Abstract
Protein synthesis by the ribosome requires the translocation of transfer RNAs and messenger RNA by one codon after each peptide bond is formed, a reaction that requires ribosomal subunit rotation and is catalyzed by the guanosine triphosphatase (GTPase) elongation factor G (EF-G). We determined 3 Å resolution x-ray crystal structures of EF-G complexed with a non-hydrolyzable GTP analogue and bound to the Escherichia coli ribosome in different states of ribosomal subunit rotation. The structures reveal that EF-G binding to the ribosome stabilizes switch regions in the GTPase active site, resulting in a compact EF-G conformation that favors an intermediate state of ribosomal subunit rotation. These structures suggest that EF-G controls the translocation reaction by cycles of conformational rigidity and relaxation preceding and following GTP hydrolysis.
Introduction
GTPases–enzymes that catalyze the hydrolysis of guanosine 5′-triphosphate–are widespread in biology, and use GTP hydrolysis as a “switch” between functional states driven by protein conformational changes (1). Protein biosynthesis by the ribosome is controlled by GTPase translation factors in all stages of translation (2). Although the GTPase catalytic core is highly conserved, translation factors have evolved unique domain architectures for separate and non-overlapping functions in translation initiation, elongation, termination and ribosome recycling. The distinctions between translation factors, both within the translation process and between different domains of life, are targets for numerous families of antimicrobial compounds (3, 4). However, the structural basis for how GTPase translation factors use a highly conserved GTP hydrolysis mechanism to control distinct steps of translation remains unclear.
During polypeptide elongation, bacterial elongation factors EF-Tu and EF-G alternate in catalyzing accurate messenger RNA (mRNA) decoding and mRNA and transfer (tRNA) translocation, respectively. The GTPase center of EF-Tu is coupled to distortions in aminoacyl-tRNA that contribute to the accuracy of mRNA decoding (5, 6). By contrast, EF-G promotes movement of mRNA and tRNA on the ribosome in steps that involve large-scale rearrangements of the ribosome (7–11). Biochemical and genetic experiments have shown that the GTPase centers of EF-Tu and EF-G, although highly conserved, are not interchangeable (12) whereas key amino acids in the GTPase active site of the eukaryotic translocase eEF2 can be mutated to those of EF-G and retain function (13). Furthermore, whereas EF-Tu hydrolyzes GTP rapidly only during accurate mRNA decoding, the GTPase activity of EF-G is greatly accelerated even by vacant ribosomes (14, 15). Together with the divergent architectures of EF-Tu and EF-G outside of the GTPase active site (16), these results indicate that EF-G has evolved considerably different means for linking GTP hydrolysis to ribosome dynamics.
Messenger RNA and tRNA translocation occurs in multiple steps (17). First, the 3’ acceptor ends of the tRNAs move with respect to the large ribosomal (50S) subunit, so that the peptidyl-site (P-site) and aminoacyl-site (A-site) tRNA termini move to the exit (E) and P sites, respectively, creating a hybrid P/E and A/P tRNA binding state (18). This hybrid state requires a rotation of the small ribosomal (30S) subunit relative to the 50S subunit (7) and an orthogonal rotation of the 30S subunit head domain (9, 19, 20) (Fig. 1A), conformational changes that are conserved in the eukaryotic ribosome (21). When complexed with GTP, EF-G binds the ribosome and favors ribosomal subunit rotation, a state associated with tRNA binding in the hybrid A/P and P/E sites (22–25). GTP hydrolysis by EF-G, subsequent ribosome dynamics (10), and phosphate release are then required to translocate mRNA and the tRNA anticodons on the small ribosomal subunit to complete the translocation reaction and to release EF-G/GDP from the ribosome, respectively (26, 27).
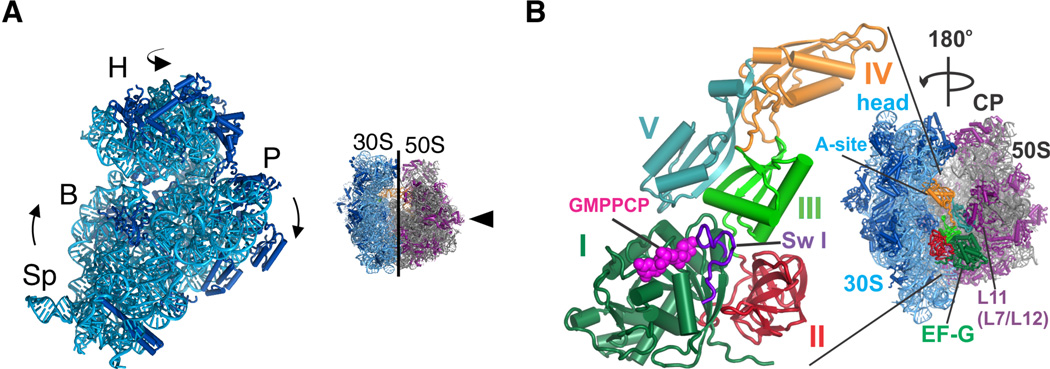
Figure 1. Global structural rearrangements in the ribosome in EF-G/GMPPCP complexes.
(A) Schematic illustrating two degrees of freedom in the 30S subunit within the 70S ribosome. 30S subunit rotation encompasses the body (B) and platform (P) domains, whereas the 30S subunit head domain (H) swivels around a nearly orthogonal rotational axis. (B) EF-G is shown bound to the 70S ribosome oriented 180° from the view shown to the left. Domains in EF-G are numbered: domain I (dark green), domain II (red), domain III (light green), domain IV (orange), and domain V (light blue). Switch I (sw I) (amino acids 38–64) in domain I is highlighted in purple spheres. The 30S subunit is in blue, and 50S subunit in grey and magenta.
The structural basis for EF-G/GTP stabilization of rotated states of the ribosome is known only at low resolution (9, 10, 28). Cryo-EM reconstructions revealed EF-G domain positions in late stages of tRNA translocation, but do not provide a molecular understanding of how the GTPase active site in EF-G, positioned by the 50S ribosomal subunit, is connected to events on the 30S subunit required for mRNA and tRNA translocation. Here, we determined structures of the ribosome in multiple states of subunit rotation, in complexes with EF-G bound to the non-hydrolyzable GTP analogue GMPPCP. These structures reveal that GTP binding rearranges switch regions in EF-G to promote EF-G interdomain packing and ribosomal subunit rotation, an allosteric mechanism reminiscent of motor proteins that use ATP hydrolysis to drive mechanical events common in biology.
Results and Discussion
Global conformations of the ribosome complexes
We determined two crystal structures of the E. coli 70S ribosome in complexes with EF-G, the non-hydrolyzable GTP analog GMPPCP and the antibiotic viomycin, to a resolution of 3 Å (Table 1) (29). Each crystal form contains four unique copies of the ribosome in the crystallographic asymmetric unit that adopt different conformations with respect to 30S subunit rotation and swiveling of the 30S subunit head domain (fig. S1). Rotation of the 30S subunit body and platform domains ranges from ~0° to ~8°, whereas the head domain of the 30S subunit is swiveled by ~6° to ~11° (table S1). EF-G, a five-domain protein, is bound to all eight copies of the ribosome with the GTPase domain (domain I or G-domain) and domains II, III and V positioned adjacent to the 50S subunit L11 arm, while domain IV projects into the 30S subunit mRNA decoding site (A site) (Fig. 1B). All of the copies of EF-G contain clear electron density for GMPPCP visible in the GTPase active site.
Table 1.
X-ray diffraction data and refinement statistics
| Crystal I | Crystal II | |
|---|---|---|
| Space group | P 21 | P 21 |
| unit cell (a, b, c in Å) | 361.60, 361.77, 433.20 | 361.14, 360.51, 429.73 |
| (α, β, γ in deg.) | 90.0, 103.566, 90.0 | 90.0, 103.217, 90.0 |
| Resolution (Å) | 70 – 3.0 | 70 – 3.0 |
| (high-resolution shell)* | (3.1 – 3.0) | (3.1 – 3.0) |
| Rmerge† | 15.2 (100.9) | 17.3 (132) |
| I/σ (I) | 6.49 (0.84) | 5.84 (0.58) |
| CC(1/2) (%) | 99.4 (42.7) | 99.3 (30.4) |
| Completeness (%) | 83.7 (60.7) | 89.6 (71.5) |
| Measurement redundancy | 3.1 (1.5) | 3.9 (2.3) |
| Unique reflections | 1,799,385 (122,106) | 1,904,514 (142,145) |
| No. crystals used | 20 | 24 |
| Refinement | ||
| Resolution (Å)† | 70–2.9 | 70–2.9 |
| No. reflections | 1,874,109 | 1,984,535 |
| Molecules per a.s.u | 4 | 4 |
| Rfree set | 8,386 | 8,583 |
| R/Rfree (%)* | 0.230/0.278 | 0.221/0.270 |
| Average B-factor | ||
| RNA | 27.4 | 31.8 |
| Protein | 30.3 | 38.5 |
| Other | 12.6 | 15.1 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.009 |
| Bond angles (°) | 1.304 | 1.376 |
Data beyond the high-resolution shell in parentheses were used for refinement and map calculation, and extend to a CC(1/2) value of about 24.5 % (54).
All statistics not in parentheses include data over the whole reported resolution range. Crystal form I contains higher occupancy for viomycin and II lower occupancy for viomycin.
Ordering of the GTPase switch regions and EF-G domain packing
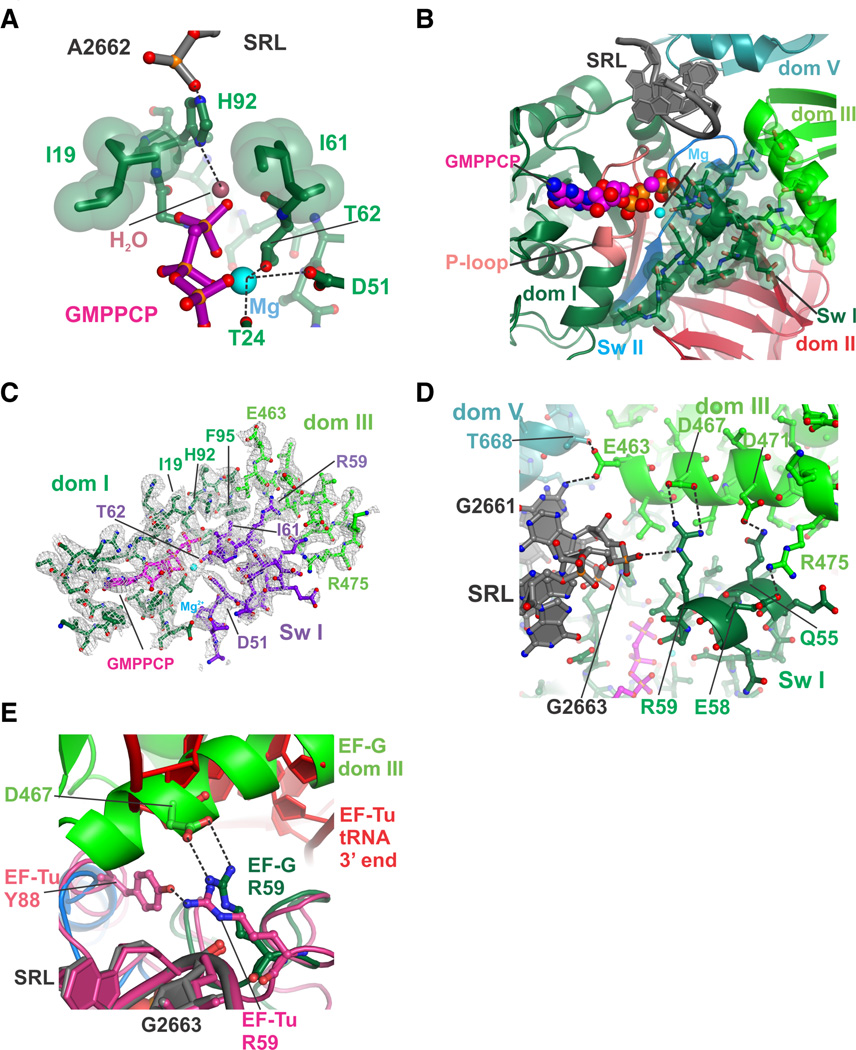
The GTPase active site in EF-G contains mobile “switch” elements termed switch I (amino acids 38–64) and switch II (amino acids 84–107), and a P loop that coordinates the triphosphate (amino acids 12–27) (30). In EF-G, the switch elements are thought to convert the free energy of GTP hydrolysis in the G-domain into the unidirectional translocation of the ribosome along an mRNA and the rapid cycling of EF-G during protein synthesis (9, 15, 27, 31). In the present structures, the G-domain of EF-G contacts the Sarcin-Ricin Loop (SRL) in 23S ribosomal RNA (rRNA) of the large ribosomal subunit. Nucleotide A2662 in the SRL coordinates a catalytic histidine in switch II (His92) in EF-G and positions its imidazole ring in the correct location to activate a nucleophilic water molecule putatively required for GTP hydrolysis, similar to the structure of the GTPase EF-Tu bound to the ribosome in the GTP state (6) (Fig. 2A). By contrast, this catalytic histidine is oriented away from the active site in a structure of the T. thermophilus 70S ribosome trapped with EF-G in a GDP state by the antibiotic fusidic acid (32). Binding of the GTP analog also orders much of switch I in EF-G (amino acids 49–64), closing the GTP binding pocket (Fig. 2B, 2C), as previously observed at low resolution by cryo-EM with a paralog of EF-G (9). During translocation, GTP hydrolysis by EF-G is rapid and is followed by a rate-limiting conformational rearrangement of the translocation complex that is coupled to EF-G in an activated GDP•Pi state (27, 33). After GTP hydrolysis and translocation, switch I becomes disordered (31), which greatly accelerates release of EF-G/GDP from the post-translocational ribosome (27, 31). In the structure of EF-G trapped on the ribosome with GDP by fusidic acid, switch I is entirely disordered (32). Thus, the current complexes likely represent either the pre-GTP hydrolysis configuration of EF-G on the ribosome, or the GDP•Pi state.
Figure 2. Compact arrangement of EF-G domains I and III in the GTP state.
(A) Position of the catalytic histidine H92. Gate residues Isoleucine 19 and Isoleucine 61 are shown with green spheres with van der Waals radii, along with the position of the proposed activated water and coordinated Mg2+ ion. (B) View of the GTPase active site, including swI, switch II (swII), P-loop, Sarcin-Ricin Loop in 23S rRNA (SRL) and the GTP nucleotide analog GMPPCP. Amino acids in swI that become ordered upon GTP binding and form salt bridges with EF-G domain III are shown as spheres with van der Waals radii. (C) Four-fold NCS averaged electron density for the GTPase domain of EF-G near the GTP binding pocket. Shown are EF-G swI, domain I, domain III, the GTP analog GMPPCP color-coded as in panel A, and a magnesium ion (light blue). Electron density (grey grid) is contoured at 1 standard deviation from the mean. (D) Closeup view of interactions between swI (dark green) and domain III (green) in the EF-G/GMPPCP complex. Contacts within hydrogen-bonding distance are indicated with dashed lines. (E) Essential salt bridge between EF-G residue R59 in swI and residue D467 in domain III. The corresponding interactions in the EF-Tu/aminoacyl-tRNA decoding complex are also shown. EF-G is color coded as in Figure 1B, while the EF-Tu/tRNA complex is colored magenta (protein, SRL) and red (tRNA).
The folding of EF-G switch I results in multiple new inter-domain contacts throughout EF-G. Arginine 59 (R59) from switch I, universally conserved in canonical translation GTPases (34), interacts with the backbone of SRL nucleotide A2663 on one side, as seen with EF-Tu (6), and forms a salt bridge with aspartate 467 (D467) in EF-G domain III (Fig. 2D), which occupies the space where the 3’-acceptor end of tRNA binds EF-Tu (6) (Fig. 2E). Although mutations of R59 in EF-Tu and EF-G do not impact GTP hydrolysis (35), mutations of R59 in EF-G decrease translocation up to 50-fold (35), similar to the rate that occurs in the absence of GTP or with a non-hydrolyzable GTP analog (35). The rate of translocation is decreased only 5-fold when R59 is replaced with lysine, showing that the salt bridge with D467 in domain III is important for EF-G function. A second salt bridge forms between Glutamate 58 and Arginine 475 (Fig. 2D), an interaction absent in EF-Tu (6). Further differences between the geometry of switch I in EF-G and EF-Tu occur in amino acids adjacent to the α-phosphate and P loop (fig. S2), which may explain the observation that replacement of EF-G switch I amino acids 44–55 with those from EF-Tu renders EF-G functionally inactive (12).
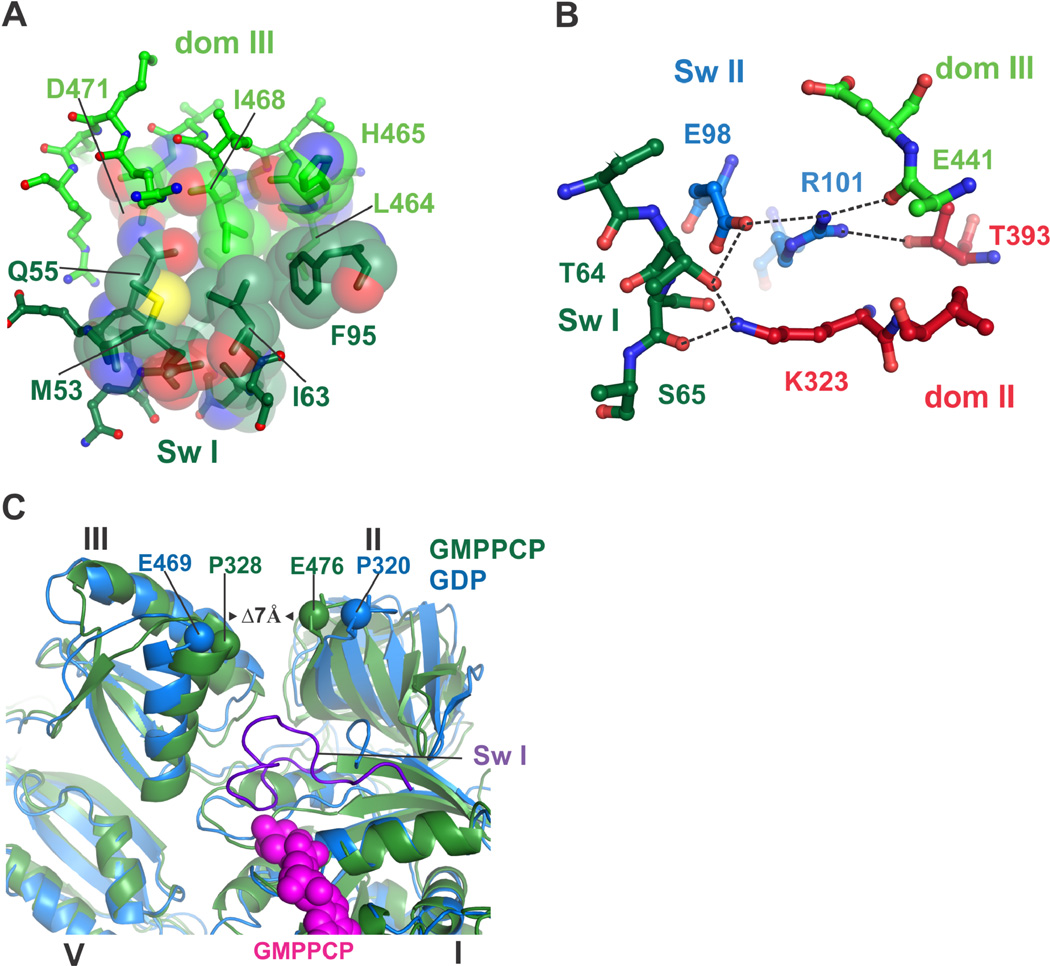
The folding of EF-G switch I further induces close hydrophobic packing between domains I, II and III, capped by the side chain of R59, and centered on switch II residue Phenylalanine 95 (F95) which is thought to contribute to GTPase activation (36) (Fig. 3A). In the ribosome complex of EF-G with GDP and fusidic acid, this interface is broken and occupied by the antibiotic, which packs against F95 (32). Notably, mutations in this interface including F95 confer fusidic acid resistance to cells (36). In some cases these mutations make EF-G an intrinsically active GTPase (36) while at the same time drastically reducing EF-G’s ability to accelerate translocation, indicating that GTP hydrolysis must be linked with ribosomal conformational changes for effective translocation (36). An additional network of polar and hydrophobic interactions surrounds salt bridges between switch II residues Arginine 101 (R101), Glutamate 98 (E98), and domain II residue Lysine 323 (K323), which are held in place by residues from switches I and II (Threonine 64 and Serine 65, and Isoleucine 97) and domains II and III (Threonine 393 and the backbone of Glutamate 441) (Fig. 3B). The contacts between these conserved residues in EF-G are lost upon GTP hydrolysis and Pi release, when switch I becomes disordered and domains II and III move apart (32).
Figure 3. A network of contacts between EF-G domains extends from the GTPase center towards the small ribosomal subunit.
(A) A new hydrophobic core forms between domains I and III in EF-G in the GTP state. Positions in swI of domain I (dark green carbons) and domain III (green carbons) are shown. (B) Polar and ionic interactions formed between swI and swII and domains II and III in the GTP state, color-coded as in Fig. 1B. Dashes indicate atoms within hydrogen-bonding distance. (C) Closure of domains II and III due to binding of the GMPPCP form of EF-G to the ribosome (7 Å) is indicated. The structure of the ribosome with EFG/ GDP/fusidic acid is shown in blue (32). The ribosome in an intermediate rotation state with EF-G/GMPPCP is in green.
EF-G coupling to the conformation of the 70S ribosome
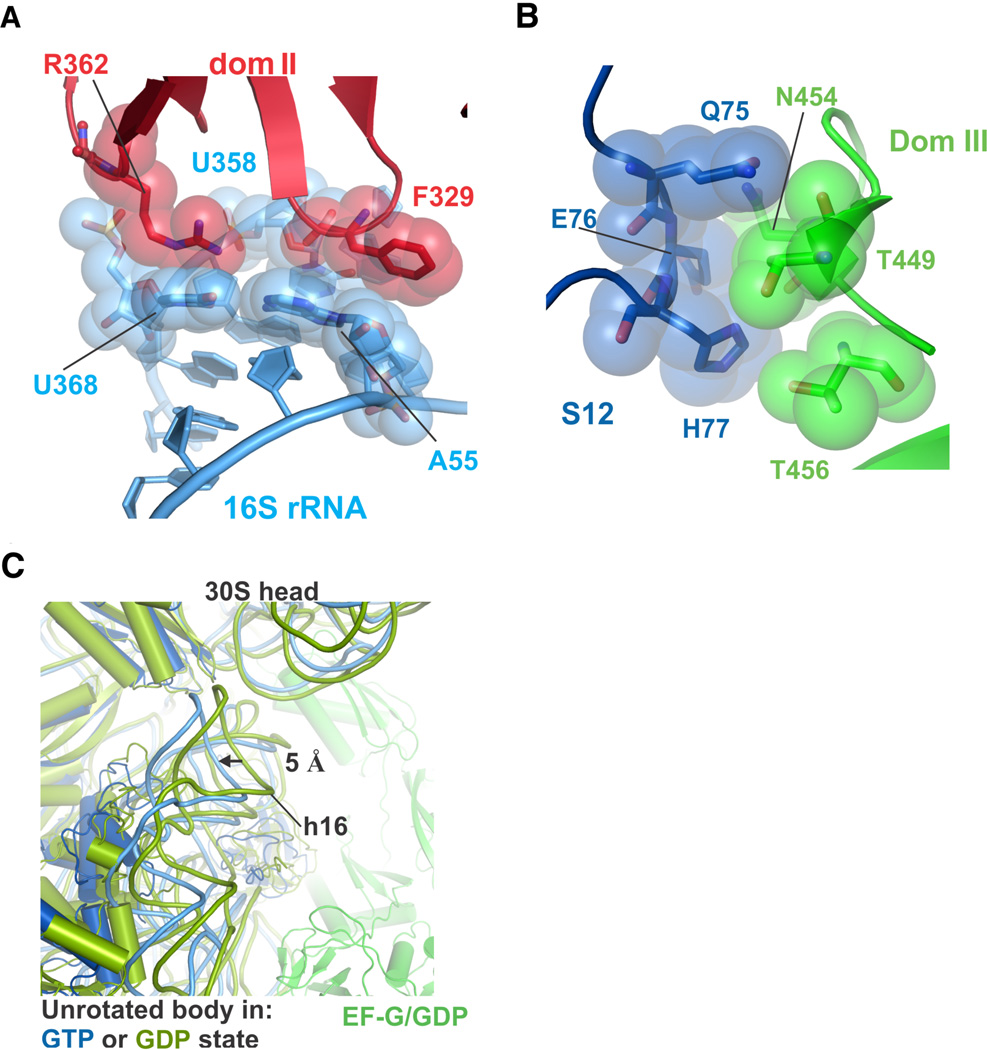
Formation of these extensive interfaces between domains I-III in EF-G in the GMPPCP complexes results in large-scale movement of domains II and III that couple to rotation of the body of the 30S ribosomal subunit. Domain II of EF-G complexed with GMPPCP moves ~7 Å closer to domain III at its extremity, when compared to the ribosome complex with EF-G, GDP and fusidic acid, which corresponds to a post-translocation state with P- and E-site tRNAs and the ribosome in an unrotated conformation (32) (Fig. 3C). EF-G domains II and III move together with the body of the 30S subunit, which is rotated by 3°–8°, and maintain contacts with 16S rRNA helices 4, 5, and 15 and ribosomal proteins S12 (Fig. 4A, 4B) (9, 10, 32).
Figure 4. Contacts between EF-G and the 30S subunit are maintained during ribosomal subunit rotation.
(A) Contacts between EF-G domain II (red) and 16S rRNA (light blue) near helices h5 (A55 or U358) and h15 (U368). (B) Contacts between EF-G domain III (green) and ribosomal protein S12 (blue), for the ribosome in an intermediate state of rotation. (C) Movement of the 30S subunit body domain away from the subunit interface in the unrotated state induced by EF-G binding in the GTP state (blue). For comparison, the structure of the ribosome with EF-G/GDP/fusidic acid in the unrotated state is shown in green and olive (32).
In the structure of the unrotated 70S ribosome in the present crystals, EF-G domain II is largely disordered, and the G-domain is partially disordered (fig. S3A), as is domain IV. Furthermore, the 30S subunit body domain is forced away from the 50S subunit interface by 5 Å when compared to EF-G bound with GDP/fusidic acid to the unrotated post-translocation state (Fig. 4C), consistent with evidence that EF-G/GTP binding favors rotated states of the ribosome (22–25). In the two structures of the ribosome in a fully-rotated state determined here, domain II and IV of EF-G are more ordered when compared to the unrotated state (fig. S3B). EF-G/GMPPCP also adopts a well-defined conformation when bound to intermediate rotated states (Fig. 5A). Thus, the interactions between EF-G and the 30S ribosomal subunit couple the nucleotide status of the GTPase center (GDP vs. GTP or GDP•Pi) to interdomain stabilization of EF-G and the rotational state of the ribosome.
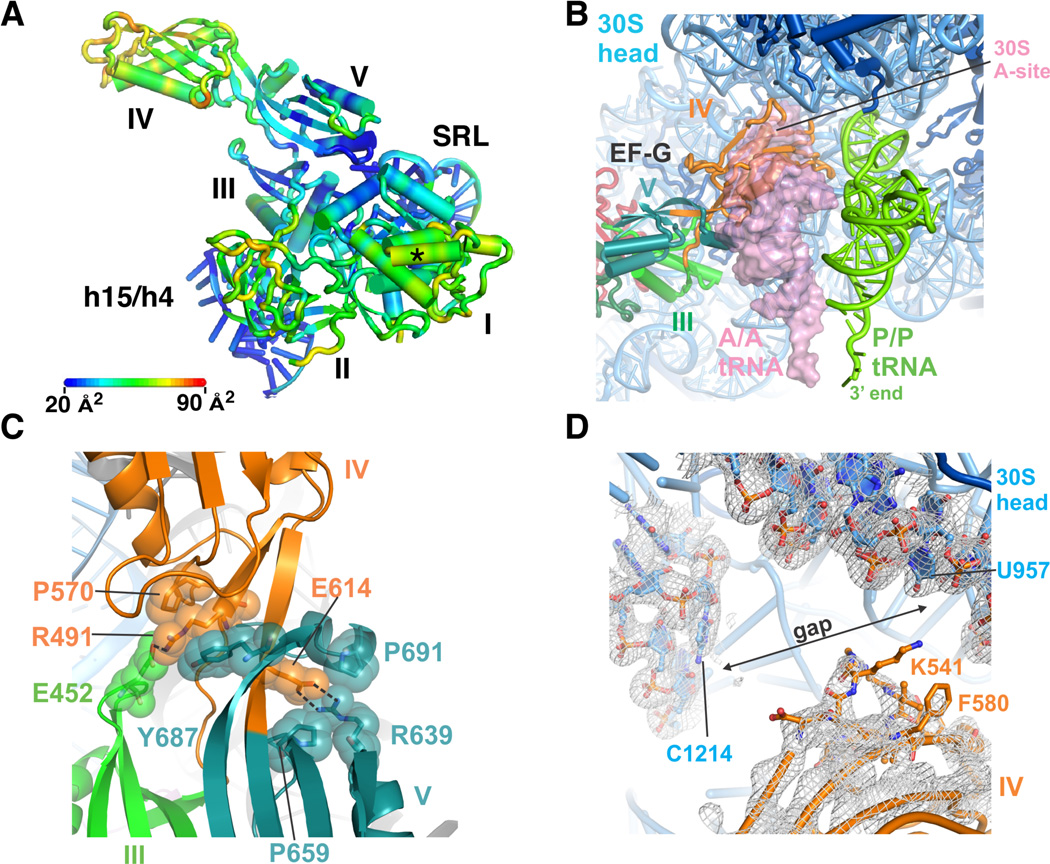
Figure 5. Conformational rigidity of EF-G/GMPPCP in the intermediate rotational state.
(A) Atomic displacement parameters (B-factors) of EF-G/GMPPCP bound to the ribosome in an intermediate state of subunit rotation. Scale bar indicates the range of color-coded B-factors. Domains in EF-G and helices in 16S and 23S rRNA are indicated. The asterisk indicates the site of interaction seen between EF-G and ribosomal proteins L7/L12 in (32). (B) Position of EF-G domain IV (orange) in the ribosomal A site. The position of tRNAs in the A site (A/A tRNA, transparent pink surface) and P site (P/P tRNA, green) are derived from the superposition of the ribosome structure in (55) with the ribosome in an intermediate state of subunit rotation, using the 30S subunit platform as a frame of reference. (C) Salt bridges between EF-G domain III (green) residue E452, domain IV (orange) residues R491 or E614 and domain V (aqua) residue R639, buttressed by hydrophobic packing. (D) Lack of contacts between EF-G domain IV and the head domain of the 30S subunit of a representative ribosome in an intermediate state of rotation. (2Fobs–2Fcalc) difference electron density is shown at a contour of 1 standard deviation from the mean.
Position of EF-G domain IV in the ribosomal A site
In the present structures, domain IV in EF-G, which is essential for tRNA and mRNA translocation (37), projects towards the 30S subunit and occupies the position of A-site tRNA (8–10, 32) (Fig. 5B). The orientation of domain IV is stabilized primarily by salt bridges with domain III and domain V (Fig. 5C), with only a few interactions occuring to the tip of h44 in 16S rRNA in the 30S subunit A site. Binding of the antibiotic viomycin in the vicinity of these weak interactions with 16S rRNA helix h44 (fig. S4) does not seem to stabilize them, since the interactions remain weak whether viomycin is bound to h44 or not (fig. S4) (29). Although the head domain of the 30S subunit adopts large swiveling angles in these structures (fig S1, table S1), there are no clear contacts between EF-G domain IV and the head domain in seven of the eight ribosomes observed here (Fig. 5D). Instead, the position of domain IV seems to function almost exclusively to preclude tRNA occupancy in the ribosomal A site (8–10, 32). In the various structures of EF-G with GMPPCP and GDP/fusidic acid, the specific interactions between EF-G and platform elements of the 30S subunit A site near helix h44 also vary in detail, and are poorly ordered in nearly all of the present structures, again suggesting that EF-G domain IV serves as a steric block to control movements of the ribosome and tRNA substrates in later steps of translocation (32, 37).
Model of mRNA and tRNA translocation
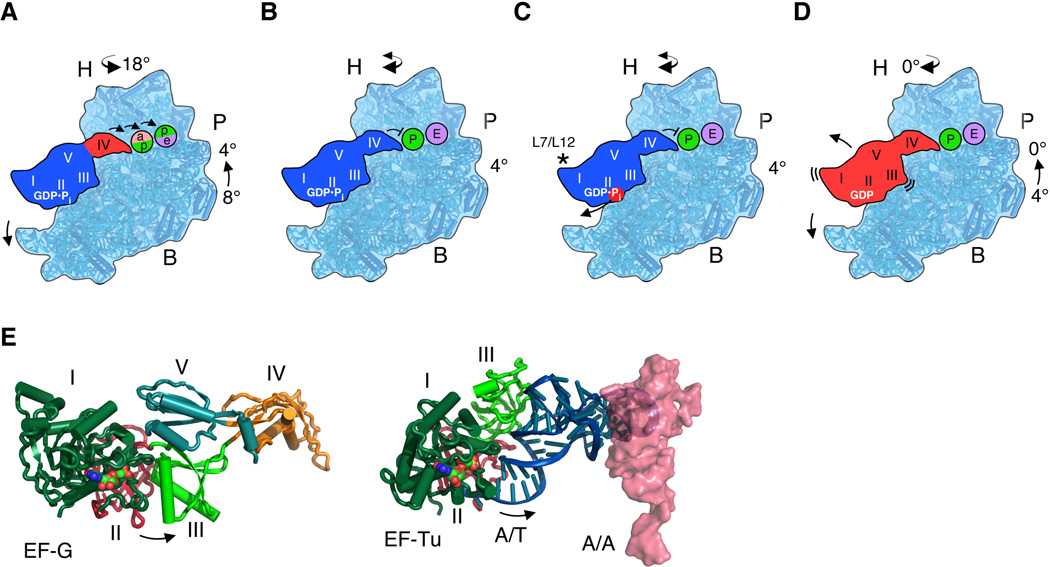
Translocation of mRNA and tRNA on the ribosome can occur in the absence of EF-G, but the rate of EF-G independent translocation is too slow to support cell growth (38, 39), and is highly reversible (40, 41). Thus, while the process of translocation is intrinsic to the ribosome, EF-G increases translocation efficiency and biases it in the forward direction. Kinetic experiments revealed that EF-G catalyzed translocation involves multiple steps, with GTP hydrolysis occurring rapidly, followed by a rate-limiting conformational change in the ribosome that precedes mRNA and tRNA translocation (27, 33, 42). However, the relationships between these kinetically-defined events and structural changes in ribosome translocation complexes remain to be determined (43–46). Several lines of evidence indicate that, while EF-G/GTP may bind the ribosome in the unrotated state (47), EF-G/GTP binds more favorably to the ribosome in an intermediate step of the translocation reaction, after tRNAs occupy hybrid A/P and P/E sites and the ribosomal subunits are in a rotated state (10, 11, 46, 48). The present structures reveal that EF-G/GMPPCP can bind to vacant ribosomes in multiple states of subunit rotation, including the unrotated state. However, the rigid arrangement of EF-G observed here is incompatible with pre-translocation complexes in which tRNA occupies the A site in the 30S subunit (49) (Fig. 5B).
The present structural data suggest that the intermediate state of rotation may be preferred before GTP hydrolysis by EF-G. However, in kinetic experiments GTP hydrolysis is rapid and precedes the rate-limiting conformational change in translocation (27, 33, 42). It is possible that the GTP and GDP•Pi states of EF-G may be in equilibrium (47), analogous to the situation with eukaryotic initiation factor eIF2 (50). Thus, the present structures of the partially and fully rotated states may represent EF-G/GDP•Pi bound to the ribosome as tRNAs move into the ap/P and pe/E sites, when the 30S head domain adopts extremely rotated positions (10, 28) (Fig. 6A). Consistent with the intermediate state of rotation observed previously by cryo-EM (10), it is likely that the activated (GDP•Pi) form of EF-G stabilizes the intermediate state of rotation (Fig. 6A) (27). However, the present structures reveal that EF-G binding to partially rotated ribosomes is independent of the position of the 30S head domain. Since EF-G domain IV only makes significant contacts to the 30S subunit head domain in the post-translocation state (30), and not when the head is swiveled (8–10, 28) (Fig. 5D), domain IV of the EF-G/GDP•Pi complex may simply act to decouple tRNA movement from the 30S subunit platform and allow the intrinsic dynamics of the 30S subunit head domain (51, 52) to translocate tRNAs into the P and E sites (10, 28). EF-G domain IV would then prevent translocated P-site tRNA from reverting its position to the A site, as suggested by previous structures (8–10, 32, 37) (Fig. 6B). Inorganic phosphate (Pi) release, accelerated by L7/L12 (32, 42) and accompanied by switch I unfolding (Fig. 6C), would then cause EF-G to relax due to loss of inter-domain contacts and allow the 30S subunit to reverse its rotation to 0°. The relaxed state of EF-G/GDP would then dissociate from the ribosome as domain III and V contacts with the ribosome are destabilized (Fig. 6D).
Figure 6. Model of EF-G controlled translocation of mRNA and tRNA.
(A) EF-G in the activated GDP•Pi state requires movement of A-site tRNA out of the decoding site (pink) of the ribosomal 30S subunit platform into the P site (green), and P-site tRNA into the E site (purple) to accommodate EF-G domain IV, as indicated by arrows. The intermediate tRNA sites in the 30S subunit and the head rotation angle are based on (10). At this point, domains I-III and V of EF-G (blue) are rigidly bound to the intermediate rotation state of the ribosome, whereas domain IV (red) moves to occupy the A site. (B) Stable interactions between EF-G domain IV and the remainder of EF-G position domain IV to prevent back-translocation of P-site tRNA. The 30S subunit head domain may remain dynamic in this post-translocation state (46, 51, 56). (C) Phosphate release from the GTPase domain of EF-G, stimulated by proteins L7/L12 (asterisk), disrupts inter-domain contacts in EF-G, allowing the ribosome to revert to the unrotated state, (D), from which EF-G/GDP dissociates from the ribosome. (E) Opening of domains II and III in EF-G after GTP hydrolysis follows the same trajectory as tRNA release from EF-Tu during mRNA decoding. The position of A-site tRNA (pink, PDB entry 3I8G) (57) was compared to that for EF-Tu in an mRNA decoding complex with the GTP analog GMPPCP (6).
Conclusion
The model of EF-G cycling between rigid and relaxed conformations is comparable to the changes in tRNA conformation that occur during mRNA decoding by EF-Tu. During mRNA decoding, GTP hydrolysis by EF-Tu releases tRNA from a bent conformation that relaxes as the tRNA is accommodated into the ribosomal A site (5, 6). The trajectory of tRNA motion after release from EF-Tu corresponds to the direction of domain II-III opening in EF-G (Fig. 6E). It will be important to determine whether similar cycles may occur with other translation GTPases. In the case of EF-G, future structural and biophysical insights will also be needed to elucidate the contribution of tRNAs to translocation, and to understand the role of EF-G in its distinct functional role in ribosome recycling (53).
Materials and Methods
Escherichia coli ribosomes lacking the C-terminus of ribosomal protein L9 (amino acids 56 to the C-terminus) were purified and used for complex formation with E. coli EF-G, the nonhydrolyzable GTP analog GMPPCP and viomycin. Purification and crystallization of the complexes are described in the Supporting Online Material. Two ribosome crystal forms, each containing four unique copies of the ribosome, were used for x-ray diffraction measurements and structure determination by molecular replacement. Details of the data measurement, structure determination, refinement, and ribosome superpositions are given in the Supporting Online Material.
Supplementary Material
Acknowledgements
We thank J. Doudna for helpful discussions and comments, J. Holton and G. Meigs for help with x-ray data collection, P. Afonine and J. Headd for advise on crystallographic refinement. This work was supported by NIH grant R01-GM65050 to J.H.D.C., by a National Cancer Institute grant CA92584 for the SIBYLS and 8.3.1 beam lines at the Advanced Light Source, and by the U.S. Department of Energy (DE-AC0376SF00098 for the SIBYLS and 8.3.1 beamlines at the ALS). Coordinates for the ribosomes have been deposited in the Protein Data Bank (16 entries TBD).
Footnotes
Supplementary Materials
Materials and Methods
Table S1
Figs. S1, S2, S3, S4
References (58–71)
References
- 1.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349:117. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 2.Clementi N, Polacek N. Ribosome-associated GTPases: the role of RNA for GTPase activation. RNA Biol. 2010 Sep-Oct;7:521. doi: 10.4161/rna.7.5.12467. [DOI] [PubMed] [Google Scholar]
- 3.Liang H. Sordarin, an antifungal agent with a unique mode of action. Beilstein J Org Chem. 2008;4:31. doi: 10.3762/bjoc.4.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sohmen D, Harms JM, Schlunzen F, Wilson DN. Enhanced SnapShot: Antibiotic inhibition of protein synthesis II. Cell. 2009 Oct 2;139:212. doi: 10.1016/j.cell.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009 Oct 30;326:688. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010 Nov 5;330:835. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000 Jul 20;406:318. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 8.Valle M, Zavialov A, Sengupta J, Rawat U, et al. Locking and unlocking of ribosomal motions. Cell. 2003 Jul 11;114:123. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 9.Connell SR, Takemoto C, Wilson DN, Wang H, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007 Mar 9;25:751. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 10.Ratje AH, Loerke J, Mikolajka A, Brunner M, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010 Dec 2;468:713. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010 Dec;17:1470. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolesnikov A, Gudkov A. Elongation factor G with effector loop from elongation factor Tu is inactive in translocation. FEBS Lett. 2002 Mar 6;514:67. doi: 10.1016/s0014-5793(02)02300-1. [DOI] [PubMed] [Google Scholar]
- 13.Bartish G, Nygard O. Importance of individual amino acids in the Switch I region in eEF2 studied by functional complementation in S. cerevisiae. Biochimie. 2008 May;90:736. doi: 10.1016/j.biochi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Rohrback MS, Bodley JW. Steady state kinetic analysis of the mechanism of guanosine triphosphate hydrolysis catalyzed by Escherichia coli elongation factor G and the ribosome. Biochemistry. 1976 Oct 19;15:4565. doi: 10.1021/bi00666a003. [DOI] [PubMed] [Google Scholar]
- 15.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997 Jan 2;385:37. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 16.Kjeldgaard M, Nyborg J, Clark BF. The GTP binding motif: variations on a theme. FASEB J. 1996 Oct;10:1347. [PubMed] [Google Scholar]
- 17.Petrov A, Kornberg G, O'Leary S, Tsai A, et al. Dynamics of the translational machinery. Curr Opin Struct Biol. 2011 Feb;21:137. doi: 10.1016/j.sbi.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989 Nov 9;342:142. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 19.Dunkle JA, Wang L, Feldman MB, Pulk A, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011 May 20;332:981. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agirrezabala X, Liao HY, Schreiner E, Fu J, et al. Structural characterization of mRNA-tRNA translocation intermediates. Proc Natl Acad Sci U S A. 2012 Apr 17;109:6094. doi: 10.1073/pnas.1201288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budkevich T, Giesebrecht J, Altman RB, Munro JB, et al. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell. 2011 Oct 21;44:214. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007 Jul 13;370:530. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 23.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008 Jun 6;30:578. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008 May 9;30:348. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010 Feb 17;29:770. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor Gdependent tRNA-mRNA translocation. J Mol Biol. 2004 Nov 5;343:1183. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 27.Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci U S A. 2006 Sep 12;103:13670. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramrath DJ, Yamamoto H, Rother K, Wittek D, et al. The complex of tmRNASmpB and EF-G on translocating ribosomes. Nature. 2012 May 24;485:526. doi: 10.1038/nature11006. [DOI] [PubMed] [Google Scholar]
- 29.Pulk A. 2013 See supplementary information. [Google Scholar]
- 30.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001 Nov 9;294:1299. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 31.Ticu C, Nechifor R, Nguyen B, Desrosiers M, Wilson KS. Conformational changes in switch I of EF-G drive its directional cycling on and off the ribosome. EMBO J. 2009 Jul 22;28:2053. doi: 10.1038/emboj.2009.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009 Oct 30;326:694. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savelsbergh A, Katunin VI, Mohr D, Peske F, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003 Jun;11:1517. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 34.Margus T, Remm M, Tenson T. A computational study of elongation factor G (EFG) duplicated genes: diverged nature underlying the innovation on the same structural template. PLoS One. 2011;6:e22789. doi: 10.1371/journal.pone.0022789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohr D, Wintermeyer W, Rodnina MV. Arginines 29 and 59 of elongation factor G are important for GTP hydrolysis or translocation on the ribosome. EMBO J. 2000 Jul 3;19:3458. doi: 10.1093/emboj/19.13.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ticu C, Murataliev M, Nechifor R, Wilson KS. A central interdomain protein joint in elongation factor G regulates antibiotic sensitivity, GTP hydrolysis, and ribosome translocation. J Biol Chem. 2011 Jun 17;286:21697. doi: 10.1074/jbc.M110.214056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J Mol Biol. 2000 Jul 21;300:951. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
- 38.Pestka S. Studies on the formation of transfer ribonucleic acid-ribosome complexes. VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. J Biol Chem. 1969 Mar 25;244:1533. [PubMed] [Google Scholar]
- 39.Gavrilova LP, Spirin AS. Stimulation of non-enzymic translocation in ribosomes by p-chloromercuribenzoate. FEBS Lett. 1971 Oct 1;17:324. doi: 10.1016/0014-5793(71)80177-1. [DOI] [PubMed] [Google Scholar]
- 40.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell. 2006 Dec 28;24:931. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konevega AL, Fischer N, Semenkov YP, Stark H, et al. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat Struct Mol Biol. 2007 Apr;14:318. doi: 10.1038/nsmb1221. [DOI] [PubMed] [Google Scholar]
- 42.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005 Dec 21;24:4316. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C, Stevens B, Kaur J, Cabral D, et al. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol Cell. 2011 May 6;42:367. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fei J, Richard AC, Bronson JE, Gonzalez RL., Jr. Transfer RNA-mediated regulation of ribosome dynamics during protein synthesis. Nat Struct Mol Biol. 2011 Sep;18:1043. doi: 10.1038/nsmb.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011 Apr;18:457. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Altman RB, Blanchard SC. Insights into the molecular determinants of EF-G catalyzed translocation. RNA. 2011 Dec;17:2189. doi: 10.1261/rna.029033.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNAbinding states in ribosomal translocation. Proc Natl Acad Sci U S A. 2008 Jul 8;105:9192. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agirrezabala X, Frank J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q Rev Biophys. 2009 Aug;42:159. doi: 10.1017/S0033583509990060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000 Feb 4;100:301. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 50.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell. 2005 Oct 28;20:251. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 51.Gabashvili IS, Agrawal RK, Grassucci R, Frank J. Structure and structural variations of the Escherichia coli 30 S ribosomal subunit as revealed by threedimensional cryo-electron microscopy. J Mol Biol. 1999 Mar 12;286:1285. doi: 10.1006/jmbi.1999.2538. [DOI] [PubMed] [Google Scholar]
- 52.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010 Jul 15;466:329. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 53.Peske F, Rodnina MV, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell. 2005 May 13;18:403. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 54.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012 May 25;336:1030. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009 May;16:528. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009 Aug 21;325:1014. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010 May;17:555. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.