Summary
The multi-copy 2 micron plasmid of Saccharomyces cerevisiae, a resident of the nucleus, is remarkable for its high chromosome-like stability. The plasmid does not appear to contribute to the fitness of the host, nor does it impose a significant metabolic burden on the host at its steady state copy number. The plasmid may be viewed as a highly optimized selfish DNA element whose genome design is devoted entirely towards efficient replication, equal segregation and copy number maintenance. A partitioning system comprised of two plasmid coded proteins, Rep1 and Rep2, and a partitioning locus STB is responsible for equal or nearly equal segregation of plasmid molecules to mother and daughter cells. Current evidence supports a model in which the Rep-STB system promotes the physical association of the plasmid with chromosomes and thus plasmid segregation by a hitchhiking mechanism. The Flp site-specific recombination system housed by the plasmid plays a critical role in maintaining steady state plasmid copy number. A decrease in plasmid population due to rare missegregation events is rectified by plasmid amplification via a recombination induced rolling circle replication mechanism. Appropriate plasmid amplification, without runaway increase in copy number, is ensured by positive and negative regulation of FLP gene expression by plasmid coded proteins and by the control of Flp level/activity through host mediated post-translational modification(s) of Flp. The Flp system has been successfully utilized to understand mechanisms of site-specific recombination, to bring about directed genetic alterations for addressing fundamental problems in biology, and as a tool in biotechnological applications.
Introduction
Selfish genetic elements (1-4), widespread in nature, are characterized by their ability to replicate efficiently and maintain themselves stably in host cell populations. A subset of these elements harbors the capacity to spread within a genome or, via horizontal transmission, between genomes. Selfish elements can also be frequently acquired by sexual transmission. The degree of selfishness can vary significantly among different elements. Some may increase the host's fitness at least under certain conditions and, in doing so, add to their own fitness in a self-serving fashion. Others may be more decidedly selfish in that they contribute little towards the host's fitness. Their long-term persistence is sustained solely by their capacity for replication and transmission during growth and division of host cells.
Selfish elements may be broadly divided into two groups: those that are integrated into the chromosome(s) of the host and those that remain extra-chromosomal (reviewed in Ref 4). The integrated class includes insertion sequences, lysogenic states of several bacteriophage, mobile DNA elements and families of repeated DNA found in eukaryotes. The extra-chromosomal class encompasses plasmids, lysogenic forms of certain bacterial viruses, RNA intermediates involved in the retro-transposition of mobile elements and epiosmes of mammalian viruses belonging to the gamma herpes and papilloma families. Plasmids, found abundantly among prokaryotes, are almost non-existent among eukaryotes, except for those encountered among members of the budding yeast (Saccharomycetaceae) lineage. The gamma herpes and papilloma viruses, whose extended latent periods of infection are characterized by their stable episomal existence, may be regarded as plasmid impostors of the eukaryotic world (5, 6).
We review here the properties of the 2 micron plasmid found nearly ubiquitously in Saccharomyces strains that justify its inclusion under the selfish DNA moniker. Furthermore, we describe the biochemical features and applications of a site-specific recombination system harbored by the plasmid. Based on genetic organization and functional attributes, it is logical to posit that the 2 micron plasmid is an authentic representative of the yeast plasmid family with respect to replication, segregation to daughter cells during cell division and maintenance and regulation of copy number (7).
The 2 micron plasmid: an optimized and miniaturized selfish DNA element
The 2 micron plasmid is a relatively small double stranded circular DNA genome (~6.3 kbp) present in the yeast nucleus at an average copy number of 40-60 per haploid cell (7-9). The stability of the plasmid is remarkably similar to that of the host chromosomes, the loss rate being as low as 10−5 to 10−4 per cell division. Four protein coding regions together with the cis-acting sequences important for replication, partitioning and copy number control leave little room to spare in the compactly designed plasmid genome (Figure 1A). Even minor disruptions of the genetic organization of the plasmid lead to prominent deleterious effects on its physiology. 2 micron based artificial plasmids routinely used for genetic manipulations in yeast are two to three orders of magnitude less stable than the native plasmid. The 2 micron plasmid may be looked upon as a minimalist selfish DNA tailored for maximum functional efficiency.
Figure 1.
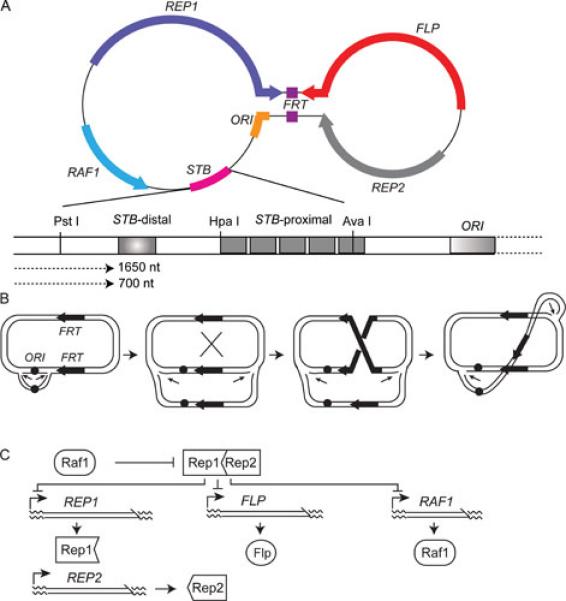
Genetic organization of the yeast plasmid and its copy number regulation. The double stranded DNA genome of the yeast 2 micron plasmid is generally represented as a dumbbell shaped molecule to highlight the 599 bp inverted repeat (the handle of the dumbbell) that separates two unique regions. The four coding regions harbored by the plasmid are REP1, REP2, FLP and RAF1. The directions in which these loci are transcribed are indicated by the arrowheads. The plasmid replication origin is indicated as ORI. Flp is a site-specific recombinase, whose target sites (FRTs) are embedded within the inverted repeat region. The plasmid partitioning locus STB can be divided into origin-proximal (STB-proximal) and origin-distal (STB-distal) segments. There are five repetitions of a 60 bp consensus sequence in STB-proximal. STB-distal, which harbors the termination signal for two origin directed plasmid transcripts (1660 nt and 700 nt long) as well as silencing element (shaded box), maintains STB-proximal as a transcription-free zone. The Rep1-Rep2-STB system ensures equal plasmid segregation. The (Flp-FRT)-Raf1 system is responsible for the maintenance of steady state plasmid copy number. B. The mechanism proposed for copy number correction of the plasmid by amplification (12) invokes a Flp mediated recombination event that changes the direction of one of the replication forks (indicated by the thin short arrows) with respect to the other during bidirectional replication of the plasmid. The ensuing dual uni-directional mode of replication amplifies the plasmid as a concatemer of tandem plasmid units. There is a marked asymmetry in the location of the FRT sites (thick arrows) with respect to the replication origin (ORI). The consequent difference in their replication status, one duplicated and the other not, is responsible for the relative inversion of the replication forks as a result of recombination between them C. Efficient amplification without the danger of unregulated increase in plasmid copy number is prevented by a transcriptional regulatory network. The putative [Rep1-Rep2] repressor negatively regulates FLP, RAF1 and REP1 expression. Raf1 is thought to antagonize the action of the [Rep1-Rep2] repressor.
The plasmid replication origin (ORI) is functionally equivalent to a chromosomal origin, so that each plasmid molecule replicates once (and only once) during S phase by utilizing the host replication machinery (10). The duplicated plasmid population is distributed equally (or almost equally) to mother and daughter cells by the plasmid partitioning system comprised of the Rep1 and Rep2 proteins and the STB locus (7, 11). Doubling of copy number followed by equal (or almost equal) segregation marks the normal steady state life style of the plasmid.
The copy number control system comes into play only when there is a reduction in plasmid population due to a rare missegregation event. The restoration of copy number is mediated by a site-specific recombination system consisting of the Flp protein and its target sites (FRTs) embedded within a 599 bp inverted repeat region. The Raf1 protein positively regulates amplification. According to the currently accepted model (12, 13), plasmid amplification is triggered by a recombination event early during bidirectional replication when the ORI-proximal FRT site, but not the distal one, has been duplicated (Figure 1B). The DNA inversion resulting from a crossover between the unreplicated FRT and a copy of the duplicated one will cause the replication forks to travel in the same direction around the circular template. This non-standard mode of replication by the two uni-directional forks spins out multiple tandem copies of the plasmid without the need for ORI to fire more than once. Amplification can be terminated by a second recombination event that restores bidirectional fork movement. The amplified plasmid DNA can be resolved into single plasmid molecules by recombination mediated by Flp or by the host homologous recombination machinery.
The mechanism for dealing with a higher-than-steady state copy number caused by missegregation, if such a mechanism exists, is not clear. It is possible that copy number may be lowered by under-replication, although there is no experimental evidence to support this notion. Very high plasmid copy numbers have a strong negative effect on cellular function, leading to premature lethality of such cells (14-16). Thus, cells bearing an undue plasmid burden would be eliminated from a growing population over time.
In sum, the 2 micron plasmid genome is comprised of three functional modules devoted to the sole purpose of self-perpetuation. They ensure (a) precise duplication of the plasmid population during a cell cycle, (b) equal plasmid segregation at the time of nuclear division and (c) preservation of plasmid copy number in individual cells.
Copy number control: plasmid and host mediated regulatory mechanisms
There is an intricate communication between the partitioning and amplification systems by which an untoward increase in plasmid copy number is avoided (17-19) (Figure 1C). The Rep1 and Rep2 proteins are thought to form a bipartite negative regulator that represses the expression of the FLP, RAF1 and REP1 genes. REP2 appears to be constitutively expressed. The level of Rep1 not only provides a measure of the plasmid copy number but also determines the effective concentration of the Rep1-Rep2 repressor. When the plasmid copy number falls below the steady state value, the corresponding drop in Rep1 reduces the repressor concentration below the threshold required for turning off FLP and RAF1 expression. As a result, the amplification system is activated. The Raf1 protein is thought to antagonize the repressor, thereby augmenting plasmid amplification. When steady state copy number is restored, the increase in Rep1 raises Rep1-Rep2 concentration above the threshold, thus turning off the amplification system. The design of this genetic regulatory circuit quickly triggers the amplification response when called for without incurring the danger of a runaway increase in the plasmid population.
In addition to the plasmid instituted control of copy number at the level of gene expression, there is also host mediated post-translational regulation of Flp (20, 21). Flp is modified by sumoylation, which may regulate its activity and/or its stability. Sumoylation could provide the signal for the secondary modification of Flp by ubiquitination and its subsequent degradation by the proteasome pathway. Under conditions that disrupt sumoylation homeostasis, the 2 micron plasmid copy number rises to very high levels causing cell lethality. A plausible cause for this aberrant plasmid amplification is the conversion of a Flp induced stand nick at the FRT site into a double strand DNA break as a result of replication (Figure 2). The invasion by the broken end of a circular plasmid template, promoted by the host's homologous recombination machinery, and repeated rounds of replication would result in an amplified plasmid concatemer. The mechanism is analogous to that proposed for the generation of copy number variations in eukaryotic genomes and the maintenance of telomere length by a telomerase independent alternative pathway (22-25).
Figure 2.
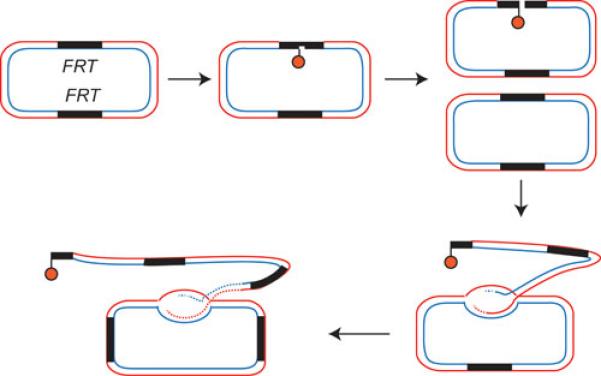
Aberrant plasmid amplification as a result of misregualtion of Flp. Strand nicks formed at FRT by the action of Flp, if unrepaired, will give rise to double strand breaks when they encounter replication forks. Such a broken end can invade an intact circular plasmid and trigger aberrant plasmid amplification by repair synthesis. Post-translational modification of Flp by sumoylation is important in preventing this mode of plasmid amplification (20, 21).
The transcriptional and post-translational controls of plasmid amplification exemplify strategies by which a selfish genome and its host genome establish long-tern coexistence with minimal conflicts between them.
Chromosome coupled segregation of the 2 micron plasmid
The organization of the 2 micron plasmid partitioning system is deceptively similar to that of well characterized bacterial plasmid partitioning systems in that it is comprised of two plasmid coded proteins (Rep1 and Rep2) and a cis-acting locus (STB) that contains iterations of a consensus sequence element (7). However, there is no evidence to suggest functional similarities between the yeast and bacterial systems. For example, neither Rep1 nor Rep2 harbors sequence motifs that would typify an ATP binding/hydrolyzing activity. The fact that the 2 micron plasmid houses a tyrosine family site-specific recombinase (Flp) would be consistent with a prokaryotic origin of the plasmid, as tyrosine recombinases have not been identified in eukaryotes except in members of the budding yeast lineage. In every such case, the recombinase is coded for by a 2 micron related plasmid. Assuming that a prokaryotic DNA element acquired by an ancestral budding yeast via horizontal transmission served as the progenitor of the extant 2 micron plasmid, the Rep-STB system must represent the evolutionary transformation of the original partitioning system during adaptation to a new biological niche.
Plasmids capable of autonomous replication in yeast but lack an active partitioning system, called ARS plasmids, are rapidly lost in the absence of continuous selection. This high instability is caused by the strong tendency of plasmid molecules to stay in the mother nucleus, causing their depletion from daughter cells (26-29). A variety of observations suggest that the Rep-STB system overcomes the mother bias by coupling plasmid segregation to chromosome segregation. For example, a number of mutations that conditionally disrupt normal chromosome segregation, ipl1-1(Ts) being one, also cause the 2 micron plasmid to missegregate (Figure 3A) (30, 31). Furthermore, the plasmid tends to co-segregate with the missegregating bulk of chromosomes. Under the same conditions, an ARS plasmid does not show any coupling to chromosomes (Figure 3B). Instead, it missegregates frequently with the characteristic strong mother bias. Attempts to uncouple plasmid segregation from chromosome segregation, except by employing conditions that directly or indirectly disable the partitioning system, have been unsuccessful. The most parsimonious hypothesis consistent with currently available data is that the 2 micron plasmid physically attaches to chromosomes, and segregates by a hitchhiking mechanism (7, 30, 32). However, a chromosome independent mode of plasmid segregation cannot be ruled out unequivocally. A direct spindle mediated segregation of the plasmid is highly unlikely. Components of the kinetochore complex, which is responsible for linking centromeres to the mitotic spindle, have not been detected at STB. The presence of two or more STB loci within a single plasmid does not lead to instabilities normally observed when two centromere (CEN) sequences are harbored by a plasmid. Delaying spindle assembly until metaphase during a cell cycle does not affect plasmid or chromosome replication. However, upon spindle restoration, chromosomes form normal spindle attachments and segregate equally whereas the plasmid missegregates (30). This result is more readily explained by the requirement of the pre-metaphase spindle for plasmid-chromosome coupling rather than by spindle mediated plasmid segregation.
Figure 3.
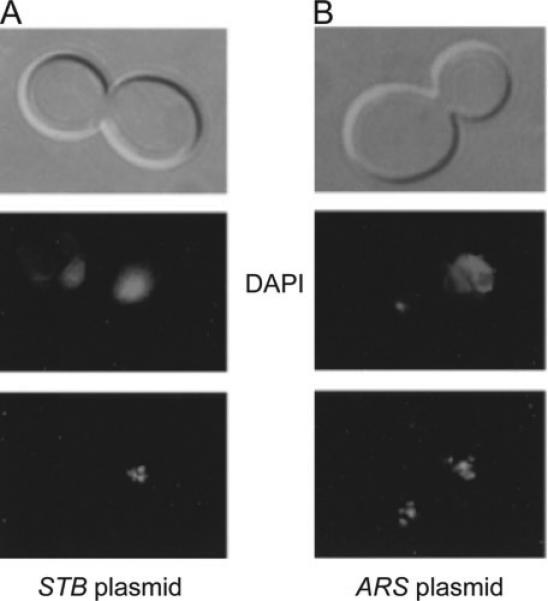
Segregation of the 2 micron plasmid in the Ipl1-1 mutant. When the Ipl1 (aurora kinase) function is inactivated by a temperature sensitive mutation, chromosomes frequently missegregate (shown here by the unequal DAPI staining in the mother and daughter compartments). A. A multi-copy 2 micron reporter plasmid, fluorescence tagged by (GFP-LacI)-LacO interaction, also shows high missegregation under this condition (31). Furthermore, the plasmid tends to be retained most often in the cell compartment that contains the bulk of the chromosomes. B. The segregation of a plasmid lacking STB (an ARS plasmid) is not coupled to that of the chromosomes. However, such a plasmid missegregates frequently regardless of the Ipl1 status.
The STB locus: a site for high-order assembly of protein factors
The Rep1 and Rep2 proteins interact with each other in vivo and in vitro, and they associate with the STB locus in vivo (33-36). The tripartite DNA protein interactions are crucial for plasmid partitioning function. Point mutations in Rep1 that disrupt its interaction with either Rep2 or STB lead to high plasmid instability (37). However direct binding of either Rep1 or Rep2 to STB in vitro has been difficult to demonstrate by standard methods such as gel mobility shift assays. Evidence has been presented for the binding of Rep2 to STB in vitro (38). Perhaps, the interactions of these proteins with STB may be nucleated or stabilized by a host factor or factors (34). Alternatively, the chromatin organization of STB may be important for the recruitment of the Rep proteins.
The STB locus can be divided arbitrarily into two regions of approximately equal size with respect to their distances from the plasmid replication origin (39) (Figure 1A). The STB-proximal, consisting of five copies of a 60 bp consensus element (the iteron unit), appears to engender the partitioning function. The STB-distal likely plays a secondary role in partitioning by helping to maintain STB-proximal in its active state. A transcription terminator sequence present within STB-distal serves to keep STB-proximal a transcription-free zone. STB-distal also harbors a silencer element that can turn down the activity of a promoter when placed adjacent to it. The potential contributions of the transcription terminator and the silencer towards plasmid partitioning have not been rigorously analyzed.
A number of host factors that associate with centromeres and contribute to normal chromosome segregation are also recruited at STB and promote equal plasmid segregation. These include the RSC2 chromatin remodeling complex, the Kip1 nuclear motor protein, the yeast cohesin complex and the centromere-specific histone H3 variant Cse4 (40-44). At least a subset of these factors may affect plasmid stability indirectly through their effects on chromosome segregation. The lack of Rsc2 causes a significant increase in plasmid loss without obvious deleterious effects on chromosome segregation (44). Presumably the RSC1 chromatin remodeling complex, in which Rsc2 is replaced by its functional homologue Rsc1, can satisfy the requirements for chromosome segregation. The association of the cohesin complex as well as Cse4 at STB is highly sub-stoichiometric (42, 45), raising concerns about their functional relevance to plasmid partitioning. However, the assembly of multiple plasmid molecules into one unit may help these factors to act in a catalytic manner by recycling among members of the unit. Such group behavior may be advantageous to a selfish genetic element by limiting its dependence on factors that are critical for the host's wellbeing but are in short supply.
The substoichiometric nature of Cse4 at STB also calls into question whether it is an authentic constituent of a specialized nucleosome assembled at STB. Cse4-STB association has been demonstrated by chromatin immunoprecipitation (ChIP) assays utilizing DNA-protein crosslinking or, alternatively, by plasmid pull-down assays in the absence of crosslinking (41, 42). Both types of analyses also reveal Cse4-centromere association, with the distinction that the efficiency of centromere (CEN) recovery in the plasmid pull-down assays is several fold higher relative to STB. A Cse4 directed antibody can specifically bring down CEN DNA corresponding to a single nucleosome from micrococcal nuclease digested chromatin (46). In this assay, the immunoprecipitated CEN DNA is detected by Southern analysis, but STB DNA is not (Sue Biggins, personal communication). In analogous assays, we are able to visualize both CEN and STB in the immunoprecipitated DNA using PCR but detect neither sequence by Southern analysis (42) (unpublished data). Whether Cse4-STB association is in the form of a nucleosome or whether this association is non-nucleosomal remains open. Nevertheless, the differences between CEN and STB in the requirements for Cse4 recruitment (41, 42), as well as the distinct life-times of Cse4-CEN and Cse4-STB associations during the cell cycle (41), suggest that the interaction between Cse4 and STB is authentic.
The localization of Cse4 to non-centromeric regions of yeast chromosomes has been established by ChIP followed by hybridization to DNA microarrays and also by ChIP in conjunction with DNA sequencing (ChIp-Seq) (47, 48). In one of the assays (47), the regions enriched in Cse4 association include repeated loci such as Ty transposable elements, telomeres and ribosomal DNA. In the other assay (48), the enriched loci, termed centromere-like regions (CLRs), map near centromeres, and at least a subset of these sequences enhance the segregation potential of reporter plasmids and chromosomes. It is possible that CLRs might denote ancient centromere sequences that became diverged or lost during the steps leading to the evolution of the present day budding yeast centromeres. As described in the succeeding section, the STB locus might have played a significant role in this evolutionary process.
The budding yeast centromere (CEN) and STB: a shared evolutionary ancestry?
The extremely short, and genetically defined, point centromere of the budding yeast stands out in sharp contrast to the much larger epigenetically specified regional centromeres common to nearly all eukaryotes (49). Interestingly, there is a strong correlation between the time of emergence of the point centromere and the partial or nearly complete loss of siRNA and heterochromatin machineries, which are central to the establishment and maintenance of regional centromeres, in the budding yeast lineage (50). Furthermore, this is the only lineage among eukaryotes whose members (though not all) harbor plasmids related to the 2 micron circle. Based on these circumstantial pieces of evidence, it has been speculated that an ancestral STB locus was domesticated by chromosomes as the point centromere to meet the evolutionary exigency that negated the assembly and function of regional centromeres (49). With the acquisition of a new type of centromere, the defunct centromeres would have diverged or become lost.
If the above model for the evolutionary transition to the point centromere is correct, it follows that the yeast chromosomes and the 2 micron plasmid utilized the same segregation mechanism at one point in their evolutionary history. The cost of bearing the plasmid burden, however small, would have provided the stimulus for the chromosome segregation machinery to evolve away from the plasmid segregation mechanism. The plasmid, in turn, might have evolved counter strategies to exploit indirectly the logic of chromosome segregation to bolster its own propagation. The present day spindle based chromosome segregation and chromosome coupled plasmid segregation might thus represent two divergent solutions to the problem of achieving equal segregation arrived at from a common start point.
The association of chromosome segregation factors at STB would be consistent with a shared evolutionary history between STB and CEN. While a subset of these associations may be relevant to the current plasmid segregation pathway, others may be evolutionary vestiges with little physiological significance. A recent analysis revealed that the functional states of STB and S. cerevisiae CEN engender an unusual positive supercoil (51, 52). It has been argued that DNA is wrapped in a non-standard right handed fashion around the specialized Cse4-containing nucleosome core present at CEN (51). The net positive supercoiling contributed by STB also requires conditions that foster the association of Cse4 with STB. The sub-stoichiometric nature of Cse4-STB interaction rules out the presence of a Cse4 containing nucleosome at every STB. Perhaps the short lived assembly of a Cse4 containing nucleosome at STB or a transient non-nucleosomal interaction between Cse4 and STB may be sufficient to induce a positive supercoil. This DNA topology may then be stably entrapped by other proteins present at STB. We cannot strictly exclude the possibility that the positive supercoiling conferred by STB results from the loss of one or two standard nucleosomes. Nevertheless, the potential presence of a positive supercoil at CEN and STB chromatin adds another tantalizing piece of circumstantial evidence to a growing list suggesting a common ancestry for the partitioning loci of the 2 micron plasmid and the chromosomes of its host.
Organization of the 2 micron plasmid and the functional plasmid unit during segregation
Fluorescence tagged STB reporter plasmids tend to form 3 to 5 foci in individual haploid nuclei. Given the mean copy number of 40-60 of the native plasmid, it has been assumed that each fluorescent focus comprises several coalesced plasmid molecules The plasmid foci are often seen in close proximity to each other and appear to segregate in time lapse assays, as a close-knit cluster (31). It is now clear that plasmid segregation as a single clustered unit is a misimpression conveyed by the small size of the haploid nucleus and the resolution limits of the microscopy assays (32) (unpublished data). Rather, each plasmid focus appears to function as an independent entity in segregation. This notion is further supported by the analysis of diploid cells with larger nuclei (and containing approximately double the plasmid copy number than haploid cells) going through meiotic cell divisions (unpublished data). In chromosome spreads prepared from cells at the pachytene stage of meiosis I, the 8-10 plasmid foci on average observed per cell are well resolved from each other.
Single copy fluorescence tagged reporter plasmids
One of the problems with assayng plasmid segregation precisely is the multi-copy nature of STB plasmids and the attendant uncertainties in quantitating plasmids in mother and daughter nuclei by standard microscopy tools. This impediment has been solved by using fluorescence tagged single copy reporter plasmids (32, 53). The general designs of such plasmids, placed in strains expressing GFP-LacI, are illustrated in Figure 4. In one strategy, the plasmid copy number is reduced to one or close to one by incorporating a CEN into it (Figure 4A). The CEN can be conditionally inactivated by driving copious transcription through it from a galactose inducible promoter. In an alternative strategy, the reporter plasmid is excised from its integrated state in a haploid chromosome (Figure 4B). In a single cell cycle, the segregation of plasmid sisters formed during S phase can be followed as 1:1 (equal), 2:0 (unequal with a mother bias) and 0:2 (unequal with a daughter bias) at the anaphase stage (Figure 4C). Under this experimental regimen, equal plasmid segregation under CEN control is 85-90% whereas that under STB control is ~70%. By contrast, the corresponding value for an ARS plasmid is ~25% with the missegregation biased strongly (90%) towards the mother (2:0).
Figure 4.

Single copy reporter plasmids for segregation assays. Two types of single copy reporter plasmids have been developed. A. The copy number of a plasmid is maintained as one or nearly one by incorporating a CEN into it. The CEN can be conditionally inactivated by driving transcription through it from the inducible GAL promoter. B. The reporter plasmid bordered by the target sites (shown by the arrowheads) for the R site-specific recombinase is integrated into a chromosome to keep the copy number strictly as one. The plasmid is excised by the inducible expression of the recombinase. C. Following inactivation of the plasmid-borne CEN (A) or recombination mediated plasmid excision (B) in G1 arrested cells, they are released into the cell cycle and plasmid segregation assayed at the anaphase stage. In either experimental scheme, the plasmid is visualized by operator-fluorescent repressor interaction.
Analyses using two single copy reporter plasmids, one tagged by green fluorescence [(GFP-LacI)-LacO interaction] and the other by red fluorescence [(TetR-RFP)-TetO interaction] in a strain expressing the two hybrid repressors, revealed that STB plasmids segregate in a sister-from-sister fashion (red from red and green from green) at a frequency significantly higher than that predicted for random association and segregation of the replicated plasmid molecules (Figure 5) (53). This finding necessitates the following refinement of the hitchhiking model. If the model is valid, STB plasmid sisters must hitchhike on sister chromatids in order to segregate in a one-to-one fashion.
Figure 5.

Segregation of two single copy STB reporter plasmids. Two single copy STB reporter plasmids cohabiting a nucleus are distinguished by tagging one with green fluorescence [(GFPLacI)-LacO] and the other with red fluorescence [(TetR-RFP)-TetO). Under a functional Rep-STB system, the red and green plasmid sisters segregate most of the time in a one-to-one fashion to yield individual cells containing one red plasmid and one green plasmid (53),
Tests of the refined hitchhiking model: do sister plasmids segregate by associating with sister chromatids?
One way to test the hitchhiking model is to force the co-segregation of sister chromatids to the same cell pole during mitosis, and scrutinize the behavior of plasmid sisters under this manipulation. The co-segregation of sister chromatids, which is antithetical to normal mitosis, and the separation of homologues is the norm during the first division of meiosis. This reductional mode of chromosome segregation is accomplished by the monopolin complex, which clamps down sister kinetochores with assistance from Sgo1 (shugoshin) and Ipl1 (aurora kinase) (54-59). By inappropriately expressing the monopolin complex early during a mitotic cell cycle, it is possible to direct sister chromatids to stay in the mother or migrate to the daughter (57) (Figure 6A). The monopolin directed co-segregation of sister chromatids occurs at roughly 30% frequency without mother-daughter bias (Figure 6B, right). STB plasmid sisters mimic sister chromatids in co-segregating under the influence of monopolin in a bias-free fashion (32) (Figure 6B, left and 6C). By contrast, ARS plasmids behave very differently from sister chromatids or STB plasmid sisters, and retain their strong mother bias. When monopolin expression is combined with the inactivation of Ipl1, a small but distinct daughter bias is imparted to sister chromatid co-segregation. A similar bias is also observed for STB plasmid sisters. Conditions that uncouple STB plasmid segregation from chromosome segregation, postponing spindle assembly until metaphase (60), for example, abolish the correlation between sister chromatids and STB plasmid sisters in their segregation patterns during monopolin driven mitosis (Figure 6D).
Figure 6.
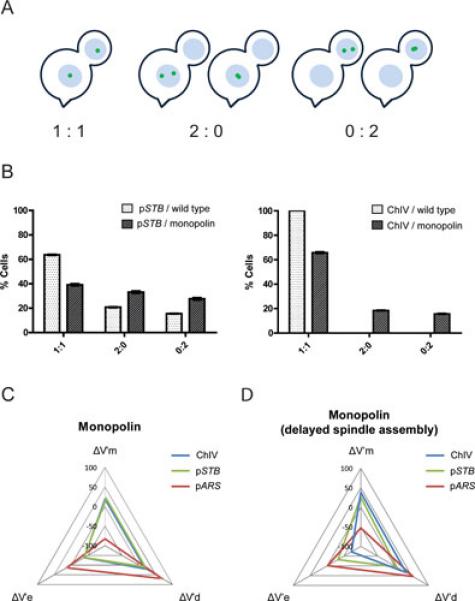
Segregation of a single copy reporter plasmid during monopolin directed mitosis. A, B. The segregation of a fluorescence tagged chromosome or a single copy reporter plasmid (pSTB) is scored as 1:1 (equal segregation) or 2:0 (mother biased co-segregation) or 0:2 (daughter biased co-segregation) during a normal or a monopolin directed mitotic cell cycle. A subset of the missegregated plasmid sisters is often seen as coalesced or overlapping foci. The normal 1:1 segregation of sister chromatids is perturbed by monopolin towards 2:0 or 0:2 segregation. A similar effect is seen for pSTB as well. C, D. In these radar plots, the degree of correlation between a reporter plasmid and a chromosome in their segregation patterns under the influence of monopolin is represented in terms of three variables: deviation from equal segregation (V’e), tendency towards mother segregation (V’m) and tendency towards daughter segregation (V’d). C. The strong correlation between an STB plasmid and a chromosome in their bias-free co-segregation under the influence of monopolin is conveyed by the near congruence of the blue and green triangles. The absence of such correlation between an ARS plasmid (lacking STB) and a chromosome is evinced by the non-overlapping disposition of the red triangle. D. Under conditions that uncouple the segregation of the 2 micron plasmid from that of the chromosomes, the tight correlation between an STB reporter plasmid and a reporter chromosome breaks down during monopolin directed mitosis. In the example shown here, G1 to G2/M phase of the cell cycle is contrived to proceed in the absence of a functional spindle (by treatment with nocodazole). The metaphase arrested cells are washed free of nocodazole to permit spindle assembly and continuation of the cell cycle. Further details can be found in Ref 32.
In more recent experiments, blocking the disassembly of the cohesin complex, which holds pairs of sister chromatids together until the onset of anaphase (61-64), has been utilized as the means for driving all chromosomes to the same cell compartment (unpublished data). Under this regimen, chromosome retention occurs in either the mother or the daughter. Even though spindle organization and separation of the duplicated spindle pole bodies are not normal under this condition, entry of the nucleus into the bud and nuclear division are not blocked. The nuclei lacking chromosomes are often malformed, and have a collapsed appearance. Despite the perturbation of the mitotic cell cycle and nuclear segregation, this analysis clearly demonstrates the near perfect correlation between chromosomes and STB plasmid sisters in their co-segregation. No such correlation is observed for ARS plasmid sisters.
The segregation results obtained by enforcing limited or total missegregation of chromosomes strongly favor the hitchhiking model in which sister copies of the 2 micron plasmid adhere to sister chromatids in a one-to-one fashion. Whether this mechanism operates in the native context of a segregation unit comprising several plasmid molecules is not clear. One possibility is that a high-order organization within a plasmid group distributes each pair of plasmid sisters formed by replication into each of two sister plasmid groups. In other words, the plasmid group is duplicated in a precisely templated fashion. Although the segregation pattern of the single copy green and red fluorescence tagged STB plasmids, referred to earlier, is consistent with this possibility, a more direct test of the model is beyond our present technical capability.
The plasmid amplification system: in vivo analyses
The Flp site-specific recombination system, responsible for the maintenance of the 2 micron plasmid copy number, has been studied only to a limited extent with respect to its in vivo physiological function. The absolute requirement of the recombination system for plasmid amplification has been demonstrated unequivocally (13). However, direct proof for the generally accepted amplification mechanism proposed by Futcher (12) is lacking. The replication intermediates predicted by the Futcher model have not been experimentally observed. An alternative pathway for amplification is the resolution of a plasmid dimer by Flp recombination while in the act of bidirectional replication (Figure 7). Pince-nez structures, two unit length 2 micron circles interconnected by DNA chains of variable length, observed by electron microscopy under DNA elongation arrest by the cdc8 mutation would be consistent with an amplifying plasmid dimer (65). Finally, the proposed over-amplification of the 2 micron plasmid under misregualtion of Flp, initiated by a linear form of the plasmid (21) (Figure 2), may represent a normal copy number control mechanism gone awry by perturbing post-translational modification(s) of Flp. The available genetic and biochemical evidence, though rather limited, highlight the critical role of self-imposed and host-imposed safeguards against unrestricted increase in plasmid population, which would be harmful to the host and, in turn, to the plasmid.
Figure 7.

A possible alternative mechanism for 2 micron plasmid amplification. A plasmid dimer may be formed by Flp mediated recombination between two monomers. Resolution of the dimer by Flp during the act of replication will give rise to two interconnected circles being replicated iteratively by unidirectional replication forks. Such structures, named pince-nez molecules, have been observed by electron microscopy (65).
Flp site-specific recombination system: in vitro analyses
In contrast to the paucity of the in vivo analysis of Flp function, Flp mediated recombination has been studied extensively in vitro as a model for phosphoryl transfer reactions in nucleic acids. Flp belongs to the tyrosine family of site-specific recombinases, whose members utilize an active site tyrosine residue as the nucleophile to break the scissile phosphate ester bond during the strand cleavage step of recombination. The molecular-genetic, biochemical and structural information derived from Flp and mechanistically related recombinases have revealed the conformational, mechanistic and dynamic attributes that underlie tyrosine family site-specific recombination (66-72). While tyrosine recombinases follow the typical type IB topoisomerase chemical mechanism, they do so in the context of a recombination synapse containing two DNA partners, each bound by a pair of recombinase monomers (Figure 8A). The reaction is completed in a carefully orchestrated sequence of two temporally separated steps of single strand exchanges. The first step generates a Holliday junction as an obligatory intermediate; the second step resolves the junction into reciprocal recombinants
Figure 8.
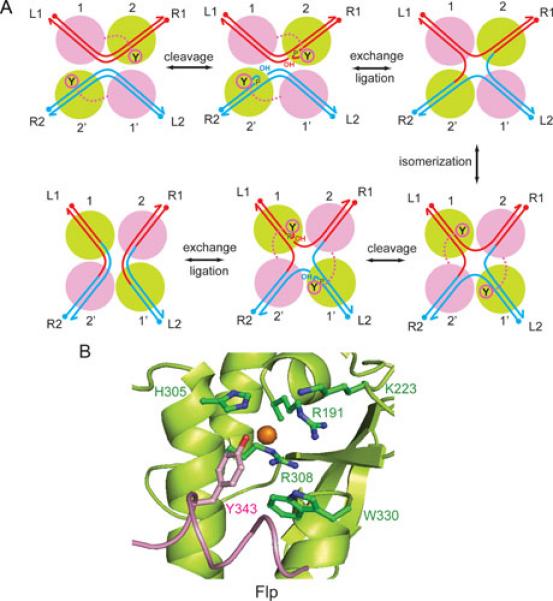
Flp mediated site-specific recombination. A. The recombination reaction is initiated by the synapsis of two FRT sites L1-R1 and L2-R2; L = left; R = right), each bound by two monomers of Flp (1, 2; 1’, 2’) across from the strand exchange region. The antiparallel arrangement of sites (left to right at the top and right to left at the bottom) within the recombination synapse is consistent with most (but not all) published data. The first pair of strand cleavage-exchange reactions gives rise to a Holliday junction intermediate; the second pair of analogous reactions resolves the junction into recombinant products, L1-R2 and L2-R1. The active Flp monomers, those adjacent to the scissile phosphates that are targeted by the active site tyrosine nucleophiles during a cleavage-exchange step, are shown in green. The switch between the active and inactive (magenta) pairs of Flp monomers accompanies the isomerization of the Holliday junction intermediate. B. The organization of key catalytic residues within the Flp active site is shown (75, 89). The conserved catalytic pentad of the tyrosine family corresponds to Arg-191, Lys-223, His-305, Arg-308 and Trp-330 in Flp. The active site tyrosine (Tyr-343) is delivered by a second Flp monomer.
Flp active site: organization and mechanism
The canonical active site of a tyrosine recombinase is characterized by an invariant tyrosine nucleophile utilized for strand cleavage as well as a highly conserved catalytic pentad (Arg-Lys-His-Arg-His/Trp) that helps stabilize the transition states involved in the strand scission and strand union reactions (72, 73). In addition, a conserved glutamic/aspartic acid reside appears to contribute structurally to the functional state of the active site (74). In Flp, the pentad residues of Arg-191, Lys-223, His-305, Arg-308 and Trp-330, assisted by Asp-194, serve to balance the negative charge on the scissile phosphate as well as to activate and orient the Tyr-343 nucleophile (75-78) (Figure 8B). The strand cleavage step results in the covalent attachment of the tyrosine to the 3’-phopshate end and the exposure of the 5’-hydroxyl group adjacent to it. This 5-hydroxyl group is the nucleophile that attacks the phosphotyrosyl intermediate during the strand joining reaction. When the joining reactions are directed across DNA partners, rather than within them, the result is the formation of reciprocal recombinant products.
While the active site mechanism described above is common to the tyrosine family, the organization of the Flp active site differs from that of other well characterized members of this family such as phage λ integrase, phage P1 Cre and E. coli XerC-XerD recombinase. Whereas the norm is the assembly of a fully functional active site within a monomer (79-83), the Flp active site is assembled at the interface of two neighboring Flp monomers (75, 84). One Flp monomer provides the pro-active site (shown in green in Figure 8A, B) comprised of the catalytic pentad and Asp-194, while the second monomer (shown in magenta in Figure 8A, B) donates the tyrosine nucleophile in trans. The shared active site may be a common feature of the sub-family of tyrosine recombinases related to Flp housed by budding yeast plasmids (85). However, allosteric interaction with a neighboring monomer may be important for active site activation even in recombinases that assemble their active sites in cis. The unusual active site design of Flp has been particularly helpful in probing its active site mechanism using experimental strategies that are not applicable to recombinase active sites with a cis configuration. For example, exogenous nucleophiles that mimic the tyrosine nucleophile, in conjunction with the catalytically inactive Flp(Y343F), can recapitulate the mechanistic features of the normal strand cleavage reaction (86, 87).
Within the recombination synapse, the interactions among the four Flp monomers permit only two of the four Tyr-343 residues to be oriented in their reactive configuration (88). This half-of-the sites activity accounts for the cleavage/exchange of the DNA strands two at a time (88, 89). The formation of the Holliday junction intermediate is accomapnied by the isomerization of its arm configuration, which results in the inactivation of the original pair of active sites and the activation of the new pair. The resulting cleavage/exchange of the second pair of strands resolves the junction. The majority of biochemical and topological evidence, as well as structural data, is consistent with the anti-parallel arrangement of the target sites within the recombination synapse (Figure 8A).
Probing Flp active site mechanism using chemical modifications of the scissile phosphate group
Modifications of the non-bridging oxygen atoms of the phosphate group to alter its electronegativity and/or stereochemistry and of the bridging oxygen atoms to modulate leaving group potential (Figure 9A, B) have provided a useful tool for analyzing mechanistic features of strand breakage and union reactions in nucleic acids. Shuman and colleagues have successfully exploited the replacement of one of the non-bridging oxygens on the scissile phosphate by sulfur (phosphorothioate) or the methyl group (methylphosphonate; MeP) as well as the 5’-bridging oxygen by sulfur (5’-thiolate) to unveil several active site attributes of vaccinia type IB topoisomerase (90-93). Analogous strategies have been employed for analyzing mechanistic features of the Flp active site (94, 95). Such studies have been performed predominantly using half-site substrates containing a single scissile phosphate or a modified scissile phosphate (Figure 9C) together with Flp variants harboring specific active site mutations.
Figure 9.
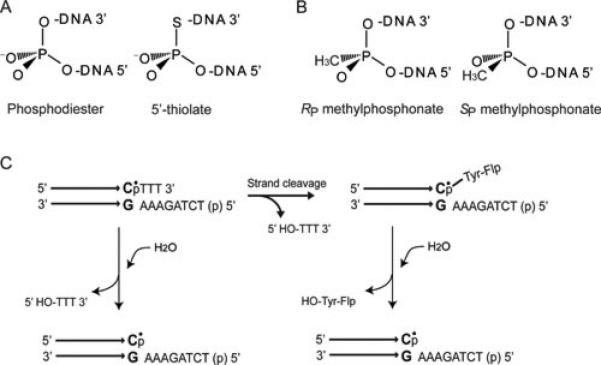
Chemical substitutions at the scissile phosphate position and half-site substrates for probing recombination mechanisms. A. When the 5’ bridging oxygen atom of the phosphodiester bond is replaced by sulfur, the significantly lower pKa of the 5’-thiol (compared to the 5’-hydroxyl) makes it a stronger leaving group. The 5’ thiolate substitution, in conjunction with active site mutants, can shed light on the general acid and/or accessory residues involved in facilitating leaving group departure. B. When one of the non-bridging oxygen atoms of the phosphate group is replaced by a methyl group, the resulting methylphosphonate (MeP) has no negative charge in the ground state. Furthermore, the methyl substitution introduces chirality at the phosphate center. The MeP substitution is useful for the analysis of electrostatic and stereochemical features of the recombination reaction. C. Half-site substrates simplify the recombination reaction while retaining its intrinsic chemical features. A half-site contains a single Flp binding element and a single scissile phosphate (or MeP) on the cleavable strand followed by three (or two) nucleotides of the strand exchange region. The modified scissile phosphate in the MeP half-site is indicated by the dot placed over ‘p’. The other (non-cleavable) strand contains all eight nucleotides of the strand exchange region. When the 5’-hydroxyl group of this strand is phosphorylated, it is blocked from partaking in a strand joining reaction. Tyr-343 mediated cleavage of the MeP bond will give rise to the Flp-linked DNA intermediate, which can be hydrolyzed slowly over time. Cleavage within a half-site is nearly irreversible, as the short tri- or dinucleotide product diffuses away from the reaction center. Since Flp bound half-sites can associate to form dimers, trimers and tetramers, Tyr-343 can be donated in trans as the cleavage nucleophile. In principle, the same hydrolysis product can also be formed by direct attack of water on the MeP bond (shown at the left). However, such a reaction is not observed during the action of Flp on native DNA substrates containing an unmodified phosphate at the scissile position.
Reactions with a 5’-thiolate substituted DNA substrate have revealed a subtle role for Trp-330 of Flp in facilitating the departure of the 5’-hydroxyl group during the strand cleavage step (77). The primary function of Trp-330 is in promoting the proper alignment of the Tyr-343 nucleophile donated in trans through hydrophobic/van der Waals interactions (77, 96). Flp(W330H) is strongly compromised in its cleavage potential, while Flp(W330A) has barely detectable cleavage activity. However, both mutants show vastly improved activity on a substrate containing the 5’-thiolate modification. The thiol is a better leaving group than hydroxyl because of its lower pKa. Thus, the catalytic deficiency caused by the absence of Trp-330 can be significantly ameliorated by a DNA modification that facilitates leaving group departure.
The reactivity of Flp variants on MeP substrates demonstrate that neutralization of the phosphate negative charge in its ground state permits transition state stabilization in the absence of one of the two conserved arginines (Arg-191 or Arg-308) of the Flp active site (94, 95). Flp(R191A) and Flp(R308A) are reactive on half-site substrates containing MeP substitution at the scissile position (Figure 10A, B) whereas both these variants are almost completely inactive on phosphate containing DNA substrates. The electrostatic suppression of the lack of a positive charge in the recombinase active site by a compensatory charge substitution in the scissile phosphate of the DNA substrate has also been demonstrated for the Cre recombinase (97, 98).
Figure 10.

Activities of Flp(R191A) and Flp(R308A) on an MeP substituted half-site substrate; difference between cis- and trans- active sites in protecting the scissile phosphate from abortive hydrolysis. The MeP half-site reactions are analyzed by electrophoresis in SDS-polyacrylamide gels (top panels) to detect the Flp linked tyrosyl-DNA intermediate or in urea-polyacrylamide gels (bottom panels) to visualize the hydrolysis product (HP). The unreacted substrate band is indicated by ‘S’. The asterisk marks the 32P label placed at the 5’ end of the cleavable strand of the half-site. A. Flp(R191A) forms the tyrosyl intermediate, which then undergoes slow hydrolysis. B. Flp(R308A) does not form the tyrosyl intermediate. Instead, it promotes direct hydrolysis of the MeP bond. C. When a cis-acting recombinase (Cre, for example) monomer binds to its target site, the tyrosine nucleophile is oriented within the active site to engage the scissile phosphate. The binding of a second monomer allosterically activates this active site to trigger tyrosine mediated strand cleavage. Thus, the scissile phosphate is not prone to attack by water nucleophile. D. Binding of a Flp monomer activates the adjacent scissile phosphate even though the tyrosine nucleophile is not oriented within the active site. The engagement of the scissile phosphate by tyrosine must await the binding of a second Flp monomer. The time lag between the two binding events renders the scissile phosphate susceptible to direct hydrolysis Electrostatic repulsion of water by Arg-308 appears to prevent this abortive reaction.
An extra-catalytic role for Arg-308 of Flp revealed by the MeP reaction
Flp(R308A) promotes direct hydrolysis of the MeP substrate without forming the tyrosyl intermediate (Figure 10B) (94). In the absence of Arg-308, the abundant water nucleophile gains access to the reaction center, outcompeting Tyr-343 in the cleavage reaction to give a dead-end hydrolytic product. By contrast, Flp(R191A) does not mediate direct hydrolysis of the MeP bond; rather, it utilizes Tyr-343 as the cleavage nucleophile to yield the tyrosyl intermediate (95) (Figure 10A). The absence of the tyrosyl intermediate during the breakage of the MeP bond by Flp(R308A) has been verified by the activity of the double mutant Flp(R308A,Y343F), which also yields the hydrolysis product with similar kinetics and Vmax as Flp(R308A). Thus, in addition to its catalytic role in balancing the phosphate negative charge, Arg-308 appears to protect the normal reaction course from abortive hydrolysis, presumably by electrostatic repulsion of water. Unlike Arg-308 of Flp, the corresponding Arg-292 of Cre does not seem to perform a protective function against direct hydrolysis of the scissile phosphate. Cre(R292A) yields the MeP-tyrosyl intermediate by utilizing the active site tyrosine nucleophile (Tyr-324) (97, 98). This difference between Flp and Cre may be rationalized by the trans versus cis organization of their respective sites. When a Cre monomer binds to one of the two binding sites of a target site, the tyrosine nucleophile concomitantly engages the adjacent scissile phosphate (Figure 10C). As a result, there is little danger from direct hydrolysis. By contrast, binding of a Flp monomer activates the adjacent scissile phosphate without immediate engagement by the tyrosine nucleophile. The donation of Tyr-343 in trans must await the binding of a second Flp monomer to the binding element across the strand exchange region. The potential time delay between the two binding events gives water the opportunity to attack the activated phosphate group (Figure 10D). This untoward reaction is avoided by utilizing the positive charge on the Arg-308 side chain to repel water (which is electrically a dipole).
As the methyl substitution of one of the non-bridging oxygen atoms turns the normally symmetric phosphate group into an asymmetric center, an additional utility of the MeP substrates is in unveiling the stereochemical course of the recombination reaction. Stereochemically pure RP and SP forms of the MeP substrates are currently being used to dissect the individual stereochemical contributions of Arg-191 and Arg-308 and to probe how other members of the catalytic pentad might influence these contributions.
Flp reaction pathway: single molecule analysis
Single molecule analysis of Flp recombination using real-time tethered particle motion (TPM) analysis has provided deeper insights into the pre-chemical and chemical steps of the reaction pathway (Figure 11) (99). The results obtained reveal interesting similarities and contrasts of Flp with λ Int and phage P1 Cre, which have also been characterized by single molecule TPM or by single molecule TPM as well as TFM (tethered fluorophore motion) (100-102). The binding of recombinase to target sites and the pairing of bound sites is quite fast in all three cases, ruling out intrinsic barriers to synapsis during tyrosine family site-specific recombination, at least in vitro. There is a strong commitment to recombination following the association of Flp with the FRT sites. The formation of non-productive complexes (those that do not synapse) and wayward complexes (those that do not form the Holliday junction intermediate or complete recombination after synapsis) constitute only minor detractions from the productive pathway. The stability of the synapse is enhanced by strand cleavage in the case of Flp and λ Int. However, Cre forms stable synapse even in the absence of strand cleavage. Despite the chemical reversibility of the individual reactions, a round of Flp recombination proceeds in an almost unidirectional fashion, presumably aided by the conformational changes associated with each cleavage or joining step. Such irreversibility would be a desirable attribute in vivo in bringing about the desired DNA rearrangement, namely, inversion of a replication fork in the case of Flp. However, the Holliday junction formed during Cre recombination is long lived, affording the opportunity for the reaction to reverse course, at least in vitro. It is possible that the in vitro Cre reaction fails to recapitulate the native regulatory features of recombination occurring within the phage genome organized into a nucleoprotein complex.
Figure 11.
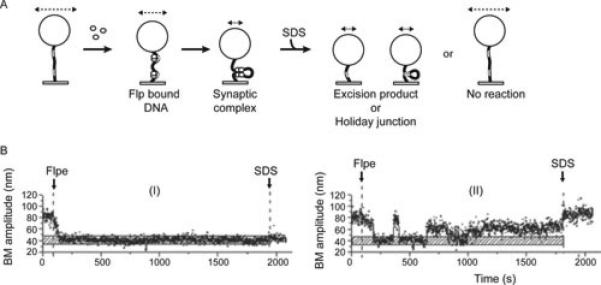
Single molecule tethered particle motion (TPM) analysis of Flp site-specific recombination. The substrate DNA molecule containing a pair of FRT sites in direct (deletion substrate) or inverted (inversion substrate) orientation is tethered to a glass surface at one end and to a polystyrene bead at the other. The effective length of the DNA changes as a result of Flp binding to FRT sites or synapsis of the bound sites. Such changes can be assayed by changes in the Brownian motion (BM) amplitude of the attached bead (indicated schematically by the dashed line with arrowheads at either end). Within the recombination synapse, the chemical steps of recombination may or may not occur. The fate of the synapsed molecules can be revealed by the BM amplitudes they display after being stripped of non-covalently associated Flp. A. The expected BM amplitudes following the addition of Flp to a deletion substrate are illustrated. Binding of Flp to the FRT sites (unfilled rectangular boxes on the tethered DNA molecule) will cause a slight reduction in the BM amplitude because of the DNA bending induced by Flp. A more marked reduction in BM amplitude follows upon synapsis of the FRT sites. This low BM amplitude will be retained by the Holliday junction intermediate and the linear product of excision even after the addition of SDS. Flp bound molecules that fail to synapse (non-productive complexes) or synapsed molecules that fail to recombine (wayward complexes) will return to the high BM amplitude of the starting DNA substrate after SDS challenge. In the case of the inversion substrate, SDS challenge cannot distinguish a wayward complex from a completed recombination event. This is because the length of the inversion product is the same as that of the parental substrate. B. Time traces of individual molecules illustrating two different states of the deletion substrate following Flp addition are shown. (I) A molecule that formed a stable synapse of the Flp bound FRT sites. The low BM amplitude of this molecule after SDS challenge indicates that it underwent Holliday junction formation or a complete recombination event. (II) A molecule that underwent Flp binding and synapsis but failed to recombine or to form the Holliday junction, as indicated by its return to the starting high BM amplitude after SDS treatment (a wayward complex). The horizontal stippled bar represents the BM amplitude of the DNA in the synapsed state of FRT sites.
Contributions of the 2 micron plasmid to basic biology and bio-engineering applications
The 2 micron plasmid provides a model for how the collaboration between an efficient partitioning system and a copy number control system can confer chromosome-like stability on an extremely simple extra-chromosomal genome (7). It exemplifies the evolutionary success of a selfish element through strategies that take advantage of the genetic endowments of its host, while at the same time moderating its selfishness to avoid harming the host (103). The plasmid partitioning system has provided the basis for propagating autonomously replicating plasmids with high stability in S. cerevisiae. Assuming that the proposed hitchhiking model for 2 micron plasmid segregation is correct, it may be possible to suitably engineer the Rep-STB system for the long-term maintenance of beneficial extra-chromosomal elements in a variety of higher eukaryotes. The Flp site-specific recombination system harbored by the plasmid has been seminal to unveiling phosphoryl transfer mechanisms in nucleic acids and to understanding conformational dynamics associated with strand exchange between DNA partners (70, 76). The simple requirements of Flp and Cre reactions have been exploited to develop an analytical tool called ‘difference topology’, which reveals the topological path of DNA within high-order DNA-protein complexes (7, 104-106). The rationale is to first assemble the complex of interest, and then utilize Flp or Cre recombination to trap the DNA supercoils sequestered within it as crossings of DNA knots formed in an inversion reaction or DNA links (catenanes) formed in a deletion reaction. Care is taken to avoid the random entrapment of supercoils by placing the recombination target sites within close proximity of the complex. The knot or catenane crossings can be counted by suitable analytical methods. A simplified description of the principles and practice of difference topology can be found in (7). The method can potentially be applied to deduce the topological aesthetics of complex DNA-protein interactions to which standard biochemical and biophysical techniques are unfortunately blind. For example, it would be interesting to know whether the interactions among promoters, enhancers and activator or repressor binding sites during transcriptional regulation engender specific DNA topologies. The targeted genetic manipulations that can be accomplished by Flp and related recombination systems (107-109) have been particularly helpful in addressing basic problems in gene regulation and developmental biology. They have also provided a powerful tool for genome engineering. The generation of altered target specificity recombinase variants (110-115), as well as the regulated and/or tissue specific expression of recombinases (108, 116-118), has vastly expanded the utility of site-specific recombination in basic research and in biotechnological applications.
Acknowledgments
Our studies on the partitioning and site-specific recombination systems have been supported by the National Institutes of Health, the National Science Foundation (MCB-1049925) and the Robert F Welch Foundation (F-1274).
References
- 1.Dawkins R. The Selfish Gene. Oxford University Press; UK: 1976. [Google Scholar]
- 2.Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 3.Orgel LE, Crick FH, Sapienza C. Selfish DNA. Nature. 1980;288:645–646. doi: 10.1038/288645a0. [DOI] [PubMed] [Google Scholar]
- 4.Rowley PA, Kachroo AH, Jayaram M. Selfish DNA. In: Malloy S, Hughes K, editors. Brenner's Encyclopedia of Genetics. Vol. 6. Elsevier; 2013. pp. 382–389. [Google Scholar]
- 5.Frappier L. Viral plasmids in mammalian cells. In: Funnell BE, Phillips G, editors. Plasmid Biology. ASM Press; Washington, DC.: 2004. pp. 325–340. [Google Scholar]
- 6.McBride AA. Replication and partitioning of papillomavirus genomes. Adv Virus Res. 2008;72:155–205. doi: 10.1016/S0065-3527(08)00404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang KM, Liu YT, Ma CH, Jayaram M, Sau S. The 2 micron plasmid of Saccharomyces cerevisiae: A miniaturized selfish genome with optimized functional competence. Plasmid. 2013;70:2–17. doi: 10.1016/j.plasmid.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Broach JR, Volkert FC. Circular DNA Plasmids of Yeasts. In: Broach JR, Pringle JR, Jones EW, editors. The Molecular Biology of the Yeast Saccharomyces. Genome Dynamics, Protein Synthesis and Energetics. Cold Spring Harbor Laboratory Press; Cold Spring harbor, New York: 1991. pp. 287–331. [Google Scholar]
- 9.Jayaram M, Yang XM, Mehta S, Voziyanov Y, Velmurugan S. The 2 micron plasmid of Saccharomyces cerevisiae. In: Funnell BE, Phillips G, editors. Plasmid Biology. ASM Press; Washington, DC.: pp. 303–324. [Google Scholar]
- 10.Zakian VA, Brewer BJ, Fangman WL. Replication of each copy of the yeast 2 micron DNA plasmid occurs during the S phase. Cell. 1979;4:923–934. doi: 10.1016/0092-8674(79)90332-5. [DOI] [PubMed] [Google Scholar]
- 11.Jayaram M, Mehta S, Uzri D, Voziyanov Y, Velmurugan S. Site-specific recombination and partitioning systems in the stable high copy propagation of the 2-micron yeast plasmid. Prog Nucleic Acid Res Mol Biol. 2004;77:127–172. doi: 10.1016/S0079-6603(04)77004-X. [DOI] [PubMed] [Google Scholar]
- 12.Futcher AB. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- 13.Volkert FC, Broach JR. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986;46:541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- 14.Dobson MJ, Pickett AJ, Velmurugan S, Pinder JB, Barrett LA, Jayaram M, Chew JS. The 2 microm plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease. Mol Cell Biol. 2005;25:4299–4310. doi: 10.1128/MCB.25.10.4299-4310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm C. Sensitivity to the yeast plasmid 2 μm DNA is conferred by the nuclear allele nib1. Mol Cell Biol. 1982;2:985–992. doi: 10.1128/mcb.2.8.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm C. Clonal lethality caused by the yeast plasmid 2 μm DNA. Cell. 1982;29:85–94. doi: 10.1016/0092-8674(82)90174-x. [DOI] [PubMed] [Google Scholar]
- 17.Murray JA, Scarpa M, Rossi N, Cesareni G. Antagonistic controls regulate copy number of the yeast 2 micron plasmid. EMBO J. 1897;6:4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds AE, Murray AW, Szostak JW. Roles of the 2 micron gene products in stable maintenance of the 2 micron plasmid of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:3566–3573. doi: 10.1128/mcb.7.10.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Som T, Armstrong KA, Volkert FC, Broach JR. Autoregulation of 2 micron circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell. 1988;52:27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- 20.Chen XL, Reindle A, Johnson ES. Misregulation of 2 micron circle copy number in a SUMO pathway mutant. Mol Cell Biol. 2005;25:4311–4320. doi: 10.1128/MCB.25.10.4311-4320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong L, Chen XL, Silver HR, Ahmed NT, Johnson ES. Deficient SUMO attachment to Flp recombinase leads to homologous recombination-dependent hyperamplification of the yeast 2 micron circle plasmid. Mol Biol Cell. 2009;20:1241–1251. doi: 10.1091/mbc.E08-06-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 24.Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat Struct Mol Biol. 2009;16:1010–1015. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gehlen LR, Nagai S, Shimada K, Meister P, Taddei A, Gasser SM. Nuclear geometry and rapid mitosis ensure asymmetric episome segregation in yeast. Curr Biol. 2011;21:25–33. doi: 10.1016/j.cub.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Khmelinskii A, Meurer M, Knop M, Schiebel E. Artificial tethering to nuclear pores promotes partitioning of extrachromosomal DNA during yeast asymmetric cell division. Curr Biol. 2011;21:R17–18. doi: 10.1016/j.cub.2010.11.034. [DOI] [PubMed] [Google Scholar]
- 28.Murray AW, Szostak JW. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- 29.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 30.Mehta S, Yang XM, Chan CS, Dobson MJ, Jayaram M, Velmurugan S. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation? J Cell Biol. 2002;158:625–637. doi: 10.1083/jcb.200204136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velmurugan S, Yang XM, Chan CS, Dobson M, Jayaram M. Partitioning of the 2-micron circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution. J Cell Biol. 2000;149:553–566. doi: 10.1083/jcb.149.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu YT, Ma CH, Jayaram M. Co-segregation of yeast plasmid sisters under monopolin-directed mitosis suggests association of plasmid sisters with sister chromatids. Nucleic Acids Res. 2013;41:4144–4158. doi: 10.1093/nar/gkt096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn YT, Wu XL, Biswal S, Velmurugan S, Volkert FC, Jayaram M. The 2micron-plasmid-encoded Rep1 and Rep2 proteins interact with each other and colocalize to the Saccharomyces cerevisiae nucleus. J Bacteriol. 1997;179:7497–7506. doi: 10.1128/jb.179.23.7497-7506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadfield C, Mount RC, Cashmore AM. Protein binding interactions at the STB locus of the yeast 2 micron plasmid. Nucleic Acids Res. 1995;23:995–1002. doi: 10.1093/nar/23.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott-Drew S, Murray JA. Localisation and interaction of the protein components of the yeast 2 micron circle plasmid partitioning system suggest a mechanism for plasmid inheritance. J Cell Sci. 1998;111:1779–1789. doi: 10.1242/jcs.111.13.1779. [DOI] [PubMed] [Google Scholar]
- 36.Velmurugan S, Ahn YT, Yang XM, Wu XL, Jayaram M. The 2 micron plasmid stability system: analyses of the interactions among plasmid- and host-encoded components. Mol Cell Biol. 1998;18:7466–7477. doi: 10.1128/mcb.18.12.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang XM, Mehta S, Uzri D, Jayaram M, Velmurugan S. Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation. Mol Cell Biol. 2004;24:5290–5303. doi: 10.1128/MCB.24.12.5290-5303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sengupta A, Blomqvist K, Pickett AJ, Zhang Y, Chew JS, Dobson MJ. Functional domains of yeast plasmid-encoded Rep proteins. J Bacteriol. 2001;183:2306–2315. doi: 10.1128/JB.183.7.2306-2315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray JA, Cesareni G. Functional analysis of the yeast plasmid partition locus STB. EMBO J. 1986;5:3391–3399. doi: 10.1002/j.1460-2075.1986.tb04655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui H, Ghosh SK, Jayaram M. The selfish yeast plasmid uses the nuclear motor Kip1p but not Cin8p for its localization and equal segregation. J Cell Biol. 2009;185:251–264. doi: 10.1083/jcb.200810130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hajra S, Ghosh SK, Jayaram M. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-micron circle partitioning locus and promotes equal plasmid segregation. J Cell Biol. 2006;174:779–790. doi: 10.1083/jcb.200603042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang CC, Hajra S, Ghosh SK, Jayaram M. Cse4 (CenH3) association with the Saccharomyces cerevisiae plasmid partitioning locus in its native and chromosomally integrated states: implications for centromere evolution. Mol Cell Biol. 2011:1030–1040. doi: 10.1128/MCB.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma CH, Cui H, Hajra S, Rowley PA, Fekete C, Sarkeshik A, Ghosh SK, Yates JR, 3rd, Jayaram M. Temporal sequence and cell cycle cues in the assembly of host factors at the yeast 2 micron plasmid partitioning locus. Nucleic Acids Res. 2013;41:2340–2353. doi: 10.1093/nar/gks1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong MC, Scott-Drew SR, Hayes MJ, Howard PJ, Murray JA. RSC2, encoding a component of the RSC nucleosome remodeling complex, is essential for 2 micron plasmid maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:4218–4229. doi: 10.1128/MCB.22.12.4218-4229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh SK, Huang CC, Hajra S, Jayaram M. Yeast cohesin complex embraces 2 micron plasmid sisters in a tri-linked catenane complex. Nucleic Acids Res. 2010;38:570–584. doi: 10.1093/nar/gkp993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci (USA) 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lefrancois P, Auerbach RK, Yellman CM, Roeder GS, Snyder M. Centromere-like regions in the budding yeast genome. PLoS Genet. 2013;9:e1003209. doi: 10.1371/journal.pgen.1003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 50.Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci (USA) 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang CC, Chang KM, Cui H, Jayaram M. Histone H3-variant Cse4-induced positive DNA supercoiling in the yeast plasmid has implications for a plasmid origin of a chromosome centromere. Proc Natl Acad Sci (USA) 2011;108:13671–13676. doi: 10.1073/pnas.1101944108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghosh SK, Hajra S, Jayaram M. Faithful segregation of the multicopy yeast plasmid through cohesin-mediated recognition of sisters. Proc Natl Acad Sci (USA) 2007;104:13034–13039. doi: 10.1073/pnas.0702996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiburz BM, Amon A, Marston AL. Shugoshin promotes sister kinetochore biorientation in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:1199–1209. doi: 10.1091/mbc.E07-06-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 56.Meyer RE, Kim S, Obeso D, Straight PD, Winey M, Dawson DS. Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Science. 2013;339:1071–1074. doi: 10.1126/science.1232518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- 59.Yu HG, Koshland D. The Aurora kinase Ipl1 maintains the centromeric localization of PP2A to protect cohesin during meiosis. J Cell Biol. 2007;176:911–918. doi: 10.1083/jcb.200609153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehta S, Yang XM, Jayaram M, Velmurugan S. A novel role for the mitotic spindle during DNA segregation in yeast: promoting 2 micron plasmid-cohesin association. Mol Cell Biol. 2005;25:4283–4298. doi: 10.1128/MCB.25.10.4283-4298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mehta GD, Rizvi SM, Ghosh SK. Cohesin: a guardian of genome integrity. Biochim Biophys Acta. 2012;1823:1324–1342. doi: 10.1016/j.bbamcr.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 62.Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol. 2011;13:1170–1177. doi: 10.1038/ncb2349. [DOI] [PubMed] [Google Scholar]
- 63.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 64.Remeseiro S, Losada A. Cohesin, a chromatin engagement ring. Curr Opin Cell Biol. 2013;25:63–71. doi: 10.1016/j.ceb.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 65.Petes TD, Williamson DH. A novel structural form of the 2 micron plasmid of the yeast Saccharomyces cerevisiae. Yeast. 1994;10:1341–1345. doi: 10.1002/yea.320101011. [DOI] [PubMed] [Google Scholar]
- 66.Azaro MA, Landy A. λ integrase and the λ int family. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington, DC.: 2002. pp. 118–148. [Google Scholar]
- 67.Barre FX, Sherratt DJ. In: Xer site-specific recombination: promoting chromosome segregation. Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. ASM Press; Washington, DC.: 2002. pp. 149–161. [Google Scholar]
- 68.Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 70.Jayaram M, Grainge I, Tribble G. Site-specific DNA recombination mediated by the Flp protein of Saccharomyces cerevisiae. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington, DC.: 2002. pp. 192–218. [Google Scholar]
- 71.Rice PA. Theme and variation in tyrosine recombinases: Structure of a Flp-DNA complex. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington, DC.: 2002. pp. 219–229. [Google Scholar]
- 72.Van Duyne GD. A structural view of tyrosine recombinase site-specific recombination. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington, DC.: 2002. pp. 93–117. [Google Scholar]
- 73.Grainge I, Jayaram M. The integrase family of recombinase: organization and function of the active site. Mol Microbiol. 1999;33:449–456. doi: 10.1046/j.1365-2958.1999.01493.x. [DOI] [PubMed] [Google Scholar]
- 74.Gibb B, Gupta K, Ghosh K, Sharp R, Chen J, Van Duyne GD. Requirements for catalysis in the Cre recombinase active site. Nucleic Acids Res. 2010;38:5817–5832. doi: 10.1093/nar/gkq384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Y, Narendra U, Iype LE, Cox MM, Rice PA. Crystal structure of a Flp recombinase-Holliday junction complex. Assembly of an active oligomer by helix swapping. Mol Cell. 2000;6:885–897. [PubMed] [Google Scholar]
- 76.Chen Y, Rice PA. New insight into site-specific recombination from Flp recombinase-DNA structures. Annu Rev Biophys Biomol Struct. 2003;32:135–159. doi: 10.1146/annurev.biophys.32.110601.141732. [DOI] [PubMed] [Google Scholar]
- 77.Ma CH, Kwiatek A, Bolusani S, Voziyanov Y, Jayaram M. Unveiling hidden catalytic contributions of the conserved His/Trp-III in tyrosine recombinases: assembly of a novel active site in Flp recombinase harboring alanine at this position. J Mol Biol. 2007;368:183–196. doi: 10.1016/j.jmb.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whiteson KL, Chen Y, Chopra N, Raymond AC, Rice PA. Identification of a potential general acid/base in the reversible phosphoryl transfer reactions catalyzed by tyrosine recombinases: Flp H305. Chem Biol. 2007;14:121–129. doi: 10.1016/j.chembiol.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arciszewska LK, Grainge I, Sherratt DJ. Action of site-specific recombinases XerC and XerD on tethered Holliday junctions. EMBO J. 1997;16:3731–3743. doi: 10.1093/emboj/16.12.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blakely GW, Davidson AO, Sherratt DJ. Binding and cleavage of nicked substrates by site-specific recombinases XerC and XerD. J Mol Biol. 1997;265:30–39. doi: 10.1006/jmbi.1996.0709. [DOI] [PubMed] [Google Scholar]
- 81.Grainge I, Sherratt DJ. Xer site-specific recombination. DNA strand rejoining by recombinase XerC. J Biol Chem. 274:6763–6769. doi: 10.1074/jbc.274.10.6763. [DOI] [PubMed] [Google Scholar]
- 82.Guo F, Gopaul DN, Van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombinase synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 83.Nunes-Duby SE, Tirumalai RS, Dorgai L, Yagil E, Weisberg RA, Landy A. Lambda integrase cleaves DNA in cis. EMBO J. 1994;13:4421–4430. doi: 10.1002/j.1460-2075.1994.tb06762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen JW, Lee J, Jayaram M. DNA cleavage in trans by the active site tyrosine during Flp recombination: switching protein partners before exchanging strands. Cell. 1992;69:647–658. doi: 10.1016/0092-8674(92)90228-5. [DOI] [PubMed] [Google Scholar]
- 85.Yang SH, Jayaram M. Generality of the shared active site among yeast family site-specific recombinases. The R site-specific recombinase follows the Flp paradigm. J Biol Chem. 1994;269:12789–12796. [PubMed] [Google Scholar]
- 86.Kimball AS, Lee J, Jayaram M, Tullius TD. Sequence-specific cleavage of DNA via nucleophilic attack of hydrogen peroxide, assisted by Flp recombinase. Biochem. 1993;32:4698–4701. doi: 10.1021/bi00069a002. [DOI] [PubMed] [Google Scholar]
- 87.Lee J, Jayaram M. Functional roles of individual recombinase monomers in strand breakage and strand union during site-specific DNA recombination. J Biol Chem. 1995;270:23203–23211. doi: 10.1074/jbc.270.39.23203. [DOI] [PubMed] [Google Scholar]
- 88.Lee J, Tonozuka T, Jayaram M. Mechanism of active site exclusion in a site-specific recombinase: role of the DNA substrate in conferring half-of-the-sites activity. Genes Dev. 1997;11:3061–3071. doi: 10.1101/gad.11.22.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Conway AB, Chen Y, Rice PA. Structural plasticity of the Flp-Holliday junction complex. J Mol Biol. 2003;326:425–434. doi: 10.1016/s0022-2836(02)01370-0. [DOI] [PubMed] [Google Scholar]
- 90.Krogh BO, Shuman S. Catalytic mechanism of DNA topoisomerase IB. Mol Cell. 2000;5:1035–1041. doi: 10.1016/s1097-2765(00)80268-3. [DOI] [PubMed] [Google Scholar]
- 91.Stivers JT, Jagadeesh GJ, Nawrot B, Stec WJ, Shuman S. Stereochemical outcome and kinetic effects of Rp- and Sp-phosphorothioate substitutions at the cleavage site of vaccinia type I DNA topoisomerase. Biochem. 2000;39:5561–5572. doi: 10.1021/bi992429c. [DOI] [PubMed] [Google Scholar]
- 92.Tian L, Claeboe CD, Hecht S, Shuman S. Guarding the genome: electrostatic repulsion of water by DNA suppresses a potent nuclease activity of topoisomerase IB. Mol Cell. 2003;12:199–208. doi: 10.1016/s1097-2765(03)00263-6. [DOI] [PubMed] [Google Scholar]
- 93.Tian L, Claeboe CD, Hecht SM, Shuman S. Mechanistic plasticity of DNA topoisomerase IB: phosphate electrostatics dictate the need for a catalytic arginine. Structure. 2005;13:513–520. doi: 10.1016/j.str.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 94.Ma CH, Rowley PA, Maciaszek A, Guga P, Jayaram M. Active site electrostatics protect genome integrity by blocking abortive hydrolysis during DNA recombination. EMBO J. 2009;28:1745–1756. doi: 10.1038/emboj.2009.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rowley PA, Kachroo AH, Ma CH, Maciaszek AD, Guga P, Jayaram M. Electrostatic suppression allows tyrosine site-specific recombination in the absence of a conserved catalytic arginine. J Biol Chem. 2010;285:22976–22985. doi: 10.1074/jbc.M110.112292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Y, Rice PA. The role of the conserved Trp330 in Flp-mediated recombination. Functional and structural analysis. J Biol Chem. 2003;278:24800–24807. doi: 10.1074/jbc.M300853200. [DOI] [PubMed] [Google Scholar]
- 97.Kachroo AH, Ma CH, Rowley PA, Maciaszek AD, Guga P, Jayaram M. Restoration of catalytic functions in Cre recombinase mutants by electrostatic compensation between active site and DNA substrate. Nucleic Acids Res. 2010;38:6589–6601. doi: 10.1093/nar/gkq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma CH, Kachroo AH, Macieszak A, Chen TY, Guga P, Jayaram M. Reactions of Cre with methylphosphonate DNA: similarities and contrasts with Flp and vaccinia topoisomerase. PLoS One. 2009;4:e7248. doi: 10.1371/journal.pone.0007248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fan HF, Ma CH, Jayaram M. Real-time single-molecule tethered particle motion analysis reveals mechanistic similarities and contrasts of Flp site-specific recombinase with Cre and λ Int. Nucleic Acids Res. 2013;41:7031–7047. doi: 10.1093/nar/gkt424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fan HF. Real-time single-molecule tethered particle motion experiments reveal the kinetics and mechanisms of Cre-mediated site-specific recombination. Nucleic Acids Res. 2012;40:6208–6222. doi: 10.1093/nar/gks274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mumm JP, Landy A, Gelles J. Viewing single lambda site-specific recombination events from start to finish. EMBO J. 2006;25:4586–4595. doi: 10.1038/sj.emboj.7601325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pinkney JN, Zawadzki P, Mazuryk J, Arciszewska LK, Sherratt DJ, Kapanidis AN. Capturing reaction paths and intermediates in Cre-loxP recombination using single-molecule fluorescence. Proc Natl Acad Sci (USA) 2012;109:20871–20876. doi: 10.1073/pnas.1211922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Velmurugan S, Mehta S, Jayaram M. Selfishness in moderation: evolutionary success of the yeast plasmid. Curr Top Dev Biol. 2003;56:1–24. doi: 10.1016/s0070-2153(03)01005-6. [DOI] [PubMed] [Google Scholar]
- 104.Grainge I, Buck D, Jayaram M. Geometry of site alignment during Int family recombination: antiparallel synapsis by the Flp recombinase. J Mol Biol. 2000;298:749–764. doi: 10.1006/jmbi.2000.3679. [DOI] [PubMed] [Google Scholar]
- 105.Harshey RM, Jayaram M. The Mu transpososome through a topniaological lens. Crit Rev Biochem Mol Biol. 2006;41:387–405. doi: 10.1080/10409230600946015. [DOI] [PubMed] [Google Scholar]
- 106.Jayaram M, Harshey RM. Difference topology: analysis of high-order DNA-protein assemblies. In: Benham CJ, Harvey S, Olson WK, Sumners DW, Swigon D, editors. Mathematics of DNA structure, function and interactions. The IMA volumes in mathematics and its applications. Vol. 150. Springer; 2009. pp. 139–158. [Google Scholar]
- 107.Garcia-Otin AL, Guillou F. Mammalian genome targeting using site-specific recombinases. Front Biosci. 2006;11:1108–1136. doi: 10.2741/1867. [DOI] [PubMed] [Google Scholar]
- 108.Turan S, Galla M, Ernst E, Qiao J, Voelkel C, Schiedlmeier B, Zehe C, Bode J. Recombinase-mediated cassette exchange (RMCE): traditional concepts and current challenges. J Mol Biol. 2011;407:193–221. doi: 10.1016/j.jmb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 109.Turan S, Zehe C, Kuehle J, Qiao J, Bode J. Recombinase-mediated cassette exchange (RMCE) - a rapidly-expanding toolbox for targeted genomic modifications. Gene. 2013;515:1–27. doi: 10.1016/j.gene.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 110.Baldwin EP, Martin SS, Abel J, Gelato KA, Kim H, Schultz P, Santoro SW. A specificity switch in selected Cre recombinase variants is mediated by macromolecular plasticity and water. Chem Biol. 2003;10:1085–1094. doi: 10.1016/j.chembiol.2003.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bolusani S, Ma CH, Paek A, Konieczka JH, Jayaram M, Voziyanov Y. Evolution of variants of yeast site-specific recombinase Flp that utilize native genomic sequences as recombination target sites. Nucleic Acids Res. 2006;34:5259–5269. doi: 10.1093/nar/gkl548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 113.Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc Natl Acad Sci (USA) 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 115.Voziyanov Y, Konieczka JH, Stewart AF, Jayaram M. Stepwise manipulation of DNA specificity in Flp recombinase: progressively adapting Flp to individual and combinatorial mutations in its target site. J Mol Biol. 2003;326:65–76. doi: 10.1016/s0022-2836(02)01364-5. [DOI] [PubMed] [Google Scholar]
- 116.Hunter NL, Awatramani RB, Farley FW, Dymecki SM. Ligand-activated Flpe for temporally regulated gene modifications. Genesis. 2005;41:99–109. doi: 10.1002/gene.20101. [DOI] [PubMed] [Google Scholar]
- 117.Kellendonk C, Tronche F, Monaghan AP, Angrand PO, Stewart F, Schutz G. Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res. 1996;24:1404–1411. doi: 10.1093/nar/24.8.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang DJ, Wang Q, Wei J, Baimukanova G, Buchholz F, Stewart AF, Mao X, Killeen N. Selective expression of the Cre recombinase in late-stage thymocytes using the distal promoter of the Lck gene. J Immunol. 2005;174:6725–6731. doi: 10.4049/jimmunol.174.11.6725. [DOI] [PubMed] [Google Scholar]


