Abstract
Background
Cholangiocarcinoma (CCA) is clinically heterogeneous; intra and extrahepatic CCA have diverse clinical presentations. Next generation sequencing (NGS) technology may identify the genetic differences between these entities and identify molecular subgroups for targeted therapeutics.
Methods
We describe successful NGS-based testing of 75 CCA patients along with the prognostic and therapeutic implications of findings. Mutation profiling was performed using either a) NGS panel of hotspot regions in 46 cancer-related genes using a 318-chip on Ion PGM Sequencer or b) Illumina HiSeq 2000 sequencing platform for 3,769 exons of 236 cancer-related genes plus 47 introns from 19 genes to an average depth of 1000X. Clinical data was abstracted and correlated with clinical outcome. Patients with targetable mutations were referred to appropriate clinical trials.
Results
There were significant differences between intrahepatic (n = 55) and extrahepatic CCA (n = 20) in regard to the nature and frequency of the genetic aberrations (GAs). IDH1 and DNA repair gene alterations occurred more frequently in intrahepatic CCA, while ERBB2 GAs occurred in the extrahepatic group. Commonly occurring GAs in intrahepatic CCA were TP53 (35%), KRAS (24%), ARID1A (20%), IDH1 (18%), MCL1 (16%) and PBRM1 (11%). Most frequent GAs in extrahepatic CCA (n = 20) were TP53 (45%), KRAS (40%), ERBB2 (25%), SMAD4 (25%), FBXW7 (15%) and CDKN2A (15%). In intrahepatic CCA, KRAS, TP53 or MAPK/mTOR GAs were significantly associated with a worse prognosis while FGFR GAs correlated with a relatively indolent disease course. IDH1 GAs did not have any prognostic significance. GAs in the chromatin modulating genes, BAP1 and PBRM1 were associated with bone metastases and worse survival in extrahepatic CCA. Radiologic responses and clinical benefit was noted with EGFR, FGFR, C-met, B-RAF and MEK inhibitors.
Conclusion
There are significant genetic differences between intra and extrahepatic CCA. NGS can potentially identify disease subsets with distinct prognostic and therapeutic implications.
Introduction
CCA represents the second most common primary liver cancer worldwide. Several recent epidemiological reports indicate that its incidence and mortality is rising, particularly in the western world [1]–[6]. The overall survival of this disease remains dismal due to delayed detection, suboptimal response to standard therapy and underlying liver disease such as non-alcoholic steatohepatitis (NASH), which can limit liver-directed therapies [7], [8]. The current clinical classification of CCA is based on its anatomic location and includes the intrahepatic, hilar and distal subgroups. The Liver Cancer Study Group of Japan has described three morphological variants of intrahepatic CCA; these include the mass-forming, intraductal and peri-hilar types, based on the patterns of disease spread [9]. These classifications have improved the understanding and surgical management of this disease. The location of the tumor (intra vs. extrahepatic) has no therapeutic implications currently in the advanced disease setting as both types receive gemcitabine-based chemotherapy. Their clinical course can vary; hilar CCA is associated with prolonged survival even in the locally advanced disease setting with liver transplantation, while the same modality in intrahepatic CCA has suboptimal results. Distal extrahepatic CCA has a clinical course that is similar to pancreatic adenocarcinoma. Underlying genetic differences between these two entities have not yet been adequately explored.
Recently, there have been several reports in regards to genomic profiling of intrahepatic CCA. Ong et al., conducted a whole exome sequencing study of patients with Opisthorchis viverrini-related CCA from East Asia. Using Sanger sequencing, they noted activating mutations in P53, KRAS, SMAD4 and 10 other newly implicated genes including MLL3, ROBO2, RNF3 and PEG3 [10]. This group further compared the genomic profile of O. viverrini-related and non-O. viverrini-related CCAs and demonstrated statistically significant differences in mutation patterns between these entities. Jiao et al. from Johns Hopkins, performed an exome sequencing analysis of 32 cases of intrahepatic CCA and demonstrated the relatively common occurrence of GAs in chromatin regulation genes [11]. Similar studies in extrahepatic CCA are lacking. Furthermore, the clinical phenotype or prognosis associated with these GAs and the effect of targeted therapies in the various CCA subsets remains to be demonstrated.
Materials and Methods
Case Selection
Surgically resected or tumor core biopsy FFPE specimens were obtained for 75 patients with CCA. All patients selected were those with pathologically confirmed CCA and a minimum of 3 months of follow-up. Patients signed an informed consent that covered review of medical records, and studies for correlated research. The study was approved by the Institutional Review Board of MD Anderson Cancer Center. Patient demographics, clinical data, survival data and treatment history were retrieved from medical records. For these cases, if adequate material was procured, somatic mutation analysis was first performed using a NGS panel of hotspot regions in 46 cancer-related genes described below. In cases where targetable mutations were not identifiable using this technology or if patients were being screened for clinical trials that required a wider NGS panel, Foundation Medicine (Boston, MA) performed additional sequencing with the IIlumina Hiseq2000 Platform.
Tumor samples
The paraffin embedded blocks were sectioned, and hematoxylin & eosin (H&E) stained slides were reviewed by surgical pathology to confirm the tumor content in each section. Ten serial sections (4 µm) were cut from selected tissue blocks and areas with tumor tissue were macrodissected from those slides using the H&E slides as templates. Clinical data were abstracted from all the patients. Approval for the study was obtained from the institutional review board at MD Anderson Cancer Center.
DNA Extraction
The pathologic diagnosis of each case of CCA was confirmed on routine H & E slides. All samples sent for DNA extraction contained a minimum of 20% DNA derived from tumor cells. DNA was extracted from 40 mm of FFPE tissue using the Maxwell 16 FFPE Plus LEV DNA Purification kit (Promega) and quantified using a standardized PicoGreen fluorescence assay (Invitrogen).
Next Generation Sequencing
An amplicon library was generated from 10 ng of DNA from each sample using the Ion Ampliseq Cancer Panel (Life Technologies). Formalin-fixed, paraffin-embedded cell pellets of the H2122 cell line diluted in the HL60 cell line were used as control. The 46 genes in the panel for detection of targetable mutations included: AKT1, BRAF, FGFR1, GNAS, IDH1, FGFR2, KRAS, NRAS, PIK3CA, MET, RET, EGFR, JAK2, MPL, PDGFRA, PTEN, TP53, FGFR3, FLT3, KIT, ERBB2, ABL1, HNF1A, HRAS, ATM, RB1, CDH1, SMAD4, STK11, ALK, SRC, SMARCB1, VHL, MLH1, CTNNB1, KDR, FBXW7, APC, CSF1R, NPM1, SMO, ERBB4, CDKN2A, NOTCH1, JAK3, and PTPN11. Primers for PCR amplification included the 190-primer pair pool provided by the vendor with an additional primer pair that was custom added to cover the ‘hotspot’ location on codon 17 of AKT1. Following PCR amplification of target sequences, barcodes were ligated to the amplicons using the Ion Xpress Barcode Adaptors Kit (Life Technologies). Library quantification was then performed using the Bioanalyzer High Sensitivity DNA Chip (Agilent Technologies, Santa Clara, CA). The library was diluted in nuclease-free water to obtain a final concentration of 16 pM. Emulsion PCR was performed manually using the Ion Xpress Template Kit (Life Technologies) followed by manual breaking of the emulsion to isolate the ion spheres (ISPs). The quality of the DNA following PCR was measured using the Qubit IonSphere Quality control kit (Life Technologies). Selective ISPs with DNA were isolated and sequenced on a Ion 316 Chip (4 samples/chip) or a Ion 318 Chip (8 samples/chip) using the vendor-provided sequencing kit (Life Technologies). Successful sequencing of a sample required at least 300 000 reads with a quality score of AQ20 (1 misaligned base per 100 bases). For a wild-type call, a minimum coverage of 250× was required. As tumor specimens were admixed with normal tissue, a minimum coverage of 500× with at least 10% frequency was used as cutoff for a variant to be considered true. All variants detected by Ion PGM with at least 10% frequency were selected for confirmation by alternate platforms. Further details regarding the methodology and analysis have been described previously [12]. NGS performed by Foundation Medicine used the Illumina Hiseq 2000 Platform for 236 targetable GA's and the methods for the same have been described previously [13], [14].
Immunohistochemistry for HER2/Neu
A standard technique for fixing tissue in formalin and embedding it in paraffin was used. The 4-µm-thick histologic sections obtained from the TMAs were deparaffinized and hydrated in decreasing alcohol concentrations. Antigens were recovered by exposure to microwaves in citrate buffer (pH 6.0) and washed in PBS (pH 7.4). The monoclonal antibody anti-ErbB2 (NCL-CB11; Novocastra) was used at a dilution of 1∶40. The primary antibody was incubated at room temperature for 60 minutes and then incubated with the complex Super Picture Polymer Detection Kit (Zymed) in a Dako autostainer. Standard criteria for HER2/Neu scoring were utilized [15].
Statistical Analysis
We investigated potential associations between genetic mutations and survival in this cohort. All patients who had at least 3 months of follow up were included for the survival analysis. Overall survival (OS) was the chosen primary endpoint for this analysis. Progression-free survival (PFS) was a secondary endpoint as in prior clinical trials in CCA, PFS correlated accurately with overall survival (OS) [16]. Patient characteristics, age, gender, primary tumor site (intra- and extrahepatic), treatments administered, GA's observed, disease course, progression date and current status were recorded. The PFS and OS were estimated by using the Kaplan–Meier method. For survival analysis, the log rank test was used for discrete variables and the Cox proportional hazard model was used for continuous variables. All variables that were significant in univariate analysis and variables of our interest were entered into a multivariate model. The Cox proportional hazards model was used to calculate hazard ratio and 95% confidence intervals. A p-value of less than 0.05 was considered significant.
Ingenuity Pathway Analysis
The web-based pathways analysis tool IPA (Ingenuity Systems, www.ingenuity.com) was used to identify signaling pathways affected in CCA. This web-based entry tool allows for the mapping of gene expression data into relevant pathways based on their functional annotation and known molecular interactions. This knowledge coming from published, peer-reviewed scientific publications is stored in the Ingenuity Pathways Knowledge Base (IPKB), and is continuously updated. The mutated genes were uploaded into IPA. Canonical pathways analysis identified molecular pathways from the IPA library of canonical pathways (part of the IPKB) that were most significant to the data set. Genes from the data set that were associated with a canonical pathway in the IPKB were considered for the analysis. The significance of the association between the genes from the dataset and the canonical pathway (in the IPKB) was measured. Fisher's exact test was used to calculate a p-value determining the probability that there is an association between the genes in the dataset and the canonical pathway that cannot be explained by chance alone.
Results
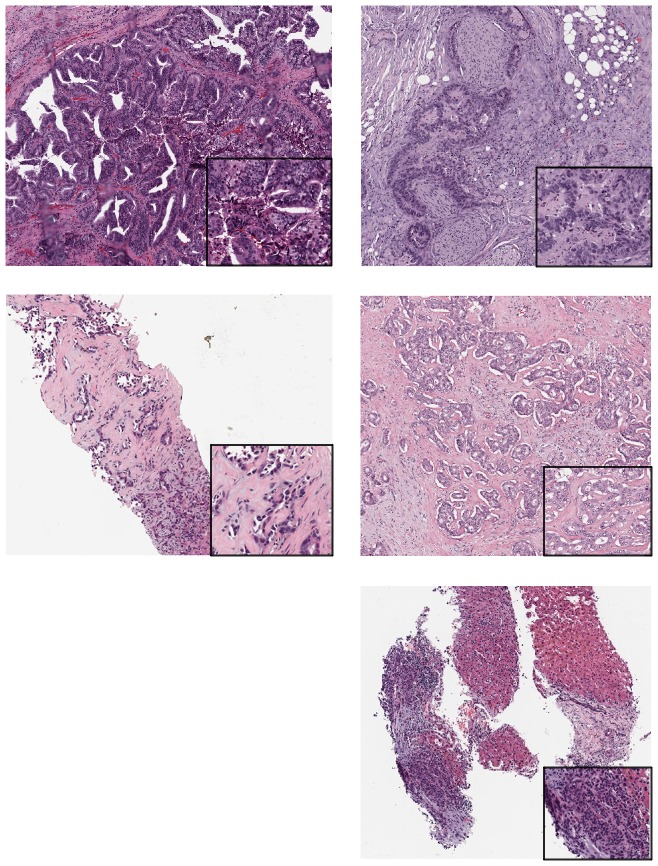
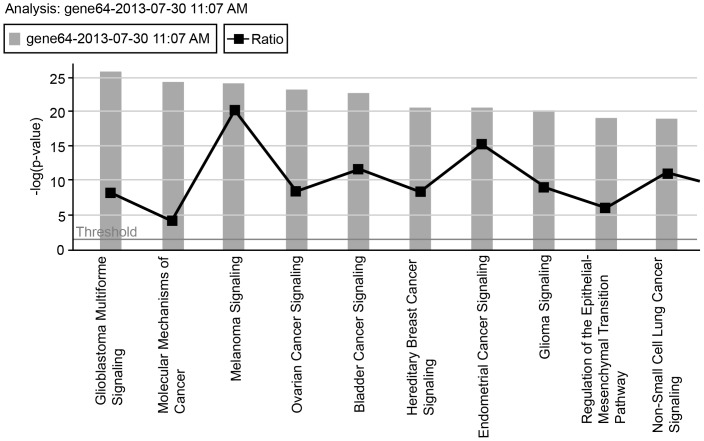
Seventy-five cases of CCA were analyzed for GAs using NGS. Tissue for NGS was obtained from the following sites: 26 samples were from surgical resections and 49 were from biopsies. Forty biopsy specimens were from the liver, 2 from retroperitoneal and peripancreatic lymph nodes, 3 from omentum nodules and 4 from peritoneum nodules. The median follow-up duration in this study was 19 months. Majority of the cases (90%) were investigated using the Illumina Hiseq Platform. Out of the 75 cases, 55 were intrahepatic CCAs and 20 were extrahepatic CCAs. For the purpose of our analysis, we considered these two entities separately. There were notable genetic differences between intra and extrahepatic CCA. IDH1 mutations occurred exclusively in intrahepatic CCA while ERBB2 mutations were seen in extrahepatic CCA. Other genetic differences between these two entities are depicted in Table 1. The allele frequency and copy number of these GAs are depicted in S1 Table. There were no morphological differences in the respective tumors associated with these GAs on light microscopy (Fig. 1). IPA canonical pathways representing these cases are represented in Fig. 2. There is close homology between the canonical pathways noted in CCA with those seen in glioblastoma, melanoma, hereditary breast cancer and bladder cancer. The affected molecular and cell functions identified on the IPA were cell cycle regulation, cellular growth, death, DNA replication and repair. We analyzed the frequency and prognostic significance of the GAs separately in intra and extrahepatic CCA.
Table 1. Genetic differences identified between Intrahepatic and Extrahepatic CCA.
| GENETIC ABNORMALITY | INTRAHEPATIC CCA (N = 55) | EXTRAHEPATIC CCA (N = 20) | ODDS RATIO | p-value | 95% CI |
| N(%) | N(%) | ||||
| GENES | |||||
| TP53 | 16 (29.1%) | 9 (45%) | 0.506 | 0.268 | 0.154–1.669 |
| KRAS | 13 (23.6%) | 8 (40%) | 0.469 | 0.244 | 0.138–1.626 |
| ARID1A | 11 (20%) | 1 (5%) | 4.678 | 0.164 | 0.599–214.83 |
| ERBB2 | 1 (1.8%) | 5 (20%) | 0.058 | 0.004 | 0.001–0.576 |
| PBRM1 | 6 (10.9%) | 1 (5%) | 2.305 | 0.667 | 0.252–112.51 |
| BAP1 | 5 (9.1%) | 2 (10%) | 0.901 | 1 | 0.132–10.26 |
| FBXW7 | 3 (5.5%) | 3 (15%) | 0.333 | 0.333 | 0.041–2.719 |
| SMAD4 | 2 (3.6%) | 5 (25%) | 0.333 | 0.333 | 0.010–0.804 |
| IDH | 13 (23.6%) | 0 (0%) | Infinite | 0.01 | 1.274 – Inf |
| PATHWAYS | |||||
| MAP-ERK | 19 (34.5%) | 11 (55%) | 0.437 | 0.121 | 0.133–1.389 |
| mTOR | 14 (25.5%) | 8 (40%) | 0.517 | 0.258 | 0.154–1.776 |
| DNA Repair | 9 (16.4%) | 8 (40%) | 0.299 | 0.058 | 0.081–1.094 |
| FGF Pathway | 7 (12.7%) | 1 (5%) | 2.741 | 0.673 | 0.316–131.27 |
| Chromatin Modification | 18 (32.7%) | 3 (15%) | 2.724 | 0.157 | 0.659–16.364 |
Figure 1. Representative histology of select tumors with the respective GA (40x, 200x).
[A. ERBB2 (S310F) B. ERBB2 (S310F) C. BAP1 (C91*) D. FGFR2-KIAA1598 fusion E. FGFR2-NOL4 fusion].
Figure 2. Ingenuity Pathway Analysis indicates canonical signaling pathways involved in CCA.
Homology noted with more commonly occurring solid tumors including melanoma and glioblastoma.
Intrahepatic CCA
Median age of the 55 patients with intrahepatic CCA was 60 years (range 24–84). Patient demographics are described in Table 2. The median PFS for these patients was 6.1 months and median OS was 10.9 months.
Table 2. Patient demographics and tumor characteristics.
| CHARACTERISTIC | INTRAHEPATIC (N = 55) | EXTRAHEPATIC (N = 20) |
| Male | 16 (29%) | 13 (65%) |
| Female | 39 (71%) | 7 (35%) |
| Asian | 2 (4%) | 1 (5%) |
| Hispanic | 8 (15%) | 0 (0%) |
| Black | 3 (5%) | 0 (0%) |
| White | 42 (76%) | 19 (95%) |
| Poorly differentiated | 28 (47%) | 7 (35%) |
| Moderate differentiated | 26 (48%) | 11 (55%) |
| Well differentiated | 1 (2%) | 0 (0%) |
| Grade N/A | 0 (0%) | 2 (10%) |
| AJCC II | 4 (7%) | 6 (30%) |
| AJCC III | 17 (31%) | 5 (25%) |
| AJCC IV | 34 (62%) | 9 (45%) |
| Surgery without adjuvant therapy | 7 (13%) | 7 (35%) |
| Radiation therapy | 10 (18%) | 1 (5%) |
| Surgery & adjuvant chemoradiation | 6 (11%) | 7 (35%) |
| Systemic Therapy | 32 (58%) | 5 (25%) |
162 GAs were identified from 55 intrahepatic CCA patient samples with an average of 2.95 GAs/patient (range 0–7). GAs identified were mutations (66%), amplifications (20%), loss/deletions (7%) and others (7%). Only 4 (7%) tumors showed no detectable GAs using the NGS. Most frequent GAs were TP53 (35%), KRAS (24%), ARID1A (20%), IDH1 (18%), MCL1 (16%) and PBRM1 (11%). FGF pathway GAs occurred in 13% of cases, these included mutations, amplifications and FGFR fusion genes. The latter were noted in 3 cases: FGFR2-KIAA1598, FGFR2-NOL4 and FGFR2-PARK2 fusions. A schematic representation of the FGFR2-KIAA1598 fusion gene is depicted in Fig. 3. Signaling pathways associated with these mutations are depicted in Table 3. MAPK, chromatin modification and mTOR pathway aberrations were relatively common (35%, 33% and 25%, respectively) followed by DNA repair (16%) and FGF signaling (13%).
Figure 3. Schematic of FGFR2-KIAA1598 fusion gene.
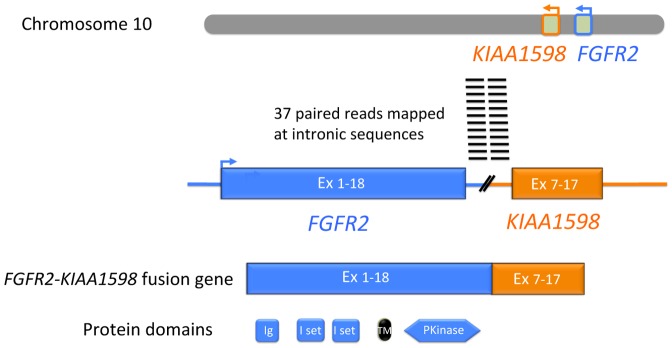
These fusions have been proven oncogenic and are potentially susceptible to FGFR inhibitors.
Table 3. Classification of select genomic variations identified based on roles in cell signaling pathways.
| PATHWAY | GENES | INTRAHEPATIC | EXTRAHEPATIC |
| FGF | FGF19, FGF3, FGFR2, FGF4, FGFR3, FGFR2-KIAA1598 fusion, FGFR2-NOL4 fusion, FGFR2-PARK2 fusion. | 13% | 5% |
| mTOR | FBXW7, PIK3CA, PTEN, NF1, NF2, PIK3R1, STK11, TSC1, TSC2 | 25% | 40% |
| MAP/ERK | KRAS, MYC, BRAF, EGFR, MAP2K1, MAP3K1, NRAS | 35% | 55% |
| DNA Repair | MSH6, BRCA1, BRCA2, BAP1, ATM, MLH1, MSH2 | 16% | 45% |
| Chromatin Modification | BAP1, ARID1A, PBRM1 | 33% | 15% |
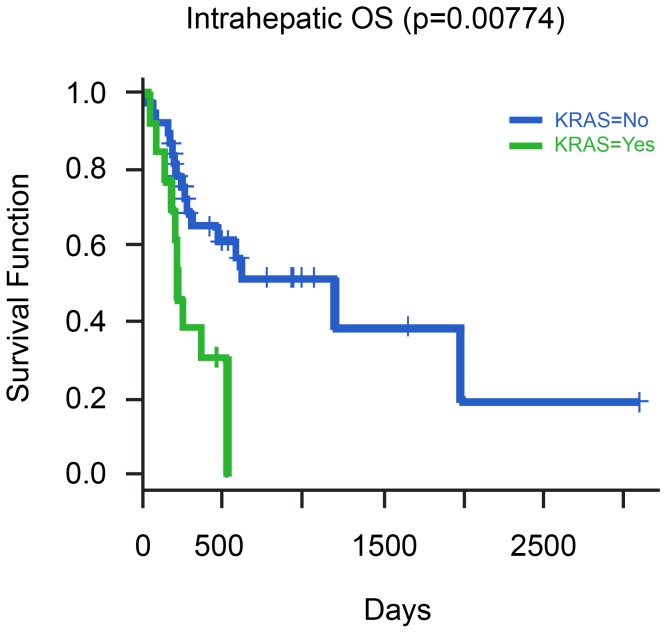
Potential associations between the mutations with PFS and overall survival OS were explored in this study. Multivariate regression analyses for PFS and OS are described in Table 4 and Table 5 respectively. In intrahepatic CCA, PFS was significantly associated with the clinical stage, KRAS mutations, and aberrations in the MAP/ERK pathway. The median PFS for patients with and without KRAS mutations was 3.4 and 10 months (p = 0.01). Median PFS for patients with and without aberrant MAP/ERK pathway genes was 3.9 and 10 months (p = 0.004). PFS was not significantly associated with age, gender, ethnicity, or presence of other mutations including IDH1.
Table 4. Multivariate regression analysis for progression free survival (PFS) in patients with intrahepatic CCA.
| RISK FACTOR | REGRESSION COEFFICIENT | HAZARD RATIO (HR) | p-value | 95% CI for HR |
| Stage III | 0.7417 | 2.0995 | 0.3531 | 0.4388–10.046 |
| Stage IV | 1.4668 | 4.3352 | 0.0593 | 0.9441–19.908 |
| KRAS | 0.3110 | 1.3648 | 0.5692 | 0.4678–3.982 |
| MAP/ERK | 0.6120 | 1.8441 | 0.2339 | 0.6732–5.052 |
Table 5. Multivariate regression analysis for overall survival (OS) in patients with intrahepatic CCA.
| RISK FACTOR | REGRESSION COEFFICIENT | HAZARD RATIO (HR) | p-value | 95% CI for HR |
| Surgery | −1.248 | 0.287 | 0.031825 | 0.09191–0.8972 |
| Male gender | 1.177 | 3.25 | 0.010583 | 1.31606–8.0071 |
| TP53 | 1.593 | 4.92 | 0.000645 | 1.97007–12.2914 |
| KRAS | 0.852 | 2.34 | 0.052791 | 0.98979–5.5519 |
| FGF | −18.842 | 6.56e-09 | 0.997732 | N/A |
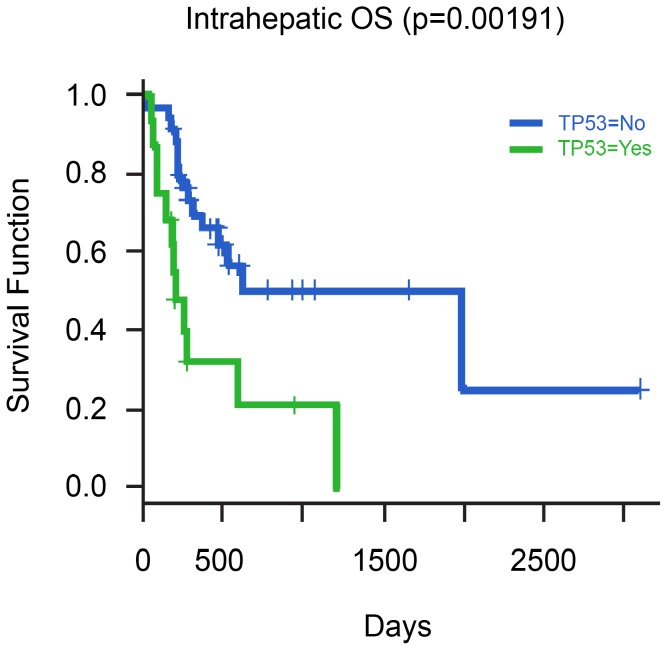
The median OS for stage II, III and IV was 20.5, 15.6 and 6 months respectively (p = 0.017). OS was significantly associated with clinical stage, surgery, gender, TP53 mutations, KRAS mutations, MAP/ERK, mTOR and FGF pathway GAs (Table 6). Kaplan-Meier curves showing the relationship between OS and GA's in KRAS, TP53 and FGF genes are depicted in Figs. 4, 5, and 6, respectively. A multivariate model was set up to investigate the effect of the clinical variables and GAs on PFS and OS. No factor was found to be significantly associated with PFS on multivariate analysis. However, male gender (HR 3.25, p = 0.01) and TP53 mutations (HR 4.92, p = 0.0006) were found to be associated with reduced OS.
Table 6. Univariate Analysis of Survival – Intrahepatic CCA.
| RISK FACTOR | MEDIAN PFS (Months) | p-value | 95% CI | MEDIAN OS (Months) | p-value | 95% CI |
| Clinical Stage | ||||||
| Stage II | 20.5 | 0.0172 | 4.0 – NA | NA | 0.01868 | NA – NA |
| Stage III | 15.6 | 3.8 – NA | 40.2 | 9.4 – NA | ||
| Stage IV | 6.0 | 3.9–10.1 | 12.3 | 8.1 - NA | ||
| Surgery | ||||||
| No | 7.3 | 0.9968 | 5.0 – NA | 12.3 | 0.0291 | 8.1 – NA |
| Yes | 6.0 | 3.9 - NA | NA | 19.5 - NA | ||
| Radiation | ||||||
| No | 7.1 | 0.6752 | 4.9 – NA | 19.5 | 10.4 – NA | |
| Yes | 6.3 | 3.0 – NA | 40.2 | 0.51 | 7.4 - NA | |
| Gender | ||||||
| Female | 10.1 | 0.09115 | 5.0–20.5 | 40.2 | 0.00329 | 20.7 – NA |
| Male | 4.9 | 3.2 – NA | 7.4 | 6.8 - NA | ||
| Ethnicity | ||||||
| Asian | 5.3 | 0.7377 | 4.6 – NA | 9.0 | 0.9659 | 9.0 – NA |
| Black | 7.3 | 1.8 – NA | 17.7 | 12.3 – NA | ||
| Hispanic | 7.7 | 7.1 – NA | 15.6 | 6.8 – NA | ||
| White | 6.3 | 4.0 – 19.4 | 20.7 | 9.4 - NA | ||
| TP53 | ||||||
| No | 7.7 | 0.5145 | 6.0–16.5 | 66.5 | 0.0005 | 17.6 – NA |
| Yes | 4.9 | 3.1 – NA | 6.6 | 4.6 - NA | ||
| KRAS | ||||||
| No | 10.1 | 0.01032 | 6.1–20.5 | 40.2 | 0.002588 | 19.5 – NA |
| Yes | 3.4 | 3.0 – NA | 7.4 | 6.1 – NA | ||
| ARID1A | ||||||
| No | 7.1 | 0.5642 | 4.2–16.5 | 19.5 | 0.7588 | 9.4 – NA |
| Yes | 14.6 | 5.0 – NA | 40.2 | 7.3 – NA | ||
| ERBB2 | ||||||
| No | 7.1 | 0.4102 | 4.6–15.6 | 20.7 | 0.09627 | 12.3 – NA |
| Yes | NA | NA – NA | 6.6 | NA – NA | ||
| PBRM1 | ||||||
| No | 7.7 | 0.1523 | 4.9–16.5 | 20.7 | 0.4427 | 12.3 – NA |
| Yes | 4.8 | 3.3 – NA | 9.4 | 9.0 – NA | ||
| BAP1 | ||||||
| No | 6.3 | 0.363 | 2.9–9.7 | 20.7 | 0.746 | 2.7 – 38.7 |
| Yes | 7.1 | 0–15.2 | 10.4 | 4.3–16.5 | ||
| FBXW7 | ||||||
| No | 7.3 | 5.0–16.5 | 20.7 | 12.3 – NA | ||
| Yes | 3.8 | 3.0 – NA | 6.1 | 4.6 – NA | ||
| SMAD4 | ||||||
| No | 7.1 | 4.9–15.6 | 20.7 | 12.3 – NA | ||
| Yes | 3.8 | NA – NA | 3.2 | 1.7 – NA | ||
| MAP/ERK Pathway | ||||||
| No | 10.1 | 0.003578 | 6.3–25.6 | 40.2 | 0.01961 | 19.5 – NA |
| Yes | 3.9 | 3.0–14.0 | 8.6 | 6.8 – NA | ||
| mTOR Pathway | ||||||
| No | 7.7 | 0.4728 | 5.0–19.4 | 66.5 | 0.0373 | 12.3 – NA |
| Yes | 4.6 | 3.8 – NA | 12.1 | 7.3 – NA | ||
| DNA Repair Pathway | ||||||
| No | 7.7 | 0.741 | 4.6–16.5 | 20.7 | 0.527 | 12.3 – NA |
| Yes | 6.1 | 3.3 – NA | 10.4 | 9.4 – NA | ||
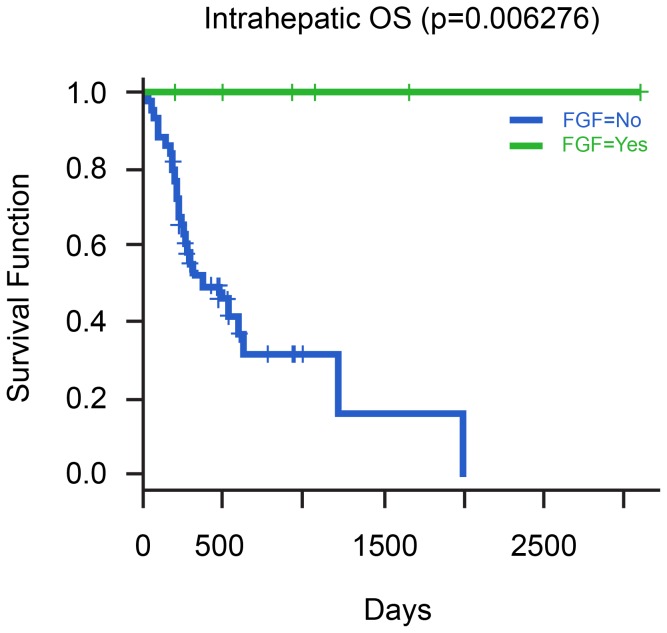
| FGF Pathway | ||||||
| No | 3.1 | 0.2031 | 4.0–16.5 | 17.6 | 0.00973 | 9.0 – NA |
| Yes | 14.0 | 6.3 – NA | NA | NA – NA | ||
| Chromatin modification Pathway | ||||||
| No | 7.3 | 0.5492 | 4.2–19.4 | 19.5 | 0.8472 | 8.6 – NA |
| Yes | 6.1 | 3.3 – NA | 40.2 | 9.4 – NA | ||
Figure 4. Relationship of Overall Survival with presence of KRAS mutation in cases of Intrahepatic CCA.
Figure 5. Relationship of Overall Survival with presence of TP53 mutation in cases of Intrahepatic CCA.
Figure 6. Relationship of Overall Survival with presence of FGF/FGFR alterations in cases of Intrahepatic CCA.
Extrahepatic CCA
Similar analyses were conducted for cases of extrahepatic CCAs. Median age of the 20 patients with extrahepatic CCA was 62.5 years (range 33–80). Patient demographics for this group are also described in Table 2. The median PFS for these patients was 7.7 months and median OS was 14 months.
A total of 78 GAs were identified from 20 extrahepatic CCA patient samples with an average of 3.9 GAs/patient (range 0–10). GAs identified were mutations (77%), amplifications (6%), loss/deletions (10%) and splices (6%). Only 1 tumor showed no detectable GA. Most frequent GAs were TP53 (45%), KRAS (40%), ERBB2 (25%), SMAD4 (25%), FBXW7 (15%), CDKN2A (15%) and CDKN2B (15%). ERBB2 GAs included 4 mutations and 1 case of amplification. One of the ERBB2 mutations was in the kinase domain (V777L) while 3 involved the extracellular domain (S310F). IHC for ERBB2 was negative in all cases with ERBB2 mutations. Signaling pathways associated with mutations in extrahepatic CCAs are depicted in Table 3. MAPK, mTOR and DNA repair pathway aberrations were relatively common (55%, 40% and 45%, respectively) followed by chromatin modification (15%) and FGF signaling (5%).
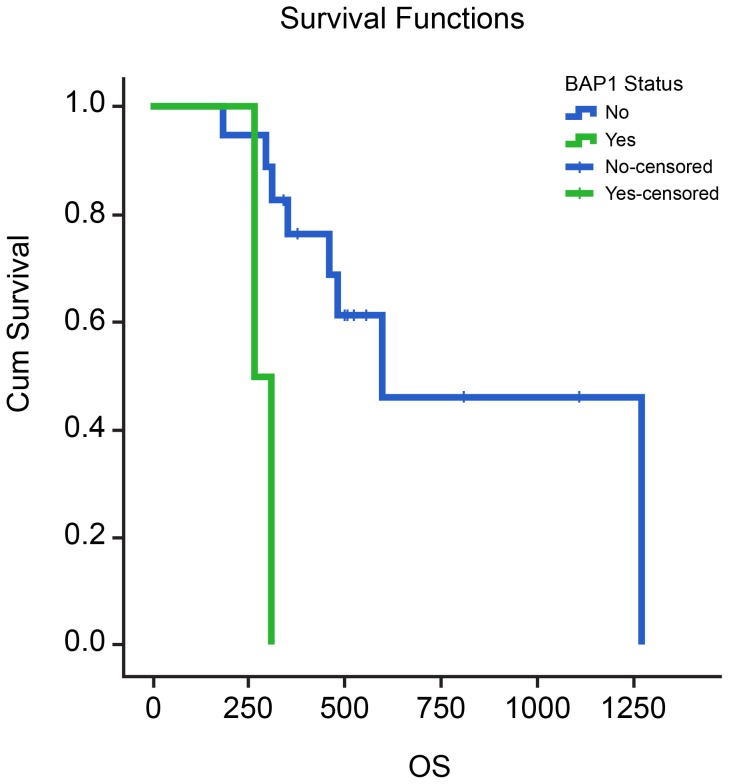
Survival analysis of extrahepatic CCA cases noted significant correlation of PFS with surgery, radiation, BAP1 mutation, and FGF pathway aberrations. OS was found to be associated with clinical stage, surgery, radiation therapy, and mutations in PBRM1 and BAP1 genes (Table 7). A Kaplan-Meier curve demonstrating the relationship between BAP1 mutations and overall survival are shown in Fig. 7. The presence of BAP1 mutations was significantly associated with reduced PFS (median: 3 vs. 8.8 months, p = 0.02) and reduced OS (median: 8.9 vs. 19.9 months, p = 0.007).
Table 7. Univariate Analysis of Survival – Extrahepatic CCA.
| RISK FACTOR | MEDIAN PFS (Months) | p-value | 95% CI | MEDIAN OS (Months) | p-value | 95% CI |
| Clinical Stage | ||||||
| Stage II | NA | 10.5 – NA | NA | 0.02864 | NA – NA | |
| Stage III | 8.6 | 3.3 – NA | 19.9 | 16.1 – NA | ||
| Stage IV | 6.1 | 0.2516 | 5.7–10.1 | 10.4 | 9.9 - NA | |
| Surgery | ||||||
| No | 5.9 | 0.0055 | 3.1 – NA | 10.4 | 0.0064 | 9.9 – NA |
| Yes | 40.0 | 8.8 - NA | 42.5 | 16.1 - NA | ||
| Radiation | ||||||
| No | 6.1 | 0.0496 | 3.3 – NA | 15.4 | 0.006144 | 10.4 – NA |
| Yes | 40.0 | 8.6 - NA | 42.5 | NA - NA | ||
| Gender | ||||||
| Female | 6.0 | 0.9625 | 3.3 – NA | 16.1 | 0.5226 | 10.4 – NA |
| Male | 8.8 | 6.1 - NA | 19.9 | 15.4 - NA | ||
| Ethnicity | ||||||
| Asian | 6.1 | 0.4504 | NA – NA | NA | 0.5673 | NA – NA |
| White | 8.8 | 6.0 – NA | 16.1 | 11.8 – NA | ||
| TP53 | ||||||
| No | 6.1 | 0.8454 | 3.3 – NA | 16.1 | 0.7181 | 10.4 – NA |
| Yes | 8.8 | 6.7 – NA | 19.9 | 15.4 – NA | ||
| KRAS | ||||||
| No | 5.9 | 0.1864 | 3.1 – NA | 11.8 | 0.213 | 10.4 – NA |
| Yes | 10.5 | 8.8 – NA | 19.9 | 15.4 - NA | ||
| ARID1A | ||||||
| No | 8.6 | 0.2948 | 6.0 – NA | 16.1 | 0.5134 | 11.8 – NA |
| Yes | NA | NA – NA | NA | NA – NA | ||
| ERBB2 | ||||||
| No | 8.6 | 0.7539 | 6.0 – NA | 42.5 | 0.1248 | 15.4 – NA |
| Yes | 8.8 | 3.1 – NA | 11.8 | 8.9 – NA | ||
| PBRM1 | ||||||
| No | 8.8 | 0.07583 | 6.1 – NA | 19.9 | 0.004671 | 15.4 – NA |
| Yes | 3.1 | NA – NA | 8.9 | NA – NA | ||
| BAP1 | ||||||
| No | 8.8 | 0.002 | 4.1–13.4 | 19.9 | 0.007 | 6.1–33.7 |
| Yes | 3.0 | NA – NA | 8.9 | NA – NA | ||
| FBXW7 | ||||||
| No | 6.7 | 5.3 – NA | 16.1 | 10.4 – NA | ||
| Yes | NA | 8.6 – NA | NA | NA – NA | ||
| SMAD4 | ||||||
| No | 6.1 | 3.3 –NA | 19.9 | 11.8 – NA | ||
| Yes | NA | 6.7 – NA | NA | 15.4 – NA | ||
| MAP/ERK Pathway | ||||||
| No | 3.3 | 0.6695 | 3.3 – NA | 16.1 | 0.5933 | 10.4 – NA |
| Yes | 8.8 | 6.7 – NA | 19.9 | 15.4 – NA | ||
| mTOR Pathway | ||||||
| No | 6.7 | 0.3786 | 3.3–19.4 | 16.1 | 0.3686 | 11.8 – NA |
| Yes | 24.3 | 6.0 – NA | 42.5 | 10.4 – NA | ||
| DNA Repair Pathway | ||||||
| No | 8.6 | 0.5778 | 6.0 – NA | 16.1 | 0.5135 | 11.8 – NA |
| Yes | 8.7 | 3.1 – NA | 19.9 | 8.9 – NA | ||
| FGF Pathway | ||||||
| No | 8.8 | 0.000013 | 6.1 – NA | 19.9 | 0.3038 | 15.4 – NA |
| Yes | 2.5 | NA – NA | 11.8 | NA – NA | ||
| Chromatin modification Pathway | ||||||
| No | 8.8 | 0.5118 | 6.1 – NA | 19.9 | 0.1392 | 15.4 – NA |
| Yes | 3.1 | 3.0 – NA | 10.4 | 8.9 – NA | ||
Figure 7. Relationship of Overall Survival with presence of BAP1 mutation in cases of Extrahepatic CCA (p = 0.007).
BAP1 Mutations
In our series, 6 cases had BAP1 mutations; 3 of these experienced early recurrence (less than six months) after potentially curative surgical resection. Bone and soft tissue metastases were common in this subpopulation. One such case is illustrated in Fig. 8; this patient experienced disease recurrence 7 weeks after surgical resection. In addition, all cases with BAP1 mutation were treated with systemic chemotherapy consisting of gemcitabine and cisplatin. No radiologic response was noted and progression after first line chemotherapy occurred in all cases within a short time frame (less than 4 months). Despite the limited number of cases, the aggressive clinical prognosis of this subgroup was very evident.
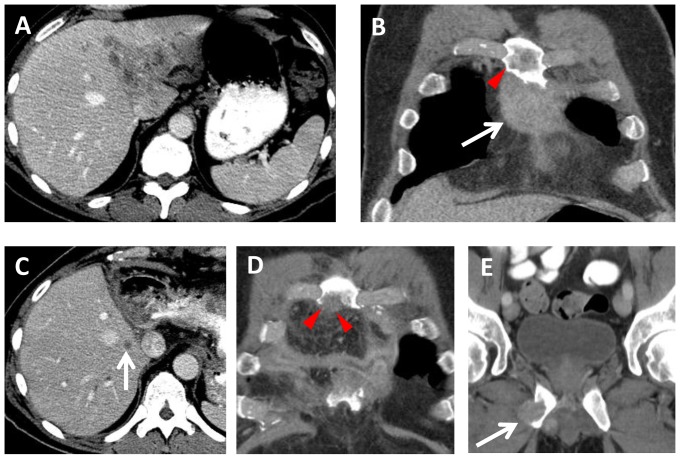
Figure 8. A 57 year old male with BAP1 mutated intrahepatic CCA.
Baseline (A) axial and (B) coronal contrast-enhanced CT images demonstrate (A) a 6.9×5.3 cm liver mass involving segments I, II, and IV, and (B) a 5.6 cm sternal metastasis (arrow). Note the sternal metastasis does not extend to the level of the right 3rd rib (arrowhead). Extended left hepatectomy and resection of the sternal metastasis was performed. (C, D) Seven weeks later, contrast-enhanced CT images show a new metastasis in the remnant liver (arrow), and a new 2 cm recurrence (arrowheads) involving the sternum adjacent to the right 3rd rib. (E) Three months later, there is a new 2.2 cm metastasis (arrow) involving the right inferior pubic ramus.
FGFR Pathway
Patients with FGFR pathway GAs appeared to have a relatively indolent prognosis in our series. FGFR Pathway GAs included mutations, fusion genes and ligand amplifications. Three patients in this subgroup had PFS>60 weeks with first line chemotherapy. These patients had FGFR2 mutation, FGFR2-PARK2 fusion and FGF19 amplification, respectively. Given the favorable prognosis of these cases, one patient with sustained response to systemic chemotherapy lasting for >2 years, received orthotopic liver transplantation with curative intent. In contrast, one patient with concomitant FGFR2-NOL4 fusion had a rapidly progressive course; this patient also had co-existing BAP1 mutation.
Response to Targeted Therapy
Based on their mutational profiles, patients were referred to appropriate clinical trials. Twelve patients were enrolled in phase I or II clinical trials. Clinical outcome from targeted therapy is illustrated below: Sustained stable disease was observed in 7 cases with KRAS wt on erlotinib. Two cases with sustained stable disease are illustrated in Fig. 9. One patient with KRAS mutation was enrolled on a clinical trial with pazopanib + trametinib and experienced stable disease after prior progression on first-line chemotherapy as illustrated in Fig. 10. Two patients with BRAF mutation were referred for a trial with BRAF inhibitor; both experienced partial responses. One of these cases with a metabolic response on FDG-PET Scan is illustrated in Fig. 11; this patient continues on therapy for 5+ months at this time. Another patient with c-met mutation was enrolled on an expansion cohort with a c-met inhibitor and experienced a metabolic response as illustrated in Fig. 12. In case of her2/neu mutations, no response was observed in two cases treated with trastuzumab and lapatinib, respectively.
Figure 9. A 69 year old female with KRAS mutated CCA metastatic to the ovaries.
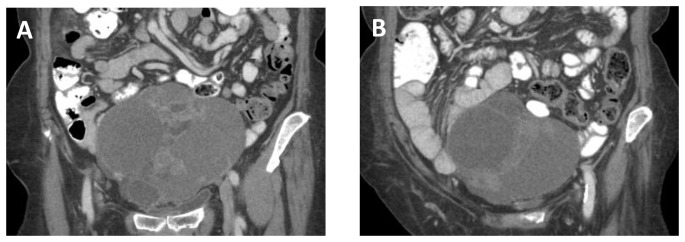
(A) Coronal contrast-enhanced CT image demonstrates a 15.4×12.6×12.2 cm (transverse × craniocaudal × AP) pelvic mass. (B) After two cycles of trametinib + pazopanib therapy, the pelvic mass is slightly decreased to 13.5×11.3×13.4 cm.
Figure 10. A 68 year old female with KRAS wt intrahepatic CCA.
Axial (A) fused PET-CT and (B) unenhanced CT images demonstrate multiple confluent FDG avid liver metastases. After 3 months of erlotinib therapy, (C) axial fused PET-CT and (D) unenhanced CT images show decreased FDG avidity and slightly decreased size of metastases.
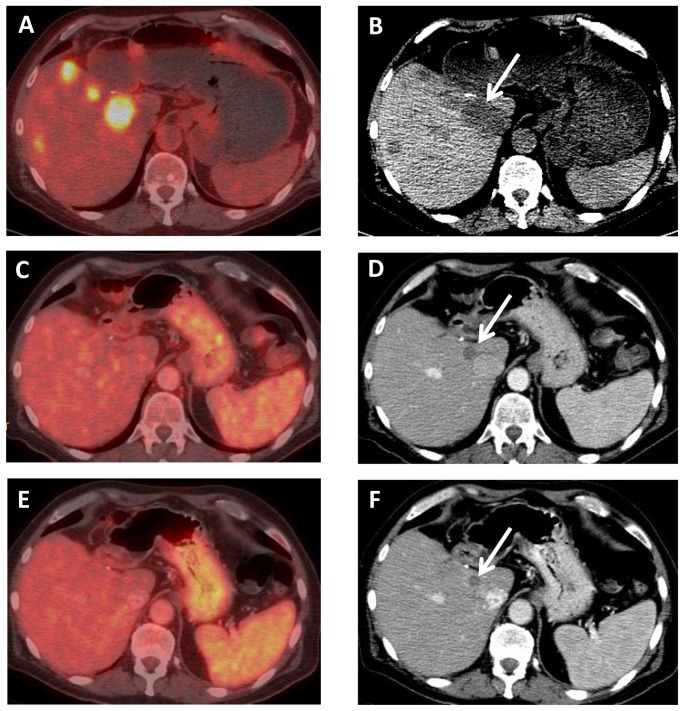
Figure 11. A 67 year old male with BRAF mutated intrahepatic CCA, who had progressed on conventional chemotherapy.
Axial (A) fused PET-CT and (B) unenhanced CT images from a PET scan demonstrate FDG avidity of multiple liver metastases. After 8 weeks of BRAF inhibitor therapy, axial (C) fused PET-CT and (D) contrast-enhanced CT images demonstrate lack of FDG avidity and decreased size of liver metastases, e.g., the dominant lesion adjacent to the IVC (arrow) decreased from 3.7 cm to 1.6 cm. After 16 weeks of therapy, axial (E) fused PET-CT and (F) contrast-enhanced CT images demonstrate continued lack of FDG avidity and further decreased size of liver metastases, e.g., the dominant lesion (arrow) now measures 1.3 cm.
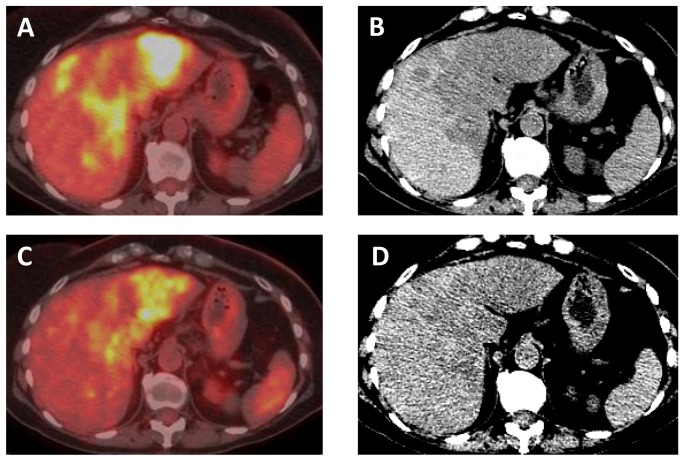
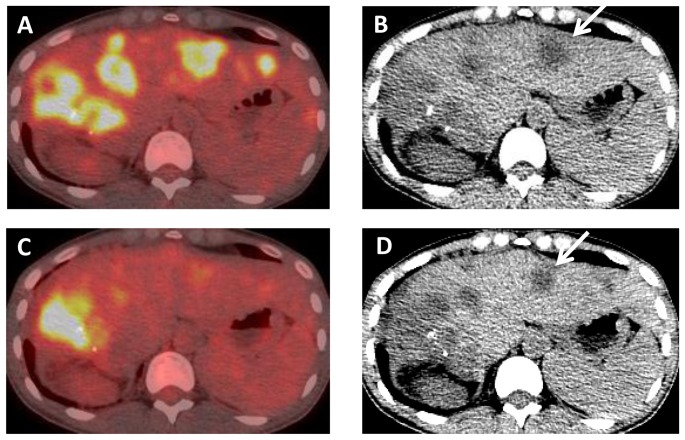
Figure 12. A 37 year old female with c-MET amplified CCA, who had progressed on conventional chemotherapy.
At baseline, axial (A) fused PET-CT and (B) unenhanced CT images demonstrate multiple FDG avid liver metastases, in this patient who is status post right hepatectomy. After 4 weeks of therapy, (C) axial fused PET-CT and (D) unenhanced CT images show decreased FDG avidity and size of metastases. A representative segment II mass (arrows) has decreased from 2.6 cm to 2.1 cm.
These cases illustrate that the mutational profile not only provided prognostic information but also provided therapeutic options.
Discussion
Recent technical advances in genomic analysis along with reduced costs of sequencing are leading to a paradigm shift in cancer management. Several recent successes of genomic directed therapies for ‘chemo-refractory’ malignancies like melanoma, renal cancer and in subgroups of non-small cell lung cancer have further incentivized research in this field. Our goal in this study was to explore the genomic landscape of CCA, an area of unmet need in oncology with a view towards identifying disease subsets that are amenable for targeted therapy. In this study, we chose NGS platforms that can identify clinically ‘actionable’ GAs from FFPE tumor specimens. The use of FFPE samples for obtaining reliable NGS data is technically challenging but has the potential of rapidly bringing the vast scope of this technology to the clinical setting. In this study, we explored a) genetic differences between intrahepatic and extrahepatic CCA, b) the prognostic importance of the GAs and c) the impact of the targeted therapy.
As noted earlier, there have been several recent exome-sequencing studies of intrahepatic CCA. There are however very limited data regarding the mutational spectrum of extrahepatic CCA and although the patient numbers in this subgroup are limited, interesting differences are highlighted by this study. Higher frequency of KRAS, P16 and SMAD4 mutations in extrahepatic CCA suggest a molecular phenotype that resembles pancreatic cancer rather than intrahepatic CCA. IDH1 and ERBB2 GAs occur exclusively in intra and extrahepatic CCA, respectively. These results contrast with the GAs seen in gallbladder cancer, where ERBB2 amplification (not mutation) occurs in 15% of the cases.
The prognostic importance of these GAs is highlighted by the association between survival and BAP1 mutation. Jiao et al., noted frequent mutations of chromatin-modulation genes including BAP1, PBRM1 and ARID1A and an adverse prognosis was noted in the BAP1 mutated cases [11]. In our study, 25 cases (33%) had GAs involving at least one of the chromatin modulating genes. These genes may behave as tumor suppressors and are frequently mutated in solid tumors. BAP1 (BRCA1-Associated Protein 1) was initially identified as a protein that binds to BRCA1 and is now recognized as a tumor suppressor that mediates its effects through chromatin modulation, ubiquitin-proteasome system and the DNA damage response pathway. Germline BAP1 mutations confer susceptibility to uveal melanoma, epithelioid atypical Spitz tumors, cutaneous melanoma, and mesothelioma [17]. Somatic BAP1 mutations are infrequent but have been reported in prostate, ovarian, colon, breast, lung cancers and in mesothelioma [18]. BAP1 loss is associated with an aggressive metastatic phenotype in uveal melanoma and in renal cancer [19]. BAP1 loss in uveal melanoma predicts increased risk of liver metastases after enucleation surgery, thereby highlighting the need for careful patient selection for this radical surgical procedure [20]. Interestingly, our study also noted early recurrence after surgery in the presence of BAP1 mutation. We propose that surgically resected CCA cases with BAP1 mutation be considered for neoadjuvant approaches, so that their disease biology can be identified before considered major hepatic resection. Several small molecule inhibitors have been developed against chromatin regulators; three (HDAC, JAK2 and DNMT inhibitors) are already approved by the Food and Drug Administration (FDA) [21]. The role of these inhibitors in CCA deserves further investigation.
Other negative prognosticators in the study included the presence of KRAS mutation. Chen et al., had previously reported a poor prognosis and presence of peri-neural invasion in the presence of KRAS mutation in CCA [22]. One case in our series with mutated KRAS experienced disease stability after prior progression on chemotherapy with trametinib + pazopanib. Response to MEK inhibitors in this setting has also been reported by other studies and this option may be worth investigating further for KRAS mutated CCA [23]–[25].
The presence of ERBB2 mutations also contrasts with the GAs reported in gallbladder cancer, wherein ERBB2 amplifications have been reported [26], [27]. ERBB2 GAs occurred in 6 cases, which included 2 amplifications and 4 somatic mutations. ERBB2 mutations have been described in non-small cell lung cancer, breast cancer, gastric cancer and colorectal cancer [28], [29]. These mutations have not been described to our knowledge in CCA. Unfortunately, these mutations may be more difficult to target than ERBB2 amplifications [30]. Yoshikawa et al., measured ERBB2 expression in 236 cases of CCA by immunohistochemistry (IHC). They reported a 0.9% and 8.5% positive ERBB2 expression rate in intrahepatic and extrahepatic CCAs, respectively [31]. In our cases with ERBB2 mutation, IHC expression of ERBB2 was not detectable in our study; one of the ERBB2 mutations was in the kinase domain (V777L) while 3 involved the extracellular domain (S310F). Based on the current knowledge of ERBB2 biology, trastuzumab or lapatinib therapy may be value in the extracellular domain mutations but could be of limited clinical benefit for kinase domain mutations and no response to either agents was noted in our cases. Bose et al., noted that V777L mutation was associated with negative ERBB2 protein expression and resistance to lapatinib [32]. Irreversible small molecule tyrosine kinase inhibitors (TKIs), including afatinib, neratinib and dacomatinib may be of value in this setting [33]. Furthermore, since ERBB2 activation can transactivate several other signaling mechanisms, combinations of ERBB2 directed TKIs with trastuzumab, mTOR or MEK inhibitors may be of value in this disease given its genomic profile.
GAs in the FGFR pathway were noted in 9% of our cases, most representing FGFR mutations. Three cases had FGFR2 fusion genes; these have been described before in CCA and are oncogenic in vitro [34]. Preclinical data indicate that these fusion proteins may indicate tumor susceptibility to targeted FGFR inhibitors, BGJ398 and PD173074. Recent data from Arai et al., indicate that FGFR2 fusion genes occur in 13.6% of intrahepatic CCA cases; Borad et al. have described responses to targeted FGFR inhibitors in this patient subgroup [35], [36]. This was also noted in our patient with FGFR fusion. The relatively good prognosis of this molecular subset is intriguing and contrasts with the poor survival of the p53 mutated subtype. Interestingly, similar findings were reported recently in bladder cancer, wherein whole genome transcriptome profiling revealed three molecular subtypes with differing prognoses [37]. The p53-subtype was resistant to chemotherapy while cases with FGFR mutation constituted a luminal subtype with good prognosis. The superior survivals of the FGFR mutated CCA subtype, if confirmed may potentially stratify these cases for liver directed therapy. One case in our series with FGFR mutation and prolonged disease stability received liver transplantation.
Other mutations identified in our study include BRAF and C-MET, both of which were associated with impressive responses seen with targeted therapy. The incidence of these mutations in CCA is low (estimated at 3% for BRAF V600E in intrahepatic CCA)[38] but the sustained responses seen with BRAF inhibitor deserve further exploration. IDH1 (R132C) mutations represented a significant number of cases that may be potentially targetable, given the advent of potent IDH inhibitors [39]–[44].
The IPA suggests genomic homology between subsets of CCA with glioblastoma, melanoma and certain categories of non-small cell lung cancer, thereby supporting the role for molecular rather than morphologic classification of human cancers. A limitation of our series is the relatively small number of cases of extrahepatic CCA and those treated on therapeutic clinical trials. Unfortunately, clinical trials are lacking for this orphan disease population.
We recognize however that the detection of candidate mutations does not necessarily indicate its relevance as a prognostic biomarker or a potential therapeutic target. An integrated approach that includes functional genomics is required to leverage the full potential of NGS and realize the promise of precision medicine. In conclusion, genomic sequencing can potentially identify distinct molecular subsets of CCA, with distinct prognostic and therapeutic implications.
Supporting Information
Detailed results of GA's including gene affected, type of alteration, allele frequency or copy number.
(XLSX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are included within the paper and its Supporting Information files.
Funding Statement
The work was supported by funds from the Cholangiocarcinoma Foundation. PO1 CA130821-01. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Shaib Y, El-Serag HB (2004) The epidemiology of cholangiocarcinoma. Semin Liver Dis 24:115–125. [DOI] [PubMed] [Google Scholar]
- 2. Shaib YH, Davila JA, McGlynn K, El-Serag HB (2004) Rising incidence of intrahepatic cholangiocarcinoma in the United States: a true increase? J Hepatol 40:472–477. [DOI] [PubMed] [Google Scholar]
- 3. Khan SA, Emadossadaty S, Ladep NG, Thomas HC, Elliott P, et al. (2012) Rising trends in cholangiocarcinoma: is the ICD classification system misleading us? J Hepatol 56:848–854. [DOI] [PubMed] [Google Scholar]
- 4. Patel T (2001) Increasing incidence and mortality of primary intrahepatic cholangiocarcinoma in the United States. Hepatology 33:1353–1357. [DOI] [PubMed] [Google Scholar]
- 5. Shin HR, Oh JK, Lim MK, Shin A, Kong HJ, et al. (2010) Descriptive epidemiology of cholangiocarcinoma and clonorchiasis in Korea. J Korean Med Sci 25:1011–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bertuccio P, Bosetti C, Levi F, Decarli A, Negri E, et al. (2013) A comparison of trends in mortality from primary liver cancer and intrahepatic cholangiocarcinoma in Europe. Ann Oncol 24:1667–1674. [DOI] [PubMed] [Google Scholar]
- 7. Parsi MA (2013) Obesity and cholangiocarcinoma. World J Gastroenterol 19:457–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tyson GL, El-Serag HB (2011) Risk factors for cholangiocarcinoma. Hepatology 54:173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamasaki S (2003) Intrahepatic cholangiocarcinoma: macroscopic type and stage classification. J Hepatobiliary Pancreat Surg 10:288–291. [DOI] [PubMed] [Google Scholar]
- 10.(2003) Opisthorchis viverrini and opisthorchiasis: The 21st Century review. Proceedings of the Congress of Opisthorchiasis and Cholangiocarcinoma. May 28–30, 2002. Khon Kaen, Thailand. Acta Trop 88:169–246. [PubMed] [Google Scholar]
- 11. Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, et al. (2013) Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 45:1470–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanagal-Shamanna R, Portier BP, Singh RR, Routbort MJ, Aldape KD, et al. (2014) Next-generation sequencing-based multi-gene mutation profiling of solid tumors using fine needle aspiration samples: promises and challenges for routine clinical diagnostics. Mod Pathol 27:314–327. [DOI] [PubMed] [Google Scholar]
- 13. Ross JS, Cronin M (2011) Whole cancer genome sequencing by next-generation methods. American journal of clinical pathology 136:527–539. [DOI] [PubMed] [Google Scholar]
- 14. Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, et al. (2009) Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nature biotechnology 27:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, et al. (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31:3997–4013. [DOI] [PubMed] [Google Scholar]
- 16. Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, et al. (2010) Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 362:1273–1281. [DOI] [PubMed] [Google Scholar]
- 17. Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, et al. (2011) Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet 43:1018–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murali R, Wiesner T, Scolyer RA (2013) Tumours associated with BAP1 mutations. Pathology 45:116–126. [DOI] [PubMed] [Google Scholar]
- 19. Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, et al. (2010) Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330:1410–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Essen TH, van Pelt SI, Versluis M, Bronkhorst IH, van Duinen SG, et al. (2014) Prognostic parameters in uveal melanoma and their association with BAP1 expression. Br J Ophthalmol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dawson MA, Kouzarides T (2012) Cancer epigenetics: from mechanism to therapy. Cell 150:12–27. [DOI] [PubMed] [Google Scholar]
- 22. Chen TC, Jan YY, Yeh TS (2012) K-ras mutation is strongly associated with perineural invasion and represents an independent prognostic factor of intrahepatic cholangiocarcinoma after hepatectomy. Ann Surg Oncol 19 Suppl 3: S675–681. [DOI] [PubMed] [Google Scholar]
- 23. Hamidi H, Lu M, Chau K, Anderson L, Fejzo M, et al. (2014) KRAS mutational subtype and copy number predict in vitro response of human pancreatic cancer cell lines to MEK inhibition. Br J Cancer [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, et al. (2008) Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 14:1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakayama N, Nakayama K, Yeasmin S, Ishibashi M, Katagiri A, et al. (2008) KRAS or BRAF mutation status is a useful predictor of sensitivity to MEK inhibition in ovarian cancer. Br J Cancer 99:2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Javle M, Rashid A, Churi C, Kar S, Zuo M, et al. (2014) Molecular characterization of gallbladder cancer using somatic mutation profiling. Hum Pathol 45:701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roa I, de Toro G, Schalper K, de Aretxabala X, Churi C, et al. (2014) Overexpression of the HER2/neu Gene: A New Therapeutic Possibility for Patients With Advanced Gallbladder Cancer. Gastrointest Cancer Res 7:42–48. [PMC free article] [PubMed] [Google Scholar]
- 28. Lee JW, Soung YH, Kim SY, Nam SW, Park WS, et al. (2006) ERBB2 kinase domain mutation in the lung squamous cell carcinoma. Cancer Lett 237:89–94. [DOI] [PubMed] [Google Scholar]
- 29. Pao W, Girard N (2011) New driver mutations in non-small-cell lung cancer. Lancet Oncol 12:175–180. [DOI] [PubMed] [Google Scholar]
- 30. Herter-Sprie GS, Greulich H, Wong KK (2013) Activating Mutations in ERBB2 and Their Impact on Diagnostics and Treatment. Front Oncol 3:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoshikawa D, Ojima H, Iwasaki M, Hiraoka N, Kosuge T, et al. (2008) Clinicopathological and prognostic significance of EGFR, VEGF, and HER2 expression in cholangiocarcinoma. Br J Cancer 98:418–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, et al. (2013) Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov 3:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bose P, Ozer H (2009) Neratinib: an oral, irreversible dual EGFR/HER2 inhibitor for breast and non-small cell lung cancer. Expert Opin Investig Drugs 18:1735–1751. [DOI] [PubMed] [Google Scholar]
- 34. Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, et al. (2013) Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 3:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arai T, Nagino M, Nimura Y (2006) [Surgical treatment for hilar cholangiocarcinoma]. Nihon Rinsho 64 Suppl 1: 476–478. [PubMed] [Google Scholar]
- 36. Borad MJ, Champion MD, Egan JB, Liang WS, Fonseca R, et al. (2014) Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet 10:e1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi W, Porten S, Kim S, Willis D, Plimack ER, et al. (2014) Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 25:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goeppert B, Frauenschuh L, Renner M, Roessler S, Stenzinger A, et al. (2014) BRAF V600E-specific immunohistochemistry reveals low mutation rates in biliary tract cancer and restriction to intrahepatic cholangiocarcinoma. Mod Pathol 27:1028–1034. [DOI] [PubMed] [Google Scholar]
- 39. Sia D, Tovar V, Moeini A, Llovet JM (2013) Intrahepatic cholangiocarcinoma: pathogenesis and rationale for molecular therapies. Oncogene 32:4861–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kipp BR, Voss JS, Kerr SE, Barr Fritcher EG, Graham RP, et al. (2012) Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum Pathol 43:1552–1558. [DOI] [PubMed] [Google Scholar]
- 41. Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, et al. (2012) Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 17:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Subbiah IM, Subbiah V, Tsimberidou AM, Naing A, Kaseb AO, et al. (2013) Targeted therapy of advanced gallbladder cancer and cholangiocarcinoma with aggressive biology: eliciting early response signals from phase 1 trials. Oncotarget 4:156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chung JY, Hong SM, Choi BY, Cho H, Yu E, et al. (2009) The expression of phospho-AKT, phospho-mTOR, and PTEN in extrahepatic cholangiocarcinoma. Clin Cancer Res 15:660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pirozzi CJ, Reitman ZJ, Yan H (2013) Releasing the block: setting differentiation free with mutant IDH inhibitors. Cancer Cell 23:570–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed results of GA's including gene affected, type of alteration, allele frequency or copy number.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are included within the paper and its Supporting Information files.