Abstract
The opportunistic pathogen Candida albicans colonizes the oral cavity and gastrointestinal tract. Adherence to host cells, extracellular matrix and salivary glycoproteins that coat oral surfaces, including prostheses, is an important prerequisite for colonization. In addition, interactions of C. albicans with commensal oral streptococci are suggested to promote retention and persistence of fungal cells in mixed-species communities. The hyphal filament specific cell wall protein Als3, a member of the Als protein family, is a major determinant in C. albicans adherence. Here, we utilized site-specific in-frame deletions within Als3 expressed on the surface of heterologous Saccharomyces cerevisiae to determine regions involved in interactions of Als3 with Streptococcus gordonii. N-terminal region amino acid residue deletions Δ166–225, Δ218–285, Δ270–305 and Δ277–286 were each effective in inhibiting binding of Strep. gordonii to Als3. In addition, these deletions differentially affected biofilm formation, hydrophobicity, and adherence to silicone and human tissue proteins. Deletion of the central repeat domain (Δ434–830) did not significantly affect interaction of Als3 with Strep. gordonii SspB protein, but affected other adherence properties and biofilm formation. Deletion of the amyloid-forming region (Δ325–331) did not affect interaction of Als3 with Strep. gordonii SspB adhesin, suggesting this interaction was amyloid-independent. These findings highlighted the essential function of the N-terminal domain of Als3 in mediating the interaction of C. albicans with S. gordonii, and suggested that amyloid formation is not essential for the inter-kingdom interaction.
Introduction
The pleiomorphic fungus Candida albicans commonly exists as a commensal of the skin, and of mucosal tissues of the oral cavity, urogenital and gastrointestinal tracts. Pathogenesis of C. albicans is facilitated by environmental changes, including alterations to the resident microbiota, modified physiology (e.g. reduced saliva flow sometimes resulting from medical intervention) and impaired immune defence. Candida species are the fourth most common micro-organisms recovered from nosocomial bloodstream infections in the USA, and infections often require long and costly hospital stays (Zaoutis et al., 2005; Moran et al., 2009).
In the oral cavity, colonization by C. albicans occurs as a result of direct contact with host tissues and with the salivary pellicle that coats most surfaces (Cannon & Chaffin, 1999). Infections by C. albicans can be enhanced by the formation of biofilms on denture materials (Minagi et al., 1985) as well as in catheters (Hawser & Douglas, 1994), and on plastics (Klotz et al., 1985). The hyphal form of C. albicans is essential for biofilm development, and hyphae express a range of specific cell wall proteins (e.g. Als3 and Hwp1) that enable adhesion and biofilm formation (Nobile et al., 2006, 2008). In addition, C. albicans is rarely detected in isolation and co-exists with a diversity of micro-organisms (Shirtliff et al., 2009). Interactions of C. albicans with Gram-positive bacteria include Staphylococcus aureus (Harriott & Noverr, 2009), Streptococcus spp. (Jenkinson et al., 1990; Xu et al., 2014a, b) and Actinomyces spp. (Grimaudo et al., 1996). Gram-negative bacteria also interact with C. albicans, and include Pseudomonas aeruginosa (Hogan & Kolter, 2002), Acinetobacter spp. (Gaddy et al., 2009) and Fusobacterium spp. (Grimaudo & Nesbitt, 1997). These associations may be deleterious to C. albicans, such as interactions with P. aeruginosa (Hogan & Kolter, 2002; Holcombe et al., 2010), or beneficial for both organisms involved (Carlson & Johnson, 1985; Bamford et al., 2009; Harriott & Noverr, 2009; Xu et al., 2014a). The relationship of C. albicans with streptococcal species is potentially synergistic (O’Sullivan et al., 2000; Bamford et al., 2009; Diaz et al., 2012; Xu et al., 2014b) as lactate production by streptococci can act as a carbon and energy source for the yeast (Jenkinson et al., 1990; Xu et al., 2014a), whilst C. albicans reduces oxygen tension to levels preferred by streptococci (O’Sullivan et al., 2000; Jenkinson & Douglas, 2002). Dual-species biofilms of C. albicans and Streptococcus gordonii have greater biomass compared with those produced by individual species (Bamford et al., 2009; Dutton et al., 2014; Xu et al., 2014a).
As a primary colonizer of the oral cavity, the bacterium Strep. gordonii attaches to mucosal or hard surfaces within the mouth (Nyvad & Kilian, 1990; Frandsen et al., 1991). Adherence of C. albicans to Strep. gordonii potentially provides an additional means for colonization of the oral cavity by C. albicans. Previous studies have identified cell surface Strep. gordonii polypeptides SspA and SspB as receptors for C. albicans hyphae (Holmes et al., 1998; Silverman et al., 2010). We have also identified several C. albicans surface proteins, including Eap1, Hwp1 and Als3, that bind Strep. gordonii cells when they are expressed on the surface of surrogate host Saccharomyces cerevisiae (Nobbs et al., 2010). A direct role for Als3 in interactions of C. albicans hyphae with Strep. gordonii was suggested by observing that an als3Δ/als3Δ (null) mutant was unable to bind Strep. gordonii cells or interact with Strep. gordonii SspB polypeptide expressed on the surface of surrogate host strain Lactococcus lactis (Silverman et al., 2010).
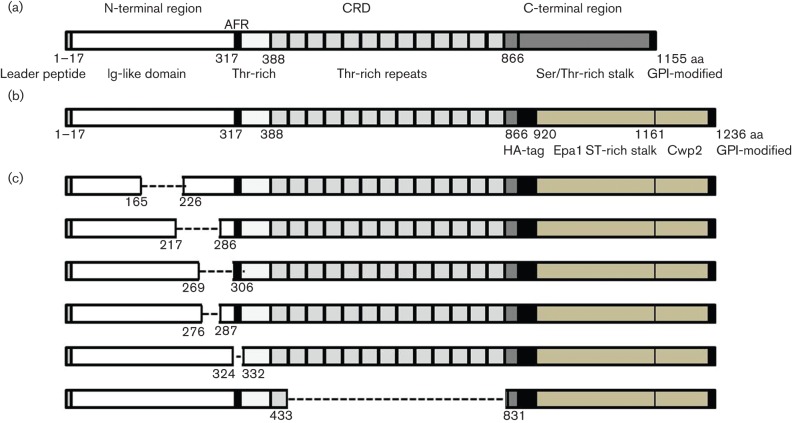
Als3 is a hypha-specific cell wall protein and a member of the Als protein family in C. albicans (Hoyer et al., 1998; Green et al., 2005). Als3 is able to bind extracellular matrix (ECM) proteins, epithelial and endothelial cells (Fu et al., 1998; Sheppard et al., 2004; Zhao et al., 2004; Liu & Filler, 2011), induce endocytosis via adherence to E- or N-cadherins (Phan et al., 2013), and mediate trafficking to the brain (Fu et al., 2013). The ability of C. albicans to form biofilms on a variety of substrata including the salivary pellicle, polystyrene and silicone elastomer is also influenced by expression of Als3 (Nobile et al., 2006, 2008; Nobbs et al., 2010). The primary structure of Als3 (Fig. 1a) contains an N-terminal domain with signal peptide, an immunoglobulin (Ig)-like region (~300 aa), a conserved Thr-rich region, a central Thr-rich repeat domain (~470 aa residues), a Ser/Thr-rich C-terminal domain and a C-terminal glycosylphosphatidylinositol modification (Gaur & Klotz, 1997; Sheppard et al., 2004; Phan et al., 2007). The central repeat domain (CRD) consists of imperfect Thr-rich tandem repeat regions, each of 36 aa residues (Frank et al., 2010). The number of repeat units varies in Als3 proteins from different C. albicans strains and the two ALS3 alleles in C. albicans SC5314 encode proteins with different numbers of repeats (Oh et al., 2005). The C-terminal Thr/Ser-rich region of Als3 is heavily glycosylated and the glycosylphosphatidylinositol modification localizes the protein to within the hyphal cell wall (Liu & Filler, 2011).
Fig. 1.
Diagrammatic representation of the Als3 polypeptide showing (a) overall structure and domains based upon primary sequence, (b) corresponding protein expressed on the surface of Sacch. cerevisiae with insertion of a HA-tag and replacement of the C-terminal region with an Epa1 Ser/Thr-rich stalk from Candida glabrata and (c) proteins expressed by als3 plasmid deletion mutants and expressed on the surface of Sacch. cerevisiae. Numbers indicate amino acid residues. Amino acid residues 325–331 corresponded to the predicted β-amyloid sequence IVIVATT (AFR). GPI, glycosylphosphatidylinositol.
The main adhesive region of Als3 has been mapped to the N-terminal domain (Zhao et al., 2006) and antibodies to N-terminal Als3 peptides were able to block adhesion of C. albicans to buccal epithelial cells. The N-terminal domain is predicted to form two Ig folds (Phan et al., 2007) with a peptide-binding cavity (PBC) for interaction with potential substrates, such as the flexible C-termini of polypeptides (Salgado et al., 2011; Lin et al., 2014). At the boundary of the Ig-like domain and conserved Thr-rich region is an amyloid-forming region (AFR) (Fig. 1a) that is implicated in cell–cell aggregation and adhesin clustering (Garcia et al., 2011; Beaussart et al., 2012), but may not be involved directly in C. albicans adherence to buccal epithelial cells (Lin et al., 2014). The CRD is believed to enhance the presentation of the adhesive N-terminal region away from the cell surface (Rauceo et al., 2006; Frank et al., 2010). In this study, we aimed to identify regions of Als3 that were necessary for binding Strep. gordonii and for interacting with the Strep. gordonii SspB cell wall adhesin. Specific amino acid sequences were deleted from within Als3 and the resulting polypeptides were surface-expressed on Sacch. cerevisiae. Our results provided evidence that regions within the N-terminal domain of Als3 were responsible for interaction of Als3 with Strep. gordonii cells and with Strep. gordonii SspB cell wall protein. However, the major AFR of Als3 appeared not to be necessary for the interaction of C. albicans Als3 protein with Strep. gordonii SspB protein – a primary mechanism for this inter-microbial association.
Methods
Microbial strains and culture conditions.
The bacterial and yeast strains used in this study are listed in Table S1 (available in the online Supplementary Material). Streptococcal cells were grown in brain heart infusion (BHI) medium (Lab M) supplemented with 5 mg yeast extract ml−1 statically in a candle jar for 16 h at 37 °C. Escherichia coli was grown in Luria–Bertani medium (Becton Dickinson) at 37 °C with vigorous shaking (200 r.p.m.). Ampicillin (100 µg ml−1) was included when necessary. L. lactis was grown in M17 medium (Difco) supplemented with 0.5 mg glucose ml−1 statically in a candle jar for 16 h at 30 °C. Erythromycin (5 µg ml−1) was added as appropriate. Sacch. cerevisiae strains were grown with shaking (200 r.p.m.) at 30 °C in complete synthetic medium (CSM) without uracil (ForMedium) supplemented with 0.67 % Yeast Nitrogen Base (Difco) and 2 % glucose (CSM-Glc). Uridine (25 µg ml−1) was added to the medium for growth of the Sacch. cerevisiae BY4742 parent strain (Brachmann et al., 1998). C. albicans was grown in YPD medium with shaking (200 r.p.m.) at 37 °C (Nobbs et al., 2010) and induced to form hyphae by transfer to YPT-Glc medium (Dutton et al., 2014).
Heterologous expression of Als3.
The ALS3 gene (larger allele) (GenBank accession number AY223552) minus the C-terminal stalk region (Fig. 1a) was amplified by PCR from C. albicans SC5314 chromosomal DNA with primers ALS3attB1 and ALS3attB2 (Gillum et al., 1984; Nobbs et al., 2010). The product was mixed with Gateway BP enzymes (Invitrogen), inserted into donor vector pDONR207 and transformed into E. coli OmniMAX 2-T1. Using Gateway LR enzymes, the C. albicans gene fragment was recombined with the destination vector pBC542 (Zupancic et al., 2008) to generate an expression vector and electroporated into Sacch. cerevisiae BY4742. To generate recombinant Als3 polypeptides with defined regions deleted, the plasmid pBC542-ALS3lg (Nobbs et al., 2010) was extracted from E. coli using a QIAquick Spin Miniprep Purification kit and used as template for inverse PCR with an Expand Long Template PCR System (Roche) and oligonucleotide primers as listed in Table S2. PCR products were purified using a QIAquick PCR Purification kit (Qiagen). Oligonucleotides were designed with BglII restriction sites at their termini to enable the coding sequence for Als3 to be religated with required deletions. Resulting plasmids were transformed into E. coli OmniMAX 2-T1 and deletions were confirmed by sequencing (Fig. S1). Plasmids with the correct sequences were linearized and transformed into Sacch. cerevisiae BY4742 by electroporation, with pBC542 and pBC542-ALS3 as controls. PCR and sequencing were again utilized to confirm successful cloning and authenticity.
Surface expression of Als3.
Sacch. cerevisiae cells were pelleted by centrifugation at 5000 g for 5 min and washed in PBS. The OD600 of the cell suspension was adjusted to 1.0 (~1×107 cells ml−1) and the cells were fixed with 4 % paraformaldehyde for 30 min at 22 °C (room temperature). After paraformaldehyde was removed, cells were blocked with heat-inactivated rabbit serum for 30 min at 22 °C. The cells were washed with PBS and then incubated for 1 h at 22 °C with mouse anti-heamagglutinin (HA) antibody (Zymed) at 10 µg ml−1. FITC-conjugated rabbit anti-mouse antibodies (Dako) diluted 1 : 100 were then added and the cells incubated for 1 h at 22 °C. Unbound antibodies were removed by washing with PBS. HA-tagged protein expression was visualized by fluorescence microscopy, and quantified by transferring portions into black 96-well microtitre plates (Greiner) and measuring relative fluorescence units (RFU) with a Molecular Devices SpectraMax M2 microtitre plate reader. The values were then adjusted to account for different levels of total protein solubilized from whole cells with 1 M NaOH and measured by the Lowry method, as described previously (Nobbs et al., 2010).
Interactions of Sacch. cerevisiae with Strep. gordonii or L. lactis expressing SspB.
Sacch. cerevisiae cells were harvested by centrifugation at 5000 g for 5 min and washed twice in CSM-Glc. Cell suspensions (2 ml) were adjusted to OD600 0.5, transferred to glass tubes and incubated with vigorous shaking (200 r.p.m.) for 3 h at 30 °C. Strep. gordonii or L. lactis cells were collected by centrifugation of cultures at 5000 g for 10 min, washed with CSM-Glc and fluorescently labelled by incubating cells with 1.5 mM FITC in 0.05 M Na2CO3 containing 0.1 M NaCl for 1 h at 22 °C. Labelled cells were collected by centrifugation, washed with CSM-Glc and adjusted to OD600 0.5 (~5×108 cells ml−1). Labelled bacteria (1 ml) were added to Sacch. cerevisiae cells and the suspensions incubated with gentle shaking (50 r.p.m.) for 1 h at 30 °C. Cells were harvested by brief centrifugation at 5000 g for 2 min, concentrated 10-fold, and imaged by transmitted light and fluorescence microscopy. Bacterial attachment to yeast cells was semi-quantified using imaging software (CellD; Olympus) and expressed as the mean number of fluorescent pixels bound per Sacch. cerevisiae cell (minimum cell count 200).
Biofilm formation on salivary pellicle.
Collection of saliva from human subjects was approved by the National Research Ethics Committee South Central Oxford C. (08/H0606/87+5). Pooled human saliva from at least six adult subjects who provided written informed consent was treated for 10 min on ice with 2.5 mM DTT. Mucins and bacteria were sedimented by centrifugation at 10 000 g for 10 min, and clarified saliva supernatant was sterilized by passage through a 0.2 µm filter and then diluted to 10 % with sterile distilled H2O. Aliquots (1 ml) were transferred to wells of a 24-well plastic plate, 13 mm diameter sterile glass coverslips were added and plates were incubated for 16 h at 4 °C. Excess saliva was removed by washing with PBS. Sacch. cerevisiae cell suspensions in CSM-Glc at OD600 1.0 were added (0.4 ml) to wells containing saliva-coated coverslips and incubated for 4 h at 30 °C with gentle rotation (50 r.p.m.). Cell suspensions were aspirated, coverslips were washed gently with PBS (1 ml), fresh CSM-Glc was added (1 ml) and then plates were incubated for 20 h at 30 °C. Coverslips were then washed carefully with PBS and removed from the wells to air-dry before staining with 0.5 % crystal violet for 2 min. Excess crystal violet was removed with distilled H2O and the biofilms were imaged by transmitted light microscopy. Alternatively, coverslips were transferred to new plate wells to release the crystal violet with 10 % acetic acid for A595 measurements, proportional to total biomass (Jakubovics et al., 2005; Nobbs et al., 2010).
Hydrophobicity assay.
Sacch. cerevisiae cells were prepared as described above, and cell suspensions (0.1 ml) added directly to wells of a 96-well microtitre plate and incubated for 4 h at 30 °C with gentle agitation (50 r.p.m.). Wells were washed thoroughly with TBS (10 mM Tris/HCl, 0.15 M NaCl, pH 8) and cells were fixed by addition of 25 % formaldehyde for 30 min at 22 °C. Biomass measurements of adherence to polystyrene, as a determinant of hydrophobicity, were then performed with crystal violet assay as described above.
Biofilm formation on silicone.
Silicone elastomer squares (1 cm2) were added to wells of a 24-well plate containing heat-inactivated horse serum (1 ml) and incubated for 16 h at 4 °C. The silicone squares were then washed with PBS, transferred to new wells and incubated with Sacch. cerevisiae cell suspension (OD600 1.0) in CSM-Glc for 4 h at 30 °C with gentle agitation (50 r.p.m.). Cell suspensions were carefully aspirated, and silicone squares were washed twice with PBS, air-dried and biomass measurements recorded using crystal violet as above.
Host protein adherence and biofilm assays.
Human fibrinogen (Calbiochem), fibronectin (Roche) or type IV collagen (Sigma) were dissolved in coating buffer (20 mM Na2CO3, 20 mM NaHCO3, pH 9.3), added to wells of a 96-well plate (10 µg ml−1) and incubated for 16 h at 4 °C. Wells were washed and blocked with 1 % gelatin in TBSC buffer (10 mM Tris/HCl, pH 7.6, 150 mM NaCl, 5 mM CaCl2) before incubation with Sacch. cerevisiae cell suspensions as described above and measurement of biomass. Values were corrected for biomass values obtained for Sacch. cerevisiae cells binding to gelatin alone.
Statistics.
For adhesion and biofilm assays, statistical analyses were performed with Student’s t-test using the Bonferroni correction for multiple comparisons.
Results
Surface expression of Als3 in heterologous host Sacch. cerevisiae BY4742
Evidence suggests that the hyphal filament-specific Als3 protein plays a major role in the binding of Strep. gordonii by C. albicans (Silverman et al., 2010). To address the question of which regions of the Als3 protein were necessary for this interaction, deletions were generated in recombinant Als3 and expressed from plasmid pBC542 (Nobbs et al., 2010) on the surface of Sacch. cerevisiae BY4742 (Fig. 1b). This strain of Sacch. cerevisiae does not bind Strep. gordonii (Nobbs et al., 2010; Silverman et al., 2010).
Specific amino acid residue sequences were selected for deletion based upon a number of criteria. Deletion of aa 218–285 and 277–286 from within the Als1 protein of C. albicans resulted in reduced adherence of expressing Sacch. cerevisiae strains to endothelial cells (Loza et al., 2004). Corresponding regions of Als3 have 85 and 90 % identity, respectively, and were therefore targeted in this study (Fig. 1c). Deletions of aa 166–225 and 270–305 were also generated (Fig. 1c) within the Ig-like N-terminal region, as detailed in Fig. S2. We also produced a deletion (aa 325–331) across the AFR sequence IVIVATT (Otoo et al., 2008) that has been proposed to induce adhesin clustering (Beaussart et al., 2012). Lastly, most of the CRD (aa 434–830) was deleted (Figs 1c and S3) to determine the role of this domain in directing adhesive interactions. The Als3 protein deletions were confirmed by sequencing (Fig. S1). Surface expression levels of proteins on Sacch. cerevisiae were measured by immunofluorescence assay using antibodies to the C-terminal HA-tag (Fig. 1) as described previously (Nobbs et al., 2010). There were small but statistically not significant differences in expression levels per microgram total protein for all the Sacch. cerevisiae strains generated (data not shown).
Effects of Als3 deletions on interactions with Strep. gordonii
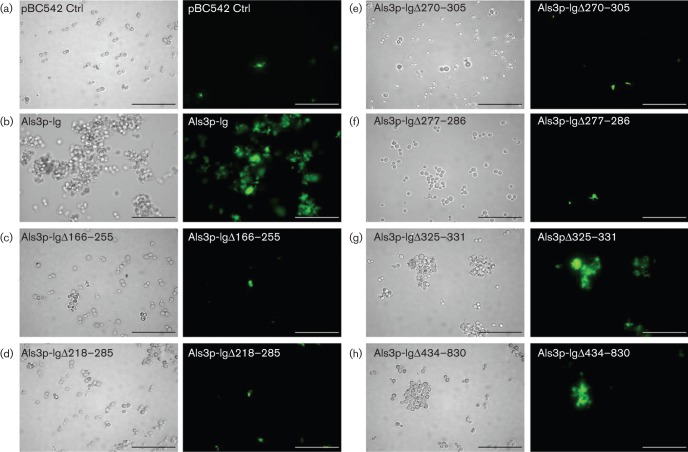
The deletions within Als3 were utilized to investigate which regions were necessary for binding Strep. gordonii. Sacch. cerevisiae cells expressing Als3 or deletion constructs were incubated with FITC-labelled Strep. gordonii cells. Coaggregation was visualized by transmitted light and fluorescence microscopy, and was quantified by calculating the mean numbers of fluorescent pixels (RFU) per Sacch. cerevisiae cell (Nobbs et al., 2010). Sacch. cerevisiae Als3+ cells showed the strongest ability to coaggregate with Strep. gordonii when compared with the vector-only control (Fig. 2a, b). All of the deletions from within the N terminus of Als3 (aa 166–225, 218–285, 270–305 and 277–286) affected binding to Strep. gordonii (Fig. 2c–f) and binding levels were reduced by >90 % compared with intact Als3 (Fig. 3). However, deletion of the AFR (aa 325–331) resulted in ~50 % reduction in binding of Strep. gordonii (Figs 2g and 3). The Als3 construct with deleted CRD (aa 434–830) also showed ~50 % reduction in ability to bind FITC-labelled Strep. gordonii cells compared with the Sacch. cerevisiae Als3+ strain (Figs 2h and 3). These results suggested that the N-terminal region of Als3 was mainly responsible for binding Strep. gordonii, but that the CRD may also have played a role, either in direct binding or stabilizing or folding of the N-terminal adhesive region.
Fig. 2.
Interactions of Sacch. cerevisiae expressing Als3 or Als3Δ polypeptides with Strep. gordonii DL1. Sacch. cerevisiae cells were incubated for 3 h at 30 °C and then with FITC-labelled Strep. gordonii for 1 h. Interactions were visualized by transmitted light microscopy and by fluorescence microscopy. (a–h) Light and corresponding fluorescence images of Sacch. cerevisiae BY4742 strains carrying pBC542 [vector-only control (Ctrl)] (a), pBC542-Als3 (b), pBC542-Als3Δ166–225 (c), pBC542-Als3Δ218–285 (d), pBC542-Als3Δ270–305 (e), pBC542-Als3Δ277–286 (f), pBC542-Als3Δ325–331 (g) and pBC542-Als3Δ434–830 (h). Bar, 50 µm.
Fig. 3.
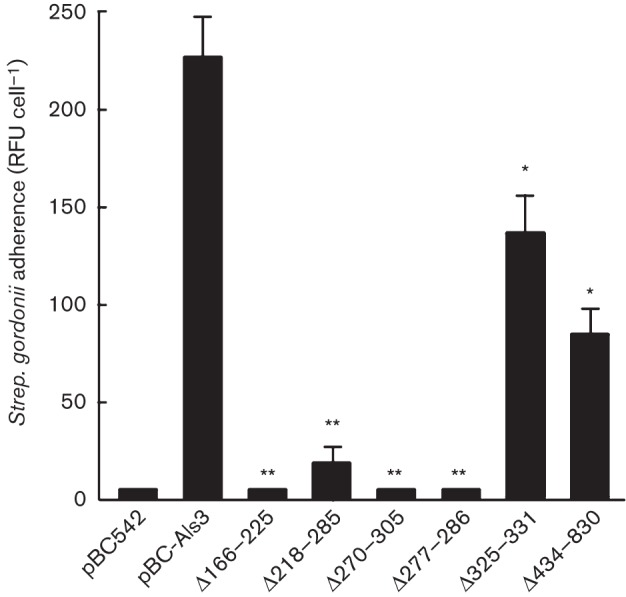
Binding levels of Strep. gordonii DL1 to Sacch. cerevisiae expressing Als3 or Als3Δ polypeptides. Sacch. cerevisiae cells were incubated for 3 h at 30 °C and then with FITC-labelled Strep. gordonii for 1 h. Strep. gordonii adherence is expressed as mean±sd (n = 3) RFU per Sacch. cerevisiae cell, as described in Methods. Statistical significance relative to pBC-Als3: *P<0.05, **P<0.005.
Effects of Als3 deletions on interactions with SspB
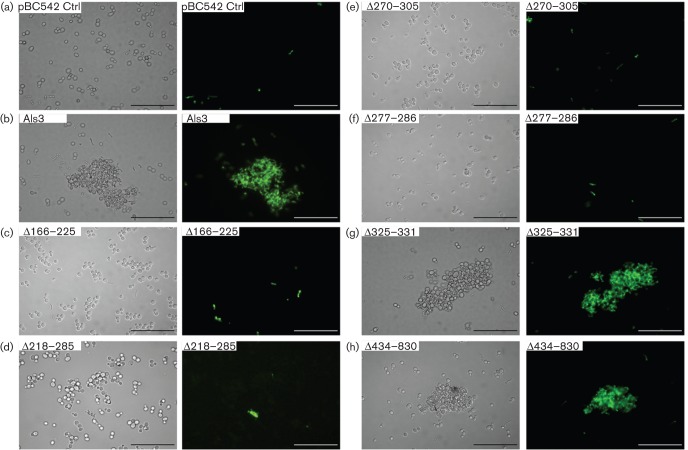
As the Strep. gordonii SspB protein specifically has been shown to interact with Als3 (Silverman et al., 2010), we then tested the ability of Sacch. cerevisiae strains to interact with L. lactis expressing SspB. L. lactis control strain containing empty vector did not interact with Sacch. cerevisiae expressing Als3 (data not shown), whilst there was extensive interaction between Sacch. cerevisiae Als3+ and L. lactis SspB+ (Fig. 4). The patterns of SspB binding by the Sacch. cerevisiae strains expressing the various Als3 deletions (Fig. 5) closely mirrored those of binding to Strep. gordonii (Fig. 3). All N-terminal region deletions, except deletion of the AFR, ablated binding to L. lactis SspB+ cells (Figs 4a–f and 5). Deletions of the AFR or CRD had no effects on binding of SspB (Figs 4g, h and 5). These results confirmed the notion that SspB is a major determinant of Strep. gordonii binding to C. albicans hyphae expressing Als3. However, the AFR and CRD, whilst involved at least in part in the Als3–Strep. gordonii interaction, did not appear to play a direct role in the Als3–SspB interaction.
Fig. 4.
Interactions of Sacch. cerevisiae expressing Als3 or Als3Δ polypeptides with surrogate host L. lactis cells expressing SspB polypeptide adhesin from Strep. gordonii. Sacch. cerevisiae cells were incubated for 3 h at 30 °C and then with FITC-labelled L. lactis for 1 h. Interactions were visualized by transmitted light microscopy and by fluorescence microscopy. (a–h) Light and corresponding fluorescence images of Sacch. cerevisiae BY4742 strains carrying pBC542 [vector-only control (Ctrl)] (a), pBC542-Als3 (b), pBC542-Als3Δ166–225 (c), pBC542-Als3Δ218–285 (d), pBC542-Als3Δ270–305 (e), pBC542-Als3Δ277–286 (f), pBC542-Als3Δ325–331 (g) and pBC542-Als3Δ434–830 (h). Bar, 50 µm.
Fig. 5.
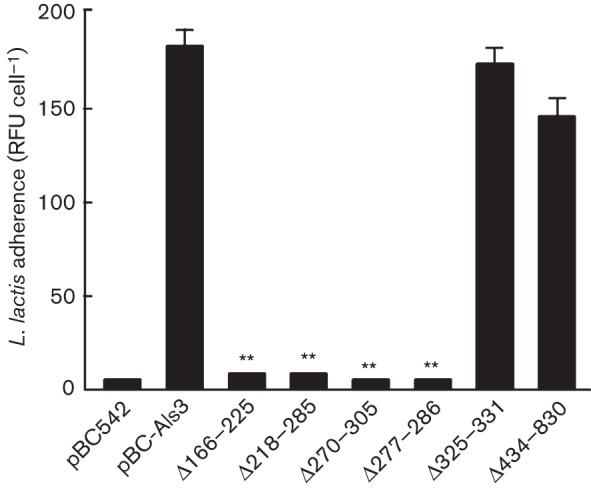
Binding levels of L. lactis expressing Strep. gordonii SspB protein to Sacch. cerevisiae expressing Als3 or Als3Δ polypeptides. Sacch. cerevisiae cells were incubated for 3 h at 30 °C and then with FITC-labelled L. lactis for 1 h. L. lactis adherence is expressed as mean±sd (n = 3) RFU per Sacch. cerevisiae cell, as described in Methods. Statistical significance relative to pBC-Als3: **P<0.005.
Biofilm formation by Sacch. cerevisiae strains expressing Als3 deletion constructs
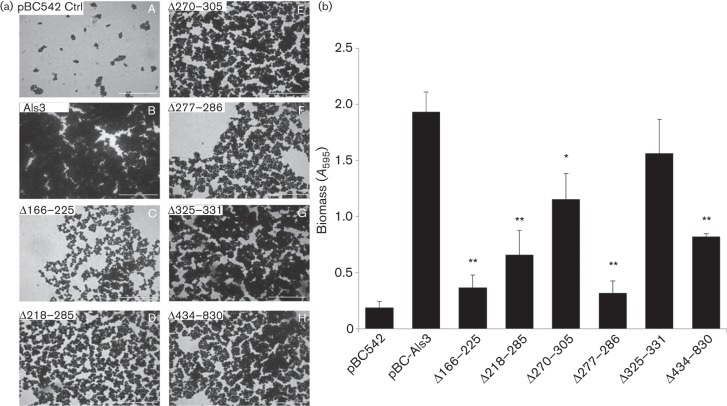
Expression of Als3 by Sacch. cerevisiae confers upon Sacch. cerevisiae the ability to form biofilms (Nobbs et al., 2010). To determine the effects of Als3 deletions on adherence and biofilm formation, saliva-coated glass coverslips were incubated with Sacch. cerevisiae strains for 24 h, washed and stained with crystal violet, and examined by microscopy or assayed for biomass (see Methods). Vector-only Sacch. cerevisiae was unable to form a biofilm (Fig. 6a), whilst Sacch. cerevisiae Als3+ cells formed a dense biofilm (Fig. 6a, b). Deletions of aa 166–225, 218–285, 277–286 and 434–830 all led to significantly reduced biofilm biomass values (60–85 % reduced) (Fig. 6b), whilst deletion of aa 270–305 had less effect (40 % reduction), and deletion of the AFR (aa 325–331) resulted in reduced, but not statistically significant, biomass accumulation (Fig. 6b).
Fig. 6.
Biofilm formation by Sacch. cerevisiae cells expressing Als3 or Als3Δ polypeptides. Sacch. cerevisiae cells were incubated with saliva-coated coverslips for 24 h at 30 °C. After washing and air-drying, cells were stained with crystal violet and imaged by light microscopy. (a) Crystal violet stained biofilms formed by Sacch. cerevisiae BY4742 strains carrying pBC542 [vector-only control (Ctrl)] (A), pBC542-Als3 (B), pBC542-Als3Δ166–225 (C), pBC542-Als3Δ218–285 (D), pBC542-Als3Δ270–305 (E), pBC542-Als3Δ277–286 (F), pBC542-Als3Δ325–331 (G) and pBC542-Als3Δ434–830 (H). Bar, 50 µm. (b) Corresponding biomass values for biofilms measured by crystal violet assay (see Methods). Values shown represent mean±sd of three experiments performed in triplicate. Significant differences in biomass from the Sacch. cerevisiae Als3-expressing strain: * P<0.05, **P<0.005.
Effects of Als3 deletions on adherence to polystyrene and silicone elastomer
A role for Als3 in adherence and biofilm formation by C. albicans on man-made materials, including polystyrene and silicone elastomer, has also been demonstrated (Nobile et al., 2008; Nobbs et al., 2010). To test the effects of Als3 deletions on adherence to polystyrene (an indirect measurement of surface hydrophobicity), Sacch. cerevisiae cells were incubated with microtitre plate wells for 4 h at 30 °C and adherence levels determined by crystal violet stain assay. Sacch. cerevisiae Als3+ cells adhered strongly to polystyrene when compared with vector-only controls (Fig. 7a). Deletion of Als3 aa 166–225 or CRD (aa 434–830) partly reduced adherence biomass (Fig. 7a), whilst the aa 277–286 deletion had no effect. Intriguingly, deletions of aa 218–285, 270–305 and 325–331 (AFR) resulted in significantly higher levels of attachment to polystyrene (Fig. 7a).
Fig. 7.
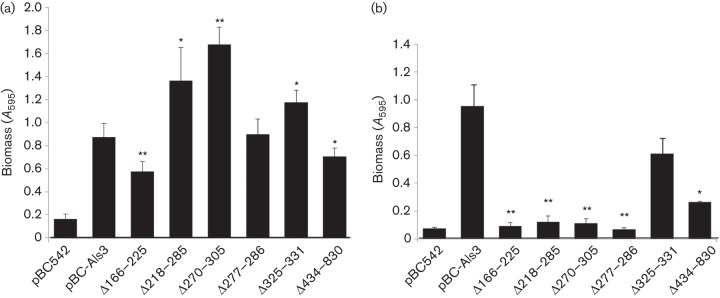
Early stage biofilm formation on (a) polystyrene or (b) serum-coated silicone elastomer by Sacch. cerevisiae cells expressing Als3 or Als3Δ polypeptides, or carrying empty vector (pBC542). (a) Sacch. cerevisiae cells were incubated in polystyrene microtitre plate wells for 4 h at 30 °C. Non-adherent cells were removed and adhered biomass measured by crystal violet staining. (b) Sacch. cerevisiae cells were incubated with serum-coated silicone elastomer squares (1 cm2) for 4 h at 30 °C. Non-adherent cells were removed and total biomass was measured by crystal violet assay. Values shown represent mean±sd of three experiments performed in triplicate. Significant differences in biomass from the Sacch. cerevisiae pBC-Als3+-expressing strain: *P<0.05, **P<0.005.
Silicone elastomer is a material commonly used for indwelling medical devices. Adherence by Als3-expressing Sacch. cerevisiae cells compared with vector-only control was measured after 4 h at 30 °C as described above. All of the Als3 deletions, except deletion of the AFR (aa 325–331), resulted in significant reductions in ability to adhere to serum-coated silicone (Fig. 7b).
Adherence to ECM proteins
Als3 interacts with fibronectin and a range of other matrix proteins (Liu & Filler, 2011). Significant reductions in adherence and biofilm formation (biomass) on collagen type IV, fibrinogen and fibronectin were observed for all Sacch. cerevisiae strains expressing Als3 deletion polypeptides as compared with Sacch. cerevisiae Als3+ (Fig. 8). In these experiments, deletion of the AFR (aa 325–331) significantly reduced the ability to form biofilms on all three matrix proteins, but the reduction was not as great as for other Als3 deletions tested.
Fig. 8.
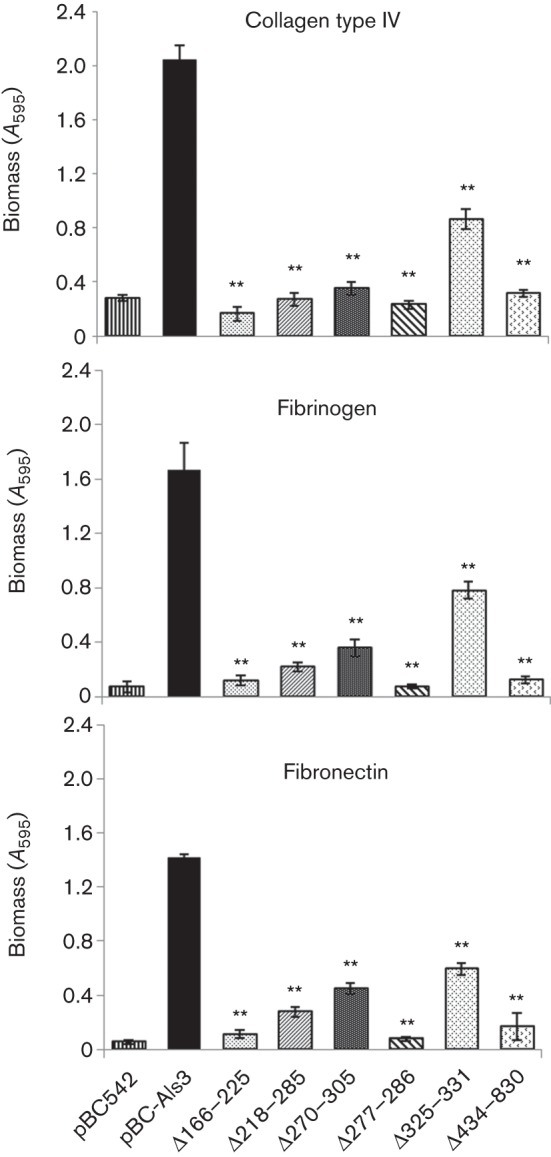
Early-stage biofilm formation on ECM proteins (collagen type IV, fibrinogen or plasma fibronectin) by Sacch. cerevisiae cells expressing Als3 polypeptides. Sacch. cerevisiae cells were incubated in microwells coated with 0.5 µg ECM protein for 4 h at 30 °C. Non-adherent cells were removed and total biomass values were estimated by crystal violet staining. Values shown represent mean±sd of three experiments performed in triplicate. Significant differences in biomass from the Sacch. cerevisiae pBC-Als3+-expressing strain: **P<0.005.
Discussion
Als3 is a key hyphal adhesin in C. albicans, and has been shown to mediate a wide range of interactive properties with host tissue proteins, mammalian cells and bacteria (Xu et al., 2014a; Liu & Filler, 2011; Peters et al., 2012). It is widely believed that the N-terminal region determines adhesive and invasive properties of the Als proteins. For example, the recombinant N-terminal region of Als3 is sufficient to mediate endocytosis by epithelial and endothelial cells (Phan et al., 2007). Cadherins can activate this pathway and in this respect are thought to function as host cell receptors for Als3 (Phan et al., 2005). Molecular modelling of the interactions of Als3 with E- or N-cadherin suggested that the Ig domains of Als3 interacted with the Ig domains of the cadherins. As the proposed interaction mimics E-cadherin molecules binding to each other, it has been suggested that Als3 functions as a molecular mimic of mammalian E-cadherin (Phan et al., 2007).
As a result of the deletion analyses, it seemed evident that the N-terminal domain was of paramount importance in the interactions of C. albicans Als3 with Strep. gordonii and with the Strep. gordonii adhesin SspB. By contrast, deletion of the AFR had less effect on binding Strep. gordonii and no effect on binding SspB. This supports previous observations that the β-amyloid-disrupting compound Congo red had little effect upon Als3 interactions with Strep. gordonii (Nobbs et al., 2010). It was noted that SspB also carried putative β-amyloid sequences (AFRs) as predicted by the tango algorithm (Silverman et al., 2010). However, the data here imply a higher specificity of interaction between SspB and Als3 than aggregate formation driven by hydrophobicity and side-chains of AFRs. Deletion of the CRD (aa 434–830) had no statistically significant effect on the binding interactions of C. albicans with L. lactis SspB+, but did significantly reduce association with Strep. gordonii cells. Thus, whilst the CRD does not seem to play a role in the Als3–SspB interaction, it may facilitate interactions with alternative streptococcal adhesins. Support for a second interactive mechanism comes from the observation that Strep. gordonii ΔsspAB is impaired but not ablated in binding surrogate host Sacch. cerevisiae expressing Als3 (Silverman et al., 2010).
Recent work describing the crystal structure of the N-terminal region of Als3 (Lin et al., 2014) has suggested the presence of a PBC that accommodates the C-terminal amino acid residues of polypeptides, thus mediating protein–protein interactions. It is proposed that differences in Als3 specificity for various substrates might be due to variations in C-terminal peptide sequences that can be accommodated in the PBC. In this model, proteolysis mediated by C. albicans secreted aspartyl proteases such as Sap9 would generate exposed C-termini of proteins on other C. albicans cells or on host tissues (Salgado et al., 2011; Dutton et al., 2014). However, in surrogate expression experiments, the specificity of Als3 (expressed on Sacch. cerevisiae) for Strep. gordonii SspB, as opposed to Strep. gordonii adhesin Hsa, was demonstrated clearly (Bamford et al., 2009). These observations suggest that the specificity of Als3 for SspB resides more than just with the PBC. Moreover, the SspB adhesin is covalently anchored to bacterial cell wall peptidoglycan via the C terminus, so there would theoretically be no C-terminal peptide ligand available. It seems more likely that interaction of SspB with Als3 occurs independently of the PBC mechanism proposed for interaction of Als3 with human epithelial cells (Lin et al., 2014). There is also the possibility that force-induced unfolding of the N-terminal domain, resulting from mechanical contact with cells or surfaces, exposes cryptic sequences involved in this protein–protein interaction (Alsteens et al., 2010; Beaussart et al., 2012; Dutton et al., 2014).
Deletions within the Als3 N-terminal region, and of the CRD, had the most significant inhibitory effects on the ability of Als3 to interact with the salivary pellicle or ECM proteins. This was also seen for attachment to serum-coated silicone, probably indicating Als3 targeting of serum ECM proteins such as fibronectin in this interaction. These data correlate with studies on Als1, for which elimination of some or all of the tandem repeats was shown to considerably reduce adhesive function (Loza et al., 2004). This suggests that the CRD may provide a secondary role in adhesion, either in presentation of the Als adhesive regions away from the cell surface or in folding of the N-terminal region. Interestingly, only deletion of the CRD, or of aa 166–225, impaired Als3 binding to polystyrene, whilst other N-terminal deletions (aa 218–285 or 270–305) actually promoted the interaction. This finding is in keeping with the evidence that the CRD (of Als5) has exposed hydrophobic surfaces that support binding to polystyrene substrata (Frank et al., 2010). Our results also suggest that CRD disruption in Als3 may concomitantly modulate presentation of the N-terminal domain structure to result in increased exposure of hydrophobic residues, thereby compensating for the loss of CRD.
The N-terminal region of Als3 comprises two IgG-type Ig domains designated N1 and N2 (Salgado et al., 2011). An invariant lysine residue (K59) within N1 (Fig. S3) is proposed to interact with the C-terminal carboxylate of peptide ligands. The PBC is formed and covered by a loop from domain N1, providing broad specificity for peptide binding. Amino acid residues in the mature protein between aa 166–172 and 294–298 have side-chains that are in close proximity to ligand within the PBC (Lin et al., 2014). All N-terminal deletions in Als3 spanning 166–305 residues resulted in major inhibitory effects upon Strep. gordonii binding, biofilm formation and adherence to human tissue proteins. These deletions would be expected to affect the overall conformation of the N-terminal region and probably disrupt formation of the PBC (Fig. S3).
The AFR in Als5 is important for efficient binding of Sacch. cerevisiae cells expressing Als5 to polystyrene (Garcia et al., 2011). Point mutation of the AFR sequence IVIVATT to INIVATT suppressed amyloidogenic potential and significantly impaired attachment to polystyrene. We observed in the present work that removal of the β-amyloid-like motif IVIVATT did not affect Als3 interactions with polystyrene, so Als3 may have additional hydrophobic residues that compensate for this deletion. As the AFR deletion also did not affect Als3 binding to SspB, it would appear to suggest that amyloid formation does not drive these adhesive interactions. We conclude that formation of amyloid-like structures may serve in some way to stabilize interactions of Als3 N-terminal functional domains with such substrata. Intriguingly, where deletion of AFR did impair Als3 function was in binding to ECM proteins, although these effects were weaker than those observed for disruption of the N-terminal region or CRD. There is precedent for interactions of ECM proteins such as fibrinogen or collagen with amyloids in neuropathies or cognitive conditions such as Alzheimer’s (Misumi et al., 2012; Muradashvili et al., 2014).
Conclusions
Our findings highlight the essential function of the N-terminal domain of Als3 in mediating binding of Strep. gordonii and of some human tissue proteins. Heterologous expression has allowed us to focus on Als3 properties in the absence of other determinants present on the C. albicans hyphal filaments that might influence interactions with Strep. gordonii. Further mutagenesis studies of Als3 in C. albicans would reveal the Strep. gordonii binding activity of Als3 in the context of other C. albicans cell wall components. The AFR plays a role in these reactions and is known to act along with the CRD in the force-induced clustering of surface adhesins to form nanodomains on the C. albicans cell surface (Alsteens et al., 2010; Chan & Lipke, 2014). Our data support the notion that the AFR could stabilize and strengthen adhesive interactions. We acknowledge that the deletion strategy that we utilized is a less precise means of determining regions of function than is, for example, site-directed mutagenesis and that deletions lead to overall conformational changes in proteins. However, our findings can now be developed to determine more accurately the residues and structure required for interactions of Als3 with SspB, and other bacterial receptors. The deletion strategy showed that the AFR and CRD both play a role in the interaction of Als3 with Strep. gordonii cells, but the AFR and CRD seem to be unnecessary for direct interaction of Als3 with SspB protein. Further understanding of these interactions will assist in the development of new drug or clinical strategies to manage C. albicans carriage and infection in association with bacteria in biofilms.
Acknowledgements
We would like to thank Lindsay Dutton, Jane Brittan, Ciara Keane and Richard Silverman for technical assistance and helpful discussions. This work was supported by National Institutes of Health (National Institute of Dental and Craniofacial Research) grant R01 DE016690.
Abbreviations:
- AFR
amyloid-forming region
- CRD
central repeat domain
- ECM
extracellular matrix
- HA
haemagglutinin
- Ig
immunoglobulin
- PBC
peptide-binding cavity
- RFU
relative fluorescence unit
Footnotes
Two supplementary tables and three supplementary figures are available with the online Supplementary Material.
References
- Alsteens D., Garcia M. C., Lipke P. N., Dufrêne Y. F. (2010). Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc Natl Acad Sci U S A 107, 20744–20749. 10.1073/pnas.1013893107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford C. V., d’Mello A., Nobbs A. H., Dutton L. C., Vickerman M. M., Jenkinson H. F. (2009). Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun 77, 3696–3704. 10.1128/IAI.00438-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaussart A., Alsteens D., El-Kirat-Chatel S., Lipke P. N., Kucharíková S., Van Dijck P., Dufrêne Y. F. (2012). Single-molecule imaging and functional analysis of Als adhesins and mannans during Candida albicans morphogenesis. ACS Nano 6, 10950–10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Cannon R. D., Chaffin W. L. (1999). Oral colonization by Candida albicans. Crit Rev Oral Biol Med 10, 359–383. 10.1177/10454411990100030701 [DOI] [PubMed] [Google Scholar]
- Carlson E., Johnson G. (1985). Protection by Candida albicans of Staphylococcus aureus in the establishment of dual infection in mice. Infect Immun 50, 655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. X., Lipke P. N. (2014). Role of force-sensitive amyloid-like interactions in fungal catch bonding and biofilms. Eukaryot Cell 13, 1136–1142. 10.1128/EC.00068-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz P. I., Xie Z., Sobue T., Thompson A., Biyikoglu B., Ricker A., Ikonomou L., Dongari-Bagtzoglou A. (2012). Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun 80, 620–632. 10.1128/IAI.05896-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton L. C., Nobbs A. H., Jepson K., Jepson M. A., Vickerman M. M., Aqeel Alawfi S., Munro C. A., Lamont R. J., Jenkinson H. F. (2014). O-mannosylation in Candida albicans enables development of interkingdom biofilm communities. MBio 5, e00911-14. 10.1128/mBio.00911-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen E. V., Pedrazzoli V., Kilian M. (1991). Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol Immunol 6, 129–133. 10.1111/j.1399-302X.1991.tb00466.x [DOI] [PubMed] [Google Scholar]
- Frank A. T., Ramsook C. B., Otoo H. N., Tan C., Soybelman G., Rauceo J. M., Gaur N. K., Klotz S. A., Lipke P. N. (2010). Structure and function of glycosylated tandem repeats from Candida albicans Als adhesins. Eukaryot Cell 9, 405–414. 10.1128/EC.00235-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Rieg G., Fonzi W. A., Belanger P. H., Edwards J. E., Jr, Filler S. G. (1998). Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect Immun 66, 1783–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Phan Q. T., Luo G., Solis N. V., Liu Y., Cormack B. P., Edwards J. E., Jr, Ibrahim A. S., Filler S. G. (2013). Investigation of the function of Candida albicans Als3 by heterologous expression in Candida glabrata. Infect Immun 81, 2528–2535. 10.1128/IAI.00013-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddy J. A., Tomaras A. P., Actis L. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77, 3150–3160. 10.1128/IAI.00096-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. C., Lee J. T., Ramsook C. B., Alsteens D., Dufrêne Y. F., Lipke P. N. (2011). A role for amyloid in cell aggregation and biofilm formation. PLoS ONE 6, e17632. 10.1371/journal.pone.0017632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur N. K., Klotz S. A. (1997). Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect Immun 65, 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum A. M., Tsay E. Y., Kirsch D. R. (1984). Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198, 179–182. 10.1007/BF00328721 [DOI] [PubMed] [Google Scholar]
- Green C. B., Zhao X., Yeater K. M., Hoyer L. L. (2005). Construction and real-time RT-PCR validation of Candida albicans PALS-GFP reporter strains and their use in flow cytometry analysis of ALS gene expression in budding and filamenting cells. Microbiology 151, 1051–1060. 10.1099/mic.0.27696-0 [DOI] [PubMed] [Google Scholar]
- Grimaudo N. J., Nesbitt W. E. (1997). Coaggregation of Candida albicans with oral Fusobacterium species. Oral Microbiol Immunol 12, 168–173. 10.1111/j.1399-302X.1997.tb00374.x [DOI] [PubMed] [Google Scholar]
- Grimaudo N. J., Nesbitt W. E., Clark W. B. (1996). Coaggregation of Candida albicans with oral Actinomyces species. Oral Microbiol Immunol 11, 59–61. 10.1111/j.1399-302X.1996.tb00337.x [DOI] [PubMed] [Google Scholar]
- Harriott M. M., Noverr M. C. (2009). Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother 53, 3914–3922. 10.1128/AAC.00657-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawser S. P., Douglas L. J. (1994). Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun 62, 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D. A., Kolter R. (2002). Pseudomonas–Candida interactions: an ecological role for virulence factors. Science 296, 2229–2232. 10.1126/science.1070784 [DOI] [PubMed] [Google Scholar]
- Holcombe L. J., McAlester G., Munro C. A., Enjalbert B., Brown A. J. P., Gow N. A. R., Ding C., Butler G., O’Gara F., Morrissey J. P. (2010). Pseudomonas aeruginosa secreted factors impair biofilm development in Candida albicans. Microbiology 156, 1476–1486. 10.1099/mic.0.037549-0 [DOI] [PubMed] [Google Scholar]
- Holmes A. R., Gilbert C., Wells J. M., Jenkinson H. F. (1998). Binding properties of Streptococcus gordonii SspA and SspB (antigen I/II family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect Immun 66, 4633–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer L. L., Payne T. L., Bell M., Myers A. M., Scherer S. (1998). Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr Genet 33, 451–459. 10.1007/s002940050359 [DOI] [PubMed] [Google Scholar]
- Jakubovics N. S., Strömberg N., van Dolleweerd C. J., Kelly C. G., Jenkinson H. F. (2005). Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol Microbiol 55, 1591–1605. 10.1111/j.1365-2958.2005.04495.x [DOI] [PubMed] [Google Scholar]
- Jenkinson H. F., Douglas L. J. (2002). Interactions between Candida species and bacteria in mixed infections. In Polymicrobial Diseases, pp. 357–373. Edited by Brogden K. A., Guthmiller J. M. Washington, DC: American Society for Microbiology; 10.1128/9781555817947.ch18 [DOI] [Google Scholar]
- Jenkinson H. F., Lala H. C., Shepherd M. G. (1990). Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun 58, 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz S. A., Drutz D. J., Zajic J. E. (1985). Factors governing adherence of Candida species to plastic surfaces. Infect Immun 50, 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Oh S. H., Jones R., Garnett J. A., Salgado P. S., Rusnakova S., Matthews S. J., Hoyer L. L., Cota E. (2014). The peptide-binding cavity is essential for Als3-mediated adhesion of Candida albicans to human cells. J Biol Chem 289, 18401–18412. 10.1074/jbc.M114.547877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Filler S. G. (2011). Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 10, 168–173. 10.1128/EC.00279-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza L., Fu Y., Ibrahim A. S., Sheppard D. C., Filler S. G., Edwards J. E., Jr (2004). Functional analysis of the Candida albicans ALS1 gene product. Yeast 21, 473–482. 10.1002/yea.1111 [DOI] [PubMed] [Google Scholar]
- Minagi S., Miyake Y., Inagaki K., Tsuru H., Suginaka H. (1985). Hydrophobic interaction in Candida albicans and Candida tropicalis adherence to various denture base resin materials. Infect Immun 47, 11–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi Y., Ueda M., Obayashi K., Jono H., Yamashita T., Ando Y. (2012). Interaction between amyloid fibril formation and extracellular matrix in the proceedings of VIIIth International Symposium on Familial Amyloidotic Polyneuropathy. Amyloid 19 (Suppl 1), 8–10. 10.3109/13506129.2012.674987 [DOI] [PubMed] [Google Scholar]
- Moran C., Grussemeyer C. A., Spalding J. R., Benjamin D. K., Jr, Reed S. D. (2009). Candida albicans and non-albicans bloodstream infections in adult and pediatric patients: comparison of mortality and costs. Pediatr Infect Dis J 28, 433–435. 10.1097/INF.0b013e3181920ffd [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muradashvili N., Tyagi R., Metreveli N., Tyagi S. C., Lominadze D. (2014). Ablation of MMP9 gene ameliorates paracellular permeability and fibrinogen-amyloid beta complex formation during hyperhomocysteinemia. J Cereb Blood Flow Metab 34, 1472–1482. 10.1038/jcbfm.2014.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs A. H., Vickerman M. M., Jenkinson H. F. (2010). Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot Cell 9, 1622–1634. 10.1128/EC.00103-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Andes D. R., Nett J. E., Smith F. J., Yue F., Phan Q. T., Edwards J. E., Jr, Filler S. G., Mitchell A. P. (2006). Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog 2, e63. 10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Schneider H. A., Nett J. E., Sheppard D. C., Filler S. G., Andes D. R., Mitchell A. P. (2008). Complementary adhesin function in C. albicans biofilm formation. Curr Biol 18, 1017–1024. 10.1016/j.cub.2008.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvad B., Kilian M. (1990). Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res 24, 267–272. 10.1159/000261281 [DOI] [PubMed] [Google Scholar]
- O’Sullivan J. M., Jenkinson H. F., Cannon R. D. (2000). Adhesion of Candida albicans to oral streptococci is promoted by selective adsorption of salivary proteins to the streptococcal cell surface. Microbiology 146, 41–48. [DOI] [PubMed] [Google Scholar]
- Oh S.-H., Cheng G., Nuessen J. A., Jajko R., Yeater K. M., Zhao X., Pujol C., Soll D. R., Hoyer L. L. (2005). Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology 151, 673–681. 10.1099/mic.0.27680-0 [DOI] [PubMed] [Google Scholar]
- Otoo H. N., Lee K. G., Qiu W., Lipke P. N. (2008). Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot Cell 7, 776–782. 10.1128/EC.00309-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. M., Ovchinnikova E. S., Krom B. P., Schlecht L. M., Zhou H., Hoyer L. L., Busscher H. J., van der Mei H. C., Jabra-Rizk M. A., Shirtliff M. E. (2012). Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology 158, 2975–2986. 10.1099/mic.0.062109-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan Q. T., Fratti R. A., Prasadarao N. V., Edwards J. E., Jr, Filler S. G. (2005). N-cadherin mediates endocytosis of Candida albicans by endothelial cells. J Biol Chem 280, 10455–10461. 10.1074/jbc.M412592200 [DOI] [PubMed] [Google Scholar]
- Phan Q. T., Myers C. L., Fu Y., Sheppard D. C., Yeaman M. R., Welch W. H., Ibrahim A. S., Edwards J. E., Jr, Filler S. G. (2007). Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol 5, e64. 10.1371/journal.pbio.0050064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan Q. T., Eng D. K., Mostowy S., Park H., Cossart P., Filler S. G. (2013). Role of endothelial cell septin 7 in the endocytosis of Candida albicans. MBio 4, e00542-e13. 10.1128/mBio.00542-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauceo J. M., De Armond R., Otoo H., Kahn P. C., Klotz S. A., Gaur N. K., Lipke P. N. (2006). Threonine-rich repeats increase fibronectin binding in the Candida albicans adhesin Als5p. Eukaryot Cell 5, 1664–1673. 10.1128/EC.00120-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado P. S., Yan R., Taylor J. D., Burchell L., Jones R., Hoyer L. L., Matthews S. J., Simpson P. J., Cota E. (2011). Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A 108, 15775–15779. 10.1073/pnas.1103496108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard D. C., Yeaman M. R., Welch W. H., Phan Q. T., Fu Y., Ibrahim A. S., Filler S. G., Zhang M., Waring A. J., Edwards J. E., Jr (2004). Functional and structural diversity in the Als protein family of Candida albicans. J Biol Chem 279, 30480–30489. 10.1074/jbc.M401929200 [DOI] [PubMed] [Google Scholar]
- Shirtliff M. E., Peters B. M., Jabra-Rizk M. A. (2009). Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett 299, 1–8. 10.1111/j.1574-6968.2009.01668.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman R. J., Nobbs A. H., Vickerman M. M., Barbour M. E., Jenkinson H. F. (2010). Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun 78, 4644–4652. 10.1128/IAI.00685-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Jenkinson H. F., Dongari-Bagtzoglou A. (2014a). Innocent until proven guilty: mechanisms and roles of Streptococcus–Candida interactions in oral health and disease. Mol Oral Microbiol 29, 99–116. 10.1111/omi.12049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Sobue T., Thompson A., Xie Z., Poon K., Ricker A., Cervantes J., Diaz P. I., Dongari-Bagtzoglou A. (2014b). Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol 16, 214–231. 10.1111/cmi.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaoutis T. E., Argon J., Chu J., Berlin J. A., Walsh T. J., Feudtner C. (2005). The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis 41, 1232–1239. 10.1086/496922 [DOI] [PubMed] [Google Scholar]
- Zhao X., Oh S. H., Cheng G., Green C. B., Nuessen J. A., Yeater K., Leng R. P., Brown A. J., Hoyer L. L. (2004). ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150, 2415–2428. 10.1099/mic.0.26943-0 [DOI] [PubMed] [Google Scholar]
- Zhao X., Daniels K. J., Oh S. H., Green C. B., Yeater K. M., Soll D. R., Hoyer L. L. (2006). Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152, 2287–2299. 10.1099/mic.0.28959-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupancic M. L., Frieman M., Smith D., Alvarez R. A., Cummings R. D., Cormack B. P. (2008). Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol Microbiol 68, 547–559. 10.1111/j.1365-2958.2008.06184.x [DOI] [PubMed] [Google Scholar]