the online Supplementary Material). As can be seen, immediately upon entry into the flow cell, S. aureusGFP rapidly and specifically adheres to the hyphae attached to the surface of the flow cell. When a higher cell density of S. aureus is flowed through the system, a more rapid and dramatic adhesion is seen resulting in development of a mature mixed-species biofilm within 5 min of initiation of the adherence process.
In order to determine the specific staphylococcal factor responsible for the hyphal binding, WT S. aureus strain JE2 and 25 different S. aureus strains mutated in previously described adhesins were individually added to SC5314 C. albicans biofilms (see Fig. 1, Table 1 for strain designations). The percentage of cells attached to the hyphae of SC5314 was determined for each mutant and compared to the hyphal attachment percentage of WT S. aureus co-cultured with either SC5314 or the SC5314 als3Δ/Δ. It was found that the percentage of WT JE2 cells binding to hyphae was that same as that seen for the staphylococcal strain M2 (84 % vs 82 % respectively) (Peters et al., 2012). The use of JE2, a S. aureus strain that is phylogenetically divergent from the M2 strain (Diep et al., 2006; Harro et al., 2013), provides broader support for the idea that this S. aureus–C. albicans interaction is species-specific and not just strain specific.
Fig. 1.
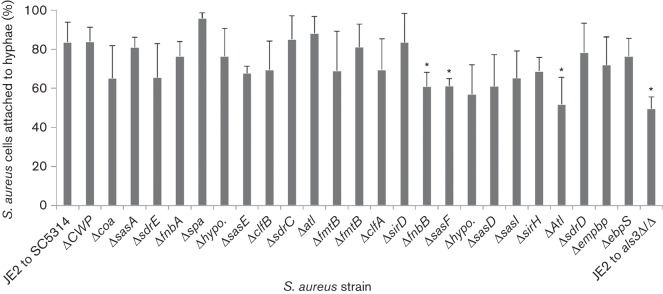
Hyphal binding assay for determining the S. aureus adhesin responsible for C. albicans hyphae binding. The WT S. aureus strain JE2 and 25 different S. aureus mutants strains in previously described adhesins, were grown in TSB, added to C. albicans hyphae, and incubated for 3 h. Non-adherent hyphae were removed by gently washing the slides in PBS, and then examined using phase-contrast microscopy under a 100× oil-immersion objective. The total number of bacterial cells per field and attached bacteria per hyphae were counted. Attachment (%) was calculated by dividing the number of attached bacteria by the total number of bacteria. A total of 10 random fields per coverslip were analysed in triplicate (n = 3). Error bars indicate sd. * denotes statistically significant (P<0.05) reductions in binding ability compared to the WT S. aureus and C. albicans attachment (JE2 to SC5314) as evaluated by a two-tailed Student’s t-test.
Several of the staphylococcal mutants tested had a statistically significant reduction in their ability to bind C. albicans, including NE728 (Δ in fibronectin binding protein B) with 62 % attachment, NE800 (Δ in sasF, a putative surface anchored protein) with 61 % attachment and NE1190 (Δ in Atl, a putative N-acetylmuramoyl-l-alanine amidase) with 52 % attachment. In addition, five other adhesin mutants showed a similarly reduced ability (52–65 % attachment) to bind C. albicans, although these differences were not statistically significant due to larger sd. The creation of single, double, triple, or octuple mutants in these adhesins has been shown to eliminate the ability of S. aureus to both form biofilms and survive in vivo (Bose et al., 2012; Chen et al., 2013; Kenny et al., 2009; Mazmanian et al., 2000; Palmqvist et al., 2005). Therefore, these S. aureus mutants are no longer able to systemically spread in cases of co-infection with C. albicans regardless of the mechanism of deep tissue delivery. Since it cannot be concluded whether a lack of systemic disease by S. aureus is a product of reduced C. albicans hyphal binding or an inability to survive within the host, subsequent studies to determine the combination of staphylococcal adhesins required for hyphal interactions in vivo were not possible.
Microscopic analysis
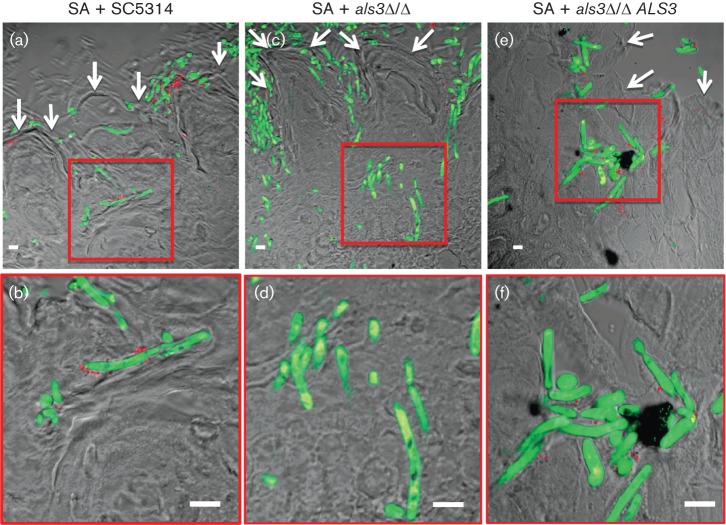
To investigate the role of Als3p in the invasive mechanism of co-colonization, experiments were performed where both micro-organisms were allowed to interact on excised tongues infected ex vivo. Similarly to the images of abiotic surfaces, S. aureus was seen strongly adhering to the C. albicans hyphae with the Als3p receptor (WT SC5314 and als3Δ/Δ ALS3; Fig. 2b, f) but only minimally to the hyphae lacking the receptor als3Δ/Δ (Fig. 2d). However, in all three C. albicans strains, S. aureus and C. albicans were seen forming a polymicrobial biofilm on the surface of the tongue (indicated by the white arrows in Fig. 2a, c, e). Importantly, in addition to mere co-colonization, this model demonstrated tissue co-penetration of S. aureus along with the invasive hyphae of C. albicans into the subepithelium (Fig. 2a, b, e, f), a process dependent on Als3p as S. aureus did not co-penetrate with als3Δ/Δ (Fig. 2c, d). We did notice that that the staphylococcal hyphal binding was markedly reduced in these studies compared to the in vitro analysis of polymicrobial biofilm development using confocal microscopy. These differences may have been due to salivary components and structural complexities of the tongue environment, as well as the tongue’s ability to produce host defence factors, such as β-defensins (Weinberg et al., 1998).
Fig. 2.
Microscopic images (low and high magnification) of sections of tongues infected and stained with probes specific to S. aureus (red) and C. albicans (green) demonstrating hyphal invasion of tissue and bacterial presence. Hyphae of Als3p-expressing C. albicans strains (WT SC5314 and complemented strain als3Δ/Δ ALS3) are seen penetrating the epithelium with S. aureus (SA) adhering to and thus co-invading the tissue with the hyphae (a, b, e, f). In contrast, although the hyphae of the C. albicans Als3p mutant (als3Δ/Δ) penetrated the tissue and SA adhered to the border of the tongues, no bacteria were seen adhering to the hyphae or detected in the subepithelium (c, d). White arrows denote tongue outer surface and scale bars represent 10 µm.
Murine in vivo model of oral co-colonization
An animal model of microbial co-colonization was developed to investigate the pathogenic implications of microbial interactions on mucosal tissue in immunocompromised hosts. Mouse oral cavities were inoculated with both S. aureus and C. albicans alone or in combination, and tongues and kidneys were harvested from euthanized animals. In addition, the C. albicans als3Δ/Δ mutant and als3Δ/Δ ALS3 complemented strain were inoculated as well, either alone or in combination with S. aureus. Clinical presentation of the harvested tongues was consistent with advanced candidiasis and diagnosis was confirmed by histopathology.
None of the animals colonized with S. aureus alone exhibited significant symptoms of disease or weight loss, and all survived until the conclusion of the experiment (Fig. 3). Mice infected with C. albicans alone exhibited symptoms of oral candidiasis and half of the mice required early euthanization due to significant weight loss (Fig. 3). However, none of these mice exhibited symptoms of systemic infection, suggesting the weight loss was solely due to the oral candidiasis, and not because of an invasive infection. There was no significant difference in the survival among all three of the C. albicans strains administered alone (data not shown).
Fig. 3.
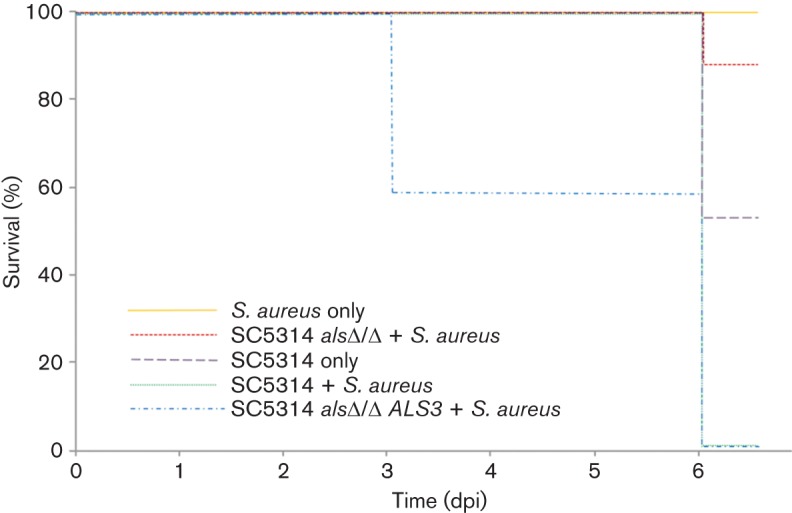
Mortality in mice orally colonized with either S. aureus or C. albicans individually or in combination. Animals were weighed 1, 3 and 6 days post-inoculation (dpi). Those animals exhibiting >20 % weight loss (defined by the University of Maryland Institutional Animal Care and Use Committee as a humane end point in rodents), were considered deceased and were euthanized. Mice co-colonized with S. aureus and C. albicans (SC5314+S. aureus) showed a significant decrease in survival vs S. aureus alone or C. albicans alone as determined by the Log Rank Test where P<0.05 is considered significant. Additionally, there was no significant difference between survival in mice colonized with S. aureus alone and mice co-colonized with the Als3p mutant (Log Rank Test P = 0.197). The dramatic weight loss and associated mortality seen in co-colonized mice was restored once Als3 expression was complemented in the als3Δ/Δ ALS3 strain. All experiments were performed with at least four mice per group. Survival in SC5314 als3Δ/Δ alone and SC5314 als3Δ/Δ ALS3 alone were not significantly different than survival with SC5314 alone (data not shown).
As opposed to the animals orally inoculated with either single species, those simultaneously inoculated with S. aureus and C. albicans containing an intact Als3p (WT SC5314 or SC5314 als3Δ/Δ ALS3 complemented) exhibited signs of systemic infection, including dehydration and lethargy. Additionally, the mice in this group showed significant loss of body weight, requiring early euthanization and halting of experiments at 6 days post-inoculation (dpi), as per the University of Maryland Institutional Animal Care and Use Committee humane end point guidelines. Significantly, all of these animals succumbed to their infection prior to the end of the experiment as determined by the humane end points (Fig. 3). In contrast, none of the animals in the group co-colonized with S. aureus and C. albicans lacking Als3p exhibited evidence of systemic infection and the majority (83 %) of these mice survived until the conclusion of the experiment (Fig. 3).
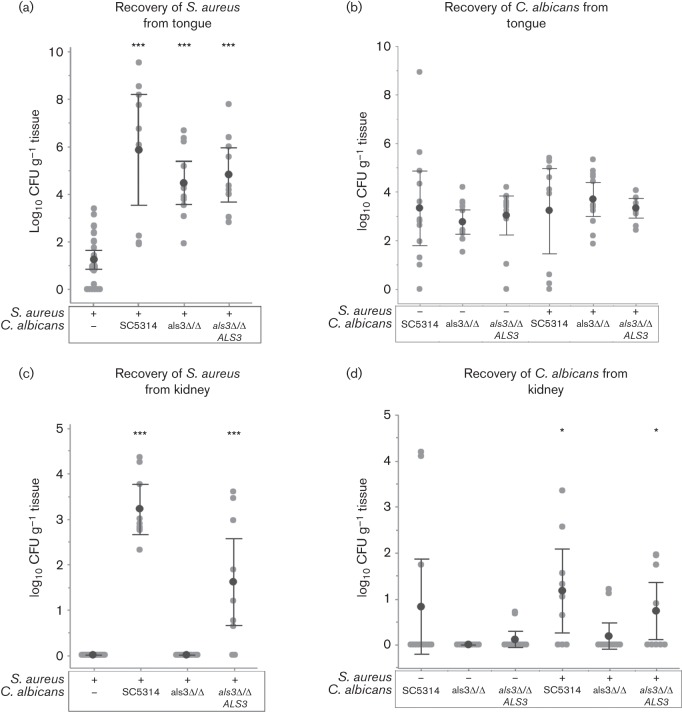
In order to diagnose the cause of morbidity in the sick animals, the tongues and kidneys were harvested and cultured to identify microbial presence. There was no significant difference in c.f.u. of C. albicans recovered from the tongues, regardless of which C. albicans strain was used and whether it was inoculated alone or co-inoculated with S. aureus, suggesting that C. albicans does not exhibit any deficiency in colonizing the tongue in the absence of Als3p (Fig. 4b). S. aureus, on the other hand, exhibited enhanced colonization of the tongue when co-inoculated with any of the C. albicans strains as compared to S. aureus given alone (Fig. 4a).
Fig. 4.
Microbial burden in the tongue and kidneys in mice orally colonized with either S. aureus or C. albicans WT (SC5314), Als3p mutant (als3Δ/Δ), and Als3p complemented (als3Δ/Δ ALS3) individually, or in combination. Three days post-infection, tissue was extracted, homogenized and cultured for assessment of microbial presence based on number of organisms recovered per gram of tissue. Number of microbes recovered is reported as log10 of c.f.u. per gram of respective tissue. (a, c) S. aureus recovered from the tongues (a) and kidneys (c) of infected mice. (b, d) C. albicans recovered from the tongues (b) and kidneys (d) of infected mice. Both microbes were recovered from tongues of mice under all conditions tested. However, S. aureus was only recovered from kidneys of mice co-colonized with either WT C. albicans or mutant C. albicans that had been complemented for Als3p expression. All experiments were performed in triplicate in groups of four mice per group. In addition to all data points, the mean of the log of c.f.u./gram is shown. Error bars indicate 95 % confidence intervals. Significance was determined using the Mann–Whitney U Test to compare each polymicrobial condition to the monomicrobial control infection. Asterisks denotes statistical significance versus the mono-microbial control infection according to the Mann–Whitney U Test. * indicates P<0.05 and *** indicates P<0.001.
However, the clinical symptoms indicating systemic disease were corroborated by the recovery of S. aureus and C. albicans from the kidneys in strains with a functional Als3p protein compared to the als3Δ/Δ (Fig. 4c, d). High numbers of S. aureus were recovered from mice co-infected with C. albicans expressing intact Als3p (WTSC5314 or SC5314 als3Δ/Δ ALS3 complemented) and no S. aureus was found in the kidneys of mice inoculated without C. albicans or with C. albicans lacking Als3p (Fig. 4c). Additionally, there were significant c.f.u. of C. albicans recovered from the kidneys of mice co-inoculated with S. aureus and the WT C. albicans. This is in contrast with mice inoculated with C. albicans alone or co-inoculated with S. aureus and C. albicans lacking Als3p, where most mice had no C. albicans present in their kidneys (Fig. 4d).
The data presented in Fig. 4 suggests that, as seen in Fig. 2, S. aureus and C. albicans are able to form a dense polymicrobial biofilm on the epithelial surface of the tongue with or without the expression of Als3p. However, S. aureus is unable to enter the bloodstream and disseminate in the absence of Als3p, ostensibly due to the lack of binding to penetrating hyphae as shown in Fig. 2. Collectively, these findings demonstrate that co-colonization of the oral mucosa by C. albicans and S. aureus in immunocompromised animals may lead to systemic S. aureus infections with high morbidity and mortality.
Discussion
Polymicrobial diseases represent the clinical manifestations induced by the presence of multiple microbial species that may act synergistically to cause complex infectious processes (Brogden & Gerberding, 2002; Pittet et al., 1993; Tuft, 2006). Yet, despite the gravity of such infections, areas of study in polymicrobial diseases and particularly those involving vastly diverse pathogens such as fungi and bacteria are in their infancy (Peleg et al., 2010; Shirtliff et al., 2009). C. albicans and S. aureus are frequent colonizers of human mucosal surfaces and exhibit a strong ability to adhere to host tissue and develop drug tolerance. However, studies investigating the implications of their interaction as they co-colonize host mucosal tissue have not been previously performed.
S. aureus avidly and specifically adheres to hyphae of C. albicans in vitro, as illustrated using static and time lapse microscopy (supplementary movie V1). Adhesion is followed by tissue penetration of S. aureus along with the invasive hyphae (Fig. 2). Although no single adhesin of S. aureus was found to be solely responsible for hyphal binding, three staphylococcal adhesin mutants (ΔfnpB, ΔsasF, and ΔAtl) were found to have significantly reduced hyphal binding compared to the WT strain (Fig. 1). FnpB (fibronectin binding protein B) is one of two important fibronectin binding proteins in S. aureus. FnpB is known to be upregulated in host tissues, and helps facilitate binding to eukaryotic cells (Menzies, 2003). Interestingly, the other structurally related S. aureus fibronectin binding protein, FnpA, did not appear to be involved in hyphal binding (Fig. 2), suggesting that the minor structural differences between FnpB and FnpA may influence the ability to bind C. albicans hyphae. SasF (staphylococcus aureus surface protein F) is a highly conserved cell surface-associated adhesin (McCarthy & Lindsay, 2010). SasF has been shown to be upregulated in response to linoleic acid (Kenny et al., 2009), which is one of the major endogenous fatty acids in C. albicans (Nigam et al., 2011). The third mutant with significantly reduced binding was a mutation of Atl, an uncharacterized protein that is annotated as a putative N-acetylmuramoyl-l-alanine amidase, and likely functions as an S. aureus autolysin (Diep et al., 2006).
In addition, five other staphylococcal mutants had similar reductions in binding (although these were not statistically significantly different from the WT strain). This was not surprising considering the redundant nature of adhesin binding partners in S. aureus (Clarke & Foster, 2006). Interestingly, neither the JE2 WT strain binding to the Als3p deficient mutant nor any of the S. aureus mutants binding to the WT C. albicans showed a reduction in binding to less than 50 %. This may be due to the expression of other proteins on C. albicans hyphae that can have minor roles in S. aureus adherence. One such protein is Als1, which was shown in an earlier study to have a role, albeit a much smaller role compared to Als3p, in binding S. aureus in vitro (Peters et al., 2012). However, the clear difference between the C. albicans als3Δ/Δ mutant and the WT strain, both in terms of hyphal binding as shown in Fig. 2 and dissemination as shown in Fig. 4, suggest that differential in vivo regulation of these other C. albicans proteins may reduce their contributions to S. aureus binding during an actual infection.
This interaction with staphylococcal adhesins and Als3p may not be surprising since a number of S. aureus adhesins are predicted to have significant 3D homology to Als3p and other Als proteins (Nobile et al., 2008; Salgado et al., 2011). This homology is also demonstrated in cross protection studies in which immunization with C. albicans Als3p has been show to provide protection from S. aureus challenge in animal models of infection (Ibrahim et al., 2013; Lin et al., 2009). Since the self–self binding and clumping due to Als proteins have been well documented, the binding of staphylococcal adhesins to Als3p may mimic this self-agglutination (Lipke et al., 2012; Ramsook et al., 2010).
In order to determine the in vivo implications of this Als3p-dependent interaction, we developed a novel murine model to demonstrate the potential progression of mucosal co-colonization into disseminated systemic disease. In our animal model, mice co-colonized with C. albicans and S. aureus succumbed to systemic bacterial infection, S. aureus was recovered from the kidneys, and Als3p was crucial for this systemic dissemination (Fig. 3 and 4). As expected, colonization with S. aureus individually did not result in death or any signs of significant morbidity, as S. aureus typically requires a breach in host surface barriers to invade (Acton et al., 2009; Smith et al., 2003; Veeh et al., 2003).
While the enhanced colonization of the tongue by S. aureus in the presence of all three C. albicans strains (Fig. 4a) may not correlate to the pattern of colonization seen in the kidney (Fig. 4c), this is likely to reflect the fact that the C. albicans factors responsible for co-colonization are different from the one required for hyphal binding. As shown in an earlier study, the co-culture of C. albicans and S. aureus together causes reciprocal changes in protein expression (Peters et al., 2010). Additionally, although S. aureus has reduced binding to C. albicans lacking Als3p, it does not show a reduction in polymicrobial biofilm formation (Peters et al., 2012). Instead, what is likely to be happening in vivo is that S. aureus and C. albicans are still able to form a dense biofilm on the epithelial surface of the tongue, but are unable to attach to penetrating hyphae. This is what is observed in Fig. 2.
It is important to note that the mutant strain lacking Als3p did not exhibit a defect in hyphal formation (Fig. 2). This is significant, since we propose that Als3p does not affect C. albicans pathogenesis when present as a mono-species infection, but instead is important for synergy with S. aureus. Also, while the mutant lacking Als3p had reduced colonization of the kidneys in co-colonization experiments, Als3p has previously been shown to be dispensable for C. albicans pathogenesis in an intravenous challenge model (Cleary et al., 2011). This suggests that the increased presence of the WT C. albicans in the kidneys in comparison to the mutant is also mediated by this synergy with S. aureus. As mentioned previously, S. aureus can produce an array of virulence factors (Bien et al., 2011; Ferry et al., 2005) that are likely to be helping to further mediate tissue lysis and facilitate dissemination of both S. aureus and C. albicans into the bloodstream. The fact that deletion of Als3p does not alter the formation of hyphae (a necessary prerequisite for S. aureus attachment; Peters et al., 2010), nor affects the virulence of C. albicans in the bloodstream, suggests that its role in our model is solely due to S. aureus attachment. Therefore, the inability of the mutant to mediate a bacterial or fungal systemic disease further confirms the specificity of Als3p in the invasion process.
As our animal model involved the establishment of a polymicrobial infection in mice receiving immunosuppression, it is most directly relevant to cases of co-colonization in immunocompromised individuals. Immunocompromised hosts represent a significant and appropriate patient population to model, as these patients frequently suffer from infections and diseases of the oral mucosa and bloodstream that involve C. albicans and S. aureus (Bassetti et al., 2012; Delorenze et al., 2013; Dongari-Bagtzoglou et al., 2009; Marukutira et al., 2014; Olczak-Kowalczyk et al., 2012; Stammler Jaliff et al., 2014; Yehia et al., 2011). Additionally, a recent pair of studies found that among HIV positive individuals, S. aureus could be isolated from the oral cavity of 92 % of individuals while C. albicans was isolated from 73 % of the study subjects (Back-Brito et al., 2009, 2011), suggesting our model is highly relevant clinically. Furthermore, due to the continued rise in the prevalence of HIV (Hall et al., 2008) and the continued increase in patients living with transplants and receiving immunosuppression (Hall et al., 2008; Port et al., 2007), the population of immunocompromised patients at risk for co-colonization with S. aureus and C. albicans continues to expand.
While this ‘hitchhiking’ phenomenon of systemic infection stemming from staphylococcal–candidal interaction has yet to be established in a human host, we propose that this may be due to a failure to look for it. It is feasible to speculate that in a significant number of the 40–50 % of cases of staphylococcal bloodstream infections with no clearly established portal of entry (del Rio et al., 2009), oral candidiasis may have been the underlying cause. Should this phenomenon occur in a vulnerable human host harbouring both species, as often occurs on mucosal surfaces, the medical consequences are likely to be significant, as reflected by the high rate of morbidity and mortality observed in our co-colonized animals (Fig. 3).
Although this study focused on C. albicans and S. aureus, it is important to note that in the oral cavity, C. albicans co-exists with the myriad of bacterial species commonly residing the various niches of this complex environment (Bagg & Silverwood, 1986; Brehm-Stecher & Johnson, 2003; Holmes et al., 1995, 1996; Jenkinson et al., 1990; Klotz et al., 2007; Silverman et al., 2010). The oral cavity is colonized by an ever-changing population (Lazarevic et al., 2010) of over 900 bacterial species and therefore, it may also be conceivable that similar phenomena with clinical relevance may also occur between C. albicans and oral bacteria (Zaura et al., 2009). This has been seen in a recent study in which Streptococcus oralis colonization of the oral and gastrointestinal tract was augmented in the presence of C. albicans and resulted in increased size and frequency of thrush lesions in a murine model of dual infection (Xu et al., 2014). As our knowledge of interspecies interactions on niches expands, the role of C. albicans in pathogenesis might prove to be more intricate than currently recognized.
A potential limitation of the study is that this interaction was only evaluated in oral cavity instead of in other body locales where these microbes, particularly S. aureus, have been isolated, including the nasopharynx, moist skin areas and the genital tract. While the model used in this study has relevance, since both of these microbes have been co-isolated in the oral cavity, we look forward to future studies where we can evaluate this interaction in the mouse nasopharynx and skin models. Also worth noting is the possibility of gastrointestinal (GI) colonization to contribute to this ‘hitchhiking’ phenomenon. Although our study focused on the oral mucosa, based on the oral administration of the microbes there was potential for the co-colonization of the GI tract to occur as well. C. albicans and S. aureus have both been shown to be able to colonize the GI tract (Acton et al., 2009; Prieto et al., 2014), and GI candidiasis can occur in immunocompromised individuals, although it is much less common than oral candidiasis (Vazquez & Sobel, 2002). Although our current study failed to examine this potential route of invasion, we feel that based on the requirement that Als3p be present in order to produce systemic S. aureus bacteraemia, any dissemination occurring through the gut mucosa is likely to be occurring through the same proposed model of Als3p binding and hitchhiking. We hope to clarify the potential role this important mucosal site may play in this phenomenon in future studies.
In summary, this study elucidates a synergistic interaction between the bacterial pathogen S. aureus and the opportunistic fungal pathogen C. albicans during polymicrobial biofilm growth in the oral cavity, for which we propose the term ‘microbial hitchhiking’. The interactions between these diverse and important commensals and opportunistic pathogens hold significant clinical implications, and therefore characterizing their complex interactions is a major step in understanding the nature of their co-existence in the host. Although retrospective and prospective clinical studies are warranted, our findings advocate screening of critically ill patients for oral candidal colonization as a measure for the prevention of systemic bacterial infections.
Acknowledgements
We would like to thank Dr Vincent Bruno for his contribution. We also graciously thank AdvanDx for the donation of several PNA-FISH probes and Timo Kreike of Westburg BV for his assistance with the Bioflux 200 and EVOS fl digital fluorescence microscope. This study was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant R01AI69568), and the National Institute of Dental and Craniofacial Research, National Institutes of Health (grant 1R01DE20939).
Abbreviations:
- d.p.i.
days post-inoculation
- PNA-FISH
peptide-nucleic acid fluorescent in situ hybridization
- TSB
trypticase soy broth
Footnotes
One supplementary video is available with the online Supplementary Material.
References
- Acton D. S., Plat-Sinnige M. J., van Wamel W., de Groot N., van Belkum A. (2009). Intestinal carriage of Staphylococcus aureus: how does its frequency compare with that of nasal carriage and what is its clinical impact? Eur J Clin Microbiol Infect Dis 28, 115–127. 10.1007/s10096-008-0602-7 [DOI] [PubMed] [Google Scholar]
- Adam B., Baillie G. S., Douglas L. J. (2002). Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J Med Microbiol 51, 344–349. [DOI] [PubMed] [Google Scholar]
- Ali S. A., Cesani F., Nusynowitz M. L., Briscoe E. G., Shirtliff M. E., Mader J. T. (1997). Skeletal scintigraphy with technetium-99m-tetraphenyl porphyrin sulfonate for the detection and determination of osteomyelitis in an animal model. J Nucl Med 38, 1999–2002. [PubMed] [Google Scholar]
- Back-Brito G. N., Mota A. J., Vasconcellos T. C., Querido S. M., Jorge A. O., Reis A. S., Balducci I., Koga-Ito C. Y. (2009). Frequency of Candida spp. in the oral cavity of Brazilian HIV-positive patients and correlation with CD4 cell counts and viral load. Mycopathologia 167, 81–87. 10.1007/s11046-008-9153-9 [DOI] [PubMed] [Google Scholar]
- Back-Brito G. N., El Ackhar V. N., Querido S. M., dos Santos S. S., Jorge A. O., Reis A. S., Koga-Ito C. Y. (2011). Staphylococcus spp., Enterobacteriaceae and Pseudomonadaceae oral isolates from Brazilian HIV-positive patients. Correlation with CD4 cell counts and viral load. Arch Oral Biol 56, 1041–1046. 10.1016/j.archoralbio.2011.02.016 [DOI] [PubMed] [Google Scholar]
- Baena-Monroy T., Moreno-Maldonado V., Franco-Martínez F., Aldape-Barrios B., Quindós G., Sánchez-Vargas L. O. (2005). Candida albicans, Staphylococcus aureus and Streptococcus mutans colonization in patients wearing dental prosthesis. Med Oral Patol Oral Cir Bucal 10 (Suppl 1), E27–E39. [PubMed] [Google Scholar]
- Bagg J., Silverwood R. W. (1986). Coagglutination reactions between Candida albicans and oral bacteria. J Med Microbiol 22, 165–169. 10.1099/00222615-22-2-165 [DOI] [PubMed] [Google Scholar]
- Bassetti M., Trecarichi E. M., Mesini A., Spanu T., Giacobbe D. R., Rossi M., Shenone E., Pascale G. D., Molinari M. P. & other authors (2012). Risk factors and mortality of healthcare-associated and community-acquired Staphylococcus aureus bacteraemia. Clin Microbiol Infect 18, 862–869. 10.1111/j.1469-0691.2011.03679.x [DOI] [PubMed] [Google Scholar]
- Bien J., Sokolova O., Bozko P. (2011). Characterization of virulence factors of Staphylococcus aureus: Novel function of known virulence factors that are implicated in activation of airway epithelial proinflammatory response. J Pathogens 2011, 601905. 10.4061/2011/601905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose J. L., Lehman M. K., Fey P. D., Bayles K. W. (2012). Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS ONE 7, e42244. 10.1371/journal.pone.0042244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady R. A., Leid J. G., Camper A. K., Costerton J. W., Shirtliff M. E. (2006). Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infect Immun 74, 3415–3426. 10.1128/IAI.00392-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady R. A., Leid J. G., Kofonow J., Costerton J. W., Shirtliff M. E. (2007). Immunoglobulins to surface-associated biofilm immunogens provide a novel means of visualization of methicillin-resistant Staphylococcus aureus biofilms. Appl Environ Microbiol 73, 6612–6619. 10.1128/AEM.00855-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady R. A., O’May G. A., Leid J. G., Prior M. L., Costerton J. W., Shirtliff M. E. (2011). Resolution of Staphylococcus aureus biofilm infection using vaccination and antibiotic treatment. Infect Immun 79, 1797–1803. 10.1128/IAI.00451-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm-Stecher B. F., Johnson E. A. (2003). Sensitization of Staphylococcus aureus and Escherichia coli to antibiotics by the sesquiterpenoids nerolidol, farnesol, bisabolol, and apritone. Antimicrob Agents Chemother 47, 3357–3360. 10.1128/AAC.47.10.3357-3360.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden K. A. (2003). Polymicrobial infections in animals and humans. In American Society for Microbiology Conference on Polymicrobial Diseases, October 19-23, Lake Tahoe, Nevada. [Google Scholar]
- Brogden K. A., Gerberding J. M. (2002). Polymicrobial Diseases, pp. 1–446. Washington: American Society for Microbiology. [Google Scholar]
- Budtz-Jłrgensen E., Mojon P., Banon-Clément J. M., Baehni P. (1996). Oral candidosis in long-term hospital care: comparison of edentulous and dentate subjects. Oral Dis 2, 285–290. 10.1111/j.1601-0825.1996.tb00239.x [DOI] [PubMed] [Google Scholar]
- Calderone R., Clancy C. (2012). Candida and Candidiasis, 2nd edn Washington, DC: American Society for Microbiology. [Google Scholar]
- Cannon R. D., Chaffin W. L. (2001). Colonization is a crucial factor in oral candidiasis. J Dent Educ 65, 785–787. [PubMed] [Google Scholar]
- Carlson E. (1983). Effect of strain of Staphylococcus aureus on synergism with Candida albicans resulting in mouse mortality and morbidity. Infect Immun 42, 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson E., Johnson G. (1985). Protection by Candida albicans of Staphylococcus aureus in the establishment of dual infection in mice. Infect Immun 50, 655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Krishnan V., Macon K., Manne K., Narayana S. V., Schneewind O. (2013). Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J Biol Chem 288, 29440–29452. 10.1074/jbc.M113.502039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. R., Foster S. J. (2006). Surface adhesins of Staphylococcus aureus. Adv Microb Physiol 51, 187–224. 10.1016/S0065-2911(06)51004-5 [DOI] [PubMed] [Google Scholar]
- Cleary I. A., Reinhard S. M., Miller C. L., Murdoch C., Thornhill M. H., Lazzell A. L., Monteagudo C., Thomas D. P., Saville S. P. (2011). Candida albicans adhesin Als3p is dispensable for virulence in the mouse model of disseminated candidiasis. Microbiology 157, 1806–1815. 10.1099/mic.0.046326-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Marrie T. J., Cheng K. J. (1985). Phenomena of Bacterial Adhesion, pp. 650–654. Edited by Savage D., Fletcher M. New York: Plenum Press. [Google Scholar]
- Cuesta A. I., Jewtuchowicz V., Brusca M. I., Nastri M. L., Rosa A. C. (2010). Prevalence of Staphylococcus spp and Candida spp in the oral cavity and periodontal pockets of periodontal disease patients. Acta Odontol Latinoam 23, 20–26. [PubMed] [Google Scholar]
- de Repentigny L., Lewandowski D., Jolicoeur P. (2004). Immunopathogenesis of oropharyngeal candidiasis in human immunodeficiency virus infection. Clin Microbiol Rev 17, 729–759. 10.1128/CMR.17.4.729-759.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio A., Cervera C., Moreno A., Moreillon P., Miró J. M. (2009). Patients at risk of complications of Staphylococcus aureus bloodstream infection. Clin Infect Dis 48 (Suppl 4), S246–S253. 10.1086/598187 [DOI] [PubMed] [Google Scholar]
- Delorenze G. N., Horberg M. A., Silverberg M. J., Tsai A., Quesenberry C. P., Baxter R. (2013). Trends in annual incidence of methicillin-resistant Staphylococcus aureus (MRSA) infection in HIV-infected and HIV-uninfected patients. Epidemiol Infect 141, 2392–2402. 10.1017/S0950268813000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep B. A., Gill S. R., Chang R. F., Phan T. H., Chen J. H., Davidson M. G., Lin F., Lin J., Carleton H. A. & other authors (2006). Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739. 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A., Dwivedi P., Ioannidou E., Shaqman M., Hull D., Burleson J. (2009). Oral Candida infection and colonization in solid organ transplant recipients. Oral Microbiol Immunol 24, 249–254. 10.1111/j.1399-302X.2009.00505.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi P., Thompson A., Xie Z., Kashleva H., Ganguly S., Mitchell A. P., Dongari-Bagtzoglou A. (2011). Role of Bcr1-activated genes Hwp1 and Hyr1 in Candida albicans oral mucosal biofilms and neutrophil evasion. PLoS ONE 6, e16218. 10.1371/journal.pone.0016218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A. M., Grossman T. J., Rudney J. D. (2006). Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infect Immun 74, 654–662. 10.1128/IAI.74.1.654-662.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry T., Perpoint T., Vandenesch F., Etienne J. (2005). Virulence determinants in Staphylococcus aureus and their involvement in clinical syndromes. Curr Infect Dis Rep 7, 420–428. 10.1007/s11908-005-0043-8 [DOI] [PubMed] [Google Scholar]
- Fidel P. L., Jr (2006). Candida-host interactions in HIV disease: relationships in oropharyngeal candidiasis. Adv Dent Res 19, 80–84. 10.1177/154407370601900116 [DOI] [PubMed] [Google Scholar]
- Gillum A. M., Tsay E. Y., Kirsch D. R. (1984). Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198, 179–182. 10.1007/BF00328721 [DOI] [PubMed] [Google Scholar]
- Goetghebeur M., Landry P. A., Han D., Vicente C. (2007). Methicillin-resistant Staphylococcus aureus: A public health issue with economic consequences. Can J Infect Dis Med Microbiol 18, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon R. J., Lowy F. D. (2008). Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46 (Suppl 5), S350–S359. 10.1086/533591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Haque A., Mukhopadhyay G., Narayan R. P., Prasad R. (2005). Interactions between bacteria and Candida in the burn wound. Burns 31, 375–378. 10.1016/j.burns.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Hall H. I., Song R., Rhodes P., Prejean J., An Q., Lee L. M., Karon J., Brookmeyer R., Kaplan E. H. & other authors (2008). Estimation of HIV incidence in the United States. JAMA 300, 520–529. 10.1001/jama.300.5.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriott M. M., Noverr M. C. (2009). Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother 53, 3914–3922. 10.1128/AAC.00657-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harro J. M., Daugherty S., Bruno V. M., Jabra-Rizk M. A., Rasko D. A., Shirtliff M. E. (2013). Draft genome sequence of the Methicillin-Resistant Staphylococcus aureus isolate MRSA-M2. Genome Announc 1, e00037-12. 10.1128/genomeA.00037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A. R., Cannon R. D., Jenkinson H. F. (1995). Interactions of Candida albicans with bacteria and salivary molecules in oral biofilms. J Ind Microbiol 15, 208–213. 10.1007/BF01569827 [DOI] [PubMed] [Google Scholar]
- Holmes A. R., McNab R., Jenkinson H. F. (1996). Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect Immun 64, 4680–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A. S., Luo G., Gebremariam T., Lee H., Schmidt C. S., Hennessey J. P., Jr, French S. W., Yeaman M. R., Filler S. G., Edwards J. E., Jr (2013). NDV-3 protects mice from vulvovaginal candidiasis through T- and B-cell immune response. Vaccine 31, 5549–5556. 10.1016/j.vaccine.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabra-Rizk M. A., Meiller T. F., James C. E., Shirtliff M. E. (2006). Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob Agents Chemother 50, 1463–1469. 10.1128/AAC.50.4.1463-1469.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz L. M., Deng D. M., van der Mei H. C., Crielaard W., Krom B. P. (2009). Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell 8, 1658–1664. 10.1128/EC.00070-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson H., Douglas L. (2002). Candida Interactions with Bacterial Biofilms, pp. 357–373. Edited by Brogden K. A., Guthmiller J. M. Washington, DC: American Society for Microbiology. [Google Scholar]
- Jenkinson H. F., Lala H. C., Shepherd M. G. (1990). Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun 58, 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamai Y., Kubota M., Kamai Y., Hosokawa T., Fukuoka T., Filler S. G. (2002). Contribution of Candida albicans ALS1 to the pathogenesis of experimental oropharyngeal candidiasis. Infect Immun 70, 5256–5258. 10.1128/IAI.70.9.5256-5258.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A. D., Otto M., Braughton K. R., Whitney A. R., Chen L., Mathema B., Mediavilla J. R., Byrne K. A., Parkins L. D. & other authors (2008). Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A 105, 1327–1332. 10.1073/pnas.0710217105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A. D., Porcella S. F., Martens C., Whitney A. R., Braughton K. R., Chen L., Craig C. T., Tenover F. C., Kreiswirth B. N. & other authors (2010). Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J Clin Microbiol 48, 4504–4511. 10.1128/JCM.01050-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny J. G., Ward D., Josefsson E., Jonsson I. M., Hinds J., Rees H. H., Lindsay J. A., Tarkowski A., Horsburgh M. J. (2009). The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS ONE 4, e4344. 10.1371/journal.pone.0004344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., Harrison L. H., Lynfield R., Dumyati G. & other authors (2007). Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298, 1763–1771. 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- Klotz S. A., Gaur N. K., De Armond R., Sheppard D., Khardori N., Edwards J. E., Jr, Lipke P. N., El-Azizi M. (2007). Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med Mycol 45, 363–370. 10.1080/13693780701299333 [DOI] [PubMed] [Google Scholar]
- Kulak-Ozkan Y., Kazazoglu E., Arikan A. (2002). Oral hygiene habits, denture cleanliness, presence of yeasts and stomatitis in elderly people. J Oral Rehabil 29, 300–304. 10.1046/j.1365-2842.2002.00816.x [DOI] [PubMed] [Google Scholar]
- Lazarevic V., Whiteson K., Hernandez D., François P., Schrenzel J. (2010). Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics 11, 523. 10.1186/1471-2164-11-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid J. G., Shirtliff M. E., Costerton J. W., Stoodley P. (2002). Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun 70, 6339–6345. 10.1128/IAI.70.11.6339-6345.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Busscher H. J., van der Mei H. C., Norde W., Krom B. P., Sjollema J. (2011). Analysis of the contribution of sedimentation to bacterial mass transport in a parallel plate flow chamber: part II: use of fluorescence imaging. Colloids Surf B Biointerfaces 87, 427–432. 10.1016/j.colsurfb.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Lin L., Ibrahim A. S., Xu X., Farber J. M., Avanesian V., Baquir B., Fu Y., French S. W., Edwards J. E., Jr, Spellberg B. (2009). Th1-Th17 cells mediate protective adaptive immunity against Staphylococcus aureus and Candida albicans infection in mice. PLoS Pathog 5, e1000703. 10.1371/journal.ppat.1000703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke P. N., Garcia M. C., Alsteens D., Ramsook C. B., Klotz S. A., Dufrêne Y. F. (2012). Strengthening relationships: amyloids create adhesion nanodomains in yeasts. Trends Microbiol 20, 59–65. 10.1016/j.tim.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch A. S., Robertson G. T. (2008). Bacterial and fungal biofilm infections. Annu Rev Med 59, 415–428. 10.1146/annurev.med.59.110106.132000 [DOI] [PubMed] [Google Scholar]
- Marukutira T., Huprikar S., Azie N., Quan S. P., Meier-Kriesche H. U., Horn D. L. (2014). Clinical characteristics and outcomes in 303 HIV-infected patients with invasive fungal infections: data from the Prospective Antifungal Therapy Alliance registry, a multicenter, observational study. HIV/AIDS–Research and Palliative Care 6, 39–47. 10.1146/annurev.med.59.110106.132000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S. K., Liu G., Jensen E. R., Lenoy E., Schneewind O. (2000). Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci U S A 97, 5510–5515. 10.1073/pnas.080520697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A. J., Lindsay J. A. (2010). Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol 10, 173. 10.1186/1471-2180-10-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies B. E. (2003). The role of fibronectin binding proteins in the pathogenesis of Staphylococcus aureus infections. Curr Opin Infect Dis 16, 225–229. 10.1097/00001432-200306000-00007 [DOI] [PubMed] [Google Scholar]
- Nigam S., Ciccoli R., Ivanov I., Sczepanski M., Deva R. (2011). On mechanism of quorum sensing in Candida albicans by 3(R)-hydroxy-tetradecaenoic acid. Curr Microbiol 62, 55–63. 10.1007/s00284-010-9666-6 [DOI] [PubMed] [Google Scholar]
- Nobile C. J., Schneider H. A., Nett J. E., Sheppard D. C., Filler S. G., Andes D. R., Mitchell A. P. (2008). Complementary adhesin function in C. albicans biofilm formation. Curr Biol 18, 1017–1024. 10.1016/j.cub.2008.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara-Nemoto Y., Haraga H., Kimura S., Nemoto T. K. (2008). Occurrence of staphylococci in the oral cavities of healthy adults and nasal oral trafficking of the bacteria. J Med Microbiol 57, 95–99. 10.1099/jmm.0.47561-0 [DOI] [PubMed] [Google Scholar]
- Olczak-Kowalczyk D., Daszkiewicz M., Krasuska-Sławińska, Dembowska-Bagińska B., Gozdowski D., Daszkiewicz P., Fronc B., Semczuk K. (2012). Bacteria and Candida yeasts in inflammations of the oral mucosa in children with secondary immunodeficiency. J Oral Pathol Med 41, 568–576. [PubMed] [Google Scholar]
- Palmqvist N., Foster T., Fitzgerald J. R., Josefsson E., Tarkowski A. (2005). Fibronectin-binding proteins and fibrinogen-binding clumping factors play distinct roles in staphylococcal arthritis and systemic inflammation. J Infect Dis 191, 791–798. 10.1086/427663 [DOI] [PubMed] [Google Scholar]
- Pate J. C., Jones D. B., Wilhelmus K. R. (2006). Prevalence and spectrum of bacterial co-infection during fungal keratitis. Br J Ophthalmol 90, 289–292. 10.1136/bjo.2005.081869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A. Y., Hogan D. A., Mylonakis E. (2010). Medically important bacterial-fungal interactions. Nat Rev Microbiol 8, 340–349. 10.1038/nrmicro2313 [DOI] [PubMed] [Google Scholar]
- Perlroth J., Choi B., Spellberg B. (2007). Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol 45, 321–346. 10.1080/13693780701218689 [DOI] [PubMed] [Google Scholar]
- Peters B. M., Noverr M. C. (2013). Candida albicans-Staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect Immun 81, 2178–2189. 10.1128/IAI.00265-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. M., Jabra-Rizk M. A., Scheper M. A., Leid J. G., Costerton J. W., Shirtliff M. E. (2010). Microbial interactions and differential protein expression in Staphylococcus aureus -Candida albicans dual-species biofilms. FEMS Immunol Med Microbiol 59, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. M., Ovchinnikova E. S., Krom B. P., Schlecht L. M., Zhou H., Hoyer L. L., Busscher H. J., van der Mei H. C., Jabra-Rizk M. A., Shirtliff M. E. (2012). Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology 158, 2975–2986. 10.1099/mic.0.062109-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan Q. T., Myers C. L., Fu Y., Sheppard D. C., Yeaman M. R., Welch W. H., Ibrahim A. S., Edwards J. E., Jr, Filler S. G. (2007). Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol 5, e64. 10.1371/journal.pbio.0050064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet D., Li N., Wenzel R. P. (1993). Association of secondary and polymicrobial nosocomial bloodstream infections with higher mortality. Eur J Clin Microbiol Infect Dis 12, 813–819. 10.1007/BF02000400 [DOI] [PubMed] [Google Scholar]
- Port F. K., Merion R. M., Finley M. P., Goodrich N. P., Wolfe R. A. (2007). Trends in organ donation and transplantation in the United States, 1996-2005. Am J Transplant 7 (s1), 1319–1326. 10.1111/j.1600-6143.2007.01778.x [DOI] [PubMed] [Google Scholar]
- Prabhakara R., Harro J. M., Leid J. G., Harris M., Shirtliff M. E. (2011a). Murine immune response to a chronic Staphylococcus aureus biofilm infection. Infect Immun 79, 1789–1796. 10.1128/IAI.01386-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakara R., Harro J. M., Leid J. G., Keegan A. D., Prior M. L., Shirtliff M. E. (2011b). Suppression of the inflammatory immune response prevents the development of chronic biofilm infection due to methicillin-resistant Staphylococcus aureus. Infect Immun 79, 5010–5018. 10.1128/IAI.05571-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto D., Román E., Correia I., Pla J. (2014). The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS ONE 9, e87128. 10.1371/journal.pone.0087128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsook C. B., Tan C., Garcia M. C., Fung R., Soybelman G., Henry R., Litewka A., O’Meally S., Otoo H. N. & other authors (2010). Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell 9, 393–404. 10.1128/EC.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Kokubu E., Inagaki S., Imamura K., Kita D., Lamont R. J., Ishihara K. (2012). Porphyromonas gingivalis entry into gingival epithelial cells modulated by Fusobacterium nucleatum is dependent on lipid rafts. Microb Pathog 53, 234–242. 10.1016/j.micpath.2012.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado P. S., Yan R., Taylor J. D., Burchell L., Jones R., Hoyer L. L., Matthews S. J., Simpson P. J., Cota E. (2011). Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A 108, 15775–15779. 10.1073/pnas.1103496108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirtliff M. E., Calhoun J. H., Mader J. T. (2002). Experimental osteomyelitis treatment with antibiotic-impregnated hydroxyapatite. Clin Orthop Relat Res 401, 239–247. 10.1097/00003086-200208000-00027 [DOI] [PubMed] [Google Scholar]
- Shirtliff M. E., Peters B. M., Jabra-Rizk M. A. (2009). Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett 299, 1–8. 10.1111/j.1574-6968.2009.01668.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman R. J., Nobbs A. H., Vickerman M. M., Barbour M. E., Jenkinson H. F. (2010). Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun 78, 4644–4652. 10.1128/IAI.00685-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. J., Robertson D., Tang M. K., Jackson M. S., MacKenzie D., Bagg J. (2003). Staphylococcus aureus in the oral cavity: a three-year retrospective analysis of clinical laboratory data. Br Dent J 195, 701–703. 10.1038/sj.bdj.4810832 [DOI] [PubMed] [Google Scholar]
- Stammler Jaliff B., Dahl-Knudsen J., Petersen A., Skov R., Benfield T. (2014). Outcome and reinfection after Staphylococcus aureus bacteraemia in individuals with and without HIV-1 infection: a case-control study. BMJ Open 4, e004075. 10.1136/bmjopen-2013-004075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P., Gow N., Berman J. (2004). The distinct morphogenic states of Candida albicans. Trends Microbiol 12, 317–324. 10.1016/j.tim.2004.05.008 [DOI] [PubMed] [Google Scholar]
- Tawara Y., Honma K., Naito Y. (1996). Methicillin-resistant Staphylococcus aureus and Candida albicans on denture surfaces. Bull Tokyo Dent Coll 37, 119–128. [PubMed] [Google Scholar]
- Timsit J. F., Cheval C., Gachot B., Bruneel F., Wolff M., Carlet J., Regnier B. (2001). Usefulness of a strategy based on bronchoscopy with direct examination of bronchoalveolar lavage fluid in the initial antibiotic therapy of suspected ventilator-associated pneumonia. Intensive Care Med 27, 640–647. 10.1007/s001340000840 [DOI] [PubMed] [Google Scholar]
- Tuft S. (2006). Polymicrobial infection and the eye. Br J Ophthalmol 90, 257–258. 10.1136/bjo.2005.084095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenza G., Tappe D., Turnwald D., Frosch M., König C., Hebestreit H., Abele-Horn M. (2008). Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros 7, 123–127. 10.1016/j.jcf.2007.06.006 [DOI] [PubMed] [Google Scholar]
- Vazquez J. A., Sobel J. D. (2002). Mucosal candidiasis. Infect Dis Clin North Am 16, 793–820, v. 10.1016/S0891-5520(02)00042-9 [DOI] [PubMed] [Google Scholar]
- Veeh R. H., Shirtliff M. E., Petik J. R., Flood J. A., Davis C. C., Seymour J. L., Hansmann M. A., Kerr K. M., Pasmore M. E., Costerton J. W. (2003). Detection of Staphylococcus aureus biofilm on tampons and menses components. J Infect Dis 188, 519–530. 10.1086/377001 [DOI] [PubMed] [Google Scholar]
- Weinberg A., Krisanaprakornkit S., Dale B. A. (1998). Epithelial antimicrobial peptides: review and significance for oral applications. Crit Rev Oral Biol Med 9, 399–414. 10.1177/10454411980090040201 [DOI] [PubMed] [Google Scholar]
- Xu H., Sobue T., Thompson A., Xie Z., Poon K., Ricker A., Cervantes J., Diaz P. I., Dongari-Bagtzoglou A. (2014). Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol 16, 214–231. 10.1111/cmi.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehia B. R., Fleishman J. A., Wilson L., Hicks P. L., Gborkorquellie T. T., Gebo K. A., HIV Research Network (2011). Incidence of and risk factors for bacteraemia in HIV-infected adults in the era of highly active antiretroviral therapy. HIV Med 12, 535–543. 10.1111/j.1468-1293.2011.00919.x [DOI] [PubMed] [Google Scholar]
- Zaremba M. L., Daniluk T., Rozkiewicz D., Cylwik-Rokicka D., Kierklo A., Tokajuk G., Dabrowska E., Pawińska M., Klimiuk A. & other authors (2006). Incidence rate of Candida species in the oral cavity of middle-aged and elderly subjects. Adv Med Sci 51 (Suppl 1), 233–236. [PubMed] [Google Scholar]
- Zaura E., Keijser B. J., Huse S. M., Crielaard W. (2009). Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9, 259. 10.1186/1471-2180-9-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli M., Widmer A. F., Dangel M., Filippi A., Frei R., Meyer J. (2009). Methicillin-resistant Staphylococcus aureus (MRSA) among dental patients: a problem for infection control in dentistry? Clin Oral Investig 13, 369–373. 10.1007/s00784-008-0244-2 [DOI] [PubMed] [Google Scholar]