Abstract
Riboswitches are structured mRNA elements that regulate gene expression in response to metabolite or second messenger binding, and are promising targets for drug discovery. Fragment-based drug discovery methods have identified weakly binding small molecule “fragments” that bind a thiamine pyrophosphate (TPP) riboswitch. However, these fragments require substantial chemical elaboration into more potent, drug-like molecules. Structure determination of the fragments bound to the riboswitch is the necessary next step. In this chapter, we describe the methods for co-crystallization and structure determination of fragment-bound TPP riboswitch structures. We focus on considerations for screening crystallization conditions across multiple crystal forms and provide guidance for building the fragment into the refined crystallographic model. These methods are broadly applicable for crystallographic analyses of any small molecules that bind structured RNAs.
1. INTRODUCTION
Riboswitches are cis-acting mRNA elements that specifically bind cellular metabolites or second messengers and modulate expression of genes in cis, typically those involved in the metabolism of their cognate ligand (Roth and Breaker, 2009; Serganov and Nudler, 2013; Zhang et al., 2010). Riboswitches are promising targets for the development of novel antibiotics, due to their specific recognition of small molecules, prevalence in bacteria, and control of genes necessary for survival or virulence in pathogens (Deigan and Ferré-D'Amaré, 2011).
Fragment-based approaches have emerged as promising methods in drug discovery (Scott et al., 2012). In a fragment-based screen, small molecules (~ 300 Da) with modest affinity for a target macromolecule are identified and then chemically elaborated into more potent compounds. Recently, application of the fragment-based method against an RNA target led to the discovery of several fragments that bind the Escherichia coli thiM thiamine pyrophosphate (TPP) riboswitch aptamer domain with Kd between 20 and 700 µM (Cressina et al., 2011). With further development, such fragments have the potential to be elaborated into ligands specific for a riboswitch from a particular organism, for use as antimicrobial compounds or chemical tools. However, rational elaboration requires structural information describing the interactions between the fragment and the riboswitch.
The method of choice to obtain high-resolution structural information on fragment binding to macromolecules is X-ray crystallography (Blow, 2002; Drenth, 2007; Rupp, 2010). The process for solving a crystal structure of a riboswitch bound to a fragment is conceptually straightforward when the structure of either the riboswitch bound to its cognate ligand or an empty riboswitch with a pre-formed ligand binding site has been determined previously. A prerequisite is the growth of well-ordered co-crystals of the riboswitch-fragment complex of interest. Fragment co-crystals can be obtained in one of two ways. First, if the empty structure has been solved and is believed to contain a pre-organized ligand binding site, the small molecules can be soaked into these empty crystals (Klein and Ferré-D'Amaré, 2006). This requires that the ligand binding site be accessible to the outside of the crystal through solvent channels. Second, if the structure of the riboswitch bound to its cognate ligand has been solved, or if the empty structure is thought to be different than the folded structure (Baird and Ferré-D'Amaré, 2010), crystals of the RNA-fragment complex, formed in solution prior to crystallization, can be grown. Generally, RNA-fragment co-crystallization conditions must be optimized, but can be guided by the co-crystallization conditions used for the cognate complex.
In this chapter, we describe the co-crystallization approach to solving structures of the E. coli thiM TPP riboswitch bound to different fragments. Application of this method has yielded structural insight into the binding mode of fragments to the E. coli thiM TPP riboswitch and visualization of fragment-induced reorganization of the ligand binding site (Warner et al., 2014). While the method is described for fragments that bind the TPP riboswitch, it is broadly applicable to any small molecules that bind structured RNAs when pertinent crystal forms have been described.
2. METHODS
2.1. Growth of riboswitch-fragment co-crystals
To increase the chance of successful fragment co-crystal growth, multiple crystal forms should be examined, if available. Starting with a known crystal form eliminates RNA sequence as a variable and allows the screening and optimization to focus on crystallization conditions.
Although crystallization conditions are generally reported in the literature as a single set or narrow range of conditions, optimization of crystallization conditions is almost always required for the growth of crystals of sufficient quality for structure determination, even when attempts are made to reproduce cognate complex co-crystals from literature conditions. Generally, single, well-ordered crystals of sufficient size are required for useful resolution, and variations in conditions may impact the presence of parasitic crystals, size and crystalline order.
When working to employ published crystals unfamiliar to the experimenter, initial screens of crystallization conditions for growing co-crystals of the RNA bound to its cognate ligand are useful to ensure that the RNA and the particular crystal form being reproduced are "well-behaved", and can offer insight into the development of efficient screens for co-crystallization of fragment complexes. The selection of a tractable number of well-behaved fragments also increases the likelihood of success. If information is available on the selectivity of the fragments, this can be used, in conjunction with practical considerations such as solubility, to define a subset of candidate fragments to initially attempt to co-crystallize. Once candidate crystal forms and fragments have been evaluated, fragment co-crystallization conditions can be screened.
1. Considerations in transcription template and RNA construct design
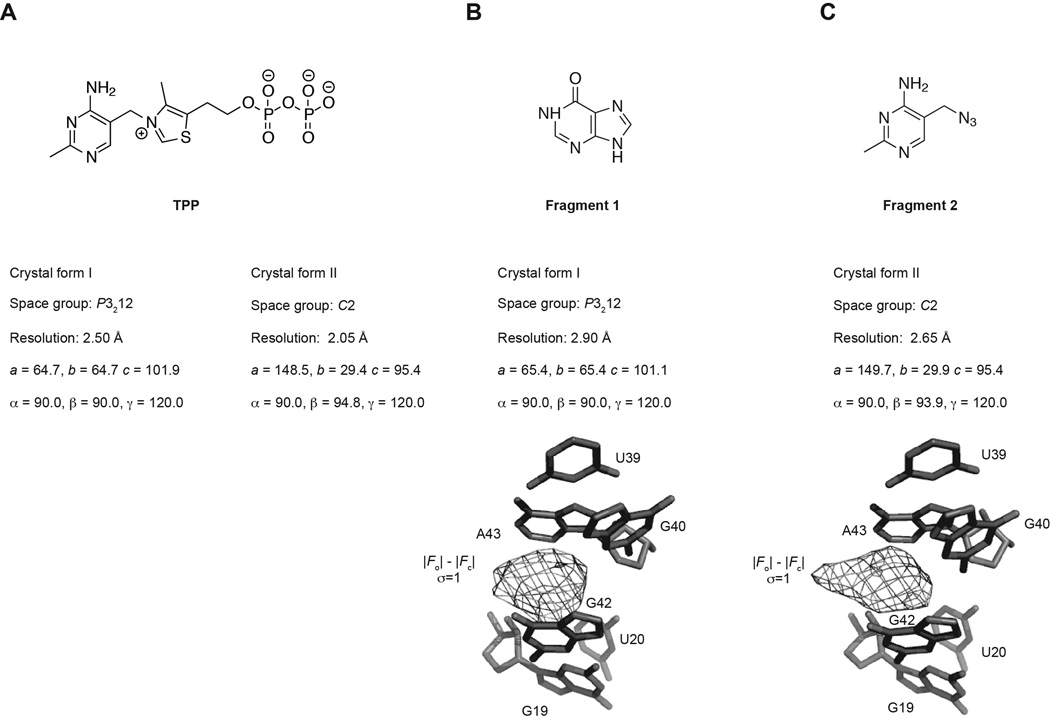
If available, multiple crystal forms should be screened to increase the chances of success. Four crystal forms of the TPP riboswitch have been described: three of the E. coli thiM TPP riboswitch (Edwards and Ferré-D'Amaré, 2006; Kulshina et al., 2010; Serganov et al., 2006), and one of the Arabidopsis thaliana thiC TPP riboswitch (Thore, 2006). For the TPP riboswitch-binding fragments, three crystal forms were examined, and fragment structures were solved using two of these (crystal forms I and II, Figure 1). We found that certain fragments only grew crystals of acceptable quality in certain crystal forms.
Figure 1.
Two crystal forms were used for growth of fragment co-crystal structures. (A) Two crystal forms of the riboswitch in complex with its cognate ligand, TPP, were used to grow fragment crystals. (B) Fragment 1 co-crystals were grown in crystal form I. Pre-ligand unbiased residual |Fo| - |Fc| electron density at 3.0 s.d. contoured around the final refined ligand is shown (bottom). (C) Fragment 2 co-crystals were grown in crystal from II. |Fo| - |Fc| electron is as in (B).
When using a previously described crystal form, it is key to replicate exactly the RNA sequence described in the original conditions. In addition, it is good practice to pay special care to other tricks that may have been employed to facilitate crystallization, such as a bimolecular construct or the use of ribozymes to produce homogenous 5’ and 3’ ends (Ferré-D'Amaré and Doudna, 1996).
If 5’ or 3’ ribozymes are employed, and the ribozyme is close in length to that of the desired riboswitch RNA (within ~ 10 nt for ~ 100 nt RNA), additional nucleotides can be added to the end of the ribozyme to allow for more efficient purification of the riboswitch RNA by polyacrylamide gel electrophoresis (PAGE). These additional nucleotides can be incorporated into the DNA template.
For crystal form I (Edwards and Ferré-D'Amaré, 2006), a hammerhead ribozyme was encoded 5' of the thiM TPP riboswitch sequence and a Varkund Satellite (VS) ribozyme was encoded 3' of the sequence. The resultant riboswitch RNA has a 5'-OH and a 2',3' cyclic phosphate.
For crystal form II (Serganov et al., 2006), a hepatitis delta virus (HDV) ribozyme was encoded 3' of the thiM TPP riboswitch sequence to yield an RNA with a 5' triphosphate and a 2',3' cyclic phosphate. Additional nucleotides were added to the 3' end of the HDV ribozyme to allow for more efficient separation of the riboswitch from the ribozyme during gel electrophoresis.
2. Considerations in fragment selection
Fragments should first be evaluated by selectivity information, if available. In this study, only fragments selective for a TPP riboswitch over a lysine riboswitch (Cressina et al., 2011) were considered. Solubility is also a factor. If fragments are only soluble in solvents other than water, the effect of that solvent on crystal growth should be considered. Fragments that were not soluble in water and were not soluble in DMSO to at least ~ 25x Kd were not considered for crystallization trials, to prevent high levels of DMSO in crystallization, which may adversely affect crystal growth.
3. In vitro transcription of TPP riboswitch RNA
TPP riboswitch RNA (Edwards and Ferré-D'Amaré, 2006; Serganov et al., 2006; Thore, 2006) was transcribed in vitro using DNA templates, which encoded the appropriate ribozymes and were produced either by restriction digestion of a plasmid or by PCR, and recombinant T7 RNA polymerase as described (Milligan et al., 1987) and purified by electrophoresis on polyacrylamide, 8 M urea, 1x TBE gels (29:1 acrylamide:bisacrylamide), where the percentage of polyacrylamide was selected to give sufficient separation between the TPP riboswitch RNA and any ribozymes used, typically 8–12%. RNA was electroeluted from gel slices using a Whatman Elutrap system, concentrated, washed once with 1 M KCl and desalted extensively through serial dilution with water by ultrafiltration using Amicon Ultra centrifugal filters (10 kDa molecular weight cutoff), and stored at 4°C in water or in 0.1 mM EDTA prior to use. We find that if proper care is taken to avoid RNase contamination, RNA stored in this manner lasts for at least six months.
4. Initial screens of crystallization conditions with the cognate ligand
In general, crystallization involves the simultaneous optimization of a number of solution variables, in order to achieve the appropriate level of supersaturation for nucleation and crystal growth (McPherson, 1999). While optimization can be carried out employing large-scale screening and powerful analytical methods (e.g. (Carter, 1997)), in the majority of cases, small-scale, qualitative screens are constructed to evaluate the effect and interdependence of a handful of variables at a time.
To evaluate the behavior of a specific crystal form in the presence of the cognate ligand, initial screens are performed, typically consisting of a small sparse matrix in which a limited number of conditions are varied. For crystal form I (Edwards and Ferré-D'Amaré, 2006), the conditions described for the cognate co-crystals indicate incubation of 150 µM RNA with 0.5 mM TPP in 5 mM Tris-HCl (pH 8.1), 3 mM MgCl2, 10 mM NaCl, 100 mM KCl, and 0.5 mM spermine at 37°C for 30 min, with co-crystals grown by vapor diffusion of 3 µL drops by mixing the RNA solution 1:1 with a reservoir solution of 26–30% polyethyleneglycol (PEG) 2000, 0.2 M NH4Cl, 10 mM CaCl2 (or MnCl2) and 50 mM Na cacodylate (pH 6.0). Patterns of behavior in crystal growth were observed while varying the percentage of the precipitant (20–30% PEG 2000), identity of the precipitant (PEG 2000 vs. PEG 3350), concentration of NH4Cl (0.05 – 0.5 M), and the ratio of RNA solution to reservoir solution. The RNA folding conditions were not varied (although this might be essential in some cases), and drops were set up as described above, except with a final drop volume of 1 µL. In crystal form I, higher concentrations of PEG 2000 were found to yield larger and more single cognate ligand co-crystals, while crystals were tolerant of variation in NH4Cl concentration. Equivalent trials were carried out for crystal form II. For a third crystal form, poor growth of the cognate ligand co-crystals was observed in trials, and the crystal form was abandoned.
5. Initial screens for fragment co-crystals
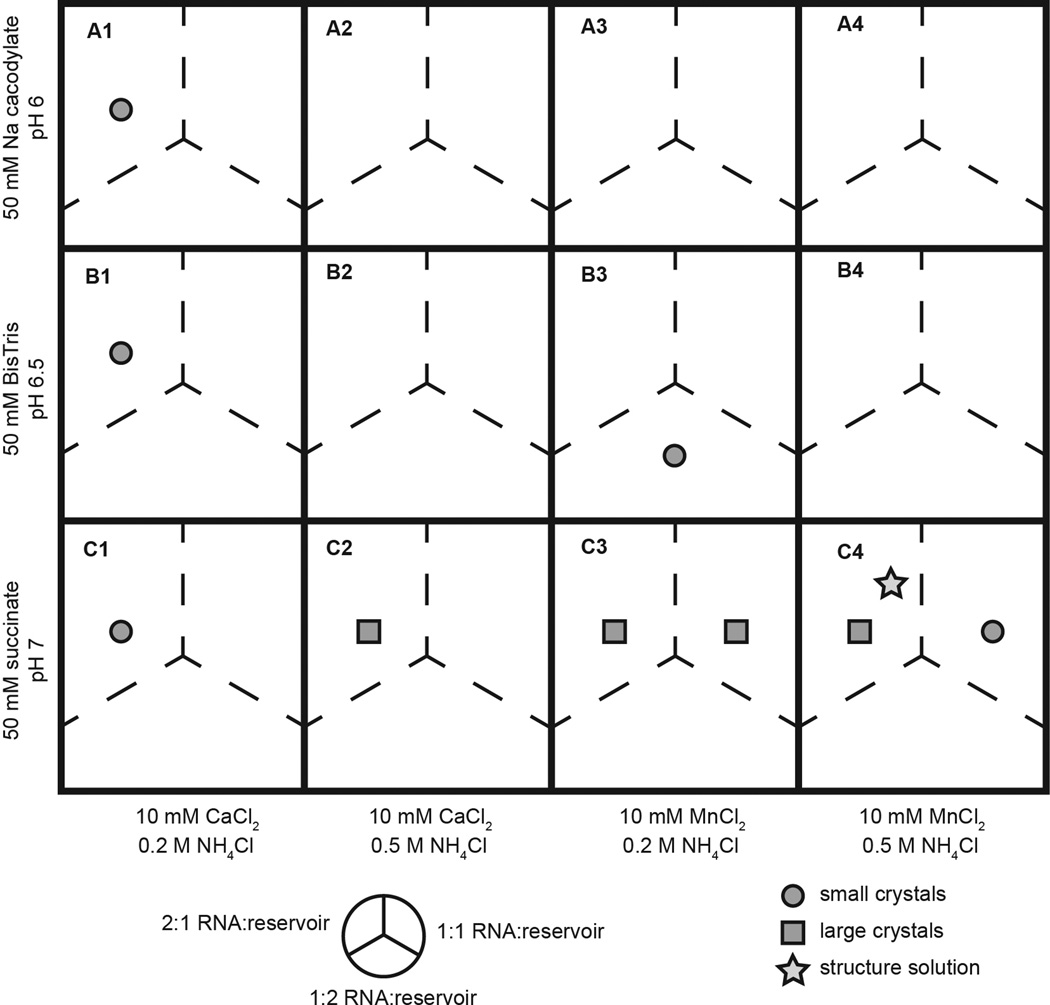
For growth of fragment co-crystals, initial crystal screens are guided by patterns observed in the cognate ligand screens. For screens with fragment 1 and crystal form I, the percentage of PEG 2000 was kept constant at 30%, the concentration of NH4Cl was either 0.2 M or 0.5 M, and the buffer identity, buffer pH, divalent cation identity, and ratio of RNA solution to reservoir solution were varied. Figure 2 shows the initial screen that resulted in good fragment co-crystal growth under a range of conditions and eventual structure solution for fragment 1 in crystal form I. While the co-crystal structure of fragment 1 was solved from a condition in the initial screen, other fragments required more extensive variation of conditions. In our study, we found that crystal growth for different RNA-fragment complexes varied substantially, without correlating with binding affinity of the fragment for the RNA.
Figure 2.
A representation of a screen of co-crystallization conditions for fragment 1 co-crystals. Each cell in the matrix represents a well on a crystal screening plate. Buffer identity, pH, divalent cation identity, and NH4Cl concentration are varied while the precipitant concentration (30% PEG 2000) and RNA folding conditions are held constant. Each well contained three drops, in which the ratio of RNA solution to reservoir solution was varied, represented by the three dashed line divisions in each cell. Circles represent conditions that yielded crystals of small size or poor quality. Squares represent conditions that yielded large crystals (greater than 100 µm in at least 2 dimensions). A star indicates the crystal growth condition for the crystal from which the fragment co-crystal structure was solved.
6. Development of cryoprotectant solutions for vitrification of fragment co-crystals
Once co-crystals are obtained, cryoprotection conditions must be determined. Ideally, cryoprotection of a crystal should allow for vitrification of the water surrounding and inside the crystal, without damaging the crystal (Garman, 2003; Rodgers, 1997). Screens for optimal cryoprotectant conditions can begin with the reported cryoprotection conditions from the cognate ligand co-crystals, and trial and error on cognate ligand co-crystals is helpful. Conditions can initially be screened for ability to vitrify, by screening mother liquor supplemented with ~ 15% cryoprotection candidate. Upon cooling, a successful cryoprotectant will yield a transparent drop and an X-ray diffraction pattern free from ice rings (water powder diffraction).
Additionally, various methods of cryoprotection schemes should be considered. Robust crystals may survive a quick dip in neat cryoprotectant. Crystals can be transferred to an artificial mother liquor, supplemented with cryoprotectant. Crystals that are sensitive to large changes in mother liquor can be serially transferred through drops of increased cryoprotection concentration. Some crystals can survive cryoprotectant addition directly to the original drop. Alternatively, particularly fragile crystals may require growth in mother liquor that contains sufficient levels of cryoprotectant for direct freezing from the crystal growth drop.
Regardless of the cryoprotection scheme employed, crystals vitrified in various cryoprotectants must be screened to assay for effects on crystal quality. Cryoprotection schemes that reproducibly yield X-ray data of poorer quality should be discarded. More extensive discussion of cryoprotection methods can be found elsewhere (Alcorn and Juers, 2010; Berejnov et al., 2006).
Fragment co-crystals in crystal form I were initially vitrified in mother liquor supplemented with 15% Ficoll or sucrose and 1 mM fragment. For fragment-TPP riboswitch co-crystals, ~10–20% glycerol, ethylene glycol, or sucrose were found to be the most useful cryoprotectants. Co-crystals were transferred to drops containing artificial mother liquor supplemented with cryoprotectant, either directly or through a series of drops containing increasing cryoprotectant concentration, in two to four increments of 2.5 – 5.0% cryoprotectant per step. Equilibration time in cryoprotectant-supplemented drops was less than one minute.
It should be noted that the concentration of ligand in the cryoprotection solution can be an important experimental variable. Moreover, the precise composition of the solution may have an important impact on whether fragments bind the RNA with sufficient occupancy to allow crystallographic visualization. For instance, for crystals of the glmS ribozyme-riboswitch, we have found that the pH of the cryoprotection conditions determines whether the cognate ligand yields detectable electron density in its binding site (Klein et al., 2007).
2.2. Structure solution by molecular replacement
After data collection and reduction, assuming fragments do not promote complete rearrangement of the RNA, fragment co-crystal structures can be solved either by molecular replacement with the structure of an empty or cognate ligand co-crystal or, if fragment co-crystals are isomorphous with the known crystal form, by inspection of a difference Fourier map, possibly after refinement of the RNA model. Here we will discuss the molecular replacement method. After molecular replacement, strong positive density in the difference (|Fo|-|Fc|) map should indicate fragment position even in the early stages of refinement. The ligand binding site and the ligand itself should only be built after all other regions of the model have been rebuilt as appropriate and refined.
1. Structure solution by molecular replacement
Fragments co-crystal structures can be solved by molecular replacement using structures of the crystal form employed for fragment co-crystal growth. Ions, ligand, and water molecules should first be removed from the search model. In this study, molecular replacement was carried out with PHASER (McCoy et al., 2007); various molecular replacement strategies are described elsewhere (Scapin, 2013). PHASER yielded results with high log-likelihood gain values (LLG; the difference between the model and a random solution) and translation function Z-scores (TFZ; the number of standard deviations above the mean of translation function search solutions): for fragment 1, LLG = 1092, TFZ = 29.8; for fragment 2, LLG = 2270, TFZ = 12.1.
2. Model building and refinement
Refinement of the fragment co-crystal structures is initially focused on all regions of the RNA except for the binding site. After all other major issues (such as significant peaks in the |Fo|-|Fc| map) outside of the binding site are accounted for, the binding site can be solved.
At this stage, a region of strong positive density in the difference (|Fo|-|Fc|) map indicates the location and, provided the resolution is sufficient, the binding orientation of the fragment (Figure 1). For the TPP riboswitch-fragment co-crystals, the electron density corresponding to the fragments was the most significant peak in the difference map, even in the first rounds of refinement. Careful inspection of the ligand binding site is necessary to account for any fragment-induced reorganization of the binding site. The fragments in this study promoted a previously unobserved rotation of a guanine (G72) into a region of the binding pocket previously occupied by a pyrophosphate of the cognate ligand TPP (Warner et al., 2014).
3. Building the fragment into the model
Before a fragment can be built into a purported fragment binding site, a series of energetic and geometric parameters for the ligand must be defined. A range of utilities exist that define these parameters for various refinement programs (Lebedev et al., 2012; Moriarty et al., 2009; Schüttelkopf and van Aalten, 2004; Winn et al., 2011). Once the ligand is built into the model, care should be taken to ensure that the correct orientation of the fragment is chosen. Factors such as geometry, real-space R factor, post-refinement |Fo|-|Fc| peak size, and anomalous signal can be used to guide ligand orientation, but in structures solved to modest resolution, ambiguity in fragment orientation may persist. In this case, it is helpful to report multiple possible fragment orientations.
3. CONCLUSIONS
Crystallographic analysis of fragment binding to the TPP riboswitch has delineated the binding location of fragments, as well as unexpected fragment-induced rearrangement of the ligand binding site (Warner et al., 2014). The growth of co-crystals and subsequent structure solution by molecular replacement described here are conceptually straightforward, and are broadly applicable for any small molecule ligands that bind in a well-formed binding site in an RNA. However, in practice the process can range from trivial, given a well-behaved crystal form and small molecule co-crystals that grow in very similar conditions, to as difficult as solving a novel crystal structure (Ferré-D'Amaré and Doudna, 2001; Ferré-D'Amaré, 2010), if small molecule co-crystals are found to require the development of a new crystal form.
ACKNOWLEDGEMENTS
We thank the staff at beamlines 24-ID-C of APS and 5.0.1 and 5.0.2 of ALS for crystallographic data collection support; and N. Baird, M. Chen, C. Jones, M. Lau, A. Serganov, M. Warner, and J. Zhang for discussions. This work is partly based on research conducted at ALS on the BCSB beamlines and at APS on the NE-CAT beamlines, all of which are supported by the National Institute for General Medical Sciences, NIH. Use of ALS and APS was supported by the US Department of Energy. This work was supported in part by the NIH-Oxford/Cambridge Research Scholars program and the intramural program of the National Heart, Lung and Blood Institute (NHLBI), NIH.
REFERENCES
- Alcorn T, Juers DH. Progress in rational methods of cryoprotection in macromolecular crystallography. Acta Crystallogr D. 2010;66:366–373. doi: 10.1107/S090744490903995X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Ferré-D'Amaré AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010;16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berejnov V, Husseini NS, Alsaied OA, Thorne RE. Effects of cryoprotectant concentration and cooling rate on vitrification of aqueous solutions. J Appl Crystallogr. 2006;39:244–251. [Google Scholar]
- Blow D. Outline of Crystallography for Biologists. Oxford: Oxford University Press; 2002. [Google Scholar]
- Carter CW. Response surface methods for optimizing and improving reproducibility of crystal growth. Methods in Enzymology. 1997;276:74–99. doi: 10.1016/S0076-6879(97)76051-8. [DOI] [PubMed] [Google Scholar]
- Cressina E, Chen L, Abell C, Leeper FJ, Smith AG. Fragment screening against the thiamine pyrophosphate riboswitch thiM. Chem Sci. 2011;2:157–165. [Google Scholar]
- Deigan KE, Ferré-D'Amaré AR. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res. 2011;44:1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenth J. Principles of Protein X-Ray Crystallography. New York: Springer; 2007. [Google Scholar]
- Edwards TE, Ferré-D'Amaré AR. Crystal Structures of the Thi-Box Riboswitch Bound to Thiamine Pyrophosphate Analogs Reveal Adaptive RNA-Small Molecule Recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Doudna JA. Use of cis- and trans-ribozymes to remove 5' and 3' heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Doudna JA. Methods to crystallize RNA. In: Beaucage SL, Bergstrom DE, Glick GD, Jones RA, editors. Curr Protoc Nucleic Acid Chem. New York: John Wiley & Sons; 2001. pp. 7.6.1–7.6.10. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR. Use of the spliceosomal protein U1A to facilitate crystallization and structure determination of complex RNAs. Methods. 2010;52:159–167. doi: 10.1016/j.ymeth.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garman E. “Cool” crystals: macromolecular cryocrystallography and radiation damage. Curr Opin Struct Biol. 2003;13:545–551. doi: 10.1016/j.sbi.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Ferré-D'Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Wilkinson SR, Been MD, Ferré-D'Amaré AR. Requirement of helix P2.2 and nucleotide G1 for positioning the cleavage site and cofactor of the glmS ribozyme. Journal of Molecular Biology. 2007;373:178–189. doi: 10.1016/j.jmb.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshina N, Edwards TE, Ferré-D'Amaré AR. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA. 2010;16:186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, Murshudov GN. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr D. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson A. Crystallization of biological macromolecules. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers DW. Practical Cryocrystallography. Methods in Enzymology. 1997;276:183–203. doi: 10.1016/S0076-6879(97)76059-2. [DOI] [PubMed] [Google Scholar]
- Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp B. Biomolecular Crystallography. New York: Garland Science; 2010. [Google Scholar]
- Scapin G. Molecular replacement then and now. Acta Crystallogr D. 2013;69:2266–2275. doi: 10.1107/S0907444913011426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Scott DE, Coyne AG, Hudson SA, Abell C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry. 2012;51:4990–5003. doi: 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- Serganov A, Nudler E. A Decade of Riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S. Structure of the Eukaryotic Thiamine Pyrophosphate Riboswitch with Its Regulatory Ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Warner KD, Homan P, Weeks KM, Smith AG, Abell C, Ferré-D'Amaré AR. Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically. Chem Biol. 2014;21:591–595. doi: 10.1016/j.chembiol.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lau MW, Ferré-D'Amaré AR. Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry. 2010;49:9123–9131. doi: 10.1021/bi1012645. [DOI] [PMC free article] [PubMed] [Google Scholar]