Abstract
The Wnt/β-catenin signaling pathway controls key aspects of embryonic development and adult tissue homeostasis, including the formation and maintenance of bone. Recently, mutations in the OSTM1 gene were found to be the cause of severe autosomal recessive osteopetrosis in both the mouse and humans. This disorder is characterized by increased bone mass resulting from a defect in osteoclast maturation. The possible role of OSTM1 in signaling of the Wnt/β-catenin “canonical” pathway was investigated in totipotent mouse F9 embryonal teratocarcinoma cells. Overexpression of OSTM1 in F9 cells increased Wnt3a-responsive β-catenin accumulation and Lef/Tcf-sensitive transcription. Similarly, knockdown of endogenous OSTM1 attenuated the ability of Wnt3a to stimulate the canonical signaling pathway. An OSTM1 mutant (detected in humans with osteopetrosis) was expressed in F9 cells and found to inhibit Wntstimulated β-catenin stabilization, gene transcription, and primitive endoderm formation. Expression of this OSTM1 C-terminal deletion mutant attenuated Lef/Tcf-sensitive gene transcription, even when transcription was activated by expression of a constitutively-active form of β-catenin. However, expression of this OSTM1 C-terminal deletion mutant was unable to alter Lef/Tcf-sensitive gene transcription when transcription was activated by expression of a β-catenin/Lef chimeric protein. From the standpoint of protein-protein interactions, expression of wild-type OSTM1 stimulated whereas mutant OSTM1 inhibited, the Wnt-dependent association of β-catenin and Lef1. On the foundation of these experiments, we propose that the human mutations in OSTM1 such as the C-terminal deletion mutant studied herein provoke dysregulation of the canonical Wnt/β-catenin signaling pathway, providing a molecular basis for severe autosomal recessive osteopetrosis.
Keywords: β-catenin, osteopetrosis, OSTM1, Wnt
Introduction
The canonical Wnt signaling cascade imposes tight regulation on the transcriptional coactivator β-catenin through a complex of proteins including glycogen synthase kinase 3β (GSK3β), Axin, and the product of the adenomatous polyposis coli gene (APC) [1–3]. Phosphorylation of β-catenin by protein kinases in this complex stimulates recognition and destruction of β-catenin by the proteasome [4], ensuring low intracellular levels of “free” β-catenin in the absence of pathway activation. The Wnt/β-catenin pathway is triggered by binding of a Wnt ligand to Frizzled (Fz), a seven-transmembrane G-protein coupled receptor [5], and subsequent downstream disruption of the β-catenin degradation complex through the action of heterotrimeric G-proteins and the phosphoprotein Dishevelled (Dvl) [6–9]. This allows β-catenin to accumulate in the cytosol, bind to members of the Lef/Tcf family of transcription factors, and activate transcription of genes necessary for both growth and differentiation [10]. The Wnt/β-catenin pathway is required for a plethora of steps in the development of multicellular organisms and regulation of adult tissue homeostasis [11, 12], including the growth and renewal of bone [13]. Several distinct steps in the accumulation of bone mass are regulated by Wnt signaling, including stem cell self-renewal, inhibition of osteocyte apoptosis, and stimulation of osteoblast differentiation and proliferation [13]. Defects in Wnt pathway components cause skeletal abnormalities, e.g., the osteoporosis-pseudoglioma disorder associated with mutations in Wnt co-receptor low-density lipoprotein receptor-related protein 5 (LRP5)[14, 15].
Severe autosomal recessive osteopetrosis is a genetic disease caused by deficiencies in bone resorption, resulting in increased bone mass. Patients with autosomal recessive osteopetrosis suffer from frequent fractures, hepatosplenomegaly and bone marrow obliteration, leading to early mortality unless treated with bone marrow transplantation [16, 17]. Mutations in two human genes, TCIRG1 and CLCN7 (which encodes the ClC-7 protein), together are known to cause ~60% of autosomal recessive osteopetrosis cases. A spontaneous mouse model of ARO allowed the identification of a third candidate gene, osteopetrosis associated transmembrane protein 1 (OSTM1) [18]. Subsequently, several human mutations in OSTM1 have been uncovered in ARO patients [16, 17]. OSTM1 is a type I transmembrane protein which localizes to intracellular vesicles (mainly endosomes and lysosomes) and homologs have been identified broadly, from humans to Caenorhabditis elegans and Drosophila melanogaster [18]. Two biological roles for OSTM1 have been suggested: serving as a cofactor necessary for ClC-7 function [19]; and, functioning as an E3 ubiquitin ligase for the heterotrimeric G-protein Gαi3 [20]. As Wnt signaling and OSTM1 are both intimately involved in bone remodeling, we explored the hypothesis that OSTM1 plays a role in Wnt/β-catenin signal transduction.
Materials and Methods
Plasmids and antibodies
An expression vector encoding human OSTM1 was obtained from OriGene (Rockville, MD). Expression vectors containing Fz1 and M50 were provided by Dr. Randall Moon (Department of Pharmacology, HHMI, University of Washington, Seattle, WA). Expression vectors containing SAβ-cat and β-cat/Lef were provided by Dr. Ken-Ichi Takemaru (Department of Pharmacology, SUNY Stony Brook, Stony Brook, NY) and an expression vector harboring VP16/Lef was provided by Dr. Howard Crawford (Department of Pharmacology, SUNY Stony Brook, Stony Brook, NY). Expression vectors harboring HA-tagged OSTM1 and OSTM1ΔC were provided by Dr. Thomas Jentsch (FMP/MDC, Berlin, Germany). The monoclonal TROMA-1 antibody was generated by the University of Iowa Developmental Studies Hybridoma Bank (Iowa City, IA). The rabbit antibody against β-catenin and the mouse antibody against actin were obtained from Sigma (Saint Louis, MO) and the anti-HA affinity matrix and anti-HA antibody were obtained from Roche (Indianapolis, IN).
Cell culture and transfection
Mouse F9 teratocarcinoma cells were obtained from the ATCC (Manassas, VA) and grown in Dulbecco’s modified Eagle’s medium supplemented with 15% fetal bovine serum, penicillin (60 µg/ml) and streptomycin (100 µg/ml) in a humidified atmosphere chamber supplied with 5% CO2 at 37° C. Cells were transfected by using Lipofectamine (Invitrogen, Carlsbad, CA), according to the manufacturer’s suggested protocol. Pre-designed siRNA oligonucleotides targeting OSTM1 were obtained from Ambion (Austin, TX). The sequences are as follows: Sense: ccaaaaaauuacuccgaagtt; Antisense: cuucggaguaauuuuuuggtg. The annealed siRNA sequences were transfected into F9 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to the manufacturer’s suggested protocol. RNA was collected 72h after transfection and knockdown efficiency measured by RT-PCR.
Reverse-transcription polymerase chain reaction
Specific primers for amplification of regions of OSTM1 and Cyclophilin A were obtained from Operon (Huntsville, AL). The sequences are as follows: OSTM1: atggctcgggacgcggag, aggcaatttgcgcagttcgcc; CycA: agcactggagagaaaggatttg, cacaatgttcatgccttctttc. RNA was isolated from wild-type F9 cells using the RNA STAT-60 reagent (Tel-Test, Friendswood, TX) according to the manufacturer’s suggested protocol. The reverse transcription reaction was performed using the Invitrogen Superscript system according to the manufacturer's suggested protocol. Primer sets (0.5 µM) were mixed with dNTPs (1 mM), F9 cDNA (100 ng), 10X Pfu buffer and Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). The PCR reaction was performed using the GeneAmp PCR System 2400 (Applied Biosystems, Foster City, CA) with the following protocol: 95° C, 5 minutes; (95° C, 1 minute; 60° C, 1 minute; 72° C, 3 minutes) 30 cycles; 72° C, 15 minutes. Reaction products were resolved electrophoretically upon 1% agarose gels and stained with ethidium bromide.
Gene transcription assay
F9 cells were seeded into 12-well plates and transiently transfected as described above. After incubation for 48 hours at 37° C, cells were serum-starved overnight, then treated with purified Wnt3a (R&D Systems, Minneapolis, MN). Cells were lysed through addition of diluted Cell Culture Lysis Reagent 5X (Promega, Madison, WI). Lysates were collected and centrifuged at 15,000 × g for 5 minutes. The supernatant was directly assayed as described below. A sample (20 µl) of lysate was added to 100 µl of luciferase assay buffer (20 mM Tricine, pH 7.8, 1.07 mM MgCO3, 4 mM MgSO4, 0.1 mM EDTA, 0.27 mM coenzyme A, 0.67 mM luciferin, 33.3 mM DTT, 0.66 µM ATP) and a luminometer (Berthold Lumat LB 9507) was used to measure luciferase activity.
Cytosolic β-catenin assay
In order to separate cytosolic from membrane-associated β-catenin, samples were treated with concanavalin A (Con A). Con A (covalently linked to Sepharose) was obtained from Amersham Biosciences (Upsala, Sweden). F9 cells were washed once with PBS and lysed in RIPA buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100) plus leupeptin (10 µg/mL), aprotinin (10 µg/mL) and PMSF (200 µM). Lysates were rotated at 4° C for 20 minutes and then centrifuged for 25 minutes at 15,000 rpm. The protein concentration of the supernatant fraction was determined and lysates were diluted with RIPA buffer to a protein content of 2.5 mg/ml. Sixty εl of Con A-Sepharose was added to each sample which then was incubated at 4° C for 1 hour with rotation. The supernatant was then removed and the protein concentration determined. The samples were subjected to SDS-PAGE on 10% acrylamide gels, the separated proteins transferred to nitrocellulose blots, and the blots probed with the anti-β-catenin antibody.
Indirect immunofluorescence
F9 cells grown in 24-well plates were fixed in 3% paraformaldehyde, then washed three times in MSM-PIPES buffer (18 mM MgSO4, 5 mM CaCl2, 40 mM KCl, 24 mM NaCl, 5 mM PIPES, 0.5% Triton X-100, 0.5% NP40). Cells were incubated at 37°C for 30 minutes with the TROMA antibody, washed with MSM-PIPES, and then incubated at 37°C for 30 minutes with an anti-mouse antibody coupled to Alexa Flour 488 (Invitrogen). Cells were then washed in blotting buffer (560 mM NaCl, 10 mM KPO4, 0.1% Triton X-100, 0.02% SDS) and imaged with a Zeiss LSM510 inverted fluorescence microscope.
Immunoprecipitation
F9 cells grown in 100 mm plates were lysed in a buffer composed of 50 mM TrisHCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, and 0.5% NP-40. 500 µg protein was incubated with 50 µl of resuspended Anti-HA Affinity Matrix overnight at 4°C. The matrix was washed three times in lysis buffer and the bound proteins were eluted with sample buffer. The samples were subjected to SDS-PAGE on 10% acrylamide gels, the separated proteins transferred to nitrocellulose blots, and the blots probed with the appropriate antibodies.
Results
OSTM1 mRNA is expressed in F9 cells
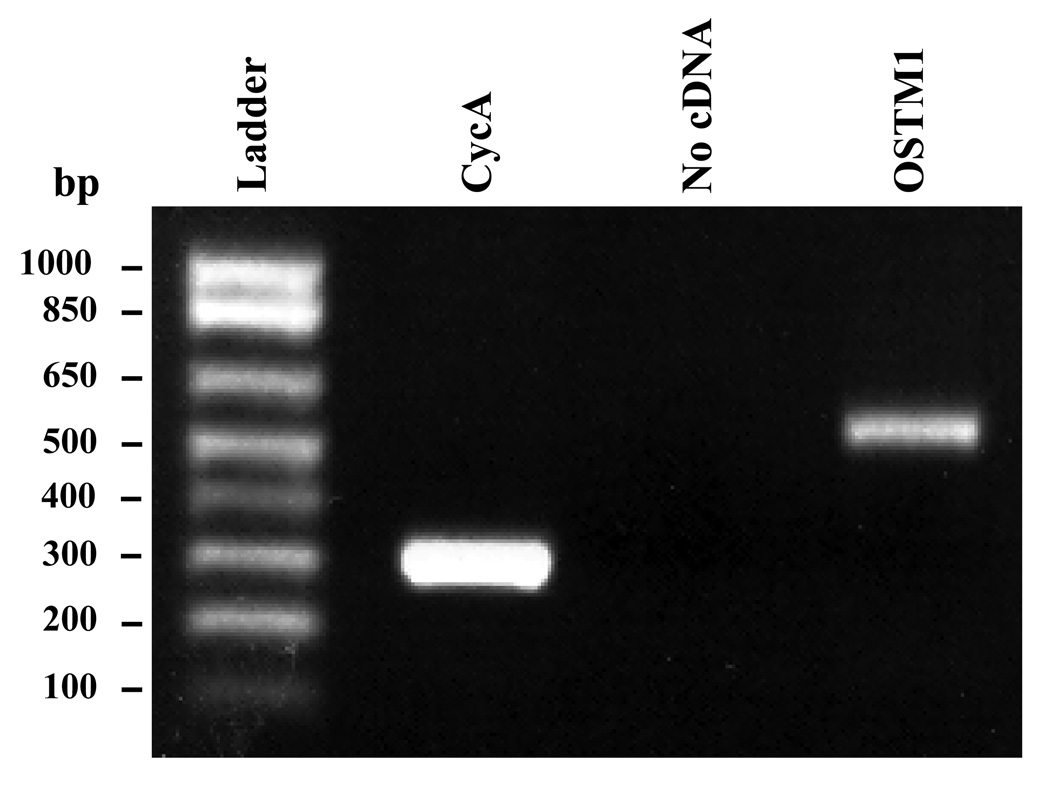
We chose to investigate the putative role of OSTM1 in Wnt/β-catenin signaling in mouse teratocarcinoma cells (F9). These pluripotent cells have been used as a model system to study signaling events in early mouse development and can be differentiated to primitive endoderm (PE) by stimulation with Wnt3a [6]. The lack of suitable antibodies precluded determination by Western blotting of OSTM1 protein expression in the mouse F9 cells. Alternatively, we sought confirmation at the level of mRNA, using reverse-transcription polymerase chain reaction (RT-PCR). OSTM1 mRNA was positively detected, being expressed in these cells (Fig. 1).
Figure 1. OSTM1 mRNA is expressed in F9 cells.
RT-PCR was performed on cDNA generated from RNA isolated from wild-type F9 cells. Primers specific for OSTM1 and Cyclophilin A were utilized in the PCR reaction. PCR products were separated by electrophoresis on a 1% agarose gel and stained with ethidium bromide. The lane labeled “No cDNA” shows that the PCR reaction is not contaminated with exogenous DNA.
OSTM1 potentiates Wnt/β-catenin signaling
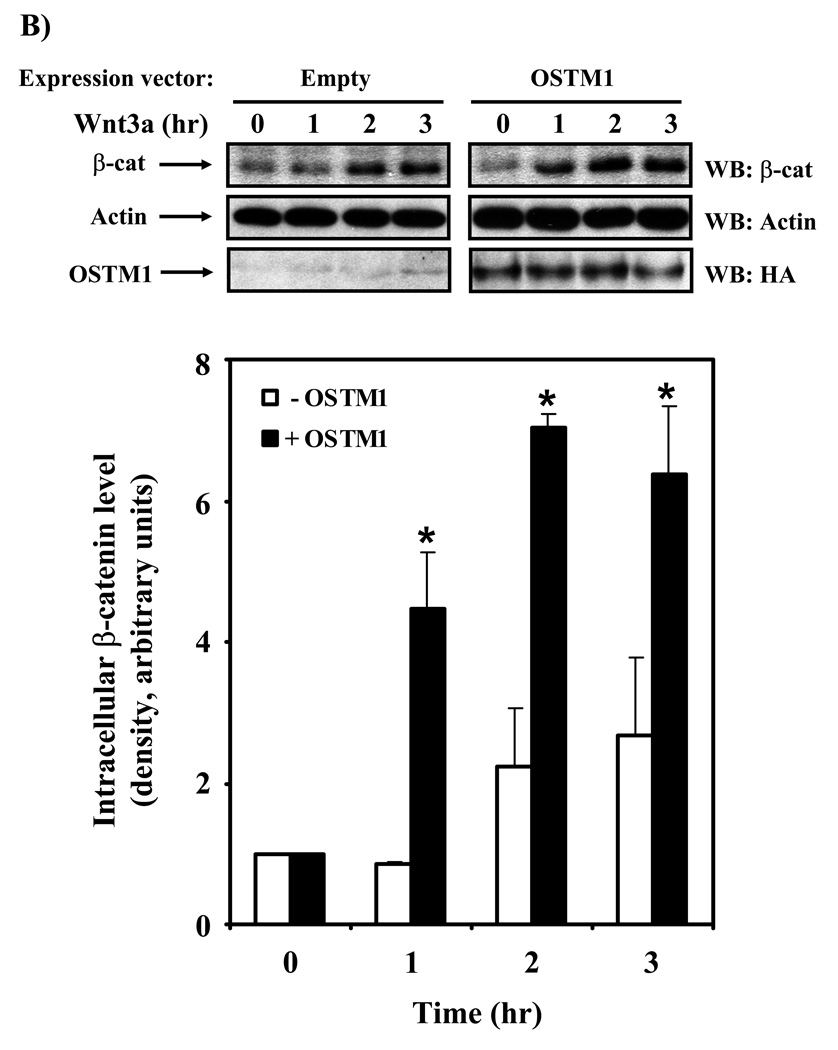
The transcriptional response to Wnt/β-catenin signaling can be measured through the use of an Lef/Tcf-responsive promoter driving luciferase expression (M50) [21]. Treatment of wild-type F9 cells with purified Wnt3a stimulated a ~10-fold increase in gene transcription, as measured by luciferase activity (Fig. 2A). We overexpressed OSTM1 in F9 cells in order to probe whether or not OSTM1 has a functional impact on Wnt/β-catenin signaling. Overexpression of OSTM1 increased the Wnt3a-induced Lef/Tcf response in a dose-dependent manner (Fig. 2A). In all conditions, constant levels of DNA were maintained through the use of an empty vector. The potentiation of Wnt3a-stimulated activity was more than double in those cells transfected with the highest amount of expression vector harboring OSTM1. The basal levels of reporter activation also were potentiated by overexpression of OSTM1. Cytosolic accumulation of β-catenin and subsequent nuclear translocation are required for Wnt-responsive Lef/Tcf-mediated gene transcription [22]. Therefore, we probed the intracellular β-catenin levels in Wnt3a-treated cells, comparing those cells that were overexpressing OSTM1 to those transfected with an empty vector (Fig. 2B). Intracellular levels of β-catenin were low in unstimulated F9 cells (Fig. 2B; 0 hr time points). Upon Wnt3a stimulation of cells transfected with an empty vector, accumulation of β-catenin was seen within 2 hours, remaining constant through 3 hours. In cells expressing OSTM1, basal levels of intracellular β-catenin were unchanged as compared with the control condition. However, in cells transfected with OSTM1 and treated with Wnt3a, accumulation of β-catenin occurred rapidly (within 1 hour), and was maintained at an elevated level past 3 hours, compared with cells expressing an empty vector.
Figure 2. OSTM1 potentiates Wnt/β-catenin signaling.
(A) F9 cells transiently transfected with Fz1, M50 and the indicated amounts of OSTM1 were treated with Wnt3a for 7 hr, then collected and subjected to luciferase assay. * denotes P < 0.05 for the difference between wildtype, Wnt3a-treated cells and those expressing OSTM1 and Wnt3a-treated for the individual doses. The data shown are mean values (+/− s.e.) of at least three separate experiments. Lower panel: Western blot analysis was used to measure the expression of transiently transfected HA-tagged OSTM1. (B) F9 cells transiently transfected with Fz1 and either empty vector or OSTM1 were treated with Wnt3a for the indicated lengths of time. Lysates were collected and treated with concanavalin A to separate cytosolic from membrane associated β-catenin. Cleared samples were subjected to SDS-PAGE on an 11% acrylamide gel, transferred to nitrocellulose and probed with an antibody against β-catenin. The results displayed are from a single representative experiment. Lower panel: Bands from multiple gels were quantified by densitometry. * denotes P < 0.05 for the difference between wild-type cells and those expressing OSTM1 for the individual time points.
OSTM1 knockdown attenuates Wnt/β-catenin signaling
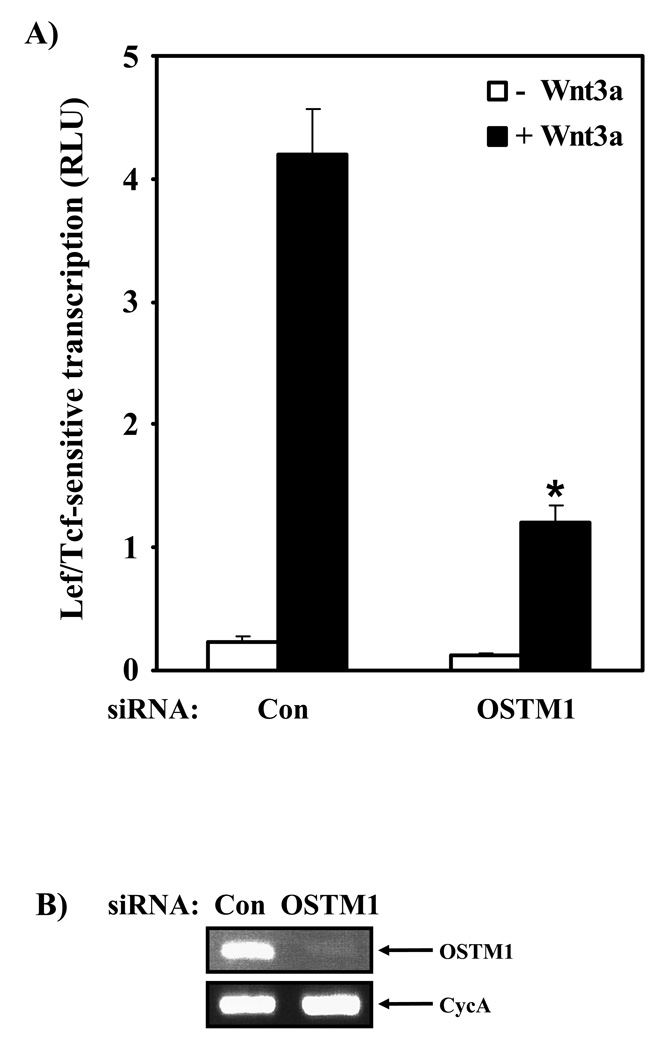
If OSTM1 acts to potentiate Wnt3a stimulation of the canonical pathway, conventional wisdom would suggest that the absence of OSTM1 would also impact signaling. To probe this role of OSTM1 in Wnt/β-catenin signaling, specific siRNA sequences were utilized to knock down OSTM1 expression in mouse F9 cells. Cells transfected with a control (scrambled) siRNA sequence responded to Wnt3a stimulation with a normal ~10-fold increase in Lef/Tcf-mediated gene transcription (Fig. 2A, Fig. 3A). In the absence of Wnt3a, basal levels of gene transcription were equivalent in control and OSTM1 knockdown conditions (Fig. 3A). The Lef/Tcf-mediated transcriptional response to Wnt3a, in sharp contrast, was attenuated by more than 70% in cells treated with an siRNA targeting mouse OSTM1 (Fig. 3A). OSTM1 appears to play an essential role in Wnt/β-catenin signaling in response to Wnt3a. RT-PCR analysis of the levels of OSTM1 mRNA demonstrated that the siRNA reagents effectively knocked down mouse OSTM1 mRNA (Fig. 3B). Two distinct siRNA sequences designed by the commercial supplier to target mouse OSTM1 were utilized in these experiments. Treatment with either of these siRNAs sharply attenuated the ability of Wnt3a to activate Lef/Tcf-sensitive transcription (data not shown).
Figure 3. OSTM1 knockdown attenuates Wnt/β-catenin signaling.
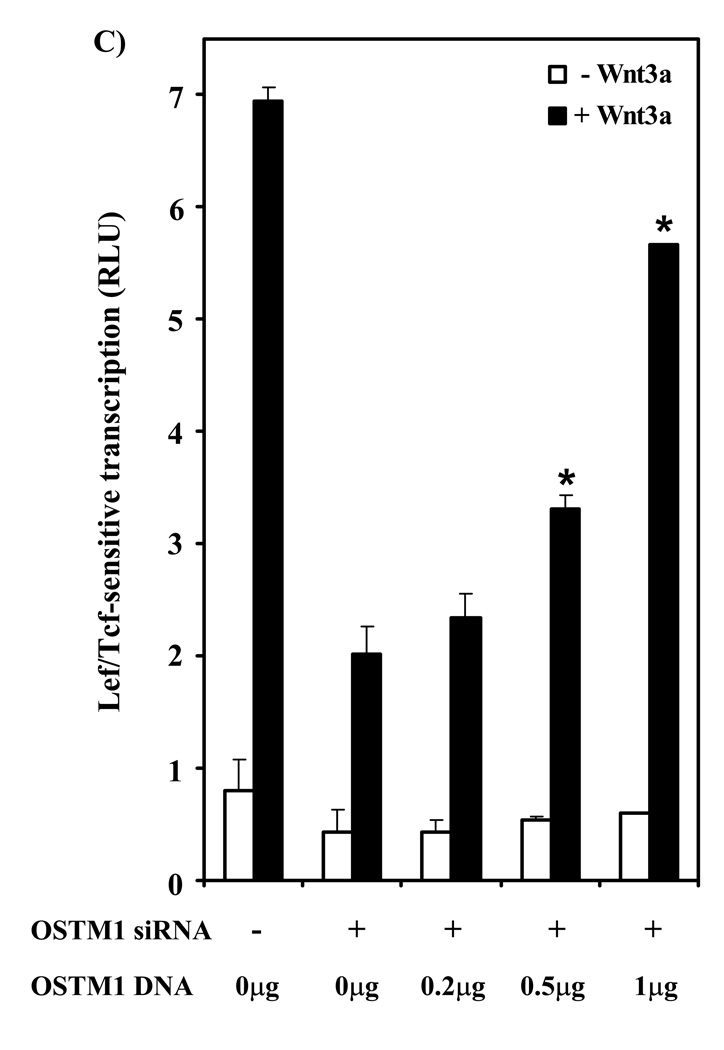
(A) F9 cells transiently transfected with siRNA sequences targeting OSTM1 (or a control sequence) and expression vectors encoding Fz1 and M50 were treated with or without Wnt3a for 7 hr, then collected and subjected to luciferase assay. * denotes P < 0.05 for the difference between cells transfected with the control siRNA and those transfected with the siRNA targeting mouse OSTM1. (B) Reverse-transcription PCR analysis was used to measure the effect of siRNA treatment on OSTM1 mRNA levels. The expression of cyclophilin A was used as a loading control. (C) F9 cells transiently transfected with siRNA sequences targeting mouse OSTM1 and expression vectors encoding Fz1, M50 and the indicated amounts of human OSTM1 were treated with or without Wnt3a for 7 hr, then collected and subjected to luciferase assay. * denotes P < 0.05 for the difference between cells transfected with the mouse OSTM1 siRNA and those transfected with the siRNA targeting mouse OSTM1 and human OSTM1 cDNA.
A rescue experiment was performed to test the specificity of the OSTM1 knockdown response. As described above, knockdown of mouse OSTM1 with siRNA sharply attenuated Wnt3a-mediated transcription (Fig. 3C). Overexpression of human OSTM1 cDNA (which is resistant to degradation by the specific siRNAs utilized in this study due to sequence divergence) in these siRNA knockdown cells lacking endogenous mouse OSTM1 successfully rescued the ability of these cells to respond to Wnt3a. The rescue of the Wnt3a-induced canonical pathway in OSTM1-depleted cells by overexpression of human OSTM1 was dose-dependent with regard to the expression vector harboring OSTM1 (Fig. 3C).
OSTM1ΔC mutant acts as a dominant negative in Wnt/β-catenin signaling
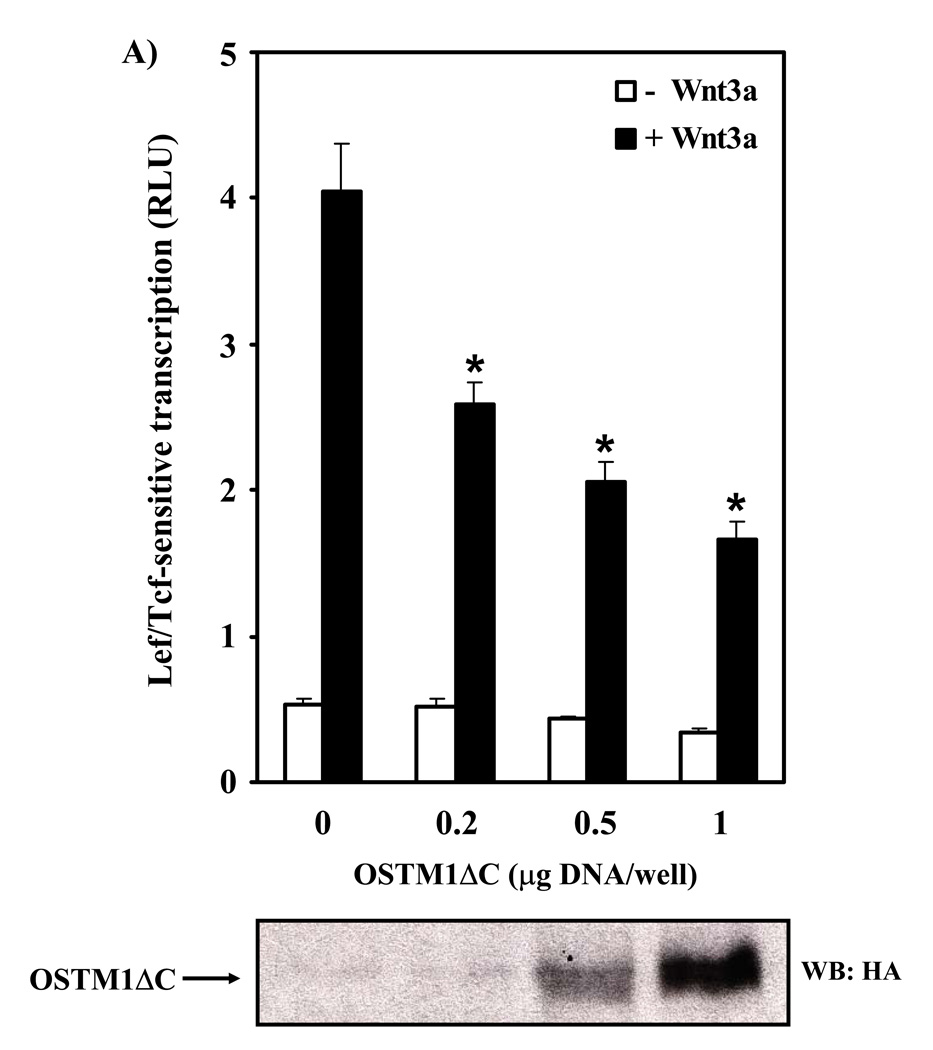
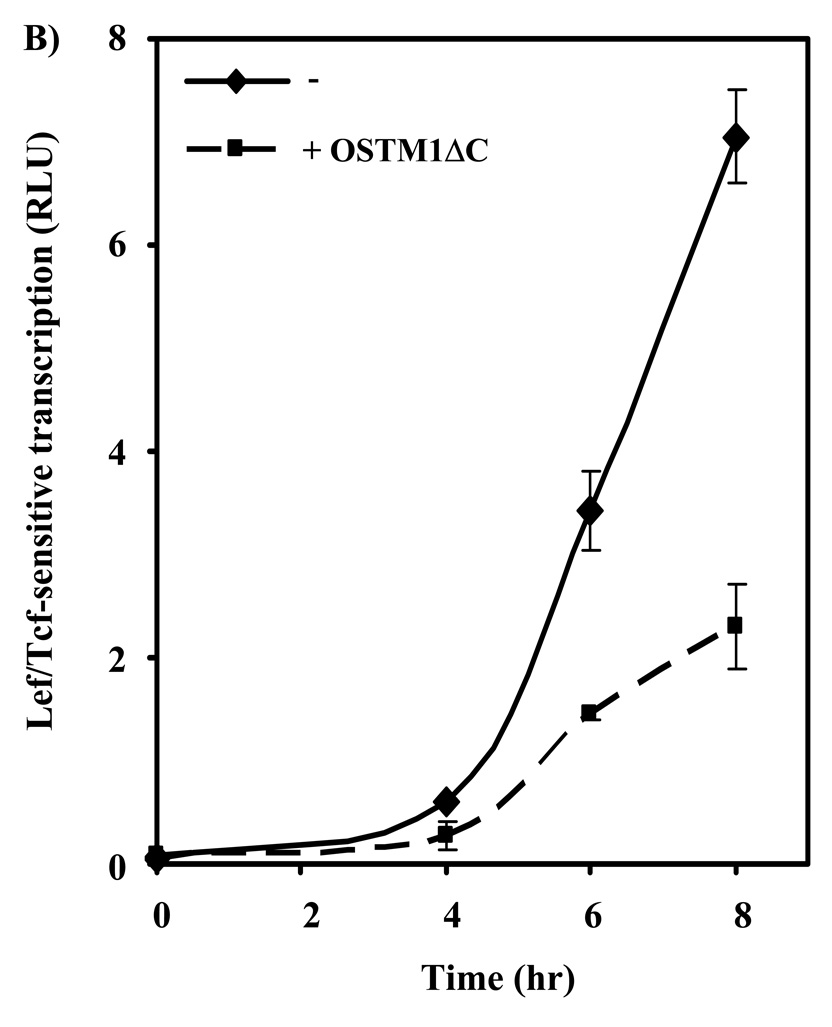
Having demonstrated a functional role of OSTM1 in Wnt/β-catenin canonical signaling we were able to focus on the effects of a known OSTM1 mutation that causes osteopetrosis in humans. There are multiple mutations of OSTM1 that cause autosomal recessive osteopetrosis [18]. We investigated the effect of a well-known human OSTM1 mutation [19] on Wnt/β-catenin signaling. The OSTM1 C-terminal deletion mutant (deletion of the final 72 amino acids; OSTM1ΔC) was constructed and we tested its ability to modulate Wnt/β-catenin canonical signaling. Overexpression of OSTM1ΔC was found to attenuate Wnt3a-induced transcription in a dose-dependent manner (Fig. 4A). In all conditions, constant levels of DNA were maintained through the use of an empty vector. Basal levels of Lef/Tcf-sensitive transcription were influenced to a lesser extent. Next, a time-course for Wnt3a-mediated gene transcription was undertaken (Fig. 4B). Treatment of F9 cells with Wnt3a induced an increase in Lef/Tcf-sensitive transcription by four hours, rising sharply at five through eight hours. Expression of OSTM1ΔC attenuated the amplitude of Wnt3a-induced transcriptional response by more than 50% (Fig. 4B).
Figure 4. OSTM1ΔC mutant attenuates Wnt/β-catenin signaling.
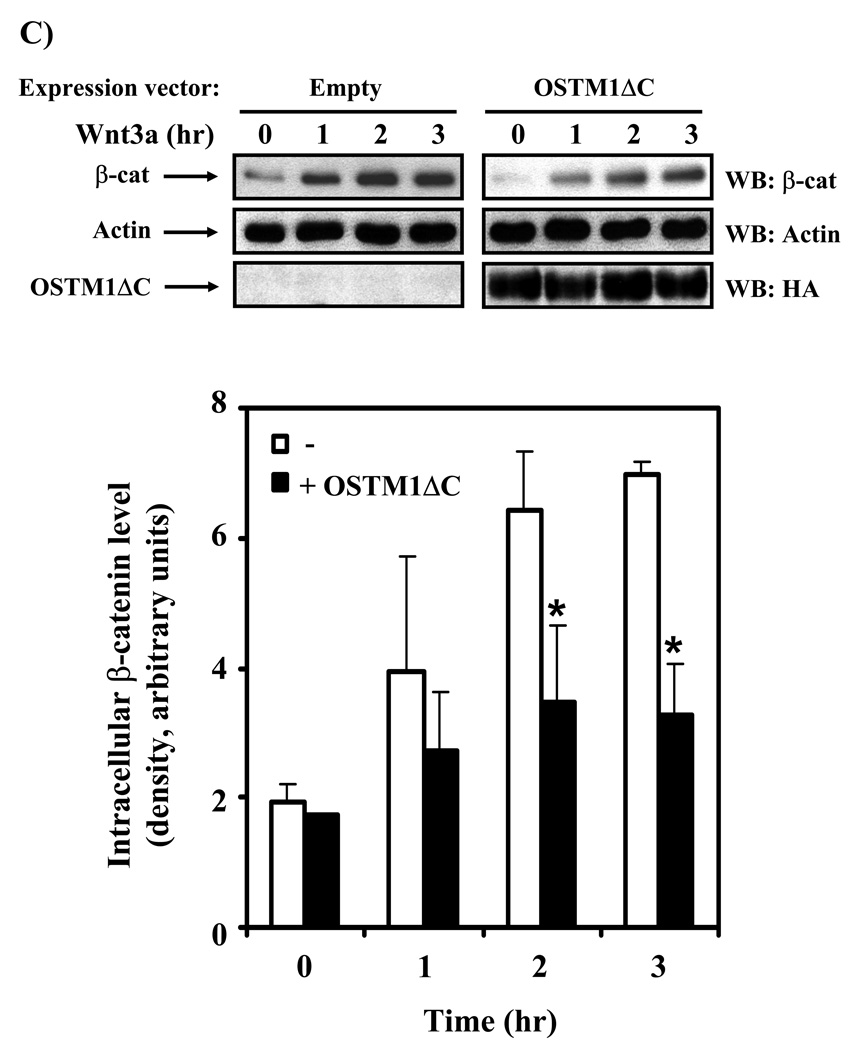
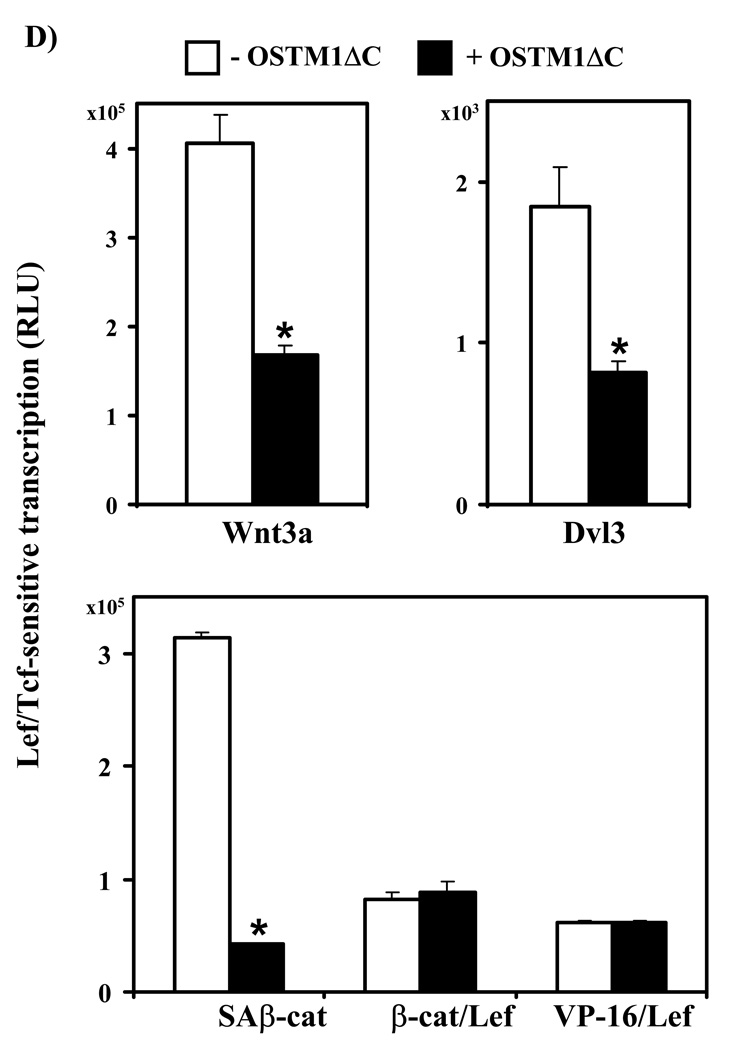
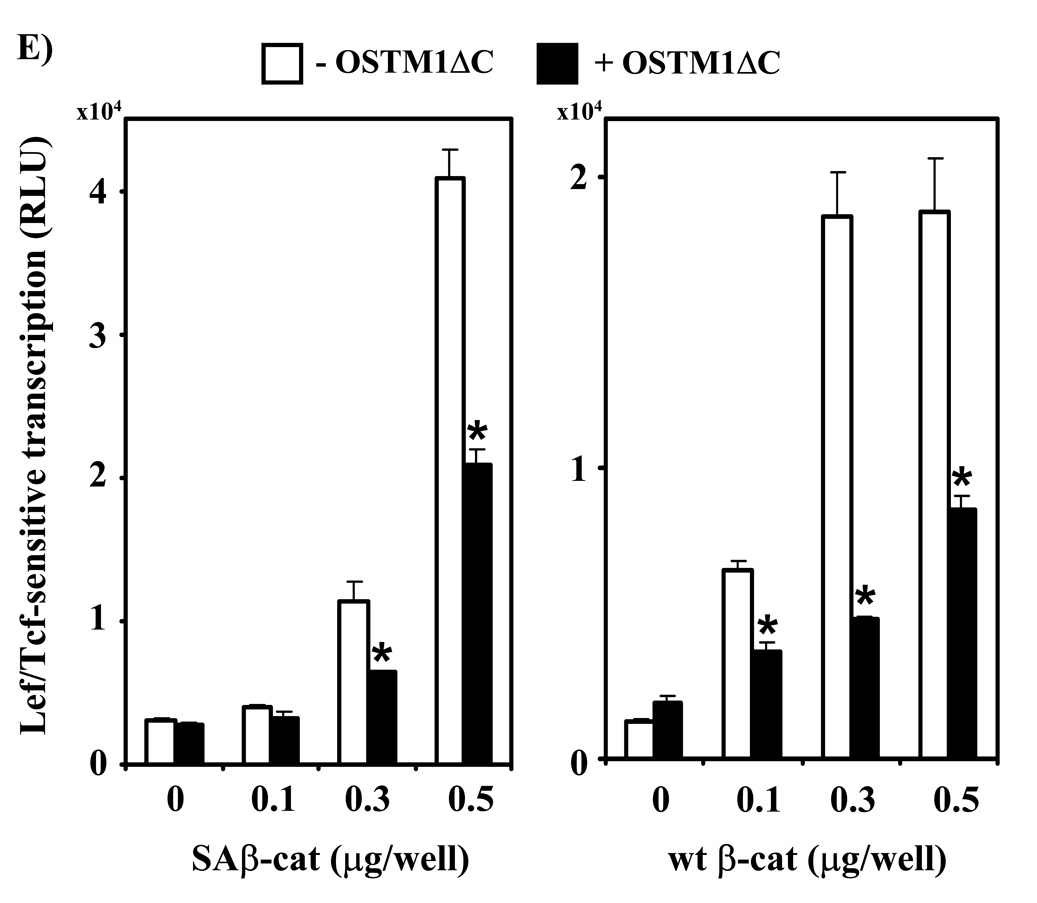
(A) F9 cells transiently transfected with Fz1, M50 and either empty vector or the indicated amounts of OSTM1ΔC were treated with Wnt3a for 7 hr, then collected and subjected to the luciferase assay. * denotes P < 0.05 for the difference between wild-type, Wnt3a-treated cells and those expressing OSTM1ΔC and Wnt3a-treated for the individual doses. Lower panel: Western blot analysis was used to measure the expression of transiently transfected HA-tagged OSTM1ΔC. (B) F9 cells transiently transfected with Fz1, M50 and either empty vector or OSTM1ΔC were treated with Wnt3a for the indicated lengths of time, then collected and subjected to luciferase assay. (C) F9 cells transiently transfected with Fz1 and either empty vector or OSTM1ΔC were treated with Wnt3a for the indicated lengths of time. Ly sates were collected and treated with concanavalin A to separate cytosolic from membrane-associated β-catenin. Cleared samples were subjected to SDS-PAGE on an 11% acrylamide gel, transferred to nitrocellulose and probed with an antibody against β-catenin. The results displayed are from a single experiment representative of more than three independent tests. Lower panel: Bands from multiple gels were quantified by densitometry. * denotes P < 0.05 for the difference between wild-type cells and those expressing OSTM1ΔC for the individual time points. (D) F9 cells transiently transfected with Fz1, M50 and the indicated constructs (or treated with Wnt3a for 7 hr) were collected and subjected to luciferase assay. * denotes P < 0.05 for the difference between Wnt/β-catenin pathway activated cells and those expressing OSTM1ΔC. (E) F9 cells transiently transfected with Fz1, M50, OSTM1ΔC and the indicated amounts of SAβ-catenin or wild-type β-catenin were collected and subjected to luciferase assay. * denotes P < 0.05 for the difference between cells transfected with SAβ-catenin or wild-type β-catenin and an empty vector and those transfected with SAβ-catenin or wild-type β-catenin and OSTM1ΔC.
To focus on the point at which the OSTM1ΔC mutant impacts the Wnt-induced transcriptional pathway, we measured β-catenin levels in cells expressing OSTM1ΔC or an empty vector and treated with Wnt3a (Fig. 4C). Wild-type F9 cells treated with Wnt3a initiated accumulation of cytosolic β-catenin within 1 hour, and reached a peak of β-catenin around 2–3 hours. Cells expressing OSTM1ΔC displayed little change in basal levels of cytosolic β-catenin. Expression of OSTM1ΔC clearly attenuated the ability of Wnt3a to induce β-catenin accumulation (Fig. 4C).
The effects of expression of OSTM1ΔC on Lef/Tcf-mediated transcription were measured in response to activation of the canonical pathway by Wnt3a and downstream elements in order to determine the epistatic level of OSTM1ΔC action on Wnt signaling (Fig. 4D). Expression of OSTM1ΔC inhibited Wnt3a-induced transcription by more than 50% (Fig. 4A,B,D). Overexpression of Dvl3 induces Lef/Tcf-mediated gene transcription [23]. OSTM1ΔC expression inhibited Dvl3-stimulated transcriptional activation by 50% (Fig. 4C). Expression of a constitutively active β-catenin mutant (lacking four N-terminal phosphorylation sites necessary for proteasome recognition; SAβ-cat) was utilized to activate Lef/Tcf gene transcription. Overexpression of OSTM1ΔC attenuated SAβ-cat-induced transcriptional activation by more than 70% (Fig. 4D). Expression of a β-catenin/Lef fusion protein (β/Lef) [24] as well as a constitutively-active Lef mutant (VP16/Lef) [25] were examined to explore further downstream signaling events in the Wnt/β-catenin pathway. In contrast to its ability to attenuate canonical signaling in response to activation by Wnt3a, overexpression of Dvl3, and expression of SAβ-catenin, expression of OSTM1ΔC was unable to attenuate Lef/Tcf-mediated signaling induced by expression of either of the Lef mutants (Fig. 4D).
To further explore the ability of OSTM1ΔC to inhibit Wnt signaling, we tested the effects of OSTM1ΔC expression on cells with increasing expression of SAβ-catenin or wild-type β-catenin (Fig. 4E). In the absence of expression of SAβ-catenin or wild-type β-catenin, OSTM1ΔC expression had no obvious effect on Lef/Tcf-sensitive transcription, when compared with cells transfected with an empty vector. At higher levels of expression of SAβ-catenin or wild-type β-catenin, however, OSTM1ΔC-induced inhibition was observed. Expression of OSTM1ΔC inhibited transcription ~50%, even at the highest tested levels of SAβ-catenin or wild-type β-catenin.
OSTM1ΔC attenuates the ability of Wnt3a to stimulate PE formation
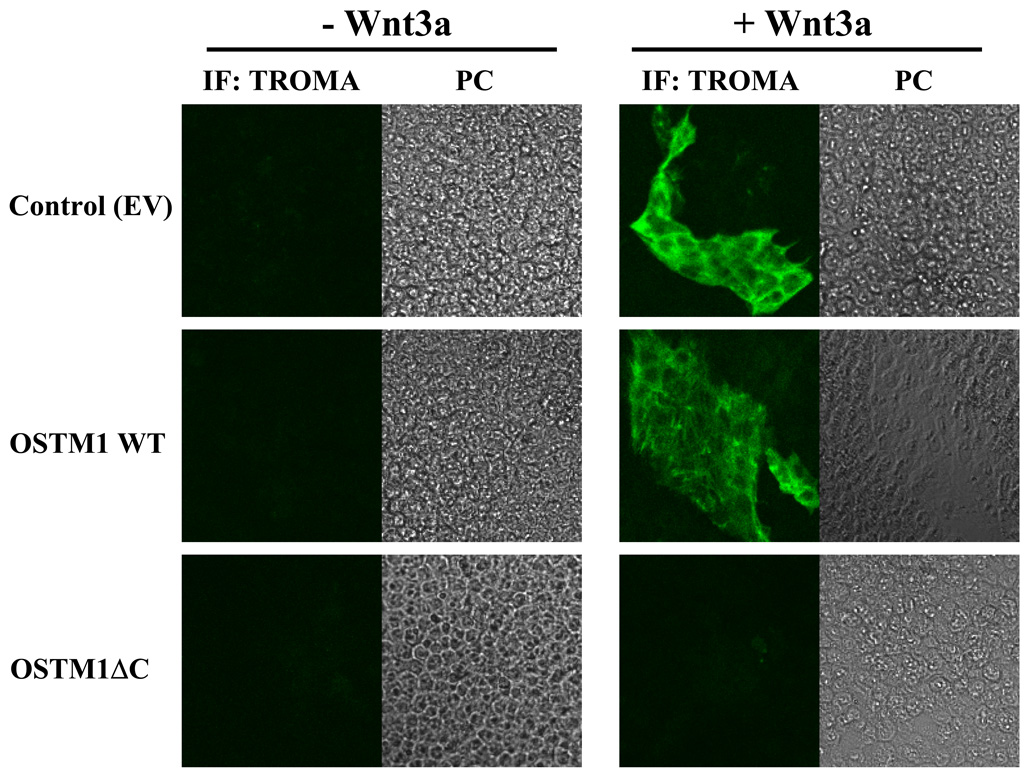
F9 cells can be induced to form PE in response to Wnt3a [6], indicated by the expression of PE-specific markers such as cytokeratin endo A. PE is a cardinal stage in early mouse development and can be measured using the mouse TROMA antibody to endo A [26]. Indirect immunofluorescence was utilized to detect expression of cytokeratin endo A in F9 cells treated with Wnt3a (Fig. 5). Untreated F9 cells do not express cytokeratin endo A, i.e., no positive staining by the TROMA antibody. Expression of OSTM1ΔC blocked the ability of F9 cells to form PE in response to Wnt3a. In the absence of Wnt3a, overexpression of OSTM1 or OSTM1ΔC had no effect on TROMA staining. In the presence of Wnt3a, cells transfected with an empty vector or OSTM1 stained positive for TROMA, indicating PE formation. Thus, expression of the OSTM1ΔC mutant blocked the ability of Wnt3a to signal to a major embryonic response, PE formation.
Figure 5. OSTM1ΔC attenuates Wnt3a-induced PE formation.
F9 cells transiently transfected with OSTM1, OSTM1ΔC or an empty vector were treated with or without Wnt3a for 4 days. Cells were stained with an antibody against cytokeratin endo A and imaged by indirect immunofluorescence. PC, phase contrast.
OSTM1 regulates β-catenin/Lef1 interaction
The epistasis experiments (Fig. 4D) implicated 0STM1ΔC action on Wnt/β-catenin signaling, downstream of β-catenin accumulation, but upstream of β-catenin/Lef1 interaction. We compared the ability of OSTM1 and OSTM1ΔC to regulate the Wnt3a-induced binding of β-catenin to Lef1 (Fig. 6). In the absence of Wnt3a, immunoprecipitated HA-Lef1 was able to pull down a small amount of endogenous β-catenin. This effect was unaltered in unstimulated cells expressing OSTM1 or OSTM1ΔC. β-catenin binding to Lef1 increased in cells treated with Wnt3a (Fig. 6, compare lanes 1 and 2). The ability of β-catenin to bind Lef1 was increased in Wnt3a-treated cells expressing OSTM1. In Wnt3a-treated cells expressing OSTM1ΔC, in sharp contrast, no detectable binding of β-catenin was observed.
Figure 6. OSTM1 regulates β-catenin/Lef1 interaction.
F9 cells transiently transfected with Lef1-HA and either OSTM1, OSTM1ΔC or an empty vector were treated with or without Wnt3a for 3 hours. Cell lysates were collected and subjected to immunoprecipitation with an anti-HA antibody. The amount of endogenous β-catenin pulled down under each condition was measured through Western blot analysis using an anti-β-catenin antibody. Lower panel: Bands from multiple gels were quantified by densitometry. * denotes P < 0.05 for the difference between untreated cells and those treated with Wnt3a.
Discussion
The Wnt/β-catenin pathway controls key steps in both the development and maintenance of multicellular organisms [11, 12]. Mutations in several Wnt pathway components lead to a number of human diseases, including cancer [12]. For example, patients with one defective APC allele present with Familiar Adenomatous Polyposis (FAP), characterized by the development of colon polyps which frequently progress to malignant adenocarcinoma [27, 28]. As many as 90% of patients with spontaneous colorectal cancer are found to have APC loss-of-function mutations [29]. However, alterations in Axin2 and β-catenin have also been reported in colon cancers with wild-type APC [30, 31]. Activating mutations in β-catenin have been identified in spontaneous pilomatricomas [32], and inactivating Lef1 mutations in sebaceous tumors [33]. Additionally, Wnt signals modulate bone formation through regulation of stem cell self-renewal, osteocyte apoptosis and osteoblast differentiation and proliferation [13]. Mutations in several Wnt signaling components lead to dramatic bone defects, including osteoporosis-pseudoglioma in patients with LRP5 defects [14]. In the current work, we focus not on a mutation that causes cancer, but one that leads to osteopetrosis.
Severe autosomal recessive osteopetrosis is a genetic disorder caused by mutations in several genes affecting osteoclast function, leading to fragile bones and increased infection rates [16, 17]. Recently, defects in the OSTM1 gene have been found to generate ARO, both in humans and in the spontaneous grey-lethal mouse model. While several functions for OSTM1 have been proposed [19, 20], the exact mechanism whereby defects in this protein lead to ARO warrant further investigation. As Wnt signaling is critically important for bone maintenance, in the present study we tested the hypothesis that OSTM1 regulates Wnt/β-catenin signaling. We found that OSTM1 overexpression potentiated, and OSTM1 knockdown suppressed, Wnt3a-mediated responses in F9 cells. Furthermore, we determined the mechanism whereby OSTM1 regulates Wnt signaling: through modulation of the β-catenin/Lef1 interaction, a crucial downstream event in Wnt/β-catenin signal transduction.
In order to assess the effect of OSTM1 overexpression or suppression on Wnt/β-catenin signaling we utilized a luciferase-based Lef/Tcf-sensitive reporter construct which can be activated in response to Wnt3a or transfection with constitutively active Wnt signaling components [21]. Overexpression of OSTM1 intensified Wnt signaling in a dose-dependent manner. In contrast, knockdown of OSTM1 attenuated Wnt3a-induced gene transcription, an effect which was rescued upon overexpression of human OSTM1 cDNA. Intracellular accumulation of β-catenin in response to Wnt3a is a hallmark of the canonical Wnt signaling pathway [34] and can be used as a reliable marker of Wnt3a signaling activity. Overexpression of OSTM1 intensified β-catenin accumulation in response to Wnt3a. This result is consistent with the transcriptional readout and suggestive of a positive role for OSTM1 in Wnt/β-catenin signaling.
Importantly, expression of an OSTM1 mutant (modeled after a known ARO human mutation) suppressed Wnt3a-mediated gene transcription in a time and dose-dependent manner, suggesting a possible Wnt signaling defect in ARO patients. Similarly, expression of OSTM1ΔC attenuated β-catenin accumulation in response to Wnt3a. In order to determine the mechanism of action of OSTM1 in the Wnt signal transduction pathway, knowledge of the epistatic level of OSTM1 was first required. Epistasis experiments revealed that OSTM1ΔC blocked Wnt/β-catenin signaling downstream of β-catenin accumulation, yet upstream of the transcriptionally active β-catenin/Lef1 protein complex. Therefore, we tested the ability of OSTM1 and OSTM1ΔC to regulate the β-catenin/Lef1 interaction. Overexpression of OSTM1 intensified, while 0STM1ΔC inhibited, the Wnt3a-stimulated interaction of β-catenin with Lef1, suggesting that OSTM1 may function as a mediator of this critical protein complex. It will be informative to probe the β-catenin/Lef1 interaction in cells from grey-lethal mice or ARO patients with OSTM1 deficiency.
Finally, expression of OSTM1ΔC was shown to inhibit the formation of primitive endoderm in response to Wnt3a. This data clearly indicates that a known human genetic lesion in OSTM1 can impact cellular differentiation and function. Our results suggest that mutations in OSTM1 may lead to severe autosomal recessive osteopetrosis through disruption of Wnt/β-catenin signaling.
Acknowledgements
The authors would like to thank the members of the Malbon lab for helpful discussions and Dr. Randall Moon (Department of Pharmacology, HHMI, University of Washington, Seattle, WA), Dr. Ken-Ichi Takemaru (Department of Pharmacology, SUNY Stony Brook, Stony Brook, NY), Dr. Howard Crawford (Department of Pharmacology, SUNY Stony Brook, Stony Brook, NY) and Dr. Thomas Jentsch (FMP/MDC, Berlin, Germany) for reagents. This work was supported by United States Public Health Service Grant DK30111 from NIDDK, National Institutes of Health (to C.C.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Embo J. 1998;17(5):1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, Kikuchi A. J Biol Chem. 1998;273(18):10823–10826. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- 3.Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL, 3rd, Lee JJ, Tilghman SM, Gumbiner BM, Costantini F. Cell. 1997;90(1):181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 4.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. Embo J. 1997;16(13):3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malbon CC. Front Biosci. 2004;9:1048–1058. doi: 10.2741/1308. [DOI] [PubMed] [Google Scholar]
- 6.Liu T, DeCostanzo AJ, Liu X, Wang H, Hallagan S, Moon RT, Malbon CC. Science. 2001;292(5522):1718–1722. doi: 10.1126/science.1060100. [DOI] [PubMed] [Google Scholar]
- 7.Yanagawa S, van Leeuwen F, Wodarz A, Klingensmith J, Nusse R. Genes Dev. 1995;9(9):1087–1097. doi: 10.1101/gad.9.9.1087. [DOI] [PubMed] [Google Scholar]
- 8.Katanaev VL, Ponzielli R, Semeriva M, Tomlinson A. Cell. 2005;120(1):111–122. doi: 10.1016/j.cell.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Rubin JS, Kimmel AR. Curr Biol. 2005;15(22):1989–1997. doi: 10.1016/j.cub.2005.10.050. [DOI] [PubMed] [Google Scholar]
- 10.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86(3):391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 11.Nusse R. Cell Res. 2005;15(l):28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 12.Clevers H. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Krishnan V, Bryant HU, Macdougald OA. J Clin Invest. 2006;116(5):1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung WM, Jin LY, Smith DK, Cheung PT, Kwan EY, Low L, Kung AW. Bone. 2006;39(3):470–476. doi: 10.1016/j.bone.2006.02.069. [DOI] [PubMed] [Google Scholar]
- 15.Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De Paepe A, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Juppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van Hul W, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML. Cell. 2001;107(4):513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 16.Quarello P, Forni M, Barberis L, Defilippi C, Campagnoli MF, Silvestro L, Frattini A, Chalhoub N, Vacher J, Ramenghi U. J Bone Miner Res. 2004;19(7):1194–1199. doi: 10.1359/JBMR.040407. [DOI] [PubMed] [Google Scholar]
- 17.Ramirez A, Faupel J, Goebel I, Stiller A, Beyer S, Stockle C, Hasan C, Bode U, Kornak U, Kubisch C. Hum Mutat. 2004;23(5):471–476. doi: 10.1002/humu.20028. [DOI] [PubMed] [Google Scholar]
- 18.Chalhoub N, Benachenhou N, Rajapurohitam V, Pata M, Ferron M, Frattini A, Villa A, Vacher J. Nat Med. 2003;9(4):399–406. doi: 10.1038/nm842. [DOI] [PubMed] [Google Scholar]
- 19.Lange PF, Wartosch L, Jentsch TJ, Fuhrmann JC. Nature. 2006;440(7081):220–223. doi: 10.1038/nature04535. [DOI] [PubMed] [Google Scholar]
- 20.Fischer T, De Vries L, Meerloo T, Farquhar MG. Proc Natl Acad Sci U S A. 2003;100(14):8270–8275. doi: 10.1073/pnas.1432965100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Curr Biol. 2003;13(8):680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 22.Noordermeer J, Klingensmith J, Perrimon N, Nusse R. Nature. 1994;367(6458):80–83. doi: 10.1038/367080a0. [DOI] [PubMed] [Google Scholar]
- 23.Feigin ME, Malbon CC. J Cell Sci. 2007;120(Pt 19):3404–3414. doi: 10.1242/jcs.011254. [DOI] [PubMed] [Google Scholar]
- 24.Filali M, Cheng N, Abbott D, Leontiev V, Engelhardt JF. J Biol Chem. 2002;277(36):33398–33410. doi: 10.1074/jbc.M107977200. [DOI] [PubMed] [Google Scholar]
- 25.Aoki M, Hecht A, Kruse U, Kemler R, Vogt PK. Proc Natl Acad Sci U S A. 1999;96(1):139–144. doi: 10.1073/pnas.96.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strickland S, Mahdavi V. Cell. 1978;15(2):393–403. doi: 10.1016/0092-8674(78)90008-9. [DOI] [PubMed] [Google Scholar]
- 27.Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hedge P, McKechnie D, et al. Science. 1991;253(5020):661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 28.Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, Koyama K, Utsunomiya J, Baba S, Hedge P. Science. 1991;253(5020):665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 29.Kinzler KW, Vogelstein B. Cell. 1996;87(2):159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Dong X, Mai M, Seelan RS, Taniguchi K, Krishnadath KK, Halling KC, Cunningham JM, Boardman LA, Qian C, Christensen E, Schmidt SS, Roche PC, Smith DI, Thibodeau SN. Nat Genet. 2000;26(2):146–147. doi: 10.1038/79859. [DOI] [PubMed] [Google Scholar]
- 31.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 32.Chan EF, Gat U, McNiff JM, Fuchs E. Nat Genet. 1999;21(4):410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 33.Takeda H, Lyle S, Lazar AJ, Zouboulis CC, Smyth I, Watt FM. Nat Med. 2006;12(4):395–397. doi: 10.1038/nm1386. [DOI] [PubMed] [Google Scholar]
- 34.Peifer M, Sweeton D, Casey M, Wieschaus E. Development. 1994;120(2):369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]