Abstract
Significant progress has been made to identify the cells at the foundation of tumorigenesis, the cancer cell of origin (CCO). The majority of data points towards resident adult stem cells (ASCs) or primitive progenitors as the CCO for those cancers studied, highlighting the importance of stem cells not only as propagators but also as initiators of cancer. Recent data suggest tumor initiation at the CCOs can be regulated through both intrinsic and extrinsic signals and that the identity of the CCOs and their propensity to initiate tumorigenesis is context dependent. In this review, we summarize some of the recent findings regarding CCOs and solid tumor initiation and highlight its relation with bona fide human cancer.
Keywords: cancer cells of origin, genetic hits, tumor initiation, niche
Decoding the cell of origin in cancer
Cancer is a complex disease due to the wide variety of cellular and molecular mechanisms associated with its initiation and progression. It is accepted that cancer cells divide and proliferate uncontrollably because of the accumulation of somatic mutations in normal tissue, which confers a selective growth advantage in the mutated progeny [1]. However, the cells that make up a tumor are heterogeneous; often making it difficult to determine the CCO, which is the normal cell that acquires the mutational load necessary to first initiate cancerous proliferation. Furthermore, since cancer is a transformative process, the cells composing advanced cancers may no longer contain morphological or molecular characteristics of the CCO [2]. The identity of the CCO could be critical to the generation of more effective treatments and preventative strategies. If CCOs can be identified and targeted specifically, it would be possible to stop cancer before it has a chance to undergo expansion. Molecular or physiological attributes specific to CCOs could be exploited to slow or block progression, thus avoiding treatments that simply kill dividing cells. This has led to significant recent efforts to define CCOs for all types of cancers, and numerous lines of evidence point towards ASCs as possible CCOs [3].
It is worth noting that CCOs are likely different from cancer stem cells. CCOs are the first cells to initiate a tumor, but cancer stem cells exist within a growing tumor and are defined by their ability to propagate tumors when serially transplanted [4]. Although cancer stem cells have many properties and gene expression patterns similar to ASCs, it is not clear whether there is a direct relation between the CCOs and cancer stem cells. It is possible and probable that cancer stem cells evolve from cells other than CCOs after tumor initiation. Cancer stem cells are covered elsewhere in several important reviews [4–6]. Here, we focus on CCOs and discuss the intrinsic and extrinsic mechanisms that regulate their ability to initiate various cancers.
ASCs and CCOs: is there a link?
ASCs make for a compelling target of tumorigenesis because of several basic properties. First, they are long lived, and thus capable of persisting long enough to accumulate DNA damage. Second, ASCs in general are multipotent (sometimes unipotent), and this could explain the variability of cell types found within most tumors. Third, ASCs, while normally quiescent, do have significant self-renewal potential, which could be critical for tumor expansion. ASCs are also capable of giving rise to a limited number of cell types [7–11]. For example, intestinal stem cells are able to differentiate into all the various secretory cell types of the villus, but not brain or muscle cells. Finally, experimental evidence from lineage tracing suggests that ASCs may in fact be the CCOs in various solid tumors [3]. However, numerous exceptions to this theory have also been identified, where various environmental insults appear to influence the nature of CCOs, and these are also discussed below.
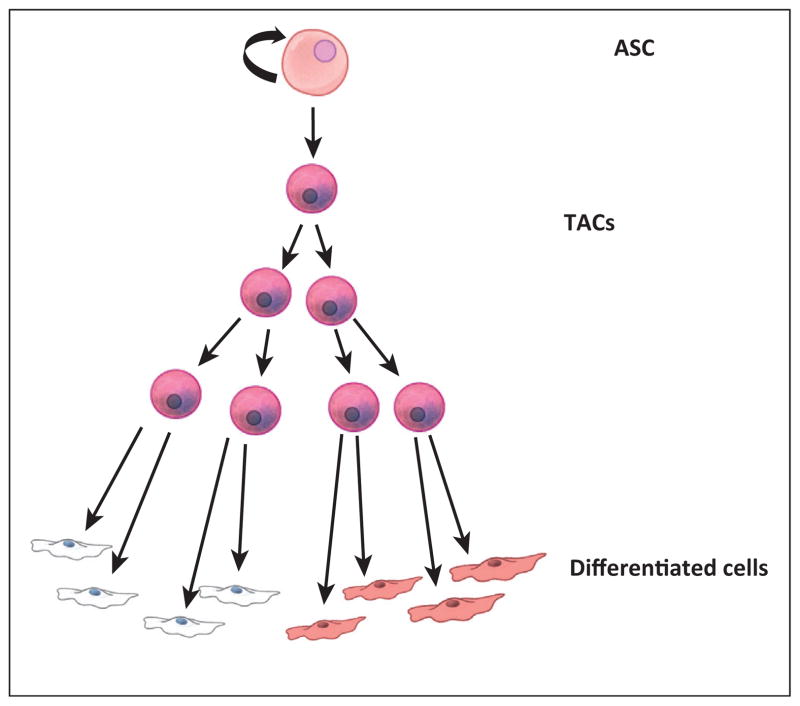
ASCs are found in many of the major adult organs and are essential for tissue homeostasis as well as regeneration in response to injury [12–17]. Most ASCs were discovered on the basis of their relative quiescence and their ability to reconstitute differentiated cell lineages of the tissue or organ in which they reside [8,18–22] (Figure 1). Either upon activation by natural turnover/cycling or in the case of regeneration due to injury, ASCs give rise to multilineage restricted progenitors or, as they are often called, transit amplifying cells (TACs) (Figure 1). These cells divide rapidly and then differentiate to generate the bulk of cells required for tissue turnover or regeneration. Due to their rapid division, TACs are also targeted by chemotherapeutics that act on cell division pathways to kill cancer cells, most obviously manifested as loss of hair and intestinal cells. In most cases, TACs quickly give rise to terminally differentiated cells that then perform the basic functions of the tissue or organ [23]. This type of hierarchy is present in most tissues, although tissues such as the epidermis and intestine experience tissue turnover and stem cell cycling with higher frequency [24]. While the identity of ASCs has not been confirmed in all tissues, most tissues are thought to possess them, with a few notable and controversial exceptions (liver and pancreas). These tissues are thought to regenerate by dedifferentiation of a differentiated cell type back to a proliferative state. However, this is thought to only happen in cases of regeneration in response to tissue injury; an example being the liver, where mature hepatocytes revert to a proliferative state in response to hepatectomy [25–30]. Cellular hierarchies based on developmental potential (ability to make more differentiated progeny) exist in all tissues, with stem cells and terminally differentiated cells at opposite ends of the spectrum.
Figure 1.
Typical ASC hierarchy. Within most mature tissues, a hierarchy exists where cell turnover is controlled first by a self-renewing ASC. These relatively quiescent cells give rise to TACs. TACs go through several rounds of division and then immediately differentiate to form the various specialized cells of the tissue. The delicate balance of cell fate decisions summarized in this figure are typically controlled by well-known signaling pathways (such as Wnt, Tgf, Bmp, Shh, and Fgf) acting through both autocrine and paracrine mechanisms. ASC, adult stem cell; TAC, transient amplifying cell.
ASCs appear to be regulated by intrinsic and extrinsic mechanisms. ASCs are intrinsically distinguished from their progeny on the basis of epigenetic, transcriptional, and potentially metabolic modes of regulation [14,31–34]. Dysregulation of these intrinsic factors such as the introduction of oncogenic mutations can result in cancer initiation [35]. Moreover, the extrinsic environment in which ASCs reside also regulates their identity and activity. ASCs live in specialized niches, which are often made up of several different cell types, frequently from different embryonic germ layers [19,36]. ASCs send and receive signals from their niche, such as growth factor signaling, extracellular matrix association, and mechanical regulation. Disruption of this signaling crosstalk or changes in the makeup of the niche can affect various aspects of ASC homeostasis such as the induction of ASC proliferation. Many of the pathways important for ASC to niche crosstalk are pathways also often aberrantly regulated in human cancer [9,37–39]. Below, we provide evidence for the role of ASCs as the CCOs of epithelial cancers including skin, intestinal, and prostate cancer.
Developmental hierarchy and cancer initiation
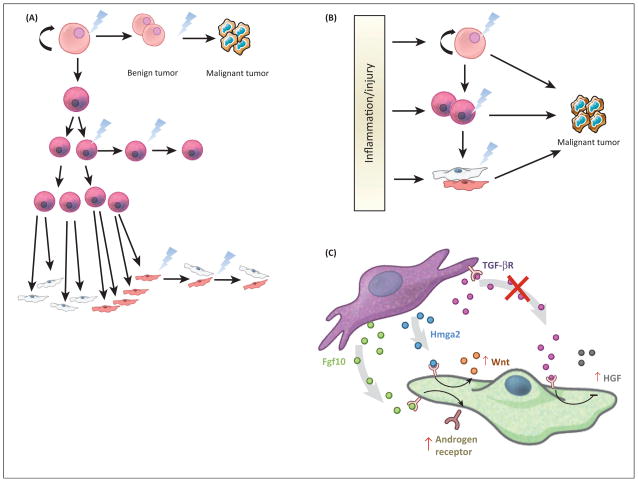
Tumors are heterogeneous and can display distinct phenotypic profiles such as morphology, gene expression, and proliferation. It has been assumed that the final morphology of the cells within a tumor can determine the CCO; however, cancer cells undergo a myriad of changes during tumor initiation to progression, suggesting that the final tumor cell may bear little resemblance to the CCO. Therefore, a priori, several scenarios are possible for tumor initiation (Figure 2A). With this realization, new lineage tracing methods have sought to uncover the origin of cancer from many tissues. Cell-type-specific promoters driving inducible Cre recombinase alleles has allowed for the prospective introduction of oncogenes or removal of tumor suppressors in postnatal murine models in intact tissue. These models are preferable to in vitro models or reconstitution/xenograft models as they contain the appropriate organization of the tissue and the presence of the native stromal, immune, lymphatic, nervous, and vascular systems. Taking advantage of lineage tracing mechanisms (CreER/CrePR) [40] and knock-in alleles [41] of oncogenes or floxed tumor suppressors [42], one can now initiate oncogenesis from particular cell types within an adult tissue by injection of an estrogen/progesterone antagonist. These experiments have suggested that pathological, retrospective studies on existing tumor tissue from human or mouse could be misleading when trying to identify the CCO.
Figure 2.
Tumor initiation scenarios and factors that can affect them. (A) Based on the existing literature, there are several scenarios by which tumor initiation could occur in the cell types of the stem cell hierarchy. Retrospective pathological studies have suggested that differentiated cells can initiate cancers, while prospective approaches to the study of cancer initiation using molecular genetics suggest that either stem or transit-amplifying cells are more relevant. (B) Data in various tissues proposes models whereby tumor initiation in unperturbed tissue follows scenario A. However, upon induction of dramatic changes to the microenvironment of tumor initiation, CCOs do not necessarily follow a typical stem cell hierarchy. There are several examples where terminally specified cells can dedifferentiate to create a cell that adopts stem cell properties and is thus able to become a CCO. (C) Changes to the signaling microenvironment can also affect tumor initiation. The same signaling pathways that are known to drive development of tissues are also implicated in tumor initiation and progression. For example, in the prostate, when stromal Tgfβ signaling is reduced, HGF is induced in the epithelium, leading to proliferation. In addition, upregulated expression of Fgf10 and Hmga2 in the meschyme can lead to increased androgen receptor signaling and Wnt signaling in the adjacent epithelium (respectively), again leading to abnormal epithelial proliferation. These cases are demonstrative of microenvironmental aberrations that can potentially enhance or induce epithelial cancer. CCO, cancer cell of origin; Fgf10, fibroblast growth factor 10; HGF, hepatocyte growth factor; Hmga2,
The simplest interpretation of the data produced by these new prospective approaches is that ASCs are more likely to serve as CCOs in many cancers [3], such as those of the skin, prostate, intestine, and brain. Since ASCs are continuously available to maintain tissue homeostasis and to repopulate cellular compartments lost during injury in tissue, it has been speculated that only ASCs are present in the tissue for a sufficient length of time to accumulate the necessary genetic mutations for tumorigenic transformation and cancer initiation (Figure 2). Below, we discuss the current understanding of the CCOs of these cancers, which represent a variety of solid tumors from well-described tissues with defined hierarchies of differentiation potential. We propose that the CCO is context dependent and can change depending on intrinsic (genetic mutation and cell of origin) and extrinsic (homeostasis or injury/inflammation) stimuli.
Intrinsic factors influence CCOs
The developmental origins for each hierarchy could yield insight into the mechanisms by which tumors arise from ASCs, because the same dominant signaling pathways that specify cell fate also play important roles in ASC homeostasis [7,35]. Indeed, developmental pathways including Wnt, Tgfβ, Bmp, Shh, Fgf, and Notch signaling, have all been implicated in the development of epithelial tissues, and for many, also in the homeostasis and proportion of ASCs and their progeny [9,37,43–53]. Gain or loss of function in these pathways often disrupts the balance between ASCs and their progeny and can act as drivers of tumor initiation. ASCs from epithelial tissue share similar regulatory schemes and routes to tumor initiation, therefore, it could be that each of them also shares defense mechanisms to prevent aberrant growth, and that lessons learned in one could be applicable to all. The degree to which authentic tumor initiation is caused by an imbalance of these pathways to maintain homeostasis versus more dramatic genetic alterations (activation of oncogenes, loss of tumor suppressors) has only been explored experimentally in murine models. However, correlative evidence from genome sequencing in human tumors suggests the possibility that disruption of these pathways could lead to excess proliferation that is then exacerbated by oncogene expression or loss of tumor suppressors [54–64].
We discuss several examples of how the accumulation of oncogenic mutations and aberrant signaling of developmental pathways can promote tumor formation in a cell-type-dependent manner. Furthermore, we discuss the emerging concept of stem cell quiescence as a barrier to tumorigenesis suggesting intrinsic cell cycle dependent changes may also regulate tumor initiation.
Oncogenic mutations in ASCs initiate cutaneous squamous cell carcinoma (SCC)
Conflicting retrospective pathological studies and experimental evidence have made it difficult to define the CCO of cutaneous SCC. Since it is pathologically defined by the presence of squamous cells, or terminally differentiated cells from the interfollicular epidermis, and not from the hair follicle, it was assumed that SCC arose from differentiated cells of the interfollicular epidermis, and not from the ASC population nor from hair follicles. By contrast, experimental evidence implicated cells of the hair follicle in SCC initiation [65–67]: the rate of tumor formation was affected by depilation, or hair removal [68]; and deletion of a hair follicle stem cell specific gene (CD34) affected tumor initiation [69].
Therefore, the question remained, does oncogenic mutation in ASCs or TACs of the hair follicle result in SCC? First, it is important to point out the convention that it takes multiple genetic hits to create bona fide cancer [70–72]. In murine models, the consensus is 2–3 hits, while in human settings, it is thought that at least three hits are required to transform cells. Of course, genetic background (sensitizing alleles [73]), injury, and inflammation can all affect the number of hits that lead to individual tumors, but regardless, cells do implement defense mechanisms to avoid transformation. Loss of defense mechanisms such as tumor suppressors alone is insufficient to drive cancer, and typically, a proliferative stimulus is also required [74]. Recent lineage tracing revealed that the expression of oncogenic Kras (KrasG12D) and the conditional deletion of tumor suppressor p53 in hair follicle ASCs (through the use of the K15-CrePR allele) resulted in the initiation of tumors, while no tumors were observed in its descendent TACs (through the use of the Shh–CreER allele), which were also exposed to the same oncogenic mutation [75,76]. Furthermore, the coupling of DMBA/TPA chemical carcinogenesis to lineage tracing demonstrated that the vast majority of SCC tumors derived from hair follicle ASCs, even when all cells of the epidermis received a carcinogenic insult [77]. Together, these data suggest not only that SCC can arise from the hair follicle, but that the cells with the greatest developmental capacity, in this case ASCs, can serve as CCOs.
Mutation of developmental pathways in ASCs results in basal cell carcinoma (BCC)
BCC is often characterized by aberrant Sonic Hedgehog (SHH) signaling. Indeed, the loss of SHH pathway component Patched (Ptch1) is found in 30–60% of human BCCs, while activated Smoothened (Smo) is found in 10–20% of human BCCs [78]. SHH signaling has been observed to regulate ASCs and is involved in the maintenance of tissue homeostasis. The binding of SHH to the receptor Ptch1 results in its repression and activation of Smo, which promotes the nuclear translocation of Gli transcription factors, consequently increasing the self-renewal capacity of some ASCs [10].
BCC was thought to derive from the hair follicle due its cellular similarity to the hair follicle [78]. However, using an inducible Cre allele, driven by either a basal keratinocyte promoter (Keratin 14), a hair follicle TAC promoter (Shh), or a hair follicle stem cell promoter (Keratin 15) to drive expression of activated Smo, it was found that cells adjacent to the follicle but within the interfollicular epidermis were the CCOs (Table 1) [79]. Coupled with careful temporal analysis, it was demonstrated that BCC tumors first initiate at the junction between follicles and the interfollicular epidermis. By contrast, another group suggested that hair follicle stem cells are the CCO for BCC in a model that used heterozygous loss of function in Ptch1 via either the Keratin 15 or Keratin 14 Cre and ionizing radiation as a mutagen [80]. However, the additional loss of p53 in the interfollicular epidermis facilitated initiation of BCC from this site. Additionally, a recent finding suggested that both interfollicular and hair follicle stem cells could give rise to BCC using a variety of lineage drivers (K15, Lgr5, K5, and K14) and a different oncogenic allele [81]. A constitutively active allele of the Gli2 transcription factor, which mimics the activation of the Shh pathway, led to the formation of two different BCC subtypes from either the interfollicular epidermis or the hair follicle [81]. While it is possible that the differences lay in the sensitivity of various cell types to the dose of pathway activation, these results, coupled with those from SCC experiments demonstrate that mutation in an appropriate cell of origin can lead to specific cancer subtypes. Whether the same is true for murine models of tumor initiation in other tissues is not yet clear, as so far, most models have only shown one particular cell type to be the cell of origin in a given tissue.
Table 1.
Experimental protocols and results for two well-explored CCO models
| Tumor type | Cre driver Cell type | Oncogenes/tumor suppressors lineage tracer | CCO | Refs |
|---|---|---|---|---|
| BCC | K14Cre-ER ShhCre-ER K15Cre-ER |
Rosa-LSL-SmoM2 | Interfollicular epidermis | Youssef et al Nature Cell Biology 2010 |
| K15Cre-PR K14Cre-ER |
Ptch1+/1 + Ionizing Radiation+ LSY-YFP +p53 KO |
HFSCs HFSCs and interfollicular epidermis |
Wang et al Cancer Cell 2011 |
|
| K15Cre-PR Lgr5Cre-ER K5Cre-ER |
Rosa-LSL-rtTA tetO-Gli2deltaN |
HFSCs = nodular BCC Interfollicular epidermis = superficial BCC |
Grachtchouk et al JCI 2011 |
|
| K15Cre-PR | Rosa-LSL-SmoM2 + wounding |
HFSCs | Wong et al PNAS 2011 |
|
| K5Cre-tTA K5Cre-PR Lgr5Cre-ER |
TRE-Gli1 Ptch1 flox/flox Ptch1 flox/flox |
Interfollicular epidermis HFSCs only when wounded |
Kasper et al PNAS 2011 |
|
| Intestinal Adenoma | Cd133Cre-ER | Activated β-catenin | Crypt stem cells | Zhu et al Nature 2009 |
| Lgr5Cre-ER AH-Cre |
APC KO | Crypt stem cells | Barker et al Nature 2009 |
|
| Xbp1sCre-ER | Activated β-catenin and IkB-α KO or KrasG12D |
Differentiated cells via dedifferentiation | Schwitalla et al Cell 2013 |
Two populations of CCOs in the intestine
Recent lineage tracing experiments in the small intestine have produced a variety of models for the identity of ASCs. There appear to be at least two candidates for ASCs in the intestinal crypt; one at the base between Paneth cells, called the crypt base columnar cells (CBCs), and the other roughly four cells up from the bottom of the crypt, named the +4 cells. The CBCs are marked by Lgr5, whereas the +4 cells are labeled by Bmi1. A variety of lineage tracing and lineage ablation protocols have been used to resolve the stem cell hierarchy in these crypts (Table 1) [32,33,82–91]. It seems that the Lgr5/CD133+ CBCs are important in normal homeostasis of the villus, dividing once a day, whereas the +4 Bmi1+ cells are a reserve stem cell population that can replenish the CBCs after injury [33,90]. Furthermore, ASCs residing in the base of the crypt give rise to a TAC population found higher up in the crypt, which in turn gives rise to a variety of differentiated cell types that populate the villus. With regard to tumorigenesis, both Lgr5/CD133+ CBCs and Bmi1+ +4 cells are capable of acting as CCOs in adenoma initiation, when Apc is conditionally deleted, whereas TACs residing further up the villus appear to be less sensitive to transformation, at least in undisturbed intestinal epithelium [33,82,87,92,93]. As APC is a critical scaffold for components of the Wnt signaling pathway, this is also further evidence where cancer can exploit a developmental signaling or cell fate pathway to drive transformation [94].
Prostate cancer: luminal or basal origin?
Models for both murine and human prostate cancer have produced conflicting conclusions within the field as to whether the CCO is of basal or luminal origin. Debate has arisen as to whether the stem cells of the prostate reside in either the basal or luminal populations [31,95–101]. Using a broader range of lineage tracing alleles, it was suggested that a multipotent population arises from the basal population, while separate unipotent progenitors populate the neuroendocrine and luminal pools [102]. The lack of a consensus on the identity of ASCs of the prostate has also clouded the interpretation of CCO studies for the prostate.
Similar to the discrepancies observed for SCC/BCC, much of the debate regarding CCOs for prostate cancer centers on the fact that prostate tumors typically adopt a morphology consistent with a luminal origin, while experimental data often point towards a basal source for CCOs. Human prostatic epithelial transplantation studies, which do not include a native stromal and immune component, indicated a basal CCO with MYC, AKT or ERG as oncogenic drivers. By contrast, genetically modified mouse models that used Pten deletion implicated both basal and luminal cells as CCOs, depending on the targeting alleles and tumorigenic strategies used [100–103]. In addition, one study showed that initiation from human basal cells generates transformed luminal-like cells that are able to propagate the tumor [95]. Together, these results suggest that the identity of the CCO for prostate cancer could be dependent on cellular, genetic, and environmental contexts, and further work will be needed to address whether differences exist between human and mouse models systems or whether the differences are caused by nonequivalent cell-intrinsic and cell-extrinsic stimuli.
Heterogeneity of tumor initiators and tumor phenotypes
The experimental models described here have proven to yield important insights into tumor initiation and CCOs. However, there are technical limitations to these models that ignore the heterogeneity of bona fide cancer initiation. Tumors are thought to be initiated in a clonal fashion as a result of mutations in individual cells surrounded by relatively normal cells, therefore, it is postulated that the heterogeneity observed in many tumors is due to either subtle variations across seemingly homogenous CCO populations; the multipotency of a single CCO; the adaptation to selective pressures within the tumor during progression, or by the resulting inflammation that typically accompanies tumor formation (discussed below). New data also suggests another possibility: interactions between individual tumor-initiating clones lead to diverse phenotypes [104]. The conditional or inducible Cre alleles used in the experimental designs described above allow for targeting of oncogenic hits to individual cells, but it is difficult to then observe the various events associated with tumor progression. In addition, if observations are not made until high grade tumors are formed, it is difficult to determine whether the tumors observed arose clonally or from several adjacent cells. Therefore, these models cannot easily identify the source of tumor heterogeneity. Alternatively, tumor initiation followed by careful lineage tracing can reveal the existence of specialized tumor-propagating cells, or cancer stem cells [105,106]. In addition, this type of protocol has also shown that tumor-propagating cells tend to expand over time in at least one malignant cancer model [107]. As murine models of cancer initiation increase in sophistication, it is likely that some combination of clonal lineage trace coupled with bar-coding could reveal the source of tumor heterogeneity. For now, the heterogeneity of phenotypes observed in many murine models could be a technical limitation that creates confounding interpretations and unnecessary controversies.
Intrinsic quiescence of ASCs as a tumor suppressor
Another key aspect of ASC biology that could potentially influence their propensity to initiate tumors relates to their inherent quiescence. In the hair follicle, ASCs go through synchronized waves of proliferation at the start of hair cycles to regenerate the bottom of the follicle and make a new hair shaft [47,108–110]. They then return to quiescence until the next hair cycle, which can take several weeks. As a result, ASCs in the hair follicle are only proliferative for brief periods. Recent work showed that when these cells are quiescent, they are unable to respond to the induction of the oncogene Ras and/or removal of the tumor suppressor p53 [111]. These data demonstrate that cellular quiescence suppresses tumor growth, which is dependent on the tumor suppressor Pten, as deletion of Pten alone had no effect on the hair cycle. Simply, activation of the hair cycle by normal mechanisms can act as an additional tumor-promoting insult to ASCs that have already accumulated genetic hits. The hair cycle is controlled by a variety of signaling pathways that converge on the stem cell niche. Some of these signals are local (Wnt, BMP, Fgf, Tgfβ), while others are systemic (hormones) [108,110,112]. This quiescent state may help explain why sun-exposed skin does not continually generate new tumors or papillomas despite the presence of Ras mutations, as regulation of these pathways serves to maintain the appropriate activation state of this stem cell population (inhibitory: Bmp and Fgf18; stimulatory: Wnt and Tgfβ) [37,113–118].
These results also suggest that the regulatory mechanisms that affect the hair cycle could, as a result, affect tumor initiation [47,108,110,118,119]. Furthermore, it is possible that studies implicating particular genes or signaling pathways in tumor initiation such as Ras, could have indirectly impacted tumor initiation by affecting the hair cycle instead. Indeed, loss of Pten by itself did not affect initiation or progression of the hair cycle, but instead affected the sensitivity of the ASCs to Ras activation [111]. Whether the dependence on hair cycle status is specific for Ras-induced tumors remains unclear because a wide variety of genetic hits have been implicated in SCC and head and neck SCC [57,58]. Whether ASCs in other tissues are also similarly refractory to transformation and tumor initiation due to tumor suppression mediated by quiescence remains to be determined.
Extrinsic factors influence CCO
While intrinsic factors such as the ones discussed above can determine the CCO of many epithelial cancers, recent evidence has suggested that extrinsic factors such as injury, inflammation, and signals from the stem cell niche can also reprogram cells to become CCOs. These findings suggest other cells beyond ASCs can also act as CCOs under varying conditions. We discuss the role of these extrinsic factors on redefining the CCO of epithelial cancer.
Does inflammation or injury reprogram TACs or differentiated cells to initiate tumors?
It is unclear whether acute or chronic inflammation is necessary for, or coincident to, tumor initiation (Figure 2B). In support of a role for inflammation, many cancers have been also associated with chronic inflammatory diseases or wounds [120–123]. If inflammation is a necessary component in tumor initiation, animal models that do not take inflammation into account might have fewer experimental variables to contend with, but could also fail to accurately model disease as it occurs in human tissues.
As previously mentioned, cells of the interfollicular epidermis are considered the CCO for BCC [79]; however, several reports suggest that the CCO for BCC can change when skin is sensitized by inflammation or injury [124–126]. It has been known for some time that hair follicle stem cells (HFSCs) can migrate out of the follicle towards interfollicular wounds to facilitate healing [127,128]. This phenomenon can complicate the interpretation of lineage tracing experiments as it impairs the ability to identify the origin of cells that reconstitute a wound site. Using strategies employing Smo activation, Ptch1 inactivation, or Gli1 overexpression in the hair follicle stem cell population, it was shown that injury can enable HFSCs to act as CCOs for BCC [124,126]. When the Shh pathway is activated during wound healing, tumorigenesis is initiated, even from HFSCs which are normally refractive to tumorigenesis at lower doses of Shh. Similarly, differentiated endocrine cells of the pancreas are able to change fate and serve as CCOs for pancreatic ductal adenocarcinoma when experimental chronic pancreatic injury is induced [129]. In the intestine, inflammation and injury also disrupt the basic hierarchy and expand the pool of possible CCOs [34,91,130–133]. In particular, a recent report demonstrated that the activation of the pro-inflammatory nuclear factor (NF)-κb pathway increased the relative dose of oncogenic Wnt stimulation by stabilizing β-catenin, the obligatory signal transducer for the Wnt pathway. This significantly enhanced downstream Wnt signaling, causing robust dedifferentiation of terminally differentiated villus cells expressing markers of stem cells, which were able to generate intestinal adenomas[132]. This study suggests inflammation could alter the CCOs by inducing cell identity changes and imparting stem cell characteristics on TACs or differentiated cells [132,134]. To support this finding, mice treated with infectious bacteria showed prostatic inflammation, which resulted in the differentiation of basal cells towards the luminal lineage [135].
The possibility that inflammation can reprogram cell fate could confound the ability to relate results from murine models to actual human tumors, where inflammation is always associated with either tumor initiation or progression. Whether authentic human CCOs are redefined by inflammation or injury will remain unclear until more suitable models are generated.
Regulation of tumorigenesis by the stem cell niche
While much of the focus on the origins of epithelial tumors has been on ASCs and cell intrinsic tumorigenic stimuli, it is important to consider the potential contribution of the niche where these cells reside (Figure 2C). It is possible that alterations in paracrine signaling from niche cells could enhance, enable, or alter the CCO during tumor initiation from ASCs. Indeed, recent studies confirm that epithelial tumors can be generated through cell-extrinsic means via the microenvironment. Significantly, both gain and loss of function experiments for various signaling modifiers within the niche has uncovered profound trans effects on epithelial tumorigenesis [136,137].
In the prostate, several studies manipulated the activity of signaling pathways both known to play roles in development and cancer in the mesenchyme surrounding the epithelia. Notably, inactivation of the transforming growth factor (TGF)-β receptor, overexpression of fibroblast growth factor ( FGF)10 or overexpression of Hmga2 in the mesenchyme can lead to benign tumorigenesis from the epithelium [138–140]. In each of these cases, perturbation of the mesenchyme led to dramatic hyperplasia of the epithelium, presumably through the secretion of factors that facilitate tumorigenesis: loss of TGFβ signaling induced hepatocyte growth factor; activation of FGF10 increased androgen receptor signaling in the adjacent epithelium; and Hmga2 overexpression in the mesenchyme induced Wnt signaling in the epithelium (Figure 2C). It is important to note that these studies did not propose that aberrant signaling from the mesenchyme altered the identity of the CCO ; only that activation of CCOs in the adjacent epithelium was enhanced. These data suggest that the niche or mesenchyme surrounding CCOs should be considered, because environmental signals emanating from these cells could shift the identity, activation, or signaling within the CCO. As a result, these data also imply that xenograft models of cancer could be influenced by the grafting location or niche into which cells are delivered. In the case of the prostate aggregation model pioneered by Witte and colleagues, the key to the experimental design is combining epithelial and mesenchymal cells into the graft. However, dissociation and transplantation of tissue likely induces a heretofore-unknown degree of inflammation. These results emphasize the importance of models that allow for investigation of cancer initiation in situ, in intact tissues that retain species-specific stromal components, but for studies of human cancers, this is a necessary trade-off.
Concluding remarks
Taken together, it is evident that ASC populations are often more susceptible to tumor initiation than the TAC and differentiated cell populations. It is also clear, however, that the choice of tumorigenic mutation used, the relative activity of the cell sustaining the tumorigenic load, and the cell extrinsic context can highly influence the CCO and course of cancer initiation and progression. As we formulate a better picture of the rules regarding how a tumor first develops using experimental variables, the future challenge will be to determine which of these processes are the most relevant or occur most often during the initiation of human cancers. These studies have been and continue to be instrumental in furthering our understanding of how cancer develops, which cannot be determined or well inferred from examination of end-point human tumors. Better understanding of cancer origins will provide the tools to develop better diagnostic tests that can detect cancer development at earlier stages, which will improve overall survival.
Current and future research is centered on how to best refine the CCO, the nature of oncogenic mutations, and the context into which tumors arise. For the skin, researchers have access to a wide variety of inducible, cell-type-specific alleles to target various cell types. However, as lineage tracing becomes more sophisticated, it may become clear that stem cell niches could contain multiple types of stem cells that may be targeted by more specific promoters [17,141–143]. In fact, it is possible that much of the confounding data generated for CCOs could be resolved with more specific targeting tools.
Furthermore, the wealth of sequencing data on human tumor samples has revealed an alarming number of mutations present in cancers [55,57,58,144–148]. Many of the newly identified mutations are related to the regulation of the epigenome or chromatin biology [148–150]. Since many of these enzymes have only recently been identified and characterized, it is not clear how these mutations directly affect tumor initiation or progression. In addition, the nature of these mutations can be difficult to discern, such as whether they represent gain or loss of function alleles. These mutations should be explored to understand human cancer by more than simple induction or deletion. These mutations will need to be introduced at the genetic level by gene replacement on various genetic backgrounds to understand whether they represent drivers or mediators of either tumor initiation or progression. These experiments could be facilitated by recent technologies that allow for facilitated gene editing over homologous recombination (e.g. Crispr/Cas9 and TALEN) [151–154].
Only by delivering particular mutations to particular cell types within a tissue will the nature of cancer initiation be truly understood. However, significant effort will be required to understand the environment into which these mutations are induced with regards to injury, inflammation, and normal turnover. Furthermore, in murine models, genetic hits are typically delivered in concert due to technical limitations and dependence on Cre mediated mechanisms. However, this is probably not the typical mode in which oncogenic hits are acquired in bona fide cancers, where genetic hits are gained over time. This last aspect could be the most difficult to tease apart, as we know little about the context of initiation of human cancers and most of our knowledge about tumor initiation in humans is correlative.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 3.Blanpain C. Tracing the cellular origin of cancer. Nat Cell Biol. 2013;15:126–134. doi: 10.1038/ncb2657. [DOI] [PubMed] [Google Scholar]
- 4.Reya T, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Kreso A, Dick JE. Evolution of the Cancer Stem Cell Model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Lowry WE, Richter L. Signaling in adult stem cells. Front Biosci. 2007;12:3911–3927. doi: 10.2741/2360. [DOI] [PubMed] [Google Scholar]
- 8.Nishikawa SI, et al. Niche required for inducing quiescent stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:67–71. doi: 10.1101/sqb.2008.73.024. [DOI] [PubMed] [Google Scholar]
- 9.Lowry WE, et al. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19:1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palma V, Ruiz i Altaba A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131:337–345. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- 11.Lim DA, et al. Noggin antagonizes BMP signaling to create a niche for adult neurogenesis. Neuron. 2000;28:713–726. doi: 10.1016/s0896-6273(00)00148-3. [DOI] [PubMed] [Google Scholar]
- 12.Quinones-Hinojosa A, et al. Cellular composition and cytoarchitecture of the adult human subventricular zone: a niche of neural stem cells. J Comp Neurol. 2006;494:415–434. doi: 10.1002/cne.20798. [DOI] [PubMed] [Google Scholar]
- 13.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 14.Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheshier SH, et al. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem F cells. Proc Natl Acad Sci USA. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaks V, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 18.Arai F, Suda T. Maintenance of quiescent hematopoietic stem cells in the osteoblastic niche. Ann N Y Acad Sci. 2007;1106:41–53. doi: 10.1196/annals.1392.005. [DOI] [PubMed] [Google Scholar]
- 19.Rezza A, et al. Adult stem cell niches: cellular and molecular components. Curr Top Dev Biol. 2014;107:333–372. doi: 10.1016/B978-0-12-416022-4.00012-3. [DOI] [PubMed] [Google Scholar]
- 20.Watt FM, Jensen KB. Epidermal stem cell diversity and quiescence. EMBO Mol Med. 2009;1:260–267. doi: 10.1002/emmm.200900033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arai F, Suda T. StemBook. Cambridge (MA): 2008. Quiescent stem cells in the niche. [PubMed] [Google Scholar]
- 22.Dhawan J, Rando TA. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005;15:666–673. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Diaz-Flores L, Jr, et al. Adult stem and transit-amplifying cell location. Histol Histopathol. 2006;21:995–1027. doi: 10.14670/HH-21.995. [DOI] [PubMed] [Google Scholar]
- 24.Barker N, et al. Tissue-resident adult stem cell populations of rapidly self-renewing organs. Cell Stem Cell. 2010;7:656–670. doi: 10.1016/j.stem.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 25.Stanger BZ, et al. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- 26.Dor Y, et al. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 27.Choi TY, et al. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146:776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorrell C, et al. Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 2011;25:1193–1203. doi: 10.1101/gad.2029411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santoni-Rugiu E, et al. Progenitor cells in liver regeneration: molecular responses controlling their activation and expansion. APMIS. 2005;113 (11–12):876–902. doi: 10.1111/j.1600-0463.2005.apm_386.x. [DOI] [PubMed] [Google Scholar]
- 31.Lawson DA, et al. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan KS, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Landeghem L, et al. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1111–G1132. doi: 10.1152/ajpgi.00519.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sancho E, et al. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- 36.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 37.Oshimori N, Fuchs E. Paracrine TGF-beta signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell. 2012;10:63–75. doi: 10.1016/j.stem.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chenn A, Walsh CA. Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in beta-catenin overexpressing transgenic mice. Cereb Cortex. 2003;13:599–606. doi: 10.1093/cercor/13.6.599. [DOI] [PubMed] [Google Scholar]
- 39.Brzoska E, et al. Regulation of muscle stem cells activation: the role of growth factors and extracellular matrix. Vitam Horm. 2011;87:239–276. doi: 10.1016/B978-0-12-386015-6.00031-7. [DOI] [PubMed] [Google Scholar]
- 40.Metzger D, et al. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alcantara Llaguno S, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carolan PJ, Melton DA. New findings in pancreatic and intestinal endocrine development to advance regenerative medicine. Curr Opin Endocrinol Diabetes Obes. 2013;20:1–7. doi: 10.1097/MED.0b013e32835bc380. [DOI] [PubMed] [Google Scholar]
- 44.Alexson TO, et al. Notch signaling is required to maintain all neural stem cell populations–irrespective of spatial or temporal niche. Dev Neurosci. 2006;28 (1–2):34–48. doi: 10.1159/000090751. [DOI] [PubMed] [Google Scholar]
- 45.Fre S, et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 46.Mutch CA, et al. Beta-catenin signaling negatively regulates intermediate progenitor population numbers in the developing cortex. PLoS ONE. 2010;5:e12376. doi: 10.1371/journal.pone.0012376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plikus MV, et al. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 2008;451:340–344. doi: 10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noles SR, Chenn A. Cadherin inhibition of beta-catenin signaling regulates the proliferation and differentiation of neural precursor cells. Mol Cell Neurosci. 2007;35:549–558. doi: 10.1016/j.mcn.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 49.Pinto D, Clevers H. Wnt, stem cells and cancer in the intestine. Biol Cell. 2005;97:185–196. doi: 10.1042/BC20040094. [DOI] [PubMed] [Google Scholar]
- 50.Zechner D, et al. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–418. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 51.Battista D, et al. Neurogenic niche modulation by activated microglia: transforming growth factor beta increases neurogenesis in the adult dentate gyrus. Eur J Neurosci. 2006;23:83–93. doi: 10.1111/j.1460-9568.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- 52.Maroof AM, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lai K, et al. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- 54.Makinen N, et al. Exomic landscape of MED12 mutation-negative and -positive uterine leiomyomas. Int J Cancer. 2014;134:1008–1012. doi: 10.1002/ijc.28410. [DOI] [PubMed] [Google Scholar]
- 55.Li S, Mao M. Next generation sequencing reveals genetic landscape of hepatocellular carcinomas. Cancer Lett. 2013;340:247–253. doi: 10.1016/j.canlet.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 56.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nikolic A, et al. Structural and functional analysis of SMAD4 gene promoter in malignant pancreatic and colorectal tissues: detection of two novel polymorphic nucleotide repeats. Cancer Epidemiol. 2011;35:265–271. doi: 10.1016/j.canep.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Durinck S, et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011;1:137–143. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sameer AS, et al. SMAD4–molecular gladiator of the TGF-beta signaling is trampled upon by mutational insufficiency in colorectal carcinoma of Kashmiri population: an analysis with relation to KRAS proto-oncogene. BMC Cancer. 2010;10:300. doi: 10.1186/1471-2407-10-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loh K, et al. Bone morphogenic protein 3 inactivation is an early and frequent event in colorectal cancer development. Genes Chromosomes Cancer. 2008;47:449–460. doi: 10.1002/gcc.20552. [DOI] [PubMed] [Google Scholar]
- 63.Chan SL, et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Lab Invest. 2007;87:644–650. doi: 10.1038/labinvest.3700547. [DOI] [PubMed] [Google Scholar]
- 64.Iwai S, et al. Mutations of the APC, beta-catenin, and axin 1 genes and cytoplasmic accumulation of beta-catenin in oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2005;131:773–782. doi: 10.1007/s00432-005-0027-y. [DOI] [PubMed] [Google Scholar]
- 65.Miller SJ, et al. Mouse skin is particularly susceptible to tumor initiation during early anagen of the hair cycle: possible involvement of hair follicle stem cells. J Invest Dermatol. 1993;101:591–594. doi: 10.1111/1523-1747.ep12366045. [DOI] [PubMed] [Google Scholar]
- 66.Lavker RM, et al. Hair follicle stem cells: their location, role in hair cycle, and involvement in skin tumor formation. J Invest Dermatol. 1993;101 (1 Suppl):16S–26S. doi: 10.1111/1523-1747.ep12362556. [DOI] [PubMed] [Google Scholar]
- 67.Andreasen E, Borum K. The influence of the mouse hair cycle on 9, 10-dimethyl-1, 2-benzanthracene-induced skin tumors. Acta Pathol Microbiol Scand Suppl. 1956;39 (Suppl 111):76–77. doi: 10.1111/j.1600-0463.1956.tb06741.x. [DOI] [PubMed] [Google Scholar]
- 68.Morris RJ, et al. Evidence that the epidermal targets of carcinogen action are found in the interfollicular epidermis of infundibulum as well as in the hair follicles. Cancer Res. 2000;60:226–229. [PubMed] [Google Scholar]
- 69.Trempus CS, et al. CD34 expression by hair follicle stem cells is required for skin tumor development in mice. CancerRes. 2007;67:4173–4181. doi: 10.1158/0008-5472.CAN-06-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whitcomb D, Greer J. Germ-line mutations, pancreatic inflammation, and pancreatic cancer. Clin Gastroenterol Hepatol. 2009;7 (11 Suppl):S29–S34. doi: 10.1016/j.cgh.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 71.Ceol CJ, et al. APC and colon cancer: two hits for one. Nat Med. 2007;13:1286–1287. doi: 10.1038/nm1107-1286. [DOI] [PubMed] [Google Scholar]
- 72.Komarova NL, Wang L. Initiation of colorectal cancer: where do the two hits hit? Cell Cycle. 2004;3:1558–1565. doi: 10.4161/cc.3.12.1186. [DOI] [PubMed] [Google Scholar]
- 73.Quigley DA, et al. Genetic architecture of mouse skin inflammation and tumour susceptibility. Nature. 2009;458:505–508. doi: 10.1038/nature07683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mondal S, et al. Two-stage chemical oncogenesis in cultures of C3H/10T1/2 cells. Cancer Res. 1976;36 (7 PT 1):2254–2260. [PubMed] [Google Scholar]
- 75.Lapouge G, et al. Identifying the cellular origin of squamous skin tumors. Proc Natl Acad Sci USA. 2011;108:7431–7436. doi: 10.1073/pnas.1012720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.White AC, et al. Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc Natl Acad Sci USA. 2011;108:7425–7430. doi: 10.1073/pnas.1012670108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li S, et al. A keratin 15 containing stem cell population from the hair follicle contributes to squamous papilloma development in the mouse. Mol Carcinog. 2012 doi: 10.1002/mc.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Donovan J. Review of the hair follicle origin hypothesis for basal cell carcinoma. Dermatol Surg. 2009;35:1311–1323. doi: 10.1111/j.1524-4725.2009.01236.x. [DOI] [PubMed] [Google Scholar]
- 79.Youssef KK, et al. Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol. 2010;12:299–305. doi: 10.1038/ncb2031. [DOI] [PubMed] [Google Scholar]
- 80.Wang GY, et al. Basal cell carcinomas arise from hair follicle stem cells in Ptch1(+/−) mice. Cancer Cell. 2011;19:114–124. doi: 10.1016/j.ccr.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grachtchouk M, et al. Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J Clin Invest. 2011;121:1768–1781. doi: 10.1172/JCI46307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 83.Walther V, Graham TA. Location, location, location! The reality of life for an intestinal stem cell in the crypt. J Pathol. 2014 doi: 10.1002/path.4370. [DOI] [PubMed] [Google Scholar]
- 84.Ritsma L, et al. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature. 2014;507:362–365. doi: 10.1038/nature12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Powell AE, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.May R, et al. Doublecortin and CaM kinase-like-1 and leucine-rich-repeat-containing G-protein-coupled receptor mark quiescent and cycling intestinal stem cells, respectively. Stem Cells. 2009;27:2571–2579. doi: 10.1002/stem.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu L, et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maria Cambuli F, et al. Brief report: musashi1-eGFP mice, a new tool for differential isolation of the intestinal stem cell populations. Stem Cells. 2013;31:2273–2278. doi: 10.1002/stem.1428. [DOI] [PubMed] [Google Scholar]
- 89.Munoz J, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tian H, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takeda N, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schepers AG, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 93.Simon E, et al. The spatial distribution of LGR5+ cells correlates with gastric cancer progression. PLoS ONE. 2012;7:e35486. doi: 10.1371/journal.pone.0035486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alman BA, et al. Increased beta-catenin protein and somatic APC mutations in sporadic aggressive fibromatoses (desmoid tumors) Am J Pathol. 1997;151:329–334. [PMC free article] [PubMed] [Google Scholar]
- 95.Stoyanova T, et al. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci USA. 2013;110:20111–20116. doi: 10.1073/pnas.1320565110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Garraway IP, et al. Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. Prostate. 2010;70:491–501. doi: 10.1002/pros.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goldstein AS, et al. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci USA. 2008;105:20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xin L, et al. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells. 2007;25:2760–2769. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- 99.Xin L, et al. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci USA. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang ZA, et al. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat Cell Biol. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ousset M, et al. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat Cell Biol. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- 103.Goldstein AS, et al. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marusyk A, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014 doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Driessens G, et al. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lapouge G, et al. Skin squamous cell carcinoma propagating cells increase with tumour progression and invasiveness. EMBO J. 2012;31:4563–4575. doi: 10.1038/emboj.2012.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Plikus MV. New activators and inhibitors in the hair cycle clock: targeting stem cells’ state of competence. J Invest Dermatol. 2012;132:1321–1324. doi: 10.1038/jid.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alonso L, Fuchs E. The hair cycle. J Cell Sci. 2006;119 (Pt 3):391–393. doi: 10.1242/jcs.02793. [DOI] [PubMed] [Google Scholar]
- 110.Paus R, Foitzik K. In search of the ‘hair cycle clock’: a guided tour. Differentiation. 2004;72 (9–10):489–511. doi: 10.1111/j.1432-0436.2004.07209004.x. [DOI] [PubMed] [Google Scholar]
- 111.White AC, et al. Stem cell quiescence acts as a tumour suppressor in squamous tumours. Nat Cell Biol. 2014;16:99–107. doi: 10.1038/ncb2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Paus R, et al. Chronobiology of the hair follicle: hunting the ‘hair cycle clock’. J Investig Dermatol Symp Proc. 1999;4:338–345. doi: 10.1038/sj.jidsp.5640241. [DOI] [PubMed] [Google Scholar]
- 113.Spencer JM, et al. Activated ras genes occur in human actinic keratoses, premalignant precursors to squamous cell carcinomas. Arch Dermatol. 1995;131:796–800. [PubMed] [Google Scholar]
- 114.Pierceall WE, et al. Ras gene mutation and amplification in human nonmelanoma skin cancers. Mol Carcinog. 1991;4:196–202. doi: 10.1002/mc.2940040306. [DOI] [PubMed] [Google Scholar]
- 115.Corominas M, et al. ras activation in human tumors and in animal model systems. Environ Health Perspect. 1991;93:19–25. doi: 10.1289/ehp.919319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van der Schroeff JG, et al. Ras oncogene mutations in basal cell carcinomas and squamous cell carcinomas of human skin. J Invest Dermatol. 1990;94:423–425. doi: 10.1111/1523-1747.ep12874504. [DOI] [PubMed] [Google Scholar]
- 117.Blanpain C, et al. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 118.Kimura-Ueki M, et al. Hair Cycle Resting Phase Is Regulated by Cyclic Epithelial FGF18 Signaling. J Invest Dermatol. 2012;132:1338–1345. doi: 10.1038/jid.2011.490. [DOI] [PubMed] [Google Scholar]
- 119.Zhang J, et al. BMP signaling inhibits hair follicle anagen induction by restricting epithelial stem/progenitor cell activation and expansion. Stem Cells. 2006 doi: 10.1634/stemcells.2005-0544. [DOI] [PubMed] [Google Scholar]
- 120.Cabodi S, Taverna D. Interfering with inflammation: a new strategy to block breast cancer self-renewal and progression? Breast Cancer Res. 2010;12:305. doi: 10.1186/bcr2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Greer JB, Whitcomb DC. Inflammation and pancreatic cancer: an evidence-based review. Curr Opin Pharmacol. 2009;9:411–418. doi: 10.1016/j.coph.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 122.Aggarwal BB. Inflammation, a silent killer in cancer is not so silent! Curr Opin Pharmacol. 2009;9:347–350. doi: 10.1016/j.coph.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 123.Quante M, Wang TC. Inflammation and stem cells in gastrointestinal carcinogenesis. Physiology. 2008;23:350–359. doi: 10.1152/physiol.00031.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kasper M, et al. Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc Natl Acad Sci USA. 2011;108:4099–4104. doi: 10.1073/pnas.1014489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mancuso M, et al. Hair cycle-dependent basal cell carcinoma tumorigenesis in Ptc1neo67/+ mice exposed to radiation. Cancer Res. 2006;66:6606–6614. doi: 10.1158/0008-5472.CAN-05-3690. [DOI] [PubMed] [Google Scholar]
- 126.Wong SY, Reiter JF. From the Cover: Wounding mobilizes hair follicle stem cells to form tumors. Proc Natl Acad Sci USA. 2011;108:4093–4098. doi: 10.1073/pnas.1013098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ito M, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 129.Gidekel Friedlander SY, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Davidson LA, et al. Alteration of colonic stem cell gene signatures during the regenerative response to injury. Biochim Biophys Acta. 2012;1822:1600–1607. doi: 10.1016/j.bbadis.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Westphalen CB, et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest. 2014;124:1283–1295. doi: 10.1172/JCI73434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schwitalla S, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152 (1–2):25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 133.Humphries A, et al. Stem cells and inflammation in the intestine. Recent Results Cancer Res. 2011;185:51–63. doi: 10.1007/978-3-642-03503-6_3. [DOI] [PubMed] [Google Scholar]
- 134.Song IY, Balmain A. Cellular reprogramming in skin cancer. Semin Cancer Biol. 2014 doi: 10.1016/j.semcancer.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kwon OJ, et al. Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin. Proc Natl Acad Sci USA. 2014;111:E592–E600. doi: 10.1073/pnas.1318157111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guasch G, et al. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nik AM, et al. Foxf2 in intestinal fibroblasts reduces numbers of Lgr5(+) stem cells and adenoma formation by inhibiting Wnt signaling. Gastroenterology. 2013;144:1001–1011. doi: 10.1053/j.gastro.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 138.Zong Y, et al. Stromal epigenetic dysregulation is sufficient to initiate mouse prostate cancer via paracrine Wnt signaling. Proc Natl Acad Sci USA. 2012;109:E3395–E3404. doi: 10.1073/pnas.1217982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Memarzadeh S, et al. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell. 2007;12:572–585. doi: 10.1016/j.ccr.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bhowmick NA, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 141.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 142.Jensen KB, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Morris RJ, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 144.Kannan K, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3:1194–1203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sanders MA, Valk PJ. The evolving molecular genetic landscape in acute myeloid leukaemia. Curr Opin Hematol. 2013;20:79–85. doi: 10.1097/MOH.0b013e32835d821c. [DOI] [PubMed] [Google Scholar]
- 146.Donley N, Thayer MJ. DNA replication timing, genome stability and cancer: late and/or delayed DNA replication timing is associated with increased genomic instability. Semin Cancer Biol. 2013;23:80–89. doi: 10.1016/j.semcancer.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huang S, Ingber DE. A non-genetic basis for cancer progression and metastasis: self-organizing attractors in cell regulatory networks. Breast Dis. 2006;26:27–54. doi: 10.3233/bd-2007-26104. [DOI] [PubMed] [Google Scholar]
- 148.Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nakagawachi T, et al. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene. 2003;22:8835–8844. doi: 10.1038/sj.onc.1207183. [DOI] [PubMed] [Google Scholar]
- 150.Pajtler KW, et al. The KDM1A histone demethylase is a promising new target for the epigenetic therapy of medulloblastoma. Acta Neuropathol Commun. 2013;1:19. doi: 10.1186/2051-5960-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang L, et al. CRISPR-Cas-Mediated Targeted Genome Editing in Human Cells. Methods Mol Biol. 2014;1114:245–267. doi: 10.1007/978-1-62703-761-7_16. [DOI] [PubMed] [Google Scholar]
- 152.Mali P, et al. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]