Runx1t1 was knocked down in hippocampal radial glial cells (RGCs) with Runx1t1-RNA interference using lentiviral vectors. Experimental evidence showed that decreased Runx1t1 expression reduced the neuronal differentiation of RGCs and increased Runx1t1 expression caused a greater number of RGCs to differentiate into neurons. Thus, Runx1t1 could be involved in the process through which RGCs differentiate into neurons.
Keywords: Runx1t1, Hippocampus, Radial glial cells, Neurogenesis, Differentiation
Abstract
The brain has the highest Runx1t1 level relative to the levels in other organs. Runx1t1 might have a regulatory function as a transcriptional corepressor in the differentiation/development of the nervous system. Neurogenesis requires factors that regulate the proliferation of progenitors and activate the neuronal differentiation process. However, the precise role of Runx1t1 in hippocampal neurogenesis is unclear. We knocked down Runx1t1 in hippocampal radial glial cells (RGCs) with Runx1t1-RNA interference using lentiviral vectors. We also used LV-Runx1t1 to induce Runx1t1 overexpression in vitro. We have provided experimental evidence that decreased Runx1t1 expression reduced the neuronal differentiation of RGCs, and increased Runx1t1 expression caused a greater number of RGCs to differentiate into neurons. We have concluded that Runx1t1 could be involved in the process through which RGCs differentiate into neurons.
Introduction
Hippocampal neurogenesis in the dentate gyrus persists throughout life [1–3]. Neurogenesis is a process through which new neurons are generated from neural stem cells (NSCs) or neural progenitor cells (NPCs). Radial glial cells (RGCs) are putative stem cells in the adult central nervous system [4–6]. They display both astroglial and neuroepithelial characteristics and have multiple differentiation potentialities. RGCs function both as progenitors and as a scaffolding onto which new neurons can migrate [7, 8]. As progenitors, RGCs can give rise to new neurons [4, 9–11). Neurogenesis is a multistep process and is regulated by many factors, both intrinsic and extrinsic [12, 13).
During the process of neurogenesis, a gene-expression cascade activated by proneural basic helix-loop-helix (bHLH) proteins plays an essential and conserved role in promoting neuronal differentiation [14]. Myeloid translocation genes (MTGs) are parts of the gene expression. MTG proteins are expressed during neuronal differentiation and can function by promoting both the transition from precursor to neuron and the expression of neuronal genes within differentiated cells [15]. A number of reports using biochemical and molecular analyses have suggested that MTG proteins function as potent transcriptional repressors [16–18]. Runx1t1 (runt-related transcription factor 1; translocated to, 1 [cyclin D-related]), a transcription factor, is a member of the MTG family. Runx1t1 is involved in the proliferation and differentiation of hematopoietic stem cells (19, 20]. However, the role of Runx1t1 in neural development has largely been unexplored. We investigated the broader role of Runx1t1 in hippocampal neurogenesis in vitro. Using RGCs from the hippocampus as cell models and Runx1t1 knockdown by small interfering RNA (siRNA), we found that Runx1t1 knockdown in hippocampal RGCs was associated with decreased neural differentiation. In contrast, Runx1t1 overexpression during the neural differentiation of hippocampal RGCs led to the differentiation of a greater number of RGCs into microtubule-associated protein 2 (MAP-2)-positive neurons. These results suggested that Runx1t1 is closely related to the neural differentiation of hippocampal RGCs in vitro.
Materials and Methods
RGC Culture and Identification
RGCs were acquired as previously described [10]. Animal experiments were conducted according to the protocols approved by the NIH Guide for the Care and Use of Laboratory Animals. Embryos were taken from pregnant rats on embryonic day 16, and the embryonic hippocampal tissues were immediately dissected and isolated. After removal of the meninges, the tissue was gently triturated into single-cell suspensions using a fire-polished pipette. After centrifugation for 2 minutes at 1,000 rpm, the cells were resuspended and maintained at a density of 1 × 105 in 10-ml Dulbecco’s modified Eagle’s medium (DMEM)/F12 containing 2% B27 and 20 ng/ml epidermal growth factor and fibroblast growth factor-2 (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com), a neurosphere expansion medium. The primary neurospheres were easily formed, and these newly formed neurospheres were passaged every week by the dissociation of bulk neurospheres using Accutase (Sigma-Aldrich). After 3 passages, the neurospheres were incubated in Accutase for about 20 minutes and triturated into single-cell suspensions, replated at a density of 1.5 × 104 cells per milliliter on poly-l-lysine (PLL)-coated coverslips in 24- or 6-well plates (adherent conditions) containing the expansion medium. Three days later, these single NSCs/NPCs exhibited elongated processes and displayed the morphological features of RGCs. Next, the cells were processed for immunocytochemistry to identify their astroglial and stem/progenitor properties.
siRNA-Mediated Knockdown and Overexpression of Runx1t1 in RGCs
Runx1t1 expression in RGCs was inhibited using specific siRNA (Silencer siRNA transfection, GV118 lentiviral expression system, LV3-RUNX1T1-RNA interference (RNAi), Shanghai GeneChem Co., Ltd., Shanghai, People’s Republic of China). A vector-based RNAi approach was used to produce intracellular short hairpin double-stranded RNA from a DNA template under the control of the porcine cytomegalovirus (pCMV) promoter. The siRNA was designed using the web-based siRNA design program on the GenScript webpage (Piscataway, NJ, http://www.genscript.com/rnai.html). The sequence used avoided the conserved LIM-homeobox domains and produced a specific hit for only RUNX1T1 in the GenBank database. Three oligonucleotide sequences were designed and were as follows: Runx1t1-RNAi (24675-2), 5′-TGAGCCTTTGCACTCAGAACATCTCGAGATGTTCTGAGTGCAAAGGCTCTTTTTTC-3′; Runx1t1-RNAi (24676-3), 5′-TAAGCAAGCGACCATGCACTATCTCGAGATAGTGCATGGTCGCTTGCTTTTTTTTC-3′; and Runx1t1-RNAi (24677-1), 5′-TCAGCGGTACAGTCCAAATAATCTCGAGATTATTTGGACTGTACCGCTGTTTTTTC-3′. The underlined letters denote the hairpin loop. The negative control (NC) sequence was 5′-CCGGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTG-3′. Vectors carrying the three oligonucleotide sequences were transfected into the hippocampal RGCs, and the most effective RUNX1T1-RNAi vector, as determined using quantitative real-time reverse transcription polymerase chain reaction (PCR), was selected for the subsequent experiments.
A GV287 lentiviral expression system (Shanghai GeneChem Co.) was used to acquire the RUNX1T1-overexpressing lentivirus LV4-Runx1t1 and the NC lentivirus LV4-NC (1 × 109 TU/ml). In brief, the full length of RUNX1T1 cDNA (NM_001108657) was obtained by oligonucleotide synthesis [21, 22]. The full length sequence was decomposed into short sequences of DNA, synthesized individually. The short sequences were then connected into the full-length sequence using T4 DNA ligase and amplified using PCR. The obtained target gene was inserted into the lentiviral vector GV287 and generated GV287-RUNX1T1-enhanced green fluorescent protein (EGFP) containing the full length of RUNX1T1. Next, GV287-RUNX1T1-EGFP was transfected into 293T cells. Finally, 48 hours later, the viral supernatants were harvested, and the titer was determined (2 × 108 TU/ml). The NC lentiviral vector LV4-NC was also purchased from Shanghai GeneChem Co.
Transfection and Stable Clones
For the transfection experiments, the cells were divided into six groups. For siRNA interference, adherent cells were cultured in expansion medium containing 30 μl of 1 × 108 TU/ml LV3-RUNX1T1-RNAi or 10 μl of 3 × 108 TU/ml LV3-NC with 8 μg/ml polybrene (Shanghai GeneChem Co.). After incubation for 12 hours, the culture medium was replaced with fresh expansion medium without lentivirus. For RUNX1T1 overexpression, adherent cells were cultured in expansion medium containing 20 μl of 2 × 108 TU/ml LV4-Runx1t1 or 4 μl of 1 × 109 TU/ml LV4-NC with 8 μg/ml polybrene for 24 hours. Next, the medium was replaced with fresh expansion medium.
In all control groups, adherent cells were cultured in expansion medium. Five days later, Runx1t1 protein and RNA expression in the RGCs was analyzed using real-time PCR, Western blot, and immunocytochemistry.
RGC Differentiation Culture
For the differentiation experiments, the adherent cells after lentiviral transfection and the cells in the control groups were transferred to DMEM/F12 medium with 2% B27 and 2% fetal bovine serum (FBS; Gibco, Grand Island, NY, http://www.invitrogen.com) to promote neuronal differentiation. Seven days later, the differentiated cells were processed for immunocytochemistry.
RNA Extraction and Real-Time PCR
Total RNA was isolated using a UNIQ-10 Spin Column RNA Purification Kit (Sangon, Shanghai, People’s Republic of China, http://www.sangon.com). First-strand cDNA was synthesized using the RevertAid First-Strand cDNA Synthesis Kit (Fermentas, Burlington, ON, Canada). First-strand cDNA was subsequently processed with the Corbett RG-6000 PCR system (Qiagen, Dusseldorf, Germany, http://www.qiagen.com) using FastStart Universal SYBR Green Master Mix (Roche Diagnostics, Basel, Switzerland, http://www.roche-applied-science.com). The reactions were optimized by varying the annealing temperatures from 48°C to 55°C. The sense and antisense primers were synthesized as follows: glyceraldehyde-3-phosphate dehydrogenase, 5′-GCAAGTTCAACGGCACAG-3′, 5′-GCCAGTAGACTCCACGACAT-3′; and RUNX1T1, 5′-CCATTGCCCACCACTA-3′, 5′-CCACTCTTCTGCCCATT-3′, respectively.
Western Blot Assay
For Western blot analysis, total protein was isolated using RIPA Lysis Buffer (Beyotime, Jiangsu, People’s Republic of China), and the protein concentration was determined using the Enhanced BCA Protein Assay Kit (Beyotime). Equal amounts of protein were resolved on sodium dodecyl sulfate polyacrylamide gel electrophoresis using 10% separation gels. The gels were transferred to polyvinylidene fluoride membranes using Bio-Rad Semi-Dry Transfer Cell (Bio-Rad, Hercules, CA, http://www.bio-rad.com) at 15 V for 45 minutes and then blocked with 5% milk in Tris-buffered saline Tween buffer. The membranes were incubated overnight with primary antibody rabbit anti-RUNX1T1 (1:300; Abcam, Cambridge, U.K., http://www.abcam.com) and mouse anti-β-actin (1:2,000; Beyotime) at 4°C. After incubation with horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibody (1:2,000), the membranes were washed and immunoreactive proteins scanned using a Chemidoc XRS system (Bio-Rad). The optical density of the membrane was measured, and the relative expression of RUNX1T1 protein in the different groups was determined semiquantitatively using Quantity One software (Bio-Rad).
Immunofluorescence Staining Analyses
The cells were fixed in 4% paraformaldehyde and incubated with primary antibodies at 4°C for 48 hours. Next, the cells were incubated overnight with secondary antibodies conjugated to fluorescein 488 and 594 at 4°C. The primary antibodies used were as follows: rabbit anti-brain lipid-binding protein (BLBP; 1:1,000), rabbit anti-glial fibrillary acidic protein (GFAP; 1:1,000), mouse anti-vimentin (1:100), mouse anti-nestin (1:100), rabbit anti-Sox2 (1:100), rat anti-RUNX1T1 (1:300), and mouse anti-MAP-2 (1:200). All primary antibodies were purchased from Millipore (Billerica, MA, http://www.millipore.com) and Abcam. The cell nuclei were counterstained with Hoechst (Sigma-Aldrich). After double-label or triple-label immunofluorescence staining for cellular markers and EGFP, the cells were observed using an Olympus laser confocal microscope (Fv10i; Olympus, Tokyo, Japan, http://www.olympus-global.com). Positively stained cells were counted in five randomly selected microscopic visual fields per well. Fluorescent intensities were determined using Leica Qwin software (Leica Microsystems, Wetzlar, Germany, http://www.leica-microsystems.com).
Statistical Analysis
Data from the experiments were subjected to one-way analysis of variance using SPSS, version 11.5, statistical software. All data are expressed as the mean ± SEM, and all experimental data were obtained from a minimum of three independent experiments.
Results
RGC Culture and Identification
Typical RGCs simultaneously express astroglial and stem/progenitor markers and present a bipolar morphology. When the hippocampal cells were cultured in stem/progenitor cell expansion medium for 3–5 days, the new-formed neurospheres (Fig. 1A) dissociated into single cells and were replated onto PLL-coated coverslips. Three days later, these cells had grown long and thin processes and displayed the typical bipolar morphology of RGCs (Fig. 1B). In the present report, we used the neural progenitor markers nestin and Sox2 and the astroglial markers vimentin, BLBP, and GFAP to identify the cells. RGCs coexpressed nestin (Fig. 1C2), Sox2 (Fig. 1D2), BLBP (Fig. 1E2), vimentin (Fig. 1F1) with GFAP (Fig. 1C1, 1D1, 1E1, 1F2). The number of double-labeled RG-like cells versus the total number of cells labeled with Hoechst was determined. The ratio of nestin- to GFAP-positive cells was 95.43% ± 3.03%, that of Sox2-to GFAP-positive cells was 94.24% ± 3.71%, that of BLBP- to GFAP-positive cells was 94.04% ± 2.95%, and that of vimentin- to GFAP-positive cells was 94.28% ± 3.33% (Fig. 1G).
Figure 1.
Hippocampal radial glial cells (RGCs) acquired from the neurosphere under adherent conditions expressed radial glial markers in vitro. The new-formed neurospheres (A) acquired from neonatal rat hippocampi were dissociated and replated under adherent conditions. Three days later, the cells had extended processes and presented a bipolar morphology (B). The RGCs coexpressed the stem/progenitor markers (green) nestin (C2) and Sox2 (D2), and the astroglial markers GFAP (red in C1, D1, and E1, but green in F2), BLBP (green, E2), and vimentin (red, F1). The ratio of nestin- to GFAP-positive cells was 95.43% ± 3.03%, that of Sox2- to GFAP-positive cells was 94.24% ± 3.71%, that of BLBP- to GFAP-positive cells was 94.04% ± 2.95%, and that of vimentin- to GFAP-positive cells was 94.28% ± 3.33% (G). Scale bars = 50 μm. Abbreviations: BLBP, brain lipid-binding protein; GFAP, glial fibrillary acidic protein.
Efficiency of siRNA-Mediated Runx1t1 Knockdown
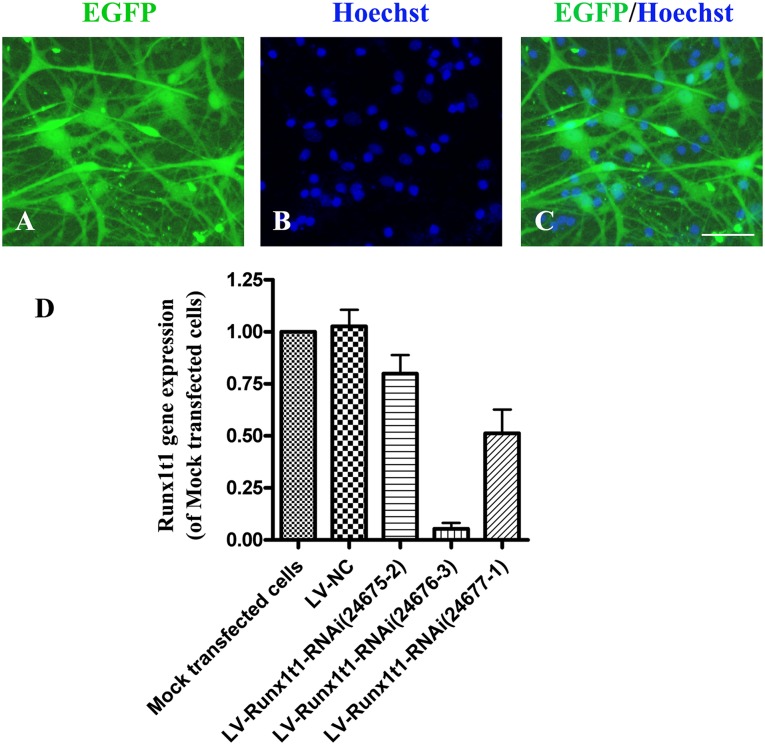
For the siRNA-mediated Runx1t1 knockdown experiments, we constructed 3 vectors carrying 3 different oligonucleotide sequences for knocking down Runx1t1 and used the vectors to transfect hippocampal RGCs. Three days later, we found that ∼70% cells expressed EGFP (Fig. 2A–2C). Real-time PCR showed that among the 3 oligonucleotide sequences, Runx1t1-RNAi (24676-3) caused obvious knockdown of the Runx1t1 gene. The RUNX1T1 gene expression level in the LV-RUNX1T1-RNAi group (24676-3) was ∼18-fold lower than that in the mock transfected cell group and ∼20-fold lower than that in the LV-NC group. The difference between the LV-RUNX1T1-RNAi (24676-3) group and the mock transfected cell/LV-NC groups was statistically significant (p < .01). However, in the LV-RUNX1T1-RNAi (24675-2) group and the LV-RUNX1T1-RNAi (24677-1) group, the slightly downregulated Runx1t1 gene expression was not significantly different from the expression in the mock transfected cells and LV-NC groups (Fig. 2D). These results have demonstrated that transfection of the hippocampal RGCs with Runx1t1-RNAi (24676-3) markedly downregulated the Runx1t1 gene. Therefore, LV-Runx1t1-RNAi (24676-3) was used in the subsequent experiments.
Figure 2.
Screening the most effective Runx1t1-RNAi vector. After transfection with LV-Runx1t1-RNAi, ∼70% cells expressed EGFP (A, C). The cell nucleus was labeled with Hoechst (B). Scale bar = 50 μm. Runx1t1 gene expression in different groups (D). Among the 3 oligonucleotide sequences, Runx1t1-RNAi (24676-3) caused obvious knockdown of the Runx1t1 gene. p < .01. Abbreviations: EGFP, enhanced green fluorescent protein; NC, negative control; RNAi, RNA interference.
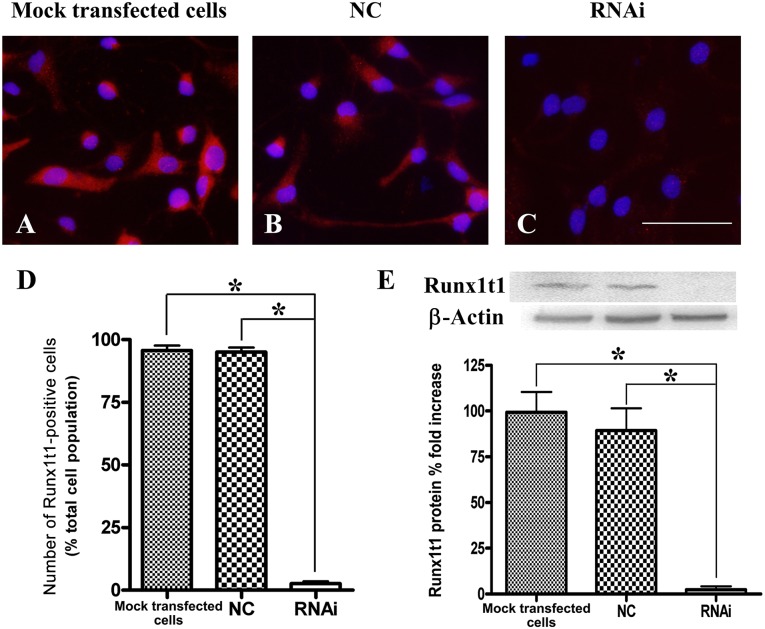
We next cocultured LV-NC and LV-Runx1t1-RNAi (Runx1t1-RNAi 24676-3), and 5 days later, the cells were processed for immunocytochemistry and total protein extraction. In order to detect the interference effect of the siRNA, we first determined the number of immunopositive cells. Almost all the cells in the mock transfected cells group and LV-NC group expressed RUNX1T1, which was seen as a red fluorescence (Fig. 3A, 3B), but not those in the LV-RUNX1T1-RNAi group (Fig. 3C), which exhibited nearly no red fluorescence. The number of RUNX1T1-immunopositive cells in the LV-RUNX1T1-RNAi group (2.63% ± 1.62%) was significantly lower than that in the mock transfected cell group (95.72% ± 3.31%) and the LV-NC group (95.03% ± 3.18%; Fig. 3D). To detect RUNX1T1 protein expression using Western blot analysis, total protein was extracted in the three groups and subjected to semiquantitative assays. The data showed that the RUNX1T1 protein level in the LV-RUNX1T1-RNAi group was significantly lower than that in the mock transfected cell group and LV-NC group (Fig. 3E). These results have indicated that LV-RUNX1T1-RNAi can effectively reduce the expression of the Runx1t1 protein and gene in RGCs.
Figure 3.
Efficiency of small interfering RNA-mediated Runx1t1 knockdown. After transfection with LV-Runx1t1-RNAi, most cells in the mock transfected cell group and LV-NC group expressed RUNX1T1 (red fluorescence; A, B) but not in the LV-RUNX1T1-RNAi group (C). Scale bar = 50 μm. The number of RUNX1T1-immunopositive cells in the LV-RUNX1T1-RNAi group (2.63% ± 1.62%) was significantly lower than that in the mock transfected cell group (95.72% ± 3.31%) and the LV-NC group (95.03% ± 3.18%) (D). The RUNX1T1 protein level in the LV-RUNX1T1-RNAi group was also significantly lower than that in the mock transfected cell and LV-NC groups (E). ∗, p < .01. Abbreviations: NC, negative control; RNAi, RNA interference.
Runx1t1 Knockdown Decreased Neuronal Differentiation of Hippocampal RGCs
To examine whether RUNX1T1 knockdown decreased the neuronal differentiation of RGCs, these cells were transfected and then transferred to DMEM/F12 medium containing 2% B27 and 2% FBS to promote neuronal differentiation. Seven days later, the differentiated cells were processed for immunocytochemistry.
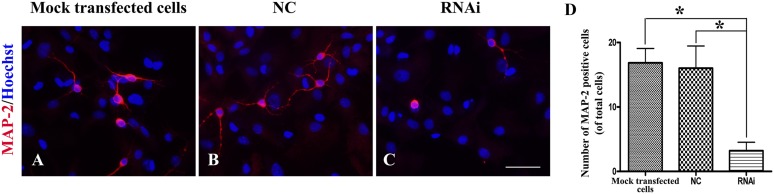
We found that in the mock transfected cell group, LV-NC group (Fig. 4A, 4B), and LV-RUNX1T1-RNAi group (Fig. 4C), the cells expressed MAP-2. The mock transfected cell group and LV-NC group showed about 17% (16.83% ± 3.88%) and 16% (16.00% ± 6.00%) MAP-2-positive cells, respectively (Fig. 4D). However, in the LV-RUNX1T1-RNAi group, fewer than ∼3.5% cells (3.20% ± 2.31%) expressed MAP-2 (Fig. 4D). The Student t test showed a significant difference between the mock transfected cell/LV-NC groups and the LV-RUNX1T1 group (p < .05; Fig. 4). In addition, we observed that the processes of the MAP-2-positive cells in the LV-RUNX1T1-RNAi group were shorter, less abundant, and even less obvious than those in the mock transfected cell and LV-NC groups.
Figure 4.
Runx1t1 knockdown decreased neuronal differentiation of hippocampal radial glial cells. Compared with the mock transfected cell group and LV-NC groups, the LV-Runx1t1-RNAi group showed fewer MAP-2-positive cells, with shorter, fewer, and less obvious processes (A–C). Scale bar = 50 μm. The number of MAP-2-positive cells differed significantly between the mock transfected cell/LV-NC groups and the LV-RUNX1T1-RNAi group (D). ∗, p < .05. Abbreviations: MAP-2, microtubule-associated protein 2; NC, negative control; RNAi, RNA interference.
Efficiency of Runx1t1 Overexpression Through LV4-Runx1t1 and Runx1t1 Overexpression Promoted the Neuronal Differentiation of Hippocampal RGCs
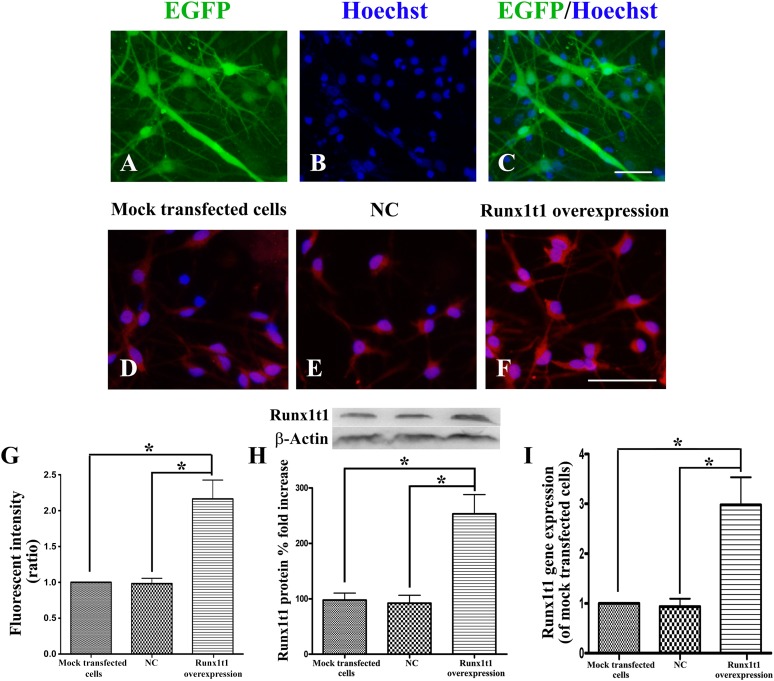
In the overexpression experiments, ∼60% cells were found to express EGFP (Fig. 5A–5C). RUNX1T1-immunopositive cells in the Runx1t1 overexpression group were stained significantly deeper than were the cells in the mock transfected cell and LV4-NC groups (Fig. 5D–5F). The number of RUNX1T1-immunopositive cells in the Runx1t1 overexpression group was also more than that in the mock transfected cell and LV4-NC groups (Fig. 5G). The RUNX1T1 protein level in the Runx1t1 overexpression group was significantly higher than that in the mock transfected cell and LV4-NC groups (Fig. 5H). The RUNX1T1 gene expression level in the Runx1t1 overexpression group was ∼20-fold higher than that in the mock transfected cell group and ∼25-fold higher than that in the LV4-NC group. The difference between the Runx1t1 overexpression group and the other groups was statistically significant (Fig. 5I). Thus, LV4-Runx1t1 markedly upregulated Runx1t1 protein and gene expression.
Figure 5.
Efficiency of Runx1t1 overexpression through LV4-Runx1t1. After transfection with LV4-Runx1t1, ∼60% cells expressed EGFP (A, C), and the cell nucleus was labeled with Hoechst (B). Almost all cells in the Runx1t1 overexpression group expressed RUNX1T1, seen as red fluorescence (F), which was significantly stronger than that in the mock transfected cell group and LV-NC group (D, E). The number of RUNX1T1-immunopositive cells in the Runx1t1 overexpression group (99.675% ± 0.32%) was higher than that in the mock transfected cell group (95.72% ± 3.31%) and the LV-NC group (95.03% ± 3.18%) (G). The RUNX1T1 protein level in the Runx1t1 overexpression group was also significantly higher than that in the mock transfected cell group and LV-NC group (H). Runx1t1 gene expression in different groups (I). ∗, p < .01. Scale bars = 50 μm. Abbreviations: EGFP, enhanced green fluorescent protein; NC, negative control.
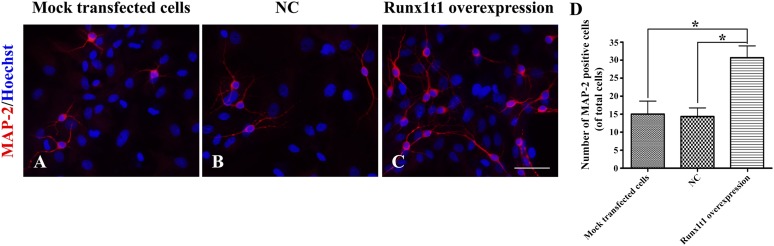
To further examine whether RUNX1T1 upregulation increased the neuronal differentiation of RGCs, these cells were also transferred to differentiation medium. We found that in the Runx1t1 overexpression group, ∼80% cells (78.31% ± 5.05%; Fig. 6) expressed MAP-2, which was significantly more than the corresponding percentages in the mock transfected cell and LV4-NC groups.
Figure 6.
Overexpression promoted the neuronal differentiation of hippocampal radial glial cells. Compared with the mock transfected cell and LV4-NC groups, the Runx1t1 overexpression group showed more MAP-2-positive cells, with more abundant and longer processes (A–C). Scale bar = 50 μm. The number of MAP-2-positive cells significantly differed between the mock transfected cell/LV4-NC groups and the Runx1t1 overexpression group (D). ∗, p < .05. Abbreviations: MAP-2, microtubule-associated protein 2; NC, negative control.
Discussion
The role of RGCs as neural progenitors and as guides for migrating neurons has been well-established. These cells exhibit a characteristic bipolar morphology and guide migrating neurons to the target location to become mature cells [4–6]. In addition to their role in radial migration, they are self-renewing and capable of differentiating into neurons. RGCs display neuroepithelial and astroglial properties. They express stem/progenitor markers, such as the intermediate filament protein nestin and the pluripotent stem cell transcription factor Sox2. They also show several astroglial markers, such as the astrocyte-specific glutamate transporter, GFAP, RC2, vimentin, the Ca+-binding protein S100b, and BLBP [4, 10, 23, 24]. In the present research, we applied adherent culture of NSCs to acquire and isolate neonatal hippocampal RGCs. These cells grew long and thin processes, presented with bipolar morphological features, coexpressed nestin, Sox2, BLBP, vimentin, and GFAP, and displayed the typical bipolar morphological features of RGCs.
The proneural bHLH proteins promote neurogenesis by inducing the changes in gene expression required for neuronal differentiation, and MTGs are a part of the different genes [14]. During the early stages of neurogenesis, MTGs are strongly induced by bHLH proteins, including XNGNR-1, Xath3, Xath5, and XNeu-roD, suggesting their role as a widely used regulator of neuronal differentiation [14, 25–27]. Inhibiting the function of MTG proteins in the developing chick spinal cord reduces the number of cells that undergo neuronal differentiation. Koyano-Nakagawa and Kintner [15] reported that MTG family members are expressed in a cascade during neuronal differentiation and perform functions required for cells to undergo terminal neuronal differentiation in the developing spinal cord. A number of studies have suggested that MTG family members act downstream of proneural proteins, as transcriptional corepressors, to promote neuronal differentiation [14, 15, 25–28].
Runx1t1, also termed ETO or MTG8, is a transcription factor and a member of the MTG family. Runx1t1 mRNA expression has been found in several human tissues, with the highest expression found in the brain and heart [28]. This high expression and the overall clues provided by the protein sequence and structure suggest that Runx1t1 could have a regulatory function in the differentiation of the nervous system. Many studies have shown that Runx1t1 is involved in the proliferation and differentiation of hematopoietic stem cells [19, 20]. Although extensive efforts have been made to understand the function of Runx1t1 proteins in the etiology of cancer, relatively less is known about their function in normal embryonic development. In the adult brain, ongoing neurogenesis was convincingly demonstrated in the subventricular zone and subgranular zone of the hippocampus. Hippocampal neurogenesis continuously generates new granule neurons, which integrate into the dentate gyrus [2, 3]. In the present study, we selected RGCs derived from the rat hippocampus under adherent conditions to investigate the relationship of Runx1t1 expression with neuronal differentiation. In different mouse and human neural cells, Runx1t1 is localized in both the nucleus and the cytoplasm and plays a role in the complex regulation of Runx1t1 in the cells of the nervous system [29, 30]. In our study, immunofluorescence staining showed that the nucleus and cytoplasm of RGCs exhibited red fluorescence, indicating Runx1t1 expression. After we used LV-RUNX1T1-RNAi to effectively knockdown Runx1t1 expression during the differentiation of hippocampal RGCs, we found that only 3.2% cells differentiated into MAP-2-positive neurons, less than the percentages in the mock transfected cell and LV3-NC groups. Moreover, the length and number of neuronal processes were also significantly reduced. In contrast, after we used LV4-Runx1t1 to upregulate Runx1t1 in the RGCs, more than 30% cells differentiated into MAP-2-positive neurons, which had more and longer processes. These results could indicate that in RGCs derived from the rat hippocampus, low Runx1t1 expression will lead to decreased neuronal differentiation, and high Runx1t1 expression will lead to increased neuronal differentiation in vitro.
Conclusion
Our findings have indicated that Runx1t1 expression is associated with the neuronal differentiation of RGCs derived from the rat hippocampus. We have deduced that Runx1t1 plays a very important role in regulating RGC differentiation.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant 31271138), the Natural Science Foundation of Jiangsu Province (Grant BK2012659), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author Contributions
Z.L.: conception, design, and manuscript writing; J.G.: conception, design, and financial support; L.H.: data analysis and interpretation; T.X.: data collection; Q.J.: provision of study material; T.M.: administrative support.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Alvarez-Buylla A, García-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- 2.Eriksson PS, Perfilieva E, Björk-Eriksson T, et al. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 3.Zou L, Jin G, Zhang X, et al. Proliferation, migration, and neuronal differentiation of the endogenous neural progenitors in hippocampus after fimbria fornix transection. Int J Neurosci. 2010;120:192–200. doi: 10.3109/00207450903464579. [DOI] [PubMed] [Google Scholar]
- 4.Anthony TE, Klein C, Fishell G, et al. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- 5.Gubert F, Zaverucha-do-Valle C, Pimentel-Coelho PM, et al. Radial glia-like cells persist in the adult rat brain. Brain Res. 2009;1258:43–52. doi: 10.1016/j.brainres.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 6.Merkle FT, Tramontin AD, García-Verdugo JM, et al. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci USA. 2004;101:17528–17532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunne B, Zhao S, Derouiche A, et al. Origin, maturation, and astroglial transformation of secondary radial glial cells in the developing dentate gyrus. Glia. 2010;58:1553–1569. doi: 10.1002/glia.21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voigt T. Development of glial cells in the cerebral wall of ferrets: Direct tracing of their transformation from radial glia into astrocytes. J Comp Neurol. 1989;289:74–88. doi: 10.1002/cne.902890106. [DOI] [PubMed] [Google Scholar]
- 9.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Jin G, Qin J, et al. Identification of neonatal rat hippocampal radial glia cells in vitro. Neurosci Lett. 2011;490:209–214. doi: 10.1016/j.neulet.2010.12.054. [DOI] [PubMed] [Google Scholar]
- 11.Noctor SC, Flint AC, Weissman TA, et al. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 12.Balu DT, Lucki I. Adult hippocampal neurogenesis: Regulation, functional implications, and contribution to disease pathology. Neurosci Biobehav Rev. 2009;33:232–252. doi: 10.1016/j.neubiorev.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doetsch F. A niche for adult neural stem cells. Curr Opin Genet Dev. 2003;13:543–550. doi: 10.1016/j.gde.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 15.Koyano-Nakagawa N, Kintner C. The expression and function of MTG/ETO family proteins during neurogenesis. Dev Biol. 2005;278:22–34. doi: 10.1016/j.ydbio.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Gelmetti V, Zhang J, Fanelli M, et al. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Hoshino T, Redner RL, et al. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Hug BA, Huang EY, et al. Oligomerization of ETO is obligatory for corepressor interaction. Mol Cell Biol. 2001;21:156–163. doi: 10.1128/MCB.21.1.156-163.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis JN, McGhee L, Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10. doi: 10.1016/s0378-1119(02)01172-1. [DOI] [PubMed] [Google Scholar]
- 20.Okumura AJ, Peterson LF, Lo MC, et al. Expression of AML/Runx and ETO/MTG family members during hematopoietic differentiation of embryonic stem cells. Exp Hematol. 2007;35:978–988. doi: 10.1016/j.exphem.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Gibson DG, Glass JI, Lartigue C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 22.Gibson DG, Smith HO, Hutchison CA, III, et al. Chemical synthesis of the mouse mitochondrial genome. Nat Methods. 2010;7:901–903. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- 23.Hartfuss E, Galli R, Heins N, et al. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- 24.Suh H, Consiglio A, Ray J, et al. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Y, Zhao H, Grunz H. XETOR regulates the size of the proneural domain during primary neurogenesis in Xenopus laevis. Mech Dev. 2002;119:35–44. doi: 10.1016/s0925-4773(02)00285-x. [DOI] [PubMed] [Google Scholar]
- 26.Logan MA, Steele MR, Van Raay TJ, et al. Identification of shared transcriptional targets for the proneural bHLH factors Xath5 and XNeuroD. Dev Biol. 2005;285:570–583. doi: 10.1016/j.ydbio.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 27.Seo S, Lim JW, Yellajoshyula D, et al. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Tümer Z, Møllgård K, et al. Characterization of a t(5;8)(q31;q21) translocation in a patient with mental retardation and congenital heart disease: Implications for involvement of RUNX1T1 in human brain and heart development. Eur J Hum Genet. 2009;17:1010–1018. doi: 10.1038/ejhg.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sacchi N, Tamanini F, Willemsen R, et al. Subcellular localization of the oncoprotein MTG8 (CDR/ETO) in neural cells. Oncogene. 1998;16:2609–2615. doi: 10.1038/sj.onc.1201824. [DOI] [PubMed] [Google Scholar]
- 30.Theriault FM, Nuthall HN, Dong Z, et al. Role for Runx1 in the proliferation and neuronal differentiation of selected progenitor cells in the mammalian nervous system. J Neurosci. 2005;25:2050–2061. doi: 10.1523/JNEUROSCI.5108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]