Abstract
Despite suffering from the major disadvantage of low sensitivity, microscopy of direct smear with the Ziehl–Neelsen stain is still broadly used for detection of acid-fast bacilli and diagnosis of tuberculosis. Here, we present a unique detection method of Mycobacterium tuberculosis (MTB) using surface functionalized magnetic microspheres (MMSs) coupled with quantum dots (QDs), conjugated with various antibodies and phage display-derived peptides. The principle is based upon the conformation of the sandwich complex composed of bacterial cells, MMSs, and QDs. The complex system is tagged with QDs for providing the fluorescent signal as part of the detection while magnetic separation is achieved by MMSs. The peptide ligand H8 derived from the phage display library Ph.D.-7 is developed for MTB cells. Using the combinations of MMS-polyclonal antibody+QD-H8 and MMS-H8+QD-H8, a strong signal of 103 colony forming units (CFU)/mL H37Rv was obtained with improved specificity. MS-H8+QD-H8 combination was further optimized by adjusting the concentrations of MMSs, QDs, and incubation time for the maximum detection signal. The limit of detection for MTB was found to reach 103 CFU/mL even for the sputum matrices. Positive sputum samples could be distinguished from control. Thus, this novel method is shown to improve the detection limit and specificity of MTB from the sputum samples, and to reduce the testing time for accurate diagnosis of tuberculosis, which needs further confirmation of more clinical samples.
Keywords: Mycobacterium tuberculosis, phage display, binding peptides, magnetic microspheres, quantum dots, detection
Introduction
With an estimated 1.4 million mortality rate in 2011, tuberculosis (TB) continues to be the second leading cause of death due to infectious diseases worldwide.1 Confirmation of infection with Mycobacterium tuberculosis (MTB) by stain or culture from sputum specimens is the mainstay of TB diagnosis but could be problematic for different reasons. Conventional microscopy of direct smear with Ziehl–Neelsen (ZN) staining is still broadly used for acid-fast bacilli detection, especially in the developing countries, for its rapidness, low price, and high positive predictive value for TB.2 Whereas, this method suffers from low sensitivity, since it can only detect bacilli if there are more than 5,000 per slide or per mL of specimen.3–5 The main reason is associated with only a small amount of sputum being analyzed with its volume not well defined. The test result is also dependent on the skill of the microscopist. In a previous study by Steingart et al they found that the alleged indirect smear could improve the detection rate of MTB, compared to the direct smear, presumptively by detecting more of sputum samples being analyzed in the test procedure.6 The conventional testing of MTB requires specialized facilities and equipment such as biosafety level 3 laboratories and centrifuges, which are not readily available in many regions of the world, especially developing countries. It is therefore critical to develop sensitive, inexpensive, and field-adapted diagnostic methods for isolating and detecting MTB.
Recently, nanotechnology and nanomaterials, such as magnetic nanoparticles or quantum dots (QDs) nanocrystals, have shown tremendous potential in offering novel approaches based on point of care (POC) devices in biomedical diagnosis of MTB.7,8 Immunomagnetic separation (IMS) is a powerful technique that can be used as an alternative to filtration or centrifugation for selective separation of a series of bacteria in clinical samples. IMS has been applied to the enrichment of mycobacteria, including MTB, Mycobacterium bovis bacillus Calmette–Guérin (BCG), M. bovis, etc, in sputum,9–12 cerebrospinal fluid,13 and environmental samples.14 Coupled with microscopy, polymerase chain reaction or culture has been used as the detection end-points.15,16 The latest research reported the ligand-coated magnetic beads, TB-beads, that show major advantages over the conventional direct smear detection of MTB on sputum samples.17
A more attractive kind of fluorescent label probe, semiconductor QDs have emerged as promising imaging contrast agents, or fluorophores.18,19 The main advantages of QDs compared with conventional fluorescent dyes are the excellent stability against photo-bleaching and wide excitation spectrum. QDs can be excited effectively at any wavelength shorter than their emissions, which are narrow, symmetrical, tunable, and size dependent. With these features, QDs of different sizes may be excited by a monochromatic light with minimal interference from natural autofluorescence, and used as effective imaging contrast agent. QDs have been used in multicolor imaging of different molecular targets in various biological specimens.20 QDs have also been applied in the detection of pathogens and characterization of their toxins. Earlier studies showed detection of unamplified mycobacterial DNA and pathogenic mycobacteria by using fluorescent QDs and magnetic beads, suggesting the possibilities of similar nanotechnologies in TB diagnosis.21,22
Recently, Foddai et al reported an IMS method for Mycobacterium avium subspecies paratuberculosis.23 They obtained maximal capture of M. avium subspecies paratuberculosis from the broth, bovine milk, and fecal specimens by using the two phage display derived peptide ligands, which was initially developed by Stratmann et al.24,25 They also evaluated a series of bead types against the antibodies and peptide ligands studied, and found that capture efficiencies are related to the bead-ligand combinations.23 Antibodies and phage display derived peptide ligands for M. bovis surface antigens have been generated and evaluated for IMS of M. bovis from the veterinary diagnostic samples, but apparently not for MTB, especially not for the nanodiagnosis of TB.12
In this study, phage display derived peptide ligands for MTB cells were generated for detection of MTB with improved detection limit and specificity. These ligands were conjugated onto the magnetic and QDs for the assembly of the basic immunomagnetic detection system. The system was also functionalized with anti-MTB polyclonal, monoclonal antibody (mAb), and an mAb specific for heat shock protein 65, which were evaluated for the best detection combinations. Experiments were carried out to investigate the specificity and the limit of detection (LOD) for MTB in the sputum matrices.
Materials and methods
Ethics statement
This study was granted by the Tongji University Ethics Committee (Permit Number: 2011-fk-03). Written informed consents were obtained from all participants prior to this study.
Antibodies
The following antibodies were included in this study: a murine (mAb) and a rabbit polyclonal antibody (Pab), both against MTB along with a monoclonal antibody (mAbc) produced in murine against MTB heat shock protein 65, which reacts with related mycobacterial species, according to the manufacturer (Thermo Fisher Scientific, Waltham, MA, USA).
Bacterial strains
Thirteen reference strains from Mycobacterium and three non-mycobacterial bacteria reference strains were used for testing the effect of MTB binding peptide and the new detection method for MTB (Table 1). Briefly, the mycobacteria were incubated in Middlebrook 7H9 broth plus 10% oleic acid-albumin-dextrose-catalase (OADC) (BD Biosciences, San Jose, CA, USA) (Middlebrook 7H9/OADC broth) to logarithmic phase at 37°C, collected by centrifugation, and washed with phosphate-buffered saline (PBS) twice. Cultures were thoroughly declumped by vortex thrice with several 3 mm glass beads for 2 minutes for the purpose of breaking large clumps and left to set down for 5 minutes. The amount of bacteria in the suspension was tested with the help of the PhoenixSpec NepheloMeter (BD Biosciences) and the accurate count (CFU/mL) was determined by spreading diluted suspensions onto the Middlebrook 7H10/OADC agar plates before detection and incubating for a suitable time at an appropriate temperature, depending on the bacteria species. Cells were then subjected to a water bath at 80°C for 30 minutes to kill the bacteria and were then stored at −20°C. Colonies of three non-mycobacterial bacteria were scraped from the blood plate and processed like mycobacterium.
Table 1.
Strains used for testing the effect of MTB binding peptide and the new MTB detection method
| Name | Code | Origin | Name | Code | Origin |
|---|---|---|---|---|---|
| Mycobacterium tuberculosis (H37Rv) | ATCC27294 | Aa | Mycobacterium aurum | ATCC23366 | A |
| Mycobacterium microti | ATCCl9422 | A | Mycobacterium kansasii | ATCCl2478 | A |
| Mycobacterium neoaurum | ATCC25795 | A | Mycobacterium gilvum | ATCC43909 | A |
| Mycobacterium parafortuitum | ATCCl9686 | A | Mycobacterium aichiense | ATCC27280 | A |
| Mycobacterium terrae | ATCCl5755 | A | Mycobacterium gordonae | ATCC14470 | A |
| Mycobacterium smegmatis | ATCCl9420 | A | Pseudomonas aeruginosa | ATCC27853 | Bb |
| Mycobacterium chelonae | ATCCl9977 | A | Staphylococcus aureus | ATCC29213 | B |
| Mycobacterium fortuitum | ATCC6841 | A | Escherichia coli | ATCC25922 | B |
Notes:
National Institute for the Control of Pharmaceutical and Biological products;
Shanghai Pulmonary Hospital.
Abbreviation: MTB, Mycobacterium tuberculosis.
Materials for MMSs and QDs’ synthesization
Chloride hydrate (FeCl3⋅6H2O), sodium acetate, ethylene glycol, ammonium hydroxide (NH4OH, 28 weight %), succinic anhydride, N,N-dimethylformamide, and hydrochloric acid (37 weight % aqueous solution) were purchased from Shanghai Reagent Company (Shanghai, People’s Republic of China); 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochlo-ride (EDC) and N-hydroxy-sulfosuccinimide sodium salt (sulfo-NHS) were purchased from Medpep Co., Ltd. (Shanghai, China). Tetraethoxysilane (TEOS) was purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Aminopropyltriethoxysilane (APTES) was purchased from Alfa Aesar (Ward Hill, MA, USA) (98%). The following chemicals were used from Sigma-Aldrich Co.: selenium powder (100 mesh, 99.99%), cadmium oxide (99.5%), tri-n-butylphosphine (90%, Tokyo Chemical Industry Co., Ltd., Tokyo, Japan), tri-n-octylphosphine oxide (90%, Sigma-Aldrich Co.), octadecylamine (90%, Thermo Fisher Scientific), 1-octadecene (90%, Thermo Fisher Scientific), oleic acid (90%, Sigma-Aldrich Co.), sulfur (Sigma-Aldrich Co.), and the triblock copolymer composed of the poly(butyl acrylate), poly(ethyl acrylate), and poly(methacrylic) acid segments.
Production of H37Rv binding peptides by phage display screening
A commercially available phage display library Ph.D.-7 (New England BioLabs Inc., Ipswich, MA, USA) was used to screen inactive MTB reference strain H37Rv whole cell according to the manufacturer’s instructions. Briefly, 107 CFU/mL H37Rv resuspended with 0.1 M NaHCO3 (pH 8.6) was used to coat the surface of a 96-well enzyme-linked immunosorbent assay (ELISA) plate by incubating overnight at 4°C. Plates were blocked with 0.1 M NaHCO3 (pH 8.6), 5 mg/mL bovine serum albumin (BSA), and 0.01% sodium azide for 1 hour at 4°C, and washed six times with Tris-Buffered Saline and Tween 20 (TBST) buffer (TBS-0.1% [volume/volume (v/v)] Tween 20). Ten μL of the Ph.D.-7 phage library (2×1011 phage for a library with 2×109 clones) was diluted 100-fold with TBST and added to the plate, followed by gentle rocking for 1 hour at room temperature to allow phage binding to the surface of H37Rv cells. Unbounded phage was thrown away and the plate was washed ten times with TBST. Bound phage was released using 0.2 M glycine-hydrochloride, 1 mg/mL BSA, and neutralized with 1M Tris-hydrochloride (pH 9.1). A small amount (1 μL) of the elution was titrated and the rest was amplified in Escherichia coli ER2738. After measuring the titer of the amplified phage elute, the input volume was calculated corresponding to the titer of 2×1011 and the amplified phage was then used as the “new” phage library for three further rounds of biopanning by using 107 CFU/mL Mycobacterium smegmatis as the reverse screening molecule with increased stringency of the washing steps (TBS-0.5% [v/v] Tween 20). After four rounds of screening, phage plaques were taken from H37Rv and M. smegmatis eluted plates at random, and amplified in E. coli ER2738 followed with DNA extraction and sequencing with −96III primer performed by the Sangon Biotech Co. Ltd (Shanghai, People’s Republic of China). The amino acid sequence of the peptide was identified using the DNASTAR software.
Preliminary evaluation of binding peptide affinity and specificity
To investigate the binding capability to H37Rv of the consensus sequence phage clones, derived from the biopanning, a phage-ELISA binding assay was performed as described in the Ph.D.-7 kit instructions. In brief, 107 CFU/mL H37Rv and M. smegmatis was coated on the 96-well ELISA plate overnight at 4°C. After blocking along with another non-coating plate, which was used to detect the non-specific binding of the selected phage display derived peptides to BSA, 1010 pfu/mL of each phage particle was added to the plates and incubated with gentle rocking at room temperature for 1–2 hours. Afterward, phage particles captured by target cell were detected with horseradish peroxidase conjugated anti-M13 mAb (GE Healthcare, UK) by using the tetramethylbenzidine solution as the reaction substrate. The absorbances of 460 nm and 620 nm were measured by using the ELISA plate reader (MK3, Thermo Fisher Scientific). The first eluted phage library served as a negative control and TBST as a blank control. All samples were tested in triplicate. Phage clones, which showed ≥3 times higher affinities to the target cell than the negative control, were defined as positive.
When binding affinity was confirmed, the phage displayed peptides were chemically synthesized in native form (H8), with N-terminal biotinylation (Bio-H8) by Bootech BioScience & Technology Co. Ltd. (Shanghai, People’s Republic of China). This was carried out before any further evaluation of H37Rv affinity, specificity, cross-reactivity to other Mycobacterium spp., and application value to detection. A structurally flexible linker peptide (SGG or GGGS) was added to the N-terminal or C-terminal ends of the phage display derived peptides to acquire the functional conformations of peptides.
Kinetics analysis using Octet® bio-layer interferometry
With bio-layer interferometry (Octet® system; FortéBio, Menlo Park, CA, USA), the kinetics of peptide H8-MTB’s binding and specificity were investigated and analyzed on the Octet-QK device.26 Samples or buffer were distributed into the 96-well microtiter plates (Greiner Bio-One GmbH, Frichenhausen, Germany) with 200 μL per well. Operation temperature was maintained at 30°C. Biosensor tips coated with streptavidin (FortéBio) were pre-wetted with buffer (FortéBio) to set up a baseline before peptide immobilization. Then, 100 μg/mL of biotin coupled peptide was immobilized on the biosensor tips with agitation for 5 minutes at 1,000 rpm. After dipping the peptide-immobilized biosensor tips in TBST buffer for 3 minutes, a bacteria matrix baseline was established. The association and dissociation of the bacteria occurred for 5 minutes in buffer agitated at 1,000 rpm. Data were produced automatically by the Octet® User Software (version 3.1) and analyzed with the text files afterward. The binding outline of each sample was generalized as the “nm shift” (the wavelength/spectral shift in nanometers), representing the diversity between the start and end of the 5 minutes sample association step.27
Synthesis of COOH-Fe3O4@silica core-shell microspheres and QDs’ nanoparticles
The Fe3O4 microsphere cores were prepared according to our previous work.28 After that, 5 mL of Fe3O4 aqueous dispersion (200 mg/mL) was managed with 1 M hydrochloride aqueous solution under sonication for 20 minutes and washed with deionized (DI) water until neutrality was reached. The treated Fe3O4 microspheres were then dispersed in the ethanol/water mixture (v/v=80/20) for 5 minutes. Ammonium hydroxide was added to adjust the pH value to about 9.5. Then 0.1 mL of TEOS was added into the reactor dropwise with vigorous mechanical stirring at room temperature. The reaction continued for 24 hours. The extra reactants were removed by washing four times with ethanol. To prepare MMSs, first, amino groups were initially introduced onto the surface of Fe3O4@silica spheres by conventional sol-gel reaction with APTES as a modifying agent. Carboxyl groups were then functionalized by chemical reaction between the amino groups and succinic anhydride. In a typical process, 20 mg of Fe3O4@silica spheres were dispersed in 25 g of mixed solvent of ethanol/DI water (v/v=70/30). Fifty μL of NH4OH was added and followed by 60 μL of APTES under vigorous stirring. The reaction lasted for 24 hours and the product was washed with ethanol and DI water respectively three times. All products were dispersed in the N,N-dimethylformamide; 15 mg of succinic anhydride was used to functionalize the microsphere with adequate carboxyl groups. MMSs were obtained after washing.
The QDs nanoparticles were prepared according to our previously reported method.29
Preparation of probe-antibody or probe-peptide conjugates
The conjugates of MMSs and different peptides, Pabs or mAbs were prepared by (EDC/sulfo-NHS)-mediated amidation reaction. Typically, 5 μL EDC (0.5 g/mL) and 10 μL sulfo-NHS (0.25 g/mL) were dissolved in 500 μL of 1×BST buffer solution containing 1 mg of MMSs. After approximately 30 minutes, appropriate antibodies or peptides were added. The reaction continued for 3 hours with gentle mixing at room temperature. The resultant mixture was blocked by 500 μL 1% BSA for 30 minutes, and stored at 4°C overnight. The conjugation reactions between the different Fe3O4 particles and antibodies/peptides were carried out as previously introduced. A similar procedure was used to conjugate QDs with antibodies or peptides.
Bacterial detection and LOD
The principle of the assay is based on the conformation of the sandwich complex, composed of the following components: 1) bacterial cells, 2) MMSs coupled with H37Rv binding peptides or the Pabs, and 3) QDs conjugated with H37Rv binding peptides or the mAbs. In an aqueous PBS solution containing a given concentration of TB, magnetic and QD probes reacted with TB through specific ligand-receptor interaction. After magnetic separation and rinsing twice, the complex was re-dispersed in PBS for measurement of the fluorescent signal (Figure 1). For this purpose, 50 μL of the bacteria, comprised of positive controls or equal volumes of PBS (blank control), were incubated with 50 μL of MMSs and 50 μL of functional QDs simultaneously at room temperature for 2 hours with gentle agitation. The dilutions were then separated from the matrix by using a magnetic device (Dynal MPC-s, Sigma-Aldrich Co.), washed twice with PBS, and resuspended in 30 μL PBS. For direct visual observation of the bacteria, 10 μL samples were smeared and observed under the epifluorescence microscope with a 50 W mercury lamp (Leica Microsystems, Wetzlar, Germany) using the filter set of 460SPUV/475DCXRU/D655/40 nm (Leica Microsystems), followed by fixing overnight under ultraviolet irradiation and acid fast staining. Fluorescence was then detected with a laser induced spectrofluorometer (excitation 405 nm emission 610 nm, photomultiplier tube (PMT) 450 V), (SP-800MCE; Shanghai Spectrum Instruments Co. Ltd., Shanghai, People’s Republic of China). The interaction between MMSs and QDs served as a negative control and the single MMSs as a blank. The result of the quantitative detection was obtained and evaluated by the fluorescent signal ratio of the positive to negative sample (S/N ratio). S/N≥2.1 designates positive, while S/N<2.1 indicates negative.
Figure 1.
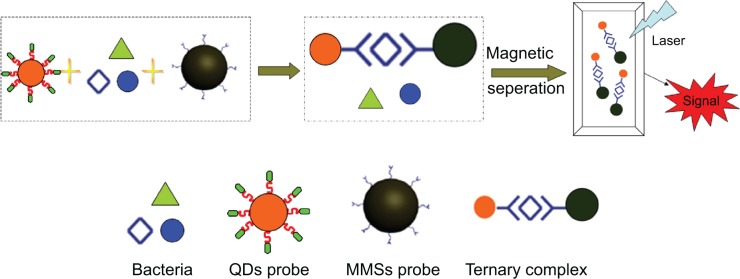
Bacteria cells are separated by MMSs coupled with specific peptides or polyclonal antibodies.
Notes: The fluorescent signal of QDs can be detected for imaging. The principle of the assay is based upon the conformation of the ternary complex, composed of bacterial cells, MMSs coupled with H37Rv binding peptides as well as the polyclonal antibodies, and QDs conjugated with H37Rv binding peptides or the monoclonal antibodies. Upon magnetic separation by MMSs, the complexes are tagged with functional QDs for detection of the fluorescent signal.
Abbreviations: MMSs, magnetic microspheres; QDs, quantum dots.
MTB reference strain H37Rv was used for the evaluation of the LOD. Two series of ten-fold dilutions ranging from 108 to 101 CFU/mL were prepared as described previously. The LOD was defined as the lowest concentration level of bacteria which was statistically different from the negative controls.
Sensitivity and specificity evaluation of MMSs and QDs conjugated with different antibodies and peptide ligands
MMSs were coupled with H8 or Pab. Independently, QDs were conjugated with H8, mAb or mAbc. These surface-conjugated MMSs and QDs were mixed in six different combinations and evaluated for the LOD on H37Rv and specificities involving 12 Mycobacterium species and three non-mycobacterial bacteria (Table 1). Cultures of each species in stationary phase were diluted to 107 CFU/mL in PBS and 50 μL of each dilution was used for the detection study. The best ligands combination was chosen based on the evaluation results.
Optimization of detection condition
MMSs and QDs were diluted to different concentrations of 50 mg/L; 100 mg/L; 150 mg/L, and 200 mg/L for the H37Rv detection. These concentrations are optimized to yield the maximum detection signal. The incubation time was also varied at different intervals: 0.5, 1, 1.5, 2, 2.5, and 3 hours for optimum detection.
Impact of sputum matrix on detection sensitivity and specificity by optimal MMSs and QDs combination
The same volume of normal sputum samples was collected from healthy individuals and spiked with 100 μL inactivated H37Rv cultured at a range of cell concentrations as before. The sputum matrixes were decontaminated by a standard N-acetyl cysteine-sodium hydroxide procedure based on the standard methodology and neutralized with PBS (pH 6.8).30 Centrifugation was carried out at 3,000× g for 20 minutes, followed by the discarding of the supernatant and re-suspending the pellet with PBS. Fifty μL of suspension was used in the detection experiment as described previously. Sputum matrixes of other Mycobacterium spp. were also processed as H37Rv.
Sputum sample detection
Sputum samples were collected from six TB patients with confirmed smear-positive pulmonary TB from Shanghai Pulmonary Hospital, Shanghai, People’s Republic of China and two healthy controls. Part of the sputum samples was used to be smeared and ZN stained directly, and the rest was disposed like before and detected by the new method.
Statistical analysis
Unpaired Student’s t-test or Mann–Whitney U-test, two-tailed, was used for data from two groups. Data from more than two groups were analyzed by one-way analysis of variance (ANOVA) and Tukey’s multiple comparison tests. P-values <0.05 were regarded as significant.
Results
Phage display biopanning to generate MTB binding peptides
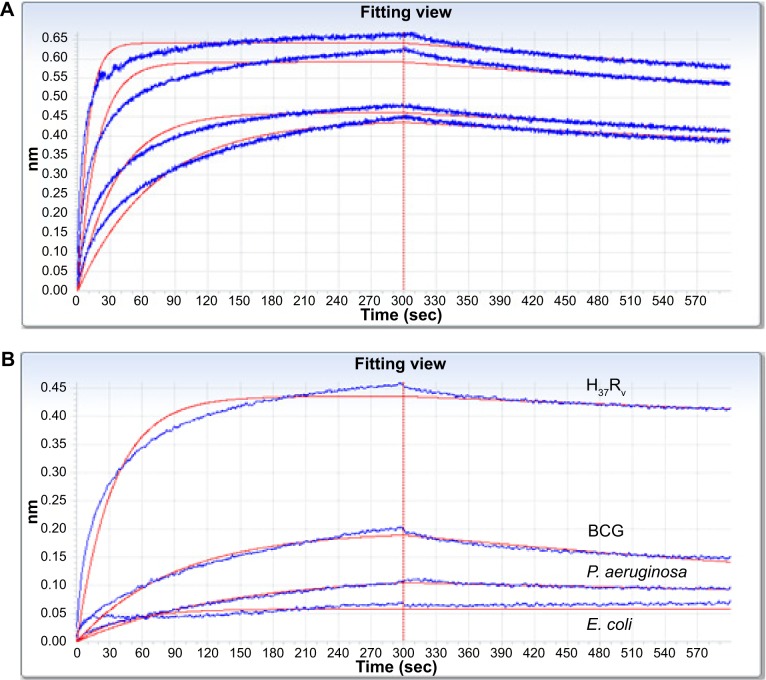
After the fourth round of panning, the amount of eluted phages bound to H37Rv gradually increased with successive screening rounds (from 5×103 to 2×104). The peptide coding region of 39 randomly selected phage clones were sequenced and five unique sequences were achieved; three were capable of binding whole H37Rv cells, of which H8 showed the highest affinity (Table 2). The corresponding peptides (designated peptide H8) were chemically synthesized in native and N-terminal biotinylation forms and evaluated for application of MTB detection. The kinetics of peptide H8-MTB binding was characterized with double diluted H37Rv and biotinylated H8 on the Octet-QK device. Given the difficulties in determining how many receptors H8 were binding on H37Rv, we used dissociation constant (Kdis) (s−1) to evaluate the binding affinity of H8 to H37Rv. As shown in the stacked graph (Figure 2A), the Kdis value of H8-MTB binding was 3.45×10−4±8.50×10−6 (R2=0.973). The binding specificity of H8 to BCG, Pseudomonas aeruginosa, and E. coli was also respectively detected with Octet-QK. The Kdis value was significantly lower than that of H37Rv (Figure 2B).
Table 2.
Outcomes of phage display biopanning
| Clone identifier(s) | Amino acid sequence | Frequencyc | Ability to bind H37Rv |
|---|---|---|---|
| Ha31,37,39; Sb3,5,15 | LHKHPTP | 6/39 | − |
| H2,5,23,24,26,27,28,30,32,38,40; S4,8,11,16 | WHTGTPH | 15/39 | ++ |
| H21; S1,2,6,7,9,10,12,13,14,18,21,22,24,29,40 | VHPHWRH | 16/39 | + |
| H8 | WHSGTPHd | 1/39 | +++ |
| S17 | CHPHTPH | 1/39 | − |
Notes:
Clones eluted by H37Rv;
clones eluted by Mycobacterium smegmatis;
number of clones with sequence/total number of clones tested after fourth round of biopanning;
H37Rv-derived peptide (highlighted in bold) capable of binding H37Rv was chemically synthesized and subsequently evaluated for detection.
Figure 2.
Kinetics of peptide H8-MTB binding and specificity were investigated and analyzed on the Octet-QK device.
Notes: (A) Kinetics analyses of H8-MTB binding with double diluted H37Rv. Kdis (s−1) was used to evaluate the binding affinity of H8 to H37Rv. Based on the stacked graph, the Kdis value of H8-MTB binding is 3.45E-04±8.50E-06 (R2=0.973). (B) Kinetics analyses of binding affinity of H8 to H37Rv, BCG, P. aeruginosa, and E. coli. The binding specificity of H8 to BCG, P. aeruginosa and E. coli was significantly lower than that to H37Rv.
Abbreviations: BCG, Mycobacterium bovis bacillus Calmette–Guérin; P. aeruginosa, Pseudomonas aeruginosa; E. coli, Escherichia coli.
Conjunction of MMSs and QDs with peptides or antibodies
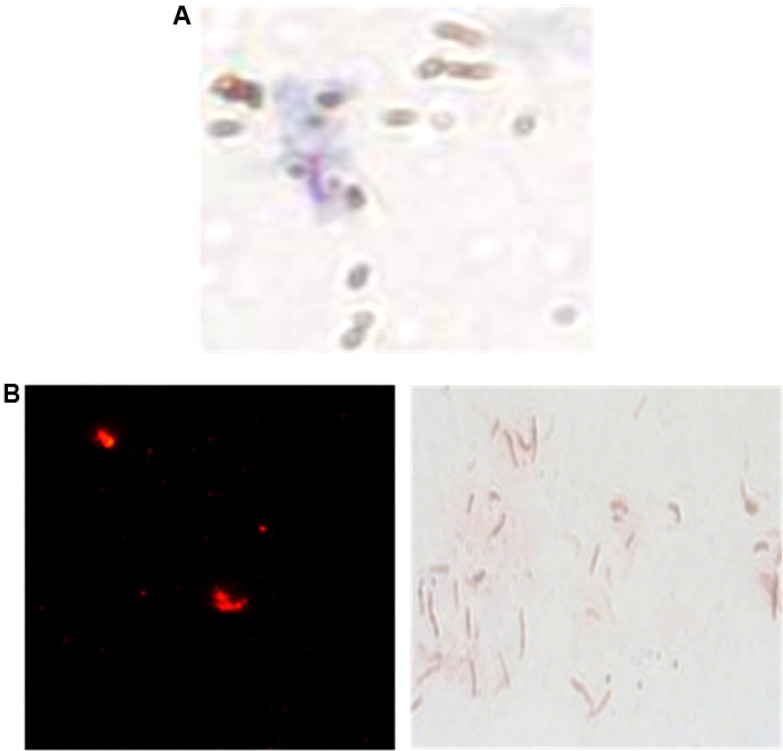
Assessment of binding affinity of functionalized MMSs and QDs to peptides or antibodies was performed by spectrophotometry at 280 nm of the peptides or antibodies solution before and after conjugation with the nanoparticles. Microscopy was also carried out on the ZN stained MMSs and QDs, with or without the peptide H8, which were used for the immunocapture of MTB cells. As shown in Figure 3, the H37Rv cells that closely associated with MMSs or QDs were observed only in the case of those coupled with H8.
Figure 3.
Microscopic observation of H37Rv bacilli, captured by the H8 conjugated MMSs.
Notes: (A) Ziehl–Neelsen stain 1,000× and QDs. (B) direct fluorescence observation 400× and Ziehl–Neelsen stain 1000×.
Abbreviations: MMSs, magnetic microspheres; QDs, quantum dots.
Bacterial detection and LOD
After incubation of H37Rv, MMSs, and QDs for 2 hours, the microscopic slide smears from different samples were prepared and examined by fluorescence microscopy. The red fluorescence from QDs and the ZN stain positive cells was respectively observed under the fluorescent and optical microscope. An approximately four-fold higher fluorescence intensity [(32.2±2.5)×103] was detected for H37Rv compared to that of the negative control [(8.3±0.9)×103]. This sharp contrast in fluorescence signal was consistent with the sandwich complex of the bacterial cells, MMSs, and QDs (Figure 4).
Figure 4.
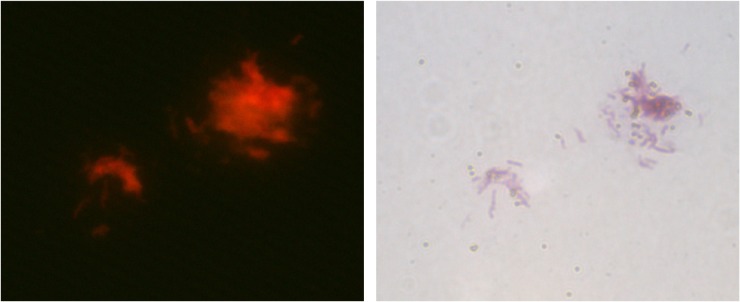
Microscopic observation of the sandwich complex of H37Rv bacterial cells, MMSs, and QDs (direct fluorescence observation 400× and Ziehl–Neelsen stain 1,000×).
Abbreviations: MMSs, magnetic microspheres; QDs, quantum dots.
Given the complexities in distinguishing the real from false signal under microscopy, the detection limit of the method was obtained primarily by measuring fluorescence intensity with the spectrofluorometer. Using this method even 103 bacterial cells/mL [(4.59±0.20)×103] generated an S/N value of more than two-fold greater than the negative control sample [(2.01±0.15)×103], which was defined as the LOD of this method.
Sensitivities and specificities evaluation of MMSs and QDs conjugated with different antibodies or peptide ligands
With MMSs coated with H8 or Pab and QDs conjugated with H8, mAb or mAbc, six combinations were evaluated for their potential to capture MTB. Preliminary evaluations were conducted using inactivated MTB H37Rv Middlebrook broth suspensions containing 108 CFU/mL which were subjected to MMSs separation monitored by the fluorescence of QDs. Different degrees of H37Rv recovery from each combination were observed, which were evidenced by different fluorescence intensity (data not shown). Figure 5 shows the LOD detection results of six combinations of MMSs coupled with H8 or Pab and QDs conjugated with H8, mAb or mAbc. As shown in this figure, the combination of MMS-Pab+QD-H8 and MMS-H8+QD-H8 provided the highest level of H37Rv capture (ratios to negative control were 4.394±0.565 and 3.956±0.525 respectively). Further trials were carried out to determine the H37Rv detection capabilities of these combinations at lower numbers of cells (101 to 108 CFU/mL). Differences in H37Rv detection capabilities between combination types were obvious when samples containing a series of H37Rv concentrations were evaluated. As also shown in Figure 5, the combinations of MMS-H8+QD-mAb and MMS-Pab+QD-mAbc produced strong signals for only 107 CFU/mL H37Rv cells, indicating relatively poor ability to detect MTB. However, the MTB detection capability was improved with the combinations of MMS-H8+QD-mAbc and MMS-Pab+QD-mAb for 105 CFU/mL or 106 CFU/mL H37Rv cells. Similarly, the combinations of MMS-Pab+QD-H8 and MMS-H8+QD-H8 yielded a significantly higher signal for 103 CFU/mL H37Rv (Figure 5) and thus demonstrated the best MTB detection capability.
Figure 5.
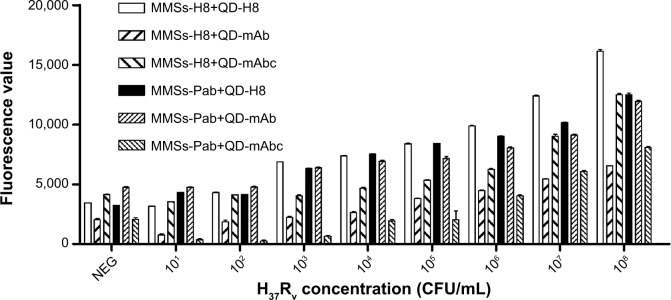
LOD detection for six combinations of MMSs coupled with H8 or Pab and QDs conjugated with H8, Mab, or mabc.
Notes: Differences in MTB detection capability between the combinations were evident for a range of MTB concentrations in the test. Combinations of MMS-H8+QD-mAb and MMS-Pab+QD-mAbc produced a strong signal for only 107 CFU/mL H37Rv cells, indicating relatively poor MTB detection capability. Combinations of MMS-H8+QD-mAbc and MMS-Pab+QD-mAb exhibited a higher but decreasing fluorescence intensity of H37Rv for 105 CFU/mL or 106 CFU/mL, showing improved MTB detection capability. Combinations of MMS-Pab+QD-H8 and MMS-H8+QD-H8 yield a significantly higher signal for 103 CFU/mL H37Rv and thus demonstrated the best MTB detection capability.
Abbreviations: LOD, limit of detection; MMSs, magnetic microspheres; QDs, quantum dots; Pab, a rabbit polyclonal antibody against MTB; mAb, a murine monoclonal antibody against MTB; mAbc, a murine monoclonal antibody against MTB heat shock protein 65 (HSP65); CFU, colony-forming unit; mAbc, .
Furthermore, the specificity of each combination was evaluated against 12 non-tuberculosis mycobacterium (NTM) and three non-mycobacterial bacteria. For all combinations, binding to the three non-mycobacterial bacteria was not observed by either microscopy or spectrofluorometer. The ligand combinations which demonstrated maximal binding of MTB cells (MMS-Pab+QD-H8 and MMS-H8+QD-H8) exhibited binding to two of the NTMs tested (M. smegmatis and Mycobacterium gilvum), less than MMS-H8+QD-mAbc and MMS-Pab+QD-mAb. Therefore, the combinations of MMS-Pab+QD-H8 and MMS-H8+QD-H8 were considered optimal for detection of MTB. Given both the feasibility of larger scale production and reduced cost of synthesizing peptides relative to Pab, we decided to use MMS-H8+QD-H8 combinations to further evaluate the detection method.
Optimization of detection concentrations for MMSs, QDs and incubation time
The concentrations of MMSs and QDs were optimized for gaining high fluorescence signals (Figure 6). Given that the values obtained using concentrations of 100 mg/L and 200 mg/L were similar (t=−1.075, P>0.05), we selected 100 mg/L as the detection concentration for MMS-H8 and QD-H8. The fluorescence signal rose to a peak at 2 hours of incubation, then began to decline (Figure 7). Thus, given that the greatest fluorescence was observed using an incubation time of 2 hours, we selected 2 hours as the incubation time for the detection assay.
Figure 6.
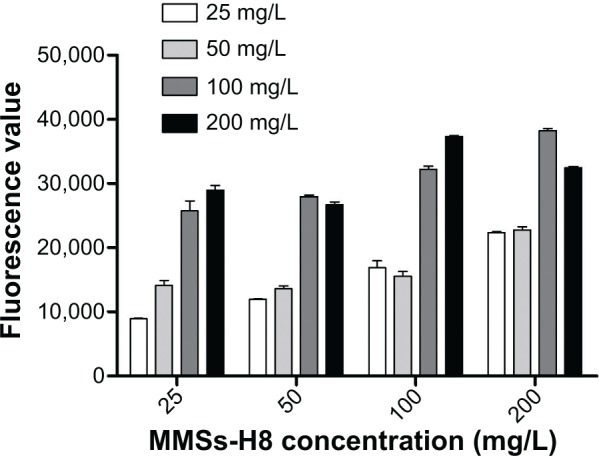
Optimization of the detection concentration of MMS-H8 and QD-H8.
Notes: MMS-H8 and QD-H8 were diluted to 50 mg/L; 100 mg/L; 150 mg/L; and 200 mg/L, respectively. The concentration was optimized for the H37Rv detection. High fluorescence was obtained for 100 mg/L as the best detection concentration for MMSs and QDs.
Abbreviations: MMSs, magnetic microspheres; QDs, quantum dots.
Figure 7.
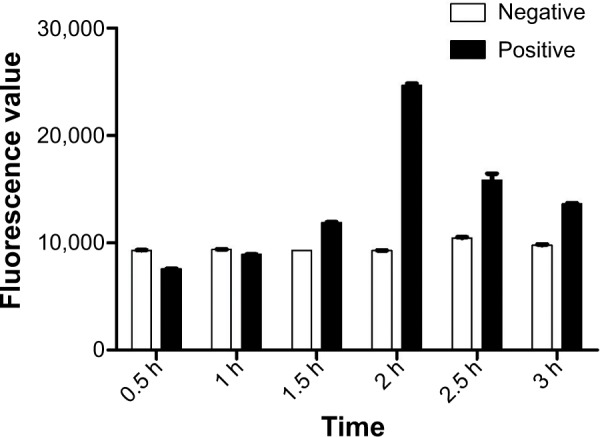
Optimization of the incubation time of bacterial cells, MMS-H8, and QD-H8.
Notes: The incubation time was set at 0.5 h, 1 h, 1.5 h, 2 h, 2.5 h, and 3 h for optimization. Highest fluorescence signal was obtained at 2 h as the optimum interaction time.
Abbreviations: MMSs, magnetic microspheres; QDs, quantum dots; h, hour.
Impact of sputum matrix on detection sensitivity and specificity of optimal MMSs and QDs combination
No significant effect of sputum matrix on detection sensitivity was found by spiking normal sputum matrices with inactivated MTB H37Rv, followed by detection using the optimal combination of MMS-H8+QD-H8. The LOD for H37Rv spiked sputum matrices was 103 CFU/mL and the specificity was similar to that observed using culture supernatant.
Sputum sample detection
Positive results were obtained for six smear-positive samples, including two one positive (1+) smear samples (S1, S2). No positive signals were detected for the two negative controls (S7, S8), seen in Figure 8.
Figure 8.
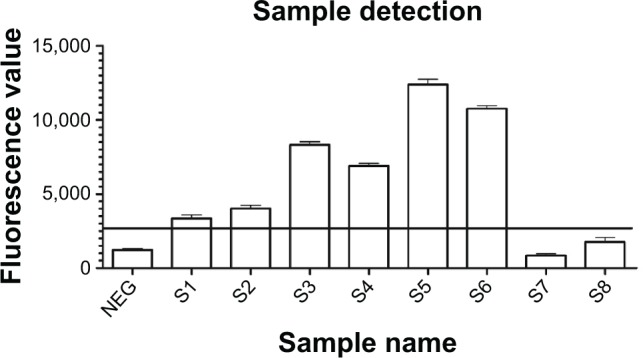
Detection of sputum samples with the new nano detection method.
Notes: Eight sputum samples were collected and detected with direct smear and the new sandwich method simultaneously. Positive results were only obtained for the six smear positive samples (S1–S6), including two one positive (1+) smear samples (S1, S2).
Discussion
Previously, Liandris et al developed a method for detection of pathogenic mycobacteria using functionalized QDs coupled with IMS.22 The major principle is based on the separation of bacteria using magnetic beads coupled with genus-specific Pab and mAb for heparin-binding hemagglutinin. These complexes were then sequentially tagged with anti-mouse biotinylated antibody and streptavidin conjugated QDs for detection of the fluorescent signal. The minimum detection limit of the assay was determined to be 104 bacteria/mL, but could be further decreased by 1 log using a spectrofluorometer. Once fully developed, this method may be applicable to clinical samples of TB. But the main issues involve unevaluated antibodies for the immune assay and rather complicated clinical procedures. Therefore, the objectives of this study were to address these limitations by: i) producing novel peptide ligands for MTB whole cells, ii) producing a new method for the detection of MTB based on MMSs coupled with functionalized QDs, and iii) evaluating the efficacy of commercial antibodies and peptide ligands for detection of MTB.
A range of magnetic beads were surface conjugated with antibodies and peptide ligands. These biomarkers were evaluated on their capacity to capture M. avium subspecies paratuberculosis and M. bovis.12,23 It was found in this study that the capturing ability depended on the bead-ligand combinations. Hence, further investigation is needed on other antibodies and alternative ligands for nano detection of MTB. Novel peptide ligands were developed in this study for MTB by a phage display biopanning approach. Phage display biopanning of specific antigens of MTB has been reported previously,31–35 although not for MTB whole cells. Therefore, in this study, the inactivated whole cells of MTB reference strain H37Rv were used as the target molecules to screen the phage display library Ph.D.-7, and to reduce or eliminate the binding to NTM, M. smegmatis was used as the reverse screening molecule. Three phage clones, capable of binding the whole H37Rv cells were produced, of which clone H8 exhibited the highest affinity based on phage-ELISA. According to the sequence results (Table 2), we believe that the replacement of the third threonine by serine of H8 may influence the structure of the peptide and cause the higher binding affinity than H2. The higher affinity and specificity to MTB of the H8 peptide, which was synthesized according to the amino acid sequence in bold in Table 2, were further confirmed by kinetics analysis (Figure 2).
In Liandris et al’s research, samples of bacteria were coupled with several biological molecules and nanoparticles, sequentially: the genus specific Pab conjugated MMSs; the heparin-binding hemagglutinin (HBHA) mAb; an anti-mouse biotinylated antibody; and finally the streptavidin-coated QDs.22 Although the final signal was amplified four times with the biotin-streptavidin reaction, these four steps render it too complex for routine and expanded use. In this study, it was found feasible to conjugate the MMSs and QDs with the ligands that can bind MTB cells directly. In this fashion, a sandwich complex was developed with bacterial cells, MMSs, and QDs coupled to MTB specific ligands. The complex structure can be magnetically separated by MMSs and viewed by fluorescence imaging with QDs (Figure 4). The method is a much more straight forward process that is easy to perform particularly in a clinical setting. In the LOD test, it was found that 103 CFU/mL MTB was detectable even in sputum matrices without biotin-streptavidin amplification, as was used in Liandris et al’s research.22
Specificity and sensitivity of MTB detection was found, in this research, to be dependent on the combinations of ligands and nanoparticles. The highest specificity and sensitivity was found for the combinations of MMS-Pab+QD-H8 and MMS-H8+QD-H8. Stewart et al achieved immunocapture of bacterium by using the phage display-derived peptides as a ligand to IMS.12 In this work, the phage display-derived peptide was used to coat MMSs/QDs simultaneously for capturing the bacteria via a sandwich complex. Although competitive combinations may exist, the results suggested an improved capturing ability of phage display-derived peptide ligands. Detection results of eight sputum samples also proved the feasible application value of the MMS-H8+QD-H8 combination for the diagnosis of TB, however, more clinical samples, especially those from patients infected by NTM are required to accurately determine the sensitivity and specificity of this new method.
It is concluded that the incubation time and the concentration of nanosystem are important factors that need to be optimized. The results of this study showed that a 2 hours incubation time is needed for the interactions among bacteria, MMSs, and QDs, which may become stabilized at 2 hours, and be adversely affected by a too short or long incubation time. Regarding the nanoparticle concentration, 100 μg/mL has been found to be optimum for sufficient signal detection.
In conclusion, we have developed phage display-derived peptide ligands that are essential for MTB detection. A new method for the detection of MTB was also developed based on functionalized MMSs coupled with QDs. The optimal ligand combinations have been found to be MMS-Pab+QD-H8 and MMS-H8+QD-H8. This optimized detection method may have considerable potential to both improve the limit and specificity of MTB detection from sputum samples and to reduce the time required for accurate diagnosis. This composite nanosystem is the subject of ongoing research to further enhance the LOD to 10–100 CFU/mL from sputum samples. Once fully developed, the method may be directly applicable on the detection of clinical samples and facilitate the early diagnosis of TB.
Acknowledgments
The authors would like to thank Dr Malcolm Duthie, from Infectious Disease Research Institute, Seattle, USA for helpful revisions and comments regarding this manuscript.
The work was supported by the grant from the National Key Project for Infectious Disease (No 2012ZX10003002-008), the Science and Technology Commission of Shanghai Municipality, Shanghai, People’s Republic of China (No 11nm0506100, 124119a1500), the National Natural Science Foundation of China (No 51173135, 81301391), and the Fundamental Research Funds for the Central Universities (No 1511219013).
Footnotes
Disclosure
The authors have no conflicts of interest to disclose.
References
- 1.World Health Organization . Global tuberculosis report (2012) Geneva: World Health Organization; 2012. [Accessed October 29, 2014]. Available from: http://apps.who.int/iris/bitstream/10665/75938/1/9789241564502_eng.pdf. [Google Scholar]
- 2.Koch ML, Cote RA. Comparison of fluorescence microscopy with Ziehl-Neelsen stain for demonstration of acid-fast bacilli in smear preparations and tissue sections. Am Rev Respir Dis. 1965;91:283–284. doi: 10.1164/arrd.1965.91.2.283. [DOI] [PubMed] [Google Scholar]
- 3.Trusov A, Bumgarner R, Valijev R, et al. Comparison of Lumin LED fluorescent attachment, fluorescent microscopy and Ziehl-Neelsen for AFB diagnosis. Int J Tuberc Lung Dis. 2009;13(7):836–841. [PubMed] [Google Scholar]
- 4.Ulrichs T, Lefmann M, Reich M, et al. Modified immunohistological staining allows detection of Ziehl-Neelsen-negative Mycobacterium tuberculosis organisms and their precise localization in human tissue. J Pathol. 2005;205(5):633–640. doi: 10.1002/path.1728. [DOI] [PubMed] [Google Scholar]
- 5.Yeager H, Jr, Lacy J, Smith LR, LeMaistre CA. Quantitative studies of mycobacterial populations in sputum and saliva. Am Rev Respir Dis. 1967;95(6):998–1004. doi: 10.1164/arrd.1967.95.6.998. [DOI] [PubMed] [Google Scholar]
- 6.Steingart KR, Ng V, Henry M, et al. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis. 2006;6(10):664–674. doi: 10.1016/S1473-3099(06)70602-8. [DOI] [PubMed] [Google Scholar]
- 7.Lee WG, Kim YG, Chung BG, Demirci U, Khademhosseini A. Nano/Microfluidics for diagnosis of infectious diseases in developing countries. Adv Drug Deliv Rev. 2010;62(4–5):449–457. doi: 10.1016/j.addr.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin D, He X, Wang K, Tan W. Using fluorescent nanoparticles and SYBR Green I based two-color flow cytometry to determine Mycobacterium tuberculosis avoiding false positives. Biosens Bioelectron. 2008;24(4):626–631. doi: 10.1016/j.bios.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 9.Albert H, Ademun PJ, Lukyamuzi G, et al. Feasibility of magnetic bead technology for concentration of mycobacteria in sputum prior to fluorescence microscopy. BMC Infect Dis. 2011;11:125. doi: 10.1186/1471-2334-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson S, Lane A, Rosedale R, Stanley C. Concentration of Mycobacterium tuberculosis from sputum using ligand-coated magnetic beads. Int J Tuberc Lung Dis. 2010;14(9):1164–1168. [PubMed] [Google Scholar]
- 11.Liang G, Chen H, Zhang S, Wu W, Kong J. Magnetic nanosensors for highly sensitive and selective detection of bacillus Calmette-Guérin. Analyst. 2012;137(3):675–679. doi: 10.1039/c1an15897j. [DOI] [PubMed] [Google Scholar]
- 12.Stewart LD, McNair J, McCallan L, Thompson S, Kulakov LA, Grant IR. Production and evaluation of antibodies and phage display-derived peptide ligands for immunomagnetic separation of Mycobacterium bovis. J Clin Microbiol. 2012;50(5):1598–1605. doi: 10.1128/JCM.05747-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazurek GH, Reddy V, Murphy D, Ansari T. Detection of Mycobacterium tuberculosis in cerebrospinal fluid following immunomagnetic enrichment. J Clin Microbiol. 1996;34(2):450–453. doi: 10.1128/jcm.34.2.450-453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweeney FP, Courtenay O, Ul-Hassan A, Hibberd V, Reilly LA, Wellington EM. Immunomagnetic recovery of Mycobacterium bovis from naturally infected environmental samples. Lett Appl Microbiol. 2006;43(4):364–369. doi: 10.1111/j.1472-765X.2006.01983.x. [DOI] [PubMed] [Google Scholar]
- 15.Ghodbane R, Drancourt M. Magnetic Bead Protocol for Culturing Mycobacterium tuberculosis from Sputum Specimens. J Clin Microbiol. 2013;51(5):1578–1579. doi: 10.1128/JCM.03428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garbaccio SG, Cataldi AA. Evaluation of an immunomagnetic capture method followed by PCR to detect Mycobacterium bovis in tissue samples from cattle. Rev Argent Microbial. 2010;42(4):247–253. doi: 10.1590/S0325-75412010000400002. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Sun ZQ, Pei H, et al. Increased case finding of tuberculosis from sputum and sputum deposits after magnetic bead concentration of mycobacteria. J Microbiol Methods. 2013;93(2):144–147. doi: 10.1016/j.mimet.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Zhu L, Ang S, Liu WT. Quantum dots as a novel immunofluorescent detection system for Cryptosporidium parvum and Giardia lamblia. Appl Environ Microbiol. 2004;70(1):597–598. doi: 10.1128/AEM.70.1.597-598.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281(5385):2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 20.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotechnol. 2003;21(1):47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 21.Gazouli M, Liandris E, Andreadou M, et al. Specific detection of unamplified mycobacterial DNA by use of fluorescent semiconductor quantum dots and magnetic beads. J Clin Microbiol. 2010;48(8):2830–2835. doi: 10.1128/JCM.00185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liandris E, Gazouli M, Andreadou M, Sechi LA, Rosu V, Ikonomopoulos J. Detection of pathogenic mycobacteria based on functionalized quantum dots coupled with immunomagnetic separation. PLoS One. 2011;6(5):e20026. doi: 10.1371/journal.pone.0020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foddai A, Elliott CT, Grant IR. Maximising capture efficiency and specificity of magnetic separation for Mycobacterium avium subsp. Paratuberculosis cells. Appl Environ Microbiol. 2010;76(22):7550–7558. doi: 10.1128/AEM.01432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stratmann J, Strommenger B, Stevenson K, Gerlach GF. Development of a peptide-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in milk. J Clin Microbiol. 2002;40(11):4244–4250. doi: 10.1128/JCM.40.11.4244-4250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stratmann J, Dohmann K, Heinzmann J, Gerlach GF. Peptide aMptD-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in bulk milk samples. Appl Environ Microbiol. 2006;72(8):5150–5158. doi: 10.1128/AEM.00590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson JL, Scott IM, McMurry JL. Optical biosensing: Kinetics of protein A-IGG binding using biolayer interferometry. Biochem Mol Biol Educ. 2010;38(6):400–407. doi: 10.1002/bmb.20442. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Schantz A, Schwegler M, Shankar G. Detection of low-affinity anti-drug antibodies and improved drug tolerance in immunogenicity testing by Octet(®) biolayer interferometry. J Pharm Biomed Anal. 2011;54(2):286–294. doi: 10.1016/j.jpba.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 28.Tang YL, Liang S, Yu SL, et al. Enhanced adsorption of humic acid on amine functionalized magnetic mesoporous composite microspheres. Colloids and Surfaces A Physicochemical and Engineering Aspects. 2012;406:61–67. [Google Scholar]
- 29.Zhang BB, Cheng J, Li DN, Liu XH, Ma GP, Chang J. A novel method to make hydrophilic quantum dots and its application on biodetection. Mater Sci Eng B. 2008;149:87–92. [Google Scholar]
- 30.Kent PT, Kubica GP. Public Health Mycobacteriology: A Guide for the Level III Laboratory. Atlanta: U.S. Department of Health and Human Services; 1985. [Google Scholar]
- 31.Gevorkian G, Segura E, Acero G, et al. Peptide mimotopes of Mycobacterium tuberculosis carbohydrate immunodeterminants. Biochem J. 2005;387(Pt 2):411–417. doi: 10.1042/BJ20041139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma A, Saha A, Bhattacharjee S, Majumdar S, Das Gupta SK. Specific and randomly derived immunoactive peptide mimotopes of mycobacterial antigens. Clin Vaccine Immunol. 2006;13(10):1143–1154. doi: 10.1128/CVI.00127-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barenholz A, Hovav AH, Fishman Y, Rahav G, Gershoni JM, Bercovier H. A peptide mimetic of the mycobacterial mannosylated lipoarabinomannan: characterization and potential applications. J Med Microbiol. 2007;56(Pt 5):579–586. doi: 10.1099/jmm.0.46920-0. [DOI] [PubMed] [Google Scholar]
- 34.Yang H, Liu ZH, Zhang LT, et al. Selection and application of peptide mimotopes of MPT64 protein in Mycobacterium tuberculosis. J Med Microbiol. 2011;60(Pt 1):69–74. doi: 10.1099/jmm.0.025098-0. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, Chen H, Liu Z, et al. A Novel B-Cell Epitope Identified within Mycobacterium tuberculosis CFP10/ESAT-6 Protein. PLoS One. 2013;8(1):e52848. doi: 10.1371/journal.pone.0052848. [DOI] [PMC free article] [PubMed] [Google Scholar]