Abstract
Staphylococcus aureus is a major cause of nosocomial and community-acquired infections and represents a significant burden on the healthcare system. S. aureus attachment to medical implants and host tissue, and the establishment of a mature biofilm, play an important role in the persistence of chronic infections. The formation of a biofilm, and encasement of cells in a polymer-based matrix, decreases the susceptibility to antimicrobials and immune defenses, making these infections difficult to eradicate. During infection, dispersal of cells from the biofilm can result in spread to secondary sites and worsening of the infection. In this review, we discuss the current understanding of the pathways behind biofilm dispersal in S. aureus, with a focus on enzymatic and newly described broad-spectrum dispersal mechanisms. Additionally, we explore potential applications of dispersal in the treatment of biofilm-mediated infections.
Keywords: biofilm, dispersal, protease, nuclease, stringent response, Staphylococcus aureus
Staphylococcus aureus biofilms and infection
Staphylococcus aureus is a Gram-positive human commensal that persistently colonizes the anterior nares of approximately 20–25% of the healthy adult population, while as many as 60% are intermittently colonized (Eriksen et al., 1995; Hu et al., 1995; Kluytmans et al., 1997; Ellis et al., 2014). Studies have linked S. aureus nasal colonization to an increased risk of infection (Dall'Antonia et al., 2005; Ellis et al., 2014). As evidence, 65% of people with S. aureus infections are colonized with the same strain, whereas the percentage jumps to 80% in nosocomial infections (Weinstein, 1959; von Eiff et al., 2001; Wertheim et al., 2004). The infections that result are quite diverse, and can include acute infections, such as bacteremia and skin abscesses, that are generally caused by planktonic cells through the production of secreted toxins and exo-enzymes (Gordon and Lowy, 2008). In contrast, chronic infections are associated with a biofilm mode of growth where S. aureus can attach and persist on host tissues, such as bone and heart valves, to cause osteomyelitis and endocarditis respectively, or on implanted materials, such as catheters, prosthetic joints, and pace makers (Parsek and Singh, 2003; Kiedrowski and Horswill, 2011; Barrett and Atkins, 2014; Chatterjee et al., 2014). Implanted materials become coated with host proteins upon insertion, and the matrix-binding proteins on the surface of S. aureus facilitate attachment to these proteins and development of a biofilm (Cheung and Fischetti, 1990; Francois et al., 1996). In cases of infected medical devices, removal of the device is often necessary to treat the infection (Darouiche, 2004).
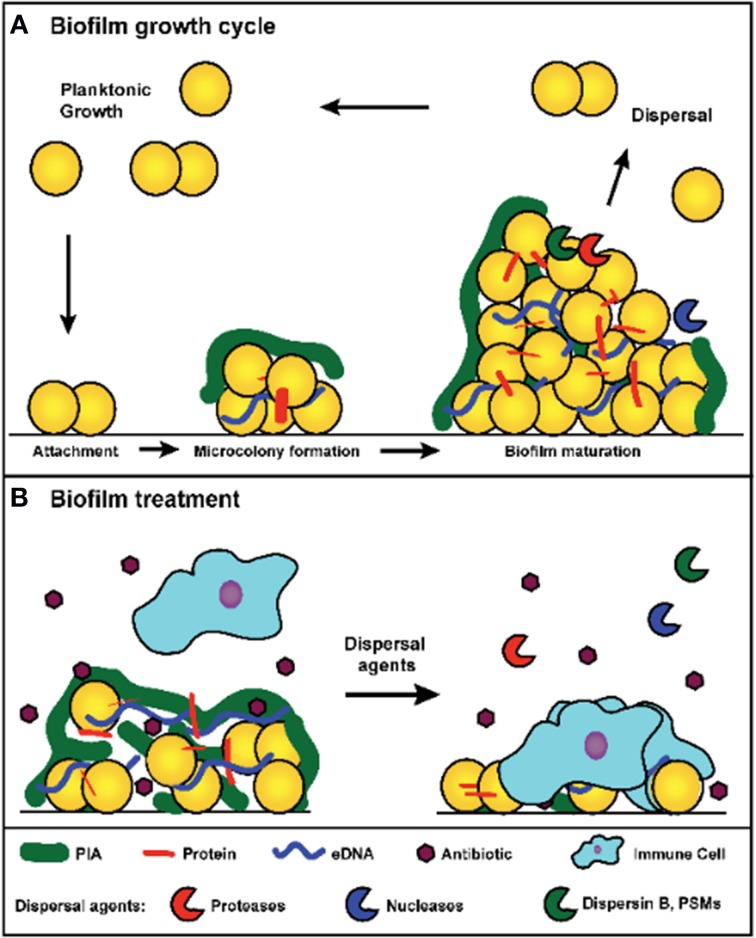
A biofilm is defined as a sessile microbial community in which cells are attached to a surface or to other cells and embedded in a protective extracellular polymeric matrix. This mode of growth exhibits altered physiologies with respect to gene expression and protein production (Parsek and Singh, 2003; Archer et al., 2011; Kiedrowski and Horswill, 2011). Biofilm developmental stages have been defined by many and can be divided into at least three major events: initial attachment, biofilm maturation, and dispersal (Figure 1A). During initial attachment, an individual planktonic cell will reversibly associate with a surface, and if the cell does not dissociate, it will bind irreversibly to the surface. Attachment is mediated through surface proteins, referred to as microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (Foster et al., 2014). During infection, these proteins play major roles in attachment to host factors such as fibrinogen, fibronectin, and collagen. Biofilm maturation occurs through cell division and the production of the extracellular polymeric matrix. The composition of the biofilm matrix varies between strains, but in general can contain host factors, polysaccharide, proteins, and extracellular DNA (eDNA) (Montanaro et al., 2011; Cue et al., 2012; Foster et al., 2014). Following biofilm accumulation, cells within the biofilm can reactivate to a planktonic state through dispersal (Boles and Horswill, 2011). The major mechanisms of S. aureus dispersal will be explored in this review.
Figure 1.
(A) Model of S. aureus biofilm growth cycle. In summary, upon coming into contact with a surface, planktonic cells attach through surface-associated proteins. Following attachment, cells divide and begin production of the extracellular matrix, which leads to the formation of a microcolony. As cell division continues, biomass accumulates and a mature biofilm is formed. Environmental signals within the biofilm trigger the activation of dispersal mechanisms, and upon dispersal, cells re-enter a planktonic growth state and can seed new sites for biofilm formation. (B) Treatment of a S. aureus biofilm. Antibiotic exposure will kill susceptible planktonic cells and metabolically active cells near the surface of the biofilm. However, persister cells and metabolically dormant cells within the biofilm survive and remain protected from immune defenses by the biofilm matrix. Treatment with dispersal agents increases the effectiveness of antibiotic penetration and promotes clearance. Antibiotic sensitive cells within the biofilm are exposed and killed after degradation of the matrix, and the antibiotic tolerant cells (such as persisters) survive and are susceptible to the immune system.
Growth in a biofilm plays an important role during infection by providing a defense against several clearance mechanisms. The biofilm matrix can impede the access of certain types of immune defenses, such as macrophages, which display incomplete penetration into the biofilm matrix and “frustrated phagocytosis” (Scherr et al., 2014). Additionally, biofilm cells display increased tolerance to antibiotics (de la Fuente-Nunez et al., 2013). In contrast to heritable antibiotic resistance mechanisms, biofilm-associated antibiotic tolerance is a transient state in which normally susceptible bacteria enter an altered physiology that decreases sensitivity. When these cells disperse and reenter a planktonic state, they regain normal antibiotic sensitivity (Singh et al., 2009). One suggested mechanism for this phenomenon is that the biofilm matrix blocks access to actively growing cells within the biofilm by decreasing antibiotic diffusion rates. However, this mechanism is dependent on the type of antibiotic, as certain antibiotics are capable of penetrating the biofilm (Singh et al., 2010). An alternative proposal is that antibiotic tolerance is due to the development of physiologically dormant persister cells that form stochastically during biofilm growth (Lewis, 2010). Due to their decreased metabolic activity, they are inherently resistant to antibiotics. Furthermore, persister cells develop at greater rates within a biofilm than within actively growing planktonic cultures (Singh et al., 2009). As such, they are thought to play a large role in the recalcitrance of biofilm-associated infections.
Beyond offering resistance to clearance mechanisms, biofilms also play an important role in the progression of chronic diseases. Following the establishment of a biofilm, individual cells can disperse from the original biofilm and either seed new sights of infection or mediate an acute infection such as sepsis (Costerton et al., 1999). The role played by the S. aureus quorum sensing system during dispersal supports this model (Boles and Horswill, 2008; Lauderdale et al., 2010). Dispersal has been the focus of many recent studies due to its importance in chronic infections and the biofilm model of growth, and an analysis of major dispersal mechanisms has led to the development of dispersal-mediated treatment options for biofilm infections (Kaplan, 2010; Boles and Horswill, 2011). This review discusses the major mechanisms for S. aureus biofilm dispersal. In addition, it analyzes the potential for developing dispersal-mediated treatments for biofilm infections (Figure 1B).
The Staphylococcus aureus biofilm matrix
The S. aureus biofilm matrix is a complex glue that encases all of the cells in the mature structure, and it is thought to be composed of host factors, secreted and lysis-derived proteins, polysaccharide, and eDNA. The contribution of each of these factors depends heavily on the strain background and on environmental conditions (Fitzpatrick et al., 2005; Abraham and Jefferson, 2012). Furthermore, the effectiveness of many dispersal mechanisms is dependent on the matrix composition (Chaignon et al., 2007; Izano et al., 2008). A brief background on the major components of the biofilm matrix and factors involved in generating these components will be provided.
A major constituent of the biofilm matrix is polysaccharide intercellular adhesin (PIA), also known as polymeric N-acetyl-glucosamine (PNAG) (O'Gara, 2007). PIA is an important component in both S. aureus and S. epidermidis biofilms that is produced by enzymes encoded in the icaADBC locus. PIA is composed of β-1,6-linked N-acetylglucosamine polymer, and the proteins encoded in the ica locus are responsible for the synthesis, export, and modification of PIA. The PIA polymer plays an important role in the structural integrity of biofilms in vitro and in vivo, although numerous studies have identified S. aureus strains capable of forming ica-independent biofilms (Beenken et al., 2003; Fitzpatrick et al., 2005; Toledo-Arana et al., 2005; Lauderdale et al., 2009; Brooks and Jefferson, 2014). The matrix components of these biofilms were later identified as proteins and eDNA (O'Neill et al., 2007, 2008; Rhode et al., 2007; Boles et al., 2010), which function as intercellular adhesins in the absence of PIA.
Many proteins have been implicated as important components in attachment and biofilm matrix development. These include surface-associated proteins such as protein A, fibrinogen-binding proteins (FnBPA and FnBPB), S. aureus surface protein (SasG), biofilm-associated protein (Bap), and clumping factor B (ClfB) (Cucarella et al., 2001; Corrigan et al., 2007; O'Neill et al., 2008; Merino et al., 2009; Geoghegan et al., 2010; Abraham and Jefferson, 2012). Many of these factors play a role both in attachment and accumulation. In addition, secreted proteins such as extracellular adherence protein (Eap), and beta toxin (Hlb) play a role in biofilm maturation (Huseby et al., 2010; Sugimoto et al., 2013). However, the importance of individual proteins varies largely between strains (Artini et al., 2013). For example, Bap-dependent biofilms have not been identified in any human isolates, and as such it is more likely that Bap plays a role in bovine mastitis (where it was originally identified) than in human diseases (Lasa and Penades, 2006). In addition to dedicated matrix proteins, intracellular proteins have been identified within the biofilm matrix. These proteins are likely released by cell lysis and nonspecifically incorporated into the matrix (Foulston et al., 2014). The relative importance of lysis-derived proteins is not yet understood.
The most recently identified and appreciated biofilm matrix component is eDNA. Due to the negative charge of the DNA polymer, eDNA potentially acts as an electrostatic polymer that anchors cells to a surface, to host factors, and to each other. Early biofilms are most sensitive to DNase treatment, suggesting that eDNA may be important during attachment (Mann et al., 2009). eDNA is produced through the autolysis of a subpopulation of the biofilm cells (Thomas and Hancock, 2009), and this altruistic suicide is mediated through the activity of murein hydrolases, encoded by the atl and lytM genes. Murein hydrolases degrade peptidoglycan and typically play an important role during cell wall rearrangements and cell division. Increased expression of these enzymes allows for autolysis in S. aureus. Autolysis is regulated through the activity of two operons, cidABC and lrgAB, that function together in a manner similar to bacteriophage holin/antiholin systems (Sadykov and Bayles, 2012). CidA, the holin in this system, oligomerizes in the cell membrane and results in the formation of a pore that is utilized for the transport of the murein hydrolase. LrgAB functions as the antiholin and prevents the activity of CidA. Studies have indicated that the regulation of autolysis is tied to micro-environmental niches that form within a biofilm, such as the hypoxic conditions found near the base of the biofilm (Moormeier et al., 2013).
There are some reported examples of interactions between eDNA and specific proteins within the biofilm. The best characterized example in S. aureus is beta toxin (Huseby et al., 2010), which is a secreted neutral sphingomyelinase capable of lysing erythrocytes and lymphocytes. However, it is structurally related to the DNaseI superfamily of proteins and is able to bind DNA. Beta toxin forms insoluble oligomers upon binding DNA that could serve as a bridge to hold the biofilm structure together. Deletion of the hlb gene correlates with a reduction in biofilm formation in both in vitro and in vivo models. Additional studies have implicated that proteins with non-specific DNA-binding activity may be important matrix components in multiple bacterial species, as antibodies against IHF, a common member of the DNABII family of proteins, are capable of disrupting existing biofilms in in vitro and in vivo models (Goodman et al., 2011; Novotny et al., 2013).
Biofilm dispersal mechanisms
The primary biofilm dispersal strategy utilized by S. aureus is the production of various exo-enzymes and surfactants to degrade the extracellular polymeric matrix. The effectiveness of individual mechanisms is highly dependent on the matrix composition of the S. aureus strain in question (Chaignon et al., 2007; Kiedrowski et al., 2011). In general, mechanisms utilizing the enzymatic self-destruction of either protein and/or eDNA in the matrix are less effective at dispersing polysaccharide-dependent biofilms. In contrast, the mechanisms specifically targeting PIA are ineffective against polysaccharide-independent biofilms. In this review, the dispersal mechanisms targeting each matrix component will be discussed, with an emphasis on self-targeting enzymatic mechanisms (Table 1), and two recently described fundamental processes with biofilm dispersing activity will also be covered. Non-specific mechanisms, such as the surfactant activity of phenol-soluble modulins (PSMs), are effective against most S. aureus biofilms and are reviewed elsewhere (Peschel and Otto, 2013).
Table 1.
Biofilm dispersal mechanisms.
| Dispersal agent | Mechanism | References | Specific factor |
|---|---|---|---|
| Proteases | Degradation of proteinaceous matrix components | McGavin et al., 1997; O'Neill et al., 2008 | V8 protease (SspA) |
| Mootz et al., 2013 | Staphopains (Cysteine Proteases) | ||
| Abraham and Jefferson, 2012 | Aureolysin (Aur) | ||
| Marti et al., 2010 | Aur, SspA | ||
| Lauderdale et al., 2010; Shukla and Rao, 2013 | Proteinase K | ||
| Beenken et al., 2003; Trotonda et al., 2005; Tsang et al., 2008; Zielinska et al., 2012 | sarA regulation | ||
| Lauderdale et al., 2009 | sigB, agr regulation | ||
| agr activation by AIP | Expression of agr regulated factors (proteases and PSMs) | Yarwood et al., 2004; Boles and Horswill, 2008; Lauderdale et al., 2010 | AIP |
| Phenol-soluble modulins | Surfactant-mediated dispersal | Peschel and Otto, 2013 | PSMs |
| S. epidermidis Esp | Degradation of proteinaceous matrix components; inhibition of autolysis through Atl degradation | Iwase et al., 2010; Chen et al., 2013; Sugimoto et al., 2013 | Esp |
| Nucleases | Degradation of eDNA | Kiedrowski et al., 2011 | Nuc |
| Kiedrowski et al., 2014 | Nuc2 | ||
| Dispersin B | Degradation of polysaccharide matrix components | Kaplan et al., 2004; Donelli et al., 2007 | DisB |
| D-amino acids | Protein synthesis inhibition in B. subtilis, unknown in S. aureus | Kolodkin-Gal et al., 2010; Hochbaum et al., 2011; Leiman et al., 2013; Sanchez et al., 2013 | D-amino acids |
| Stringent response inhibition | Unknown | de la Fuente-Nunez et al., 2014; Reffuveille et al., 2014 | Peptide 1018 |
Enzymatic dispersal mechanisms
Protease-mediated dispersal
S. aureus produces 10 secreted proteases, including seven serine proteases (SspA and SplA-F), two cysteine proteases (SspB and ScpA), and one metalloprotease (Aur) (Shaw et al., 2004). The role of proteases in biofilm dispersal was initially characterized during the analysis of S. aureus strains deficient in the global regulators sarA and sigB (Bronner et al., 2004) that were unable to form biofilm (Beenken et al., 2003; Trotonda et al., 2005; O'Neill et al., 2008). Characterization of these mutants revealed that the observed biofilm phenotypes resulted from elevated protease activity levels (Tsang et al., 2008; Lauderdale et al., 2009; Marti et al., 2010; Zielinska et al., 2012; Mootz et al., 2013). The high protease activity results in the degradation of important matrix proteins and destabilization of the biofilm (Zielinska et al., 2012). This phenotype could be reversed by the deletion of multiple protease genes or the addition of protease inhibitors (McGavin et al., 1997; Tsang et al., 2008; Mootz et al., 2013). The ability of the V8 serine protease (SspA), the staphopains (SspB and ScpA), and aureolysin (Aur) to disrupt biofilms have been demonstrated (Table 1), with the relative importance of each varying between strains and conditions. The V8 serine protease can degrade FnBPs and Bap (McGavin et al., 1997; O'Neill et al., 2008; Marti et al., 2010), and aureolysin can degrade ClfB and Bap to mediate biofilm disruption (Marti et al., 2010; Abraham and Jefferson, 2012). While the staphopains can disrupt the biofilm matrix, no target proteins have yet been characterized (Mootz et al., 2013). Additional targets such as Atl, Spa, and SasG have been proposed, but have not been linked to individual proteases (Lauderdale et al., 2009; Kolar et al., 2013). Despite the identification of some specific matrix proteins as targets for degradation, the large number of proteases and potential matrix protein targets will require proteomic analysis to dissect the complex mechanism behind protease-mediated dispersal.
The production of proteases is positively regulated through the S. aureus quorum sensing system, agr (Thoendel et al., 2011). The agr system is activated upon detection of an autoinducing peptide (AIP) that is encoded and produced by the agr operon. The AIP is detected by a two-component system that regulates virulence through the production of a regulatory RNA, RNAIII. The agr system regulates the virulence state of the cell by activating the production of secreted toxins and enzymes and the down-regulation of surface factors. The agr system induces the expression of both proteases and PSMs, which act as surfactants to disperse biofilms (Peschel and Otto, 2013). Thus, activation of the agr system can result in a shift from a biofilm state to a planktonic state of growth. This has been demonstrated through the addition of AIP to existing biofilms, which results in complete dispersal (Boles and Horswill, 2008; Lauderdale et al., 2010), and through the use of fluorescent reporters, which demonstrated that cells detach from the biofilm after agr activation (Yarwood et al., 2004).
In addition to native S. aureus proteases, recent studies have indicated that the production of non-native proteases may impact S. aureus biofilm growth in bacterial communities. The serine protease Esp produced by S. epidermidis has been shown to disperse S. aureus biofilms (Sugimoto et al., 2013). This was first identified when it was observed that S. aureus colonization rates of the human nares negatively correlate with colonization rates of esp positive S. epidermidis (Iwase et al., 2010). Following this discovery, it was shown that Esp is able to cleave an array of S. aureus proteins, including Eap, FnBPA, and Atl (Chen et al., 2013; Sugimoto et al., 2013). The mechanism of Esp-mediated dispersal is thus two-fold: Esp degrades matrix proteins important for intercellular adhesion and prevents the release of eDNA by degrading murein hydrolase.
Nuclease-mediated dispersal
S. aureus produces two extracellular nucleases, referred to here as nuclease (Nuc) and nuclease2 (Nuc2) (Tang et al., 2008). The production of the major secreted Staphylococcal nuclease, also known as micrococcal nuclease or thermonuclease, is conserved across most clinical isolates and is produced in vivo. A recent study utilized this fact and developed a nuclease-specific probe for imaging S. aureus infections (Hernandez et al., 2014). Nuclease is regulated by the global regulator sigB and the SaeRS two-component system (Kiedrowski et al., 2011; Olson et al., 2013), and the expression of nuc is greatly reduced during biofilm growth conditions, suggesting that Nuc may play a role in the biofilm growth cycle (Olson et al., 2013).
Two major roles have been proposed for Nuc during infection, the disruption of neutrophil extracellular traps (NETs) and modulating biofilm development. It has been shown that the expression of nuclease results in reduced biofilm formation in vitro, while a nuc mutant displays enhanced biofilm formation (Mann et al., 2009; Kiedrowski et al., 2011). These phenotypes correlate with levels of eDNA accumulation during biofilm growth, where lack of nuclease results in the preservation of high molecular weight eDNA (Mann et al., 2009; Kiedrowski et al., 2011). This agrees with an earlier study that found a minimum size of 11 kb fragments was necessary for biofilm integrity (Izano et al., 2008). The second role proposed for nuclease during infection is the evasion of NETs. NETs are a newly discovered killing mechanism utilized by neutrophils against bacterial infections. Activated neutrophils secrete nuclear DNA at the site of infection to entrap bacteria and enhance bacterial killing. Nuclease is able to degrade NETs and promote resistance against killing by neutrophils (Berends et al., 2010; Thammavongsa et al., 2013). The relative importance of each activity during infection has not yet been explored. Overall, in vivo studies indicate that nuc mutants are attenuated during infection (Berends et al., 2010; Olson et al., 2013). However, it is unclear whether this attenuation results from a reduced ability to disperse from a biofilm and disseminate to new sites, an increased susceptibility to killing by neutrophils, or the inability to scavenge nucleotides in the host. It is possible that all these functions of nuclease are important during infection.
In contrast to nuclease, the function of Nuc2 is still relatively unknown. This is in part due to the difficulty in studying Nuc2 in wild type backgrounds, as its activity is masked by Nuc. A recent study has shown that Nuc2 is a membrane-bound nuclease with an extracellular catalytic domain. Nuc2 activity is detectable in a nuc mutant, but the activity is very low (Kiedrowski et al., 2014). This is likely due to low expression levels, as mechanistic studies demonstrated that the Nuc2 catalytic domain is functional. Addition of purified Nuc2 was able to partially disperse existing biofilms, suggesting that Nuc2 could play a role in localized dispersal during infection. This localized dispersal could result in the formation of channels within the biofilm or supplement Nuc activity in high flow environments (such as those seen during endocarditis) where Nuc would be unable to accumulate. However, further studies will be necessary to determine the function of Nuc2 in vivo.
Dispersin B-mediated dispersal
The enzyme dispersin B isolated from Actinobacillus actinomycetemcomitans is able to disperse polysaccharide-dependent Staphylococcus epidermidis and S. aureus biofilms (Kaplan et al., 2004). Dispersin B disrupts the biofilm by hydrolyzing the glycosidic linkages of PIA. No homolog of dispersin B has been identified in the S. aureus genome so it is unlikely the organism utilizes this mechanism for dispersal during biofilm growth. However, treatment of biofilms with dispersin B does result in increased susceptibility to antimicrobials (Donelli et al., 2007). Thus, dispersin B could be developed as a potential anti-biofilm treatment.
Broad-spectrum dispersal mechanisms
D-amino acids
It has been reported that D-amino acids produced during late stationary phase induce biofilm dispersal in multiple bacteria, including S. aureus (Kolodkin-Gal et al., 2010). The role of D-amino acids in dispersal was initially discovered in Bacillus subtilis. The proposed mechanism behind this dispersal was the incorporation of D-amino acids into the peptidoglycan, resulting in a failure to attach the major matrix protein, TasA, to the cell wall. Subsequently, this resulted in decreased intercellular adhesion via the detachment of existing TasA fibers. This dispersal mechanism was tested in additional bacterial species, including S. aureus and P. aeruginosa, where a similar phenotype were observed (Hochbaum et al., 2011).
However, a recent study has found that the effect of D-amino acids observed in B. subtilis was due to a strain specific mutation in the dtd gene (Leiman et al., 2013). dtd encodes a D-tyrosyl-tRNA deacylase and is responsible for preventing the misincorporation of D-amino acids into protein. As such, the D-amino acid biofilm dispersal effect observed in the dtd mutant was due to a growth defect caused by interference with protein synthesis. The impact of D-amino acids on S. aureus biofilm is therefore unclear and requires further investigation. However, D-amino acids may still offer clinical applications for the prevention of biofilm infections. It has been shown that pre-treatment of polymeric surfaces with D-amino acids reduces S. aureus biofilm formation in vitro (Hochbaum et al., 2011; Sanchez et al., 2013).
Stringent response
The stringent response is a general bacterial system triggered by nutrient starvation that allows cells to adapt to stressful conditions, such as those seen during infection (Srivatsan and Wang, 2008). During nutrient starvation, the alarmone ppGpp is produced by RelA/SpoT homologs and elicits regulatory changes that switch the cell to a metabolically inactive state. Studies have linked the stringent response to virulence and biofilm formation in multiple bacterial species (Lemos et al., 2004; Nguyen et al., 2011; Vogt et al., 2011; Chavez de Paz et al., 2012; He et al., 2012; Wexselblatt et al., 2012; Sugisaki et al., 2013). In S. aureus, evidence suggests the stringent response plays a role during infection (Geiger et al., 2010), but its impact on biofilm has not been extensively studied.
A recent study identified a synthetic cationic peptide capable of dispersing biofilms in a large number of clinically relevant bacterial pathogens, including S. aureus, without inhibiting planktonic growth (de la Fuente-Nunez et al., 2014). The peptide affected both Gram-negative and Gram-positive organisms, implicating that the peptide was targeting a general bacterial process. Further investigation determined that the peptide was inhibiting the stringent response through a direct interaction with ppGpp that resulted in the degradation of the alarmone. This result indicates that the metabolic state of the cell plays some role in dispersal. Additional research will be necessary to explore the role of stringent response in S. aureus biofilm dispersal.
Implications for clinical treatment of biofilm infections
Biofilm dispersal has drawn interest as a potential means of treating persistent S. aureus infections. The intentional dispersal of a biofilm coupled with antibiotic therapy would expose and kill metabolically active cells and render any remaining persister cells vulnerable to the immune system (Figure 1B). Increased antibiotic susceptibility has been observed with most dispersal agents, including many industrially produced enzymes such as dispersin B, proteinase K, and DNaseI (Lauderdale et al., 2010; Kaplan et al., 2012b; Shukla and Rao, 2013; Reffuveille et al., 2014). The efficacy of dispersal-mediated treatments could potentially be improved by the inclusion of a drug targeting persister cells (Conlon et al., 2013). In addition to the treatment of existing infections, dispersal mechanisms could be utilized in the prevention of biofilm formation associated with implanted medical devices. Several studies have found that pretreatment of polymeric surfaces with dispersing agents can reduce biofilm formation in vivo (Donelli et al., 2007; Sanchez et al., 2013). The slow release of dispersal agents from the implanted device should prevent biofilm accumulation and facilitate clearance of the bacteria by the immune system. While these approaches sound promising, there are several concerns that have not yet been thoroughly addressed. First, induced dispersal could result in acute infections if the antibiotic fails to eradicate the released cells. Sub-inhibitory concentrations of certain antibiotics have been linked to enhanced agr activation (Joo et al., 2010), which could accelerate an acute response. Sub-inhibitory concentrations of β-lactams have also been linked to the induction of eDNA release and biofilm formation (Kaplan et al., 2012a), which could be counter-productive when coupled with a dispersal agent. Embolisms resulting from the release of cell clumps embedded in matrix components represent another major concern. Studies will need to address these challenges before dispersal agents are tested in a clinical setting.
Concluding remarks and future perspectives
The ability to form a biofilm is an important virulence determinant for the persistence of S. aureus chronic infections. In this review, we focused on the strategies utilized by S. aureus to escape from a biofilm through dispersal and disseminate to other body sites. Ongoing research continues to improve our understanding of the exo-enzymes and surfactants that degrade the biofilm matrix and release cells into the surrounding environment. The enzymes that have drawn the most attention are the secreted cysteine proteases (staphopains), V8 serine protease (SspA), and nuclease (Nuc). The relative importance of each enzyme will depend on the strain-specific composition of the biofilm matrix. The proteases and surfactant molecules are under agr quorum-sensing control, and activation of this regulatory system is a known dispersing mechanism.
Going forward, additional studies are necessary to fill specific knowledge gaps. The targets of the major proteases (V8, Aur, staphopains) are still not fully described, although some candidate surface proteins, like the FnBPs and ClfB, have been identified. The function of Nuc in biofilm dispersal has not been examined in detail. It is likely other exo-enzymes, such as hyaluronidase and lipases, are also important in dispersal mechanisms, but have not been fully investigated in biofilm studies (Rosenthal et al., 2014). In addition to the matrix-degrading mechanisms, it is possible that D-amino acids and the stringent response may play a role in dispersal, but further work is needed to better characterize these mechanisms. Perhaps the area of greatest need is confirming dispersal mechanisms in relevant animal models of infection and testing the efficacy of dispersal agents in treating biofilm infections. Additionally, coupling these agents with antibiotic therapy to facilitate clearance of a recalcitrant infection has received little attention. Overall our knowledge of enzymatic dispersal mechanisms has expanded in recent years, but many details still remain unclear. Further work on the topic will allow for the development of better treatment options for biofilm-mediated diseases.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grant AI083211 from the National Institute of Allergy and Infectious Diseases.
References
- Abraham N. M., Jefferson K. K. (2012). Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology 158, 1504–1512. 10.1099/mic.0.057018-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer N. K., Mazaitis M. J., Costerton J. W., Leid J. G., Powers M. E., Shirtliff M. E. (2011). Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2, 445–459. 10.4161/viru.2.5.17724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artini M., Papa R., Scoarughi G. L., Galano E., Barbato G., Pucci P., et al. (2013). Comparison of the action of different proteases on virulence properties related to the staphylococcal surface. J. Appl. Microbiol. 114, 266–277. 10.1111/jam.12038 [DOI] [PubMed] [Google Scholar]
- Barrett L., Atkins B. (2014). The clinical presentation of prosthetic joint infection. J. Antimicrob. Chemother. 69 Suppl. 1, i25–i27. 10.1093/jac/dku250 [DOI] [PubMed] [Google Scholar]
- Beenken K. E., Blevins J. S., Smeltzer M. S. (2003). Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71, 4206–4211. 10.1128/iai.71.7.4206-4211.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berends E. T., Horswill A. R., Haste N. M., Monestier M., Nizet V., von Kockritz-Blickwede M. (2010). Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immun. 2, 576–586. 10.1159/000319909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles B. R., Horswill A. R. (2008). Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. 10.1371/journal.ppat.1000052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles B. R., Horswill A. R. (2011). Staphylococcal biofilm dissasembly. Trends Microbiol. 19, 449–455. 10.1016/j.tim.2011.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles B. R., Thoendel M., Roth A. J., Horswill A. R. (2010). Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE 5:e10146. 10.1371/journal.pone.0010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner S., Monteil H., Prevost G. (2004). Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28, 183–200. 10.1016/j.femsre.2003.09.003 [DOI] [PubMed] [Google Scholar]
- Brooks J. L., Jefferson K. K. (2014). Phase variation of poly-N-acetylglucosamine expression in Staphylococcus aureus. PLoS Pathog. 10:e1004292. 10.1371/journal.ppat.1004292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaignon P., Sadovskaya I., Ragunah C., Ramasubbu N., Kaplan J. B., Jabbouri S. (2007). Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl. Microbiol. Biotechnol. 75, 125–132. 10.1007/s00253-006-0790-y [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Maiti P., Dey R., Kundu A., Dey R. (2014). Biofilms on indwelling urologic devices: microbes and antimicrobial management prospect. Ann. Med. Health Sci. Res. 4, 100–104. 10.4103/2141-9248.126612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez de Paz L. E., Lemos J. A., Wickstrom C., Sedgley C. M. (2012). Role of (p)ppGpp in biofilm formation by Enterococcus faecalis. Appl. Environ. Microbiol. 78, 1627–1630. 10.1128/AEM.07036-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Krishnan V., Macon K., Manne K., Narayana S. V. L., Schneewind O. (2013). Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J. Biol. Chem. 288, 29440–29452. 10.1074/jbc.M113.502039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. L., Fischetti V. A. (1990). The role of fibrinogen in staphylococcal adherence to catheters in vitro. J. Infect. Dis. 161, 1177–1186. 10.1093/infdis/161.6.1177 [DOI] [PubMed] [Google Scholar]
- Conlon B. P., Nakayasu E. S., Fleck L. E., Lafleur M. D., Isabella V. M., Coleman K., et al. (2013). Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370. 10.1038/nature12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan R. M., Rigby D., Handley P., Foster T. J. (2007). The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153, 2435–2446. 10.1099/mic.0.2007/006676-0 [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- Cucarella C., Solano C., Valle J., Amorena B., Lasa I., Penades J. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183, 2888–2896. 10.1128/JB.183.9.2888-2896.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D., Lei M. G., Lee C. Y. (2012). Genetic regulation of the intercellular adhesion locus in staphylococci. Front. Cell. Infect. Microbiol. 2, 38. 10.3389/fcimb.2012.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Antonia M., Coen P. G., Wilks M., Whiley A., Millar M. (2005). Competition between methicillin-sensitive and -resistant Staphylococcus aureus in the anterior nares. J. Hosp. Infect. 61, 62–67. 10.1016/j.jhin.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Darouiche R. O. (2004). Treatment of infections associated with surgical implants. N. Engl. J. Med. 350, 1422–1429. 10.1056/NEJMra035415 [DOI] [PubMed] [Google Scholar]
- de la Fuente-Nunez C., Reffuveille F., Fernandez L., Hancock R. E. (2013). Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 16, 580–589. 10.1016/j.mib.2013.06.013 [DOI] [PubMed] [Google Scholar]
- de la Fuente-Nunez C., Reffuveille F., Haney E. F., Straus S. K., Hancock R. E. (2014). Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10:e1004152. 10.1371/journal.ppat.1004152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelli G., Francolini I., Romoli D., Guaglianone E., Piozzi A., Ragunath C., et al. (2007). Synergistic activity of dispersin B and cefamandole nafate in inhibition of staphylococcal biofilm growth on polyurethanes. Antimicrob. Agents Chemother. 51, 2733–2740. 10.1128/AAC.01249-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M. W., Schlett C. D., Millar E. V., Crawford K. B., Cui T., Lanier J. B., et al. (2014). Prevalence of nasal colonization and strain concordance in patients with community-associated Staphylococcus aureus skin and soft-tissue infections. Infect. Control Hosp. Epidemiol. 35, 1251–1256. 10.1086/678060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen N. H., Espersen F., Rosdahl V. T., Jensen K. (1995). Carriage of Staphylococcus aureus among 104 healthy persons during a 19-month period. Epidemiol. Infect. 115, 51–60. 10.1017/S0950268800058118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick F., Humphreys H., O'Gara J. P. (2005). Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 43, 1973–1976. 10.1128/JCM.43.4.1973-1976.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Geoghegan J. A., Ganesh V. K., Hook M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. 10.1038/nrmicro3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulston L., Elsholz A. K., Defrancesco A. S., Losick R. (2014). The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. MBio 5, e01667–e01614. 10.1128/mBio.01667-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois P., Vaudaux P., Foster T. J., Lew D. P. (1996). Host-bacteria interactions in foreign body infections. Infect. Control Hosp. Epidemiol. 17, 514–520. 10.2307/30141285 [DOI] [PubMed] [Google Scholar]
- Geiger T., Goerke C., Fritz M., Schafer T., Ohlsen K., Liebeke M., et al. (2010). Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 78, 1873–1883. 10.1128/IAI.01439-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J. A., Corrigan R. M., Gruszka D. T., Speziale P., O'Gara J. P., Potts J. R., et al. (2010). Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 192, 5663–5673. 10.1128/JB.00628-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman S. D., Obergfell K. P., Jurcisek J. A., Novotny L. A., Downey J. S., Ayala E. A., et al. (2011). Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 4, 625–637. 10.1038/mi.2011.27 [DOI] [PubMed] [Google Scholar]
- Gordon R. J., Lowy F. D. (2008). Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46, S350–S359. 10.1086/533591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Cooper J. N., Mishra A., Raskin D. M. (2012). Stringent response regulation of biofilm formation in Vibrio cholerae. J. Bacteriol. 194, 2962–2972. 10.1128/JB.00014-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez F. J., Huang L., Olson M. E., Powers K. M., Hernandez L. I., Meyerholz D. K., et al. (2014). Noninvasive imaging of Staphylococcus aureus infections with a nuclease-activated probe. Nat. Med. 20, 301–306. 10.1038/nm.3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum A. I., Kolodkin-Gal I., Foulston L., Kolter R., Aizenberg J., Losick R. (2011). Inhibitory effects of D-amino acids on Staphylococcus aureus biofilm development. J. Bacteriol. 193, 5616–5622. 10.1128/JB.05534-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Umeda A., Kondo S., Amako K. (1995). Typing of Staphylococcus aureus colonising human nasal carriers by pulsed-field gel electrophoresis. J. Med. Microbiol. 42, 127–132. 10.1099/00222615-42-2-127 [DOI] [PubMed] [Google Scholar]
- Huseby M. J., Kruse A. C., Digre J., Kohler P. L., Vocke J. A., Mann E. E., et al. (2010). Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 14407–14412. 10.1073/pnas.0911032107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T., Uehara Y., Shinji H., Tajima A., Seo H., Takada K., et al. (2010). Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349. 10.1038/nature09074 [DOI] [PubMed] [Google Scholar]
- Izano E. A., Amarante M. A., Kher W. B., Kaplan J. B. (2008). Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74, 470–476. 10.1128/AEM.02073-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H., Chan J. L., Cheung G. Y. C., Otto M. (2010). Subinhibitory concentrations of protein synthesis-inhibiting antibiotics promote increased expression of the agr virulence regulator and production of phenol-soluble modulin cytolysins in community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54, 4942–4944. 10.1128/AAC.00064-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J. B. (2010). Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89, 205–218. 10.1177/0022034509359403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J. B., Izano E. A., Gopal P., Karwacki M. T., Kim S., Bose J. L., et al. (2012a). Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. MBio 3, e00198–e00112. 10.1128/mBio.00198-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J. B., Lovetri K., Cardona S. T., Madhyastha S., Sadovskaya I., Jabbouri S., et al. (2012b). Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in staphylococci. J. Antibiot. 65, 73–77. 10.1038/ja.2011.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J. B., Velliyagounder K., Ragunath C., Rohde H., Mack D., Knobloch J. K., et al. (2004). Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186, 8213–8220. 10.1128/JB.186.24.8213-8220.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski M. R., Crosby H. A., Hernandez F. J., Malone C. L., McNamara J. O., 2nd., Horswill A. R. (2014). Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PLoS ONE 9:e95574. 10.1371/journal.pone.0095574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski M. R., Horswill A. R. (2011). New approaches for treating staphylococcal biofilm infections. Ann. N.Y. Acad. Sci. 1241, 104–121. 10.1111/j.1749-6632.2011.06281.x [DOI] [PubMed] [Google Scholar]
- Kiedrowski M. R., Kavanaugh J. S., Malone C. L., Mootz J. M., Voyich J. M., Smeltzer M. S., et al. (2011). Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS ONE 6:e26714. 10.1371/journal.pone.0026714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluytmans J., van Belkum A., Verbrugh H. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10, 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar S. L., Ibarra J. A., Rivera F. E., Mootz J. M., Davenport J. E., Stevens S. M., et al. (2013). Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2, 18–34. 10.1002/mbo3.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I., Romero D., Cao S., Clardy J., Kolter R., Losick R. (2010). D-amino acids trigger biofilm disassembly. Science 328, 627–629. 10.1126/science.1188628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I., Penades J. R. (2006). Bap: a family of surface proteins involved in biofilm formation. Res. Microbiol. 157, 99–107. 10.1016/j.resmic.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Lauderdale K. J., Boles B. R., Cheung A. L., Horswill A. R. (2009). Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77, 1623–1635. 10.1128/IAI.01036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauderdale K. J., Malone C. L., Boles B. R., Morcuende J., Horswill A. R. (2010). Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J. Orthop. Res. 28, 55–61. 10.1002/jor.20943 [DOI] [PubMed] [Google Scholar]
- Leiman S. A., May J. M., Lebar M. D., Kahne D., Kolter R., Losick R. (2013). D-amino acids indirectly inhibit biofilm formation in Bacillus subtilis by interfering with protein synthesis. J. Bacteriol. 195, 5391–5395. 10.1128/JB.00975-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos J. A., Brown T. A., Burne R. A. (2004). Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72, 1431–1440. 10.1128/iai.72.3.1431-1440.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. (2010). Persister cells. Annu. Rev. Microbiol. 64, 357–372. 10.1146/annurev.micro.112408.134306 [DOI] [PubMed] [Google Scholar]
- Mann E. E., Rice K. C., Boles B. R., Endres J. L., Ranjit D., Chandramohan L., et al. (2009). Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 4:e5822. 10.1371/journal.pone.0005822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M., Trotonda M. P., Tormo-Mas M. A., Vergara-Irigaray M., Cheung A. L., Lasa I., et al. (2010). Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microb Infect 12, 55–64. 10.1016/j.micinf.2009.10.005 [DOI] [PubMed] [Google Scholar]
- McGavin M. J., Zahradka C., Rice K. C., Scott J. E. (1997). Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect. Immun. 65, 2621–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino N., Toledo-Arana A., Vergara-Irigaray M., Valle J., Solano C., Calvo E., et al. (2009). Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191, 832–843. 10.1128/JB.01222-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L., Poggi A., Visai L., Ravaioli S., Campoccia D., Speziale P., et al. (2011). Extracellular DNA in biofilms. Int. J. Artif. Organs 34, 824–831. 10.5301/ijao.5000051 [DOI] [PubMed] [Google Scholar]
- Moormeier D. E., Endres J. L., Mann E. E., Sadykov M. R., Horswill A. R., Rice K. C., et al. (2013). Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Appl. Environ. Microbiol. 79, 3413–3424. 10.1128/AEM.00395-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz J. M., Malone C. L., Shaw L., Horswill A. R. (2013). Staphopains modulate Staphylococcus aureus biofilm integrity. Infect. Immun. 81, 3227–3238. 10.1128/IAI.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D., Joshi-Datar A., Lepine F., Bauerle E., Olakanmi O., Beer K., et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986. 10.1126/science.1211037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny L. A., Amer A. O., Brockson M. E., Goodman S. D., Bakaletz L. O. (2013). Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS ONE 8:e67629. 10.1371/journal.pone.0067629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gara J. P. (2007). ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270, 179–188. 10.1111/j.1574-6968.2007.00688.x [DOI] [PubMed] [Google Scholar]
- Olson M. E., Nygaard T. K., Ackermann L., Watkins R. L., Zurek O. W., Pallister K. B., et al. (2013). Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect. Immun. 81, 1316–1324. 10.1128/IAI.01242-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E., Pozzi C., Houston P., Humphreys H., Robinson D. A., Loughman A., et al. (2008). A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190, 3835–3850. 10.1128/JB.00167-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E., Pozzi C., Houston P., Smyth D., Humphreys H., Robinson D. A., et al. (2007). Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 45, 1379–1388. 10.1128/JCM.02280-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M. R., Singh P. K. (2003). Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57, 677–701. 10.1146/annurev.micro.57.030502.090720 [DOI] [PubMed] [Google Scholar]
- Peschel A., Otto M. (2013). Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 11, 667–673. 10.1038/nrmicro3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reffuveille F., de la Fuente-Nunez C., Mansour S., Hancock R. E. (2014). A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob. Agents Chemother. 58, 5363–5371. 10.1128/AAC.03163-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode H., Burandt E. C., Siemssen N., Frommelt L., Burdelski C., Wurster S., et al. (2007). Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28, 1711–1720. 10.1016/j.biomaterials.2006.11.046 [DOI] [PubMed] [Google Scholar]
- Rosenthal C. B., Mootz J. M., Horswill A. R. (2014). Staphylococcus aureus biofilm formation and inhibition, in Antibiofilm Agents, eds Rumbaugh K. P., Ahmad I. (New York, NY: Springer; ), 233–256. [Google Scholar]
- Sadykov M. R., Bayles K. W. (2012). The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr. Opin. Microbiol. 15, 211–215. 10.1016/j.mib.2011.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C. J., Jr., Prieto E. M., Krueger C. A., Zienkiewicz K. J., Romano D. R., Ward C. L., et al. (2013). Effects of local delivery of D-amino acids from biofilm-dispersive scaffolds on infection in contaminated rat segmental defects. Biomaterials 34, 7533–7543. 10.1016/j.biomaterials.2013.06.026 [DOI] [PubMed] [Google Scholar]
- Scherr T. D., Heim C. E., Morrison J. M., Kielian T. (2014). Hiding in plain sight: interplay between staphylococcal biofilms and host hmmunity. Front. Immunol. 5:37. 10.3389/fimmu.2014.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L., Golenka E., Potempa J., Foster S. J. (2004). The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150, 217–228. 10.1099/mic.0.26634-0 [DOI] [PubMed] [Google Scholar]
- Shukla S. K., Rao T. S. (2013). Dispersal of Bap-mediated Staphylococcus aureus biofilm by proteinase K. J. Antibiot. 66, 55–60. 10.1038/ja.2012.98 [DOI] [PubMed] [Google Scholar]
- Singh R., Ray P., Das A., Sharma M. (2009). Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: an in vitro study. J. Med. Microbiol. 58, 1067–1073. 10.1099/jmm.0.009720-0 [DOI] [PubMed] [Google Scholar]
- Singh R., Ray P., Das A., Sharma M. (2010). Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 65, 1955–1958. 10.1093/jac/dkq257 [DOI] [PubMed] [Google Scholar]
- Srivatsan A., Wang J. D. (2008). Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11, 100–105. 10.1016/j.mib.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Sugimoto S., Iwamoto T., Takada K., Okuda K., Tajima A., Iwase T., et al. (2013). Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation. J. Bacteriol. 195, 1645–1655. 10.1128/JB.01672-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugisaki K., Hanawa T., Yonezawa H., Osaki T., Fukutomi T., Kawakami H., et al. (2013). Role of (p)ppGpp in biofilm formation and expression of filamentous structures in Bordetella pertussis. Microbiology 159, 1379–1389. 10.1099/mic.0.066597-0 [DOI] [PubMed] [Google Scholar]
- Tang J., Zhou R., Shi X., Kang M., Wang H., Chen H. (2008). Two thermostable nucleases coexisted in Staphylococcus aureus: evidence from mutagenesis and in vitro expression. FEMS Microbiol. Lett. 284, 176–183. 10.1111/j.1574-6968.2008.01194.x [DOI] [PubMed] [Google Scholar]
- Thammavongsa V., Missiakas D. M., Schneewind O. (2013). Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science 342, 863–866. 10.1126/science.1242255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M., Kavanaugh J. S., Flack C. E., Horswill A. R. (2011). Peptide signaling in the staphylococci. Chem. Rev. 111, 117–151. 10.1021/cr100370n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas V. C., Hancock L. E. (2009). Suicide and fratricide in bacterial biofilms. Int. J. Artif. Organs 32, 537–544. [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A., Merino N., Vergara-Irigaray M., Debarbouille M., Penades J. R., Lasa I. (2005). Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187, 5318–5329. 10.1128/JB.187.15.5318-5329.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotonda M. P., Manna A. C., Cheung A. L., Lasa I., Penades J. R. (2005). SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J. Bacteriol. 187, 5790–5798. 10.1128/JB.187.16.5790-5798.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang L. H., Cassat J. E., Shaw L. N., Beenken K. E., Smeltzer M. S. (2008). Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS ONE 3:e3361. 10.1371/journal.pone.0003361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt S. L., Green C., Stevens K. M., Day B., Erickson D. L., Woods D. E., et al. (2011). The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect. Immun. 79, 4094–4104. 10.1128/IAI.00193-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eiff C., Becker K., Machka K., Stammer H., Peters G. (2001). Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344, 11–16. 10.1056/NEJM200101043440102 [DOI] [PubMed] [Google Scholar]
- Weinstein H. J. (1959). Control of nasal staphylococcal-carrier states. N. Engl. J. Med. 260, 1308–1310. 10.1056/NEJM195906252602602 [DOI] [PubMed] [Google Scholar]
- Wertheim H. F., Vos M. C., Ott A., van Belkum A., Voss A., Kluytmans J. A., et al. (2004). Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364, 703–705. 10.1016/S0140-6736(04)16897-9 [DOI] [PubMed] [Google Scholar]
- Wexselblatt E., Oppenheimer-Shaanan Y., Kaspy I., London N., Schueler-Furman O., Yavin E., et al. (2012). Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog. 8:e1002925. 10.1371/journal.ppat.1002925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood J. M., Bartels D. J., Volper E. M., Greenberg E. P. (2004). Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850. 10.1128/jb.186.6.1838-1850.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska A. K., Beenken K. E., Mrak L. N., Spencer H. J., Post G. R., Skinner R. A., et al. (2012). sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol. Microbiol. 86, 1183–1196. 10.1111/mmi.12048 [DOI] [PMC free article] [PubMed] [Google Scholar]