Abstract
Infection by the Gram-negative pathogen Burkholderia pseudomallei results in the disease melioidosis, acquired from the environment in parts of southeast Asia and northern Australia. Clinical symptoms of melioidosis range from acute (fever, pneumonia, septicemia, and localized infection) to chronic (abscesses in various organs and tissues, most commonly occurring in the lungs, liver, spleen, kidney, prostate and skeletal muscle), and persistent infections in humans are difficult to cure. Understanding the basic biology and genomics of B. pseudomallei is imperative for the development of new vaccines and therapeutic interventions. This formidable task is becoming more tractable due to the increasing number of B. pseudomallei genomes that are being sequenced and compared.
Here, we compared three B. pseudomallei genomes, from strains MSHR668, K96243 and 1106a, to identify features that might explain why MSHR668 is more virulent than K96243 and 1106a in a mouse model of B. pseudomallei infection. Our analyses focused on metabolic, virulence and regulatory genes that were present in MSHR668 but absent from both K96243 and 1106a. We also noted features present in K96243 and 1106a but absent from MSHR668, and identified genomic differences that may contribute to variations in virulence noted among the three B. pseudomallei isolates. While this work contributes to our understanding of B. pseudomallei genomics, more detailed experiments are necessary to characterize the relevance of specific genomic features to B. pseudomallei metabolism and virulence. Functional analyses of metabolic networks, virulence and regulation shows promise for examining the effects of B. pseudomallei on host cell metabolism and will lay a foundation for future prediction of the virulence of emerging strains. Continued emphasis in this area will be critical for protection against melioidosis, as a better understanding of what constitutes a fully virulent Burkholderia isolate may provide for better diagnostic and medical countermeasure strategies.
Introduction
Melioidosis, the disease caused by Burkholderia pseudomallei, presents with a wide range of non-specific signs and symptoms, including fever, pneumonia, acute septicemia, and chronic localized infection [1]–[3]. Initial infection can also be asymptomatic. Chronic stages of the disease are characterized by abscesses in various organs and tissues, most commonly occurring in the lungs, liver, spleen, kidney, prostate and skeletal muscle [1], [3], [4]. Melioidosis is community-acquired through bacterial contamination of wounds, inhalation, and ingestion [5]. Research in Thailand and Australia has provided critical information about the clinical epidemiology of the disease; the clinical presentations of melioidosis caused by Thai and Australian strains differ in several ways: 1) parotid abscesses are not prevalent in Australia, but occur in Thailand; 2) prostate abscesses are uncommon in Thailand, but are more commonly seen in Australia [3]; and 3) an encephalomyelitis syndrome is seen in tropical Australia more often than in Thailand [5]. This latter condition was associated with the illnesses caused by B. pseudomallei strains MSHR668 [6] and MSHR305 [7]. However, there is evidence that the same strain can cause different clinical presentations in different individuals, and a number of risk factors, such as diabetes have been identified for melioidosis [8]. Therefore, host factors may be important in determining the severity and duration of disease [9], [10].
The high incidences of infection in geographical areas where B. pseudomallei is endemic may be due to its resilience and ability to survive under sometimes harsh environmental conditions. B. pseudomallei can survive nutrient depletion, a wide range of pH differences, salt concentrations, and temperatures [11], detergent solutions [12] and acidic environments [13]. It seems that harsh environmental conditions may confer a selective advantage for the growth of B. pseudomallei [5]. These resilience characteristics may explain why B. pseudomallei can cause persistent infections in the human host that are difficult to cure. Also, B. pseudomallei is naturally resistant to a variety of antimicrobial agents [14], [15]. In some cases, there is a latency period before symptoms present that can last for days to years [5]. In other cases, an initial acute infection and extensive antibiotic treatment is followed by a variable period of bacterial persistence, with subsequent recrudescence of the disease months or years after the initial infection [16], [17].
Our understanding of B. pseudomallei pathogenesis is further complicated by the natural diversity of its genome. B. pseudomallei is a soil-dwelling bacterium that utilizes lateral gene transfer at a very high rate [18]. As a result, there is substantial variation among B. pseudomallei genomes, which may also contribute to differential virulence. Fortunately, as we now have access to many B. pseudomallei genomes from various geographic locations, it is possible to identify genomic features that the various strains have in common, as well as features that are unique to one or more strains.
Comparative studies of genomes from Australian and Thai B. pseudomallei isolates have revealed genomic differences that contribute to our understanding of this organism. The genomes of B. pseudomallei analyzed so far contain from 16–21 genomic islands (GIs) [7], [19], [20]. The genome of B. pseudomallei strain K96243 contains 16 GIs [19] that are variably present in other B. pseudomallei genomes [20], and each GI shows micro-evolutionary changes that generate GI diversity [20]. In addition to GIs, the genomes of Thai strains K96243 and 1106a contain a horizontally acquired Yersinia-like fimbrial (YLF) gene cluster, while the comparable region in the Australian strains (MSHR668, MSHR305, DM98, 1655 and 13177) is the B. thailandensis-like flagellum and chemotaxis (BTFC) gene cluster [21]. Previous studies showed that BTFC is dominant in Australian strains, while YLF is dominant in strains from Thailand and elsewhere [21]. In addition, clinical isolates are more likely to belong to group YLF, whereas environmental isolates are more likely to belong to group BTFC [21]. In contrast to these trends, we found that the Australian strain MSHR346 contains the YLF cluster (data not shown), and Tuanyok and colleagues reported that 406e, a clinical isolate from Thailand, has BTFC [21].
Previous studies began to address the question of why different strains show differences in virulence and disease presentation. Many studies have focused on host risk factors such as diabetes and alcoholism; but to date only one study has identified genes associated with different disease presentations [22]. This suggests that virulence factors that are variably present in B. pseudomallei strains may be important for pathogenesis. Taken together with the genomic variation, geographical distribution and differences in environmental habitats [18], [21], [23], comparative genomic studies suggest that strains associated with human melioidosis may possess an accessory genome that differs from animal and environmental strains [24]. We hypothesize that differences in virulence may be associated with variations in metabolic and regulatory capabilities among B. pseudomallei strains.
In this study we compared three B. pseudomallei genomes, from clinical strains MSHR668, K96243 and 1106a, seeking to identify metabolic characteristics that might explain why MSHR668 is more virulent than K96243 and 1106a in a mouse model of B. pseudomallei infection. Analyses focused on genomic features, including metabolic, virulence and regulatory genes that were present in MSHR668 but absent from both K96243 and 1106a. Features present in K96243 and 1106a but absent from MSHR668 were noted, and we also identified virulence-associated genes that were present in all three genomes. Here we have identified genomic features that may contribute to variations in virulence noted among B. pseudomallei isolates.
Results
Comparative Virulence of B. pseudomallei Isolates
For the purposes of this manuscript, we measured the LD50 upon intraperitoneal (IP) challenge to assess potential differences in virulence among the three B. pseudomallei strains. While studies evaluating clinical infection are complicated by a range of factors such as host risk factors, exposure routes and dose of exposure, experimental studies using inbred mice were used in an attempt to limit the number of host factors that may contribute to differences. Experiments involving infections of BALB/c and C57BL/6 mice [25] with B. pseudomallei strains K96243, MSHR668, and 1106a revealed differences in LD50 values among the B. pseudomallei strains. LD50 values were calculated after 21 and 60 days post-challenge. Differences were more pronounced in the BALB/c model, where the LD50 values of MSHR668 were 30 to 100-fold lower than those of K96243 and 1106a (Table 1). The LD50 values for MSHR668 were also lower in the C57BL/6 model, although the differences were not as great. Since K96243 and 1106a had similar virulence properties in both mouse infection models, we were interested in identifying the genomic features that these strains shared but were not common to MSHR668.
Table 1. LD50 values from intraperitoneal exposure of BALB/c and C57BL/6 mice to B. pseudomallei strains MSHR668, K96243 and 1106a.
| BALB/c | Strain | Day 21 LD50 | 95% HPD Credible Interval | Day 60 LD50 | 95% HPD Credible Interval |
| K96243 | 6.15×104 | 2.65×105–1.38×105 | 3.45×104 | 1.18×104–1.06×105 | |
| 668 | 1.34×102 | 37 −4.53×102 | 1.35×102 | 37–4.5×102 | |
| 1106a | 4.15×104 | 1.69×104–9.55×105 | 4.14×104 | 1.70×104–9.39×105 | |
| C57BL/6 | Strain | Day 21 LD50 | 95% HPD Credible Interval | Day 60 LD50 | 95% HPD Credible Interval |
| K96243 | 2.24×106 | 1.15×106–4.29×106 | 1.09×106 | 4.97×105–2.25×106 | |
| 668 | 1.70×105 | 9.93×104–3.01×105 | 3.18×104 | 1.34×104–7.24×104 | |
| 1106a | 3.47×106 | 1.48×106–8.35×106 | 1.17×106 | 4.55×105–3.12×106 |
HPD: Highest Posterior Density.
Genome Features
We performed an extensive comparative analysis of the B. pseudomallei genomes to identify genomic features that are common and unique among the various strains, and to begin to address differences in virulence and disease presentation. Because the K96243 genome that we downloaded from NCBI contained nearly 1,500 fewer CDS than the other two genomes, we re-annotated all three genomes using the RAST system [26] to ensure consistent comparisons. Table 2 compares the three complete genomes in terms of their general features. Comparisons of the CDS in each genome identified by RAST annotation compared to the original annotations showed that the number of CDS in the K96243 genome increased by 1,317 (18.7%). The numbers of CDS in the MSHR668 and 1106a genomes were also increased, but by smaller percentages (3.5% and 4.2%, respectively). These analyses provided a common annotation platform from which the ensuing comparisons were made.
Table 2. General genome features.
| Feature | MSHR668 | K96243 | 1106a |
| Genome size (bp) | 7,040,403 | 7,247,547 | 7,089,249 |
| No. chromosomes | 2 | 2 | 2 |
| Genes | 6,940 | 7,116 | 6,946 |
| Protein coding (RAST annotation) | 6,869 | 7,045 | 6,875 |
| Protein coding (original annotation) | 7,116 | 5,728 | 7,174 |
| Mobile elements | 72 | 79 | 89 |
| rRNA operons | 12 | 12 | 12 |
| tRNA genes | 59 | 59 | 59 |
| GC% | 68.3 | 68.1 | 68.3 |
| Regulatory elements | 333 | 332 | 328 |
| 2-component system | 79 | 81 | 77 |
Pseudogenes and mobile elements
The number of pseudogenes in each originally annotated genome varied depending on the resource used to identify them. Holden et al. (2004) originally reported that the genome of K96243 contains 26 pseudogenes [19], whereas the IMG system [27] identified 122 pseudogenes in K96243, 5 in MSHR668, and 8 in 1106a. The Pathway Tools [28] identified 136 pseudogenes in K96243, 10 pseudogenes in MSHR668, and 15 pseudogenes in 1106a. Because of this discrepancy, and since we re-annotated the genomes using RAST, which does not include an automatic pseudogene identification step, we identified the potential pseudogenes in each genome using the Psi Phi program [29], which is a comparative method for pseudogene identification. Psi Phi identified no additional pseudogenes in the RAST-predicted CDS of K96243, MSHR668 and 1106a. However, Psi Phi identified a few candidate pseudogenes in the intergenic regions, and there were some CDS with less than full length alignments to known protein sequences in the public databases. Since this report does not focus on pseudogenes, we did not explore these further.
The genomes of K96243 and 1106a contained more genes annotated by RAST [26] as encoding mobile elements (79 and 89, respectively) compared to MSHR668 (S1 Table). The number of genes encoding mobile elements that were identified by RAST annotation of K96243 was greater than the originally reported number of 42 mobile elements in K96243 [19]. This discrepancy is likely due to the higher number of total CDS in the RAST annotation of the K96243 genome.
Chromosome alignments
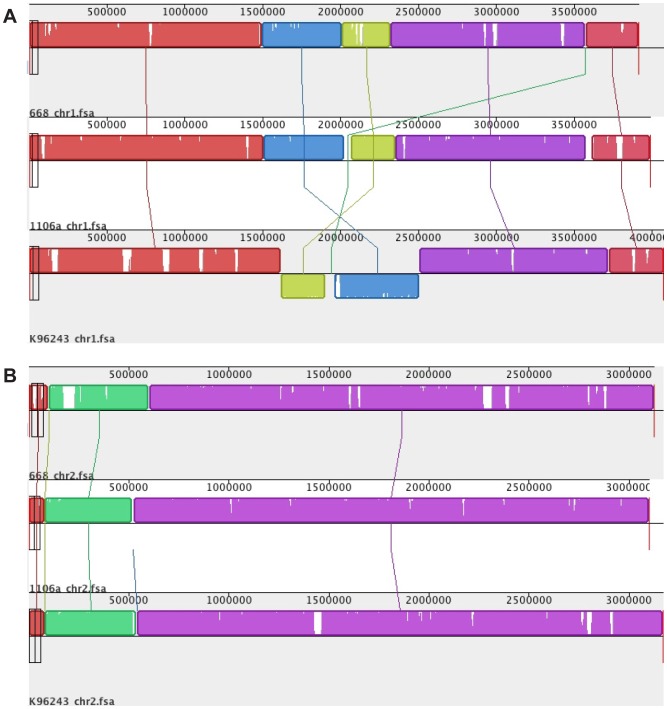
Individual chromosomes of B. pseudomallei MSHR668, 1106a and K96243 were aligned using Mauve [30], and results showed that they are largely collinear, except for an inversion of the K96243 chromosome 1 and a small gap in between the locally collinear blocks in the inverted region (Fig. 1).
Figure 1. Mauve alignment of B. pseudomallei chromosomes 1 (panel A) and 2 (panel B).
Homologous regions in the genomes are illustrated as locally collinear blocks of the same color that are linked across the chromosomes. The three genomes showed five homologous regions in chromosome 1, and three homologous blocks in chromosome 2.
Coding sequence comparisons
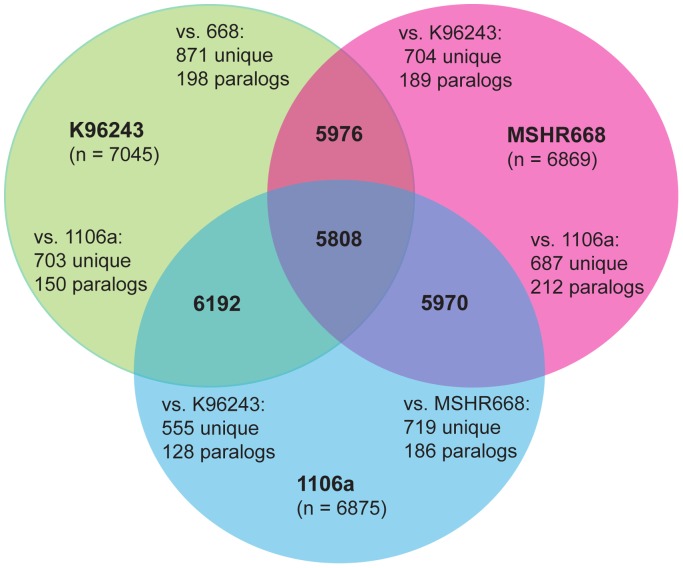
The protein coding sequences (CDS) in common among the genomes (putative homologs) were identified by a bidirectional best BLASTp hits analysis. This also enabled the identification of unique genes that were only present in each genome or group of genomes. Fig. 2 shows the results of the analyses for each pair of genomes, as well as all three genomes together. A total of 5,808 CDS were shared by all three genomes. The pairwise comparisons showed 5,976 CDS shared between K96243 and MSHR668, 6,192 CDS in common between K96243 and 1106a, and 5,970 CDS shared between MSHR668 and 1106a.
Figure 2. Venn diagram illustrating the numbers of CDS shared by B. pseudomallei strains K96243, MSHR668 and 1106a, determined by a bidirectional best BLAST hits analysis.
The number of CDS unique to each genome in each pairwise comparison and the number of putative paralogs are shown. The total number of CDS present in each genome is given below the genome name.
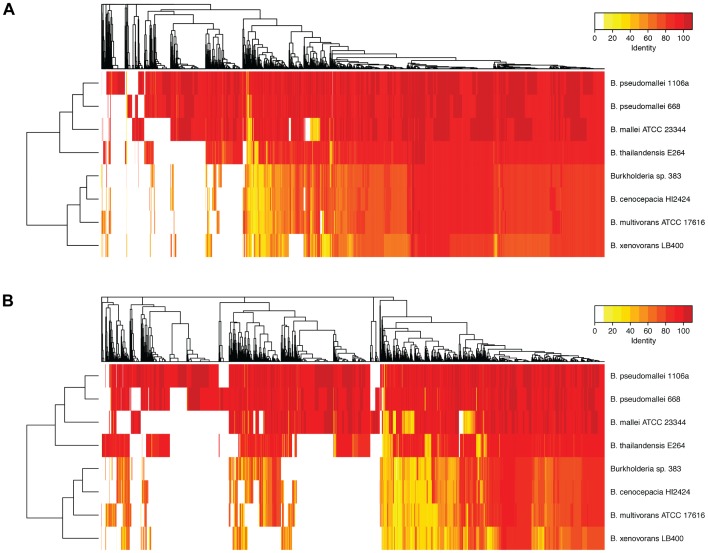
The distribution of BLASTp hits to strain K96243 is also displayed in a heatmap in Fig. 3. These comparisons included the two pseudomallei strains plus B. thailandensis, B. mallei and other near neighbors to illustrate overall similarities and differences in percent identities across the genomes. The number of best BLASTp hits in these eight Burkholderia genomes is also summarized at different percent identity cutoffs in Fig. 4.
Figure 3. Heatmap displaying best BLAST hits of protein sequences from eight Burkholderia genomes to B. pseudomallei K96243 proteins on chromosome 1 (Panel A) and chromosome 2 (panel B).
The protein BLAST was run without the filter and an E-value cutoff of 1e-15.
Figure 4. Summary of the number of best BLAST hits matching B. pseudomallei K96243 proteins at different percent identity cutoffs.
Gene content comparisons
Although genomic islands (GIs) and their gene content vary greatly among B. pseudomallei strains, a thorough comparison of the GIs in the three genomes was already performed [7]. Therefore to investigate potential virulence and metabolism-related genes, we focused on gene clusters and individual CDS (not found in GIs) that were unique to strain MSHR668 and not present in the genomes of 1106a and K96243 (Tables 3 and 4). Many of the genomic features that were present in strain MSHR668 but absent in the genomes of 1106a and K96243 were also present in the genomes of one or more of the other Australian strains, for example strain MSHR305. This result is particularly interesting because of the similar clinical presentations of disease caused by these Australian strains, involving general septicemic infections and the somewhat rare events of encephalomyelitis caused by strains MSHR668 [6] and MSHR305.
Table 3. Genes present in the MSHR668 genome that were absent in both K96243 and 1106a.
| 668 CDS (locus tag) | Function | Present in other Bp genomes? |
| BURPS668_0139 | cytidine/deoxycytidylate deaminase | MSHR1043, BDI, BEZ |
| BURPS668_0798 | multidrug ABC transporter permease | 1655, S13, MSHR1043, NAU20B-16 |
| BURPS668_0860 | CRISPR-associated RAMP Cmr1 | no |
| in RAST annotation (320373.8.peg.1061) | Beta-glucosidase (EC 3.2.1.21) | NCTC 13179, 354e, 1026ab |
| in RAST annotation (320373.8.peg.1096) | Glycine-rich cell wall structural protein 1.8 precursor | no |
| BURPS668_1498 | phage protein, possible ATP synthase | many |
| BURPS668_1596 | transposase | 576, Pakistan 9, 1710ab, MSHR6137 |
| BURPS668_1621 | trans-aconitate 2-methyltransferase | no |
| in RAST annotation (320373.8.peg.1826) | putative HIT domain protein | NCTC 13178, NCTC 13179 |
| BURPS668_2012 | gp30 | MSHR6137, Pakistan 9, MSHR346, 1710a |
| BURPS668_2112 | Multidrug resistance protein, major facilitator superfamily | NAU20B-16, MSHR511, MSHR146 |
| BURPS668_2138 | XRE family transcriptional regulator | no |
| in RAST annotation (320373.8.peg.2249) | LuxR family transcriptional regulator | 576, 1710a, MSHR1043, MSHR6137 |
| BURPS668_2839 | putative septum site-determining protein MinD | MSHR1043, 406e, MSHR346 |
| BURPS668_3493 | integrase | no |
| BURPS668_3499 | XRE family transcriptional regulator | no |
| BURPS668_A0076 | putative dienelactone hydrolase | 1026ab, MSHR346, MSHR338, 406e |
| BURPS668_A0193 | glycosyl transferase group 2 family protein | MSHR6137, MSHR305, NCTC13179, NCTC13178, MSHR511, MSHR146, NAU20B-16 |
| BURPS668_A0194 | putative queuine/archaeosine tRNA-ribosyltransferase | NCTC13178, NCTC13179, 1655, MSHR6137 |
| BURPS668_A0197 | putative sugar nucleotidyltransferase | MSHR6137, MSHR305, 406e, NCTC13179, NCTC13178, MSHR511, MSHR146, NAU20B-16, 1655 |
| BURPS668_A0198 | CDP-glycerol glycerophosphotransferase | MSHR6137, MSHR305, 406e, NCTC13179, NCTC13178, MSHR511, MSHR146, NAU20B-16, 1655, MSHR1043 |
| BURPS668_A0218 | flagellar motor switch protein FliM | MSHR305, NCTC 13179, NCTC 13178, MSHR520, MSHR511, MSHR146, NAU20B-16, 406e, 1655 |
| BURPS668_A0222 | flagellar hook-basal body protein FliE | same as above |
| BURPS668_A0227 | flagellar protein FliJ | same as above |
| BURPS668_A0230 | signal transduction histidine kinase | same as above |
| BURPS668_A0231 | flagellar hook-length control protein FliK | same as above |
| BURPS668_A0232 | flagellar basal body rod protein | same as above |
| BURPS668_A0234 | flagellar biosynthesis anti-sigma factor | same as above |
| BURPS668_A0235 | flagellar biosynthesis protein FliR | same as above |
| BURPS668_A0245 | flageller rod assembly protein | same as above |
| BURPS668_A0248 | flagellar hook associated protein | same as above |
| BURPS668_A0249 | flagellar hook-length control protein FliK | MSHR305, 406e, 1655 |
| in RAST annotation (320373.8.peg.4106) | membrane protein | no |
| BURPS668_A0981 | integrase | 1026ab, NCTC13178, MSHR5858, 576, NAu35A-3, 1710b, others |
| BURPS668_A1335 | DNA-binding protein | MSHR305, MSHR346, MSHR6137, Pasteur 52237, 1710b |
| BURPS668_A1383 | beta-lactamase class A | MSHR305, S13, Pakistan 9, MSHR346, 1710a |
| BURPS668_A1459 | response regulator of the LytR/AlgR family | 1710b, MSHR1655, MSHR146, MSHR511, MSHR305, MAU20B-16, MSHR520 |
| BURPS668_A1550 | thymidylate kinase | BPC006 |
| BURPS668_A1697 | CurM protein | no |
| BURPS668_A1836 | DGPF domain-containing protein | S13, MSHR346, MSHR305, 1710b |
| BURPS668_A1843 | LysR family transcriptional regulator | no |
| BURPS668_A2058 | endoribonuclease L-PSP | MSHR346 |
| BURPS668_A2983 | DNA repair ATPase | no |
Presence in other B. pseudomallei genomes was determined by NCBI BLAST against all genomes in GenBank.
Table 4. Genes present in both K96243 and 1106a genomes that were absent in MSHR668.
| K96243 and 1106a CDS (locus tag) | Function | Present in other Bp genomes? |
| BPSL0348/BURPS1106A_0385 | putative inclusion body protein | MSHR5858, 576, 1026b, 1710b, NCTC13179, others |
| BPSL0349/BURPS1106A_0386 | DNA-directed RNA polymerase subunit beta | MSHR5858, NAU35A-3, MSHR3865, NCTC13179, others |
| in RAST annotation 272560.34.peg.842/357348.16.peg.782 | phage integrase | 1258ab, 354ae, MSHR6137, 1026a, MSHR520, MSHR338, MSHR346, MSHR1043 |
| BPSL0763/357348.16.peg.3555 | helicase | no |
| BPSL0764/357348.16.peg.3554 | putative restriction enzyme | no |
| BPSL0765/357348.16.peg.3551 | helicase | MSHR5855 |
| 272560.34.peg.1128/BURPS1106A_1060 | putative OmpA family protein | 1106b, BPC006, 576, MSHR6137, 1710b, MSHR1043 |
| BPSL1028/several | transposase | MSHR5858, NCTC13178, 576, 1710b, others |
| 272560.34.peg.1421/BURPS1106A_1350 | LysR family transcriptional regulator | NAU35A-3, BPC006, MSHR1153, 1026b, others |
| BPSL1298/BURPS1106A_1411 | histidine kinase | MSHR2243, NCTC13179, MSHR1153, others |
| BPSL1563/BURPS1106A_2170 | membrane protein | MSHR5858, MSHR2243, NAU35A-3, others |
| BPSL1564/BURPS1106A_2169 | Cro/Cl family transcriptional regulator | MSHR5855, MSHR5858, MSHR2243, NAU35A-3, others |
| 272560.34.peg.2810/BURPS1106A_2805 | putative periplasmic substrate binding protein | 1106b, 576 |
| in RAST annotation 272560.34.peg.3267/357348.16.peg.3156 | D-glycero-D-manno-heptose 7-phosphate kinase | S13, MSHR346, MDHR305, BPC006 others |
| BPSL2817/several | transposase | 1026b, MSHR1153, NCTC13179, others |
| 272560.34.peg.3457/BURPS1106A_3460 | LysM repeat protein | Pakistan 9, BPC006, 1710a |
| BPSS0121/BURPS1106A _A0164 | beta fimbrial chaperone protein | 1026b, MSHR5858, BPC006, others |
| BPSS0123/BURPS1106A _A0167 | beta fimbrial major subunit | 1026b, MSHR5858, BPC006, 1710b, others |
| 272560.34.peg.4508/BURPS1106A_A0545 | phage holin | MSHR5858, MSHR346, MSHR1655, 1026b, others |
| BPSS0395/BURPS1106A_A0542 | phage protein | MSHR5858, 1710b, MSHR146, others |
| BPSS0396/BURPS1106A_A0540 | phage protein | MSHR305, MSHR520, 576, others |
| BPSS1075/357348.16.peg.3542 | phage tail completion protein | 1026b, NCTC13179, others |
| BPSS1080/357348.16.peg.3545 | phage baseplate assembly protein | 1026b, NCTC13179, others |
| BPSS1081/357348.16.peg.3544 | phage tail fiber protein | 1026b, NCTC13179, others |
| in RAST annotation 272560.34.peg.6291/357348.16.peg.6156 | integrase | 1026b, MSHR305, MSHR520, others |
| BPSS2057/BURPS1106A _A3044 | transposase | MSHR1153, NCTC13179, others |
| in RAST annotation 272560.34.peg.6685/357348.16.peg.6527 | transposase | 576, MSHR63, MSHR2243, NAU35A-3, others |
| in RAST annotation 272560.34.peg.6695/357348.16.peg.6539 | transposase | 1026b, MSHR5855, NCTC13179, others |
| BPSS2292/BURPS1106A_A3098 | universal stress protein | 1026b, 1710b, NCTC13179, others |
| BPSS2298/BURPS1106A _A3104 | thioredoxin | 1026b, 1710b, 576, others |
Presence in other B. pseudomallei genomes was determined by NCBI BLAST against all genomes in GenBank.
Addressing differences in the gene content of MSHR668 compared to both K96243 and 1106a, Table 3 lists individual genes (CDS) that were present in the MSHR668 genome but not present in the genomes of both K96243 and 1106a. Table 4 compares the gene content of both K96243 and 1106a, listing CDS that were present in both K96243 and 1106a genomes but absent in MSHR668. Most of the individual genes listed in Table 4 have mobile-element related annotated functions.
Metabolic genes and chokepoint reactions
There were some metabolism-related genes in the MSHR668 genome that did not have putative homologs in the K96243 and 1106a genomes (Table 5). The MSHR668 genome had thirteen genes with annotated functions in metabolism that were not present in the K96243 and 1106a genomes. Only four of the functions listed in Table 5 (cytidine/deoxycytidylate deaminase family protein, beta-glucosidase, putative dienelactone hydrolase, beta-lactamase) had additional copies in the MSHR668 genome. Only one gene (BURPS668_1621, encoding trans-aconitate 2-methyltransferase) was associated with a chokepoint reaction by the Pathway Tools [28]. This enzyme transfers one-carbon groups in the reaction that produces S-adenosyl-L-homocysteine from S-adenosyl-L-methionine [31].
Table 5. Metabolic and regulatory genes in the MSHR668 genome that were not present in either K96243 or 1106a.
| 668 Gene | Function | MetaCyc Pathways | KEGG Pathways |
| Metabolic | |||
| BURPS668_0139* | cytidine/deoxycytidylate deaminase family protein (EC 3.5.4.5/EC 3.5.4.12) | pyrimidine ribonucleosides degradation I, pyrimidine ribonucleotides salvage, purine and pyrimidine metabolism, pyrimidine ribonucleosides degradation II | pyrimidine metabolism |
| not in previous annot.* (320373.8.peg.1061) | Beta-glucosidase (EC 3.2.1.21) | various sugars converted to beta-D-glucose | Starch and sucrose metabolism Phenylpropanoid biosynthesis Cyanoamino acid metabolism |
| BURPS668_1621 | trans-aconitate 2-methyltransferase (EC 2.1.1.144) # | Reaction: S-adenosyl-L-methionine+trans-aconitate = S-adenosyl-L-homocysteine+(E)-3-(methoxycarbonyl)pent-2-enedioate | same reaction as MetaCyc |
| not in previous annot. (320373.8.peg.1826) | putative HIT domain protein (nucleotide hydrolase or transferase) | NA | NA |
| BURPS668_A0076* | putative dienelactone hydrolase (EC 3.1.1.45) | Reaction: dienelactone+H2O < = >2-maleylacetate+H+ | Chlorohexane, chlorobenzene, fluorobenzene, toluene degradation |
| BURPS668_A0193 | glycosyl transferase group 2 family protein | NA | Mucin-type O-glycan biosynthesis |
| BURPS668_A0194 | putative queuine/archaeosine tRNA-ribosyltransferase (EC 2.4.2.29) | NA | NA |
| BURPS668_A0197 | putative sugar nucleotidyltransferase | NA | NA |
| BURPS668_A0198 | CDP-glycerol glycerophosphotransferase(EC 2.7.8.12) | Reaction: CDP-glycerol+(glycerophosphate)(n) = Cmp+(glycerophosphate)(n+1). | NA |
| BURPS668_A1383* | beta-lactamase class A | NA | NA |
| BURPS668_A1550 | thymidylate kinase (EC 2.7.4.9) | Reaction: ATP+dTMP< = > ADP+dTDP | Pyrimidine metabolism |
| BURPS668_A1697 | CurM protein | NA | NA |
| BURPS668_A2058 | endoribonuclease L-PSP | NA | NA |
| Regulatory | |||
| BURPS668_2138* | XRE family transcriptional regulator | NA | NA |
| not in previous annot.* (320373.8.peg.2249) | LuxR family transcriptional regulator | NA | NA |
| BURPS668_3499* | XRE family transcriptional regulator | NA | NA |
| BURPS668_A0230* | signal transduction histidine kinase | NA | NA |
| BURPS668_A1459* | response regulator of the LytR/AlgR family | NA | NA |
| BURPS668_A1843* | LysR family transcriptional regulator | NA | NA |
*MSHR668 has one or more additional genes for this function.
#candidate chokepoint.
NA: function too general or no pathway associated.
Metabolic genes of interest in the K96243 and 1106a genomes that were not present in MSHR668 (Table 6) included D-glycero-D-manno-heptose 7-phosphate kinase, which is a candidate chokepoint enzyme, a LysM repeat protein and thioredoxin. The thioredoxin function was encoded by additional copies in both K96243 and 1106a genomes. D-glycero-D-manno-heptose 7-phosphate kinase is involved in the biosynthesis of lipopolysaccharide and is a virulence factor and potential protective antigen for B. pseudomallei [32].
Table 6. Metabolic and regulatory genes in the K96243 and 1106a genomes that were not present in MSHR668.
| K96243/1106a Gene | Function | MetaCyc Pathways | KEGG Pathways |
| Metabolic | |||
| not in prev. annot. (272560.34.peg.3267) | D-glycero-D-manno-heptose 7-phosphate kinase (EC 2.7.1.167)# | ADP-L-glycero-β-D-manno-heptose biosynthesis | Lipopolysaccharide biosynthesis |
| not in prev. annot./BURPS1106A_3460 | LysM repeat protein (putative peptidoglycan hydrolase) | NA | NA |
| BPSS2298/BURPS1106A _A3104* | thioredoxin (protein disulfide reductase) | NA | NA |
| Regulatory | |||
| not in prev. annot./BURPS1106A_1350* | LysR family transcriptional regulator | NA | NA |
| BPSL1298/BURPS1106A_1411* | histidine kinase | NA | NA |
| BPSL1564/BURPS1106A_2169 | Cro/Cl family transcriptional regulator | NA | NA |
| BPSS2292/BURPS1106A_A3098* | universal stress protein | NA | NA |
* K96243 and 1106a have one or more additional genes for this function.
#candidate chokepoint.
NA: function too general or no pathway associated.
Metabolic pathways
Metabolic pathways were identified in the three B. pseudomallei genomes by Pathway Tools [28] and compared using Pathway Tools, MetaCyc [31], KEGG [33], BLAST analysis [34] and IMG [27]. Pathways comprising central carbon metabolism and the main inputs and outputs are listed in S2 Table. All three of the genomes had components of the main pathways of central carbon metabolism and genes encoding transporters systems, anapleurotic reactions, and pathways for amino acid biosynthesis. The genomes of all three strains had complete pathways to make the amino acids and vitamins that humans obtain from diet and that more fastidious host-restricted intracellular pathogens, such as F. tularensis, do not contain. These included histidine, isoleucine, leucine, lysine, methionine, cysteine, phenylalanine, tyrosine, threonine, tryptophan, valine, folate, biotin, lipoic acid, pantothenate, thiamine, riboflavin and vitamin K2 (menaquinone). All of the genomes had genes encoding the cobalamin adenosyltransferase that converts cobalamin to vitamin B12. None of the genomes had genes encoding the enzymes needed to make vitamin K1 (phylloquinone).
Bacterial gene expression is controlled by transcriptional regulators, such as transcription factors and sigma factors. The functions of these proteins in gene expression regulation were first described in Escherichia coli [35] and were previously reviewed in Pseudomonas aeruginosa [36]. There were many transcription and sigma factors, response regulators, and DNA-binding proteins identified in the B. pseudomallei genomes (Table 2). S3 and S4 Tables list the differences in regulatory gene numbers, while Tables 5 and 6 compare gene content between MSHR668 and K96243/1106a. Nearly all of regulatory functions listed in these tables were present in additional copies in the genomes, although their exact gene targets are not known.
Virulence genes and metabolism
Table 7 lists virulence genes compiled from online databases [37]–[39]and literature [19], [40] with annotated metabolic and regulatory functions that were present in all three genomes. At least twenty five of the metabolic genes in Table 7 were identified as potential chokepoints by the Pathway Tools.
Table 7. Virulence genes in B. pseudomallei K96243, 1106a and MSHR668 genomes with metabolic and regulatory functions.
| Gene | Annotated Function | Pathways (KEGG, MetaCyc) or process |
| Metabolism | ||
| BPSL0338 | non-hemolytic phospholipase C (EC 3.1.4.3) | Inositol phosphate metabolism, Glycerophospholipid metabolism, Ether lipid metabolism |
| BPSL0374 | metallo-beta-lactamase superfamily protein | NA |
| BPSL0395 | cytidylyltransferase | various |
| BPSL0413 | lipoate protein ligase B (EC 2.7.7.63) | Lipoic acid metabolism |
| BPSL0634 | oxidoreductase | various |
| BPSL0808 | peptidase; serine protease (EC 3.4.21.-) | various |
| BPSL0908 | phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) | Purine metabolism, One carbon pool by folate, Biosynthesis of secondary metabolites |
| BPSL1103 | endonuclease III (EC 4.2.99.18)# | various |
| BPSL1196 | acetolactate synthase 3 catalytic subunit (EC 2.2.1.6)# | Branched chain amino acid biosynthesis, Butanoate metabolism, C5-branched dibasic acid metabolism, Pantothenate and CoA biosynthesis, Biosynthesis of secondary metabolites |
| BPSL1561 | metallo-beta-lactamase | Hydrolysis of beta-lactam antibiotics |
| BPSL1776 | L-ornithine 5-monooxygenase MbaA/PvdA (EC 1.13.12.-) | Siderophore biosynthesis |
| BPSL1777 | siderophore-related non-ribosomal peptide synthase MbaI | Siderophore biosynthesis |
| BPSL1778 | siderophore related non-ribosomal peptide synthase MbaJ | Siderophore biosynthesis |
| BPSL1876 | phospholipase; phosphoesterase | various |
| BPSL2403 | non-hemolytic phospholipase C (EC 3.1.4.3) | Inositol phosphate metabolism, Glycerophospholipid metabolism, Ether lipid metabolism |
| BPSL2433 | peptidase; Do family protease; serine protease | various |
| BPSL2519 | phosphoserine aminotransferase (EC 2.6.1.52)# | Glycine, serine and threonine metabolism, Methane metabolism, Vitamin B6 metabolism |
| BPSL2672 | epimerase/dehydratase capsule polysaccharide biosynthesis protein | various |
| BPSL2673 | undecaprenyl phosphate N-acetylglucosaminyltransferase; glycoside hydrolase family protein; UDP-D-N-acetylhexosamine:polyprenol phosphate D–N-acetylhexosamine-1-phosphate transferases (EC 2.7.8.-) | various |
| BPSL2674 | NAD-dependent epimerase/dehydratase | various |
| BPSL2675 | glycosyl transferase | various |
| BPSL2676 | glycosyl transferase | various |
| BPSL2677 | O-antigen methyl transferase (EC 2.4.1.-) | various |
| BPSL2678 | glycosyl transferase | various |
| BPSL2679 | NAD-epimerase/dehydratase | various |
| BPSL2680 | O-antigen acetylase WbiA (EC 2.3.1.-) | various |
| BPSL2683 | dTDP-4-dehydrorhamnose reductase (EC 1.1.1.133)# | Biosynthesis of secondary metabolites |
| BPSL2684 | dTDP-6-deoxy-D-glucose-3,5 epimerase (EC 5.1.3.13)# | Biosynthesis of secondary metabolites |
| BPSL2685 | glucose-1-phosphate thymidylyltransferase (EC 2.7.7.24)# | Biosynthesis of secondary metabolites |
| BPSL2686 | dTDP-glucose 4,6-dehydratase (EC 4.2.1.46)# | Biosynthesis of secondary metabolites |
| BPSL2687 | diadenosine tetraphosphatase (EC 3.6.1.41)# | Purine metabolism |
| BPSL2688 | 1-acyl-SN-glycerol-3-phosphate acyltransferase; Lysophospholipid Acyltransferases (LPLATs) of Glycerophospholipid Biosynthesis (EC 2.3.1.51)# | various |
| BPSL2786 | acetyltransferase | various |
| BPSL2787 | acyl-CoA transferase; 8-amino-7-oxononanoate synthase (EC 2.3.1.47)# | Biotin metabolism |
| BPSL2788 | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (EC 3.5.1.108)# | various |
| BPSL2789 | capsular polysaccharide biosynthesis fatty acid synthase; type I polyketide synthase WcbR | various |
| BPSL2790 | capsular polysaccharide biosynthesis transmembrane protein; sulfatase (EC 3.1.6.-) | various |
| BPSL2791 | capsular polysaccharide biosynthesis dehydrogenase/reductase; short chain dehydrogenase/reductase family oxidoreductase | various |
| BPSL2792 | capsule polysaccharide biosynthesis/export protein KpsS | various |
| BPSL2793 | D-glycero-d-manno-heptose 1,7-bisphosphate phosphatase (EC 3.1.3.82)# | various |
| BPSL2794 | D-glycero-d-manno-heptose 1-phosphate guanosyltransferase (EC 2.7.7.71) | various |
| BPSL2795 | phosphoheptose isomerase (EC 5.3.1.28)# | Lipopolysaccharide biosynthesis |
| BPSL2796 | sugar kinase; D-glycero-D-manno-heptose 7-phosphate kinase; related to galactokinase and mevalonate kinase (EC 2.7.7.70)# | Lipopolysaccharide biosynthesis |
| BPSL2797 | GDP sugar epimerase/dehydratase; GDP-6-deoxy-D-lyxo-4-hexulose reductase (EC 1.1.1.281)# | Fructose and mannose metabolism, Amino sugar and nucleotide sugar metabolism |
| BPSL2798 | capsular polysaccharide biosynthesis protein; NAD-dependent epimerase/dehydratase | various |
| BPSL2799 | capsular polysaccharide biosynthesis protein | various |
| BPSL2800 | glycosyl transferase | various |
| BPSL2801 | capsular polysaccharide biosynthesis protein | various |
| BPSL2802 | capsular polysaccharide biosynthesis protein | various |
| BPSL2803 | glycosyltransferase | various |
| BPSL2808 | capsular polysaccharide glycosyltransferase biosynthesis protein | various |
| BPSL2810 | GDP-mannose pyrophosphorylase; mannose-1-phosphate guanylyltransferase (EC 2.7.7.13/EC 2.7.7.22) | Fructose and mannose metabolism, Amino sugar and nucleotide sugar metabolism, Biosynthesis of secondary metabolites |
| BPSL2818 | phosphoribosylaminoimidazole synthetase (EC 6.3.3.1)# | Purine metabolism, Biosynthesis of secondary metabolites |
| BPSL2825 | hypothetical protein BPSL2825; para-aminobenzoate synthase, component I PabB (EC 2.6.1.85)# | tetrahydrofolate biosynthesis and salvage, superpathway of chorismate metabolism, superpathway of tetrahydrofolate biosynthesis, 4-aminobenzoate biosynthesis |
| BPSL3051 | anthranilate synthase component II (EC 4 1.3.27) | Phenylalanine, tyrosine and tryptophan biosynthesis, Biosynthesis of secondary metabolites |
| BPSL3133 | imidazole glycerol phosphate synthase subunit HisF (EC 4.1.3.−/EC 2.4.2.-)# | Histidine biosynthesis, Purine biosynthesis |
| BPSL3168 | 3-dehydroquinate synthase (EC 4.2.3.4)# | Phenylalanine, tyrosine and tryptophan biosynthesis, Biosynthesis of secondary metabolites |
| BPSS0067 | non-hemolytic phospholipase C (EC 3.1.4.3) | Inositol phosphate metabolism, Glycerophospholipid metabolism, Ether lipid metabolism |
| BPSS0419 | glucose-1-phosphate cytidylyltransferase (EC 2.7.7.33)# | Starch and sucrose metabolism, Amino sugar and nucleotide sugar metabolism, Biosynthesis of secondary metabolites |
| BPSS0420 | CDP-glucose 4,6-dehydratase (EC 4.2.1.45)# | Amino sugar and nucleotide sugar metabolism, Biosynthesis of secondary metabolites |
| BPSS0421 | lipopolysaccharide biosynthesis protein rfbH | Lipopolysaccharide biosynthesis |
| BPSS0422 | aminotransferase | various |
| BPSS0424 | glycosyl transferase group 2 | various |
| BPSS0425 | heptosyltransferase (O-antigen related) | Lipopolysaccharide biosynthesis |
| BPSS0426 | heptosyltransferase (O-antigen related) | Lipopolysaccharide biosynthesis |
| BPSS0427 | O-acetyl transferase; galactoside O-acetyltransferase | Lipopolysaccharide biosynthesis |
| BPSS0428 | glycosyl transferase (O-antigen related) | Lipopolysaccharide biosynthesis |
| BPSS0581 | salicylate biosynthesis isochorismate synthase (EC 5.4.4.2)# | Ubiquinone biosynthesis, Biosynthesis of siderophore group nonribosomal peptides, Biosynthesis of secondary metabolites |
| BPSS0582 | isochorismate-pyruvate lyase (EC 4.2.99.21) | Siderophore biosynthesis |
| BPSS0583 | pyochelin biosynthetic protein PchC (EC 3.1.2.-) | Siderophore biosynthesis |
| BPSS0584 | salicyl-AMP ligase; 2,3-dihydroxybenzoate-AMP ligase (EC 2.7.7.58)# | Siderophore biosynthesis |
| BPSS0586 | pyochelin synthetase | Siderophore biosynthesis |
| BPSS0587 | pyochelin synthetase | Siderophore biosynthesis |
| BPSS0588 | pyochelin biosynthetic protein | Siderophore biosynthesis |
| BPSS0666 | peptidase; collagenase (EC 3.4.24.3) | Digestion of native collagen |
| BPSS0885 | N-acylhomoserine lactone synthase; autoinducer synthase BpsI (EC 2.3.1.184) | Quorum sensing |
| BPSS0946 | beta-lactamase precursor | Hydrolysis of beta-lactam antibiotics |
| BPSS1180 | N-acylhomoserine lactone synthase; autoinducer synthetase | Quorum sensing |
| BPSS1570 | N-acylhomoserine lactone synthase; autoinducer synthetase BpmI | Quorum sensing |
| BPSS1705 | 3-isopropylmalate dehydrogenase (EC 1.1.1.85)# | Branched chain amino acid biosynthesis, Butanoate metabolism, C5-branched dibasic acid metabolism, Biosynthesis of secondary metabolites |
| BPSS1825 | glycosyltransferase | various |
| BPSS1826 | glycosyltransferase | various |
| BPSS1828 | glycosyltransferase group 1 protein | various |
| BPSS1829 | glycosyltransferase | various |
| BPSS1830 | exopolysaccharide capsular polysaccharide biosynthesis-like tyrosine-protein kinase | capsule biosynthesis |
| BPSS1831 | exopolysaccharide (EPS) capsular polysaccharide biosynthesis related polysaccharide lipoprotein | capsule biosynthesis |
| BPSS1832 | exopolysaccharide (EPS) capsular polysaccharide biosynthesis-like; low molecular weight protein-tyrosine-phosphatase | capsule biosynthesis |
| BPSS1833 | UDP-glucose 6-dehydrogenase 2 (EC 1.1.1.22)# | Pentose and glucuronate interconversions, Ascorbate and aldarate metabolism, Starch and sucrose metabolism, Amino sugar and nucleotide sugar metabolism,Biosynthesis of secondary metabolites |
| BPSS1834 | lipopolysaccharide biosynthesis-like protein; undecaprenyl-phosphate glucose phosphotransferase (EC 2.7.8.31) | NA |
| BPSS1915 | metallo-beta-lactamase | NA |
| BPSS1993 | serine metalloprotease precursor | NA |
| BPSS1997 | class D beta-lactamase | Hydrolysis of beta-lactam antibiotics |
| Regulation | NA | |
| BPSL0812 | TetR family regulatory protein; multidrug efflux pump repressor protein BpeR | NA |
| BPSS0887 | N-acylhomoserine lactone dependent regulatory protein; autoinducer-binding transcriptional regulator BpsR | NA |
| BPSS1176 | N-acyl-homoserine lactone dependent regulatory protein; ATP-dependent transcriptional regulator LuxR | NA |
| BPSS1569 | N-acylhomoserine lactone-dependent regulatory protein; autoinducer-binding transcriptional regulator BmpR | NA |
| BPSL1787 | extracytoplasmic-function sigma-70 factor | NA |
| BPSL1805 | TetR family regulatory protein; multidrug efflux operon transciptional regulator AmrR | NA |
| BPSL2347 | LuxR family transcriptional regulator | NA |
| BPSL2434 | sigma E factor regulatory protein | NA |
| BPSL2435 | sigma E factor negative regulatory protein, RseA family | NA |
| BPSL2866 | oxidative stress regulatory protein OxyR; LysR family transcriptional regulator | NA |
| BPSS0312 | LuxR family transcriptional regulator | NA |
| BPSS0585 | AraC family transcriptional regulator PchR | NA |
| BPSS1391 | AraC family transcriptional regulator | NA |
| BPSS1520 | AraC family transcriptional regulator | NA |
| BPSS1522 | two-component response regulator; LuxR family DNA-binding response regulator | NA |
While K96243 GenBank locus tags are listed, genes are present in all three genomes.
NA: no pathway associated with the enzyme.
Various: enzyme may participate in multiple pathways or annotation too general to identify pathways by EC number.
# Candidate chokepoint.
All of the genes in this table were present in various other B. pseudomallei genomes, as determined by NCBI BLAST against all genomes in GenBank.
Discussion
Experimental infection of mouse models with the three B. pseudomallei strains showed that the K96243 and 1106a strains from Thailand had similar LD50 values in both BALB/c (more susceptible) and C57BL/6 (more resistant) murine infection models, while the Australian strain MSHR668 was more virulent as measured by LD50. Given the incredible amount of genomic diversity among B. pseudomallei strains, we sought to identify candidate genomic differences that may correlate with variations in virulence. We conducted whole genome comparisons focusing on virulence, metabolism and regulation and identified genes in common among all three genomes. We also identified genes that were present in MSHR668 but absent in K96243 and 1106a (and vice versa). Our findings and the implications on our understanding of melioidosis as a disease are discussed below.
Comparison of the three B. pseudomallei genomes revealed genomic differences that included the previously reported variability in GIs [7], [19], which were likely acquired by horizontal transfer [19], as evidenced by their proximity to transposases, integrases, tRNA genes, and the presence of phage-related genes within the GI. This variability in the GI regions may contribute to virulence potential, particularly because these regions can encode a broad array of functions [20]. The intracellular life cycle and adaptation of a pathogen to the host cell environment depends on the expression of virulence factors, which is controlled by regulatory elements, and may be affected by the metabolic state of the pathogen [41]. The genomes of B. pseudomallei MSHR668, K96243, and 1106a contained complete gene sets for the core pathways comprising carbon metabolism. They also contained gene sets encoding transporters and utilization pathways for a wide range of carbon substrates, anapleurotic reactions and fatty acid degradation products (S2 Table), providing many potential targets for metabolic regulation.
An important outcome of the metabolic pathway analysis was identification of chokepoint reactions in the three genomes by the Pathway Tools software [28]. Inhibition of an enzyme that consumes a unique substrate might cause accumulation of the substrate and be potentially toxic to the cell. Conversely, inhibition of an enzyme that produces a unique product might result in starvation for that product, which could cripple essential cell functions. Thus, chokepoint enzymes may be essential to the pathogen and therefore represent potential drug targets. We identified two chokepoint reactions among the lists of genes in Tables 5 and 6, which were differentially present in the three genomes. Among the genes in Table 7, we identified twenty-five candidate chokepoint enzymes in common among the three genomes, involved in a variety of metabolic functions. The complete list of chokepoint reactions, including candidates, totals approximately 1,200−1,300 reactions for each genome (data not shown) and requires additional curation and more extensive comparative analysis to determine which ones are the most promising targets. While our findings indicate that there are only a few metabolic differences among the B. pseudomallei genomes, it is becoming increasingly apparent that virulence and metabolism are linked together by complex regulatory interactions occurring between intracellular pathogens and their host cells [41]–[45]. We did find a few differences in regulatory gene content between MSHR668 and the other two genomes, in particular the K96243 and 1106a genomes contained more predicted sigma factor encoding genes than MSHR668 (S4 Table). Also the MSHR668 genome encoded additional transcriptional regulators, specifically two XRE family, two LysR family and one LuxR family, that were not present in the other genomes (Table 3). The genomes of both 1106a and K96243 encoded one LysR family and one Cro/CI family regulator that were not present in the MSHR668 genome (Table 4). These results suggest that differences in regulation may contribute to the differences in virulence observed among these strains. Although further work is needed to test this hypothesis, the observed differences in transcriptional regulatory genes may contribute to the differential virulence observed in this study.
Increasing evidence indicates that virulence gene expression is regulated by nutrients in the environment surrounding B. pseudomallei [46]. The expression of pathogen genes involved in transport and utilization of nutrients containing carbon and nitrogen is controlled by transcriptional regulators that are activated by the presence of nutrients [47]–[51]. For example, RpoS is involved in the response of B. pseudomallei to carbon starvation, heat shock, osmotic stress and oxidative stress. The expression of metabolic pathway genes involved in central carbon metabolism is controlled by RpoS, and by RpoS and BpsI co-regulation [52]. Therefore, the inter-regulation of stress response and metabolic genes by RpoS and BpsI may play an important role in B. pseudomallei survival and virulence [52]. RpoS has been reported to play a role in virulence gene expression in Salmonella typhimurium [53], and may also influence host macrophage responses to B. pseudomallei infection [54], [55]. The polyamines spermidine and putrescine regulate gene expression at the transcriptional level by affecting regulatory protein binding to DNA. The Fur protein is a positive regulator of peroxidase and iron-containing superoxide dismutase expression, but in response to increased iron concentrations, Fur reduces the transcription of iron-regulated promoters [56].
Several studies have examined transcriptional profiles of B. pseudomallei during infection [46], [57]–[61]. Results of these efforts support the idea that some virulence functions leading to infection and disease are linked to pathogen metabolism through regulation of gene expression. In some cases, metabolic enzymes may act as virulence factors through their role in providing nutrients to the pathogen during infection. For instance, phosphoserine aminotransferase, encoded by serC, is involved in serine and pyridoxal-5-phosphate synthesis, and may be a virulence factor in B. pseudomallei, as it is co-expressed with other virulence genes and auxotrophic mutation attenuates virulence [59]. Several studies have shown that some genes involved in metabolic processes and virulence are upregulated in B. pseudomallei during infection, while other metabolic genes are downregulated [46], [57], [58], [60], [61]. In spite of the increasing evidence linking metabolism and virulence, further work is needed to thoroughly characterize the overlapping roles of virulence factors, regulators and metabolic pathways in B. pseudomallei pathogenicity. Comparative genomic approaches such as those described here can be a key first step in generating hypotheses with respect to the roles of various bacterial factors in virulence.
Bacterial pathogens have evolved strategies to alter their lifestyle depending on whether they are in their natural environment or infecting a host, shifting resources from normal cell functions to the production of virulence factors, and altering metabolism to take advantage of the nutrients provided by host cells to facilitate survival and growth [62]. This should be especially true for B. pseudomallei given its presence and survival in a range of soil samples [63]–[68] and ability to cause severe disease in humans. Our comparison of the genomic features of two B. pseudomallei strains from Thailand (K96243 and 1106a) to one strain from Australia (MSHR668) revealed that the genomes are very similar in the repertoires of metabolic and virulence genes that they contain, leading to the conclusion that differential virulence studies on a larger scale are warranted. Detailed experiments will be necessary to characterize the relevance of specific genomic features to B. pseudomallei metabolism and virulence, and particular attention should be focused on the regulatory mechanisms influencing gene expression. Continued emphasis in this area will be critical to protection against melioidosis, as a better understanding of what constitutes a fully virulent Burkholderia isolate may inform better diagnostic and medical countermeasure strategies. The comparative genomic analysis that we present in this report, combined with more detailed functional analyses of metabolic networks, virulence and regulation, shows promise for examining the effects of B. pseudomallei and other intracellular pathogens on host cell metabolism and will lay a foundation for future prediction of the virulence of emerging strains.
Materials and Methods
Animal Challenges
Mouse challenges and statistical analyses were performed to establish LD50 values for each strain of B. pseudomallei. The United States Army of Medical Research Institute of Infectious Diseases is compliant with all federal and Department of Defense regulations pertaining to the use of Select Agents. Cultures were initiated by inoculating Glycerol Tryptone broth (GTB-10g/L tryptone, 5 g/L NaCl, 40 ml/L glycerol) with defrosted freezer stock of B. pseudomallei. The cultures were grown at 37°C while shaking at 200 RPM for approximately 8–10 hours in order to harvest cells at late logarithmic phase of growth. Challenge doses were prepared according to OD620 nm values and cultures were plated on sheep blood agar plates to confirm the number of colony forming units per milliliter (CFU/ml). At least 5 dose groups were used and 10 mice were included in each group. BALB/c and C57BL/6 mice were ordered from the National cancer Institute-NCI Frederick and were approximately 7–10 weeks of age at time of challenge. Mice were challenged intraperitoneally with bacterial doses suspended in 200 µl of GTB. Mice were observed at least daily for signs of illness or distress and monitoring frequency increased as indicated by the advancement of clinical signs. Challenged mice were observed at least twice daily for 60 days for clinical signs of illness. Humane endpoints were used during all studies, and mice were humanely euthanized when moribund according to an endpoint score sheet. Animals were scored on a scale of 0–12∶0–2 = no clinical signs; 3–7 = clinical symptoms; increase monitoring; greater than or equal to 8 = distress; euthanize. Those animals receiving a score of 8 or greater were humanely euthanized by CO2 exposure using compressed CO2 gas followed by cervical dislocation. However, even with multiple checks per day, some animals died as a direct result of the infection. Animal research at The United States Army of Medical Research Institute of Infectious Diseases was conducted and approved under an Institutional Animal Care and Use Committee in compliance with the Animal Welfare Act, PHS Policy, and other Federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
A Bayesian probit analysis was performed for each Burkholderia strain to estimate the lethal dose response curve. Prior distributions for each parameter were assumed to be independent, weakly informative Cauchy distributions with center 0 and scale 10. Using samples from the posterior distributions of the slope and intercept parameters from the probit analysis, the median and 95% credible intervals of the range of dose responses are estimated. Direct comparisons of the posterior samples of the LD50s of each strain permit us to make probabilistic statements about how likely it is that one strain is more or less potent than any other strain, given the observed data.
Genome Analysis
Whole genome sequences were obtained from NCBI (accession numbers NC_006351.1, NC_006350.1, NC_009074.1, NC_009075.1, NC_009076.1, NC_009078.1). To facilitate consistency in genome comparisons, genomes were annotated with RAST [26]. The GenBank format files for the RAST-annotated genomes are included in S1–S3 Files. The numbers of pseudogenes in each genome were obtained through the software package Psi Phi, which was kindly provided by Prof. Lerat [29]. In preparation for running Psi Phi, annotated protein sequences from each query genome were obtained from NCBI and used to query the nucleotide sequences of the other target genomes using tblastn. To identify potential pseudogenes, the Psi Phi software compares protein sequence matches in a query genome to the GenBank file of the target genome. We identified matches having a blast score with E-value <10−10 and a minimal percentage of protein identity of 80% Matches with 80% to 100% protein sequence identity to the query protein were retained. If a query sequence had two matches in close proximity in the target genome (as might result from frameshifts or insertion), the matches were merged if they were <300 nt apart [69].
Mobile genetic elements, transcription factors, sigma factors, response regulators, DNA binding proteins and two-component signal transduction systems were identified in each genome by searching the annotated genomes in the SEED [70]. Functional analysis was accomplished through the RegPrecise database [71].
Whole genome alignments were performed with Mauve [30]. To identify putative homologs among the genomes of B. pseudomallei strains K96243, 668 and 1106a, we performed a bidirectional best hits analysis, using BLASTp with an E-value cutoff of 1e−5 to obtain liberal best hits for the proteins of each genome compared to the others. Genes x and y from genomes 1 and 2 are considered as homologs if y is the best BLASTp hit for x and vice versa. We used the blast2gi program from the Seals package [72] to format the BLAST results in tabular form. Each pair of genomes was subjected to this analysis. To obtain the CDS shared by all three genomes, the sequences in common to each pair of genomes were compared to generate a list of CDS present in all three genomes. Sequences unique to each genome were identified by comparison of the total number of CDS in each genome to the common sequences from each pairwise comparison. We gathered the sequences that were unique to MSHR668 and not found in either K96243 and 1106a, and those that were unique to both K96243 and 1106a but not found in MSHR668. These sets of sequences were compared to the originally annotated genomes from GenBank, to determine whether RAST annotation predicted similar CDS to the previously annotated genomes in GenBank. Predicted CDS were not included in the unique set if there were high identity hits (>95%) in the original annotation. The locus_tags in Tables 3–5 refer to CDS present in both the RAST and original annotations. To create heatmaps comparing CDS from each B. pseudomallei genome to other Burkholderias, we used protein BLAST version 2.2.26+ to compare B. pseudomallei K96243 protein translations against eight other Burkholderia proteomes that we also annotated using RAST. We disabled filtering and set the E-value cutoff to 1e−15 and then saved the best hit to each subject protein. The best hits were binned into groups based on percent identity (100%, 90–99.9%, 80–89.9%, etc) and then displayed as a heatmap (Fig. 3), which was created in R using complete linkage hierarchical clustering with euclidean distances. A matrix showing the numbers of CDS shared in each pairwise comparison and percent identity was created by counting the number of best hits in each bin (Fig. 4).
Virulence gene lists were compiled from [19], [37]–[40], [73]. Blast analysis was used to compare the virulence gene sequences among the three genomes and between the original and RAST annotations. Metabolic pathways of the original and RAST-annotated B. pseudomallei genomes were analyzed using the Pathway Tools version 18.0 [28]. Chokepoint reactions were identified in B. pseudomallei MSHR668, K96243 and 1106a using the chokepoint reaction finder, with human reactions excluded.
Ethics Statement
Animal research at The United States Army Medical Research Institute of Infectious Diseases (USAMRIID) was conducted and approved under an Institutional Animal Care and Use Committee in compliance with the Animal Welfare Act, PHS Policy, and other Federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011. The USAMRIID IACUC approved this animal care and use protocol. USAMRIID policy does not allow approved animal protocol numbers to be published.
Supporting Information
Number of mobile element genes in B. pseudomallei genomes.
(DOCX)
Metabolic Pathway Comparison among B. pseudomallei genomes.
(DOCX)
Transcriptional regulatory genes in B. pseudomallei genomes.
(DOCX)
Sigma factor genes in B. pseudomallei genomes.
(DOCX)
RAST-annotated MSHR668 genome in GenBank file format.
(ZIP)
RAST-annotated 1106a genome in GenBank file format.
(ZIP)
RAST-annotated K96243 genome in GenBank file format.
(ZIP)
Acknowledgments
Disclaimer: Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U. S. Army.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was funded by the Defense Threat Reduction Agency (CCAR# CB3846 PPE-1 Burkholderia and CBS119924543-7049-BASIC). The funder had no role in study design, data collection and analysis, or decision to publish the manuscript. DNW contributed to the writing of the manuscript.
References
- 1. Currie BJ, Ward L, Cheng AC (2010) The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis 4:e900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Inglis TJJ, Sousa AQ (2009) The public health implications of melioidosis. Braz J Infect Dis 13:59–66. [DOI] [PubMed] [Google Scholar]
- 3. White NJ (2003) Melioidosis. Lancet 361:1715–1722. [DOI] [PubMed] [Google Scholar]
- 4. Currie BJ (2003) Melioidosis: an important cause of pneumonia in residents of and travellers returned from endemic regions. Eur Respir J 22:542–550. [DOI] [PubMed] [Google Scholar]
- 5. Cheng AC, Currie BJ (2005) Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev 18:383–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nandi T, Ong C, Singh AP, Boddey J, Atkins T, et al. (2010) A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence. PLoS Pathogens 6:e1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tuanyok A, Leadem B, Auerbach R, Beckstrom-Sternberg S, Beckstrom-Sternberg J, et al. (2008) Genomic islands from five strains of Burkholderia pseudomallei . BMC Genomics 9:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Limmathurotsakul D, Peacock SJ (2011) Melioidosis: a clinical overview. Br Med Bull 99:125–139. [DOI] [PubMed] [Google Scholar]
- 9. Choy JL, Mayo M, Janmaat A, Currie BJ (2000) Animal melioidosis in Australia. Acta Tropica 74:153–158. [DOI] [PubMed] [Google Scholar]
- 10. Currie BJ, Mayo M, Anstey NM, Donohoe P, Haase A, et al. (2001) A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am J Trop Med Hyg 65:177–179. [DOI] [PubMed] [Google Scholar]
- 11. Robertson J, Levy A, Sagripanti J-L, Inglis TJJ (2010) The Survival of Burkholderia pseudomallei in Liquid Media. Am J Trop Med Hyg 82:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gal D, Mayo M, Smith-Vaughan H, Dasari P, McKinnon M, et al. (2004) Contamination of hand wash detergent linked to occupationally acquired melioidosis. Am J Trop Med Hyg 71:360–362. [PubMed] [Google Scholar]
- 13. Dejsirilert S, Kondo E, Chiewsilp D, Kanai K (1991) Growth and survival of Pseudomonas pseudomallei in acidic environments. Jpn J Med Sci Biol 44:63–74. [DOI] [PubMed] [Google Scholar]
- 14. Jenney AW, Lum G, Fisher DA, Currie BJ (2001) Antibiotic susceptibility of Burkholderia pseudomallei from tropical northern Australia and implications for therapy of melioidosis. Int J Antimicrob Agents 17:109–113. [DOI] [PubMed] [Google Scholar]
- 15. Thibault FM, Hernandez E, Vidal DR, Girardet M, Cavallo JD (2004) Antibiotic susceptibility of 65 isolates of Burkholderia pseudomallei and Burkholderia mallei to 35 antimicrobial agents. J Antimicrob Chemother 54:1134–1138. [DOI] [PubMed] [Google Scholar]
- 16. Mays EE, Ricketts EA (1975) Melioidosis: recrudescence associated with bronchogenic carcinoma twenty-six years following initial geographic exposure. Chest 68:261–263. [DOI] [PubMed] [Google Scholar]
- 17. Ngauy V, Lemeshev Y, Sadkowski L, Crawford G (2005) Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II. J Clin Microbiol 43:970–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, et al. (2009) Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol 7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holden MT, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, et al. (2004) Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei . Proc Natl Acad Sci USA 101:14240–14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tumapa S, Holden MT, Vesaratchavest M, Wuthiekanun V, Limmathurotsakul D, et al. (2008) Burkholderia pseudomallei genome plasticity associated with genomic island variation. BMC Genomics 9:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tuanyok A, Auerbach R, Brettin T, Bruce D, Munk A, et al. (2007) A horizontal gene transfer event defines two distinct groups within Burkholderia pseudomallei that have dissimilar geographic distributions. J Bacteriol 189:9044–9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarovich DS, Price EP, Webb JR, Ward LM, Voutsinos MY, et al. (2014) Variable Virulence Factors in Burkholderia pseudomallei (Melioidosis) Associated with Human Disease. PLoS One 9:e91682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng AC, Godoy D, Mayo M, Gal D, Spratt BG, et al. (2004) Isolates of Burkholderia pseudomallei from Northern Australia are distinct by multilocus sequence typing, but strain types do not correlate with clinical presentation. J Clin Microbiol 42:5477–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sim SH, Yu Y, Lin CH, Karuturi RK, Wuthiekanun V, et al. (2008) The core and accessory genomes of Burkholderia pseudomallei: implications for human melioidosis. PLoS Pathog 4:e1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ulett GC, Ketheesan N, Hirst RG (2000) Cytokine gene expression in innately susceptible BALB/c mice and relatively resistant C57BL/6 mice during infection with virulent Burkholderia pseudomallei . Infection and immunity 68:2034–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, et al. (2006) The Integrated Microbial Genomes (IMG) system. Nucleic Acids Res 34:D344–D348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karp PD, Paley S, Romero P (2002) The Pathway Tools software. Bioinformatics 18 Suppl 1: S225–S232. [DOI] [PubMed] [Google Scholar]
- 29. Lerat E, Ochman H (2004) Psi-Phi: exploring the outer limits of bacterial pseudogenes. Genome Res 14:2273–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Darling AC. B M, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, et al. (2010) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 38(Database issue): D473–479. [DOI] [PMC free article] [PubMed]
- 32. DeShazer D, Brett PJ, Woods DE (1998) The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol Microbiol 30:1081–1100. [DOI] [PubMed] [Google Scholar]
- 33. Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- 35. Reznikoff WS, Siegele DA, Cowing DW. Gross CA (1985) The regulation of transcription initiation in bacteria. Annu Rev Genet 19:355–387. [DOI] [PubMed] [Google Scholar]
- 36. Potvin E, Sanschagrin F, Levesque RC (2008) Sigma factors in Pseudomonas aeruginosa . FEMS Microbiol Rev 32:38–55. [DOI] [PubMed] [Google Scholar]
- 37.Brinkac LM, Davidsen T, Beck E, Ganapathy A, Caler E, et al. (2010) Pathema: a clade-specific bioinformatics resource center for pathogen research. Nucleic Acids Res 38(Database issue): D408–414. [DOI] [PMC free article] [PubMed]
- 38. The UniProt Consortium (2014) Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res 42:D191–D198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiang Z, Tian Y, He Y (2007) PHIDIAS: a pathogen-host interaction data integration and analysis system. Genome Biol 8:R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Galyov E, Brett P, DeShazer D (2010) Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu Rev Microbiol 64:495–517. [DOI] [PubMed] [Google Scholar]
- 41. Eisenreich W, Dandekar T, Heesemann J, Goebel W (2010) Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol 8:401–412. [DOI] [PubMed] [Google Scholar]
- 42. Abu Kwaik Y, Bumann D (2013) Microbial quest for food in vivo: ‘nutritional virulence’ as an emerging paradigm. Cell Microbiol 15:882–890. [DOI] [PubMed] [Google Scholar]
- 43. Zhang YJ, Rubin EJ (2013) Feast or famine: the host-pathogen battle over amino acids. Cell Microbiol 15:1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fuchs TM, Eisenreich W, Heesemann J, Goebel W (2012) Metabolic adaptation of human pathogenic and related nonpathogenic bacteria to extra- and intracellular habitats. FEMS Microbiol Rev 36:435–462. [DOI] [PubMed] [Google Scholar]
- 45. Barbier T, Nicolas C, Letesson JJ (2011) Brucella adaptation and survival at the crossroad of metabolism and virulence. FEBS Lett 585:2929–2934. [DOI] [PubMed] [Google Scholar]
- 46. Ooi W, Ong C, Nandi T, Kreisberg J, Chua H, et al. (2013) The Condition-Dependent Transcriptional Landscape of Burkholderia pseudomallei . PLoS Genet 9:e1003795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fujita Y (2009) Carbon catabolite control of the metabolic network in Bacillus subtilis . Biosci Biotechnol Biochem 73:245–259. [DOI] [PubMed] [Google Scholar]
- 48. Deutscher J, Francke C, Postma PW. (2006) How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mao XJ, Huo YX, Buck M, Kolb A, Wang YP (2007) Interplay between CRP-cAMP and PII-Ntr systems forms novel regulatory network between carbon metabolism and nitrogen assimilation in Escherichia coli . Nucleic Acids Res 35:1432–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Atkinson MR, Wray LVJ, Fisher SH (1990) Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis . J Bacteriol 172:4758–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fisher SH (1999) Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence. Mol Microbiol 32:223–232. [DOI] [PubMed] [Google Scholar]
- 52. Wongtrakoongate P, Tumapa S, Tungpradabkul S (2012) Regulation of a quorum sensing system by stationary phase sigma factor RpoS and their co-regulation of target genes in Burkholderia pseudomallei . Microbiol Immunol 56:281–294. [DOI] [PubMed] [Google Scholar]
- 53. Fang F, Libby SJ, Buchmeier NA, Loewen PC, Switala J, et al. (1992) The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA 89:11978–11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lengwehasatit I, Nuchtas A, Tungpradabkul S, Sirisinha S, Utaisincharoen P (2008) Involvement of B. pseudomallei RpoS in apoptotic cell death in mouse macrophages. Microb Pathog 44:238–245. [DOI] [PubMed] [Google Scholar]
- 55. Utaisincharoen P, Arjcharoen S, Limposuwan K, Tungpradabkul S, Sirisinha S (2006) Burkholderia pseudomallei RpoS regulates multinucleated giant cell formation and inducible nitric oxide synthase expression in mouse macrophage cell line (RAW 264.7). Microb Pathog 40:184–189. [DOI] [PubMed] [Google Scholar]
- 56. Loprasert S, Sallabhan R, Whangsuk W, Mongkolsuk S (2000) Characterization and mutagenesis of fur gene from Burkholderia pseudomallei . Gene 254:129–137. [DOI] [PubMed] [Google Scholar]
- 57. Chieng S, Carreto L, Nathan S (2012) Burkholderia pseudomallei transcriptional adaptation in macrophages. BMC Genomics 13:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chin CY, Monack DM, Nathan S (2010) Genome wide transcriptome profiling of a murine acute melioidosis model reveals new insights into how Burkholderia pseudomallei overcomes host innate immunity. BMC Genomics 11:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rodrigues F, Sarkar-Tyson M, Harding SV, Sim SH, Chua HH, et al. (2006) Global map of growth-regulated gene expression in Burkholderia pseudomallei, the causative agent of melioidosis. J Bacteriol 188:8178–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tuanyok A, Tom M, Dunbar J, Woods DE (2006) Genome-wide expression analysis of Burkholderia pseudomallei infection in a hamster model of acute melioidosis. Infect Immun 74:5465–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Velapatiño B, Limmathurotsakul D, Peacock SJ, Speert DP (2012) Identification of differentially expressed proteins from Burkholderia pseudomallei isolated during primary and relapsing melioidosis. Microbes Infect 14:335–340. [DOI] [PubMed] [Google Scholar]
- 62. Yang J, Tauschek M, Robins-Browne RM (2011) Control of bacterial virulence by AraC-like regulators that respond to chemical signals. Trends Microbiol 19:128–135. [DOI] [PubMed] [Google Scholar]
- 63. Berg G, Eberl L, Hartmann A (2005) The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol 7:1673–1685. [DOI] [PubMed] [Google Scholar]
- 64. Kaestli M, Schmid M, Mayo M, Rothballer M, Harrington G, et al. (2012) Out of the ground: aerial and exotic habitats of the melioidosis bacterium Burkholderia pseudomallei in grasses in Australia. Environ Microbiol 14:2058–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663. [DOI] [PubMed] [Google Scholar]
- 67. Trung TT, Hetzer A, Göhler A, Topfstedt E, Wuthiekanun V, et al. (2011) Highly sensitive direct detection and quantification of Burkholderia pseudomallei bacteria in environmental soil samples by using real-time PCR. Appl Environ Microbiol 77:6486–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Suebrasri T, Wang-ngarm S, Chareonsudjai P, Sermswan RW, Chareonsudjai S (2013) Seasonal variation of soil environmental characteristics affect the presence of Burkholderia pseudomallei in Khon Kaen, Thailand. African J Microbiol Res 7:1940–1945. [Google Scholar]
- 69. Lerat E, Ochman H (2005) Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res 33:3125–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Aziz RK, Devoid S, Disz T, Edwards RA, Henry CS, et al. (2012) SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS One 7:e48053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Novichkov PS, Laikova ON, Novichkova ES, Gelfand MS, Arkin AP, et al. (2010) RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res 38:D111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Walker DR, Koonin EV (1997) A system for easy analysis of lots of sequences. Intell Syst Mol Biol 5:333–339. [PubMed] [Google Scholar]
- 73.Chen L, Xiong Z, Sun L, Yang J, Jin Q (2012) VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res 40(Database issue): D641–645. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of mobile element genes in B. pseudomallei genomes.
(DOCX)
Metabolic Pathway Comparison among B. pseudomallei genomes.
(DOCX)
Transcriptional regulatory genes in B. pseudomallei genomes.
(DOCX)
Sigma factor genes in B. pseudomallei genomes.
(DOCX)
RAST-annotated MSHR668 genome in GenBank file format.
(ZIP)
RAST-annotated 1106a genome in GenBank file format.
(ZIP)
RAST-annotated K96243 genome in GenBank file format.
(ZIP)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.