Abstract
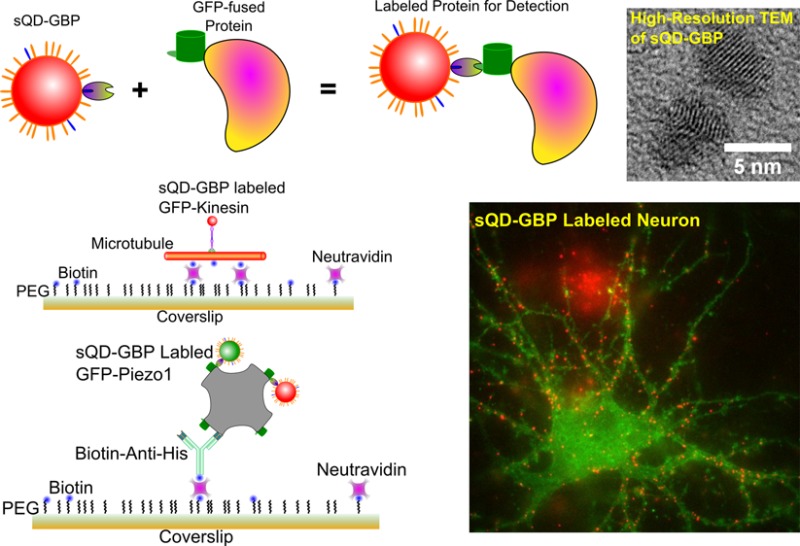
Immunofluorescence, a powerful technique to detect specific targets using fluorescently labeled antibodies, has been widely used in both scientific research and clinical diagnostics. The probes should be made with small antibodies and high brightness. We conjugated GFP binding protein (GBP) nanobodies, small single-chain antibodies from llamas, with new ∼7 nm quantum dots. These provide simple and versatile immunofluorescence nanoprobes with nanometer accuracy and resolution. Using the new probes we tracked the walking of individual kinesin motors and measured their 8 nm step sizes; we tracked Piezo1 channels, which are eukaryotic mechanosensitive channels; we also tracked AMPA receptors on living neurons. Finally, we used a new super-resolution algorithm based on blinking of (small) quantum dots that allowed ∼2 nm precision.
Introduction
Immunofluorescence is a powerful technique to detect specific targets using fluorescently labeled antibodies. It has been widely used in both scientific research and clinical diagnostics. This technique makes use of the specificity of antibodies to their antigens and allows visualization of target molecules in vivo and in vitro via fluorescence. Examples of immunofluorescence include immunostaining, immunohistochemistry, and immunoprecipitation. More recently, immunofluorescence has been utilized in superaccuracy and super-resolution microscopies.1−5
Brighter and more photostable fluorophores are clearly desirable, so we replaced the fluorophores with quantum dots (QDs).6 QDs are ∼100× brighter than organic fluorophores, and are highly resistant to photobleaching. However, the hydrodynamic diameter of commercial (biologically functionalized) QDs is 15–20 nm, a fairly large size in sterically constrained situations.7 We recently developed small quantum dots (sQDs), which are ∼7 nm in diameter (or ∼9 nm when functionalized with streptavidin (SA)), and about 1/3 the brightness of commercial QDs.8
Reducing the size of antibodies would also assist in sterically limited environments. In conventional immunofluorescence, a full IgG antibody (with two binding sites) is usually used, at ∼150 kDa and ∼14.5 × 8.5 × 4 nm3 in size, with a binding affinity that ranges from nanomolar to picomolar. In contrast, the single binding fragments derived from llama antibodies, often called “nanobodies”, are much smaller. A nanobody to GFP (or YFP), known as GFP binding protein (GBP), is only ∼13 kDa and 1.5 × 2.5 nm2 with subnanomolar affinity.9,10
In this Article we report conjugates of our new small quantum dots to GBP, thereby extending immunofluorescence to any GFP-labeled protein. The hydrodynamic diameter of our sQD-GBP conjugates is smaller than the size of any QD-antibody or QD-nanobody conjugates reported in the literature.11,12 We first applied the new probes to track the walking of individual kinesin motors and measured their ∼8 nm step sizes. Then we utilized the new probes in super-resolution imaging for measuring the size of Piezo1 proteins in cells. Piezo1 is a recently cloned cation selective eukaryotic mechanosensitive ion channel,13−16 containing 4 identical subunits. The distances between subunits are generally too large for more conventional techniques (FRET, for example), yet sufficiently small that the size of the probes may be important: hence, using a GBP-sQD is ideal. In addition, we used a new super-resolution algorithm based on blinking of QD that allowed unusually good (∼2 nm) precision. Last, we used the probes to label and track AMPA receptors in the synaptic cleft on the membrane of neurons. Here, the size of the probes is critical because of the constrained volume surrounding the receptors.
Results and Discussion
We prepared sQD-GBP conjugates following a protocol from ref (8) described in the Supporting Information. Briefly, organic CdSe/ZnS QDs (emission = 580 nm, or 620 nm) were mixed with a mixture of commercially available PEGylated alkanethiol (HSC11(EG)4-OH, 97.5%) and carboxyl PEGylated alkanethiol (HSC11(EG)4-COOH, 2.5%) in H2O/toluene with tetraethylammonium hydroxide (TEAH; 20 wt % in H2O) as base. Similar thiol-ligands (but longer) have been utilized in the literature.17−19 The reaction went on for 4 h under nitrogen at 60 °C, resulting in the transfer of QDs from organic phase into aqueous phase, monitored by fluorescence under UV. After washing with chloroform three times, negatively charged QDs (i.e., COOH-sQD) were purified from the aqueous solution using a self-packed DEAE anion exchange column. The carboxylated sQDs can then be conjugated to streptavidin or GBP nanobodies via coupling by EDC, which cross-links carboxyl groups on the sQDs to amine groups on the proteins. Unconjugated proteins were removed by a 100 kDa cutoff centrifugal filter unit, while aggregates were removed by centrifugal filter units with 0.2 μm pore-size. Conjugated sQD-GBPs were stored in PBS buffer at 4 °C for later use.
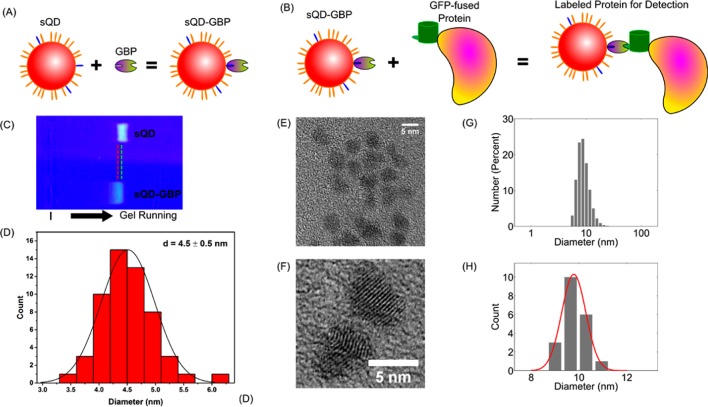
The conjugation of GBP to sQD was confirmed by several controls. For example, agarose gel electrophoresis (1%, 10 mM sodium phosphate, pH 8.0) was used since the fluorescence of sQDs can be detected directly.17,18 As shown in Figure 1C, the unconjugated sQDs show a single band with higher mobility while the sQD-GBP conjugates show a slightly retarded band, caused by the increased overall size after GBP conjugation.
Figure 1.
Synthesis and characterization of sQD-GBP conjugates. (A) CdSe/ZnS QDs (Red) were first coated with a mixture of PEGylated alkanethiol [HSC11(EG)4-OH] (orange) and carboxyl PEGylated alkanethiol [HSC11(EG)4-COOH] (blue) under the described conditions.8 The resultant COOH-functionalized QDs (sQD) were further conjugated to GBP via EDC coupling to produce GBP functionalized sQDs (sQD-GBP). (B) sQD-GBP binds to a GFP-fused protein, allowing for the detection of the protein of interest. (C) Agarose gel electrophoresis was used to confirm the success of sQD-GBP conjugation. Unconjugated sQDs show a band with higher mobility while sQD-GBP conjugates ran slightly more slowely, due to the increased size after the attachment of GBP proteins. (D) High resolution TEM shows that the size distribution of sQD-GBP conjugates is monodispersed, with a peak around 4.5 ± 0.5 nm (mean ± SD, n = 55). (E) Typical high resolution TEM image of sQD-GBP conjugates (scale bar = 5 nm). (F) Subtle lattice of the CdSe/ZnS qdots can be observed from the high resolution TEM images (scale bar = 5 nm). (G) An example of a DLS measurement shows the hydrodynamic size (diameter) of sQD-GBP ≈ 9.2 nm. (H) Twenty DLS measurements of the sQD-GBP size give an average diameter of 9.8 ± 0.4 (Mean ± SD).
The size of the sQD-GBP conjugates was characterized by both high resolution transmission electron microscope (TEM) and dynamic light scattering (DLS). For TEM imaging, the conjugates were loaded on an ultrathin carbon film TEM grid and imaged on a JEOL 2010 LaB6 high resolution TEM operating at 200 kV. The size (diameter) distribution of the conjugates, measured from TEM, showed a monodispersion distribution, with a peak center at 4.5 ± 0.5 nm (mean ± SD, n = 55, Figure 1D). A representative sample of sQD-GBP conjugates is shown in Figure 1E, which also proves that quantum dots were properly stabilized by the protein conjugates. In addition, subtle lattice of the CdSe/ZnS qdots can be observed from the high resolution TEM images (Figure 1F). Note that TEM mainly measures the size of the core and inorganic shell of the nanocrystals. The hydrodynamic size (diameter) of the sQD-GBP conjugates was also measured by DLS. A typical DLS measurement is shown in Figure 1G, with a hydrodynamic size of 9.2 nm. Multiple DLS measurements give a monodispersion distribution for the hydrodynamic size, centered at 9.8 ± 0.4 (mean ± SD, n = 20, Figure 1H). Compared to bare sQD, the hydrodynamic size of the sQD-GBP conjugate is increased by ∼4 nm; compared to the hydrodynamic size of carboxylated sQD, the hydrodynamic size of sQD-GBP increases by ∼1.5 nm. This increase in the size after conjugation to GBP once again confirms the success of conjugation.
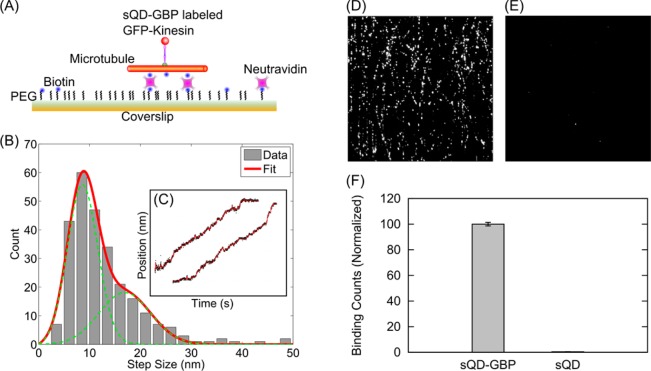
We first applied sQD-GBP to measure the in vitro step-size of kinesin, a molecular motor which walks on microtubules and plays a key role in many cellular transport processes.20 It has been shown that kinesin moves in 8.3 nm center-of-mass steps and that kinesin “walks” in a hand-over-hand fashion with ∼16.6 nm stride length.21,22 We attached the sQD-GBP to a GFP fused to the heavy chain of truncated kinesin560, and watched the motor walking at a low ATP concentration of 400 nM (Figure 2A). The high spatial resolution is achieved by FIONA (fluorescence imaging with one nanometer accuracy).2 Two example traces of kinesin labeled with a GBP-sQD are shown in Figure 2C. Traces were tested for steps with Students’ t test and the resulting step sizes were plotted in a histogram (Figure 2B). The mean step size was determined by fitting the distribution with
where
which takes into account the asymmetric distance distribution due to noise.23I0 is the modified Bessel function of integer order zero, x is the measured distance, σi is the localization error, and μ is the true distance. Note that the second term [A2p(x, 2μ, σ2)] in the fitting function originates from motors with a stepping rate faster than the experimental time resolution. The major population (65%) gives a step size of 8.1 ± 0.2 nm (mean ± SEM), consistent with previous reports.24,25
Figure 2.
Measurement of the step size of kinesin using sQD-GBP as probes. (A) Scheme of the experiment. (B) Distribution of step sizes. (C) Two example traces of kinesin labeled with a sQD-GBP. In the context of immunofluorescence, microtubules were decorated using kinesins as primary antibodies and (D) sQD-GBP conjugates or (E) unconjugated sQD as secondary antibodies. We observed (D) a lot of binding with sQD-GBP conjugates and (E) almost no binding with carboxylated sQD. Microtubules and kinesins were present in both samples and experiments were run in parallel. (F) Quantification of the binding count shows that less than 1% of the binding is due to nonspecific binding (100.0 ± 1.4% vs 0.45 ± 0.06% [Mean ± SD, n = 6]).
The system of microtubule and kinesin was also used as a third control to confirm the success of sQD-GBP conjugation. In the context of immunofluorescence, we decorated microtubules by using kinesins as primary antibodies (as kinesin binds to microtubule under appropriate conditions) and sQD-GBP conjugates (or unconjugated sQD) as secondary antibodies. We observed a lot of binding with sQD-GBP conjugates and almost no binding with unconjugated sQD (100.0 ± 1.4% vs 0.45 ± 0.06%), as shown in Figure 2D,E,F.
We then labeled Piezo1 proteins, mechanosensitive channels recently identified in mammalian cells,13,14 with sQD-GBP. Mutations of this protein are associated with human diseases such as xerocytosis which disrupts red blood cell volume regulation.15 It has been proposed that Piezo1 proteins assemble as homotetramers in vitro and may do so in vivo (with a total molecular weight of 4 × 0.3 MDa ≈ 1.2 MDa). A crystal structure is not yet available and FRET measurements of monomer spacing have yet to be made—and in most cases, they are likely too large to be measured. Consequently we measured the monomer/monomer distances via single-molecule high-resolution co-localization (SHREC), a two-color form of FIONA with <10 nm resolution.26 Here, we used sQD-GBP conjugates with two colors (sQD580-GBP, emission at 580 nm; and sQD620-GBP, emission at 620 nm) to co-localize subunits by measuring the N-to-N and C-to-C distances.
We isolated single His-tagged Piezo homotetramers for fluorescent imaging using single-molecule pull-down (SiMPull).27 In brief, GFP-fused Piezo1 proteins were expressed in HEK293 cells and collected after cell lysis, followed by labeling with sQD580-GBP and sQD620-GBP conjugates (Supporting Information Figure 1a). The labeled Piezo1 proteins were immobilized on a microscope coverslip. The coverslip was functionalized with PEG and biotinylated PEG to facilitate the binding of neutravidin, on top of which biotinylated anti-His antibodies are bound (Supporting Information Figure 1b). The anti-His antibodies bind to the proteins and immobilize them on the surface for imaging.
Each color was imaged with an Opto-Splitter on an EMCCD. Nanoholes were used for co-localization and correction for achromatic aberration.28,29 We achieved ∼1.1 nm for the target registration error for mapping transformation between the two color probes (Supporting Information notes and Supporting Information Figure 2b and c), three times better than that obtained by using a fluorescent bead and moving the microscope stage,26 or by using nonuniform nanoholes.29
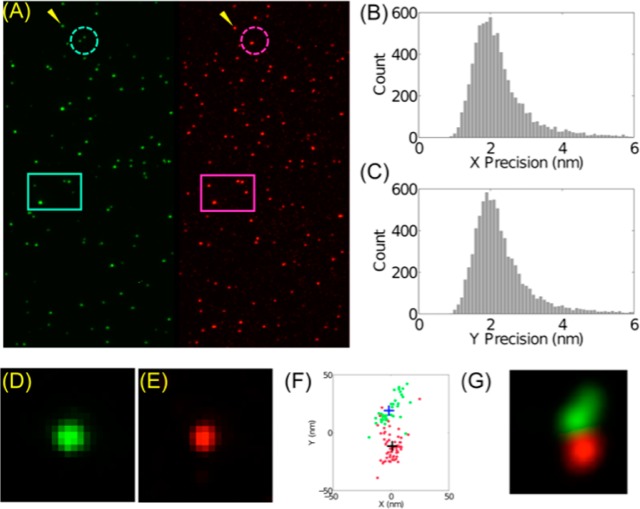
Figure 3A shows images of Piezo1 labeled with sQD-GBP conjugates. Some Piezo1 spots show co-localization of two labels, indicating that these individual Piezo1 channels were labeled by at least one sQD580-GBP and one sQD620-GBP (boxed area in Figure 3A). On the other hand, due to the stochastic nature of labeling, some Piezo1 channels were labeled with only one color (circled area in Figure 3A).
Figure 3.
Measurement of the distance between N-termini of monomers of Piezo1 channels. (A) Piezo1 proteins labeled with sQD-GBP in two colors (Left: ∼580 nm, Right: ∼620 nm). The rectangles show example spots with co-localization in the two channels, while the circles indicate proteins labeled by sQD-GBP of only one color, which is expected, as the labeling is stochastic. Panels B and C show localization of spots using a super-resolution algorithm based on QD-blinking30,31 for both colors. The precision of localization in (B) x and (C) y directions is 2.0 ± 0.5 nm (Mean ± SD). (D, E) Enlarged images of the spots indicated by yellow arrow in (A). (F) Super-resolution analysis shows multiple localizations of sQD580-GBP (green dots) and sQD620-GBP (red dots). Their centers of masses (blue and black + signs) are used to calculate the distance between the two sQDs. (G) A different representation of (F) by rendering each dot as a Gaussian function with σ = 10 nm.
Instead of localizing the spots using FIONA directly, we performed super-resolution analysis using an algorithm based on QD-blinking for both channels30,31 (Supporting Information notes) to localize sQD-GBP for multiple times, with localization precision ∼2.0 ± 0.5 nm (Mean ± SD, Figure 3B,C; or 2.7 nm using a more theoretical accurate formula29,32). An advantage is that the super-resolution algorithm allows us to have multiple localization events for the same sQD-GBP and thus to reduce errors in distance calculations. A dual-labeled Piezo1 cluster was indicated by the yellow arrows shown in Figure 3A and enlarged in Figure 3D and E. Super-resolution analysis, with correction for stage drift using QDs in the same field of view, gives multiple localizations of sQD580-GBP (green dots in Figure 3F) and sQD620-GBP (red dots in Figure 3F). Their centers of masses (blue and black + signs in Figure 3F) were used to calculate the distance between the two QDs.
We measured the distance between N-termini of subunits of human Piezo1 (ΔNN) and the distance between C-termini (ΔCC) (Figure 4A and B, respectively). With the assumption that Piezo1 assembles as a homotetramer, the labels can be on adjacent monomers or opposite monomers. Simple geometric considerations predict the two distances to differ from each other by √2. Thus, we fit the distribution with a sum of two Gaussians whose centers differ by a factor of √2: f(x) = A1 exp[−(x – μ)2/(2σ12)] + A2 exp[−(x – √2 μ)2/(2σ2)]. The fitted curves give μ = 36 ± 10 nm (mean ± SD) for ΔNN and μ = 22 ± 5 nm (mean ± SD) for ΔCC.
Figure 4.
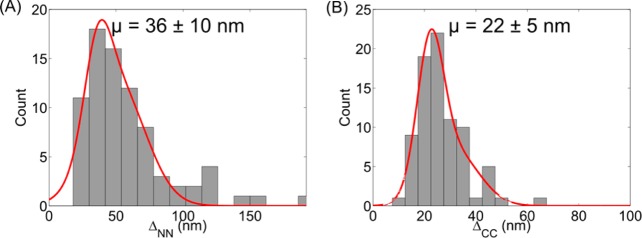
Distributions of (A) the distances between N-terminus of subunits of human Piezo1 (ΔNN) and (B) the distance between C-terminus (ΔCC). Fitting curves f(x) = A1 exp[−(x – μ)2/(2σ12)] + A2 exp[−(x – √2 μ)2/(2σ2)] give (A) μ = 36 ± 10 nm (Mean ± SD) for ΔNN and (B) μ = 22 ± 5 nm (Mean ± SD) for ΔCC.
Last, we labeled AMPA receptors (AMPARs) with sQD-GBP expressed in neurons (Figure 5). AMPARs are ligand-gated ion channels that mediate fast synaptic transmission in the central nervous system. Trafficking of AMPARs in and out of synapses is one of the effects of synaptic plasticity, which underlies the molecular mechanism of learning and memory.33
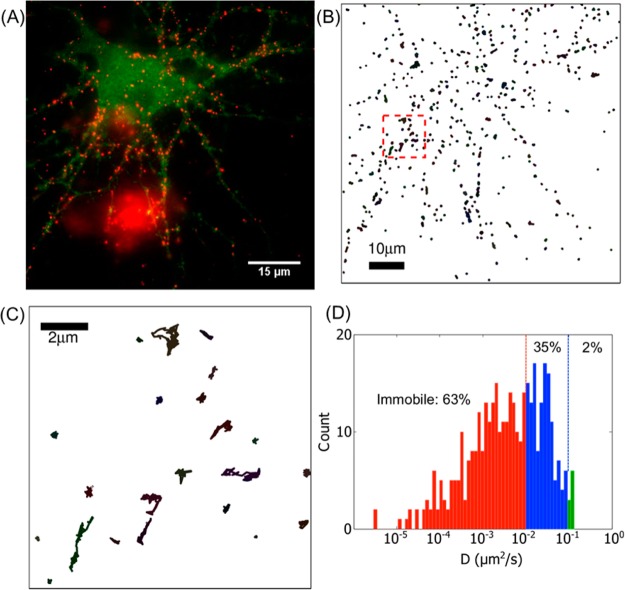
Figure 5.
(A) Specific labeling of AMPA receptors on neurons. Green: GluA2-pHluorin; Red: sQD-GBP. High labeling specific is demonstrated by sQD-GBP only labeling neurons which expressed GluA2-pHluorin but not untransfected neurons. (B) Diffusion trajectories of individual AMPA receptors from (A). (C) Enlargement of the red square area of (B). (D) Distribution of diffusion coefficient D for all trajectories shown in (B).
To study AMPAR lateral diffusion at the synapse, quantum dots are sometimes used as fluorescent probes due to their brightness and photostability.8,34 However, the big size of commercially available QDs (15–20 nm in diameter) can lead to problems when labeling the AMPARs inside the synaptic cleft, a 20–40 nm gap. Here we used sQD620-GBP to track AMPAR movements on the surface of neurons, and to see whether the sQD-GBP could enter the synapse. AMPAR subunit GluA2 was genetically fused with pHluorin,35 a pH-sensitive GFP, and expressed in dissociated cortical neuron cultures. The neurons were labeled with sQD620-GBP in a 5 min incubation. As shown in Figure 5A, the, sQD-GBP (red) specifically bound to the neuron that expressed GluA2-pHluorin (green), but not to untransfected neurons nearby or to the coverslip. (These untransfected neurons are not fluorescently visible.) Note that the big red cluster on the lower part of the image is not due to nonspecific labeling but to sQDs trapped in the membrane of a dead cell.
We used single particle tracking to study the diffusion of the sQD-labeled AMPARs. Figure 5B shows the trajectories of individual AMPARs, with an enlarged image shown in Figure 5C. We calculated the diffusion coefficients for the AMPARs labeled with sQD-GBP and found that 63% of the receptors are immobile (defined as the diffusion coefficient, D, <0.01 μm2/s). The percentage is much higher than using commercial QDs (11%) and also higher than using sQD-SA (37%).8 This indicates that the sQD-GBP labeled AMPARs have a greater chance of diffusing into the synapse and being trapped there. Another advantage of the sQD-GBP is that GBP only has one binding site to GFP, whereas SA has 4 binding sites to biotins. Therefore, using sQD-GBP instead of sQD-SA can reduce the possibility of cross-linking when labeling target proteins.
To conclude, the small quantum dots conjugated to nanobodies are versatile immunofluorescence probes. We demonstrated its applications by tracking the walking of individual kinesin motors, detecting and measuring the size of Piezo1 channels, and tracing AMPA receptors on neurons. The hydrodynamic diameter of our sQD-GBP conjugates is 9.8 nm, ∼20% smaller in diameter than the size of the recently reported QD-nanobody conjugates.12 In fact, the sQD-GBP probes reported here are smaller than full antibodies labeled with organic fluorophores.36 However, due to the use of QDs, the probes are much brighter than the conventional fluorophores. The new probes can be applied to many GFP-fused (and eventually other FP-) proteins. We expect that the current conjugation method could be used with other nanobodies or single domain antibodies/fragments.
Acknowledgments
This work was funded in part by NSF 1063188, NSF 1430124, and NIH NS087413, HL054887, NS090903, and GM108578.
Supporting Information Available
Experimental procedures and characterization of co-localization and localization precision. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Huang B.; Wang W.; Bates M.; Zhuang X. (2008) Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toprak E., Kural C., and Selvin P. R. (2010) Super-Accuracy and Super-Resolution: Getting Around the Diffraction Limit, in Methods in Enzymology (Walter N. G., Ed.) pp 1–26, Chapter One, Academic Press. [DOI] [PubMed] [Google Scholar]
- Betzig E.; Patterson G. H.; Sougrat R.; Lindwasser O. W.; Olenych S.; Bonifacino J. S.; Davidson M. W.; Lippincott-Schwartz J.; Hess H. F. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645. [DOI] [PubMed] [Google Scholar]
- Shroff H.; Galbraith C. G.; Galbraith J. A.; Betzig E. (2008) Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5, 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M.; Huang B.; Dempsey G. T.; Zhuang X. (2007) Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner K. D.; Lindén S.; Jin Z.; Jennings T. L.; el Khoulati R.; van Bergen en Henegouwen P. M. P.; Hildebrandt N. (2013) Nanobodies and nanocrystals: highly sensitive quantum dot-based homogeneous FRET immunoassay for serum-based EGFR detection. Small 10, 734–740. [DOI] [PubMed] [Google Scholar]
- Sperling R. A.; Liedl T.; Duhr S.; Kudera S.; Zanella M.; Lin C.-A. J.; Chang W. H.; Braun D.; Parak W. J. (2007) Size determination of (bio)conjugated water-soluble colloidal nanoparticles: a comparison of different techniques. J. Phys. Chem. C 111, 11552–11559. [Google Scholar]
- Cai E.; Ge P.; Lee S. H.; Jeyifous O.; Wang Y.; Liu Y.; Wilson K. M.; Lim S. J.; Baird M. A.; Stone J. E.; Lee K. Y.; Davidson M. W.; Chung H. J.; Schulten K.; Smith A. M.; Green W. N.; Selvin P. R. (2014) Stable small quantum dots for synaptic receptor tracking on live neurons. Angew. Chem., Int. Ed. 53, 12484–12488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U.; Zolghadr K.; Tillib S.; Nowak D.; Schermelleh L.; Gahl A.; Backmann N.; Conrath K.; Muyldermans S.; Cardoso M. C.; Leonhardt H. (2006) Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3, 887–889. [DOI] [PubMed] [Google Scholar]
- Ries J.; Kaplan C.; Platonova E.; Eghlidi H.; Ewers H. (2012) A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584. [DOI] [PubMed] [Google Scholar]
- Clarke S.; Pinaud F.; Beutel O.; You C.; Piehler J.; Dahan M. (2010) Covalent monofunctionalization of peptide-coated quantum dots for single-molecule assays. Nano Lett. 10, 2147–2154. [DOI] [PubMed] [Google Scholar]
- Sukhanova A.; Even-Desrumeaux K.; Kisserli A.; Tabary T.; Reveil B.; Millot J.-M.; Chames P.; Baty D.; Artemyev M.; Oleinikov V.; Pluot M.; Cohen J. H. M.; Nabiev I. (2012) Oriented conjugates of single-domain antibodies and quantum dots: toward a new generation of ultrasmall diagnostic nanoprobes. Nanomed. Nanotechnol. Biol. Med. 8, 516–525. [DOI] [PubMed] [Google Scholar]
- Coste B.; Mathur J.; Schmidt M.; Earley T. J.; Ranade S.; Petrus M. J.; Dubin A. E.; Patapoutian A. (2010) Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B.; Xiao B.; Santos J. S.; Syeda R.; Grandl J.; Spencer K. S.; Kim S. E.; Schmidt M.; Mathur J.; Dubin A. E.; Montal M.; Patapoutian A. (2012) Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 483, 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae C.; Gnanasambandam R.; Nicolai C.; Sachs F.; Gottlieb P. A. (2013) Xerocytosis is caused by mutations that alter the kinetics of the mechanosensitive channel PIEZO1. Proc. Natl. Acad. Sci. U. S. A. 110, e1162–e1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo I.; Alper S. L.; Franceschi L. D.; Auriemma C.; Russo R.; Falco L. D.; Vallefuoco F.; Esposito M. R.; Vandorpe D. H.; Shmukler B. E.; Narayan R.; Montanaro D.; D’Armiento M.; Vetro A.; Limongelli I.; Zuffardi O.; Glader B. E.; Schrier S. L.; Brugnara C.; Stewart G. W.; Delaunay J.; Iolascon A. (2013) Multiple clinical forms of dehydrated hereditary stomatocytosis arise from mutations in PIEZO1. Blood 121, 3925–3935. [DOI] [PubMed] [Google Scholar]
- Zhu Z.-J.; Yeh Y.-C.; Tang R.; Yan B.; Tamayo J.; Vachet R. W.; Rotello V. M. (2011) Stability of quantum dots in live cells. Nat. Chem. 3, 963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M.; Liu W.; Puthenveetil S.; Zheng Y.; Marshall L. F.; Schmidt M. M.; Wittrup K. D.; Bawendi M. G.; Ting A. Y. (2008) Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nat. Methods 5, 397–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.-C.; Patra D.; Yan B.; Saha K.; Miranda O. R.; Kim C. K.; Rotello V. M. (2011) Synthesis of cationic quantum dotsvia a two-step ligand exchange process. Chem. Commun. 47, 3069–3071. [DOI] [PubMed] [Google Scholar]
- Vale R. D.; Milligan R. A. (2000) The way things move: looking under the hood of molecular motor proteins. Science 288, 88–95. [DOI] [PubMed] [Google Scholar]
- Yildiz A.; Tomishige M.; Vale R. D.; Selvin P. R. (2004) Kinesin walks hand-over-hand. Science 303, 676–678. [DOI] [PubMed] [Google Scholar]
- Svoboda K.; Schmidt C. F.; Schnapp B. J.; Block S. M. (1993) Direct observation of kinesin stepping by optical trapping interferometry. Nature 365, 721–727. [DOI] [PubMed] [Google Scholar]
- Stirling Churchman L.; Flyvbjerg H.; Spudich J. A. (2006) A non-Gaussian distribution quantifies distances measured with fluorescence localization techniques. Biophys. J. 90, 668–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer M. J.; Block S. M. (1997) Kinesin hydrolyses one ATP per 8-nm step. Nature 388, 386–390. [DOI] [PubMed] [Google Scholar]
- Yardimci H.; Duffelen M. van; Mao Y.; Rosenfeld S. S.; Selvin P. R. (2008) The mitotic kinesin CENP-E is a processive transport motor. Proc. Natl. Acad. Sci. U. S. A. 105, 6016–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman L. S.; Ökten Z.; Rock R. S.; Dawson J. F.; Spudich J. A. (2005) Single molecule high-resolution co-localization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc. Natl. Acad. Sci. U. S. A. 102, 1419–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A.; Liu R.; Ramani B.; Arauz E.; Ishitsuka Y.; Ragunathan K.; Park J.; Chen J.; Xiang Y. K.; Ha T. (2011) Probing cellular protein complexes using single-molecule pull-down. Nature 473, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baday M.; Cravens A.; Hastie A.; Kim H.; Kudeki D. E.; Kwok P.-Y.; Xiao M.; Selvin P. R. (2012) Multicolor super-resolution DNA imaging for genetic analysis. Nano Lett. 12, 3861–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertsinidis A.; Mukherjee K.; Sharma M.; Pang Z. P.; Park S. R.; Zhang Y.; Brunger A. T.; Südhof T. C.; Chu S. (2013) Ultrahigh-resolution imaging reveals formation of neuronal SNARE/Munc18 complexes in situ. Proc. Natl. Acad. Sci. U. S. A. 110, E2812–E2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson P. D.; Rothenberg E.; Selvin P. R. (2011) Single-molecule-based super-resolution images in the presence of multiple fluorophores. Nano Lett. 11, 5090–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Fruhwirth G.; Cai E.; Ng T.; Selvin P. R. (2013) 3D super-resolution imaging with blinking quantum dots. Nano Lett. 13, 5233–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen K. I.; Churchman L. S.; Spudich J. A.; Flyvbjerg H. (2010) Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat. Methods 7, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huganir R. L.; Nicoll R. A. (2013) AMPARs and synaptic plasticity: the last 25 years. Neuron 80, 704–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groc L.; Lafourcade M.; Heine M.; Renner M.; Racine V.; Sibarita J.-B.; Lounis B.; Choquet D.; Cognet L. (2007) Surface trafficking of neurotransmitter receptor: comparison between single-molecule/quantum dot strategies. J. Neurosci. 27, 12433–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby M. C.; Rue S. A. D. L.; Ralph G. S.; Uney J.; Collingridge G. L.; Henley J. M. (2004) Removal of AMPA receptors (AMPARs) from synapses is preceded by transient endocytosis of extrasynaptic AMPARs. J. Neurosci. 24, 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbmann U.; Connah M.; Fish B.; Varley P.; Gee C.; Mulot S.; Chen J.; Zhou L.; Lu Y.; Shen F.; Yi J.; Harding S. E. (2007) Dynamic light scattering as a relative tool for assessing the molecular integrity and stability of monoclonal antibodies. Biotechnol. Genet. Eng. Rev. 24, 117–128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.