Abstract
Background
Preference for fatty foods is a risk factor for obesity. It is a complex behaviour that involves the brain reward system and is regulated by genetic and environmental factors, such as the opioid receptor mu-1 gene (OPRM1) and prenatal exposure to maternal cigarette smoking (PEMCS). We examined whether OPRM1 and PEMCS interact in influencing fat intake and whether exposure-associated epigenetic modifications of OPRM1 may mediate this gene–environment interaction.
Methods
We studied adolescents from a French Canadian genetic founder population, half of whom were exposed prenatally to maternal cigarette smoking. Fat intake was assessed with a 24-hour food recall in the form of a structured interview conducted by a trained nutritionist. The OPRM1 variant rs2281617 was genotyped for the whole sample with the Illumina Human610-Quad and HumanOmniExpress BeadChips. Methylation of blood DNA was assessed at 21 CpGs across OPRM1 in a subset of the sample using the Illumina HumanMethylation450 BeadChip.
Results
We included 956 adolescents in our study. In the whole sample, OPRM1 (T carrier in rs2281617) was associated with lower fat intake (−1.6%, p = 0.017), and PEMCS was associated with higher fat intake (+1.6%, p = 0.005). OPRM1 and PEMCS interacted with each other (p = 0.003); the “protective” (fat intake–lowering) allele of OPRM1 was associated with lower fat intake in nonexposed (−3.2%, p < 0.001) but not in exposed individuals (+0.8%, p = 0.42). Further, PEMCS was associated with lower DNA methylation across multiple CpGs across OPRM1 in exposed versus nonexposed individuals (p = 0.031).
Limitations
A limitation of our study was its cross-sectional design.
Conclusion
Our study suggests that PEMCS may interact with OPRM1 in increasing fat preference. Silencing of the protective OPRM1 allele in exposed adolescents might be related to epigenetic modification of this gene.
Introduction
Obesity is a major health problem with many severe consequences, including the leading causes of morbidity and mortality: cardiovascular disease and type 2 diabetes mellitus.1 Despite increased public awareness of the health risks associated with obesity and the benefits of exercise and healthy eating, the rates of obesity have not decreased.
Research in humans and experimental animals suggests that excess dietary fat contributes to the development of obesity,2,3 as fats compared with other macronutrients (i.e., carbohydrates, proteins) are of higher energy density and efficiency.4 Fats contain more than twice the amount of energy per gram than carbohydrates and proteins,4 and almost all calories eaten as fats are stored, whereas a substantial proportion of calories eaten as carbohydrates and proteins are lost during their absorption, processing and storage.4 Finally, fat is highly palatable, and dietary preference for fat is a behaviour regulated in part by reward-related mechanisms that process the hedonic properties of food independently of the body’s energy status.5 Such mechanisms may overlap with those mediating the addictive properties of drugs of abuse5 and may involve dopaminergic and opioidergic signalling.6–8
Preference for fatty foods is a complex trait regulated by genetic and environmental factors, but only a few such factors have been identified in human populations.9–14 These include the opioid receptor mu-1 gene (OPRM1)13 and prenatal exposure to maternal cigarette smoking (PEMCS).14 OPRM1 encodes a receptor that is highly expressed in reward-processing regions of the brain and that is known to modulate fat preference in animals;6,15,16 it has been associated with dietary intake of fat and risk for obesity in a genome-wide association study.13 Prenatal exposure to maternal cigarette smoking is a well-established risk factor for obesity in the exposed offspring17–19 and has been associated with enhanced dietary preference for fat.14 Effects of genetic and environmental factors may be independent from each other, or the 2 types of factors may interact. Environment-induced epigenetic modifications of genes and their regulatory sequences may, at least in part, mediate such interactions and thus influence phenotypes.20,21
One well-understood epigenetic modification is DNA methylation (DNAm), which is the addition of a methyl group at the 5’ position of cytosines, occurring most frequently in the context of CpG dinucleotides. It plays an important role in the regulation of gene transcription — while DNAm at gene promoters has mainly been linked to lower gene expression,22 DNAm at gene bodies has been associated with higher gene expression.23 Recent research suggests that gene-body DNAm may also play a key role in the regulation of alternative transcription.24–26
Cigarette smoke is considered one of the most powerful environmental modifiers of DNAm; this holds for self-exposure to cigarette smoking as well as PEMCS.27–31 Cigarette smoke contains a large number of chemicals, such as carcinogens, nicotine and carbon monoxide, which can modify DNAm (see the review by Lee and Pausova31). Critical for the present study, many chemicals contained in cigarette smoke can easily pass from a smoking pregnant woman to the developing embryo.32 Acting during early phases of embryogenesis, when global erasure and re-establishment of DNAm occur in undifferentiated cells (before their commitment into specific tissues), these chemicals may induce lasting soma-wide modifications of DNAm in the exposed offspring that would be detectable later in life in peripheral lymphocytes.20,30,31,33 The DNAm modifications induced during embryogenesis are maintained by the action of maintenance DNA methyltransferases that copy these modifications from cell to cell during successive cell divisions throughout life.34 Further, as the DNAm machinery is not sequence-specific (i.e., DNA methyl-transferases do not have specific target DNA sequences34), these modifications may spread over larger segments of DNA. Consistent with this, it has been observed that DNAm is correlated across neighbouring CpGs.35
Previous research suggests that OPRM113 and PEMCS14 modulate dietary preference for fat. It is unclear, however, whether these 2 factors interact, and, if so, whether their interaction could involve PEMCS-associated modifications of DNAm in OPRM1. The present study investigated these questions in a population-based sample of adolescents recruited from the French Canadian founder population of Saguenay–Lac-Saint-Jean, Canada, as part of the Saguenay Youth Study (SYS).36
Methods
The Saguenay Youth Study and prenatal exposure to maternal cigarette smoking
The SYS is a population-based cross-sectional study investigating the long-term consequences (and genetic modifiers) of PEMCS on cardiovascular, metabolic, brain and mental health in adolescence.36 It is a family-based study that recruited sibling pairs from the genetic founder population of the Saguenay–Lac-Saint-Jean region in the province of Quebec, Canada. The power of genetic analyses is expected to be higher in founder than in regular (outbred) populations owing to more homogeneous genetic and environmental backgrounds. All participants were recruited in high schools. At recruitment, adolescents who were exposed and nonexposed prenatally to maternal cigarette smoking were matched by school attended and by maternal education to minimize the potentially confounding influence of socioeconomic status. Being exposed was defined as having a mother who smoked more than 1 cigarette per day during the second trimester; being nonexposed was defined as having a mother who did not smoke for at least 1 year before — and throughout — the pregnancy. This information was reported by the mothers during a structured telephone interview with a research nurse at the time of our study and was subsequently validated against medical records in a subset of 260 adolescents; κ statistics with a value of 0.69 ± 0.04 indicated good agreement (> 0.6–0.8)37 in this subset.
The main exclusion criteria of the SYS were premature birth (< 35 wk) or detached placenta; maternal alcohol abuse during pregnancy; positive medical history of the participant for type 1 diabetes, heart disease requiring surgery or sustained medication; and contraindication to MRI. Additional study details have been described elsewhere.36 Written consent of the parents and assent of the adolescents were obtained. The research ethics committee of the Chicoutimi Hospital approved the SYS study protocol.
Dietary intake of fat
Dietary intake of fat was assessed using 24-hour food recall. It is a well-established method of assessing diet used, for example, in the U.S. National Health and Nutrition Examination Surveys, which are the only nationally representative dietary surveys in the United States.38 This method has been validated for Quebec youth.39 In the SYS, all 24-hour food recalls took place in the form of a structured interview conducted by a trained nutritionist who collected information about all foods and drinks consumed in the preceding 24 hours and then analyzed this information using the Recipe File (U.S. Department of Agriculture) to obtain quantitative data on energy and macronutrient (fat, carbohydrate, protein) intake. All 24-hour food recalls took place on a Saturday during a hospital session of the phenotyping protocol.36 Therefore, for all participants, the assessed 24-hour periods occurred on Fridays. In the present study, we analyzed fat, carbohydrate and protein intake (percent of energy consumed as fat, carbohydrate and protein, respectively) and energy intake.
OPRM1 genotype
Here we studied the OPRM1 single nucleotide polymorphism (SNP) previously associated with dietary intake of fat (rs2281617)14 and other reward-related behaviours.40 Genotypes for this SNP were obtained as follows: in the first 570 participants (the first wave of genotyping), the SNP was genotyped with the Illumina Human610-Quad BeadChip at the Centre National de Génotypage in Paris, France. In the remaining 415 participants (the second wave of genotyping), it was genotyped with the Illumina HumanOmniExpress BeadChip at the Genome Analysis Center of the Helmholtz Research Center for Environmental Health in Neuherberg, Germany. On both genotyping platforms, the SNP passed the quality control criteria of call rate over 95% and minor allele frequency over 0.01. Owing to a low number of TT homozygotes in our sample (n = 12), TT homozygotes were pooled together with CT heterozygotes and analyzed as T carriers, as done previously.14
DNAm of OPRM1
We assessed DNAm in OPRM1 using the Infinium Human-Methylation450 BeadChip (Illumina) in a subset of unrelated participants of the SYS (nonexposed matched to exposed by sex, age and maternal education). The exposed participants (half male, half female) were selected based on medians of exposure (10 cigarettes per day in both sexes).
Bisulfite conversion of DNA extracted from peripheral blood cells (800 ng) was performed with the EZ-96 DNA Methylation Kit (Zymo Research), and subsequent hybridization of this DNA was carried out with the Infinium HumanMethylation450 BeadChip. Both these procedures were performed at the Genome Analysis Center of the Helmholtz Research Center for Environmental Health. Samples were loaded onto the array in a random order with respect to PEMCS and sex and processed by the same technician simultaneously to minimize batch effects. The chip interrogates DNAm at more than 485 000 CpGs, providing coverage to more than 99% of RefSeq genes, targeted across their promoters, 5′-untranslated regions (UTRs), first exons, gene bodies and 3’UTRs.41 The DNAm score at each CpG, described as the DNAm-β value, ranges between 0 and 1 and is derived from the fluorescent intensity ratio (β = intensity of the methylated allele ÷ [intensity of the unmethylated allele + intensity of the methylated allele + 100]). Methylation values were normalized using the preprocess Illumina algorithm implemented in the Minfi R package.42 Parameters were set to mimic the Illumina Genome Studio normalization procedure. The CpGs within transcriptional start sites (TSS), 5′UTRs and first exons were classified as promoter CpGs, and CpGs within gene bodies and 3′UTRs were classified as gene-body CpGs.41
We considered CpGs located within the OPRM1 region with a detection p value less than 0.05 and no cross-reactivity.43 We excluded any probes that demonstrated abnormal distribution of DNAm-β values as well as any outliers from further analysis. In the present study, 23 CpGs located within the OPRM1 region were considered (Table 1). Two of them (cg06649410 and cg11881038) demonstrated abnormal distribution of DNAm-β values, suggestive of strong confounding genetic influences (see the Appendix, Fig. S1, available at jpn.ca); the studied OPRM1 SNP (rs2281617), however, was not associated with DNAm-β at either of these CpGs (Appendix, Table S1). In addition, a DNAm-β value at cg14262937 of 1 participant was a distinct outlier (Appendix, Fig. S1) and was excluded.
Table 1.
Probed CpGs within OPRM1
| PCA loading matrix‡ | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| CpG no. | Illumina ID | DNAm-β | UCSC RefGene Group* | Modified position category† | PC1, 16.2% | PC2, 13.3% |
| 1 | cg12944573 | 0.56 | TSS | Promoter | 0.42§ | 0.52§ |
| 2 | cg17256711 | 0.24 | TSS | Promoter | 0.32§ | 0.23 |
| 3 | cg13245264 | 0.68 | 1st Exon (Exon 11) | Promoter | 0.12 | 0.44§ |
| 4 | cg10143581 | 0.88 | TSS | Promoter | 0.04 | 0.36§ |
| 5 | cg22370006 | 0.15 | TSS | Promoter | 0.44§ | −0.09 |
| 6 | cg14262937 | 0.16 | TSS | Promoter | 0.41§ | −0.11 |
| 7 | cg06649410 | 0.19 | 1st Exon (Exon 1) | Promoter | — | — |
| 8 | cg05215925 | 0.13 | 1st Exon (Exon 1) | Promoter | 0.74§ | −0.22 |
| 9 | cg12838303 | 0.13 | 1st Exon (Exon 1) | Promoter | 0.74§ | −0.21 |
| 10 | cg22719623 | 0.44 | 1st Exon (Exon 1) | Promoter | 0.70§ | 0.02 |
| 11 | cg15085086 | 0.21 | 1st Exon (Exon 1) | Promoter | 0.38§ | −0.47 |
| 12 | cg05017309 | 0.88 | Body | Body | 0.00 | 0.12 |
| 13 | cg22366819 | 0.91 | Body | Body | 0.37§ | 0.50§ |
| 14 | cg12466324 | 0.79 | Body | Body | −0.61 | 0.45§ |
| 15 | cg23384428 | 0.93 | Body | Body | 0.16 | 0.58§ |
| 16 | cg11881038 | 0.78 | Body | Body | — | — |
| 17 | cg04170440 | 0.85 | Body | Body | 0.20 | 0.59§ |
| 18 | cg07813322 | 0.85 | 3’UTR | Body | −0.31 | 0.13 |
| 19 | cg15658985 | 0.86 | Body | Body | −0.08 | −0.52 |
| 20 | cg26415516 | 0.78 | 3’UTR | Body | −0.27 | 0.08 |
| 21 | cg27296341 | 0.68 | Body | Body | 0.44§ | 0.09 |
| 22 | cg10276116 | 0.92 | Body | Body | −0.01 | 0.47§ |
| 23 | cg07356123 | 0.87 | Body | Body | 0.21 | 0.38§ |
DNAm-β = DNA methylation β; OPRM1 = opioid receptor mu-1 gene; PC = principal component; PCA = principal component analysis; TSS = transcription start site; UTR = untranslated region; UCSC = University of California, Santa Cruz.
Annotations according to the UCSC database (http://genome.ucsc.edu/)
Modified position categorizations, as adopted from Sandoval and colleagues.41
Percentages represent the proportion of variance among CpG DNA methylation explained by the principal component.
Positive PC loading ≥ 0.30, as suggested previously for data with sample sizes ≥ 100.45
Statistical methods
We first examined whether OPRM1 and PEMCS were independently associated with fat intake. We then examined whether OPRM1 interacted with PEMCS in their associations with fat intake. In these analyses, we used multivariate regression, with OPRM1 genotype (T carriers v. CC homozygotes at rs2281617) and/or PEMCS (exposed v. nonexposed) as the main factors and age, sex and perinatal variables previously shown to be associated with PEMCS (birth weight [adjusted for gestation duration], breastfeeding duration)44 as potentially confounding factors. We also tested an additional model that included current cigarette smoking by adolescents as a potentially confounding factor.
Next, we examined whether PEMCS was associated with differential DNAm of OPRM1. Since DNAm correlated across neighbouring CpGs,35 we first used principal component analysis (PCA) to identify independent components of shared variance among the tested OPRM1 CpGs and then examined whether any of the identified components were associated with PEMCS. Principal component analysis is a multivariate statistical technique used to extract shared variance from correlated data. It transforms a number of possibly correlated variables into a number of uncorrelated variables, the so-called principal components (PCs). Each PC represents a different linear combination of the original correlated variables. The original variables are first normalized to their respective means and then used to generate a correlation matrix. Principal component analysis is then performed by eigenvalue decomposition of the correlation matrix. In the present study, PCA was performed on 21 residuals of DNAm-β values adjusted for age, sex and perinatal variables (birth weight [adjusted for gestation duration], breastfeeding duration). The PCs with an eigenvalue over 1 were considered significant, as suggested previously for data on samples of more than 100 participants.45 Associations between PEMCS and significant PCs were tested with 2-sided t tests. Normality of the dependent variables (i.e., significant PCs) was assessed and statistical outliers (mean ± 3 standard deviations) were excluded (PC1 contained 2 outliers and PC2 contained 1 outlier). An additional model including current cigarette smoking by adolescents as a potentially confounding factor was also tested using multivariate regression analysis with PEMCS as the main factor. All analyses were performed using JMP version 9.0 (SAS Institute Inc.).
Results
Participants
The SYS included a total of 1028 adolescents. In the present study, all individuals with missing 24-hour food recall (n = 29), OPRM1 genotype (n = 14), and perinatal data (birth weight, gestation duration, breastfeeding duration; n = 17) were excluded. Statistical outliers (mean ± 3 standard deviations) for perinatal variables were also excluded (n = 12). Thus, the final analyzed sample was 956 adolescents (457 exposed). Descriptive characteristics of these individuals are provided in Table 2. In the present study, average exposure to maternal smoking was 10.7 ± 6.7 (range 1–35) cigarettes per day throughout gestation.
Table 2.
Basic characteristics of all studied adolescents
| Group; mean ± SD* | |||
|---|---|---|---|
|
|
|||
| Characteristic | Nonexposed | Exposed | p value† |
| Sex, no. male:female | 258:241 | 208:249 | 0.06 |
| Age, yr | 14.5 ± 1.8 | 14.6 ± 1.8 | 0.61 |
| Family income, $CAD/yr | 51 424 ± 17 446 | 50 518 ± 19 417 | 0.53 |
| Current smoking, yes:no‡ | 36:459 | 54:395 | 0.013 |
| PEMCS, cigarettes/d | 0 | 10.6 ± 6.7 | N/A |
| Gestational duration, wk | 39.3 ± 1.5 | 39.2 ± 1.5 | 0.65 |
| Birth weight, g | 3511 ± 457 | 3265 ± 475 | < 0.001 |
| Breastfeeding duration, wk | 10.5 ± 13.8 | 6.3 ± 11.2 | < 0.001 |
N/A = not applicable; PEMCS = prenatal exposure to maternal cigarette smoking; SD = standard deviation.
Unless otherwise indicated.
We use a t test for continuous variables and a χ2 test for categorical variables.
Adolescents were classified as currently smoking if they reported to have smoked at least 1 cigarette in the last 30 days. Data were missing from 12 adolescents.
We assessed DNAm in OPRM1 in 132 unrelated participants (66 nonexposed and 66 exposed). The exposed participants comprised 33 boys and 33 girls who were prenatally exposed to maternal cigarette smoking of 8.9 ± 3.9 (range 5–20) cigarettes per day. Descriptive characteristics of the participants in this subset are provided in the Appendix, Table S2.
OPRM1 × PEMCS interaction and fat intake
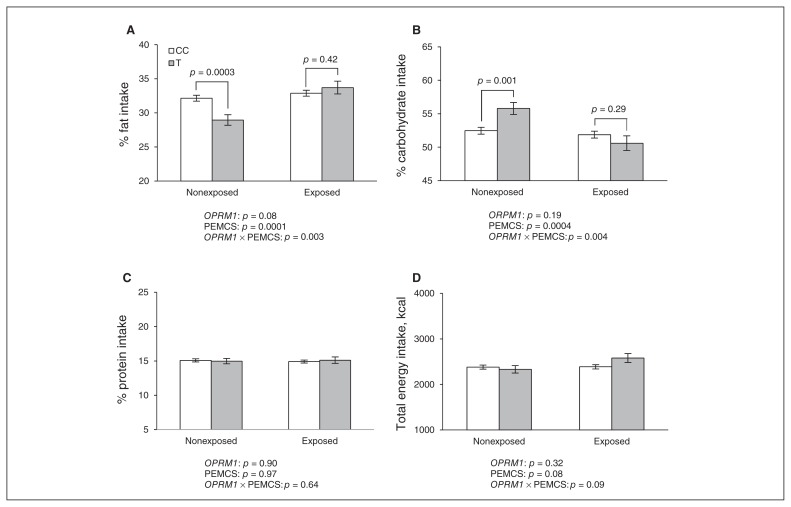
The present study (n = 956) confirms associations of OPRM113 and PEMCS14 with fat intake, as observed previously in smaller subsets of the sample, and identifies a new interaction between these 2 factors. Thus, the OPRM1 T carriers (n = 196) demonstrated lower fat intake than CC homozygotes (n = 754; −1.6%, p = 0.017), and adolescents exposed to PEMCS (n = 453) showed higher fat intake than nonexposed adolescents (n = 497; +1.6%, p = 0.005). The “protective” T allele of OPRM1 was associated with lower fat intake in nonexposed (−3.2%, p = 0.0003) but not in exposed (by +0.8%, p = 0.42) adolescents (OPRM1 × PEMCS interaction, p = 0.003; Fig. 1). This T allele was also associated with higher carbohydrate intake in nonexposed but not exposed adolescents (Fig. 1). No differences were seen in protein intake and overall energy intake (Fig. 1). These results are consistent with those of a previous animal study suggesting that the mu-opioid receptor may exert opposite effects on fat and carbohydrate intake independently of energy status (being satiated or hungry).16 All the above results remained virtually unchanged after adjusting for the potentially confounding effects of birth weight, breastfeeding and current smoking by the adolescents (Appendix, Table S2). Finally, neither PC1 nor PC2 were significantly associated with rs2281617.
Fig. 1.
Association of the opioid receptor mu-1 gene (OPRM1; rs2281617) and prenatal exposure to maternal cigarette smoking (PEMCS) with fat, carbohydrate, protein, and energy intake. Means ± standard errors of the mean, adjusted for age, sex and perinatal variables (birth weight, adjusted for gestation duration and breastfeeding duration) are shown.
PEMCS association with DNAm of OPRM1
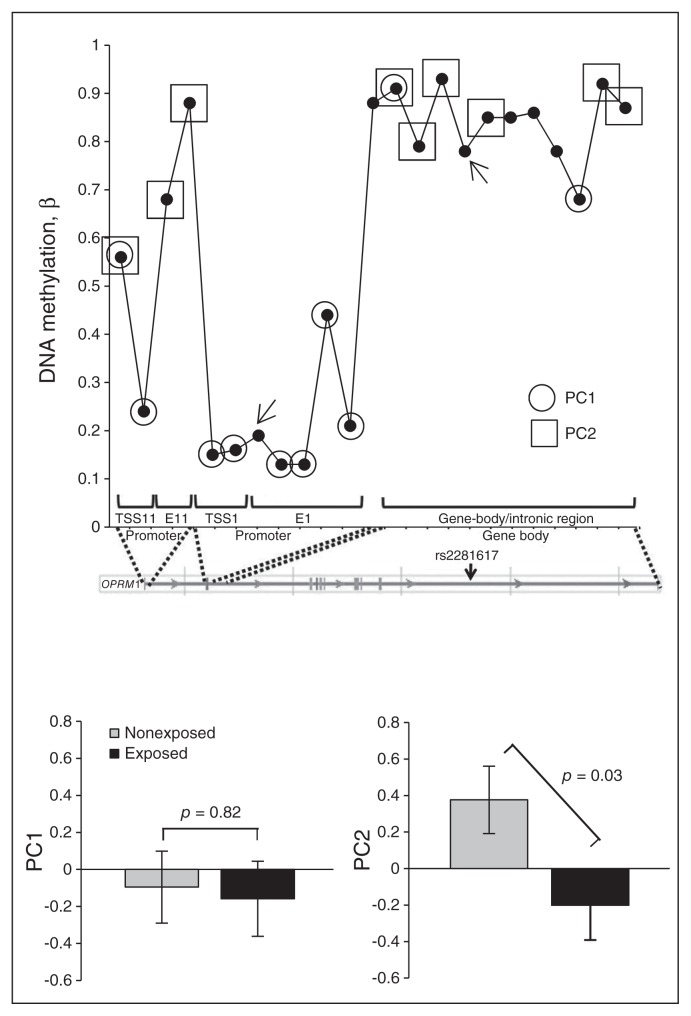
Next, we examined whether DNAm across the tested OPRM1 CpGs (n = 21) differed between 66 exposed and 66 nonexposed adolescents matched by sex, age and maternal education. The PCA of DNAm at the 21 CpGs identified 2 independent PCs. The first, PC1, explained 16.2% of shared variance (p < 0.001) and was strongly positively loaded by CpGs within promoter regions (Table 1, Fig. 2). The second, PC2 explained 13.3% of shared variance (p < 0.001) and was positively loaded mainly by CpGs within the gene body (Table 1, Fig. 2). We found that PC1 did not differ between exposed and nonexposed adolescents (p = 0.82), whereas PC2 was lower in exposed than in nonexposed adolescents (p = 0.031; Fig. 2), suggesting that PEMCS may modulate gene-body rather than promoter DNAm. These results remained virtually unchanged after adjusting for the potentially confounding effects of current cigarette smoking by the adolescents (PC1: p = 0.63 and PC2: p = 0.025).
Fig. 2.
Studied opioid receptor mu-1 gene (OPRM1) CpGs and their association with prenatal exposure to maternal cigarette smoking (PEMCS). (Top) A total of 23 CpGs located within OPRM1 were considered, and their position and mean DNA methylation level (β, DNAm-β) across 132 investigated adolescents are shown. In addition, significant contributions (loading ≥ 0.30) of individuals’ CpGs to principal component 1 (PC1) and principle component 2 (PC2) are indicated by circles and squares, respectively. Arrows indicate excluded CpGs with abnormal distribution (see the Appendix, Fig. S1, available at jpn.ca). (Bottom) Associations between PEMCS and PC1 or PC2, with means ± standard errors of the mean adjusted for age, sex and perinatal variables (birth weight, adjusted for gestation duration, and breastfeeding duration) are shown. E = exon; TSS = transcription start site.
Discussion
The results of the present study suggest that PEMCS may interact with OPRM1 to enhance dietary intake of fat, and that the mechanisms of this gene-environment interaction might involve exposure-associated decreases of gene-body DNAm in OPRM1.
OPRM1 encodes the mu-opioid receptor for endogenous (β-endorphin, enkephalin) and exogenous (morphine, heroin, methadone) opioids. The gene is highly expressed in brain regions processing reward from drugs of abuse as well as palatable foods, such as dietary fat.6,15,16 Previous research suggests that a specific variant of OPRM1 (T allele of rs2281617) may confer a “protective” effect — it was associated with lower fat intake and lower body adiposity in adolescence.14 Interestingly, the same variant of OPRM1 was associated with lower amphetamine-induced euphoria in an independent study of adults.40 Here, we found a significant interaction between this variant and PEMCS — the protective T allele was associated with lower fat intake in the nonexposed but not in the exposed adolescents (Fig. 1). One possible cellular mechanism underlying this gene–environment interaction could be PEMCS-associated modulations of DNAm in OPRM1 that would silence the protective T allele in the exposed individuals.
Cigarette smoke is considered a powerful environmental modifier of DNAm.27–31 Chemicals contained in cigarette smoke may alter the expression or activity of DNAm machinery or may induce DNA lesions that recruit this machinery to the site of repair and enhance DNAm there (see the review by Lee and Pausova31). When exposure to cigarette smoke takes place during early embryogenesis, namely during the process of global erasure and re-establishment of DNAm in undifferentiated cells (before their commitment into specific tissues), it may induce soma-wide modifications of DNAm present in diverse tissues, such as brain and peripheral blood cells. Consistent with this possibility, it has been demonstrated recently that, despite a substantial degree of between-tissue variation in DNAm, interindividual differences in DNAm are highly correlated across brain and peripheral blood cells (cerebellum v. blood: r = 0.76, cortex v. blood: r = 0.66).33 Epigenetic changes occurring early in development (before complete tissue differentiation) have been suggested as a possible cause of these between-tissue correlations.33 Further, these early environment-induced soma-wide modifications of DNAm may be long-lasting, as DNAm patterns established during embryogenesis are maintained during life by copying DNAm marks during successive cell divisions by maintenance DNA methyltransferases.34 In the present study, we observed that PEMCS is associated, in adolescence, with lower DNAm across multiple CpGs located mainly in the gene body of OPRM1 (Fig. 2). Recent research suggests that gene-body DNAm plays a critical role in regulating the use of alternative splice sites.24–26 OPRM1 is a complex gene with multiple 5′ and 3′ alternative splice sites that can generate more than 20 alternative transcripts.46,47 Further, gene-body methylation may be involved in suppression of gene expression by, for example, inhibiting alternative promoters embedded in gene bodies48 or by impeding RNA-polymerase transit and transcription elongation.49,50 Whether the observed lower gene-body methylation of OPRM1 modulates its expression requires further experimental research.
In previous studies, PEMCS and OPRM1 were associated not only with fat intake but also with adiposity and volume of amygdala (a brain structure involved in reward processing).13,14 Therefore, in the present study, we also tested adiposity and amygdala volume. Adiposity showed a pattern of group differences suggestive of a possible role of the PEMCS × OPRM1 interaction in regulating adiposity (PEMCS × OPRM1 interaction: p = 0.17) in a similar way as fat intake (Appendix, Fig. S2). Further studies, however, are needed to confirm this possibility. In contrast, amygdala volume demonstrated clear evidence for the absence of PEMCS × OPRM1 interaction (p = 0.99), suggesting that amygdala volume is not involved in mediating the effect of this interaction on fat preference.
Limitations
The present study was a cross-sectional rather than longitudinal study of adolescents and, as such, could not examine developmental trajectories of the observed effects of genetic variants and early environment-induced epigenetic modifications of those variants from birth to adolescence. In addition, DNAm of OPRM1 was assessed only in a subset (n = 132) of all analyzed individuals (n = 956). Finally, based on the results of the present study, we do not know whether gene-body methylation of OPRM1 is associated with its higher or lower expression, but we presented several examples from previous research indicating a critical role of gene-body methylation in regulating expression of alternative transcripts and/or gene expression.
Conclusion
The present study identifies a novel gene–environment interaction that may be increasing dietary preference for fatty foods in adolescents. It also provides some initial evidence for a possible underlying mechanism of this interaction: PEMCS-associated epigenetic modifications of OPRM1.
Acknowledgments
The authors thank Ms. Celine Bourdon for her contribution to statistical analyses. The Canadian Institutes of Health Research (D. Gaudet, T. Paus, Z. Pausova) funds the Saguenay Youth Study. The McLaughlin Centre at the University of Toronto (L. Bouchard, T. Paus, Z. Pausova) funds the DNA methylation study. M. Abrahamowicz is a James McGill Professor of Biostatistics at McGill University. T. Paus is the Tanenbaum Chair in Population Neuroscience at the Rotman Research Institute, University of Toronto.
Footnotes
Competing interests: None declared.
Contributors: Z. Pausova designed the study. G.T. Leonard, L. Richer, M. Perron, S. Veillette, E. Reischl, D. Gaudet, T. Paus and Z. Pausova acquired the data, which K.W.K. Lee, M. Abrahamowicz, L. Bouchard, T. Paus and Z. Pausova analyzed. K.W.K. Lee and Z. Pausova wrote the article, which all authors reviewed and approved for publication.
References
- 1.Must A, Spadano J, Coakley EH, et al. The disease burden associated with overweight and obesity. JAMA. 1999;282:1523–9. doi: 10.1001/jama.282.16.1523. [DOI] [PubMed] [Google Scholar]
- 2.Klesges RC, Klesges LM, Haddock CK, et al. A longitudinal analysis of the impact of dietary intake and physical activity on weight change in adults. Am J Clin Nutr. 1992;55:818–22. doi: 10.1093/ajcn/55.4.818. [DOI] [PubMed] [Google Scholar]
- 3.Hill JO, Peters JC. Environmental contributions to the obesity epidemic. Science. 1998;280:1371–4. doi: 10.1126/science.280.5368.1371. [DOI] [PubMed] [Google Scholar]
- 4.Tappy L. Thermic effect of food and sympathetic nervous system activity in humans. Reprod Nutr Dev. 1996;36:391–7. doi: 10.1051/rnd:19960405. [DOI] [PubMed] [Google Scholar]
- 5.Kenny PJ. Common cellular and molecular mechanisms in obesity and drug addiction. Nat Rev Neurosci. 2011;12:638–51. doi: 10.1038/nrn3105. [DOI] [PubMed] [Google Scholar]
- 6.Taha SA. Preference or fat? Revisiting opioid effects on food intake. Physiol Behav. 2010;100:429–37. doi: 10.1016/j.physbeh.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manabe Y, Matsumura S, Fushiki T. Preference for high-fat food in animals. In: Montmayeur JP, le Coutre J, editors. Fat detection: taste, texture, and postingestive effects. Boca Raton (FL): CRC Press; 2010. [PubMed] [Google Scholar]
- 8.Baldo BA, Pratt WE, Kelley AE. Control of fat intake by striatal opioids. In: Montmayeur JP, le Coutre J, editors. Fat detection: taste, texture, and postingestive effects. Boca Raton (FL): CRC Press; 2010. [PubMed] [Google Scholar]
- 9.Bauer F, Elbers CC, Adan RA, et al. Obesity genes identified in genome-wide association studies are associated with adiposity measures and potentially with nutrient-specific food preference. Am J Clin Nutr. 2009;90:951–9. doi: 10.3945/ajcn.2009.27781. [DOI] [PubMed] [Google Scholar]
- 10.Qi L, Kraft P, Hunter DJ, et al. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum Mol Genet. 2008;17:3502–8. doi: 10.1093/hmg/ddn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka T, Ngwa JS, van Rooij FJ, et al. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr. 2013;97:1395–402. doi: 10.3945/ajcn.112.052183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lussana F, Painter RC, Ocke MC, et al. Prenatal exposure to the Dutch famine is associated with a preference for fatty foods and a more atherogenic lipid profile. Am J Clin Nutr. 2008;88:1648–52. doi: 10.3945/ajcn.2008.26140. [DOI] [PubMed] [Google Scholar]
- 13.Haghighi A, Melka MG, Bernard M, et al. Opioid receptor mu 1 gene, fat intake and obesity in adolescence. Mol Psychiatry. 2014;19:63–8. doi: 10.1038/mp.2012.179. [DOI] [PubMed] [Google Scholar]
- 14.Haghighi A, Schwartz DH, Abrahamowicz M, et al. Prenatal exposure to maternal cigarette smoking, amygdala volume, and fat intake in adolescence. JAMA Psychiatry. 2013;70:98–105. doi: 10.1001/archgenpsychiatry.2012.1101. [DOI] [PubMed] [Google Scholar]
- 15.Pecina S, Smith KS. Hedonic and motivational roles of opioids in food reward: implications for overeating disorders. Pharmacol Biochem Behav. 2010;97:34–46. doi: 10.1016/j.pbb.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, Gosnell BA, Kelley AE. Intake of high-fat food is selectively enhanced by mu opioid receptor stimulation within the nucleus accumbens. J Pharmacol Exp Ther. 1998;285:908–14. [PubMed] [Google Scholar]
- 17.Al Mamun A, Lawlor DA, Alati R, et al. Does maternal smoking during pregnancy have a direct effect on future offspring obesity? Evidence from a prospective birth cohort study. Am J Epidemiol. 2006;164:317–25. doi: 10.1093/aje/kwj209. [DOI] [PubMed] [Google Scholar]
- 18.Oken E, Levitan EB, Gillman MW. Maternal smoking during pregnancy and child overweight: systematic review and meta-analysis. Int J Obes (Lond) 2008;32:201–10. doi: 10.1038/sj.ijo.0803760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Syme C, Abrahamowicz M, Mahboubi A, et al. Prenatal exposure to maternal cigarette smoking and accumulation of intra-abdominal fat during adolescence. Obesity (Silver Spring) 2010;18:1021–5. doi: 10.1038/oby.2009.354. [DOI] [PubMed] [Google Scholar]
- 20.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–7. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 21.Szyf M, Bick J. DNA methylation: a mechanism for embedding early life experiences in the genome. Child Dev. 2013;84:49–57. doi: 10.1111/j.1467-8624.2012.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–68. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 23.Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–8. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shukla S, Kavak E, Gregory M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li-Byarlay H, Li Y, Stroud H, et al. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc Natl Acad Sci U S A. 2013;110:12750–5. doi: 10.1073/pnas.1310735110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gelfman S, Ast G. When epigenetics meets alternative splicing: the roles of DNA methylation and GC architecture. Epigenomics. 2013;5:351–3. doi: 10.2217/epi.13.32. [DOI] [PubMed] [Google Scholar]
- 27.Breitling LP, Yang R, Korn B, et al. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–7. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shenker NS, Polidoro S, van Veldhoven K, et al. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22:843–51. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 29.Zeilinger S, Kuhnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joubert BR, Haberg SE, Nilsen RM, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120:1425–31. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee KW, Pausova Z. Cigarette smoking and DNA methylation. Front Genet. 2013;4:132. doi: 10.3389/fgene.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambers DS, Clark KE. The maternal and fetal physiologic effects of nicotine. Semin Perinatol. 1996;20:115–26. doi: 10.1016/s0146-0005(96)80079-6. [DOI] [PubMed] [Google Scholar]
- 33.Davies MN, Volta M, Pidsley R, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. ChemBioChem. 2011;12:206–22. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 35.Bell JT, Pai AA, Pickrell JK, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pausova Z, Paus T, Abrahamowicz M, et al. Genes, maternal smoking, and the offspring brain and body during adolescence: design of the Saguenay Youth Study. Hum Brain Mapp. 2007;28:502–18. doi: 10.1002/hbm.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74. [PubMed] [Google Scholar]
- 38.Thompson FE, Subar AF. Dietary assessment methodology. In: Bousky C, Coulston AM, editors. Nutrition in the prevention and treatment of disease. 2nd ed. Waltham (MA): Academic Press; 2008. [Google Scholar]
- 39.Berthiaume P, Lavalee C, Villeneuve M, et al. Volet nutrition. Québec (QC): Institute de la Statistique du Quebec; 2004. [accessed 2012 Nov. 12]. Enquête sociale et de santé auprès des enfants et des adolescents québécois; pp. 19–33. Available: www.stat.gouv.qc.ca/statistiques/sante/enfants-ados/alimentation/enfants-ados-nutrition.pdf. [Google Scholar]
- 40.Dlugos AM, Hamidovic A, Hodgkinson C, et al. OPRM1 gene variants modulate amphetamine-induced euphoria in humans. Genes Brain Behav. 2011;10:199–209. doi: 10.1111/j.1601-183X.2010.00655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 42.Hansen KD, Aryee M. R package version 1.2.0. Minfi: Analyze Illumina 450k Methylation Arrays. [Google Scholar]
- 43.Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium Human-Methylation450 microarray. Epigenetics. 2013;8:203–9. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cnattingius S. The epidemiology of smoking during pregnancy: smoking prevalence, maternal characteristics, and pregnancy outcomes. Nicotine Tob Res. 2004;6(Supple 2):S125–40. doi: 10.1080/14622200410001669187. [DOI] [PubMed] [Google Scholar]
- 45.Woolston A, Tu YK, Baxter PD, et al. A comparison of different approaches to unravel the latent structure within metabolic syndrome. PLoS ONE. 2012;7:e34410. doi: 10.1371/journal.pone.0034410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shabalina SA, Zaykin DV, Gris P, et al. Expansion of the human mu-opioid receptor gene architecture: novel functional variants. Hum Mol Genet. 2009;18:1037–51. doi: 10.1093/hmg/ddn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersen S, Baar C, Fladvad T, et al. The N-terminally truncated micro3 and micro3-like opioid receptors are transcribed from a novel promoter upstream of exon 2 in the human OPRM1 gene. PLoS ONE. 2013;8:e71024. doi: 10.1371/journal.pone.0071024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zilberman D, Gehring M, Tran RK, et al. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–9. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 50.Deaton AM, Webb S, Kerr AR, et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–86. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]