Abstract
Exploiting genotyping, DNA sequencing, imputation and trans-ancestral mapping, we used Bayesian and frequentist approaches to model the IRF5–TNPO3 locus association, now implicated in two immunotherapies and seven autoimmune diseases. Specifically, in systemic lupus erythematosus (SLE), we resolved separate associations in the IRF5 promoter (all ancestries) and with an extended European haplotype. We captured 3230 IRF5–TNPO3 high-quality, common variants across 5 ethnicities in 8395 SLE cases and 7367 controls. The genetic effect from the IRF5 promoter can be explained by any one of four variants in 5.7 kb (P-valuemeta = 6 × 10−49; OR = 1.38–1.97). The second genetic effect spanned an 85.5-kb, 24-variant haplotype that included the genes IRF5 and TNPO3 (P-valuesEU = 10−27–10−32, OR = 1.7–1.81). Many variants at the IRF5 locus with previously assigned biological function are not members of either final credible set of potential causal variants identified herein. In addition to the known biologically functional variants, we demonstrated that the risk allele of rs4728142, a variant in the promoter among the lowest frequentist probability and highest Bayesian posterior probability, was correlated with IRF5 expression and differentially binds the transcription factor ZBTB3. Our analytical strategy provides a novel framework for future studies aimed at dissecting etiological genetic effects. Finally, both SLE elements of the statistical model appear to operate in Sjögren's syndrome and systemic sclerosis whereas only the IRF5–TNPO3 gene-spanning haplotype is associated with primary biliary cirrhosis, demonstrating the nuance of similarity and difference in autoimmune disease risk mechanisms at IRF5–TNPO3.
INTRODUCTION
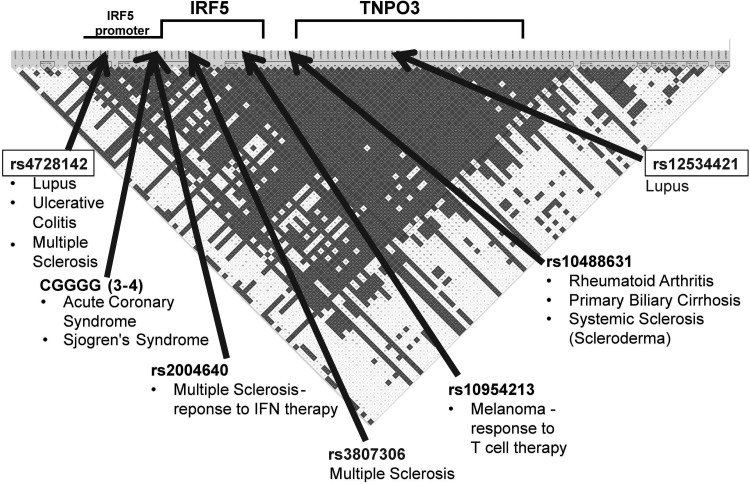
Interferon regulatory factor 5 (Irf5) mediates interferon activation and apoptosis with complex transcriptional regulation and is critical in a number of inflammatory signaling pathways. Variants in or near IRF5 are associated with lupus, rheumatoid arthritis, ulcerative colitis, systemic sclerosis, Sjögren's syndrome, primary biliary cirrhosis, multiple sclerosis and responses to interferon therapy in multiple sclerosis and to T-cell therapy in metastatic melanoma (Fig. 1) (1–11). IRF5 association with lupus is convincing and is found in every major ancestral group tested (1–8,11–29) (Supplementary Material, Fig. S1). However, at this time, multiple sets of proposed functional variants have been nominated to explain the biological mechanisms that drive this association.
Figure 1.
Reported IRF5–TNPO3 associations. Disorders and immunotherapies purported to be associated with IRF5 and TNPO3. The most closely associated variant reported is identified. Disequilibrium as assessed by r2 in European-Americans controls is presented. The positions presented are not on a nucleotide scale.
Biologically, Irf5 is activated by pattern recognition receptors such as Toll-like receptor 7 and 9 (TLR7 and TLR9) and is a critical regulator of the immune response to infection (30). Multiple studies have identified significantly attenuated disease in IRF5-deficient mice relative to wild-type mice in mouse models of lupus (31–33). Many of the original candidate studies of the genetic association of variants around IRF5 fail to identify the neighboring gene that also contains lupus-associated variants: transportin-3 gene (TNPO3). Tnpo3 is a nuclear importer, which is required for the infection of several lentiviruses including HIV (34).
The genetic analyses in lupus that have been reported from the IRF5–TNPO3 locus have focused mainly on subjects of European origin and incomplete identification of the genomic region containing the associated causal genetic variants (Supplementary Material, Fig. S1). Previous regression analyses have identified multiple genetic effects, but to date, there is no genetic model explaining the genetic association of variants in IRF5 and TNPO3 across the major human ancestries. We obtained comprehensive genetic variant coverage in a large trans-ancestral cohort of cases and controls to identify a unifying model to help explain the genetic risk of two genetic effects in lupus. Furthermore, we demonstrate that this model at IRF5–TNPO3 is also, at least in part, consistent with data showing an association at this locus in three other autoimmune diseases.
Our strategy included two different analytical approaches. The frequentist approach computes a probability against the null hypothesis that there is no enrichment of an allele or haplotype in the cases relative to the controls. The Bayesian method calculates a Bayes factor (BF) that provides the increased likelihood that an allele is actually enriched in the cases compared with the control population (reviewed in 35). Under specific assumptions, the posterior probability that a polymorphic variant is driving the association signal can be calculated from the BF (35). For both of the component genetic effects at IRF5–TNPO3, we used this method to identify the likelihood that variants were causal.
Previous reports have identified biologically functional variants across the IRF5–TNPO3 locus that affect transcription factor binding, RNA stability and the DNA-binding domain of IRF5 (3,5,7,8,22,26,36–39). For the IRF5–TNPO3 locus, we performed careful statistical modeling to clarify the biological mechanisms driving a statistical association of genetic variants with disease risk in the context of an overabundance of potential candidates. We demonstrate that the minor allele of rs4728142, a variant tagging the IRF5 promoter effect and not previously included in lupus-risk models (16), leads to the increased binding of the transcription factor ZBTB3 and increased expression of the IRF5 transcript. Very little is known about ZBTB3, a zinc finger transcription factor. We conclude that there are many biologically functional variants at the IRF5–TNPO3 locus, and not all of these are equally likely to be responsible for the genetic associations identified through the statistical models generated herein. We present ZBTB3 as a candidate causal transcription factor responsible for disease risk, realizing that this is only a candidate at this point and that there may be other causal elements, either in addition to or instead of ZBTB3, left yet to be identified.
RESULTS
Experimental design
We genotyped 112 genetic variants spanning the IRF5–TNPO3 locus and in subsets of our sample collected genotyping from the IRF5 promoter copy number variant, CGGGG(3-4), and performed sequencing across the associated region. The data are evaluated across five ethnicities in 8395 systemic lupus erythematosus (SLE) cases and 7367 controls. We imputed against 1000 genomes, adding 2815 high-quality variants with minor allele frequency (MAF) of >0.01. DNA sequencing from the IRF5–TNPO3 region in European (EU) and African American (AA) ancestry imputation identified an additional 303 variants with MAF of >0.01 that were then incorporated. These 3230 variants, all with MAF of > 0.01 (Supplementary Material, Table S1), constitute the most complete assessment of relevant variation for SLE at the IRF5–TNPO3 locus reported to-date and form the basis for model construction of this association under the assumption that common variants (MAF > 0.01) are responsible for the association observed.
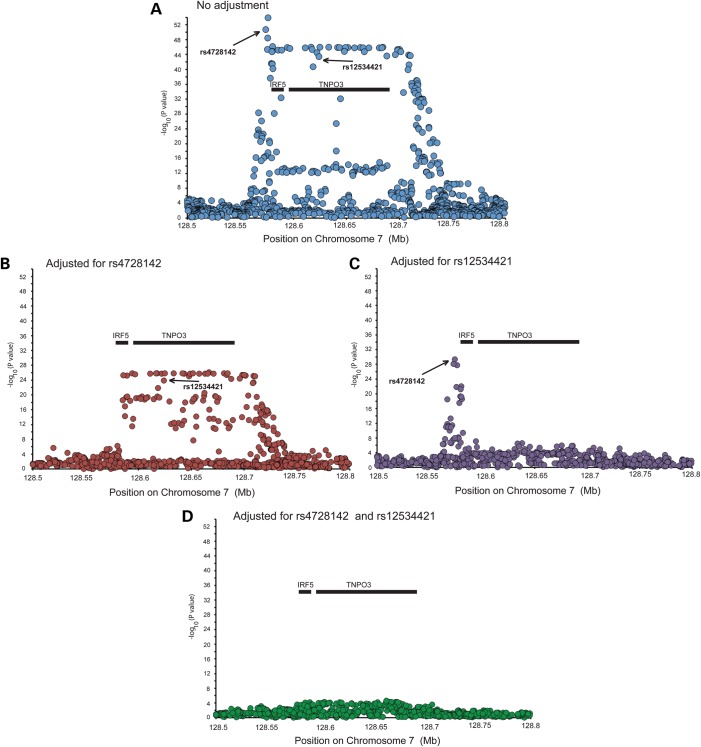
We separated the subjects by ancestry, tested each single-nucleotide polymorphism (SNP) for association with SLE and performed a corresponding step-wise logistic regression analysis, first in each ethnicity (Fig. 2) and then in a meta-analysis to identify independent genetic loci (Figs 3 and 4). In a meta-analysis of four ancestral data sets, adjusting for rs4728142 (IRF5 promoter region) and any variant in high linkage disequilibrium (LD) (r2 > 0.9) with rs12534421 (located in a haplotype of SNPs spanning the IRF5 and TNPO3 genes) was sufficient to remove nearly all residual association (Fig. 3). Adjusting for either variant separately resulted in residual association at the other variant, either at the IRF5 promoter (upon adjustment on rs12534421) or at the IRF5–TNPO3 gene-spanning region (upon adjustment on rs4728142). Based upon the results of the regression analysis, we concluded that these genetic effects made independent contributions to disease risk. Our conclusion was further supported by the LD between rs12534421 and rs4728142 being low in all ancestries (e.g. European LD: r2 = 0.087, D’ = 0.714). We found no statistical interaction among the associations marked by rs4728142 and rs12534421.
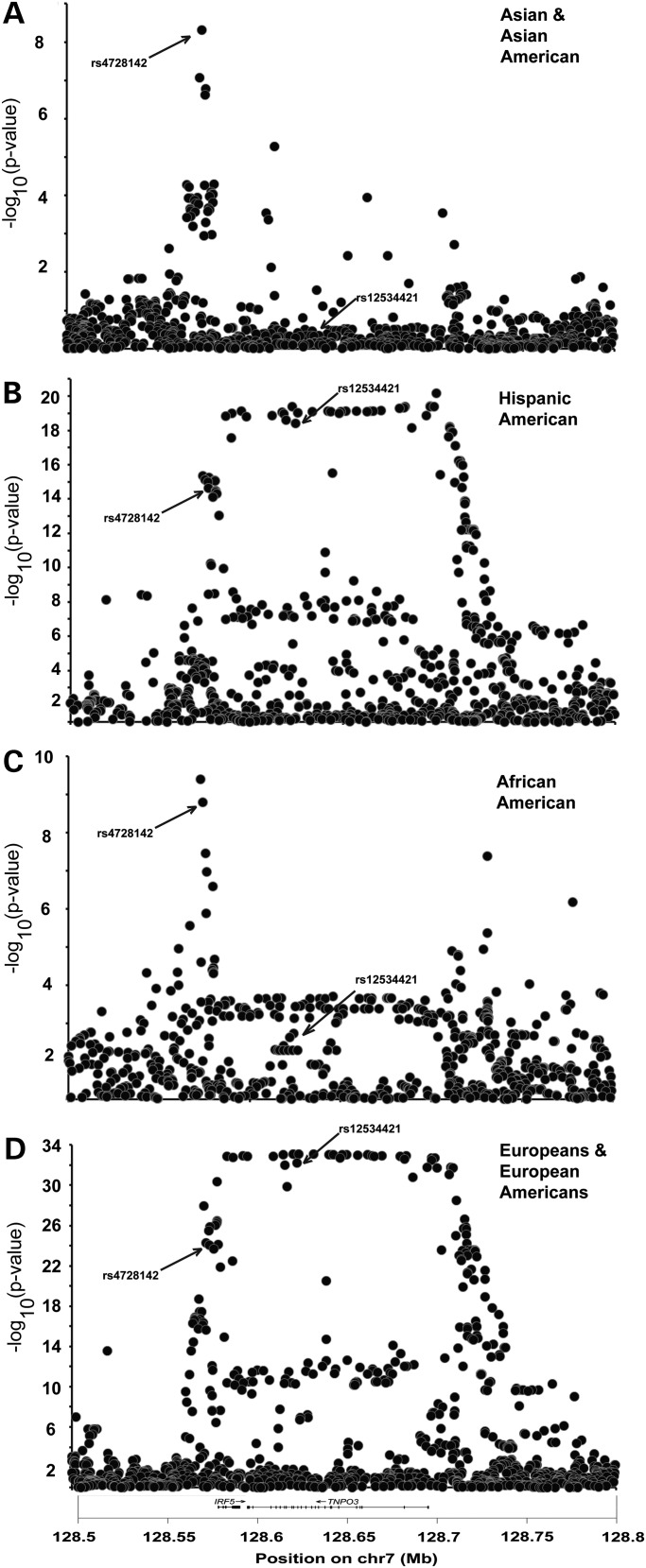
Figure 2.
IRF5- and TNPO3-imputed genetic variants demonstrate different patterns of lupus association in four distinct ethnicities. The association of genotyped and imputed variants in cohorts of SLE in European Americans (EU) (3936 cases and 3491 controls), AAs (AA) (1527 cases and 1811 controls), Asians and AS (1265 cases and 1260 controls) and HAs (HA) (1225 cases and 614 controls) was assessed in a logistic regression with an adjustment for admixture. While the association of SNPs in the promoter of IRF5 is present in all of the cohorts, the association of SNPs spanning the IRF5 and TNPO3 genes is only seen in the cohorts with European admixture.
Figure 3.
Trans-ancestral meta-analysis of genotyped and imputed variants. A meta-analysis of five ancestral cohorts contained 8395 SLE cases and 7367 controls identifies the IRF5–TNPO3 association and its boundaries. For each meta-analysis, the adjusted P-values from the step-wise regression of each ancestry were combined. Panels 1b and 1c present the association of each SNP in the region in a logistic regression adjusting for admixture covariants and rs4728142 (Panel 1a) in the IRF5 promoter and rs12534421 in a haplotype that spans IRF5 and TNPO3 (Panel 1c). After adjusting for these two variants and admixture, no other variant provides convincing evidence of association.
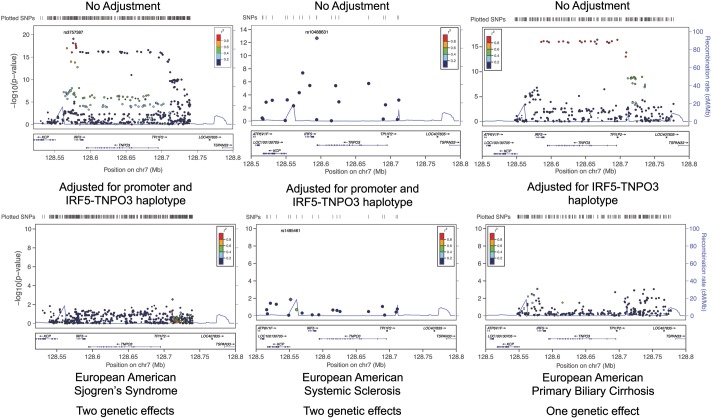
Figure 4.
The IRF5–TNPO3 association across other inflammatory diseases. Genomic position is given with Human Genome Build 37 coordinates. An analysis of European ancestry Sjögren's syndrome and systemic sclerosis using a different set of initially genotyped markers reproduces both components of the model we present herein for the European ancestry in SLE. For the IRF5–TNPO3 association with primary biliary cirrhosis, the IRF5 promoter association identified in SLE is absent, whereas the IRF5–TPO3 haplotype association is present. The full step-wise logistic regression analysis for Sjögren's syndrome, systemic sclerosis and primary biliary cirrhosis is presented in Supplementary Material, Figures S3, S4 and S5, respectively.
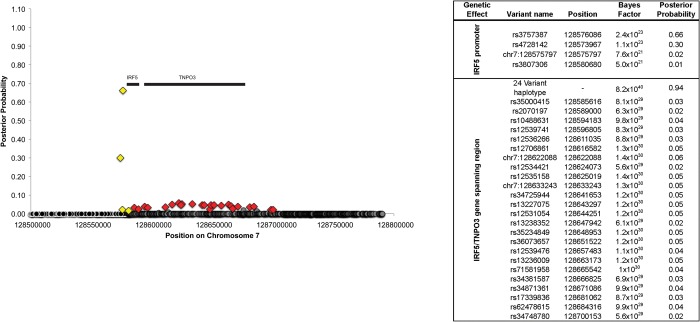
A meta-analysis of the 5 ancestral cohorts containing 8395 cases and 7367 controls clearly identified a group of SNPs in the IRF5 promoter including rs4728142 [P-valuemeta = 1.8 × 10−51; OR range (the range of the OR from the 4 analyses in individual ethnicities) = 1.38–1.97]. We chose rs4728142 to represent this group because it was genotyped and in nearly perfect LD with the most highly associated variant rs3757397 (P-valuemeta = 1.3 × 10−54). The group of variants in the IRF5–TNPO3 gene-spanning haplotype, which can be tagged by rs12534421, were also highly significant in the meta-analysis (P-valuemeta = 3.9 × 10−44; OR range = 1.66–2.44) (Fig. 3). Again, we chose rs12534421 to tag this variant group because it was genotyped and always present on the risk haplotype of imputed variants with stronger association.
The IRF5 promoter association tagged by rs4728142 is present in each population sample evaluated, whereas the association of variants in the IRF5–TNPO3 gene-spanning region, tagged by rs12534421, is polymorphic only in the EU samples or those with admixed European ancestry [AA, Native American (NA) and Hispanic American (HA)] (Fig. 2). In the EU cohort (3936 cases, 3491 controls), we identified both association effects (rs12534421: P-valueEU = 1.1 × 10−32, OREU = 1.701; rs4728142: P-valueEU = 9.34 × 10−26, OREU = 1.381) (Fig. 2D).
The haplotype of 24 variants defining the European IRF5–TNPO3 gene-spanning association was not found in Africans in the HapMap3 or 1000 Genomes controls or in a group of African lupus patients from South Africa (40). The observed frequency of 2.3% in AA controls is close to the 1.6% predicted by admixture alone (Supplementary Material, Table S2). Local admixture analysis showed increased European admixture in the IRF5–TNPO3 region in the admixed AA cases relative to controls. This means that while the AA lupus cases and controls have similar global European admixture, the AA lupus cases have increased local European admixture at the IRF5–TNPO3 locus, supporting the conclusion that the genetic variation of the associated variants in the region spanning the IRF5 and TNPO3 genes is a risk factor for lupus (Supplementary Material, Fig. S2).
Perhaps, some of the diseases and immunotherapies associated with IRF5 use the same mechanism of phenotypic risk that is developed herein for SLE. An analysis of European ancestry in Sjögren's syndrome and systemic sclerosis using a different set of initially genotyped markers reproduces both components of the model we present herein for the European ancestry in SLE (Fig. 6 and Supplementary Material, Figs S3–S4). In contrast, only the IRF5–TNPO3 gene-spanning haplotype is associated with a cohort of subjects with primary biliary cirrhosis (10), an overwhelmingly European disease (41) (Fig. 6 and Supplementary Material, Fig. S5). This is a finding consistent with there being no IRF5–TNPO3 association in Asians with primary biliary cirrhosis (42) (Asians do not have this haplotype by our analysis). These results suggest powerful fundamental similarities and nuanced differences in disease risk mechanisms (Supplementary Material, Figs S3–S5) in the four autoimmune diseases evaluated. What the mechanistic similarities are for how variants at IRF5 contribute to other autoimmune disorders and immunotherapies remains an open but compelling question.
Figure 6.
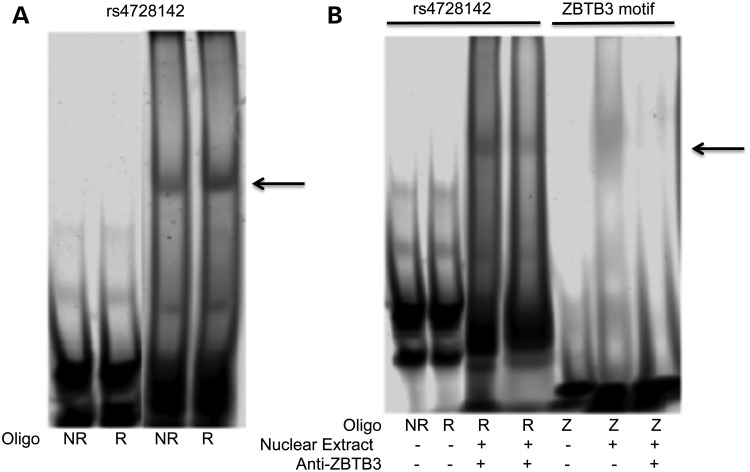
Anti-ZBTB3 reduces the shifted band intensity at rs4728142 suggesting ZBTB3 binding. Labeled oligonucleotides corresponding to the non-risk (NR) and risk (R) variants at rs4728142. (A) Oligonucleotides NR and R at rs4728142 were used to probe nuclear extract from B cells. The four lanes shown are from the same blot. (B) Anti-ZBTB3 antibody was used to compete with the labeled oligonucleotides for binding of the nuclear extract. An oligonucleotide corresponding to the consensus ZBTB3 binding site was used as a positive control. The arrow indicates the specific band.
Lupus-risk haplotypes in IRF5 are associated with a type I Interferon (IFN) signature in lupus patients. Clinically, the response and production of type I IFNs are hypothesized to mediate autoimmune inflammation (40,43–45). Studies in subjects of European ancestry have also identified lupus-associated IRF5 haplotypes that affect mRNA expression, splicing and stability (16,19,21,22,26,38,44,46,47). Other studies have demonstrated that specific IRF5-associated variants affect IRF5 DNA-binding activity and hyper-responsiveness to microbial stimuli (16,39,48,49). Consider the predicted change in Sp1 transcription factor binding at CGGGG(3−4), the differential splicing at rs2004640, the Exon 9 indel at rs10954213, which changes the rate of IRF5 mRNA degradation, and the Exon 6 indel that changes the IRF5 DNA-binding domain. Each of these has a clear biological basis to be a strong functional candidate for causing increased disease risk (16). Our data support the lupus association for all of these variants except for the Exon 6 indel. However, after adjusting for the three variants with association (CGGGG(3-4), rs2004640 and rs10954213), significant residual association remains both at rs4728142 (P < 10−8) in the meta-analysis and at both rs12534421 and the 24-member IRF5–TNPO3 haplotype (residual P < 10−15 for single marker and haplotype). The residual association after adjustment for these variants is also apparent in individual population analyses (Supplementary Material, Table S3).
By assessing the Akaike Information Criterion (AIC), we formally compared the previously reported three-SNP risk-haplotype model (16) (rs2004640, the Exon 6 indel and rs10954213) with our two-SNP model (rs4728142 and rs12534421) in all four ancestries. We found that the AIC for our two-SNP model presented herein was substantially smaller than the three-SNP risk haplotype model (16) (i.e. had significantly more empirical support in all three ancestries): EU: ΔAIC = 45.2, HA: ΔAIC = 8.9, AA: ΔAIC = 3.7, AS: ΔAIC = 10.4 (Supplementary Material, Table S4). Furthermore, using an analysis of haplotypic and genotypic probabilities in Europeans, we adjusted for the risk haplotype of the three-SNP model and found significant residual association with the risk variant of rs12534421 and separately with the risk variant of rs4728142. As previously discussed, the risk haplotype of the three-SNP haplotype model and that of rs12534421 are not polymorphic (MAF < 0.01) in the AS cohort. Owing to the high degree of co-linearity between rs12534421 and the risk haplotype of the previously suggested (16) three-SNP haplotype model, the analysis did not succeed in completely distinguishing the two-variant model presented herein from the three-component haplotype presented previously (16) in the AA or HA samples in our possession. On the other hand, in the EU cohort, we had a sufficient sample size to support the inference that the two-SNP model alone (rs4728142 and rs12534421) explains residual variation beyond what is explained by the three-SNP haplotype. Furthermore, the three-SNP haplotype does not account for the variation explained by rs4728142 in any of the four ancestral cohorts (Supplementary Material, Table S4). Our conclusion is that the three-SNP haplotype model and rs12534421 associations are reflecting the action of the same underlying causal variant(s) underlying the association of the IRF5-TPNO3 haplotype, whereas the three-SNP haplotype model (16) is a much less robust reflection of the association in the promoter tagged by rs4728142.
Using a Bayesian approach, we identified variants at both genetic effects with large posterior probabilities that represent those most likely to be causal among the SNPs typed (Fig. 5) (35) constitute the credible set that could reasonably be considered as plausible candidate causal variants for the IRF5–TNPO3 haplotype association (Fig. 5). Importantly, the Bayesian analysis fails to include the four previously identified functional variants (16), as they were not in the credible set of variants accounting for 95% of the posterior probability in the region for either genetic effect (35).
Figure 5.
Bayesian association plot showing the signal strength in the promoter of IRF5 (position < 128 585 000) and in the IRF5–TNPO3 gene-spanning region (position > 128 585 000) as the posterior probability of each SNP. Genomic position is given with Human Genome Build 37 coordinates; European American data are shown. Single-nucleotide polymorphisms are colored according to membership in credible sets: yellow diamonds, 95% credible set in the promoter region of IRF5; red diamonds, 95% credible set in the region spanning the IRF5 and TNPO3 genes; gray outline, neither set. The position of IRF5 and TNPO3 are labeled. Variants with larger posterior probabilities (>0.01) represent those most likely to be causal among the variants genotyped (yellow or red). Variants with relatively low posterior probabilities (<0.01) are unlikely to be causal (gray or black).
We systematically assessed every possible pair of variants in the imputed data set of all 3230 variants to identify all two-variant combinations that could account for all of the lupus-associated variability of P < 0.01 in the EU, AA and HA cohorts. We confirm that the only possible models include variants that are included in the credible set of 4 variants in the promoter of IRF5 and 24 variants spanning the IRF5 and TNPO3 genes (Fig. 5).
If we have succeeded in considering all relevant variants with MAF of >1% for the IRF5 relationship to SLE risk and if our analysis of this sample reflects the behavior in the population of lupus cases and appropriate controls, then, under these assumptions, two (the two associations detected are independent with no evidence of interaction) or more of the variants identified are likely to contribute to the risk of lupus and would therefore contribute to the cause of disease.
Our genetic analysis demonstrated that rs4728142 is one of four variants that explain 95% of the posterior probability in the IRF5 promoter and is a critical part of our genetic model. We, therefore, examined the biological function of this tagging variant. The location of this variant in the promoter of IRF5 led us to suspect that disease risk at this genetic effect may be mediated by IRF5 expression. We used publically available SNP-mRNA expression data to identify a strong eQTL of rs4728142 with IRF5 using six independent probes across two microarray platforms and three independent groups of subjects (10−4 > P > 10−16) (Supplementary Material, Table S5). The eQTL of other lupus-associated variants in the IRF5 promoter has been previously published (16,21,22,37,47,50), Owing to the linkage-disequilibrium in the region, it is not possible to identify a single variant that is causal in driving the differential expression from this analysis. Indeed, each of the variants in the credible set that could be driving the association in the promoter of IRF5 demonstrates a strong eQTL with IRF5 expression [(16,21,22,37,47,50) and Supplementary Material, Table S5].
To predict which specific transcription factors might have binding sites affected by this genetic variant, we compiled a library of transcription factor motifs utilizing data from the Transfac (51), JASPAR (52) and UniProbe databases (53), as well as HT-SELEX-derived motifs from a recent study (54) and motifs obtained from ENCODE ChIP-seq data (55). The four variants in the credible set for the IRF5 promoter were evaluated with these tools. The most promising candidate was ZBTB3, which was strongly predicted to bind the risk allele of rs4728142. We tested the hypothesis that ZBTB3 differentially bound an oligo of the sequence including the risk or non-risk allele of rs4728142 using electrophoretic mobility shift assays (EMSAs) (Fig. 6A) and confirmed the previously reported differential binding of the risk allele of rs4728142 to nuclear lysate from B-cell lines (7). Using antibodies against ZBTB3, we demonstrated that anti-ZBTB3 was able to inhibit the binding of nuclear lysate to the labeled oligo containing the risk variant of rs4728142 (Fig. 6B), providing evidence for a specific interaction at a plausible candidate causal variant. Taken together, these results suggest that the presence of the rs4728142 risk allele results in increased binding of ZBTB3 in B cells, proving a plausible potential mechanism in which the genotypically elevated expression of IRF5 in lupus patients increases lupus risk.
DISCUSSION
The genotyping data from the substantial sample of SLE cases and controls allow resolution of the IRF5 association with SLE into two separate and independent associations: one in the promoter of IRF5 that is found in all human ancestries and one in a group of variants spanning the IRF5 and TNPO3 genes constituting a haplotype that is present only in the European ancestry. Our approach to this problem at IRF5 has been greatly aided by the differences in the posterior probability of variants in the context of multiple ancestries and aided by the inclusion of variants that we obtained after sequencing and imputation.
A recent study (56) identifies rs12531711 as a variant that originated in Neanderthals. The strong disequilibrium between rs12531711 and rs12534421 (European r2 = 0.78) [while not present between rs12531711 and rs4728142 (European r2 = 0.053)] is consistent with the possibility that the IRF5–TNPO3 spanning haplotype has a Neanderthal origin. The gene flow from Neanderthals into Europeans, but not into Africans, would explain its presence and, given this event in recent evolutionary time, probably the size of the haplotype.
At least two variants, a minimum of one for each component of the risk model we derive, and possibly more than two variants from the two sets of plausible causal candidate variants, are responsible for disease risk at IRF5 in SLE. This raises challenges for explaining why the previously identified functional variants that clearly alter, or are predicted to alter, IRF5 gene product activity (3,5,7,8,22,26,36–39,50) do not also alter disease risk. These formerly identified variants, responsible for changing gene expression or function, are not included among the plausible candidate causal variants in the model we have constructed.
At the moment, our perception of the relevance to disease risk of the binding of ZBTB3 to the risk variant at rs4728142 is completely dependent upon how robust our model of genetic association proves to be. In any case, the most that can be advocated is that the binding of ZBTB3 is a candidate mechanism to explain the association of SLE with the promoter of IRF5. There are an unknown large number of possible genetic models, given the data in our possession that could be constructed to explain lupus risk at this locus. We present a model that is preferred over those previously described (3,5,7,8,22,26,36–39,50). It remains possible that other models could be constructed that cannot be statistically distinguished from the one presented herein.
If the model we present is correct, then its failure to include the previously identified functional variants (3,5,7,8,22,26,36–39,50) suggests four possibilities: the level of IRF5 gene product activity in the cell cannot be a simple function of the level of gene expression; the nature of the mechanism generating disease risk in the IRF5 promoter region is far more nuanced and complex than is accommodated by current concepts; our idea that IRF5 gene expression is involved in disease risk is wrong at its base or there is some other now unknown function that is important to disease risk, perhaps, even at a gene other than IRF5 in cis with this element. After all, association experiments with DNA variants are both powerful and weak because they are agnostic with regard to mechanistic hypotheses; they are associations with a piece of DNA that have no a priori structural or functional requirements. Consequently, and until specific experiments require the conclusion that a particular function is explanatory and causal, the relationship of a particular identified function with disease causality is little more than an unsupported assumption. We argue that attempts to identify the biological functionality of genetic associations often lead to the premature attribution of causality.
The 3-SNP haplotype of functional variants identified by Graham et al. (16) identifies the IRF5–TNPO3 haplotype from our analysis. Graham et al. did not have a complete representation of variants from this association. Consequently, given the data available at that time and the evidence that expression is related to disease risk, their formulation is logical and reasonable. Now that we have a complete data sets of variants and data from four ethnicities, we learn that the formulation that Graham et al. provide reflects the association of the IRF5–TNOP3 (tagged by rs12534421) but not with the IRF5 promoter (tagged by rs4728142). Thus, our work improves on their discovery and separates the second effect that we find at the IRF5 promoter. The variants identified by Graham et al., either individually (Supplementary Material, Table S3) or collectively as a haplotype (Supplementary Material, Table S4), only partially explain the variation at the IRF5 locus whereas the model that uses the two critical sets identified herein, tagged by rs4728142 and rs12534421, completely explains all of the variation at the IRF5 locus, including the variation left unexplained by the 3-SNP model presented by Graham et al. Consequently, we cannot and do not eliminate the possibility that these variants contribute.
Indeed, recent studies and the mechanistic studies herein demonstrating the functionality of rs4728142 suggest that all of the biologically functional variants in this locus have not yet been identified, not to mention how they might relate to disease risk. In general, we now have a growing overabundance of functionally relevant variants at a locus known to create a gene product (IRF5) that mediates pathways that are central for disease etiology, which we suspect will make the convincing identification of disease mechanism more difficult. This is not a problem isolated to the promoter of IRF5, but is and will remain the major impediment to progress for the vast majority of the now known >10 500 associations found by genome-wide association studies and is a sobering assessment of the magnitude of the work remaining, not to mention the incisive experimental creativity that will be required to establish plausible causality.
Physical mapping of genetic associations is limited by the sample evaluated and its population history. In the case of the IRF5 promoter, the capacity of the model-building exercise to limit the set of plausible causal variants to four members is an enormous benefit to experimental efficiency. As we had started with 3230 possible variants, the reduction of those under consideration by almost a 1000-fold is an important contribution to subsequent conceptual construction. Evaluating the IRF5–TNPO3 haplotype set with its 24 members stretched over 85 kb is not so straightforward. A different experimental orientation for identifying candidate causal mechanisms is required for such extended haplotypes than it is for much smaller elements, such as we find in the promoter of IRF5.
Our results underscore the perplexing and, as yet, unanswered question of why so many biologically functional variants are present at the IRF5–TNPO3 locus that do not appear to contribute to the statistical models for lupus disease risk. Perhaps, these variants contribute to risk for other properties of the SLE cases, such as phenotypic variation or disease outcome, or to one of the other associated disorders not yet evaluated using this approach (Fig. 1). Imputed variants were used in this analysis, and to address this limitation, we performed a two-step imputation to a 1000-genome and lupus-specific reference panel and did not use imputed variants in our genetic modeling. We used a trans-ancestral cohort instead of a classical discovery-replication cohort design, and future studies will assess the genetic model presented herein in a cohort of independent subjects.
Characteristics of the variation we model in disease risk for SLE are consistent, at least in part, with the data available from Sjögren's syndrome, systemic sclerosis and primary biliary cirrhosis, raising the possibility that particular IRF5–TNPO3 variants operate to alter disease risk in similar ways across these different disease phenotypes.
We have presented an approach to identify the possible causal variants at a locus associated with disease etiology. All of the variants at each independent association were assessed for their potential to drive the association in order to be confident that no plausible candidate is unintentionally removed from consideration because the data are missing. We calculated the posterior probability from the BF for each variant. This analysis identified the variants most likely to cause the genetic association based upon the relative posterior probabilities. Once the variants with the highest likelihood of driving the genetic associations were identified, they were assessed individually and in combination for function using both bioinformatic and experimental approaches.
In conclusion, we performed a large trans-ancestral fine mapping and disease risk modeling study of the IRF5–TNPO3 locus to identify genetic variants that increase lupus risk. Using frequentist and Bayesian methods, we clearly identified two independent genetic effects: one in the promoter of IRF5 which is present in all ancestries and one that spans the IRF5 and TNPO3 genes which is European, perhaps really Neanderthal in origin. Posterior probabilities identify the variants most likely to drive the association signals. Of these, we identified an allele-specific novel biological function at rs4728142 in the promoter of IRF5. After assessing the lupus model of IRF5 disease risk to three other autoimmune diseases, we found striking similarities and differences in the disease risk derived from association studies at the IRF5–TNPO3 locus. Finally, we present a specific application of a general approach for extracting as much information as is available from population history in the service of evaluating allele-associated differences in biological function for their potential to explain phenotype, herein as disease phenotypes.
MATERIALS AND METHODS
Subjects and study design
We used a large collection of samples from case–control subjects from multiple ethnic groups. These samples were from the collaborative Large Lupus Association Study 2 (LLAS2) (57) and were contributed by participating institutions in the USS, Asia and Europe. According to genetic ancestry, subjects were grouped into five ethnic groups including European American (EU), AA, Asian and Asian American (AS), NA and HA. All SLE patients met the American College of Rheumatology criteria for the classification of SLE (58).
Genotyping of genetic variants and sample quality control
We genotyped 112 SNPs covering the entire IRF5–TNPO3 region (Supplementary Material, Table S1, 128–129 MB on Chr 7, Build 37), as part of a larger custom genotyping study. The variants were chosen based upon the results of a genome-wide association study of 720 women of European ancestry and 2337 controls in the LLAS2 (11). Specifically, the variants were chosen to span the association interval identified with the Infinium HumanHap330 array. Genotyping of SNPs was completed with Infinium chemistry on an Illumina iSelect custom array according to the manufacturer's protocol. The following quality-control procedures were implemented to identify SNPs for analysis: well-defined clusters for genotype calling, call rate of >90% across all samples genotyped, MAF of >1% (except for the rare variant analysis as described later) and P < 0.05 for differential missingness between cases and controls (the total proportion missing was <5%). Markers with evidence of a departure from Hardy–Weinberg proportion expectation (P < 0.0001 in controls) were removed from the initial analysis.
We removed samples with a call rate of <90% or excess heterozygosity. The remaining individuals were examined for excessive allele sharing as estimated by identity-by-descent (IBD). In sample pairs with excessive relatedness (IBD > 0.4), one individual was removed from the analysis on the basis of the following criteria: (1) remove the sample with the lower call rate, (2) remove the control and retain the case, (3) remove the male sample before the female sample, (4) remove the younger control before the older control and (5) in a situation with two cases, remove the case with the less complete phenotype data available. Discrepancies between self-reported and genetically determined gender were evaluated.
Ascertainment of population stratification
Genetic outliers from each ethnic and/or racial group were removed from further analysis as determined by principal component (PC) analysis and admixture estimates [Fig. 1 of ref. 59 (60) and ref. 61 and 62 (63,64)]. We used 347 ancestral informative markers (AIMs) from the same custom genotyping study that passed quality control in both EIGENSTRAT (64) and ADMIXMAP (59,61) to distinguish the four continental ancestral populations, allowing identification of the substructure within the sample set (62,65). The AIMs were selected to distinguish four continental ancestral populations: Africans, Europeans, American Indians and East Asians. We utilized PCs from EIGENSTRAT outputs to identify outliers of each of the first three PCs for the individual population clusters with visual inspection.
Statistical analysis: workflow
The analysis was initiated by assessing the association of genotyped variants in each of the four ancestral cohorts individually. Strategically, we analyzed the genotyped, then imputed variants, performed full haplotype analysis, executed an analysis of LD and finally built statistical models to account for the lupus-associated variability in each ancestry. In building the two-SNP models of association in each ancestry, we comparatively evaluated every possible combination of two variants for their ability to better account for the lupus-associated genetic variation. A meta-analysis was finally performed to combine the association results across ancestries (see Meta-analysis).
Statistical analysis: - frequentist approach
We tested each SNP for association with SLE using logistic regression models that included gender and three admixture proportion estimates as covariates as implemented in PLINK v 1.07 and SNPTEST (66,67). The additive genetic model is the primary model of inheritance. Other models are subsequently considered, but only if they are substantially superior.
Step-wise logistic regression was performed to identify those SNPs independently associated with the development of lupus in PLINK and SNPTEST. For these analyses, the allelic dosage(s) of specific variant(s) are added to the logistic model as covariates in addition to the admixture estimates and gender. Haplotypic associations were assessed using logistic regression and incorporating admixture measurements and gender as covariates. For the 24-SNP haplotype spanning the IRF5 and TNPO3 genes, logistic regression was used to determine the residual association of the haplotype after accounting for individual SNPs, such as rs12534421.
Linkage disequilibrium and haplotypes were determined with HAPLOVIEW v 4.2 (68–70). We calculated haplotype blocks for those haplotypes present at >3% frequency using the four gamete rule algorithms with a minimum r2-value of 0.8. Haplotypic associations were performed in PLINK using both a sliding window approach and by assessing the association of haplotypes defined using logistic regression, as described earlier.
Statistical analysis: Bayesian approach
Using SNPTEST, we calculated the BF for each SNP: the probability of the genotype configuration at that SNP in cases and controls under the alternative hypothesis that the SNP is associated with disease status divided by the probability of the genotype configuration at that SNP in cases and controls under the null hypothesis that disease status is independent of genotype at that SNP as previously described (we used the methods developed and introduced in ref. 35). We used three admixture estimates as covariates, as we did for the frequentist approach. Large values of the BF correlate to robust evidence for association, as small P-values correlate to strong evidence in a frequentist approach. For well-powered studies, the BFs of relatively common variants are highly correlated with the P-values [reviewed in ref. 71 (72)]. We used the additive model. The linear predictor is log(pi/(1 − pi)) = µ + ßGi, and the prior is µ∼N(0,12), ß∼N(0,0.22) (variables are defined in the supplementary note in ref. 35).
To identify the variants most likely to be driving the statistical association, we calculated a posterior probability under the assumption that any of the variants within a single genetic effect could be causal and that only one of these variants is causal for each genetic effect. Variants with a low posterior probability are highly unlikely to be causal regardless of the allele frequency or presence of the actual causal variant in the analysis, following the procedure as presented (35). Regardless of whether the causal variants have been genotyped in this experiment, variants with a low posterior probability are unlikely to be causal (35).
Meta-analysis
The meta-analyses were conducted using METAL (http://www.sph.umich.edu/csg/abecasis/Metal/) with inverse normal approach. This tool allowed P-values across ancestral cohorts to be combined [taking sample size and direction of effect (odds ratio) into account]. For the meta-analysis of adjusted data from the step-wise regression, the adjusted P-values of each ancestry were combined.
Re-sequencing
We re-sequenced the IRF5–TNPO3 region as described previously (73). DNA from AA and European American subjects included in the current genotyping experiment was sequenced. To assess the accuracy of sequence-based SNP calling, we cross-referenced the sequenced and genotyped allele calls. We observed ∼99% concordance between genotypes and sequence-based variant detection, suggesting high-quality sequence data. We manually inspected the samples with 5% of variants differing between sequencing and genotyping to determine where sequence quality was poor (for example at CGGGG(3-4) see below).
Briefly, 3–5 micrograms of whole genomic DNA from each sample was sheared and prepared for sequencing with an Illumina Paired-End Genomic DNA Sample Prep Kit. Targeted regions of interest from each sample were then enriched with a SureSelect Target Enrichment System utilizing a custom-designed bait pool (Agilent Technologies). Post-sequence data were processed with Pipeline software v.1.7 (Illumina). All samples were sequenced to minimum average fold coverage of 253. Variant detection and quality control were also performed as previously described (73).
CGGGG(3-4) is a known lupus-associated functional insertion in the promoter of IRF5 (21,22,26,39). CGGGG(3-4) repeat is in public databases but is poorly covered in HapMap and 1000 genomes owing to the variable repeat. Our deep sequencing data include CGGGG(3-4), which we concluded was not reliable. We consequently used polymerase chain reaction product size differences to genotype this variant in 654 cases of AA ancestry and 346 EU cases whose data are included in the genotyping reported (bringing the total markers genotyped in the study to 113) and were used to impute the CGGGG(3-4) genotype in the remaining subjects. Specifically, a file set containing only European Americans with and without the CGGGG(3-4) was created, and the CGGGG(3-4) variant was imputed. This process was repeated in AAs. For Asian and HAs, all four ancestries were combined and European American and AA subjects with direct CGGGG(3-4) were used to impute the insertion/deletion. For this imputation, concordance of 500 directly genotyped AA and EU individuals with the imputed CGGGG(3-4) when the variant was masked was found to be 99.2%.
Imputation to composite 1000 genomes reference panel
To detect associated variants that were not directly genotyped, we imputed the IRF5-TNP03 region with IMPUTE2 and using a composite imputation reference panel based on 1000 Genomes Project sequence data freezes from March 2012 (21,67,74). Imputed genotypes were required to meet or exceed a probability threshold of 0.5, and information measure of >0.4 and the same quality-control criteria threshold described for the genotyped markers. In the statistical analyses, the probability threshold from each imputed value was incorporated into the statistical analysis using SNPTEST. We assessed 2015 subjects for the concordance of the imputed SNPs with 400 high-quality variants genotyped on the ImmunoChip as part of a separate study. The overall genotype-imputed variant concordance rate was >99%.
Electrophoretic mobility shift assay
Pairs of single-stranded 5′ IRDye infrared dye-labeled and -unlabeled 27 or 53 base oligonucleotides (obtained from IDT, Inc., Coralville, Iowa, USA) were annealed to generate double-stranded probes. 25–50 fmoles of labeled probes were incubated with 8 or 10 μg of nuclear extract prepared from Epstein–Barr virus-transformed B-cell lines, 6 ug poly (dI-dC) and 1 μl salmon sperm provided along with the buffers and protocols supplied with the Odyssey Infrared EMSA kit (LI-COR Biosciences, Lincoln, Nebraska, USA). Competition experiments were performed using 1 μg of anti-ZBTB3 antibody (Novus Biologicals, Cambridge, UK, Cat # NBP1-82079). The binding reactions were analyzed using electrophoresis on 6% TBE polyacrylamide gels and detected by an infrared fluorescent procedure using the Odyssey Infrared Imaging System (LI-COR Biosciences).
ZBTB3 motif:
5′ AAGCTGCTATTGCAGTGCCTGCAAGAA 3′
5′ TTCTTGCAGGCACTGCAATAGCAGCTT 3′
Non-risk:
5′ GGTCACACCCCAAAAAGCTCTGAGCCGGTGTTAGTAAGAAATGGGGAGGAAGG3′
5′ CCTTCCTCCCCATTTCTTACTAACACCGGCTCAGAGCT TTTTGGGGTGTGACC 3′
Risk:
5′ GGTCACACCCCAAAAAGCTCTGAGCCAGTGTTAGTAAGAAATGGGGAGGAAGG 3′
5′ CCTTCCTCCCCATTTCTTACTAACACTGGCTCAGAGCTTTTTGGGGTGTGACC 3′
Imputation to custom reference panel
GATK Readbacked algorithm was used to determine variant phase. An imputation reference panel was derived from the sequencing data using the VCFtools analysis suite. IMPUTE2 was used to impute variants captured in the targeted resequencing of the IRF5–TNPO3 region. A second imputation procedure, BEAGLE, was also used to determine variant phase with virtually identical association results. Phased genotypes with a phred-scaled phasing quality score of <10 were not used (indicating a 1 of 10 chances that the genotype was inaccurately phased). The results from the three imputation strategies were combined for a final data set that captures the common variation (MAF > 0.01) in the region.
Local admixture analysis
The local admixture assessments were made with SNPs from the IRF5–TNPO3 locus. LAMP is an algorithm that computes the ancestry structure for overlapping windows of contiguous SNPs and combines the results with a majority vote. The algorithm is based on a window-based processing combined within a hierarchical Hidden Markov Model, which can process 2–5 mixing populations.
For local ancestry estimation, we used the software LAMP in LAMPANC mode providing allele frequencies for the HGDP West Africans and European ancestral populations. A total of 1460 SNPs were included in the analysis, and configuration parameters were set as follows: mixture proportions (alpha) = 0.25, 0.75; number of generations since admixture (g) = 7; recombination rate (r) = 1 × 10−8; fraction of overlap between adjacent windows (offset) = 0.2; number of EM iterations = 2000 and r2 threshold (ld cut-off) = 0.1 (70).
Non-lupus data sets
Data for Sjögren's syndrome and controls were taken from an genome-wide association study using the data imputed from the OMNI-1 array (Illumina) led by Chris Lessard and Kathy Moser Sivils at the Oklahoma Medical Research Foundation following data quality criteria similar to those presented earlier (71). Data for primary biliary cirrhosis were taken from a candidate gene study using the ImmunoChip (Illumina) and being led by Kathy Siminovitch of the University of Toronto, again following similar data quality standards (21). Data for systemic sclerosis were from a genome-wide association study of European and North American subjects with systemic sclerosis and controls genotyped on the Human CNV370K and 550K BeadChips (Illumina) being led by Javier Martin, Maureen Mayes and Timothy Radstake (75). No sequencing data were available for systemic sclerosis or primary biliary cirrhosis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. The contents are the sole responsibility of the authors and do not necessarily represent the official views of NIAMS or NIH.
FUNDING
This work has been supported by National Institutes of Health grants and contracts (AI024717, AR042460, AI031584, DE015223, AR057172, AI083194, AR043418, AR065626, AR049084, AI082714, AR052300, AR062277, AR060366, AI094377, GM103510, RR020143, AR062755, AR30692, AR048940, RR027190, RR026314, RR029882, 1RR025741, TR000165, AR 002138, AI070304, AR43727, AR0608040, DE015223, AR058959, AR049084, AR053483 AI082714, DE018209-02, DE018209, RR020143, AI083194, RR027190, AI101934, and GM103510); the U.S. Department of Defense (PR094002); the U.S. Department of Veterans Affairs (IMMA 9); the General Center Research Center (RR-000079); Alliance for Lupus Research; the Korea Healthcare technology R&D Project; Ministry for Health and Welfare, Republic of Korea (HI12C1834); Mary Kirkland Scholar (J.B.H. and L.A.C.); the Swedish Rheumatism Association, American College of Rheumatology Research and Education Foundation/Abbott Healthy Professional Graduate Student Preceptorship Award 2009; Oklahoma Medical Research Foundation, Sjögren's Syndrome Foundation (4434); Phileona Foundation, French Ministry of Health (PHRC N°2006-AOM06133); The Strategic Research Program at Helse Bergen, Western Norway Regional Health Authority and The Broegelmann Foundation (KFO 250, TP03, WI 1031/6-1″, "KFO 250, Z1); Medical Research Council (UK G0800629); Northumberland, Tyne & Wear CLRN; the Canadian Institutes for Health Research (MOP74621); the Ontario Research Fund (REO-061); the PBC Society of Canada; Canadian Institutes of Health Research; and the Canada Research Chair and the Sherman Family Chair in Genomic Medicine.
Supplementary Material
REFERENCES
- 1.Hirschfield G.M., Liu X., Han Y., Gorlov I.P., Lu Y., Xu C., Chen W., Juran B.D., Coltescu C., Mason A.L., et al. Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nat. Genet. 2010;42:655–657. doi: 10.1038/ng.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson C.A., Boucher G., Lees C.W., Franke A., D'Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A., et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin J.E., Broen J.C., Carmona F.D., Teruel M., Simeon C.P., Vonk M.C., van't Slot R., Rodriguez-Rodriguez L., Vicente E., Fonollosa V., et al. Identification of CSK as a systemic sclerosis genetic risk factor through Genome Wide Association Study follow-up. Hum. Mol. Genet. 2012;21:2825–2835. doi: 10.1093/hmg/dds099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stahl E.A., Raychaudhuri S., Remmers E.F., Xie G., Eyre S., Thomson B.P., Li Y., Kurreeman F.A., Zhernakova A., Hinks A., et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miceli-Richard C., Gestermann N., Ittah M., Comets E., Loiseau P., Puechal X., Hachulla E., Gottenberg J.E., Lebon P., Becquemont L., et al. The CGGGG insertion/deletion polymorphism of the IRF5 promoter is a strong risk factor for primary Sjogren's syndrome. Arthritis Rheum. 2009;60:1991–1997. doi: 10.1002/art.24662. [DOI] [PubMed] [Google Scholar]
- 6.Fan J.H., Gao L.B., Pan X.M., Li C., Liang W.B., Liu J., Li Y., Zhang L. Association between IRF-5 polymorphisms and risk of acute coronary syndrome. DNA Cell Biol. 2010;29:19–23. doi: 10.1089/dna.2009.0929. [DOI] [PubMed] [Google Scholar]
- 7.Kristjansdottir G., Sandling J.K., Bonetti A., Roos I.M., Milani L., Wang C., Gustafsdottir S.M., Sigurdsson S., Lundmark A., Tienari P.J., et al. Interferon regulatory factor 5 (IRF5) gene variants are associated with multiple sclerosis in three distinct populations. J. Med. Genet. 2008;45:362–369. doi: 10.1136/jmg.2007.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vosslamber S., van der Voort L.F., van den Elskamp I.J., Heijmans R., Aubin C., Uitdehaag B.M., Crusius J.B., van der Pouw Kraan T.C., Comabella M., Montalban X., et al. Interferon regulatory factor 5 gene variants and pharmacological and clinical outcome of Interferonb therapy in multiple sclerosis. Genes Immun. 2012;13:443. doi: 10.1038/gene.2011.18. [DOI] [PubMed] [Google Scholar]
- 9.Uccellini L., De Giorgi V., Zhao Y., Tumaini B., Erdenebileg N., Dudley M.E., Tomei S., Bedognetti D., Ascierto M.L., Liu Q., et al. IRF5 gene polymorphisms in melanoma. J. Transl. Med. 2012;10:170. doi: 10.1186/1479-5876-10-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juran B.D., Hirschfield G.M., Invernizzi P., Atkinson E.J., Li Y., Xie G., Kosoy R., Ransom M., Sun Y., Bianchi I., et al. Immunochip analyses identify a novel risk locus for primary biliary cirrhosis at 13q14, multiple independent associations at four established risk loci and epistasis between 1p31 and 7q32 risk variants. Hum. Mol. Genet. 2012;21:5209–5221. doi: 10.1093/hmg/dds359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harley J.B., Alarcón-Riquelme M.E., Criswell L.A., Jacob C.O., Kimberly R.P., Moser K.L., Tsao B.P., Vyse T.J., Langefeld C.D. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunninghame Graham D.S., Manku H., Wagner S., Reid J., Timms K., Gutin A., Lanchbury J.S., Vyse T.J. Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation. Hum. Mol. Genet. 2007;16:579–591. doi: 10.1093/hmg/ddl469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demirci F.Y., Manzi S., Ramsey-Goldman R., Minster R.L., Kenney M., Shaw P.S., Dunlop-Thomas C.M., Kao A.H., Rhew E., Bontempo F., et al. Association of a common interferon regulatory factor 5 (IRF5) variant with increased risk of systemic lupus erythematosus (SLE) Ann. Hum. Genet. 2007;71:308–311. doi: 10.1111/j.1469-1809.2006.00336.x. [DOI] [PubMed] [Google Scholar]
- 14.Ferreiro-Neira I., Calaza M., Alonso-Perez E., Marchini M., Scorza R., Sebastiani G.D., Blanco F.J., Rego I., Pullmann R., Jr, Pullmann R., et al. Opposed independent effects and epistasis in the complex association of IRF5 to SLE. Genes Immun. 2007;8:429–438. doi: 10.1038/sj.gene.6364407. [DOI] [PubMed] [Google Scholar]
- 15.Gateva V., Sandling J.K., Hom G., Taylor K.E., Chung S.A., Sun X., Ortmann W., Kosoy R., Ferreira R.C., Nordmark G., et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham R.R., Kyogoku C., Sigurdsson S., Vlasova I.A., Davies L.R., Baechler E.C., Plenge R.M., Koeuth T., Ortmann W.A., Hom G., et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proci. USA. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. Natl. Acad. Sc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han J.W., Zheng H.F., Cui Y., Sun L.D., Ye D.Q., Hu Z., Xu J.H., Cai Z.M., Huang W., Zhao G.P., et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 18.Jarvinen T.M., Hellquist A., Zucchelli M., Koskenmies S., Panelius J., Hasan T., Julkunen H., D'Amato M., Kere J. Replication of GWAS-identified systemic lupus erythematosus susceptibility genes affirms B-cell receptor pathway signalling and strengthens the role of IRF5 in disease susceptibility in a Northern European population. Rheumatology (Oxford) 2012;51:87–92. doi: 10.1093/rheumatology/ker263. [DOI] [PubMed] [Google Scholar]
- 19.Kawasaki A., Kyogoku C., Ohashi J., Miyashita R., Hikami K., Kusaoi M., Tokunaga K., Takasaki Y., Hashimoto H., Behrens T.W., et al. Association of IRF5 polymorphisms with systemic lupus erythematosus in a Japanese population: support for a crucial role of intron 1 polymorphisms. Arthritis Rheum. 2008;58:826–834. doi: 10.1002/art.23216. [DOI] [PubMed] [Google Scholar]
- 20.Kelly J.A., Kelley J.M., Kaufman K.M., Kilpatrick J., Bruner G.R., Merrill J.T., James J.A., Frank S.G., Reams E., Brown E.E., et al. Interferon regulatory factor-5 is genetically associated with systemic lupus erythematosus in African Americans. Genes Immun. 2008;9:187–194. doi: 10.1038/gene.2008.4. [DOI] [PubMed] [Google Scholar]
- 21.Lofgren S.E., Yin H., Delgado-Vega A.M., Sanchez E., Lewen S., Pons-Estel B.A., Witte T., D'Alfonso S., Ortego-Centeno N., Martin J., et al. Promoter insertion/deletion in the IRF5 gene is highly associated with susceptibility to systemic lupus erythematosus in distinct populations, but exerts a modest effect on gene expression in peripheral blood mononuclear cells. J. Rheumatol. 2010;37:574–578. doi: 10.3899/jrheum.090440. [DOI] [PubMed] [Google Scholar]
- 22.Nordang G.B., Viken M.K., Amundsen S.S., Sanchez E.S., Flato B., Forre O.T., Martin J., Kvien T.K., Lie B.A. Interferon regulatory factor 5 gene polymorphism confers risk to several rheumatic diseases and correlates with expression of alternative thymic transcripts. Rheumatology (Oxford) 2012;51:619–626. doi: 10.1093/rheumatology/ker364. [DOI] [PubMed] [Google Scholar]
- 23.Qin L., Lv J., Zhou X., Hou P., Yang H., Zhang H. Association of IRF5 gene polymorphisms and lupus nephritis in a Chinese population. Nephrology (Carlton) 2010;15:710–713. doi: 10.1111/j.1440-1797.2010.01327.x. [DOI] [PubMed] [Google Scholar]
- 24.Reddy M.V., Velazquez-Cruz R., Baca V., Lima G., Granados J., Orozco L., Alarcon-Riquelme M.E. Genetic association of IRF5 with SLE in Mexicans: higher frequency of the risk haplotype and its homozygozity than Europeans. Hum. Genet. 2007;121:721–727. doi: 10.1007/s00439-007-0367-6. [DOI] [PubMed] [Google Scholar]
- 25.Shin H.D., Sung Y.K., Choi C.B., Lee S.O., Lee H.W., Bae S.C. Replication of the genetic effects of IFN regulatory factor 5 (IRF5) on systemic lupus erythematosus in a Korean population. Arthritis Res. Ther. 2007;9:R32. doi: 10.1186/ar2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sigurdsson S., Goring H.H., Kristjansdottir G., Milani L., Nordmark G., Sandling J.K., Eloranta M.L., Feng D., Sangster-Guity N., Gunnarsson I., et al. Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Hum. Mol. Genet. 2008;17:872–881. doi: 10.1093/hmg/ddm359. [DOI] [PubMed] [Google Scholar]
- 27.Sigurdsson S., Nordmark G., Goring H.H., Lindroos K., Wiman A.C., Sturfelt G., Jonsen A., Rantapaa-Dahlqvist S., Moller B., Kere J., et al. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am. J. Hum. Genet. 2005;76:528–537. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siu H.O., Yang W., Lau C.S., Chan T.M., Wong R.W., Wong W.H., Lau Y.L., Alarcon-Riquelme M.E. Association of a haplotype of IRF5 gene with systemic lupus erythematosus in Chinese. J. Rheumatol. 2008;35:360–362. [PubMed] [Google Scholar]
- 29.Yang W., Shen N., Ye D.Q., Liu Q., Zhang Y., Qian X.X., Hirankarn N., Ying D., Pan H.F., Mok C.C., et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cham C.M., Ko K., Niewold T.B. Interferon regulatory factor 5 in the pathogenesis of systemic lupus erythematosus. Clin. Dev. Immunol. 2012;2012:780436. doi: 10.1155/2012/780436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng D., Yang L., Bi X., Stone R.C., Patel P., Barnes B.J. Irf5-deficient mice are protected from pristane-induced lupus via increased Th2 cytokines and altered IgG class switching. Eur. J. Immunol. 2012;42:1477–1487. doi: 10.1002/eji.201141642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tada Y., Kondo S., Aoki S., Koarada S., Inoue H., Suematsu R., Ohta A., Mak T.W., Nagasawa K. Interferon regulatory factor 5 is critical for the development of lupus in MRL/lpr mice. Arthritis Rheum. 2011;63:738–748. doi: 10.1002/art.30183. [DOI] [PubMed] [Google Scholar]
- 33.Yang L., Feng D., Bi X., Stone R.C., Barnes B.J. Monocytes from Irf5-/- mice have an intrinsic defect in their response to pristane-induced lupus. J. Immunol. 2012;189:3741–3750. doi: 10.4049/jimmunol.1201162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larue R., Gupta K., Wuensch C., Shkriabai N., Kessl J.J., Danhart E., Feng L., Taltynov O., Christ F., Van Duyne G.D., et al. Interaction of the HIV-1 intasome with transportin 3 protein (TNPO3 or TRN-SR2) J. Biol. Chem. 2012;287:34044–34058. doi: 10.1074/jbc.M112.384669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wellcome Trust Case Control Consortium, Maller J.B., McVean G., Byrnes J., Vukcevic D., Palin K., Su Z., Howson J.M., Auton A., Myers S., et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dideberg V., Kristjansdottir G., Milani L., Libioulle C., Sigurdsson S., Louis E., Wiman A.C., Vermeire S., Rutgeerts P., Belaiche J., et al. An insertion–deletion polymorphism in the interferon regulatory factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Hum. Mol. Genet. 2007;16:3008–3016. doi: 10.1093/hmg/ddm259. [DOI] [PubMed] [Google Scholar]
- 37.Graham R.R., Kozyrev S.V., Baechler E.C., Reddy M.V., Plenge R.M., Bauer J.W., Ortmann W.A., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat. Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 38.Feng D., Stone R.C., Eloranta M.L., Sangster-Guity N., Nordmark G., Sigurdsson S., Wang C., Alm G., Syvanen A.C., Ronnblom L., et al. Genetic variants and disease-associated factors contribute to enhanced interferon regulatory factor 5 expression in blood cells of patients with systemic lupus erythematosus. Arthritis Rheum. 2010;62:562–573. doi: 10.1002/art.27223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hedl M., Abraham C. IRF5 risk polymorphisms contribute to interindividual variance in pattern recognition receptor-mediated cytokine secretion in human monocyte-derived cells. J. Immunol. 2012;188:5348–5356. doi: 10.4049/jimmunol.1103319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niewold T.B., Kelly J.A., Kariuki S.N., Franek B.S., Kumar A.A., Kaufman K.M., Thomas K., Walker D., Kamp S., Frost J.M., et al. IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Ann. Rheum. Dis. 2012;71:463–468. doi: 10.1136/annrheumdis-2011-200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farrell G.C. Primary biliary cirrhosis in Asians: less common than in Europeans, but just as depressing. J. Gastroenterol. Hepatol. 2008;23:508–511. doi: 10.1111/j.1440-1746.2008.05379.x. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura M., Nishida N., Kawashima M., Aiba Y., Tanaka A., Yasunami M., Nakamura H., Komori A., Nakamuta M., Zeniya M., et al. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am. J. Hum. Genet. 2012;91:721–728. doi: 10.1016/j.ajhg.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niewold T.B., Kelly J.A., Flesch M.H., Espinoza L.R., Harley J.B., Crow M.K. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58:2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rullo O.J., Woo J.M., Wu H., Hoftman A.D., Maranian P., Brahn B.A., McCurdy D., Cantor R.M., Tsao B.P. Association of IRF5 polymorphisms with activation of the interferon alpha pathway. Ann. Rheum. Dis. 2010;69:611–617. doi: 10.1136/ard.2009.118315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guthridge J.M., Clark D.N., Templeton A., Dominguez N., Lu R., Vidal G.S., Kelly J.A., Kauffman K.M., Harley J.B., Gaffney P.M., et al. Effects of IRF5 lupus risk haplotype on pathways predicted to influence B cell functions. J. Biomed. Biotechnol. 2012;2012:594056. doi: 10.1155/2012/594056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malarstig A., Sigurdsson S., Eriksson P., Paulsson-Berne G., Hedin U., Wallentin L., Siegbahn A., Hamsten A., Syvanen A.C. Variants of the interferon regulatory factor 5 gene regulate expression of IRF5 mRNA in atherosclerotic tissue but are not associated with myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 2008;28:975–982. doi: 10.1161/ATVBAHA.108.163733. [DOI] [PubMed] [Google Scholar]
- 47.Alonso-Perez E., Suarez-Gestal M., Calaza M., Kwan T., Majewski J., Gomez-Reino J.J., Gonzalez A. Cis-regulation of IRF5 expression is unable to fully account for systemic lupus erythematosus association: analysis of multiple experiments with lymphoblastoid cell lines. Arthritis Res. Ther. 2011;13:R80. doi: 10.1186/ar3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozyrev S.V., Lewen S., Reddy P.M., Pons-Estel B., Witte T., Junker P., Laustrup H., Gutierrez C., Suarez A., Francisca Gonzalez-Escribano M., et al. Structural insertion/deletion variation in IRF5 is associated with a risk haplotype and defines the precise IRF5 isoforms expressed in systemic lupus erythematosus. Arthritis Rheum. 2007;56:1234–1241. doi: 10.1002/art.22497. [DOI] [PubMed] [Google Scholar]
- 49.Wen F., Ellingson S.M., Kyogoku C., Peterson E.J., Gaffney P.M. Exon 6 variants carried on systemic lupus erythematosus (SLE) risk haplotypes modulate IRF5 function. Autoimmunity. 2011;44:82–89. doi: 10.3109/08916934.2010.491842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alonso-Perez E., Fernandez-Poceiro R., Lalonde E., Kwan T., Calaza M., Gomez-Reino J.J., Majewski J., Gonzalez A. Identification of three new cis-regulatory IRF5 polymorphisms: in vitro studies. Arthritis Res. Ther. 2013;15:R82. doi: 10.1186/ar4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wingender E., Dietze P., Karas H., Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newburger D.E., Bulyk M.L. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G., et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 55.Skipper M., Dhand R., Campbell P. Presenting ENCODE. Nature. 2012;489:45. doi: 10.1038/489045a. [DOI] [PubMed] [Google Scholar]
- 56.Sankararaman S., Mallick S., Dannemann M., Prufer K., Kelso J., Paabo S., Patterson N., Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rasmussen A., Sevier S., Kelly J.A., Glenn S.B., Aberle T., Cooney C.M., Grether A., James E., Ning J., Tesiram J., et al. The lupus family registry and repository. Rheumatology (Oxford) 2011;50:47–59. doi: 10.1093/rheumatology/keq302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hochberg M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 59.Hoggart C.J., Parra E.J., Shriver M.D., Bonilla C., Kittles R.A., Clayton D.G., McKeigue P.M. Control of confounding of genetic associations in stratified populations. Am. J. Hum. Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lessard C.J., Adrianto I., Kelly J.A., Kaufman K.M., Grundahl K.M., Adler A., Williams A.H., Gallant C.J., Anaya J.M., Bae S.C., et al. Identification of a systemic lupus erythematosus susceptibility locus at 11p13 between PDHX and CD44 in a multiethnic study. Am. J. Hum. Genet. 2011;88:83–91. doi: 10.1016/j.ajhg.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoggart C.J., Shriver M.D., Kittles R.A., Clayton D.G., McKeigue P.M. Design and analysis of admixture mapping studies. Am. J. Hum. Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith M.W., Patterson N., Lautenberger J.A., Truelove A.L., McDonald G.J., Waliszewska A., Kessing B.D., Malasky M.J., Scafe C., Le E., et al. A high-density admixture map for disease gene discovery in African Americans. Am. J. Hum. Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKeigue P.M., Carpenter J.R., Parra E.J., Shriver M.D. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann. Hum. Genet. 2000;64:171–186. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- 64.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 65.Halder I., Shriver M., Thomas M., Fernandez J.R., Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum. Mutat. 2008;29:648–658. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- 66.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 68.Barrett J.C. Haploview: visualization and analysis of SNP genotype data. Cold Spring Harb. Protoc. 2009 doi: 10.1101/pdb.ip71. 2009. http://cshprotocols.cshlp.org/content/2009/10/pdb.ip71.abstract . [DOI] [PubMed] [Google Scholar]
- 69.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 70.Sankararaman S., Sridhar S., Kimmel G., Halperin E. Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lessard C.J., Li H., Adrianto I., Ice J.A., Rasmussen A., Grundahl K.M., Kelly J.A., Dozmorov M.G., Miceli-Richard C., Bowman S., et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjogren's syndrome. Nat. Genet. 2013;45:1284–1292. doi: 10.1038/ng.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stephens M., Balding D.J. Bayesian statistical methods for genetic association studies. Nat. Rev. Genet. 2009;10:681–690. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 73.Lessard C.J., Adrianto I., Ice J.A., Wiley G.B., Kelly J.A., Glenn S.B., Adler A.J., Li H., Rasmussen A., Williams A.H., et al. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am. J. Hum. Genet. 2012;90:648–660. doi: 10.1016/j.ajhg.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Bonnen P.E., de Bakker P.I., Deloukas P., Gabriel S.B., et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Radstake T.R., Gorlova O., Rueda B., Martin J.E., Alizadeh B.Z., Palomino-Morales R., Coenen M.J., Vonk M.C., Voskuyl A.E., Schuerwegh A.J., et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat. Genet. 2010;42:426–429. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.