Abstract
Bacteria have branched aerobic respiratory chains that terminate at different terminal oxidases. These terminal oxidases have varying properties such as their affinity for oxygen, transcriptional regulation and proton pumping ability. The focus of this study was a quinol oxidase encoded by cyoABCD. Although this oxidase (Cyo) is widespread among bacteria, not much is known about its role in the cell, particularly in bacteria that contain both cytochrome c oxidases and quinol oxidases. Using Rhizobium etli CFN42 as a model organism, a cyo mutant was analysed for its ability to grow in batch cultures at high (21 % O2) and low (1 and 0.1 % O2) ambient oxygen concentrations. In comparison with other oxidase mutants, the cyo mutant had a significantly longer lag phase under low-oxygen conditions. Using a cyo :: lacZ transcriptional fusion, it was shown that cyo expression in the wild type peaks between 1 and 2.5 % O2. In addition, it was shown with quantitative reverse transcriptase PCR that cyoB is upregulated approximately fivefold in 1 % O2 compared with fully aerobic (21 % O2) conditions. Analysis of the cyo mutant during symbiosis with Phaseolous vulgaris indicated that Cyo is utilized during early development of the symbiosis. Although it is commonly thought that Cyo is utilized only at higher oxygen concentrations, the results from this study indicate that Cyo is important for adaptation to and sustained growth under low oxygen.
Introduction
Bacteria have remarkable adaptability to environmental changes, such as fluctuations in oxygen concentration. Presumably, an important aspect of coping with variation in oxygen concentration is that aerobic bacteria have a variety of terminal oxidases (Poole & Cook, 2000; Bueno et al., 2012; Morris & Schmidt, 2013). Terminal oxidases are the enzymes that catalyse oxygen reduction during aerobic respiration. In many bacteria this step is achieved by cytochrome c oxidases, which catalyse electron transfer from cytochrome c to oxygen. Prior to this reaction, cytochrome c is reduced by quinol through the action of ubiquinol–cytochrome c oxidoreductase (Fbc), also known as the bc1 complex (Fig. 1). Quinol oxidation is a key branch point in aerobic respiration. Electrons from quinol flow either through the aforementioned Fbc or directly to oxygen via terminal oxidases known as quinol oxidases (Fig. 1). Because oxygen is a substrate for both quinol and cytochrome c oxidases, oxygen is expected to be a major factor in how each of these oxidases is regulated and utilized within bacteria.
Fig. 1.

Predicted aerobic respiratory chains of R. etli CFN42. Electrons from quinol to oxygen can be transferred through Fbc (square), ultimately leading to cytochrome c oxidases (circles): FixN_P, Cta and CoxM_P. Independent of the Fbc pathway, R. etli can transfer electrons directly from quinol to oxygen via Cyo (triangle). In parentheses, below each of the oxidases and Fbc complex, are the indicated operons that encode each of the oxidases followed by their NCBI reference numbers of the encoded proteins. Not included in the diagram are two putative cytochrome c oxidases, RHE_CH00981-85 and RHE_PB00063-66.
The quinol oxidase encoded by cyoABCD (Cyo) is widespread among aerobic bacteria. Much of what is known about Cyo has been concluded from studies performed in Escherichia coli, where Cyo was used at high oxygen concentrations and a high-affinity quinol oxidase (Cyd), encoded by cydAB, was utilized at lower oxygen concentrations (D’Mello et al., 1995, 1996; Tseng et al., 1996). However, E. coli does not contain the Fbc pathway. Therefore, the regulation and utilization of Cyo may be very different in bacteria that also contain cytochrome c oxidases. Currently, Cyo is classified as a low-affinity oxidase along with the cytochrome c aa3 oxidase (Cta), encoded by ctaCDGE (García-Horsman et al., 1994, Morris & Schmidt, 2013).
In this study, Rhizobium etli CFN42 was used as a model organism. Like other rhizobia, R. etli contains both the quinol oxidase and the Fbc pathway that contains multiple cytochrome c oxidases. This enables a direct comparison between quinol oxidases and cytochrome c oxidases within the same organism. This strain of R. etli only contains one quinol oxidase, Cyo, making it an ideal organism to study this particular enzyme.
Much of what is known about aerobic respiration in R. etli is limited to the necessary components involved in the symbiotic interaction with Phaseolus vulgaris (Delgado et al., 1998). It has been well established that a high-affinity cbb3 cytochrome c oxidase (Preisig et al., 1996), encoded by fixNOQP, is required for nitrogen fixation in P. vulgaris. For this reason, the regulation of this oxidase (FixN_P in Fig. 1) has been studied extensively in R. etli. It is tightly regulated and expressed at low oxygen (Michiels et al., 1998; Girard et al., 2000; Lopez et al., 2001; Moris et al., 2004; Granados-Baeza et al., 2007). How the other oxidases in R. etli, such as Cyo, are utilized and regulated in R. etli CFN42 is unclear. Mutants, with altered levels of Cyo, have been isolated and examined in the symbiotic interaction (Soberón et al., 1989, 1990). However, to our knowledge these mutations are either not in the cyo genes or they have not been genetically defined.
In this study, the oxygen conditions in which Cyo was utilized in liquid culture were determined. Initially, the ability of a cyo mutant to grow at various oxygen concentrations in comparison to other oxidase mutants was analysed. In addition, the activity of the cyo promoter was measured at various oxygen conditions. Lastly, the impact of cyo mutation on symbiosis with P. vulgaris was examined.
Methods
Bacterial strains and growth conditions.
R. etli strains were derived from strain CE3, a streptomycin-resistant derivative of wild-type strain CFN42 (Noel et al., 1984), whose genome nucleotide sequence has been determined (González et al., 2006). R. etli strains were grown at 30 °C on a rotating shaker in TY liquid medium [0.5 %, w/v, tryptone (Difco); 0.3 %, w/v, yeast extract (Difco); 10 mM CaCl2]. E. coli strains were grown in LB liquid medium (1.0 %, w/v, tryptone; 0.5 %, w/v, yeast extract; 0.5 %, w/v, NaCl) at 37 °C on a rotating shaker (Sambrook et al., 1989). Agar medium contained 1.5 % (w/v) Bacto agar (Difco). To analyse growth at low-oxygen conditions, fully grown cultures were diluted 1 : 200 into 5 ml of TY medium resting in 60 ml serum vials. The serum vials were then capped and the headspace was flushed with nitrogen gas. Using a sterile syringe needle, ambient air (assumed 21 % O2) was injected back into the headspace to make it 1 and 0.1 % O2. For growth at 21 % O2, the vials were covered with aluminium foil. Cultures were then grown at 30 °C on a rotating shaker. To follow growth, 400 µl was removed from the cultures using a sterile syringe needle and the OD600 was analysed. In addition, for some cultures the c.f.u. ml−1 was enumerated. To ensure oxygen was not reintroduced into the cultures while sampling for growth, nitrogen gas was aspirated from a separate vial prior to sampling.
Materials and techniques for DNA isolation.
Genomic DNA was isolated from R. etli strains using the GenElute Bacterial Genomic DNA kit (Sigma) for use in cloning. E. coli NEB 5-α (Invitrogen) competent cells were transformed (Hanahan, 1983), and plasmids were isolated using QIAprep spin miniprep kit (Qiagen). DNA was recovered from agarose gels using Gel/PCR DNA Fragments Extraction kit (IBI Scientific) and modified with restriction enzymes (NEB). Custom primers were synthesized by Eurofins MWG Operon.
Cloning and site-directed mutagenesis.
The fixN_P mutant was generously provided by L. Girard (Girard et al., 2000). Using PCR, cyoA, cyoB, ctaC and coxN were amplified separately from R. etli CE3 genomic DNA (primer sequences are listed in Table S1, available with the online Supplementary Material). PCR products were inserted into a TA cloning vector, pCR2.1. Plasmids were then digested using restriction enzymes, and DNA fragments were then inserted into plasmid pEX18Tc (Hoang et al., 1998). Either the gentamicin-resistant cassette from plasmid pUCGm, the kanamycin-resistant cassette from plasmid pBSL86 or the omega chloramphenicol resistant cassette from pBSL119 was inserted (Schweizer, 1993; Alexeyev, 1995; Alexeyev et al., 1995). Plasmids carrying the mutated ORFs were transferred into R. etli CE3 by using the plasmid-mobilizer strain MT616 on TY agar plates (Finan et al., 1986; Glazebrook & Walker, 1991). CE3 transconjugants containing these constructs were selected and purified as previously described (Ojeda et al., 2010). Double-crossover recombinants were screened on TY agar plates supplemented with 1 µg of tetracycline ml−1. Of the recombinants that were sensitive to 1 µg tetracycline ml−1 and resistant to 8 % (w/v) sucrose on TY agar, it was then verified by PCR that the colonies contained only the mutant allele and the wild-type allele was absent. The resulting strains are listed in Table 1.
Table 1. Strains and plasmids used in study.
| Bacterial strain or plasmid | Description, genotype or phenotype | Reference or Source |
| Strains | ||
| R. etli | ||
| CE3 | Wild-type strain, str-1 | Noel et al. (1984) |
| CE3/pZL39 | CE3 carrying pZL39; TcR | This study |
| CE119 | CE3 derivative, str-1 fbcF :: Tn5 | This study |
| CE426 | CE3 with mTn5SSgusA11 at unknown site | Duelli et al. (2001) |
| CE574 | CE3 derivative, str-1 cyoA :: Km; KmR | This study |
| CE574/pZL34 | CE574 carrying pZL34; Tcr | This study |
| CE582 | CE3 derivative, str-1 coxN :: CmΩ; CmR | This study |
| CE583 | CFNX641 derivative, str-1 cyoB :: Gm fixNf :: Sp fixNd :: Km; GmR KmR | This study |
| CE598 | CE3 derivative, str-1 ctaC :: Gm; GmR | This study |
| CE607 | CE574 with mTn5SSgusA11 at unknown site | This study |
| CFNX641 | CE3 derivative; str-1 fixNf :: Sp fixNd :: Km; SpR KmR | Girard et al. (2000) |
| E. coli | ||
| NEB-5α | Competent strain used for cloning | NEB |
| MT616 | pro thi endA hsdR supE44 recA-J6 pRK2013Km :: Tn9 | Finan et al. (1986) |
| Plasmids | ||
| pBSL86 | nptII gene cassette; KmR | Alexeyev (1995) |
| pBSL119 | CmΩ gene cassette; CmR | Alexeyev et al. (1995) |
| pCAM111 | Carries mTn5SSgusA11; SmR SpR ApR | Wilson et al. (1995) |
| pCR2.1 | T-A cloning vector for PCR products; ApR KmR | Invitrogen |
| pEX18Tc | Suicide plasmid; Tcr oriT sacB | Hoang et al. (1998) |
| pFAJ1708 | Expression vector with nptII promoter | Dombrecht et al. (2001) |
| pMP220 | Transcriptional lacZ fusion vector; TcR | Spaink et al. (1987) |
| pSY6 | cyoA with Km inserted at SalI site in pEX18Tc | This study |
| pUCGm | aaC1 gene cassette; GmR | Schweizer (1993) |
| pZL3 | cyoB with Gm inserted at SalI site in pEX18Tc; GmR | This study |
| pZL12 | coxN with CmΩ inserted at PstI site; CmR | This study |
| pZL31 | Gm cassette replacing 798 bp of ctaC ORF; GmR | This study |
| pZL22 | cyoB qRT-PCR fragment cloned into pCR2.1 for standard curve | This study |
| pZL34 | 1.3 kb BamHI, PstI fragment with cyoA in pFAJ1708 | This study |
| pZL39 | pMP220 derived, 350 bp KpnI, XbaI fragment upstream of cyoA fused with lacZ | This study |
ApR, ampicillin resistance; CmR, chloramphenicol resistance; GmR, gentamicin resistance; KmR, kanamycin resistance; SmR, streptomycin resistance; SpR, spectinomycin resistance; TcR, tetracycline resistance.
Isolation of the fbc mutant.
The fbc mutant (CE119) was isolated via random Tn5 mutagenesis as a fix mutant that still developed nodules (K. D. Noel, J. Richmond & P. Pachori, unpublished data). The genomic location of the transposon was determined to be in the fbcF gene of the fbcFBC operon (K. D. Noel & E. Rosado, unpublished data). The cytochrome c reduction activity was deficient in this mutant, but activity was restored after the addition of the wild-type fbcFBC operon (K. D. Noel & K. J. Ojeda, unpublished). In addition, an independent mutant was constructed by inserting a gentamicin cassette in the fbcB gene. This mutant (CE581) displayed the same phenotypic characteristics as CE119.
Quinol oxidase activity measurements.
Membranes were prepared and solubilized by modifying a protocol previously described (Ludwig, 1986). Cells were either grown in 500 ml Erlenmeyer flasks containing 250 ml of TY medium or 120 ml serum vials containing 10 ml of TY medium. Cells in the exponential growth phase in 250 ml of TY medium were harvested and washed with 50 mM K-phosphate buffer (pH 7.5). The washed cells were suspended in 15 ml of buffer and broken by sonication. Crude extracts were obtained by centrifugation of the sonicate for 15 min at 6 000 g. The supernatant (crude extract) was removed and centrifuged at 75 000 g for 2 h 15 min. The pellet (membranes) was suspended in 3 ml of buffer. Triton-X was added to the membranes at a final concentration of 2 % and allowed to incubate on a rotator for 2 h at 4 °C to solubilize the membranes. The solubilized membranes were kept at −80 °C until tested. Measurements of quinol oxidase activity were essentially performed as previously described (Richter et al., 1994). The activity of solubilized membranes was determined by measuring the change of absorbance at 275 nm. The membranes were incubated in a 1 ml reaction containing 50 mM of K-phosphate buffer (pH 7.5) and 30 µM antimycin A. The substrate, 75 µM quinol (reduced according to the method described by Rieske, 1967), was added to the mixture and the absorbance was immediately detected. The specific activity was then calculated by using the molar absorption coefficient of quinone (12,500 M−1 cm−1) and the total protein (BCA assay; Thermo Scientific) used in the reaction. To inhibit quinol oxidase activity, 1 mM potassium cyanide was added to the reaction prior to the addition of quinol.
lacZ fusion and beta-galactosidase measurements.
To generate a cyoA :: lacZ transcriptional fusion, a 350 bp fragment of the promoter region of cyoA was amplified from R. etli CE3 genomic DNA by PCR (primer sequences in Table S1). The PCR product was then inserted into the TA cloning vector, PCR 2.1. The fragment was then inserted into the pMP220 plasmid (Spaink et al., 1987) at the KpnI, XbaI restriction sites resulting in the plasmid pZL39. The plasmid pZL39 was transferred into CE3 using the MT616 plasmid-mobilizer strain. The empty vector, pMP220, was introduced into CE3 as a negative control. At different oxygen conditions (0.1–21 % O2), 1 ml of culture was withdrawn and washed with cold Z-buffer (Sambrook et al., 1989). The beta-galactosidase assay was performed as previously described (Sambrook et al., 1989).
Quantitative reverse transcriptase PCR (qRT-PCR).
Cultures were pelleted and immediately frozen in dry ice and stored at −80 °C. When ready for testing, cells were thawed on ice and RNA was extracted using NucleoSpin RNA II kit (Macherey–Nagel). The RNA concentration was measured by NanoDrop and 1 µg RNA was converted to cDNA using an EasyScript cDNA synthesis kit (MidSci) with the specific reverse primer for the gene of interest. As a negative control, water was added instead of the reverse-transcriptase. cDNA products were quantified by real-time PCR using EvaGreen qPCR Mastermix (MidSci), gene specific primers and the Bio-Rad iCycler. For analysing cyo expression, primers were designed to detect a 118 bp fragment in cyoB (Table S1). Samples were initially denatured at 95 °C for 10 min followed by a 40-cycle amplification protocol (95 °C for 15 s, 60 °C for 60 s). After the PCR, a melt curve was performed to ensure only one amplification product was present. The expression of the 16S rRNA gene was analysed using the same approach. Results for cyoB expression were normalized to the expression of the 16S rRNA gene.
Western blotting of BacS.
R. etli CE3 was grown at various oxygen concentrations (0.1–21 % O2) as described above. At full growth, 1 ml of cells were pelleted and resuspended in 100 µl 1× SDS buffer. The sample was then boiled for 6 min and stored at −20 °C until testing. The extracts were separated by SDS-PAGE (Laemmli, 1970) with 15 % acrylamide in the separating gel. The gel was stained with Coomassie blue (Dzandu et al., 1984) to visualize protein amounts in each sample or the contents were electroblotted onto a nitrocellulose membrane. The blot was incubated with rabbit polyclonal antiserum against the BacS protein (Jahn et al., 2003), and bound antibodies were detected with goat alkaline-phosphatase-conjugated anti-rabbit IgG (Sigma) that was developed with 5-bromo-4-chloro-3-indolyl phosphate.
Nodule assays.
Inoculation of bean seeds (P. vulgaris) with R. etli was performed as described previously (Box & Noel, 2011). For nodule staining, the wild-type and cyo mutants were tagged with beta-glucuronidase (GUS) by introducing pCAM111 via mating (Wilson et al., 1995; Duelli et al., 2001). Slicing and staining of the nodules were performed as previously described (Box & Noel, 2011). Nitrogenase activity of nodules was measured as acetylene reduction. Shoots were removed, and intact roots were incubated with acetylene in serum-capped vials. The production of ethylene was measured by gas chromatography on a Porapak N column (Noel et al., 1982). The ratio of ethylene : acetylene was normalized to the collective weight of the nodules on the root to give an overall specific activity.
Results
Predicted aerobic respiratory branches of R. etli CFN42 and sequence-directed mutant construction
The genome nucleotide sequence of R. etli CFN42 has been determined (González et al., 2006). With blast (Altschul et al., 1990) searches, the terminal oxidases encoded by the genome of this strain were predicted (Figs 1 and S1). The only quinol oxidase revealed in this way was a potential Cyo quinol oxidase, encoded by contiguous cyoABCD genes. On the other hand, the R. etli CFN42 genome revealed sequences for several cytochrome c oxidases: FixN_P (two copies), Cta and an alternative aa3-oxidase (CoxM_P), encoded by coxMNOP. Two additional operons that encode putative cytochrome c oxidases were also revealed in the genome (RHE_CH00981-85 and RHE_PB00063-66; Fig. S1).
To gain insight into the physiological function of the oxidases, strains were constructed with mutations in the essential subunits I and/or II of the oxidases using antibiotic-resistance cassettes (Table 1). The fixN_P mutant, strain CFNx641, was previously constructed by Girard et al. (2000). In this strain both fixNOQP operons have been mutated. An fbc mutant, carrying a Tn5 in the iron–sulfur cluster gene (fbcF), was also studied. Despite repeated attempts, a double mutant (fbc, cyo) was not attained. Specific mutations in RHE_CH00981-85 and RHE_PB00063-66 putative cytochrome c oxidases were not constructed.
Quinol oxidase activity
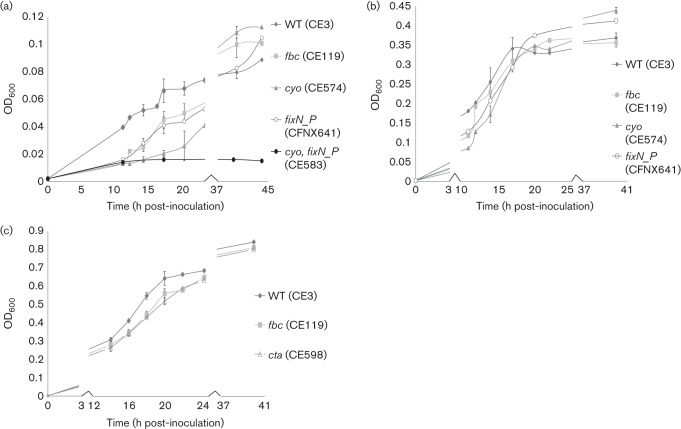
To confirm that the cyo genes encoded active proteins, and that Cyo was the only quinol oxidase present under aerobic conditions in R. etli CFN42, quinol oxidase activity was measured in the wild-type (CE3), fbc (CE119) and cyo (CE574) mutants during exponential growth (Fig. 2). Quinol oxidase activity was undetectable in the cyo mutant, supporting the notion that Cyo is the only quinol oxidase present under these conditions. Addition of the wild-type cyo gene (pZL34) restored activity similar to that of the wild-type. The fbc mutant had increased quinol oxidase activity compared with the wild-type.
Fig. 2.
Quinol oxidase activities of solubilized membranes. Strains were grown under aerobic conditions in 250 ml of TY medium in 500 ml Erlenmeyer flasks, and solubilized membranes were prepared as described in Methods. The addition of KCN depleted activity in all strains. Reaction mixtures with no substrate (quinol) and no membranes were used as separate negative controls (data not shown) that gave no apparent quinol oxidase activity. Mean±sd values were calculated from measured activities from three separate experiments and cultures.
Growth at varying oxygen concentrations
To determine the growth of mutants at low oxygen, the headspace in sealed batch cultures was adjusted to 1 or 0.1 % O2. To ensure that low oxygen conditions had been met, BacS (Jahn et al., 2003) was used as a physiological biomarker. BacS is a protein that is regulated by NifA and is expressed only at low oxygen conditions. As the oxygen concentration was lowered, the amount of BacS increased (Fig. S2).
In general, growth was monitored by measuring the OD600 of the cultures. However, to assess whether OD600 was a valid indicator of growth, c.f.u. were measured in some cases as well. It was determined at low oxygen (0.1 % O2) that an increase in OD600 correlated with an increase in c.f.u. (Fig. S3). As expected, the growth of the high-affinity fixN_P mutant was deficient in low-oxygen conditions (Fig. 3a, b). The fbc mutant was defective similarly to the fixN_P mutant in low-oxygen conditions.
Fig. 3.
Growth curves at (a) 0.1, (b) 1.0 and (c) 21 % O2. Strains were initially grown in TY liquid under a gas phase with 21 % O2. At full growth they were subcultured 1 : 200 into 5 ml of TY medium in 60 ml serum vials. As described in Methods, nitrogen and air were added to the headspaces in the vials above the liquid to give the indicated concentrations of oxygen. Growth was followed by measuring the OD600. Error bars indicate sd from at least three separate experiments.
Unexpectedly, the cyo mutant was the strain that had the longest lag phase compared with the wild-type and other respiratory mutants under both 1 and 0.1 % O2 conditions (Fig. 3a, b). This effect was alleviated after transferring the wild-type copy of cyoA to the cyo background (Fig. S4). Furthermore, a cyo/fixN_P mutant (CE583) was unable to grow under the 0.1 % condition, whereas the single cyo or fixN_P mutant recovered to grow after a delay (Fig. 3a). The fbc mutant, presumably completely dependent on Cyo activity, not only suffered less delay than cyo but sustained its growth at 0.1 % comparably to the cyo, which could use FixN_P. The cta (CE598) and the coxM_P (CE582) displayed growth similar to the wild-type at low oxygen conditions (data not shown).
Under fully aerobic conditions, the cta mutant reproducibly had a slight growth defect (Fig. 3c). The fbc mutant had a similar growth defect at high-oxygen conditions. The other oxidase mutants grew similarly to the wild-type at high-oxygen conditions (data not shown).
cyo expression at various oxygen concentrations
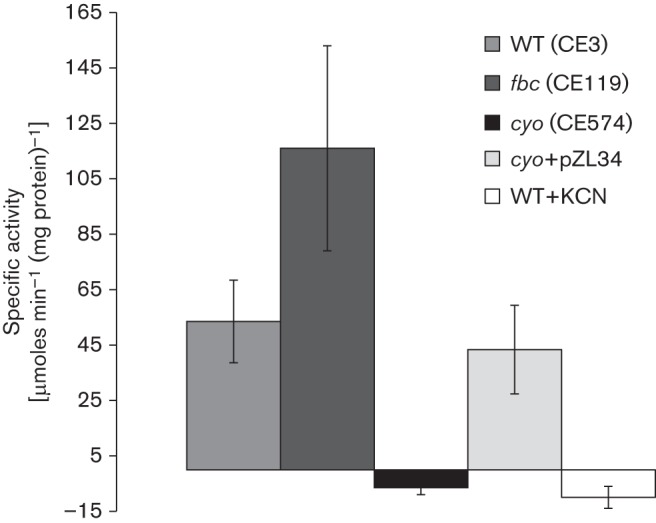
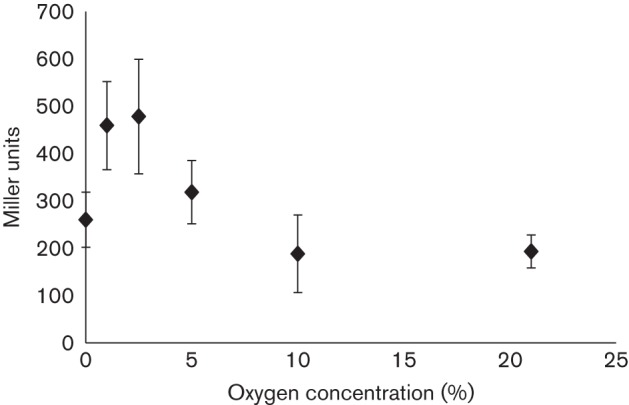
A transcriptional fusion containing the promoter region of cyo and the lacZ reporter (Pcyo :: lacZ) was used to analyse cyo expression at various oxygen concentrations ranging from 0.1 to 21 % O2 in the wild-type (Fig. 4). Expression of cyo gradually increased as oxygen was lowered, peaking at 1–2.5 % O2. The beta-galactosidase activity was approximately 2.5-fold higher from cells grown at 1 % O2 compared with 21 % O2 conditions (Fig. 5a). As the oxygen concentration was further lowered to 0.1 % O2, the expression decreased (Fig. 4) even though analysis of mutant growth had shown a more obvious importance of Cyo at 0.1 % O2 (Fig. 3a). Expression of cyo at 1 and 21 % O2 was investigated more closely using qRT-PCR, which showed that cyo expression was upregulated approximately fivefold at 1 % O2 compared with fully aerobic conditions. Depicted in Fig. 5(b) are data from one of the qRT-PCR experiments performed. Furthermore, the quinol oxidase activity in the wild-type was greatly increased at 1 % O2 compared with 21 % O2 (Fig. 5c).
Fig. 4.
Impact of oxygen concentration on cyo promoter activity. Wild-type (CE3) cells, carrying the PcyoA :: lacZ transcriptional fusion (pZL39), were harvested from exponentially growing cultures at different oxygen concentrations (0.1–21 % O2), washed in cold Z-buffer and beta-galactosidase assay was performed. Specific activity is given in Miller units. Mean±sd values were calculated from three or more separate lacZ assays from two different cultures.
Fig. 5.
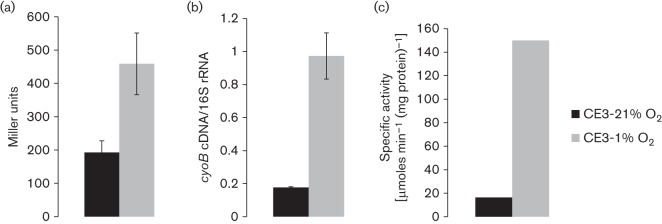
Comparison of cyo expression and quinol oxidase activity at low (1 %) versus high (21 %) oxygen. Wild-type cells were grown either in 21 or 1 % O2. Protein, RNA and membranes were all extracted from cells in the exponential phase. (a) Beta-galactosidase activity of wild-type cells carrying the transcriptional fusion plasmid, pZL39 (cyoA :: lacZ). Mean±sd values were calculated from three or more separate lacZ assays from two different cultures. (b) qRT-PCR of cyoB. RNA was extracted and converted to cDNA using the reverse gene-specific primer. As described in Methods, cDNA was then quantified by qPCR. The amount of cyoB cDNA (ng) was then normalized to the amount of 16S rRNA cDNA (pg) from the original RNA sample. Mean±sd values were calculated from three separate qPCR assays. (c) Specific quinol oxidase activity of wild-type solubilized membranes. Wild-type cells were grown in six separate 120 ml serum vials containing 10 ml of TY medium. These cultures were pooled together and the membranes were prepared and solubilized as described in Methods. In these experiments, growth was in serum vials rather than in Erlenmeyer flasks as described in Fig. 2.
Impact of cyo on development of symbiosis
To analyse the symbiotic role of Cyo, nodules harbouring the wild-type were compared to nodules harbouring the cyo mutant. Although cyo mutant and wild-type nodules displayed nitrogenase activity on the same day [8 days post-inoculation (p.i.)], nodules harbouring the cyo mutant had significantly less activity compared with the wild-type (Fig. 6a). At 9 days p.i., the nitrogenase activity was similar to the wild-type (data not shown).
Fig. 6.
Symbiotic phenotype of cyo. (a) Nitrogenase activity of intact nodulated roots whose nodules harboured wild-type or cyo at 8 days p.i. Error bars indicate sd (n = 8; P<0.05). Acetylene reduction activity = [(ethylene peak/acetylene peak)/nodule weight]. (b) Gus-staining of nodules harbouring gus-tagged wild-type (CE426) and gus-tagged cyo mutants (CE607) at different days p.i. The gusA gene was constitutively expressed at equivalent levels (in TY culture) in both strains under low (0.1 % O2) and high (21 % O2) oxygen conditions. Therefore, staining reflects the relative density of bacterial cells. Bars in the lower right corner of each image represent 500 µm. Samples are from 6, 7, 8 and 10 days p.i.
As a more direct measure of infection, the bacterial content within nodules was examined by tagging the bacteria with the gus gene (Fig. 6b), assuming that the intensity of GUS staining was proportional to the bacterial content within the nodule. The GUS activities of the studied gus-tagged strains were equivalent in cells grown in TY liquid at various oxygen conditions (Fig. S5). In the early days post-inoculation (6–7 days p.i.), the staining of nodules harbouring the gus-tagged wild-type (CE426) was more intense compared with the nodules harbouring the gus-tagged cyo mutant (CE607). By 8 days p.i., the cyo nodules had comparable staining to the wild-type.
Discussion
In this study we sought to determine the oxygen conditions in which Cyo is utilized in an Fbc-containing organism such as R. etli CFN42. This strain is specifically useful for studying Cyo, as inspection of the genome indicated that Cyo is the only terminal oxidase independent of the Fbc pathway. The inability to obtain a double cyo, fbc mutant under aerobic conditions supports this notion. In addition, quinol oxidase activity was undetectable in the cyo mutant under aerobic conditions. On the other hand, the quinol oxidase activity was increased in an fbc mutant, as might be predicted, given that Cyo is the only viable respiratory option in this case. These results indicate that the quinol oxidase assay is specific for Cyo in R. etli CFN42, and presumably enables measurement of the activity of Cyo directly under different physiological conditions. However, it is still possible that a cryptic quinol oxidase might be induced under conditions not yet studied.
To begin to understand the oxygen condition under which Cyo is utilized, we analysed the ability of the cyo mutant to grow at low oxygen in comparison to the wild-type and other oxidase mutants. Since the inoculant is from an aerobic culture, the results can be interpreted as the bacterium’s ability to adapt to a sudden decrease in oxygen concentration. Surprisingly, the cyo mutant had the greatest growth defect under low-oxygen conditions but no growth defect at high oxygen, even though Cyo is classified as a low-affinity oxidase. This result is supported by a previously reported observation in R. etli CFN42 that cyo mutants were slow to grow on minimal medium plates under micro-aerobic conditions compared with the wild-type (Landeta et al., 2011). In addition, the fbc mutant was able to reach a similar final OD600 as rapidly as the cyo at 0.1 % O2. Interestingly, both fbc and cyo reproducibly reached an OD600 higher than that of the wild-type. The basis for this observation is unknown. These growth results, along with the upregulation of cyo in the wild-type, indicate that Cyo is utilized and important for adapting to and sustaining growth under low oxygen.
As predicted, the high-affinity fixN_P mutant had a growth defect at low oxygen and the low-affinity cta mutant had a growth defect at high oxygen. The fbc mutant had similar growth defects to the FixN_P mutant at low oxygen and similar growth defects to the cta mutant under fully aerobic conditions. This is consistent with the literature in that the Fbc pathway terminates with the aa3 oxidase under aerobic conditions and the cbb3 oxidase under micro-aerobic conditions (Bott et al., 1990; Preisig et al., 1996). The coxM_P mutant had no a distinct observable growth defect under any of the conditions tested. The two other putative cytochrome c oxidases (RHE_CH00981-85 and RHE_PB00063-66) were not specifically addressed in this study.
The symbiotic process from infection to bacteroid differentiation requires R. etli to adapt and eventually to respire at very low oxygen concentrations. Based on the results from growth in liquid cultures, it was hypothesized that Cyo may have a role during infection, because it is required for the bacterium to grow while adapting to lower levels of oxygen. The onset of nitrogen fixation was initially tested, as it was assumed that the more efficiently the bacteria infected, the faster they would differentiate into bacteroids and fix nitrogen. Although wild-type nodules and cyo nodules started to fix nitrogen at the same time, the wild-type nodules showed significantly more nitrogenase activity compared with the cyo nodules at the earliest time investigated. A similar result was reported in Bradyrhizobium japonicum, where a mutant defective for a Cyo homologue (coxWXYZ) displayed an approximately 30 % decrease in nitrogenase activity (Surpin and Maier, 1999). However, in the present study there was no significant difference in nitrogenase activity after 8 days p.i. Using gus-tagged bacteria, the results show that there was significantly less bacterial content in early-developed cyo nodules (6–7 days p.i.). As nodules matured, the staining was similar between the wild-type and cyo nodules. Taken together, these results indicate that Cyo is advantageous during the early stages of symbiosis but has little role in the later stages. A logical inference is that Fbc-dependent oxidases are not sufficient to provide optimal bacterial growth during the infection phase.
A potential reason for observing such gross phenotypes associated with Cyo is the fact that R. etli CFN42 does not contain the high-affinity Cyd quinol oxidase. However, studies in other rhizobial species have revealed that Cyo may be utilized at lower oxygen conditions regardless of the presence of Cyd. Two separate studies in Sinorhizobium meliloti, using DNA microarray and transcriptional fusion respectively, indicated that cyo is transcriptionally upregulated (2–4-fold) at lower oxygen concentrations (1–2 % O2) (Trzebiatowski et al., 2001; Bobik et al., 2006). In B. japonicum, based on cyanide inhibitor titration patterns of cell membranes, the Cyo homologue (coxWXYZ) is predicted to be expressed under micro-aerobic conditions (1 % O2) (Surpin et al., 1996). In addition, a mutation in coxWXYZ had a growth defect at low oxygen levels under chemolithotrophic conditions (Surpin & Maier, 1998). To our knowledge these organisms contain a Cyd quinol oxidase based on blast searches. It has yet to be ruled out whether the absence of Cyd has had an impact on the manner in which Cyo is utilized and expressed in R. etli CFN42. Nevertheless, our results indicate that Cyo and Cta oxidases have distinct physiological roles in regard to oxygen. Based on our growth studies, Cyo enhances the ability to respire at lower oxygen concentrations in comparison with the Cta oxidase. Furthermore, it appears that Cyo is capable of functioning in batch culture at 0.1 % O2 as effectively as the FixN_P oxidase.
We propose that Cyo is a versatile oxidase that can function under a broad range of oxygen concentrations based on the growth results of the fbc mutant, in which it is assumed that Cyo is the only functional oxidase. With only slight deficiency in both instances, the fbc mutant was able to grow under both fully aerobic and low oxygen conditions. On the other hand, Cta and FixN_P seem more specialized with respect to the oxygen concentrations at which they support growth. The cyo, fixN_P mutant was unable to grow at 0.1 % O2, indicating that Cta is unable to function at this oxygen concentration. Under fully aerobic conditions, a cyo, cta double mutant was unattainable indicating that other cytochrome c oxidases are in sufficient for growth under fully aerobic conditions. A recent study on the alphaproteobacterium Gluconobacter oxydans also indicates that Cyo is capable of functioning under a wide range of oxygen conditions as a cyo mutant had a defect in growth and oxygen consumption under both free oxygen and oxygen-limiting conditions (Richhardt et al., 2013). This bacterium is an obligate aerobe yet it contains neither cytochrome c oxidases nor a high-affinity Cyd quinol oxidase, indicating that it may be taking advantage of the versatility of Cyo that we have shown in the present study.
Although it is capable of functioning under a wide range of oxygen concentrations, Cyo may be most important at intermediate oxygen levels. Based on our results, cyo expression peaks at approximately 1–2.5 % O2. Perhaps at these concentrations, oxygen is too low for Cta to be adequately effective but not low enough to induce FixN_P. Having an oxidase such as Cyo that is capable of functioning at various oxygen concentrations would be of great benefit for many bacteria, particularly soil bacteria that frequently have to adjust to wide ranges of oxygen conditions.
Acknowledgements
We would like to thank Dr Lourdes Girard for providing the R. etli strain, CFNx641. We acknowledge Jodi Richmond, Pappi Pachori and Eric Rosado for the isolation and genomic identification of CE119 and Sihui Yang for the construction of pSY6. Funds from NIH grant no. 1 R15 GM087699-01A1 helped support this research.
Abbreviations:
- Cyo
cytochrome c oxidase
- Fbc
ubiquinol–cytochrome c oxidoreductase
- GUS
beta-glucuronidase
Footnotes
Five supplementary figures and one supplementary table are available with the online Supplementary Material.
References
- Alexeyev M. F. (1995). Three kanamycin resistance gene cassettes with different polylinkers. Biotechniques 18, 52–56, 54, 56. [PubMed] [Google Scholar]
- Alexeyev M. F., Shokolenko I. N., Croughan T. P. (1995). Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene 160, 63–67. 10.1016/0378-1119(95)00108-I [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bobik C., Meilhoc E., Batut J. (2006). FixJ: a major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti. J Bacteriol 188, 4890–4902. 10.1128/JB.00251-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott M., Bolliger M., Hennecke H. (1990). Genetic analysis of the cytochrome c-aa3 branch of the Bradyrhizobium japonicum respiratory chain. Mol Microbiol 4, 2147–2157. 10.1111/j.1365-2958.1990.tb00576.x [DOI] [PubMed] [Google Scholar]
- Box J., Noel K. D. (2011). Controlling the expression of rhizobial genes during nodule development with elements and an inducer of the lac operon. Mol Plant Microbe Interact 24, 478–486. 10.1094/MPMI-07-10-0155 [DOI] [PubMed] [Google Scholar]
- Bueno E., Mesa S., Bedmar E. J., Richardson D. J., Delgado M. J. (2012). Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid Redox Signal 16, 819–852. 10.1089/ars.2011.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Mello R., Hill S., Poole R. K. (1995). The oxygen affinity of cytochrome bo’ in Escherichia coli determined by the deoxygenation of oxyleghemoglobin and oxymyoglobin: Km values for oxygen are in the submicromolar range. J Bacteriol 177, 867–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Mello R., Hill S., Poole R. K. (1996). The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology 142, 755–763. 10.1099/00221287-142-4-755 [DOI] [PubMed] [Google Scholar]
- Delgado M. J., Bedmar E. J., Downie J. A. (1998). Genes involved in the formation and assembly of rhizobial cytochromes and their role in symbiotic nitrogen fixation. Adv Microb Physiol 40, 191–231. 10.1016/S0065-2911(08)60132-0 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Vanderleyden J., Michiels J. (2001). Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol Plant Microbe Interact 14, 426–430. [DOI] [PubMed] [Google Scholar]
- Duelli D. M., Tobin A., Box J. M., Kolli V. S. K., Carlson R. W., Noel K. D. (2001). Genetic locus required for antigenic maturation of Rhizobium etli CE3 lipopolysaccharide. J Bacteriol 183, 6054–6064. 10.1128/JB.183.20.6054-6064.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzandu J. K., Deh M. E., Barratt D. L., Wise G. E. (1984). Detection of erythrocyte membrane proteins, sialoglycoproteins, and lipids in the same polyacrylamide gel using a double-staining technique. Proc Natl Acad Sci U S A 81, 1733–1737. 10.1073/pnas.81.6.1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M., Kunkel B., De Vos G. F., Signer E. R. (1986). Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol 167, 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Horsman J. A., Barquera B., Rumbley J., Ma J., Gennis R. B. (1994). The superfamily of heme-copper respiratory oxidases. J Bacteriol 176, 5587–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard L., Brom S., Dávalos A., López O., Soberón M., Romero D. (2000). Differential regulation of fixN-reiterated genes in Rhizobium etli by a novel fixL-fixK cascade. Mol Plant Microbe Interact 13, 1283–1292. 10.1094/MPMI.2000.13.12.1283 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Walker G. C. (1991). Genetic techniques in Rhizobium meliloti. Methods Enzymol 204, 398–418. 10.1016/0076-6879(91)04021-F [DOI] [PubMed] [Google Scholar]
- González V., Santamaría R. I., Bustos P., Hernández-González I., Medrano-Soto A., Moreno-Hagelsieb G., Janga S. C., Ramírez M. A., Jiménez-Jacinto V. & other authors (2006). The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc Natl Acad Sci U S A 103, 3834–3839. 10.1073/pnas.0508502103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Baeza M. J., Gómez-Hernández N., Mora Y., Delgado M. J., Romero D., Girard L. (2007). Novel reiterated Fnr-type proteins control the production of the symbiotic terminal oxidase cbb3 in Rhizobium etli CFN42. Mol Plant Microbe Interact 20, 1241–1249. 10.1094/MPMI-20-10-1241 [DOI] [PubMed] [Google Scholar]
- Hanahan D. (1983). Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166, 557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Jahn O. J., Davila G., Romero D., Noel K. D. (2003). BacS: an abundant bacteroid protein in Rhizobium etli whose expression ex planta requires nifA. Mol Plant Microbe Interact 16, 65–73. 10.1094/MPMI.2003.16.1.65 [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Landeta C., Dávalos A., Cevallos M. Á., Geiger O., Brom S. C., Romero D. (2011). Plasmids with a chromosome-like role in rhizobia. J Bacteriol 193, 1317–1326. 10.1128/JB.01184-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez O., Morera C., Miranda-Rios J., Girard L., Romero D., Soberón M. (2001). Regulation of gene expression in response to oxygen in Rhizobium etli: role of FnrN in fixNOQP expression and in symbiotic nitrogen fixation. J Bacteriol 183, 6999–7006. 10.1128/JB.183.24.6999-7006.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig B. (1986). Cytochrome c oxidase from Paracoccus denitrificans. Methods Enzymol 126, 153–159. 10.1016/S0076-6879(86)26017-6 [DOI] [PubMed] [Google Scholar]
- Michiels J., Moris M., Dombrecht B., Verreth C., Vanderleyden J. (1998). Differential regulation of Rhizobium etli rpoN2 gene expression during symbiosis and free-living growth. J Bacteriol 180, 3620–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris M., Dombrecht B., Xi C., Vanderleyden J., Michiels J. (2004). Regulatory role of Rhizobium etli CNPAF512 fnrN during symbiosis. Appl Environ Microbiol 70, 1287–1296. 10.1128/AEM.70.3.1287-1296.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R. L., Schmidt T. M. (2013). Shallow breathing: bacterial life at low O2. Nat Rev Microbiol 11, 205–212. 10.1038/nrmicro2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel K. D., Carneol M., Brill W. J. (1982). Nodule protein synthesis and nitrogenase activity of soybeans exposed to fixed nitrogen. Plant Physiol 70, 1236–1241. 10.1104/pp.70.5.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel K. D., Sanchez A., Fernandez L., Leemans J., Cevallos M. A. (1984). Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J Bacteriol 158, 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda K. J., Box J. M., Noel K. D. (2010). Genetic basis for Rhizobium etli CE3 O-antigen O-methylated residues that vary according to growth conditions. J Bacteriol 192, 679–690. 10.1128/JB.01154-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole R. K., Cook G. M. (2000). Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv Microb Physiol 43, 165–224. 10.1016/S0065-2911(00)43005-5 [DOI] [PubMed] [Google Scholar]
- Preisig O., Zufferey R., Thöny-Meyer L., Appleby C. A., Hennecke H. (1996). A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol 178, 1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richhardt J., Luchterhand B., Bringer S., Büchs J., Bott M. (2013). Evidence for a key role of cytochrome bo3 oxidase in respiratory energy metabolism of Gluconobacter oxydans. J Bacteriol 195, 4210–4220. 10.1128/JB.00470-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter O. M. H., Tao J. S., Turba A., Ludwig B. (1994). A cytochrome ba3 functions as a quinol oxidase in Paracoccus denitrificans. Purification, cloning, and sequence comparison. J Biol Chem 269, 23079–23086. [PubMed] [Google Scholar]
- Rieske J. S. (1967). Preparation and properties of reduced coenzyme Q-cytochrome c reductase (complex III of the respiratory chain). Methods Enzymol 10, 239–245. 10.1016/0076-6879(67)10047-5 [DOI] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Schweizer H. D. (1993). Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15, 831–834. [PubMed] [Google Scholar]
- Soberón M., Williams H. D., Poole R. K., Escamilla E. (1989). Isolation of a Rhizobium phaseoli cytochrome mutant with enhanced respiration and symbiotic nitrogen fixation. J Bacteriol 171, 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberón M., Membrillo-Hernández J., Aguilar G. R., Sánchez F. (1990). Isolation of Rhizobium phaseoli Tn5-induced mutants with altered expression of cytochrome terminal oxidases o and aa3. J Bacteriol 172, 1676–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink H. P., Okker R. J. H., Wijffelman C. A., Pees E., Lugtenberg B. J. J. (1987). Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol 9, 27–39. 10.1007/BF00017984 [DOI] [PubMed] [Google Scholar]
- Surpin M. A., Maier R. J. (1998). Roles of the bradyrhizobium japonicum terminal oxidase complexes in microaerobic H2-dependent growth. Biochim Biophys Acta 1364, 37–45. 10.1016/S0005-2728(98)00003-6 [DOI] [PubMed] [Google Scholar]
- Surpin M. A., Maier R. J. (1999). Symbiotic deficiencies associated with a coxWXYZ mutant of bradyrhizobium japonicum. Appl Environ Microbiol 65, 339–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M. A., Lübben M., Maier R. J. (1996). The Bradyrhizobium japonicum coxWXYZ gene cluster encodes a bb3-type ubiquinol oxidase. Gene 183, 201–206. 10.1016/S0378-1119(96)00559-8 [DOI] [PubMed] [Google Scholar]
- Trzebiatowski J. R., Ragatz D. M., de Bruijn F. J. (2001). Isolation and regulation of Sinorhizobium meliloti 1021 loci induced by oxygen limitation. Appl Environ Microbiol 67, 3728–3731. 10.1128/AEM.67.8.3728-3731.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C. P., Albrecht J., Gunsalus R. P. (1996). Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol 178, 1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. J., Sessitsch A., Corbo J. C., Giller K. E., Akkermans A. D. L., Jefferson R. A. (1995). β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology 141, 1691–1705. 10.1099/13500872-141-7-1691 [DOI] [PubMed] [Google Scholar]