Abstract
Autophagy is a lysosomal degradation pathway that acts as a dynamic regulator of tumorigenesis. Specifically, autophagy has been shown to impede early cancer development while facilitating advanced tumor progression. Recent studies have uncovered several tumor-promoting functions for autophagy; these include the maintenance of multiple metabolic pathways critical for aggressive tumor growth and the promotion of tumor cell survival downstream of the unfolded protein response. Furthermore, autophagy supports anoikis resistance and cancer cell invasion. At the same time, because autophagy cargo receptors, which are essential for selective autophagy, lie upstream of diverse cancer-promoting signaling pathways, they may profoundly influence how alterations in autophagy affect tumor development. This review focuses on how these tumor cell autonomous functions of autophagy broadly impact tumorigenesis.
Keywords: autophagy, metabolism, unfolded protein response, anoikis, invasion, cancer
Overview of autophagy and tumorigenesis
Macroautophagy (hereafter called autophagy) is an evolutionarily conserved pathway of lysosomal-mediated cellular self-digestion. It involves the formation of a double-membrane vesicle, the autophagosome, which engulfs cytoplasmic components and delivers them to the lysosome for degradation (Box 1). Landmark studies in yeast have identified over 30 autophagy-related genes (atgs); in response to stress and starvation, numerous signaling pathways impinge on these ATGs to induce autophagy. The resulting lysosomal digestion and recycling of cellular contents is proposed to refuel cells with metabolic building blocks that are critical for survival during stress [1–3]. Additionally, during normal cellular homeostasis, autophagy functions as a primary route of degradation for damaged organelles and protein aggregates [4]. Because of these conserved functions in eukaryotic cells, autophagy has been proposed to act as a crucial cellular adaptation pathway that promotes tumorigenesis by facilitating the survival of cancer cells under duress [5–7].
Box 1. Autophagosome formation and maturation in mammals.
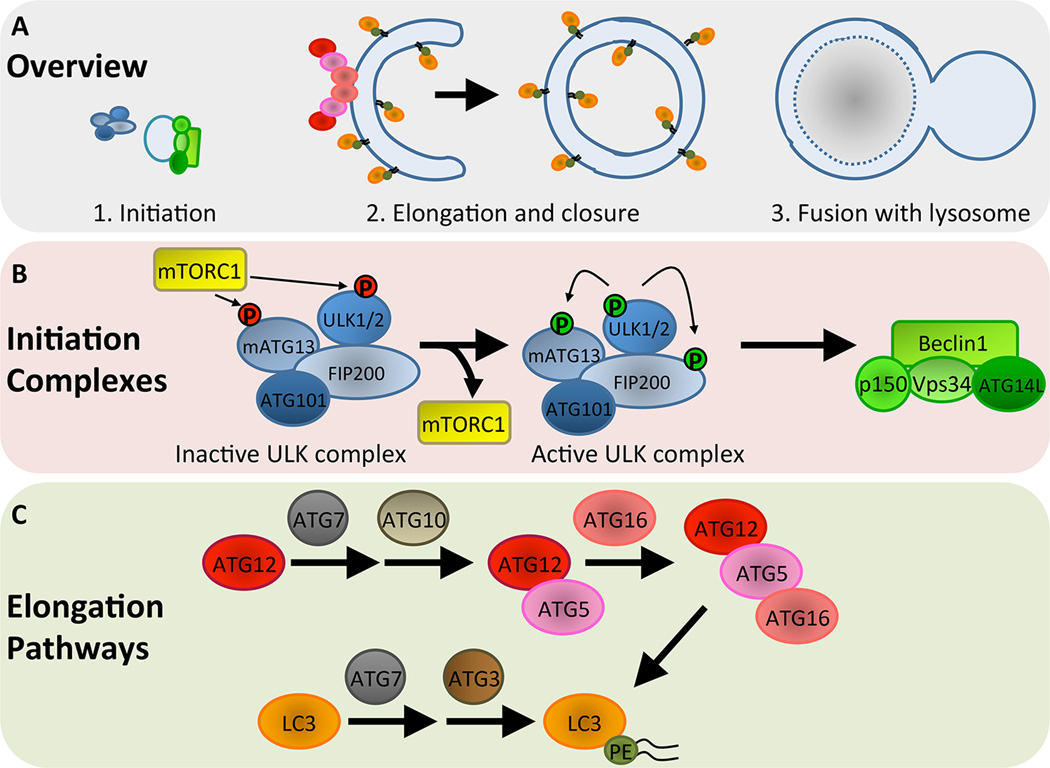
Autophagosome formation and maturation is a highly regulated process that occurs through a series of distinct steps controlled by ATGs (Fig IA). Initiation of autophagosome formation is regulated by the unc-51-like kinase (ULK) and class III phosphatidylinositol (PI3K) complexes (Fig IB). The ULK complex consists of ULK1/2, which are the mammalian orthologues of ATG1, as well as mATG13, FIP200 (ATG17), and ATG101 [82]. Under nutrient-rich conditions, the ability of the ULK complex to initiate autophagy is inhibited by mTORC1, which phosphorylates and inactivates ULK1/2 [83]. Upon starvation, mTORC1 activity is suppressed, leading to disassociation from and activation of the ULK complex. The class III PI3K complex, consisting of the lipid kinase Vps34, Beclin1 (ATG6), ATG14L, and p150, is then activated by the ULK complex [84]. The PI3K complex functions to generate phosphatidylinositol 3-phosphate (PI3P) at the site of early autophagosome formation for recruitment of additional ATGs that will subsequently mediate elongation and closure of the autophagosome membrane.
Elongation and closure is controlled by two ubiquitin-like conjugation pathways that conjugate ATG12 to ATG5 and LC3 to the lipid phosphatidylethanolamine (PE) [85] (Fig IC). Conjugation of ATG12 to ATG5 is regulated by the E1- and E2-like activities of ATG7 and ATG10, respectively. The ATG12-ATG5 complex then associates with ATG16 to form a multimeric complex that localizes to the outer surface of the autophagosomal membrane. LC3 is conjugated to PE by ATG7 and the E2-like enzyme ATG3, and the ATG12-ATG5-ATG16 complex functions in an E3-like fashion to promote LC3 lipidation by PE. PE is inserted into the autophagosome membrane, and LC3-PE is localized to both the inner and outer membranes. Importantly, these core ATGs that directly control elongation of the autophagosome membrane are commonly targeted for experimental purposes, either by genetic deletion or RNAi-mediated depletion, to conduct functional studies of autophagy during tumorigenesis. Additionally, LC3-PE (also termed LC3-II) is commonly used as a marker of autophagosomes to monitor the induction or inhibition of autophagy [86].
Ultimately, the autophagosome fuses with endocytic and lysosomal compartments, leading to formation of the autolysosome (Fig IA). Autophagic cargo is then degraded through the activity of lysosomal proteases. The mechanisms underlying these late stage maturation steps are only beginning to emerge, but studies aimed at identifying essential regulators of autolysosome formation have unveiled roles for common mediators of cellular membrane fusion, including Rab and SNARE proteins [87–88]. Further elucidation of genes involved in these late stages will facilitate more comprehensive functional analyses of the autophagy pathway in cancer.
Nonetheless, over the last decade, studies of how autophagy impacts cancer development have yielded conflicting results. Initial studies demonstrating that loss of the essential autophagy regulator beclin1 (atg6) results in increased tumorigenesis in mice provided genetic evidence that autophagy serves tumor suppressive functions [8–9]. Additionally, BECN1 was proposed to be a haploinsufficient tumor suppressor in humans, but a recent analysis of human cancer sequencing data questions these original findings [10–12]. Further support for a role for autophagy in limiting tumorigenesis came from elegant studies in which the deletion of atg5 or atg7 led to spontaneous development of premalignant liver tumors due to accumulation of oxidative stress and activation of genome damage responses [13–14]. However, as tumor progression requires cancer cells to thrive in untoward environments, tumor-supporting functions for autophagy have also been uncovered [7]. Now, we appreciate that autophagy serves dual roles during tumorigenesis; its homeostatic function limits genome-damaging events that would otherwise favor tumor initiation, while its ability to help cells mitigate stress facilitates advanced tumor progression [5–6]. Importantly, studies in genetically engineered mouse models (GEMMs) of cancer have provided additional support for these opposing functions of autophagy; during oncogene activation, genetic deletion of ATGs enhances early tumor development but impairs advanced tumorigenesis [15–18] (Table 1). Additionally, novel insight into how autophagy controls tumor cell fate and regulates cell phenotypes other than growth and survival has emerged. Here, we focus on how GEMMs have improved our understanding of how autophagy controls tumor cell metabolism and cell survival as well as highlight new cell biological functions for autophagy in tumor cells during cancer progression.
Table 1.
Effects of atg deletion on cancer progression and metabolism in GEMMs of cancer
| GEMM | Atg deletiona | Phenotype upon autophagy inhibition | Ref | ||
|---|---|---|---|---|---|
| Cancer type | Genotype | Tumor progressionb | Tumor cell metabolismc |
||
| Mammary carcinoma |
MMTV-PyMT |
FIP200 (MMTV-Cre) |
Decreased initiation and progression |
Impaired glycolysis | [21] |
| Non-small cell lung cancer |
lox-stop-lox-KrasG12D; Tp53flox/flox |
Atg7 (intranasal adenoviral Cre) |
Decreased progression, oncocytoma formation |
Impaired mitochondrial metabolism and fatty acid oxidation, lipid accumulation |
[24] |
| Non-small cell lung cancer |
lox-stop-lox-KrasG12D |
Atg5 (intranasal adenoviral Cre) |
Increased initiation, decreased progression, oncocytoma formation |
Impaired mitochondrial metabolism |
[15] |
| Non-small cell lung cancer |
frt-stop-frt-KrasG12D; Tp53frt/frt |
Atg7 (Ubc-CreERT2) |
Decreased progression, oncocytoma formation |
Lipid accumulation | [25] |
| Non-small cell lung cancer |
BrafV600E; Tp53flox/flox |
Atg7 (intranasal adenoviral Cre) |
Increased initiation, decreased progression, oncocytoma formation |
Impaired mitochondrial glutamine metabolism |
[18] |
| Pancreatic cancer | lox-stop-lox-KrasG12D; Pdx-cre |
Atg5 or Atg7 (Pdx-Cre) |
Increased initiation, decreased progression |
ND | [16] |
| Pancreatic cancer |
lox-stop-lox-KrasG12D; Tp53flox/flox; Pdx-cre |
Atg5 or Atg7 (Pdx-Cre) |
Increased progression | Increased glycolysis | [16] |
| Pancreatic cancer |
lox-stop-lox-KrasG12D; Tp53flox/+; Pdx-cre |
Atg5 (Pdx-Cre) |
Increased initiation, decreased progression |
ND | [17] |
Method of cre-recombinase mediated deletion is indicated in parentheses. MMTV-Cre expression is mammary epithelial cell specific, Ubc-CreERT2 expression is ubiquitous and tamoxifen-inducible, and Pdx-Cre is exocrine and endocrine pancreatic specific.
Effects on tumor initiation are related to tumor onset and development of early stage tumors. Effects on progression are related to advanced tumorigenesis.
ND indicates metabolic phenotype was not determined in the context of atg deletion.
Control of tumor cell metabolism by autophagy
Rapidly proliferating tumor cells have increased anabolic demands, which are met by metabolic changes induced upon activation of oncogenes and loss of tumor suppressors [19]. At its most fundamental level, autophagy couples catabolic breakdown of cellular content with anabolic pathways of macromolecule synthesis by supplying the cell with intracellular metabolites generated via lysosomal-mediated degradation. Despite this salient feature of autophagy, its importance in tumor cell metabolism was not appreciated until recently.
Studies of oncogenic Ras-transformation were the first to demonstrate a role for autophagy in supporting tumor cell proliferation and in maintaining metabolic function in the context of oncogene activation. In mouse embryonic fibroblasts (MEFs) transformed with oncogenic HRas and MDA-MB-231 human breast carcinoma cells, which harbor oncogenic KRas, genetic autophagy inhibition reduced anchorage-independent transformation, slowed proliferation, and decreased glycolysis [20]. Similar results were obtained in a transgenic model of breast tumorigenesis driven by the polyoma middle T (PyMT) oncogene; deletion of FIP200, which is essential for autophagy initiation, impaired glycolysis in these tumor cells in vitro and reduced mammary tumorigenesis in vivo [21]. An additional requirement for autophagy in cancer cell metabolism was subsequently shown using HRas-transformed immortalized baby mouse kidney (iBMK) cells and pancreatic ductal adenocarcinoma (PDAC) cell lines with activated Ras [22–23]. Remarkably, these studies described an increase in autophagy with oncogenic activation of Ras, suggesting that sustained autophagy allows Ras-transformed tumor cells to meet their high metabolic demands. Accordingly, inhibiting autophagy in these models led to multiple defects in mitochondrial metabolism, including decreased production of TCA cycle intermediates, reduced mitochondrial respiration, and diminished ATP production. Although these various studies of Ras-transformation uncovered different requirements for autophagy during glycolysis versus mitochondrial metabolism, collectively, they demonstrated that autophagy is important for supporting the diverse metabolic demands of different tumor types.
The role of autophagy in sustaining Ras-regulated metabolism has also been explored in lung and pancreatic cancer GEMMs driven by oncogenic KRas or the Ras effector, Braf (Table 1). Deletion of atg5 or atg7 in an oncogenic KRas-induced lung cancer model led to diminished overall lung tumor burden; notably, autophagy-deficient tumors exhibited oncocytic differentiation, marked by the accumulation of abnormal mitochondria within tumor cells [15, 24–25]. In the absence of the tumor suppressor p53, this loss of mitochondrial homeostasis resulted in defective fatty acid oxidation and, consequently, impaired lipid metabolism when atg7 was deleted [24]. Based on these results, the authors concluded that reduced lipid catabolism compromises the ability of autophagy-deficient tumor cells to cope with nutrient deprivation. In a Braf-driven lung cancer model, advanced tumor progression was similarly reduced by atg7 deletion [18]. Autophagy deficient cell lines derived from these tumors harbored aberrant mitochondria and addition of the metabolite glutamine rescued defects in mitochondrial metabolism, suggesting that autophagy-inhibited tumor cells exhibit slowed growth due to increased metabolic stress associated with a lack of intermediates that drive mitochondrial metabolic pathways. Defects in lipolysis were not observed here as in the KRas lung model, but these studies nonetheless corroborated the importance of autophagy in regulating metabolic homeostasis by broadly controlling proper mitochondrial function. Furthermore, autophagy inhibition also reduced growth and survival of central nervous system tumor cells with activated Braf. While the impact of autophagy on metabolism was not investigated here, the results obtained in the Braf lung model suggest an underlying mechanism by which autophagy may impact Braf-driven growth across multiple tumor types [26].
Studies of pancreatic cancer GEMMs driven by mutant KRas have revealed a seemingly complex and varying role for autophagy in controlling tumor cell metabolism. In the context of embryonic p53 deletion in the pancreas, genetic and pharmacological inhibition of autophagy actually accelerates pancreatic tumor progression [16]. Cells isolated from these tumors lacking both ATG7 and p53 exhibited increased rates of glycolysis and increased levels of metabolites in the pentose phosphate pathway, a key side branch of glucose metabolism that facilitates tumor growth. In contrast, when p53 inactivation occurred by somatic loss of heterozygosity (LOH), autophagy inhibition resulted in impaired PDAC progression, and pharmacological targeting of autophagy using the lysosomal inhibitor chloroquine led to defects in mitochondrial respiration across a panel of human PDAC cell lines, regardless of p53 status [17].
Much remains to be learned with regard to the precise mechanisms through which autophagy controls metabolism. While specific enzymes involved in glycolysis have been shown to regulate autophagy, no such regulation by autophagy on particular steps of the glycolytic pathway has been uncovered [27–28]. Additionally, although accumulation of abnormal mitochondria due to decreased mitophagy may explain the defects associated with mitochondrial metabolism upon autophagy inhibition, impaired mitophagy was not observed in autophagy-deficient PDAC cell lines that exhibited diminished oxidative phosphorylation [23]. This discrepancy suggests that there are mitophagy-independent pathways through which autophagy controls mitochondrial metabolism and that engagement of these various regulatory mechanisms may be context-dependent. Moreover, in most studies, these metabolic defects have been characterized using tumor cell lines in culture. While this method certainly facilitates a detailed analysis of metabolic parameters, it may not accurately recapitulate the metabolic state of tumors in vivo. In vivo application of established NMR-based technologies to assay glycolysis during tumor formation or use of methods to measure metabolism in freshly isolated mitochondria from tumors will provide further insight into the role of autophagy in cancer metabolism [29–30].
Autophagy and the unfolded protein response in cancer
In addition to being critical for metabolic adaptation, autophagy has other functions in helping tumors cope with oncogene, environmental, and therapy-induced stresses, particularly during induction of the unfolded protein response (UPR) (Fig 1). The UPR is a cytoprotective pathway that alleviates stress associated with accumulation of misfolded proteins in the endoplasmic reticulum (ER) [31]. It is regulated by three sensors, including the ER kinase, PERK, which phosphorylates the translation regulatory protein, eIF2α, ultimately leading to a block in translation to prevent further accumulation of unfolded proteins. Recently, oncogenic activation of c-Myc, which promotes increased translation, was shown to activate the UPR to accommodate this increase in protein synthesis [32]. Knockout of PERK led to cell death in the context of activated c-Myc, and this cytoprotective function was due to PERK-mediated activation of autophagy. Similar results were obtained in a Drosophila model of Myc overexpression in which induction of the UPR and PERK led to an autophagy-dependent increase in cell growth [33].
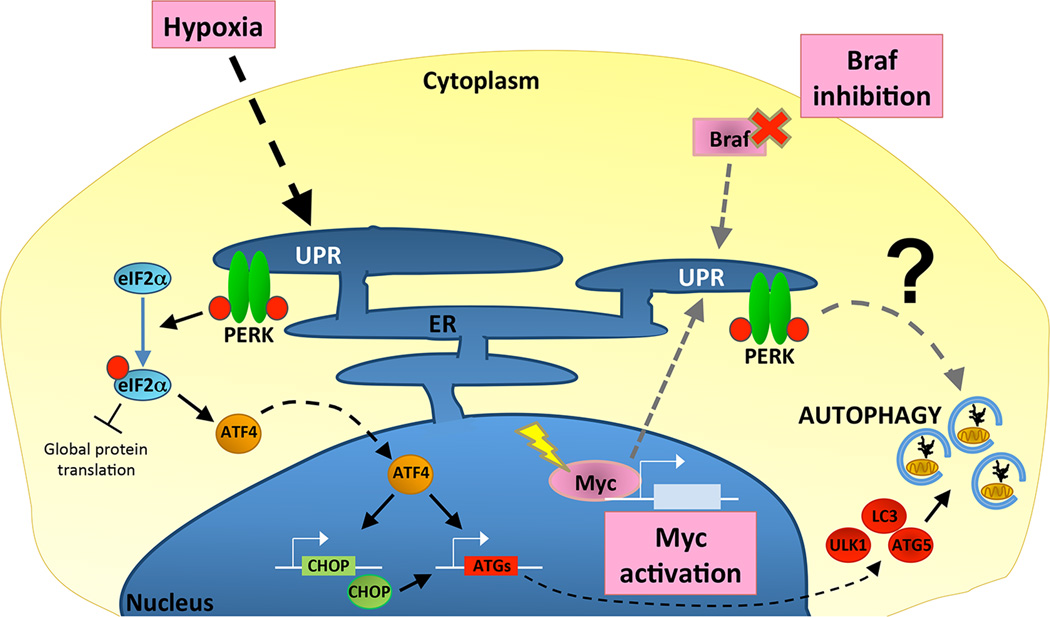
Figure 1. UPR-mediated induction of cytoprotective autophagy supports tumor cell survival and growth.
Multiple stresses promote the unfolded protein response (UPR) in tumor cells, such as hypoxia, activation of the oncogenic transcription factor Myc, and inhibition of the growth-promoting kinase Braf. In response to these stimuli, UPR-dependent activation of the ER-resident kinase, PERK has been most strongly implicated in the induction of autophagy as a cytoprotective pathway. Hypoxia activates PERK, which phosphorylates eIF2α to suppress global translation and to selectively induce ATF4. ATF4 is a transcription factor that promotes expression of the transcription factor CHOP. Together, ATF4 and CHOP drive expression of multiple core autophagy machinery genes, including ULK1, MAP1LC3B (LC3), and ATG5, which collectively promote autophagosome formation (see Box 1). Although the mechanisms by which PERK induces autophagy during Myc activation and Braf inhibition have not been determined, activation of ATF4 and CHOP downstream of PERK may similarly induce the transcription of core autophagy regulators.
While the mechanism of autophagy induction by the UPR during Myc-transformation remains unclear, hypoxia can also induce UPR-dependent upregulation of autophagy downstream of PERK via transcriptional mechanisms [34–35]. During hypoxia-induced UPR, increased expression of the transcription factors ATF4 and CHOP leads to enhanced expression of ULK1, which is required for initiation of autophagy, and MAP1LC3B and ATG5, which are both essential for autophagosome formation. Increased expression of essential ATGs downstream of the UPR is also seen during other stresses, such as extracellular-matrix (ECM) detachment, suggesting transcriptional upregulation of ATGs may be a general route of autophagy upregulation by the UPR [36]. Moreover, because hypoxia occurs in multiple tumor types, this mechanism of cytoprotective autophagy induction may be common across many cancers. Similarly, inhibition of Braf by targeted therapy in melanoma activates autophagy downstream of PERK, and this induction mediates resistance of tumor cells to Braf inhibitors [37]. Thus, like hypoxia, activation of autophagy by therapy-induced UPR and PERK may be another route of cytoprotective autophagy induction in various cancers.
While autophagy may support tumor cells during hypoxia by diverse mechanisms, one can speculate that the ability of autophagy to facilitate glycolysis and supply metabolites, as has been observed during Ras transformation, may also be critical for metabolic adaption to oxygen deprivation. A similar requirement for autophagy during Myc-induced metabolic changes may also exist, since Myc transformation has been associated with enhanced glycolysis and glutamine metabolism [38]. Overall, an important outcome of UPR-mediated activation of autophagy may be to sustain tumor cell metabolism. Future studies interrogating connections between the various requirements for autophagy during diverse stresses and in the context of different oncogenes may uncover conserved mechanisms for control of tumorigenesis by autophagy.
Regulation of cellular invasion and metastasis by autophagy
Metastasis, the process by which tumor cells spread to foreign sites throughout the body, involves phenotypic changes that allow tumor cells to gain entry into and out of the vasculature and to survive stresses associated with traversing the circulation and growing in a foreign microenvironment [39–40]. In many cancer patients, metastasis is the primary cause of mortality, primarily because limited treatments for metastatic disease exist. Interestingly, autophagy impacts cell biological phenotypes that regulate metastasis, such as resistance to anoikis and invasion (Fig. 2). Resistance to anoikis allows cells to survive stress associated with ECM detachment, which may occur while tumor cells are in the circulation or at the metastatic site where they cannot fully engage the foreign ECM [41]. Invasion allows tumor cells to access the vasculature for dissemination and to exit the circulation at metastatic sites [42–43].
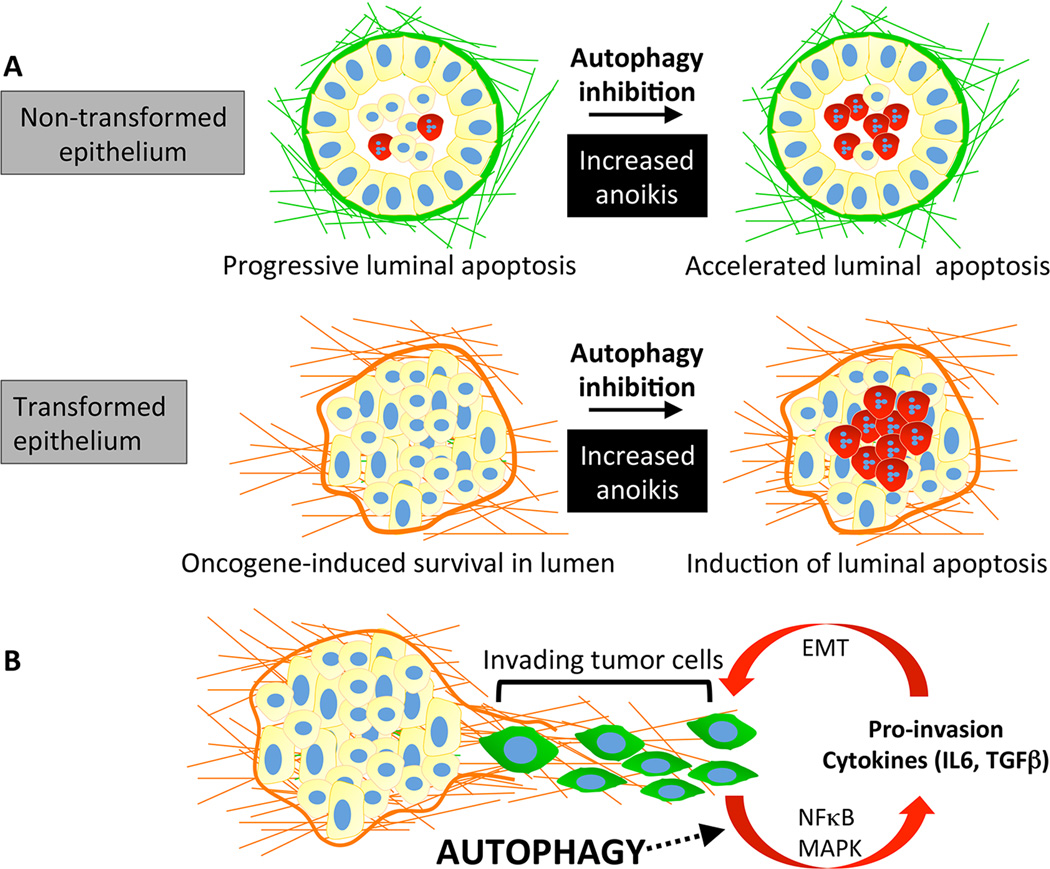
Figure 2. Autophagy promotes anoikis resistance and tumor cell invasion.
(A) Three-dimensional (3D) culture of MCF10A cells leads to the formation of acini with hollow lumens. Luminal clearance occurs through anoikis of central cells (depicted in red) lacking extracellular matrix (ECM) contact. Oncogene activation protects luminal cells from anoikis, leading to the formation of structures with filled lumens. Autophagy promotes the survival of both normal and transformed epithelial cells deprived of ECM-contact; therefore, inhibiting autophagy leads to increased anoikis. (B) Autophagy promotes tumor cell invasion by facilitating the secretion of multiple pro-invasive cytokines. Activation of the NFκB and MAPK pathways by autophagy has been shown to contribute to the increased production of these secreted factors. In turn, these cytokines may augment a pro-invasive gene signature program through the induction of epithelial-to-mesenchymal transition (EMT).
Autophagy was first shown to promote the survival of non-transformed mammary epithelial cells during ECM detachment; subsequent studies revealed that detachment-induced autophagy is critical for adhesion-independent transformation [20, 44]. Multiple Ras-transformed human cancer cell lines upregulate autophagy upon detachment, and autophagy inhibition compromises adhesion-independent growth and survival of cells harboring activated Ras. Similarly, when oncogenic PI3K-transformed MCF10A cells were grown in three-dimensional (3D) culture, autophagy inhibition led to increased apoptosis of luminal cells deprived of ECM contact [45]. Recently, this requirement for autophagy during anoikis resistance was shown to be necessary for metastasis of hepatocellular carcinoma (HCC) cells [46]. Autophagy inhibition attenuated pulmonary metastasis of HCC cells following orthotopic transplantation into nude mice, and this defect correlated with increased anoikis of autophagy deficient HCC cells.
Autophagy has also emerged as a regulator of cellular invasion and migration. In an organotypic model of invasion through a collagen matrix, knockdown of the essential autophagy regulator, ATG12, decreased invasive capacity of glioma cells [47]. Although this study did not delineate the mechanism of autophagy-mediated invasion, other studies have demonstrated multiple routes by which autophagy controls invasion. For example, in glioblastoma (GBM) stem cells, autophagy inhibition or knockdown of the autophagy regulator, p62, decreased invasion and migration in vitro and led to metabolic defects [48]. Based on previous evidence indicating that glycolysis is important for GBM invasion, the authors proposed a model in which p62-dependent autophagy impacts metabolism to control invasion [49].
Further roles for autophagy in regulating invasion have been uncovered in other models. Invasion of HCC cells during starvation was shown to be autophagy-dependent, due to the ability of autophagy to stimulate TGFβ and promote epithelial-to-mesenchymal transition (EMT), a well established transcriptional program that supports metastasis [50]. A similar requirement for autophagy in controlling invasion was observed in Ras-transformed epithelial cells in 3D culture [51]. Autophagy inhibition attenuated invasion and caused a partial reversion of EMT. Additionally, impaired self-eating led to decreased secretion of multiple pro-invasive cytokines, including interleukin-6 (IL6). Notably, decreased invasion upon autophagy inhibition was partly restored with IL6 re-addition, demonstrating a specific need for the autophagy pathway in controlling secretion of this cytokine. Furthermore, autophagy-deficient Ras-transformed cells exhibited reduced pulmonary metastases. Overall, these findings uncovered a new role for autophagy during cancer cell invasion by promoting secretion and suggested that autophagy-dependent secretion may be important for metastasis in vivo. An additional report showed that induction of autophagy by toll-like receptors (TLRs) promotes secretion of pro-invasive factors, including IL6, in lung cancer cells, further corroborating a role for autophagy as a determinant of pro-invasive secretion [52].
The mechanism by which autophagy controls secretion during invasion remains poorly defined. Although these phenotypes may be secondary to autophagic turnover of secretory regulators, autophagy has been directly implicated in promoting both conventional and unconventional secretion in other contexts [53]. During TLR mediated invasion, autophagy upregulates signaling pathways, such as NFκB and MAPK, that promote secretion, but how autophagy controls these pathways in this model is unclear [52]. Furthermore, IL6 can promote EMT and stimulate TGFβ signaling, which argues that autophagy-dependent secretion may also be important for HCC cell invasion [54].
Lastly, in contrast to the studies above, autophagy has been described as a suppressor of metastasis by preventing p62-dependent stabilization of the EMT-promoting transcription factor, Twist1 [55]. Because EMT is regulated by multiple signaling pathways and transcription factors, these varying roles of autophagy in regulating EMT and invasion point to a complex relationship between self-eating and metastasis [56]. Hence, additional work is needed to establish the cellular functions for autophagy during in vivo cancer invasion and metastasis.
Regulation of tumorigenesis by autophagy cargo receptors
Autophagy cargo receptors mediate selective degradation of autophagy substrates by binding ubiquitinated targets and recruiting autophagosomes to this cargo. This recruitment generally occurs through interaction of receptors with LC3 via an LC3 interacting region (LIR), but the recent identification of the ferritin receptor nuclear receptor coactivator 4 (NCOA4), which lacks a canonical LIR, points to additional mechanisms by which cargo receptors interact with autophagosomes [57–58]. Importantly, because these receptors are themselves degraded during selective autophagy, inhibition of autophagy promotes their accumulation and results in aberrant regulation of their downstream pathways. This may have crucial implications for tumorigenesis, as illustrated by studies demonstrating tumor-promoting functions for the archetypal autophagy cargo receptor p62/SQSTM1 [59–60].
p62/SQSTM1 is a versatile, multi-domain adapter that regulates several signaling pathways to promote tumorigenesis [59–60] (Fig. 3). Among these, NFκB-mediated control of pro-inflammatory signaling and regulation of the antioxidant response by Nrf2 have been most strongly linked to tumorigenesis in the context of autophagy inhibition. Through interaction with TRAF6, p62 promotes NFκB signaling, and this p62-mediated activation of NFκB is required for Ras-induced lung and pancreatic tumorigenesis [60–62]. Indeed, atg5 knockout or overexpression of p62 in tumorigenic iBMK cells enhances tumor growth by modulating NFκB signaling [63]. Additionally, p62 regulates tumorigenesis of transformed MEFs during autophagy inhibition resulting from knockout of FIP200 [64]. When p62 is depleted in FIP200 null tumor cells, tumor growth is inhibited. Conversely, when p62 is re-expressed in p62 knockout cells also deleted for FIP200, tumor growth is enhanced and NFκB signaling is upregulated. Similarly, regulation of the transcription factor Nrf2 by p62 is also important for tumorigenesis. p62 inhibits degradation of Nrf2, a key regulator of the oxidative stress response, by binding to Keap1, an adaptor for the E3 ubiquitin ligase that promotes Nrf2 degradation [65]. The ability of p62 to support tumor growth by activating Nrf2 is crucial for the spontaneous development of liver tumors due to atg5 or atg7 knockout [13–14, 66].
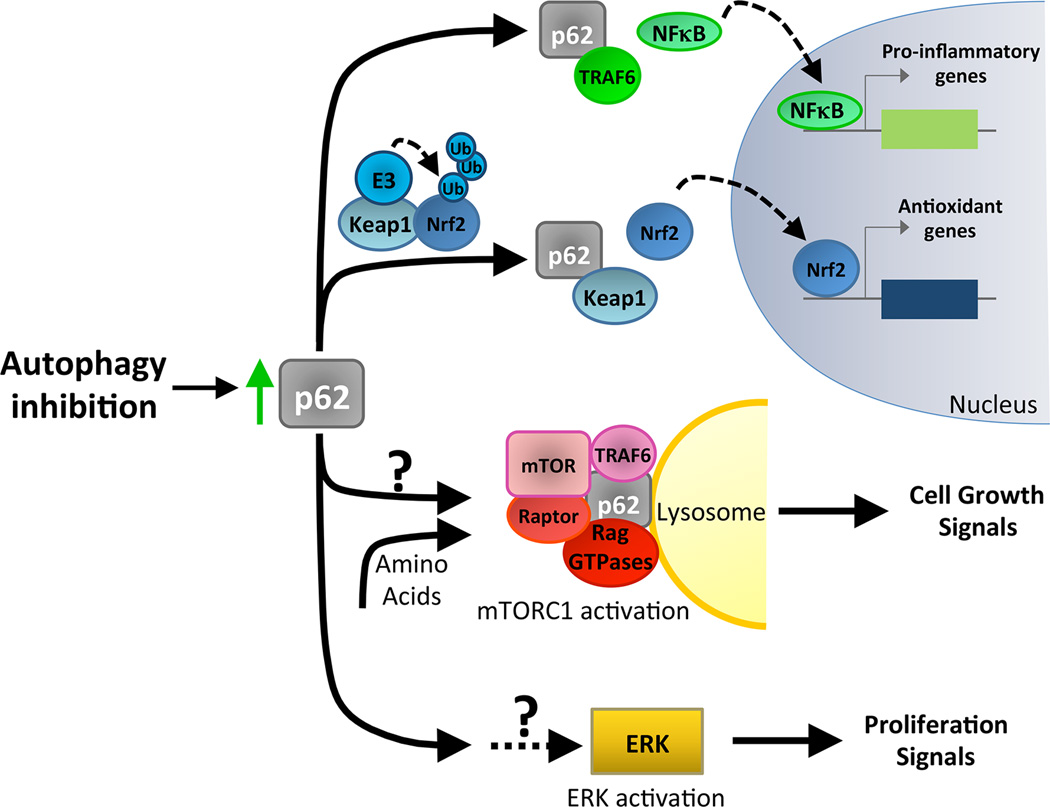
Figure 3. p62/SQSTM1 activates multiple signaling pathways that support tumorigenesis.
The selective autophagy cargo receptor p62/SQSTM1 functions as a positive regulator of multiple tumor promoting pathways. Because p62 accumulates with autophagy inhibition, impairment of autophagy has been correlated with increased activation of multiple pathways and with enhanced tumor cell growth. First, Atg deletion and p62 have been shown to promote NFκB activity. p62 promotes NFκB function by interaction with TRAF6. The increased expression of pro-inflammatory genes by NFκB supports tumorigenesis. Second, Atg deletion promotes Nrf2 stability and activity by increasing levels of p62, which interacts with and sequesters Keap1. Keap1 is an adaptor for the E3-ubiquitin ligase that promotes Nrf2 ubiquitination and proteosomal degradation. Nrf2-mediated expression of antioxidants promotes tumor growth. Furthermore, in response to nutrient signaling, p62 complexes with regulators of mTOR to activate mTORC1 at the lysosome and promote cell growth. The impact of p62 on mTORC1 pathway activation during autophagy inhibition has not been established. Finally, RNAi-mediated depletion of ATGs or p62 overexpression enhances ERK signaling to enable the proliferation of transformed cells. The mechanism underlying this increase in ERK activity is not understood.
p62 also controls additional pro-tumorigenic pathways, including those regulated by mTORC1 and ERK. mTORC1 critically regulates tumor cell growth, and p62 activates mTORC1 by potentiating its ability to complex with Rag GTPases and TRAF6 and by facilitating its recruitment to lysosomes; this regulation of mTORC1 by p62 supports tumor growth in vivo and cell proliferation in vitro [67–68]. Autophagy inhibition or p62 overexpression can also enhance the growth of PI3K-transformed MCF10A cells in 3D culture [45]. In this model, p62-induced proliferation correlates with activation of mitogenic ERK signaling. These studies collectively point to a pro-tumorigenic function for p62 and highlight the varied regulatory roles of p62 during tumorigenesis.
In addition to p62, other cargo receptors, such as NDP52, OPTN, and NBR1, mediate selective autophagy [57]. Similar to p62, NDP52 has been implicated in regulation of NFκB signaling in lung cancer cells [69]. Although this regulation was proposed to occur through NDP52-mediated selective autophagy, the precise mechanism is unknown. In lung cancer cells, the ubiquitylation of OPTN increases autophagy, promotes the degradation of p62 and damaged proteins, and suppresses lung tumorigenesis in nude mice [70]. Moreover, OPTN can inhibit NFκB signaling, but how this regulation affects tumor development has not been investigated [71]. While formal evidence for NBR1 in mediating tumor progression is lacking, it is noteworthy that NBR1 can support Nrf2 antioxidant signaling to potentially impact cancer [72]. Additionally, NBR1 regulates selective autophagic clearance of midbodies that form during cell division, and midbody accumulation contributes to increased growth of tumor cells in vitro [73]. Overall, further establishing the contribution of these selective autophagy regulators to cancer progression remains an important and exciting topic for future study.
Concluding remarks
Given the diversity of tumor types and the numerous oncogenic drivers involved in cancer, the role of autophagy during cancer progression will likely continue to remain complex and intensely debated. Thus, going forward it will be imperative to use appropriate models of disease to accurately determine the clinical contexts in which autophagy inhibition or activation should be considered therapeutically. Moreover, the controversial and context-dependent role of autophagy during initiation and advanced progression of tumors emphasizes the need for a comprehensive understanding of the cell biological processes regulating autophagy during tumorigenesis. Certainly, the highly dynamic nature of autophagy and its regulation by many evolutionarily conserved genes and diverse signaling pathways suggests there are countless avenues by which cancer cells can modulate the pathway to their benefit.
Although research to date has identified many stress pathways in tumor cells that induce autophagy, better elucidating the relationship between autophagy and these pathways in cancer GEMMs remains an important goal. For example, understanding if there is a defined repertoire and coordination of stress-induced pathways that drive autophagy in particular tumor types may reveal unexpected routes for therapeutic modulation of autophagy in a tumor-specific manner. Such knowledge may also lead to development of therapeutic alternatives to hydroxychloroquine (HCQ)-mediated lysosomal inhibition, which is currently the major clinical option for inhibiting autophagy in patients [74–81]. Furthermore, autophagy has traditionally been viewed as a pathway that promotes tumor cell survival during stress; nevertheless, the aforementioned studies of invasion, secretion and EMT have begun to illuminate new functions for autophagy in tumor cells. As these novel roles for autophagy during tumorigenesis continue to emerge, understanding the mechanisms by which self-eating controls these processes may expose opportunities for specific targeting of autophagy-dependent phenotypes in tumor cells.
Finally, this review specifically focuses on the functions of autophagy in tumor cells. However, because the effects of pharmacological inhibitors like HCQ are not confined to cancer cells, their long-term use may be limited by adverse effects associated with autophagy and lysosomal inhibition in normal cells. Indeed, a recent study of inducible systemic atg7 deletion in adult mice demonstrates that acute autophagy ablation elicits the rapid regression of KRas lung tumors; however, extended periods of autophagy deficiency causes the deterioration of multiple tissues and lethal neurodegeneration [25]. To identify new strategies that selectively target autophagy in cancer cells without harming normal tissue, we must continue to define the cellular and metabolic functions of autophagy in both normal and tumor cells.
Figure I.
Autophagosome formation and maturation in mammals
Highlights.
Autophagy sustains metabolic pathways required for aggressive tumor growth.
The unfolded protein response induces cytoprotective autophagy in stressed tumors.
Autophagy facilitates anoikis resistance and tumor cell invasion.
Selective autophagy cargo receptors control multiple pro-tumor signaling pathways.
Acknowledgements
We are grateful to Srirupa Roy for helpful discussions. CMK is supported by NIH F31CA167905. JD is supported by the NIH (CA126792 and CA188404), the DOD BCRP (W81XWH-11-1-0130 and W81XWH-12-1-0505), and the Samuel Waxman Cancer Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo JY, et al. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–1219. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yue Z, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aita VM, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 11.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 12.Laddha SV, et al. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Mol Cancer Res. 2014;12:485–490. doi: 10.1158/1541-7786.MCR-13-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inami Y, et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao S, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun. 2014;5:3056. doi: 10.1038/ncomms4056. [DOI] [PubMed] [Google Scholar]
- 16.Rosenfeldt MT, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 17.Yang A, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strohecker AM, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E–driven lung tumors. Cancer Discov. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lock R, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei H, et al. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–1527. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang S, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo JY, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karsli-Uzunbas G, et al. Autophagy is Required for Glucose Homeostasis and Lung Tumor Maintenance. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy JM, et al. Autophagy Inhibition Improves Chemosensitivity in BRAFV600E Brain Tumors. Cancer Discov. 2014;4:773–780. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts DJ, et al. Hexokinase-II positively regulates glucose starvation-induced autophagy through TORC1 inhibition. Mol Cell. 2014;53:521–533. doi: 10.1016/j.molcel.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z, et al. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med. 2013;210:2119–2134. doi: 10.1084/jem.20130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu S, et al. 13C–pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab. 2011;14:131–142. doi: 10.1016/j.cmet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Graham BH. Measurement of mitochondrial oxygen consumption using a Clark electrode. Methods Mol Biol. 2012;837:63–72. doi: 10.1007/978-1-61779-504-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardner BM, et al. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5:a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hart LS, et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. J Clin Invest. 2012;122:4621–4634. doi: 10.1172/JCI62973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy P, et al. Myc-driven overgrowth requires unfolded protein response-mediated induction of autophagy and antioxidant responses in Drosophila melanogaster. PLoS Genet. 2013;9:e1003664. doi: 10.1371/journal.pgen.1003664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouschop KM, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pike LR, et al. Transcriptional up-regulation of ULK1 by ATF4 contributes to cancer cell survival. Biochem J. 2013;449:389–400. doi: 10.1042/BJ20120972. [DOI] [PubMed] [Google Scholar]
- 36.Avivar-Valderas A, et al. PERK integrates autophagy and oxidative stress responses to promote survival during extracellular matrix detachment. Mol Cell Biol. 2011;31:3616–3629. doi: 10.1128/MCB.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma XH, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller DM, et al. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sethi N, Kang Y. Unravelling the complexity of metastasis -molecular understanding and targeted therapies. Nat Rev Cancer. 2011;11:735–748. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paoli P, et al. Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta. 2013;1833:3481–3498. doi: 10.1016/j.bbamcr.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 42.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 43.Reymond N, et al. Crossing the endothelial barrier during metastasis. Nat Rev Cancer. 2013;13:858–870. doi: 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- 44.Fung C, et al. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen N, et al. Autophagy restricts proliferation driven by oncogenic phosphatidylinositol 3-kinase in three-dimensional culture. Oncogene. 2013;32:2543–2554. doi: 10.1038/onc.2012.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng YF, et al. Autophagy inhibition suppresses pulmonary metastasis of HCC in mice via impairing anoikis resistance and colonization of HCC cells. Autophagy. 2013;9:2056–2068. doi: 10.4161/auto.26398. [DOI] [PubMed] [Google Scholar]
- 47.Macintosh RL, et al. Inhibition of autophagy impairs tumor cell invasion in an organotypic model. Cell Cycle. 2012;11:2022–2029. doi: 10.4161/cc.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galavotti S, et al. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene. 2013;32:699–712. doi: 10.1038/onc.2012.111. [DOI] [PubMed] [Google Scholar]
- 49.Beckner ME, et al. Identification of ATP citrate lyase as a positive regulator of glycolytic function in glioblastomas. Int J Cancer. 2010;126:2282–2295. doi: 10.1002/ijc.24918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, et al. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis. 2013;34:1343–1351. doi: 10.1093/carcin/bgt063. [DOI] [PubMed] [Google Scholar]
- 51.Lock R, et al. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov. 2014;4:466–479. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhan Z, et al. Autophagy facilitates TLR4- and TLR3-triggered migration and invasion of lung cancer cells through the promotion of TRAF6 ubiquitination. Autophagy. 2014;10:257–268. doi: 10.4161/auto.27162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deretic V, et al. Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. Trends Cell Biol. 2012;22:397–406. doi: 10.1016/j.tcb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Reilly S, et al. Interleukin-6 (IL-6) trans signaling drives a STAT3-dependent pathway that leads to hyperactive transforming growth factor-beta (TGF-beta) signaling promoting SMAD3 activation and fibrosis via Gremlin protein. J Biol Chem. 2014;289:9952–9960. doi: 10.1074/jbc.M113.545822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiang L, et al. Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc Natl Acad Sci U S A. 2014;111:9241–9246. doi: 10.1073/pnas.1322913111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogov V, et al. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 58.Mancias JD, et al. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puissant A, et al. When autophagy meets cancer through p62/SQSTM1. Am J Cancer Res. 2012;2:397–413. [PMC free article] [PubMed] [Google Scholar]
- 60.Komatsu M, et al. p62/SQSTM1/A170: physiology and pathology. Pharmacol Res. 2012;66:457–462. doi: 10.1016/j.phrs.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 61.Duran A, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 62.Ling J, et al. KrasG12D–induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei H, et al. p62/SQSTM1 synergizes with autophagy for tumor growth in vivo. Genes Dev. 2014;28:1204–1216. doi: 10.1101/gad.237354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 66.Ichimura Y, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 67.Duran A, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Linares JF, et al. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51:283–296. doi: 10.1016/j.molcel.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Newman AC, et al. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-kappaB signalling. PLoS One. 2012;7:e50672. doi: 10.1371/journal.pone.0050672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Z, et al. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell. 2014;26:106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu G, et al. Optineurin negatively regulates TNFalpha- induced NF-kappaB activation by competing with NEMO for ubiquitinated RIP. Curr Biol. 2007;17:1438–1443. doi: 10.1016/j.cub.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 72.Rubio N, et al. p38(MAPK)-regulated induction of p62 and NBR1 after photodynamic therapy promotes autophagic clearance of ubiquitin aggregates and reduces reactive oxygen species levels by supporting Nrf2-antioxidant signaling. Free Radic Biol Med. 2014;67:292–303. doi: 10.1016/j.freeradbiomed.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 73.Kuo TC, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol. 2011;13:1214–1223. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amaravadi RK, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barnard RA, et al. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy. 2014:10. doi: 10.4161/auto.29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mahalingam D, et al. Combined autophagy and HDAC inhibition: A phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy. 2014:10. doi: 10.4161/auto.29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rangwala R, et al. Combined MTOR and autophagy inhibition: Phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014:10. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rangwala R, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014:10. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rosenfeld MR, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014:10. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vogl DT, et al. Combined autophagy and proteasome inhibition: A phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014:10. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolpin BM, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19:637–638. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 83.Chan EY. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci Signal. 2009;2:pe51. doi: 10.1126/scisignal.284pe51. [DOI] [PubMed] [Google Scholar]
- 84.Itakura E, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shen HM, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci. 2014;39:61–71. doi: 10.1016/j.tibs.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 88.Ao X, et al. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348–358. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]