Abstract
There is a lack of tools to ease the integration and ontology based semantic queries in biomedical databases, which are often annotated with ontology concepts. We aim to provide a middle layer between ontology repositories and semantically annotated databases to support semantic queries directly in the databases with expressive standard database query languages. We have developed a semantic query engine that provides semantic reasoning and query processing, and translates the queries into ontology repository operations on NCBO BioPortal. Semantic operators are implemented in the database as user defined functions extended to the database engine, thus semantic queries can be directly specified in standard database query languages such as SQL and XQuery. The system provides caching management to boosts query performance. The system is highly adaptable to support different ontologies through easy customizations. We have implemented the system DBOntoLink as an open source software, which supports major ontologies hosted at BioPortal. DBOntoLink supports a set of common ontology based semantic operations and have them fully integrated with a database management system IBM DB2. The system has been deployed and evaluated with an existing biomedical database for managing and querying image annotations and markups (AIM). Our performance study demonstrates the high expressiveness of semantic queries and the high efficiency of the queries.
Keywords: Semantics, ontology, biomedical databases
1. Introduction
Biomedical ontologies have proliferated in biomedical domains to support semantic queries, semantic interoperability and data integration [1, 2]. The National Center for Biomedical Ontology (NCBO) BioPortal [3–5] alone hosts nearly five million terms for about 329 ontologies. Example ontologies include NCI Thesaurus (NCIt) for cancer, RadLex [6] for radiology image annotations, GO [7] for genes, etc.
Increasingly, biomedical databases are becoming semantic enabled through semantically annotated data models, i.e. data objects are described through links to ontological concepts. Examples include AIM model for NCI Image Markup and Annotation project [8], and the pathology analytical imaging standards (PAIS) project [9–11], which use the following attributes to describe semantically linked concepts: codeValue — the unique identification code for the concept, codeName — the meaning of the concept code, codingSchemeDesignator — the ontology or controlled vocabulary where the data element depends on, and codingSchemeVersion — the version of the codingSchemeDesignator. While storing data in RDF triples is attractive, in reality, re-implementing an existing database with different data stores and query languages may not be realistic. Instead, annotated biomedical databases often take the standard SQL table based data representation, with additional attributes for the ontology based semantic annotations. Recently XML has become a popular data model and XML databases can be used to support similar annotated data through adding additional attributes.
Such semantically annotated databases provide the opportunity to support ontology based semantic queries. For example, a concept may be relaxed to provide more semantically related results: one may return descendant terms “gliosarcoma” and “giant cell glioblastoma” when a term “astrocytoma (WHO grade IV)” is posed in a query. Such operations need interplay between ontology queries and database queries, and require querying an ontology repository and integrating the results into database queries for further processing. Custom coding to support such queries is possible, but requires major programming on manually translating queries back and forth between databases and ontology repositories. Such an approach is not generic either, and repeated development is needed for similar queries for each new database. Meanwhile, database users often prefer writing queries with a declarative query language, such as the structured query language (SQL) for tabular data and the XML query language (XQuery) for XML data. For example, for above concept relaxation query, a user may want to specify a SQL query with a simple extended function like getHyponym (term), without any additional programming. A declarative query interface based approach is generic: once developed, the interface can be used for different queries and databases. This will save major development effort, and provides high usability for end users.
The gap in support for convenient ontology based queries in biomedical databases is exacerbated by the limitations of current biomedical repositories, including complex interfaces, primitive query operations, and overhead of network communications. While biomedical repositories such as NCBO BioPortal provide the management and query capabilities for ontologies, the query interfaces are normally designed for machine consumption and are cumbersome for humans. For example, for the getHyponym query to retrieve a list of descendant terms, it can be supported by writing codes to submit queries to an ontology repository, e.g. NCBO BioPortal. The results returned from NCBO BioPortal interfaces, however, are very complex XML documents that have to be parsed, filtered and aggregated before further processing. In addition, ontology repositories normally provide primitive queries. To support a complex semantic query such as getHyponym, a user has to develop his/her own application with multiple queries on the ontology repository and additional semantic reasoning on query results. For example, for getHyponym query, recursive calls have to be called until no more descendant nodes are found. Invocation of multiple queries from a remote repository also leads to inefficiency due to the network overhead. For example, it takes 162 seconds to retrieve recursively all descendant concepts of a concept with a depth of 4 based on the NCI Thesaurus ontology (NCIt) [12] hosted at BioPortal.
The limitation and mismatch of ontology repositories make it difficult to directly support the requirements of declarative, expressive and reusable semantic queries on biomedical database systems. This motivates us to develop DBOntoLink, a system to provide a middle layer between ontology repositories and semantically annotated databases to support semantic queries in the databases with declarative languages and interfaces. DBOntoLink provides the following salient features.
Expressive semantic query operators
The system generalizes a comprehensive set of ontology based semantic operators. These semantic operations include basic information of concepts, relations between concepts, as well as advanced ontology reasoning. Generalized semantic operations simplify the usage of ontology, thus extend the utility of existing ontologies.
Tight integration of semantic operators with the database engine
The semantic operators are implemented in the database management system as extended user defined functions thus expressive ontology based semantic queries can be directly specified in structural query language (SQL) or XML query language (XQuery), without any programming needed. This makes it highly expressive and convenient to run semantic queries against ontologies.
Extensible to different ontologies and databases
The configuration management provides easy customization to define the mapping between relation labels and conceptual meanings, and the mapping between ontology names and versions. This enables the system with high adaptability to support major ontologies, such as those in NCBO BioPortal. The software can be quickly deployed to different biomedical databases through simple customization.
High efficiency on semantic queries
The caching management automatically caches ontology concepts and relationships in the database from executed queries, which significantly boosts the query performance.
2. Background
2.1. Related work
An ontology represents concepts and relations between concepts in forms that can be interpreted by both humans and machines. Many biomedical ontologies have been developed in the past for different domains [1, 2, 6, 7, 12]. Most of these ontologies are included in the NCBO BioPortal [3, 5], which provides the abilities to browse, search and visualize ontologies as well as to comment on, and create mappings for ontologies. LexGrid [13] is a framework for representing, storing, and querying biomedical terminologies, and often used by ontology repositories as the backend for managing ontologies and providing query services. caDSR [14] is a database and a set of APIs and tools to create, edit, control, deploy, and find common data elements (CDEs) for use by metadata consumers.
Many applications or databases are providing semantic annotations to the data by linking data to ontology concepts. For example, NCI Annotation and Image Markup project [8] are Pathology Analytical Imaging Standards project [9–11] provide semantic enabled models to support semantic interoperability.
Lim et al. [15, 16] summarize major problems and challenges of supporting semantic queries in relational databases, such as graph based queries and vagueness of queries, and propose query-by-example (QBE) based semi-automatic approach to solve the problems. In SciPort project [17], semantic enabled authoring and queries are provided to link specific ontologies such as RadLex with structured data, through providing RESTful Web Service based interfaces. Extending to additional ontologies, however, needs development of new RESTful APIs for each ontology repository. Early work in [18–20] tried to support semantic operations in RDBMS through user-defined functions. Our work provides an effective generic framework and can support multiple different ontologies. Semantic Web based knowledge management has been an active research area [21]. DBOntoLink takes a middle layer based approach and is extended on existing database management systems and query languages.
2.2. Ontology search and semantic queries
An ontology can provide fundamental semantic reasoning capabilities, supported by an ontology repository as a set of services. Common services provided by an ontology repository includes:
Concept search: Given term(s), return matched ontology concepts with either exact match or approximate match, and the associated information.
Identity search: Given a concept id and ontology version id, return properties and concepts related to the concept (e.g. child concepts, synonym concepts).
Hierarchy search: Given a concept id, return the path from the concept to its root or leaves, usually in the form of sequences of concatenated concept ids.
These primitive operators provide the foundation for semantic operations. For example, we may combine multiple identity based searches to generate a list of descendant concepts, by parsing each search result and recursively searching on child concepts until the leaf concepts. However, such approach is extremely cumbersome and could only be implemented through programming, and each new query needs to be supported by a new program. This approach also lacks expressiveness — humans prefer to write queries in a declarative and expressive way, such as the popular standard SQL query language for relational data or XQuery query language for XML data. Next we show how BioPortal is limited on such capabilities.
2.3. BioPortal
BioPortal implements the ontology services listed above as two types of interfaces: SOAP based Web Services and RESTful based services. The latter processes HTTP URL formatted requests and responds with a set of result in the form of XML. For example, to query the properties and related concepts of “lung” in RadLex, the following URL needs to be issued. In the expression, “45137” is the ontology version id and “RID29152” is the concept id. http://rest.bioontology.org/bioportal/concepts/45137?conceptid=RID29152&light=0&apikey=YourAPIKey
The result is an XML document with 483 lines with a very complex XML schema. These interfaces are designed for machine based queries and processing, and have the following limitations for human use:
The URL formatted request is inconvenient for interpretation and editing, especially when the queries are complex or there are many terms used in a query.
They are limited as semantic operations. In many cases, users have to provide multiple requests to build a query. More advanced queries such as finding common ancestors of multiple concepts are difficult and require users to write complex queries.
The XML based query results contain complex — often redundant — information for users. Users almost always need to parse and filter information from the results. In reality, users often prefer simpler query result, for example, a list of child concept terms for a “get children” query.
BioPortal services suffer from network delay. Each request will incur a network overhead, and this could seriously hamper the performance of complex queries when multiple requests are needed, or repetitive queries from a large database.
2.4. Database user defined functions
The gap between ontologies and applications — especially databases — represents a key impediment to enabling the full potential of ontologies in biomedical databases. Our goal is to bridge the gap through database extensibility techniques provided by modern DBMSs.
User defined functions (UDFs) in DBMSs provide an opportunity for close integration of ontology repositories into the database. A UDF could return a single value or tabular value — which can be further converted into a table view for SQL operations. While its implementation could be in multiple programming languages, such as Java or C/C++, a UDF can be embedded in a SQL query as simple as an ordinary SQL function. Thus, UDFs can take full advantage of the expressive power of SQL. Comprehensive application logic, such as composing complex semantic operations on an ontology repository, can be realized as logically extended functions for SQL, and expressed in natural declarative SQL language. For an ontology reasoning operation, instead of writing complex codes to perform the operation, a user could simply submit a SQL query by embedding the corresponding UDF into the query, and have the result returned in a tabular format. For example, advance semantic reasoning, such as searching for shared descendants of multiple concepts, may need to be queried with recursive hierarchy searches, where many complex intermediate processing of XML based results have to performed. Using our system, user can invoke a single function call directly where all the reasoning is transparent to users. This is highexpressive and makes it very convenient for users. Next we discuss the overall architecture and methods of our work.
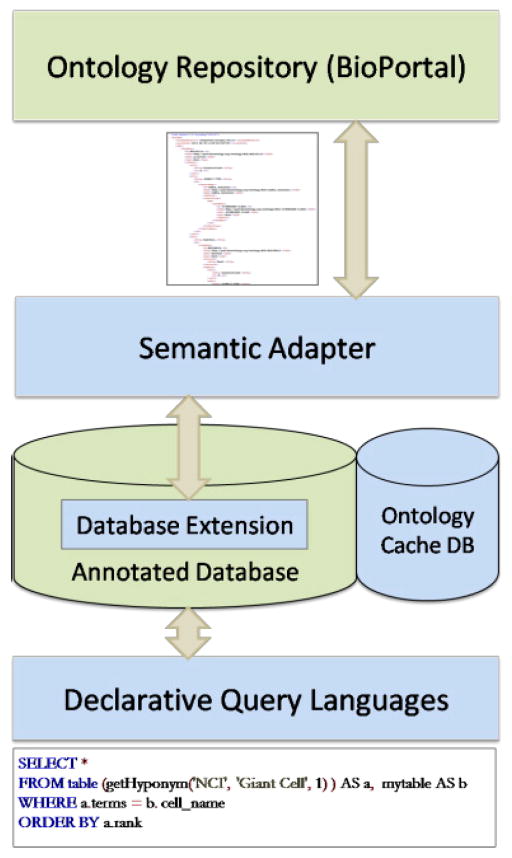
3. Architecture of DBOntoLink
DBOntoLink has three major components: ontology repository, semantic adapter, and database extension. We rely on BioPortal (with its RESTful interfaces) as the ontology repository since it is the most commonly used biomedical ontology repository. The semantic adapter provides a mediation layer between applications (via databases or RESTful web services) and the ontology repository, by supporting a comprehensive set of semantic operations. The semantic adapter sends requests to BioPortal, parses, processes and composes query results. The architecture overview is shown in Fig. 1.
Fig. 1.
Overview of DBOntoLink.
Operations implemented in the semantic adapter are consumed either by databases or applications. In the database, these semantic operations are wrapped as user defined functions to be consumed by SQL or XQuery queries.
The semantic adapter provides three major components: the semantic query engine for processing the semantic operations, caching management for caching query results to improve performance, and configuration management to configure the system for different ontologies (Fig. 2).
Fig. 2.
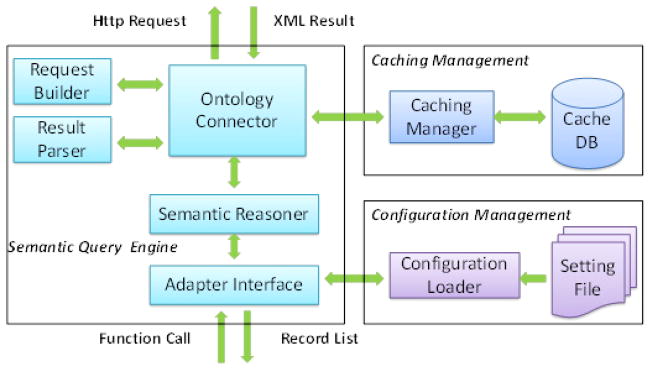
The architecture of DBOntoLink.
3.1. Semantic query engine
The semantic query engine provides semantic reasoning, query requesting and processing, and it interfaces with databases and applications. The workflow of the engine is as follows:
When an adapter interface receives a function call issued either by a UDF or HTTP request, the semantic reasoner analyzes the procedures and requests needed to answer this call. It then submits requests through the ontology connector.
Once the ontology connector receives a query request, it first checks the caching manager to see if this query has been previously issued. If the result is already cached, it will be retrieved from the cache database directly.
If the query result is not cached, the ontology connector will issue RESTful requests composed by the request builder and transfers returned XML results to result parser.
The result parser extracts the relevant information from the XML result, and passes them to the semantic reasoner via ontology connector.
After all the required information has been collected through multiple ontology repository calls, the semantic reasoner generates the final result for the adapter interface, in a form of a list of records.
This semantic adapter works as a translation engine to interpret expressive, simple semantic operations (seen by end users and applications) into a process of complex operations of ontology repository queries, result filtering and restructuring. Such process is transparent to users, thus users could enjoy writing declarative semantic queries without worrying about how the queries are realized. This approach thus significantly improves the usability of ontology repositories.
3.2. Caching management
To improve the efficiency when processing queries, we implement caching for query results to avoid the overhead of multiple query requests to remote repositories. The caching layer is realized by persisting results into relational database tables: multiple tables are created for multiple types of information. Metadata of concepts is stored in concept cache table, including description, child count and other basic information. Relationship graphs between concepts are stored in relation cache table, in the form of edges — each edge is represented as a record containing attributes of start concept, relation label and end concept. In this way, information of ontology is stored in a flexible structure for convenient usage and update. The caching management provides significant speedup of semantic query processing, as the performance study of result section demonstrates.
3.3. Configuration management
Different ontologies often have different definitions of hierarchies and relationships [22]. DBOntoLink provides customization to define the mapping between relation label and its conceptual meaning, as well as the mapping between ontology name and the used version. In this way, the label of relation can be customized conveniently if necessary. All the customization settings are defined in an XML based setting file, therefore easy to interpret and configure. An adapter loads the configuration of a given ontology via the configuration loader when the adapter is initialized.
4. Semantic Operations Based on Ontologies
The semantic adapter generalizes many semantic operations with convenient interfaces for databases and applications. Such operations include metadata queries for concepts, semantic enabled term queries and ontology relation queries.
4.1. Metadata queries for terms
This class of functions employs the search service of BioPortal to retrieve a term’s metadata.
getDescription: Retrieve description or definition of a term.
getSemanticType: Obtain the semantic type or role of a term. For example, both “Antigen Gene” and “Fusion Gene” have the same semantic type “Gene or Genome”.
getChildCount: Retrieve the count of all child terms.
getRelevantTerm: Retrieve all the relevant terms for each word in input text. For example, given the text “cancer patient”, the function returns terms such as: “Cancer cell growth”, “Patient Allergic to Contrast Media” and so forth, which are related to the terms in the input text.
4.2. Semantic enabled term queries
Operations in this set are implemented based on the term service of BioPortal, and focus on retrieving related concepts for a given concept.
getHyponym: When a user specifies a query with a term, there may be subclasses of the concept that can generate favorable results as well. For example, for “Abnormal Cell”, this function returns its hyponyms such as “Neoplastic Cell” and “Signet Ring Cell”. This function allows query in expanded domain using hyponyms.
getHypernym: In many cases, there may not be any result from a query using a certain term, but users may still want to look at the closest results or related results by relaxing the concept to a broader scope. For example, if a query with the term “Tumor Lysis Syndrome” returns no result, the user may want to see if there is any result from the relaxed term “Cancer-Related Condition”, a super class of “Tumor Lysis Syndrome”.
getSynonym: Queries using a precise term often suffer from the problem that results from its synonyms will be missing. For example, “Alcoholism” and “Alcohol Dependence” are synonyms. To support synonym detection, we send a request to the corresponding ontology and retrieve all its synonyms. With these, the final query will include a combination of all possible terms and return more accurate result.
getSibling: Retrieve concepts that belong to the same category of the input concept to expand the search domain. For example, “Copine VII”, “Mitochondrial Membrane Protein” and “Neogenin Homolog 1” are siblings sharing the parent class “Membrane Protein”.
Methods getHyponym and getHypernym can be configured with a depth limit for searching. For example, depth 2 will retrieve all terms up to two levels in an ontology hierarchy. All the operations in this set could be applied with result constraints such as child node count and semantic type.
4.3. Ontology relation queries
The operations in this set examine relations among multiple concepts.
getCommonAncestor: Given a set of terms, return ancestral classes shared by all the input terms. For example, “Abnormal Eosinophil” and “Leukemic Cell” share the same ancestral concept “Abnormal Hematopoietic and Lymphoid Cell”.
getCommonDescendant: Given a set of terms, return mutual descendants of all the input terms. For example, “Giant Cell” and “Atypical Epithelial Cell” have mutual child concept “Giant Epithelial Cell”.
getRelation: Discover relation between two concepts, check if one is the ancestor, descendant or sibling of the other. For example, given terms “Cystic Fibrosis” and “Chromosome Disorder”, the relation between them is identified as sibling.
4.4. Concept domain queries
OWL is an ontology representation language, and the property owl:intersectionOf in OWL presents a list of parent concepts for a given concept. For example, “Neoplastic Polyp” is the common child of “Polyp”, “Precancerous Condition” and “Neoplasm by Morphology”. Regard each concept as a domain, the “Neoplastic Polyp” is the intersection area shared by these three domains. Operations in this category check the intersection condition of input concepts.
getAdjunctTerm: Retrieve terms which have intersection domain with the given concept. For example, a search of adjunct terms of “Abnormal Germ Cell” will return “Neoplastic Cell”, since they share the intersection concept domain, “Neoplastic Germ Cell”.
checkIfAdjunct: Check if the input terms intersect on their domain. For example, the domain overlapping condition of “Behavior-Related Disorder” and “Psychiatric Disorder” is true, since they share the same child concept “Socialized Conduct Disorder”.
4.5. Text content annotation
The operation in this set adapts the annotation service of BioPortal.
getAnnotation: Annotate the terms in given text, return score of accuracy and other information according to configuration. For example, consider the following text in an sample pathology report: “Carcinoma of breast. Post-operative diagnosis: Same. Left UOQ breast mass”. Based on the SNOMEDCT ontology, getAnnotation will return “Carcinoma of breast”, “Mass”, “Entire breast”, “Breast structure”, and others as the result.
5. Implementation
We have implemented DBOntoLink as an open source software, which supports major ontologies hosted at BioPortal. The software is available at DBOntoLink wiki [23]. We first implement the above semantic operations as Java based APIs, and then port them into upper level UDF interface. Currently the database management system supported by DBOntoLink is IBM DB2, and it can be easily extended to support other database management systems such as Oracle, PostgreSQL, and mySQL. Once the software is deployed, the corresponding database can immediately support ontology based semantic queries, expressed as UDFs extended to the database query language, as discussed next.
Implementation of UDFs wraps the Java APIs as database user defined functions based on specific database UDF specifications. They are deployed based on database specific UDF deployment processes. We use DB2 in our current implementation, but it is easy to adapt it to other databases by following different UDF development processes and interfaces. These UDFs can be directly invoked by SQL queries just as normal SQL functions, therefore they offer great usability and convenience for database users.
According to the format of the return value, UDFs can be classified into scalar functions and table functions. A scalar function, such as getDescription, getChildCount and checkRelation, returns a single value for each record of a query. For example, the following SQL query retrieves the description for all the terms in the column “Cell Name”:
SELECT cell name, getDescription (‘NCI’,mytable.cell name) FROM myatable
A table function, such as getHyponym, getCommonAncestor and getAnnotation, can be applied in the FROM clause of a SQL query. For example, the following query selects records with Cell Name as the hyponym of “Giant Cell”, with results ordered by the relevance rank:
SELECT * FROM table (getHyponym(‘NCI’, ‘Giant Cell’, 1)) AS a, mytable AS b WHERE a.terms = b.cell name
The following query returns result with symptom “Chest Pain” from all possible synonyms (“thoracodynia”, “PAIN IN CHEST”, and “thoracalgia”):
SELECT * FROM table (getHyponym(‘NCI’, ‘Giant Cell’, 1)) AS a, mytable AS b WHERE a.terms = b.cell name
5.1. RESTful web service based semantic operations
Similar to UDFs, the RESTful server works as a layer between the semantic adapter and web client. Our RESTful Web Services are implemented as JAVA Servlets. It processes HTTP URL based request and responds with XML formatted results.
As an example, to query the hyponym of “Giant Cell”, a user can issue the following query, where the parameters are in the format “parameter name = parameter value” and separated by “&”:
http://URLprefix/?operation=getHyponym&ontology=NCI&termName=GiantCell&distance=1
The XML formatted result contains concise information, which includes term name, the relation label and the distance, as well as metadata information of the result set.
6. XQuery Based Semantic Queries for an Image Markup and Annotation Database
AIM is a NCI project [8] with the goal to provide standardization for image annotation and markup, especially for clinical trials. The model includes dozens of classes such as patient, observer, equipment, image, anatomic entities, image observations and image observation characteristics, geometric shapes, text annotations, and calculations. The AnatomicEntity, ImagingObservation, and ImagingObservationCharacteristic classes represent essential features for an annotation, and rely on an ontology (e.g. RadLex or NCI Thesaurus) or controlled vocabulary to populate the data. The representation of AIM data is an XML based format. AIM data management and sharing is through xService [24, 25], which relies on an XML based data management system — we use IBM DB2 pureXML for this case. We use LIDC dataset [26] for our examples and load the dataset into DB2 database with XML documents stored as XML type. DB2 has a hybrid approach for managing relational data and XML data, and supports both SQL and XQuery. The UDFs we develop can be used in both SQL and XQuery. Next we show some sample use cases that the XQuery/SQL queries are semantically enriched with the UDFs we developed.
QUERY 1: Retrieve all the anatomic entity concepts and their descriptions in this dataset:
declare namespace ns1=“gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM”;
for$t indb2-fn:xmlcolumn(‘XMLTABLE.XMLCOLUMN’)/
ns1:ImageAnnotation/ns1:anatomicEntityCollection/ ns1:AnatomicEntity/@codeMeaning
let$d:=db2-fn:sqlquery(“values(xmlcast(getDescription(‘RadLex’, parameter(1)) as xml))“, $t)
return<result term =“{$t}“ description =“{$d}“/>
This query returns result as a list of XML elements, and below is an example element from the result:
<result term=“lung“ description=“One of a pair of viscera occupying the pulmonary cavities of the thorax, the organs of respiration in which aeration of the blood takes place. . .“/>
QUERY 2: Retrieve all the image annotation documents that contain the descendant concepts of the term “lobular organ”:
declare namespace ns1=“gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM“; for$a indb2-fn:xmlcolumn(‘XMLTABLE.XMLCOLUMN’)/ ns1:ImageAnnotation for$hypo indb2-fn:sqlquery(“select xmlcast(terms as xml) from table(getHyponym(‘NCI’, Spatial Qualifier’, 1))“) where$a/ns1:imagingObservationCollection/ ns1:ImagingObservation/ns1:imagingObservationCharacteristicCollection/ ns1:ImagingObservationCharacteristic/@codeMeaning=$hypo return$a
QUERY 3: Retrieve common ancestors with “liver” for each of the anatomic entity:
declare namespacens1=”gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM”;
for$t indb2-fn:xmlcolumn(‘XMLTABLE.XMLCOLUMN’)/
ns1:ImageAnnotation/ns1:anatomicEntityCollection/ ns1:AnatomicEntity/@codeMeaning
for$a indb2-fn:sqlquery(”select xmlcast(terms as xml) from
table(getCommonAncestors (‘RadLex’, CONCAT (‘liver;’,parameter(1))))”,$t)
return<result term =”{$t}” ancestor =”{$a}”/>
QUERY 4: Annotate imaging observation characteristic terms with NCI Thesaurus ontology, and return the annotations as ontology concept terms if available:
declare namespace ns1=”gme://caCORE.caCORE/3.2/edu.northwestern.radiology.AIM”; for$t indb2-fn:xmlcolumn(‘XMLTABLE.XMLCOLUMN’)/ ns1:ImageAnnotation/ ns1:imagingObservationCollection/ ns1:ImagingObservation/ ns1:imagingObservationCharacteristicCollection/ ns1:ImagingObservationCharacteristic/@codeMeaning for$a indb2-fn:sqlquery(“select xmlcast(terms as xml)
7. System Performance
We present experimental results of typical UDFs with sample test cases summarized in Table 1. The experiment is performed with the NCI Thesaurus ontology hosted at BioPortal, and the database we use is IBM’s DB2 V9.7. The machine we use for the test is a desktop with i5-760 at 2.8 GHz, 8 GB of RAM, 1 TB RAID 0 (2 × 500 GB SATA7200rpm HDDs) hard drive. The running time for each query is the average of 10 executions. The local catching database stores a subset of the original ontology.
Table 1.
UDFs used in testing.
| Test UDF | Test case description |
|---|---|
| getAnnotation | Annotate the sentence “lung cancer patient” |
| getDescription | Retrieve the description of “Abnormal Cell” |
| getRelevantTerm | Retrieve relevant terms of “patient” |
| getChildCount | Retrieve child node amount of “Abnormal Cell” |
| getHypernym | Retrieve the parent node of “Neoplastic Cell” |
| getHyponym | Retrieve the child nodes of “Neoplastic Cell” |
| checkIfAdjunct | Check if concepts “Abnormal Germ Cell” and “Neoplastic Large Cell” overlap. |
| getRelation | Retrieve the relation of “Abnormal Cell” and “Circulating Tumor Cell”. |
| getCommonAncestors | Retrieve the common ancestors of concept “Malignant Cell” and “Neoplastic Germ Cell” |
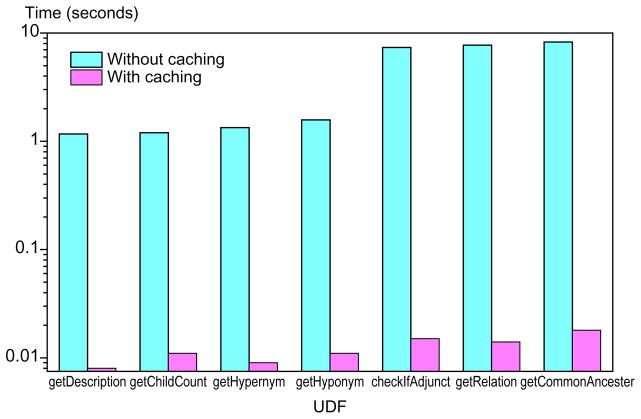
For each query in Table 1, Fig. 3 shows performance comparison between methods without caching and with local caching. The experiment of querying without catching shows that simple queries take around 1 second as only one or two HTTP requests are needed. However, complex queries take much longer. This is because many complex queries are reasoned as queries with recursive operations that result in multiple repository requests. Such performance is unacceptable when a query runs across many terms. With caching, most queries run within 0.1 second, a significant performance improvement. Performance of caching may decrease as the size of caching grows. In reality, the execution of previous queries may also affect the efficiency of caching because of the caching mechanism of local caching database.
Fig. 3.
Sample query performance without and with local cache (in seconds).
In addition, we also compared the performance enhancement of UDF getHyponymat different query depths. This query searches the descendants of the concept “Abnormal Cell”. Table 2 shows the number of concepts need to be checked and the total number of result concepts returned in the recursive processes, as well as the performance before and after local caching. The experiment shows that caching provides relatively constant performance, and could be several orders of magnitude faster for high depth queries.
Table 2.
Running time without and with local cache for getHyponym UDF at different depths (in seconds).
| Depth | Examined concepts | Retrieved concepts | Time without cache | Time with cache |
|---|---|---|---|---|
| 1 | 1 | 15 | 1.21 | 0.019 |
| 2 | 16 | 83 | 7.7 | 0.027 |
| 3 | 84 | 267 | 34.8 | 0.083 |
| 4 | 268 | 503 | 162.2 | 0.192 |
7.1. Visualization of local ontology in a database
In practice, a local biomedical database only uses a subset of one or multiple ontologies to make annotations of the data, and the subset of concepts consist of fragments of ontologies and can be difficult to visualize. By taking advantage of DBOntoLink, we can generate a graph view of a local ontology. To build the graph, each distinct concept used in the local database is treated as a leaf and the hierarchy is recursively built until the root using DBOntoLink functions. Intermediate nodes with same concepts are merged together. Figure 4 shows an example local ontology graph, for a small set of local concepts “Giant Cell”, “Malignant Cell”, “Liver” and “Lung”.
Fig. 4.
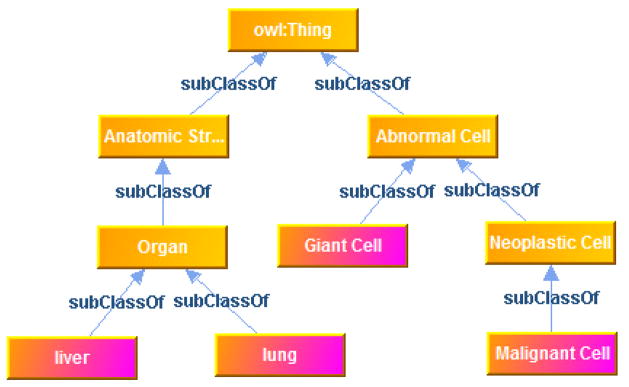
Local ontology graph generated by system.
We can also link data objects which contain a concept into the graph, for example, an AIM XML document for “Lung” can be attached to the “lung” node in the ontology graph. This will enable an ontology based navigation of instances in the database.
8. Discussions and Future Work
While DBOntoLink provides an architecture that supports semantic operations in the database, more semantic operations can be extended in the future, such as complex semantic operations. This could be implemented by extending current set of semantic operators, and providing corresponding translation in the semantic query engine and UDFs in the database.
Another future work is to extend the system to support more database management systems. Since different DBMSs have their own UDF frameworks, we can port current UDFs to databases such as Oracle, PostgreSQL and mySQL by implementing the UDF interfaces, where the internal Java codes can be reused across DBMSs.
9. Conclusion
While ontologies are proliferating in biomedical domains, most biomedical data are available as structured data managed in relational DBMS or XML DBMS. Using ontologies to enrich the semantics of data for queries and interoperability is becoming increasingly important, but there is a lack of tools to ease the integration and use of ontologies in databases using standard query languages or query interfaces. DBOntoLink provides a bridge between ontology repositories and databases to support semantic operations directly inside a database based on standard database query languages. Semantically annotated biomedical databases thus can be easily extended with powerful and expressive semantic enabled queries with DBOntoLink to use major ontologies hosted at NCBO BioPortal, with high query efficiency achieved through caching management.
Acknowledgments
This research is supported in part by PHS Grant UL1RR025008 from the CTSA program, R01LM009239 from the NLM, and NCI Contract No. HHSN261200800001E.
Contributor Information
Shuai Zheng, Email: shuai.zheng@emory.edu, Department of Mathematics and Computer Science, Emory University Atlanta, Georgia, USA.
Fusheng Wang, Email: fusheng.wang@emory.edu, Department of Biomedical Informatics, Emory University Atlanta, Georgia, USA.
James Lu, Email: jlu@mathcs.emory.edu, Department of Mathematics and Computer Science, Emory University Atlanta, Georgia, USA.
References
- 1.Cimino JJ, Zhu X. The practical impact of ontologies on biomedical informatics. Methods Inf Med. 2006;45:124–35. [PubMed] [Google Scholar]
- 2.Rubin DL, Shah NH, Noy NF. Biomedical ontologies: A functional perspective. Brief Bioinform. 2007 doi: 10.1093/bib/bbm059. [DOI] [PubMed] [Google Scholar]
- 3.NCBO BioPortal. [accessed Jul. 2013]; bioportal.bioontology.org/
- 4.Noy NF, Shah NH, Whetzel PL, et al. BioPortal: Ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009:170–3. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whetzel PL, Noy NF, Shah NH, et al. BioPortal: Enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.RadLex: A Lexicon for Uniform Indexing and Retrieval of Radiology Information Resources. [accessed Jul. 2013]; http://radlex.org/
- 7. [accessed Jul. 2013];Gene Ontology. http://www.geneontology.org/
- 8.Channin D, Mongkolwat P, Kleper V, et al. The caBIG annotation and image markup project. Journal of Digital Imaging. 2009 doi: 10.1007/s10278-009-9193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. [accessed Jul. 2013];Pathology Analytical Imaging Standards (PAIS) https://web.cci.emory.edu/confluence/display/PAIS.
- 10.Wang F, Kurc T, Widener P, et al. High-performance systems for in silico microscopy imaging studies. Proc of Seventh International Conference on Data Integration in the Life Sciences. 2010:25–27. [Google Scholar]
- 11.Wang F, Kong J, Cooper L, et al. A data model and database for high-resolution pathology analytical image informatics. Journal of Pathology Informatics. 2011;2(32) doi: 10.4103/2153-3539.83192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. [accessed Jul. 2013];NCI Thesaurus. http://ncit.nci.nih.gov/
- 13.Pathak J, Solbrig HR, Buntrock JD, et al. LexGrid: A framework for representing, storing, and querying biomedical terminologies from simple to sublime. J Am Med Inform Assoc. 2009;16(3):305–15. doi: 10.1197/jamia.M3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Data Standards Registry and Repository (caDSR) [accessed Jul. 2013]; https://cabig.nci.nih.gov/concepts/caDSR/
- 15.Lim L, Wang H, Wang M. Semantic queries in databases: Problems and challenges. CIKM 2009. :1505–1508. [Google Scholar]
- 16.Lim L, Wang H, Wang M. Semantic data management: Towards querying data with their meaning. ICDE 2007. :1438–1442. [Google Scholar]
- 17.Wang F, Liu P, Pearson J. Database Technology for Life Sciences and Medicine. World Scientific; 2010. SciPort: An extensible data management platform for biomedical research. [Google Scholar]
- 18.Stoffel K, Saltz J, Hendler J, et al. Semantic indexing for complex patient grouping. AIMIA. 1997 [Google Scholar]
- 19.Andrade HCM, Saltz J. Towards a knowledge base management system KBMS: An ontology-aware database management system DBMS. SBBD. 1999 [Google Scholar]
- 20.Stoffel K, Davis JD, Rottman G, et al. A graphical tool for ad hoc query generation. AMIA. 1998 [PMC free article] [PubMed]
- 21.Davies J, Fensel D, Harmelen FV. Towards the Semantic Web: Ontology-driven Knowledge Management. Wiley; 2003. [Google Scholar]
- 22.Smith B, Ceusters W, Klagges B, et al. Relations in biomedical ontologies. Genome Biol. 2005;6(5):R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DBOntoLink wiki. [accessed Jul. 2013];2012 https://web.cci.emory.edu/confluence/display/DBOntoLink.
- 24. [accessed Jul. 2013];caGridxService. https://web.cci.emory.edu/confluence/display/xmlds.
- 25.Wang F, Pan T, Sharma A, et al. Managing and querying image annotation and markup in XML. Proc of SPIE Medical Imaging. 2010 doi: 10.1117/12.843606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armato SG, McNitt-Gray MF, Reeves AP, et al. The Lung Image Database Consortium (LIDC): An evaluation of radiologist variability in the identification of lung nodules on CT scans. Acad Radiol. 2007;14(11):1409–21.s. doi: 10.1016/j.acra.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]