Abstract
Autotransporter (AT) proteins provide a diverse array of important virulence functions to Gram-negative bacterial pathogens, and have also been adapted for protein surface display applications. The “autotransporter” moniker refers to early models that depicted these proteins facilitating their own translocation across the bacterial outer membrane. Although translocation is less autonomous than originally proposed, AT protein segments upstream of the C-terminal transmembrane β-barrel have nevertheless consistently been found to contribute to efficient translocation and/or folding of the N-terminal virulence region (the “passenger”). However, defining the precise secretion functions of these AT regions has been complicated by the use of multiple overlapping and ambiguous terms to define AT sequence, structural, and functional features, including “autochaperone”, “linker” and “junction”. Moreover, the precise definitions and boundaries of these features vary among ATs and even among research groups, leading to an overall murky picture of the contributions of specific features to translocation. Here we propose a unified, unambiguous nomenclature for AT structural, functional and conserved sequence features, based on explicit criteria. Applied to 16 well studied AT proteins, this nomenclature reveals new commonalities for translocation but also highlights that the autochaperone function is less closely coupled with a conserved sequence element than previously believed.
Introduction
The autotransporter (AT, or type Va) secretion system is the most common and arguably the simplest of all known mechanisms to secrete proteins to the surface of Gram-negative bacterial pathogens (Dalbey and Kuhn, 2012; Dautin and Bernstein, 2007; Henderson et al., 2004). AT proteins perform a wide variety of crucial virulence-related functions (Henderson and Nataro, 2001) and have been adapted for cell surface display of heterologous proteins (Jose and Meyer, 2007; Jong et al., 2010). Yet despite its simplicity, many features of the AT secretion mechanism remain unclear (Braselmann & Clark, 2012; Dautin & Bernstein, 2007; Leyton et al., 2012). This has sparked intense interest in determining the precise mechanism of secretion and the contributions of specific regions of an AT protein to this mechanism.
To facilitate their secretion, all AT proteins follow a specific organizational pattern: at the extreme N-terminus, a signal sequence directs export of the AT across the inner (cytoplasmic) membrane via the Sec translocon. The signal sequence is followed by a secreted domain (the “passenger”), a linker, and the C-terminal 12-stranded transmembrane β-barrel (Henderson et al., 1998). The primary function of the linker and β-barrel is to facilitate translocation of the passenger across the outer membrane (OM) (Fig. 1), whereas the passenger represents the extracellular, functional part of an AT virulence protein. After OM translocation, some ATs are cleaved within the linker to separate the passenger from the β-barrel. After cleavage, a passenger may be released from the cell surface or remain attached to the membrane. Most but not all AT passengers include β-helical structure (Bradley et al., 2001; Junker et al., 2006; Kajava et al. 2001; Kajava et al., 2006 Oomen et al., 2004). The distinct organizational pattern of these structural features can be used to identify putative AT genes in bacterial genomes (Celik et al., 2012; Junker et al., 2006; Loveless and Saier 1997; Wells et al., 2010 Yen et al., 2002). Yet despite these common traits, AT proteins have quite diverse sequences and functions. Even among conserved regions, there appears to be much mixing and matching between different sequence motifs (Celik et al., 2012).
Figure 1.
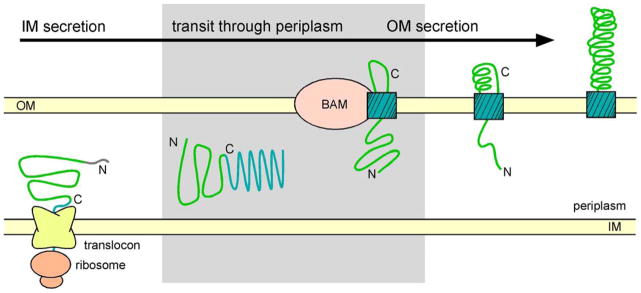
Overview of the autotransporter (AT) secretion mechanism. ATs are synthesized as precursors with an N-terminal signal sequence (grey) for export across the inner membrane (IM). Following IM translocation, the AT passenger (green), linker (red and magenta) and C-terminal β-barrel (cyan), cross the periplasm and the β-barrel forms a pore that spans the outer membrane (OM). The passenger is translocated across the OM, presumably through the pore formed within the β-barrel. Studies of the B. pertussis AT pertactin (Prn) have shown that the passenger is translocated across the OM from C- to N-terminus (Junker et al., 2009). The shaded area indicates early steps in the AT secretion mechanism that remain unclear, including transit of the precursor through the periplasm, the conformation of the passenger in the periplasm, the role of periplasmic chaperones in facilitating OM secretion, the insertion of the β-barrel domain into the OM, and translocation of the first β-strands of the passenger across the OM. The β-barrel assembly machinery (BAM) plays a role in some early AT OM translocation steps, including insertion of the transmembrane β-barrel into the OM (Ieva and Bernstein, 2009; Ruiz-Perez et al., 2009; Sauri et al., 2009). Figure adapted from (Braselmann and Clark, 2012).
The organization of AT proteins suggests a deceptively simple picture of their secretion mechanism (Fig. 1). Initially it was thought that after the signal sequence directs transport across the inner membrane, the C-terminal β-barrel forms a pore in the outer membrane through which the passenger is translocated (Klauser et al., 1990). However, it is now known that the Bam complex facilitates assembly of AT and other OM β-barrels (Gessmann et al., 2014; Hagan et al., 2011). Bam has also been implicated in the translocation of the passenger (Ieva and Bernstein, 2009), although its precise role remains an active area of investigation. Many other important questions also remain unresolved: Where does the energy come from to drive secretion across the outer membrane, given the absence of ATP or an ion concentration gradient across the outer membrane (Leyton et al., 2014; Roman-Hernandez et al., 2014; Thanassi et al., 2005)? Given that the folded passenger is rather wide (≥4 nm), what conformation does it adopt during OM translocation through a narrow pore, and how is passenger folding coordinated with secretion? What are the minimum requirements for secretion, and how can they be manipulated to display heterologous proteins on the bacterial cell surface (Jose & Meyer, 2007; van Ulsen et al., 2014)?
Thanks to efforts from many laboratories, insights have been gained into several of these questions (see (Grijpstra et al., 2013; Leo et al., 2012; Leyton et al., 2012) for recent reviews). However, each laboratory tends to focus on a specific aspect of the AT secretion mechanism, using a specific model system, and often using a distinct experimental approach. While this diversity of models and approaches could accelerate our understanding of AT secretion determinants, it can also make it challenging to identify common mechanistic features. Exacerbating this challenge is the often vague and inconsistent nomenclature used to describe AT protein features.
To facilitate the identification of common elements of the AT secretion mechanism, we analyzed published studies of well-characterized AT proteins, and used these results to develop a unified nomenclature to unambiguously label identifiable structural, functional, and conserved sequence features. This analysis revealed that the majority of our understanding of AT secretion has been developed from studies of proteins from a closely related clade within the AT family. Moreover, several features often represented as common to all AT proteins have been well-characterized for only one or a few AT proteins. Nevertheless, organizing published results within this unified nomenclature revealed common themes that heretofore have received little attention. Our hope is that, going forward, adoption of this unified nomenclature will help the AT community improve our understanding of the translocation mechanism, and facilitate direct comparisons across model systems, revealing common characteristics of AT secretion as well as ’variations on a theme’.
The need for unambiguous definitions of common autotransporter secretion features
Two particularly frustrating aspects of the current ad hoc AT nomenclature are that names used to describe structural, functional, and conserved sequence features are often used interchangeably, and several of these features have been assigned more than one name (see Table 1 for a short glossary of historically and/or ambiguously used terms). Most of this ambiguity occurs in the region between the C-terminus of the stably folded passenger and the N-terminus of the transmembrane β-barrel (grey shaded areas in Figs 1&2), which for many ATs includes sequence segments that are relatively poorly defined in terms of their structure and specific function(s) but play a crucial role in the early steps of OM translocation (Berthiaume et al., 2007; Ivie et al., 2010; May and Morona, 2008; Ohnishi et al., 1994; Oliver et al., 2003a,b; Velarde and Nataro, 2004). For example, “linker” has been used to describe a structurally flexible segment within this region (Besingi et al., 2013), but this same name is also used to define a larger sequence segment that includes the α-helix that spans the OM through the center of the β-barrel (Oliver et al., 2003b), or used as a functional term to denote the entire region N-terminal to the β-barrel that is required for efficient OM translocation of heterologous proteins and can include the C-terminus of the folded passenger (Velarde and Nataro, 2004). Similarly, “autochaperone” has been defined as a region that functions to promote extracellular folding of an AT passenger, even when supplied in trans (Dutta et al., 2003; Oliver et al., 2003b). An autochaperone function has been identified for only a handful of AT proteins, but this term is also used to refer to a weakly conserved sequence element at the C-terminus of many AT passengers (Rutherford et al., 2006). As a result, it can be difficult to compare the specific functional roles of different portions of AT proteins, and ambiguous statements like “mutations in the linker region disrupt passenger folding” or “the autochaperone is required for efficient OM translocation” can generate considerable confusion. Of course, as in any protein, structural, functional and sequence features can be closely related and occasionally overlap. Nevertheless, developing a truly comprehensive understanding of AT secretion will require that we (a) avoid using the same names interchangeably to describe different features, (b) explicitly define the criteria used to designate a particular feature of an autotransporter without having to trace each term back to the early literature pertaining to a specific AT protein, and (c) apply this nomenclature consistently across all autotransporters. To achieve these goals, we present below a consistent, unified nomenclature that explicitly defines our current understanding of the components of an autotransporter protein and distinguishes whether a particular component is defined by structural, functional, and/or sequence conservation criteria.
Table 1.
Glossary of names used historically and sometimes ambiguously to describe structural and/or functional features of AT proteins, listed from N- to C-terminus.
| Feature name | Description | Preferred term |
|---|---|---|
| α-domaina | Historically used to refer to the passenger. For IgAP, the term “α-peptide” is used to refer to one small portion of the passenger. | Passenger |
| Autochaperoneb | Functional unit at C-terminus of the passenger, capable of catalyzing passenger folding and efficient OM translocation in trans. Historically also used to refer to any conserved sequence at the passenger C-terminus. | (Avoid – see text for details) |
| Helperc | Early historical term used to describe the entire translocator unit. | Translocator (if known) |
| Hydrophobic secretion facilitation domain (HSF)d | A seldom-used term that describes a conserved hydrophobic region near the C-terminus of the passenger, found to be functionally required for secretion of EspP. Overlaps with the PL and autochaperone regions. | PL region |
| Junctione | Loosely refers to residues connecting the passenger and the β-barrel. Occasionally used to refer to a conserved sequence near the C-terminus of the passenger. | Linker or PL region (see text for distinction) |
| Linkerf | Segment connecting the folded passenger to the central α-helix. May also include the α-helix. Historically also used as a synonym for “junction”. | Linker or PL region (see text for distinction) |
| Translocator, Translocation unitg | C-terminal part of the AT protein, responsible for facilitating OM translocation. Includes, but is not limited to, the β-barrel domain and the central α-helix. Historically sometimes defined as the entire AT sequence C-terminal to the passenger cleavage site(s) or to the N-terminus of the α-helix. | Translocator |
| β-domainh | Historically used to refer to the β-barrel structural domain, or entire AT sequence C-terminal to the passenger cleavage site(s), or the entire translocator functional unit. | β-barrel or Translocator (if known; see text for distinction) |
Figure 2.
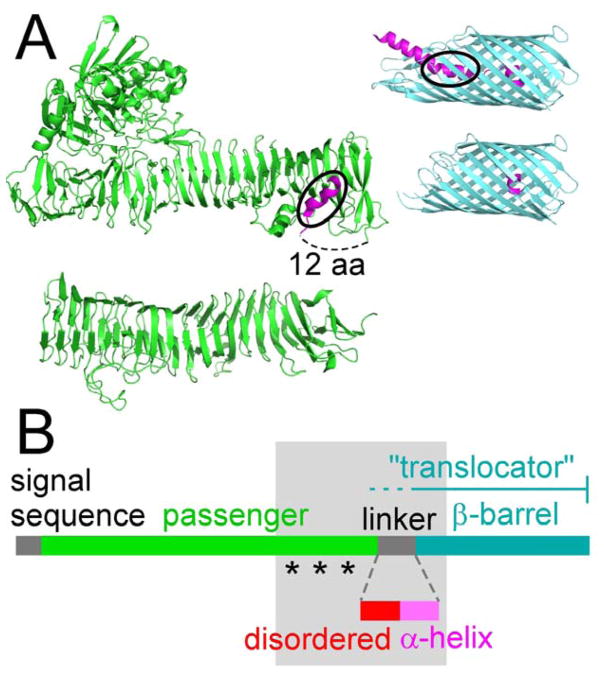
Application of a unified nomenclature to explicitly describe typical structural, functional, and sequence conservation features important for AT secretion. (A) Ribbon diagrams of representative AT structures. Passengers are shown in green, α-helices in pink, and β-barrels in cyan. The passengers are from E. coli EspP (top, PDB ID 3SZE (Khan et al., 2011)) and B. pertussis pertactin (Prn, bottom, PDB ID 1DAB (Emsley et al., 1996)). The EspP β-barrel and α-helix are shown in the post-cleavage (bottom, PDB ID 2QOM (Barnard et al., 2007)) form and in a form designed to represent a pre-cleavage intermediate (top, PDB ID 3SLO (Barnard et al., 2012)). C-terminal to the EspP passenger β-helix is a short α-helix that lies at the surface of the passenger. Note that these residues also appear in the structure of the EspP translocator in the pre-cleavage form (black ovals indicate the same sequence). (B) Sequence annotation of universal and other common AT structural, functional and sequence features. Lengths are drawn to scale using the B. pertussis pertactin sequence. As for many AT proteins, the N-terminus of the translocator unit (the minimal unit required for OM translocation) is not clearly defined for pertactin (dotted line). Asterisks indicate the approximate locations of three features found in the C-terminus of some AT passengers (PL region, stable core, and autochaperone). The shaded area indicates features that are often inaccurately or ambiguously annotated in the AT literature, but are known to play an important role in AT biogenesis.
The nomenclature described below is confined to terms related to AT secretion, as the secretion mechanism and its sequence and structural requirements are the common and defining features of all autotransporter proteins. Specific virulence functions, which are reflected primarily in passenger diversity, are not discussed. For clarity, we have organized the features of AT proteins from N- to C-terminus, describing first the four major features found in all AT proteins: signal sequence, passenger, linker and β-barrel. We recommend that the readily identified boundaries of these four universal features be used as reference points in all studies of AT secretion and folding. Following the descriptions of these features, we describe alternative terms and more specialized sub-features identified in some AT proteins. To avoid ambiguity and misattribution, we distinguish whether each feature has been defined by structural, functional, and/or sequence conservation criteria, and caution against assuming these terms to represent general features of AT secretion until unambiguously established using a broader, more representative set of AT proteins.
Signal sequence
This short (<60 aa) N-terminal sequence is defined as the sequence required to direct transport of the AT protein across the inner membrane. It is typically cleaved after translocation across the inner membrane. In isolation, signal sequences lack a well-defined structure (Briggs et al., 1986) but are often accurately predicted from the amino acid sequence (Hiller et al., 2004; Petersen et al., 2011). Interestingly, some AT signal sequences have an N-terminal extension of 15–25 residues, the function of which is still unclear (Desvaux et al., 2007; Jong & Luirink, 2008; Szabady et al., 2005). Although little controversy surrounds the function and boundaries of the signal sequence, it is nevertheless a universal feature of the AT secretion mechanism and thus we include it here for the sake of completeness.
Passenger
We define the passenger as following the signal sequence and consisting of one or more extracellular domains that adopt a stable, defined structure. The C-terminal boundary of the passenger can therefore be defined by x-ray crystallography (Emsley et al., 1997) or limited proteolytic digestion and mass spectrometry (Junker et al., 2006; Renn & Clark, 2008). >95% of passengers are predicted to include a right-handed β-helical structure, although unusually short passengers often deviate from this pattern (Junker et al., 2006; Celik et al., 2012; Otto et al., 2005; van den Berg, 2010). The β-helical passenger of pertactin from B. pertussis has a C-terminal cap that inhibits aggregation (Bryan et al., 2011), and an N-terminus that cannot fold independently, which may contribute to efficient secretion (Renn et al., 2012). Although not the focus of this analysis, the passenger also includes the virulence activity of the AT protein (Henderson and Nataro, 2001).
Extreme caution must be exercised when sub-dividing the passenger further into regions with more specific features. In particular, the terms “PL (pertactin-like) region”, “stable core” and “autochaperone”, which respectively describe a sequence, structural and functional feature within the C-terminus of some AT proteins, have often been used ambiguously. See below for a complete description of these terms and a set of rigorous criteria for determining their presence within an AT protein.
Linker
All AT proteins possess a linker, defined as the region connecting the C-terminus of the passenger to the N-terminus of the transmembrane β-barrel (Oliver et al., 2003b; Velarde and Nataro, 2004). The linker includes an α-helix within the center of the transmembrane β-barrel, which for some AT proteins can span the width of the OM and even extend further into the extracellular space (Barnard et al., 2007; Oomen et al., 2004; Tajima et al., 2010; van den Berg, 2010). In many ATs the linker also includes a disordered region N-terminal to the α-helix. If the passenger is cleaved from the β-barrel after OM translocation, this cleavage site can be located within the linker and after OM translocation will be positioned either on the cell surface (Kühnel and Diezmann, 2011; Meng et al., 2011; Pohlner et al., 1987) or within the folded β-barrel structure (Dautin et al., 2007; Tajima et al., 2010). Some passengers have additional cleavage sites that separate different passenger domain structures after secretion (Fink et al., 2001; Nguyen et al., 2001; Ohnishi and Horinouchi, 1996; Pohlner et al., 1987). Many ATs perform these cleavage reactions autocatalytically, while others rely on proteases present on the cell surface (Shere et al., 1997; van Ulsen et al., 2003). Functionally, the linker region is strictly required for secretion in all AT proteins where its role has been tested (Berthiaume et al., 2007; Brockmeyer et al., 2009; Dutta et al., 2003; Maurer et al., 1999; Oliver et al., 2003a; Suzuki et al., 1995; Yang et al., 2004).
β-barrel
The last universal, readily identified AT feature is the C-terminal 12-stranded β-barrel structure that spans the OM and forms a hydrophilic pore through the center of the barrel (Barnard et al., 2007; Oomen et al., 2004; Tajima et al., 2010; van den Berg, 2010; Zhai et al., 2011). Upon completion of OM translocation, this pore is blocked by the central α-helix of the linker. Both the N- and C-terminal ends of the barrel reside in the periplasm. AT transmembrane β-barrel domains share sufficient sequence conservation that they have been identified as conserved domains (either Pfam 03797 or the similar but distinct smart00869). Note, however, that there is still a significant degree of variability and apparent swapping of the order of β-strands within the β-barrels from different AT proteins (Celik et al., 2012).
Alternative terms: What about “translocator”? Or “β-domain”?
The term “translocator” was originally devised as part of early models, when it was thought that autotransporters were completely self-contained secretion systems (Konieczny et al., 2000; Loveless and Saier, 1997). In some studies, an AT translocator or β-domain was defined as all portions of the AT sequence that are C-terminal to an intramolecular cleavage site within the linker. But not all AT proteins undergo cleavage (Besingi et al., 2013; van den Berg 2010), so this nomenclature cannot be universally applied. Adding to this ambiguity, other studies define translocator or β-domain as the minimal C-terminal portion of the AT protein required for OM translocation of the passenger. By this definition, the translocator or β-domain always includes the β-barrel domain and the α-helix of the linker but often extends further to include the disordered portion of the linker (Maurer et al., 1999; Oliver et al., 2003a; Ramesh et al., 2012; Sevastsyanovich et al., 2012; Suzuki et al., 1995; Yang et al., 2004). In some cases, the translocator extends still further, well into the folded domain of the passenger (Fischer et al., 2001; Ivie et al., 2010; Velarde and Nataro, 2004). There can therefore be overlap between the passenger and the translocator functional unit (Fig. 2B), which can create considerable confusion. Note also that within a single AT protein or close homologs the location of the N-terminal boundary of the minimal translocation unit may vary, depending on whether what is being translocated is a bone fide passenger, which are often quite large (>500 aa), or a smaller heterologous protein (Jose & Meyer, 2007). For Pet, the minimal construct capable of mediating the secretion and folding of a heterologous passenger consisted of the β-barrel plus a short N-terminal extension that included 9 aa N-terminal of the cleavage site within the linker (Sevastsyanovich et al., 2012). Yet for the closely related Pet homolog EspP, hydrophobic residues within the C-terminus of the passenger have been shown to be important for OM translocation of the endogenous EspP passenger and are considered part of the translocator (Velarde and Nataro, 2004). Despite these potential ambiguities, we recognize that it is useful to define a term that indicates the N-terminal boundary of the AT region demonstrated via quantitative functional studies to be required for efficient OM translocation of a passenger. Hence we recommend the term “translocator” be used exclusively in this way. In the absence of functional studies to determine the N-terminus of the translocator, we recommend using only the terms “passenger”, “linker” and “β-barrel”, the boundaries of which are more readily determined by sequence comparisons and structural studies.
We also appreciate that, for the limited number of AT proteins that have a single, well defined cleavage site (such as SPATES), “β-domain“ may be a convenient term to describe all portions of the AT protein that are C-terminal to this cleavage site. However, we discourage use of “β-domain“ for the following reasons: (1) to avoid confusion with the similar but non-synonymous “translocator”, (2) because, unlike “translocator”, it is not particularly meaningful in terms of the OM translocation process, (3) because it cannot be defined for uncleaved AT proteins, and (4) because it is challenging to define unambiguously for those AT proteins with multiple cleavage sites.
Back to the passenger: What about more specialized terms: “PL region”, “stable core”, and “autochaperone”?
As mentioned above, the terms “PL region”, “stable core”, and “autochaperone” respectively describe a sequence, structural and functional feature found within the C-terminus of a minor fraction of AT passengers. Because these terms are neither synonymous nor universal to all (or even most) ATs (see Fig. 3), they should be avoided unless their existence has been confirmed by the following specific criteria.
Figure 3.
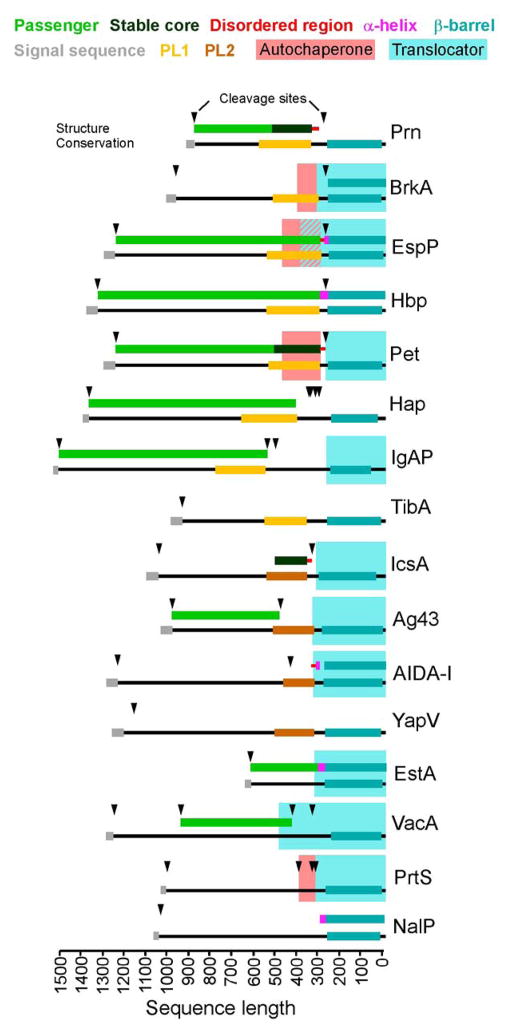
Regions of known structure (top), sequence conservation (bottom), and functional features (background shading) for 16 well characterized AT proteins. Sequences were aligned at their C-termini in order to highlight the widespread conservation of C-terminal sequence features. The conserved β-barrel region (cyan, bottom line) corresponds to smart00869 in YapV and Ag43 and Pfam 03797 in all other proteins shown. Note that solved structures of β-barrel domains are also colored in cyan (top line). The conserved PL region (cd00253) at the C-terminus of many passengers corresponds to subgroup PL1 (yellow; cd01343) or subgroup PL2 (brown; cd01344). Conserved domains were identified using NCBI Conserved Domain Search (Marchler-Bauer et al., 2013). Structural features correspond to regions resolved within PDB structures and/or defined biophysically. Functional features were identified biochemically. Overlap between the EspP autochaperone and translocator is shown as a hatched area. See Table 2 for specific boundaries of highlighted features.
The PL (pertactin-like) region is a conserved sequence feature named after B. pertussis pertactin (Prn), the first crystal structure determined for an AT passenger (Emsley et al., 1996), and is classified in the NCBI Conserved Domain Database as cl00185 (Marchler-Bauer et al., 2013). Based on sequence similarity, there are two distinct sub-families of PL regions classified as cd01343 and cd01344, which for brevity we designate PL1 and PL2, respectively (Figs 4&5). There is no clear functional connection between the PL region and autochaperone functional feature, and hence these terms must not be used interchangeably. For example, PrtS (also known as SSP), which is one of only a few AT proteins that contains a well-characterized autochaperone, lacks a PL region (Ohnishi et al., 1994) (Fig. 3).
Figure 4.
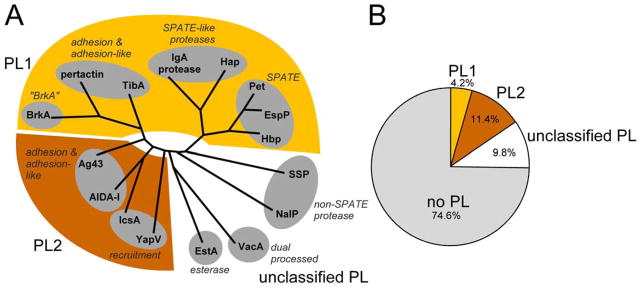
Passenger sequence conservation amongst the 16 well-characterized AT proteins shown in Figure 3. (A) AT passenger sequences were aligned using Muscle, and a pylogenetic tree was calculated using PROTML from the PHYLIP phylogeny package (Felsenstein, 2005). The overall passenger phylogeny corresponds well to the distinction between the three types of PL regions (PL1 (cd01343; yellow), PL2 (cd01344; brown), and no PL region (no shading)). (B) PL domain assignments for a nonredundant set of 1882 AT proteins (maximum identity = 60%) collected from the NCBI Reference Sequence Database. An AT is assigned as unclassified PL if it had a PL superfamily assignment (cl00185) but did not have an assignment to a specific PL subset (PL1 or PL2). Domains were assigned using NCBI Conserved Domain BLAST.
Figure 5.
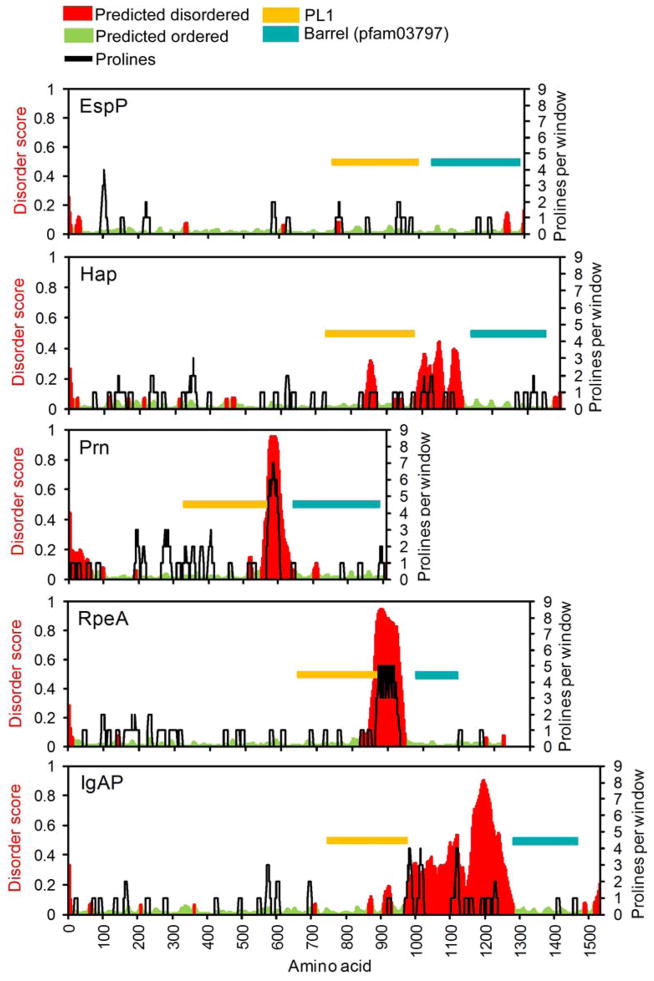
AT linkers exhibit a diverse range of proline content and predicted disorder, even within the sub-family of PL1-containing AT proteins. Disorder was predicted using Espritz (Walsh et al., 2012). Conserved domains are colored according to the assignments in Figure 3. AT linkers may entirely lack a disordered region (EspP), have a disordered region without high proline content (Hap), or contain a polyproline region that is also predicted to be disordered (Prn). There is wide variety in the length of these disordered regions, with some ATs having regions of predicted disorder that are hundreds of residues in length (IgAP).
The stable core is a structural feature, an ~25 kDa portion of the passenger C-terminus that is more stable than the rest of the passenger, as determined by an increased resistance to protease digestion and chemical denaturation. Stable cores have been reported in the C-terminal regions of three β-helical passengers (Junker et al., 2006; Kühnel and Diezmann, 2011; Renn and Clark, 2008) (Fig. 3). The higher stability of this core relative to the remainder of the passenger has been shown to promote efficient OM translocation (Renn et al., 2012). However, at least one AT passenger is translocated efficiently across the OM without a canonical stable core (Besingi et al., 2013), and hence the extent to which a stable core represents a widespread functional feature of AT secretion remains to be determined. The stable core and PL region are not synonymous, as one is a structural feature and the other a sequence feature: YapV, which contains a PL2 region, lacks a stable core (Besingi et al., 2013) (Fig. 3). Likewise, the stable core and autochaperone are not synonymous: the stable core is a structural feature that is confined entirely within the passenger structure, whereas the autochaperone is a functional feature that spans approximately half the length of a stable core.
An autochaperone is a C-terminal portion of the passenger that promotes correct folding of the entire passenger at the cell surface. Crucially, a bone fide autochaperone will promote efficient passenger folding and OM translocation even when supplied in trans (as a separate protein, anchored to the OM) (Ohnishi et al., 1994; Oliver et al., 2003b). This functional feature has been characterized for only three AT proteins, BrkA, EspP/Pet, and PrtS (SSP). We have lumped the closely related EspP and Pet AT proteins together here because what was shown is that the C-terminus of EspP can rescue the folding of some Pet mutants at the cell surface when supplied in trans (Dutta et al., 2003). Crucially, while mutations in the C-terminal regions of several other AT passengers have been shown to impair passenger folding in vivo (Berthiaume et al., 2007; Ivie et al., 2010; May & Morona, 2008), these regions are not considered autochaperones because it is not known whether their expression in trans is capable of rescuing passenger folding.
Common themes (and variations) for characterized autotransporters
Application of our unified, unambiguous nomenclature to 16 well-characterized ATs (Fig. 3, Table 2) revealed several common themes. The first is that some features considered to be broadly common have been well characterized in only a few AT proteins. For example, a stable core has been found in pertactin (Prn) (Junker et al., 2006), Pet (Renn and Clark, 2008) and IcsA (Kühnel and Diezmann, 2011), and is known to be absent in YapV (Besingi et al., 2013), but its existence has not been tested in any other AT protein. Conversely, the minimal translocator unit has never been determined for pertactin or Hbp, but is well characterized for IgAP (Klauser et al., 1993), Ag43 (Ramesh et al., 2012), IcsA (Suzuki et al., 1995), and AIDA-1 (Maurer et al., 1999). As a result of the limited results available, direct mapping of functional features onto structural units across the diverse AT family is challenging and subject to bias. As an alternative approach, we used sequence conservation as a bridge to connect structural and functional units identified from diverse AT proteins (Fig. 3).
Table 2.
Sequence locations of well-characterized autotransporter secretion features included in this analysis.
| Autotransporter Organism | Signal sequence | Passenger | Stable core | Disordered region | α-helix | β-barrel | PDB ID | Auto- chaperone | Translocator |
|---|---|---|---|---|---|---|---|---|---|
| Prna Bordetella pertussis |
1 34 |
35 573 |
385 573 |
574 ~601 |
1DAB | ||||
| BrkAb B. pertussis |
1 42 |
745 1010 |
3QQ2 | 601 692 |
694 1010 |
||||
| EspPc Escherichia coli |
1 55 |
56 997 |
998 1013 |
1014 1030 |
1037 1300 |
3SZE, 2QOM, 3SLJ | 821 997 |
907 1300 |
|
| Hbpd E. coli |
1 52 |
53 1074 |
1075 1082 |
1083 1114 |
1115 1377 |
1WXR, 3AEH | |||
| Pete E. coli |
1 52 |
53 992 |
780 992 |
993 1018 |
4OM9 | 819 992 |
1010 1295 |
||
| Hapf Haemophilus influenzae |
1 25 |
26 977 |
3SYJ | ||||||
| IgAPg Neisseria gonorrhoeae |
1 27 |
28 989 |
3H09 | 1259 1532 |
|||||
| TibAh E. coli |
1 54 |
||||||||
| IcsA (VirG)i Shigella flexneri |
1 52 |
591 740 |
741 758 |
3ML3 | 780 1102 |
||||
| Ag43j E. coli |
1 52 |
53 552 |
4KH3 | 700 1039 |
|||||
| AIDA-Ik E. coli |
1 49 |
~950 964 |
965 980 |
1003 1286 |
4MEE | 955 1286 |
|||
| YapVl Yersinia pestis |
1 45 |
none | |||||||
| EstAm Pseudomonas aeruginosa |
1 24 |
25 330 |
none | 331 361 |
364 646 |
3KVN | 321 646 |
||
| VacAn Helicobacter pylori |
1 33 |
345 855 |
2QV3 | ~798 1287 |
|||||
| PrtS (SSP)o Serratia marcescens |
1 27 |
646 716 |
716 1045 |
||||||
| NalPp Neisseria meningitidis |
1 27 |
786 811 |
812 1084 |
1UYN |
Structural units are included only when direct experimental confirmation exists in the form of a PDB structure or resistance to protease digestion in the case of stable core. Signal sequence cleavage was typically confirmed by N-terminal sequencing of the remaining passenger. Inclusion of an autochaperone is predicated on it being experimentally shown to rescue folding-deficient passenger mutants when supplied in trans. Translocators correspond to the minimal units required to secrete a passenger, typically a heterologous one.
Barnard et al. (2007, 2012); Brockmeyer et al. (2009); Dutta et al. (2003); Khan et al. (2011); Skillman et al. (2005); Szabady et al. (2005); Velarde and Nataro (2004)
Johnson et al. (2009); Klauser et al. (1993, 1990); Pohlner et al. (1987, 1995); van Ulsen et al. (2003)
Brandon and Goldberg (2001); Kühnel and Diezmann (2011); May and Morona (2008); Suzuki et al. (1995); Teh and Morona (2013)
Benz and Schmidt (1992); Berthiaume et al. (2007); Charbonneau et al. (2006); Charbonneau and Mourez (2007); Gawarzewski et al. (2014); Maurer et al. (1999); Rutherford et al. (2006); Suhr et al. (1996)
Cover and Blaser (1992); Fischer et al. (2001); Gangwer et al. (2007); Ivie et al. (2010); Nguyen et al. (2001); Schmitt and Haas (1994)
The most conserved AT sequence elements are the N-terminal signal sequence and C-terminal transmembrane β-barrel. The β-barrel structural unit consistently correlates with conserved sequences, so these features can be considered equivalent for most practical purposes. Notably, AT β-barrels contain a highly conserved “mortise and tenon” joint between two adjacent β-strands within the barrel that appears important for positioning a portion of the AT passenger within the barrel during its insertion into the OM (Leyton et al., 2014). Interestingly, although Pfam 03797 and smart00869 do not extend into the α-helical or disordered linker segments, a conserved motif that does map to this region was identified in 100% of 1523 AT sequences analyzed (Celik et al., 2012).
Although not as broadly conserved as the signal sequence or β-barrel, the PL region (cd00253) stands out as a conserved sequence element within a subset of AT passengers. It can be divided into two distinct subgroups, PL1 and PL2, based on sequence conservation. Which of the two subgroups is present in a particular AT protein correlates with the position of the AT in a phylogenetic tree of passenger sequences (Fig. 4). Most AT secretion studies have used ATs that contain a PL1 region, however it is the rarest of all PL subgroups, and the vast majority of AT proteins lack a PL region altogether (Fig. 4B). Hence the current collection of well-studied ATs over-represents a relatively narrow range of AT proteins. Going forward, developing a comprehensive understanding of the connections between AT protein sequence features such as the PL region and AT structural and functional properties will require studies that sample a broader variety of more phylogenetically diverse AT proteins.
The PL region can overlap with other features at the C-terminus of the passenger. It appears to encode a folded structure, since it overlaps with known passenger structures — specifically the C-terminal β-helical structure — rather than with disordered linkers. Some PL regions include a stable core and/or autochaperone feature, and it might therefore be tempting to regard the PL region as synonymous with structural stability and autochaperone functionality. However, because these three features do not consistently overlap (Fig. 3), and because so few well-characterized stable cores and autochaperones have been identified, the temptation to extend structural and functional significance to a conserved sequence feature must be resisted. Similarly, although C-terminal passenger stability has also been linked to efficient OM secretion (Brockmeyer et al., 2009; Drobnak et al., 2014; Renn et al., 2012; Soprova et al., 2010), and hence it might be tempting to speculate that the PL region plays a key role in the secretion of wild type β-helical passengers. However, a similar effect of C-terminal passenger stability on secretion has also been observed in at least one AT without an identified PL region (Ivie et al., 2010). This suggests the passenger stability effect could be more widespread, not tied exclusively to the PL sequence but perhaps to a wider superfamily of sequences or to the C-termini of β-helices in general. In short, the function of the PL region is far from clear, and therefore in the absence of a detailed, quantitative structural and functional characterization its presence should not be used to infer increased structural stability, autochaperone functionality or effects on OM secretion.
We also examined connections between sequence, structure and function within the linker that connects the passenger to the β-barrel. For many ATs, the linker includes an unstructured region N-terminal to the α-helix, and in many cases this unstructured region is rich in proline residues (Charles et al., 1989; Leyton et al., 2007; Lindenthal and Elsinghorst, 1999). To investigate how widespread proline enrichment is within the linker across all AT proteins, we defined a proline-rich region as containing ≥4 prolines within a window of 11 amino acids, ≤200 aa N-terminal to the N-terminus of the conserved AT β-barrel domain (see below). A cutoff of ≥4 Pro residues was chosen because it gave a positive result for pertactin (Prn) and TibA, two well-characterized ATs previously known to have a proline-rich region region (Charles et al., 1989; Lindenthal and Elsinghorst, 1999) (Fig. 5), but was uncommon overall in the E. coli proteome (0.14% of all 11 aa windows). Across a nonredundant set of 1882 AT proteins (maximum identity = 60%) collected from the NCBI Reference Sequence Database, 18.5% of ATs contained a putative proline-rich region. Due to its low sequence complexity, it is very difficult to draw conclusions regarding evolutionary conservation of this region, but high proline content could rigidify the N-termini of these linkers (Williamson, 1994). Proline-rich regions can play an important role in binding to other macromolecules, either through direct interactions or by acting as spacers (Côté and Mourez, 2011; Williamson, 1994). The proline-rich region tends to overlap with the portion of the linker found between the passenger domain and the central α-helix, although some linkers include a predicted disordered region without having a proline-rich region (Fig. 5).
While there is strong sequence conservation in the signal sequence, translocator and C-terminal portion of some AT passengers, no similarly broadly conserved sequence feature has been identified in the N-terminal regions of AT passengers. This might reflect the need to accommodate a broad spectrum of virulence functions across the AT family, but also illustrates the versatility of the AT secretion mechanism, which can obviously be used to transport many different passenger sequences to the cell surface. Also of note is the ability of the AT translocation machinery to secrete a variety of heterologous proteins to the bacterial cell surface (Jong et al., 2010; Jose & Meyer, 2007), although it is important to keep in mind that these heterologous proteins are often significantly smaller (<300 aa) than a typical AT passenger (>500 aa) (Junker et al., 2006; Celik et al., 2012). It is tempting therefore to point to the passenger C-terminus as the crucial region that determines whether an endogenous AT passenger is compatible with AT secretion, but it should be noted that premature formation of stable folded structures at the N-terminus of an endogenous passenger can also reduce its secretion efficiency (Jong et al., 2007; Leyton et al., 2011; Renn et al., 2012; Saurí et al., 2012). Moreover, many of these less conserved passenger N-terminal sequences still adopt a β-helical fold, likely reflecting the plasticity of the β-helical fold to amino acid substitutions (Schuler et al., 2000).
Discussion
Analyzing the sequence determinants for AT OM translocation within the framework of our unified nomenclature has yielded new insights. For example, the functional distinction between the disordered portion of the linker and the PL region of the passenger was not previously widely recognized, in part because some older naming conventions lumped together these functionally distinct regions. As highlighted in Figs 2 and 4, this disordered region and the PL region are distinct units in terms of structure, function and sequence. The PL region adopts a stably folded conformation, whereas the disordered region is by definition less well structured. Not all AT proteins include this disordered portion within the linker, yet where present this portion of the linker is typically essential for OM translocation. This is consistent with the need for an estimated 30–45 residues N-terminal to the β-barrel to form a proposed early intermediate in OM translocation, consisting of an extended loop that crosses the OM twice, connecting the waiting passenger in the periplasm to the cell surface and back to the periplasmic N-terminus of the β-barrel (Fig. 1) (Drobnak et al., 2014; Ohnishi et al., 1994; Pohlner et al., 1987). While the disordered portion of the linker can be crucial for OM translocation, the PL region primarily appears to be necessary for the folding (and potentially secretion) of wild type, β-helical passengers that are also much larger than the commonly used heterologous passengers (such as cholera toxin B subunit). More work, including both bioinformatics analyses and quantitative assays of the effects of mutations on secretion efficiency, will be required to determine the precise function(s) of PL regions and how they compare to the roles that the α-helix and unstructured linker regions play in AT secretion.
An additional point of clarity achieved here surrounds the term “autochaperone”. This term was originally coined to define a C-terminal region of the AT passenger that promotes extracellular passenger folding even if present only in trans during OM translocation (Ohnishi et al., 1994; Oliver et al., 2003b). By this definition, mutations that lead to passenger misfolding are not sufficient to characterize a region as an autochaperone. Partial overlap between the BrkA autochaperone and its PL region (Oliver et al., 2003b) has unfortunately led to widespread misuse of the term “autochaperone” to describe the entire PL region, even though the two terms are not synonymous. The BrkA autochaperone (Oliver et al., 2003b) spans only half of the BrkA PL region, and one of the few other AT proteins known to contain a bone fide autochaperone, PrtS (SSP) (Ohnishi et al., 1994), lacks a PL domain (Fig. 3). Similarly, although the C-terminal rungs of the β-helix domain in VacA are required for the correct folding and secretion of the passenger (Ivie et al., 2010), no PL domain has been identified for VacA, nor has it been shown that this putative VacA autochaperone can rescue the folding-impaired phenotype when provided in trans. What is more, the stable core of IcsA, which overlaps closely with the IcsA PL region, does not appear to function as an autochaperone (Teh and Morona, 2013). Hence, while the C-terminus of a PL region can function as an autochaperone, the same or similar functionality can also be encoded in a non-PL sequence at the C-terminus of other AT passengers, whereas some PL regions may lack the autochaperone functionality. Going forward, it will be important to determine what sequence and/or structural features represent the crucial determinants of autochaperone function. Until that time, we recommend avoiding the term “autochaperone” except in cases where it has been functionally well described (currently only PrtS, BrkA and EspP/Pet).
New ATs with important virulence functions are being discovered on a monthly basis, yet much of the detailed structural and functional characterization of ATs to date has focused on a relatively narrow group of AT proteins, many of which are members of the minority fraction of PL1-containing AT proteins (Fig. 4). The AT community has developed a number of novel assays specifically to study the connections between AT sequence, structure and OM translocation (reviewed in (Henderson et al., 2004; Leyton et al., 2012)). To make the best use of these approaches and develop a truly comprehensive picture of the AT secretion mechanism, it will be important to diversify beyond our current well-characterized model AT proteins. We predict that targeted structural and functional studies using a manageable number of phylogenetically diverse AT proteins representative of the entire AT superfamily, focused on the regions from the C-terminal part of the passenger to the N-terminus of the β-barrel, will resolve many remaining ambiguities regarding the roles of specific AT protein features on OM translocation. Together with adopting the unified, unambiguous nomenclature described here, these efforts will provide a more comprehensive understanding of the AT secretion mechanism.
Methods
Sequence data
The NCBI RefSeq Protein database (Tatusova et al., 2014) was searched for keyword “autotransporter”. The resulting sequences were downloaded in FASTA format. Redundant identical protein sequences were removed.
Filtering for sequence similarity
The sequences were compared by BLASTP (e-value cutoff 10−7). BLAST results were processed to remove sequences sharing similarity above a percent identity threshold. The first query sequence was marked as included, and each subsequent sequence marked as excluded if it had an alignment to the first sequence with percent identity > percent identity threshold. This was repeated for each subsequent, non-excluded sequence in the file. Filtering was performed at percent identity cutoffs of 60% and 85%. A 60% cutoff was used to reflect the diversity in the well characterized AT set (maximum percent identity in this set is 57%, between Pet and EspP).
Conserved domains: Identifying autotransporters
Methods used for classification of proteins as ATs were based on Junker et al. (2006). Following filtering, non-redundant protein sequences were analyzed for conserved domains using the NCBI Conserved Domain Search (Marchler-Bauer et al., 2013) batch search option. Results were filtered for hits with e-values < 0.0001. A protein was considered an AT if it had a predicted β-barrel domain (pfam03797 or, in its absence, smart00869) ending < 100 aa from the C-terminus of the protein (because ATs have C-terminal barrels), and beginning > 250 aa from the N-terminus of the protein (to allow space for a passenger domain), and did not contain YadA-like domains (because this related classification could lead to false positives). Sequences meeting these criteria were also analyzed for the presence of PL domains (e-value < 0.0001). AT proteins were classified as PL1 or PL2 if they had specific hits to these domain classes (cd01343 or cd01344), or unclassified PL if they lacked a specific hit to PL1 or PL2 but did have a PL superfamily hit (cl00185).
Polyproline regions
The region of interest was defined as 200 amino acids preceding the N-terminus of the conserved barrel domain. This distance was defined because 95% of ATs with a conserved PL domain had a distance <200 amino acids between the PL C-terminus and the barrel N-terminus. This region was searched for polyproline regions and predicted disorder. An AT was considered to have a polyproline region if it contained ≥4 Pro residues in a window of 11 amino acids. This cutoff was chosen because it gave positive results for pertactin (Prn) and Ag43, the only two model ATs previously known to have polyproline regions, but was uncommon overall in the E. coli proteome (0.14% of windows).
Disorder prediction
AT protein sequences were analyzed for regions of intrinsic disorder using the Espritz web server (Walsh et al., 2012), with default settings (prediction based on X-ray training set, best SW decision threshold).
Acknowledgments
This work was supported by the NIH (R01 GM097573). E.B. was supported by a graduate fellowship from the Chemistry-Biochemistry-Biology Interface Program (T32 GM075762). I.D. was supported by an Entrepreneurial Innovation Postdoctoral Fellowship from the College of Science at the University of Notre Dame. J.C. was supported by a Clare Boothe Luce Graduate Fellowship.
References
- Barnard TJ, Dautin N, Lukacik P, Bernstein HD, Buchanan SK. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat Struct Mol Biol. 2007;14:1214–1220. doi: 10.1038/nsmb1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard TJ, Gumbart J, Peterson JH, Noinaj N, Easley NC, Dautin N, et al. Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter. J Mol Biol. 2012;415:128–142. doi: 10.1016/j.jmb.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz I, Schmidt MA. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol Microbiol. 1992;6:1539–1546. doi: 10.1111/j.1365-2958.1992.tb00875.x. [DOI] [PubMed] [Google Scholar]
- Berthiaume F, Rutherford N, Mourez M. Mutations affecting the biogenesis of the AIDA-I autotransporter. Res Microbiol. 2007;158:348–354. doi: 10.1016/j.resmic.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Besingi RN, Chaney JL, Clark PL. An alternative outer membrane secretion mechanism for an autotransporter protein lacking a C-terminal stable core. Mol Microbiol. 2013;90:1028–1045. doi: 10.1111/mmi.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P, Cowen L, Menke M, King J, Berger B. BETAWRAP: Successful prediction of parallel β-helices from primary sequence reveals an association with many microbial pathogens. Proc Natl Acad Sci USA. 2001;98:14819–14824. doi: 10.1073/pnas.251267298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon LD, Goldberg MB. Periplasmic transit and disulfide bond formation of the autotransported Shigella protein IcsA. J Bacteriol. 2001;183:951–958. doi: 10.1128/JB.183.3.951-958.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braselmann E, Clark PL. Autotransporters: The cellular environment reshapes a folding mechanism to promote protein transport. J Phys Chem Lett. 2012;3:1063–1071. doi: 10.1021/jz201654k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MS, Cornell DG, Dluhy RA, Gierasch LM. Conformations of signal peptides induced by lipids suggest initial steps in protein export. Science. 1986;233:206–208. doi: 10.1126/science.2941862. [DOI] [PubMed] [Google Scholar]
- Brockmeyer J, Spelten S, Kuczius T, Bielaszewska M, Karch H. Structure and function relationship of the autotransport and proteolytic activity of EspP from shiga toxin-producing Escherichia coli. PLoS ONE. 2009;4:e6100. doi: 10.1371/journal.pone.0006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan AW, Starner-Kreinbrink JL, Hosur R, Clark PL, Berger B. Structure-based prediction reveals capping motifs that inhibit β-helix aggregation. Proc Natl Acad Sci USA. 2011;108:11099–11104. doi: 10.1073/pnas.1017504108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik N, Webb CT, Leyton DL, Holt KE, Heinz E, Gorrell R, et al. A bioinformatic strategy for the detection, classification and analysis of bacterial autotransporters. PLoS ONE. 2012;7:e43245. doi: 10.1371/journal.pone.0043245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau MÈ, Berthiaume F, Mourez M. Proteolytic processing is not essential for multiple functions of the Escherichia coli autotransporter adhesin involved in diffuse adherence (AIDA-I) J Bacteriol. 2006;188:8504–8512. doi: 10.1128/JB.00864-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau MÈ, Mourez M. Functional organization of the autotransporter Adhesin Involved in Diffuse Adherence. J Bacteriol. 2007;189:9020–9029. doi: 10.1128/JB.01238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles IG, Dougan G, Pickard D, Chatfield S, Smith M, Novotny P, et al. Molecular cloning and characterization of protective outer membrane protein P.69 from Bordetella pertussis. Proc Natl Acad Sci USA. 1989;86:3554–3558. doi: 10.1073/pnas.86.10.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté JP, Mourez M. Structure-function analysis of the TibA self-associating autotransporter reveals a modular organization. Infect Immun. 2011;79:1826–1832. doi: 10.1128/IAI.01129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- Dalbey RE, Kuhn A. Protein traffic in Gram-negative bacteria — how exported and secreted proteins find their way. FEMS Microbiol Rev. 2012;36:1023–1045. doi: 10.1111/j.1574-6976.2012.00327.x. [DOI] [PubMed] [Google Scholar]
- Dautin N, Barnard TJ, Anderson DE, Bernstein HD. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J. 2007;26:1942–1952. doi: 10.1038/sj.emboj.7601638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautin N, Bernstein HD. Protein secretion in Gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- Desvaux M, Scott-Tucker A, Turner SM, Cooper LM, Huber D, Nataro JP, Henderson IR. A conserved extended signal peptide region directs posttranslational protein translocation via a novel mechanism. Microbiology. 2007;153:59–70. doi: 10.1099/mic.0.29091-0. [DOI] [PubMed] [Google Scholar]
- Drobnak I, Braselmann E, Clark PL. Multiple driving forces required for secretion of autotransporter virulence proteins. 2014. Manuscript under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta PR, Sui BQ, Nataro JP. Structure-function analysis of the enteroaggregative Escherichia coli plasmid-encoded toxin autotransporter using scanning linker mutagenesis. J Biol Chem. 2003;278:39912–39920. doi: 10.1074/jbc.M303595200. [DOI] [PubMed] [Google Scholar]
- Emsley P, Charles IG, Fairweather NF, Isaacs NW. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature. 1996;381:90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylip (phylogeny inference package) version 3.6. Department of Genome Sciences, University of Washington; Seattle: 2005. Distributed by the author. [Google Scholar]
- Fink DL, Cope LD, Hansen EJ, Geme JWS. The Hemophilus influenzae Hap autotransporter is a chymotrypsin clan serine protease and undergoes autoproteolysis via an intermolecular mechanism. J Biol Chem. 2001;276:39492–39500. doi: 10.1074/jbc.M106913200. [DOI] [PubMed] [Google Scholar]
- Fischer W, Buhrdorf R, Gerland E, Haas R. Outer membrane targeting of passenger proteins by the vacuolating cytotoxin autotransporter of Helicobacter pylori. Infect Immun. 2001;69:6769–6775. doi: 10.1128/IAI.69.11.6769-6775.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, Lacy DB. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc Natl Acad Sci USA. 2007;104:16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawarzewski I, DiMaio F, Winterer E, Tschapek B, Smits SH, Jose J, Schmitt L. Crystal structure of the transport unit of the autotransporter adhesin involved in diffuse adherence from Escherichia coli. J Struct Biol. 2014;187:20–29. doi: 10.1016/j.jsb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, Zaccai NR, Fleming KG. Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc Natl Acad Sci USA. 2014;111:5878–5883. doi: 10.1073/pnas.1322473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grijpstra J, Arenas J, Rutten L, Tommassen J. Autotransporter secretion: Varying on a theme. Res Microbiol. 2013;164:562–582. doi: 10.1016/j.resmic.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Hagan CL, Silhavy TJ, Kahne D. β-Barrel membrane protein assembly by the Bam complex. Ann Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Nataro JP. Virulence functions of autotransporter proteins. Infect Immun. 2001;69:1231–1243. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Nataro JP. The great escape: structure and functions of the autotransporter proteins. Trends Microbiol. 1998;6:370–378. doi: 10.1016/s0966-842x(98)01318-3. [DOI] [PubMed] [Google Scholar]
- Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, et al. The antigen 43 structure reveals a molecular velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc Natl Acad Sci USA. 2014;111:457–462. doi: 10.1073/pnas.1311592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller K, Grote A, Scheer M, Münch R, Jahn D. PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 2004;32:W375–W379. doi: 10.1093/nar/gkh378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci USA. 2009;106:19120–19125. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R, Tian P, Peterson JH, Bernstein HD. Sequential and spatially restricted interactions of assembly factors with an autotransporter β domain. Proc Natl Acad Sci USA. 2011;108:E383–E391. doi: 10.1073/pnas.1103827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivie S, McClain M, Algood H, Lacy DB, Cover T. Analysis of a β-helical region in the p55 domain of Helicobacter pylori vacuolating toxin. BMC Microbiol. 2010;10:60. doi: 10.1186/1471-2180-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TA, Qiu J, Plaut AG, Holyoak T. Active-site gating regulates substrate selectivity in a chymotrypsin-like serine protease: The structure of haemophilus influenzae immunoglobulin A1 protease. J Mol Biol. 2009;389:559–574. doi: 10.1016/j.jmb.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong WS, Luirink J. The conserved extension of the Hbp autotransporter signal peptide does not determine targeting pathway specificity. Biochem Biophys Res Commun. 2008;368:522–527. doi: 10.1016/j.bbrc.2008.01.122. [DOI] [PubMed] [Google Scholar]
- Jong WS, Saurí A, Luirink J. Extracellular production of recombinant proteins using bacterial autotransporters. Curr Opin Biotechnol. 2010;21:646–652. doi: 10.1016/j.copbio.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Jong WSP, Ten Hagen-Jongman CM, Den Blaauwen T, Jan Slotboom D, Tame JRH, Wickström D, et al. Limited tolerance towards folded elements during secretion of the autotransporter Hbp. Mol Microbiol. 2007;63:1524–1536. doi: 10.1111/j.1365-2958.2007.05605.x. [DOI] [PubMed] [Google Scholar]
- Jose J, Meyer TF. The autodisplay story, from discovery to biotechnical and biomedical applications. Microbiol Mol Biol Rev. 2007;71:600–619. doi: 10.1128/MMBR.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker M, Besingi RN, Clark PL. Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol Microbiol. 2009;71:1323–1332. doi: 10.1111/j.1365-2958.2009.06607.x. [DOI] [PubMed] [Google Scholar]
- Junker M, Clark PL. Slow formation of aggregation-resistant beta-sheet folding intermediates. Proteins. 2010;78:812–824. doi: 10.1002/prot.22609. [DOI] [PubMed] [Google Scholar]
- Junker M, Schuster CC, McDonnell AV, Sorg KA, Finn MC, Berger B, Clark PL. Pertactin β-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc Natl Acad Sci USA. 2006;103:4918–4923. doi: 10.1073/pnas.0507923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC. Beta-helix model for the filamentous haemagglutinin adhesion of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol. 2001;42:279–292. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Steven AC. The turn of the screw: variations of the abundant beta-solenoid motif in passenger domains of Type V secretory proteins. J Struct Biol. 2006;155:306–315. doi: 10.1016/j.jsb.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Khan S, Mian HS, Sandercock LE, Chirgadze NY, Pai EF. Crystal structure of the passenger domain of the Escherichia coli autotransporter EspP. J Mol Biol. 2011;413:985–1000. doi: 10.1016/j.jmb.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser T, Krämer J, Otzelberger K, Pohlner J, Meyer TF. Characterization of the Neisseria Iga β-core: The essential unit for outer membrane targeting and extracellular protein secretion. J Mol Biol. 1993;234:579–593. doi: 10.1006/jmbi.1993.1613. [DOI] [PubMed] [Google Scholar]
- Klauser T, Pohlner J, Meyer TF. Extracellular transport of cholera toxin B subunit using Neisseria IgA protease β-domain: conformation-dependent outer membrane translocation. EMBO J. 1990;9:1991–1999. doi: 10.1002/j.1460-2075.1990.tb08327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny M, Benz I, Hollinderbäumer B, Beinke C, Niederweis M, Alexander Schmidt M. Modular organization of the AIDA autotransporter translocator: The N-terminal β1-domain is surface-exposed and stabilizes the transmembrane β2-domain. Antonie van Leeuwenhoek. 2001;80:19–34. doi: 10.1023/a:1012084325728. [DOI] [PubMed] [Google Scholar]
- Konieczny MP, Suhr M, Noll A, Autenrieth IB, Alexander Schmidt M. Cell surface presentation of recombinant (poly-) peptides including functional T-cell epitopes by the AIDA autotransporter system. FEMS Immunol Med Microbiol. 2000;27:321–332. doi: 10.1111/j.1574-695X.2000.tb01446.x. [DOI] [PubMed] [Google Scholar]
- Kühnel K, Diezmann D. Crystal structure of the autochaperone region from the Shigella flexneri autotransporter IcsA. J Bacteriol. 2011;193:2042–2045. doi: 10.1128/JB.00790-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo JC, Grin I, Linke D. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Phil Trans R Soc B. 2012;367:1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyton DL, Adams LM, Kelly M, Sloan J, Tauschek M, Robins-Browne RM, et al. Contribution of a novel gene, rpeA, encoding a putative autotransporter adhesion to intestinal colonization by rabbit-specific enteropathogenic Escherichia coli. Infect Immun. 2007;75:4664–4669. doi: 10.1128/IAI.00972-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyton DL, Johnson MD, Thapa R, Huysmans GH, Dunstan RA, Celik N, et al. A mortise-tenon joint in the transmembrane domain modulates autotransporter assembly into bacterial outer membranes. Nat Commun. 2014;5:4239. doi: 10.1038/ncomms5239. [DOI] [PubMed] [Google Scholar]
- Leyton DL, Rossiter AE, Henderson IR. From self-sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol. 2012;10:213–225. doi: 10.1038/nrmicro2733. [DOI] [PubMed] [Google Scholar]
- Leyton DL, Sevastsyanovich YR, Browning DF, Rossiter AE, Wells TJ, Fitzpatrick RE, et al. Size and conformation limits to secretion of disulfide-bonded loops in autotransporter proteins. J Biol Chem. 2011;286:42283–42291. doi: 10.1074/jbc.M111.306118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenthal C, Elsinghorst EA. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect Immun. 1999;67:4084–4091. doi: 10.1128/iai.67.8.4084-4091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveless BJ, Saier MH. A novel family of channel-forming, autotransporting, bacterial virulence proteins. Mol Membr Biol. 1997;14:113–123. doi: 10.3109/09687689709048171. [DOI] [PubMed] [Google Scholar]
- Makoff AJ, Oxer MD, Ballantine SP, Fairweather NF, Charles IG. Protective surface antigen P69 of Bordetella pertussis: Its characterization and very high level expression in Escherichia coli. Nat Biotech. 1990;8:1030–1033. doi: 10.1038/nbt1190-1030. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer J, Jose J, Meyer TF. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J Bacteriol. 1999;181:7014–7020. doi: 10.1128/jb.181.22.7014-7020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May KL, Morona R. Mutagenesis of the Shigella flexneri autotransporter IcsA reveals novel functional regions involved in IcsA biogenesis and recruitment of host neural Wiscott-Aldrich syndrome protein. J Bacteriol. 2008;190:4666–4676. doi: 10.1128/JB.00093-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Spahich N, Kenjale R, Waksman G, St Geme JW. Crystal structure of the Haemophilus influenzae Hap adhesin reveals an intercellular oligomerization mechanism for bacterial aggregation. EMBO J. 2011;30:3864–3874. doi: 10.1038/emboj.2011.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza-Aguilar JD, Fromme P, Torres-Larios A, Mendoza-Hernández G, Hernandez-Chiñas U, de los Monteros RAAE, et al. X-ray crystal structure of the passenger domain of plasmid encoded toxin (Pet), an autotransporter enterotoxin from enteroaggregative Escherichia coli (EAEC) Biochem Biophys Res Commun. 2014;445:439–444. doi: 10.1016/j.bbrc.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Caprioli RM, Cover TL. Carboxy-terminal proteolytic processing of Helicobacter pylori vacuolating toxin. Infect Immun. 2001;69:543–546. doi: 10.1128/IAI.69.1.543-546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Horinouchi S. Extracellular production of a Serratia marcescens serine protease in Escherichia coli. Biosci Biotech Biochem. 1996;60:1551–1558. doi: 10.1271/bbb.60.1551. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Nishiyama M, Horinouchi S, Beppu T. Involvement of the COOH-terminal pro-sequence of Serratia marcescens serine protease in the folding of the mature enzyme. J Biol Chem. 1994;269:32800–32806. [PubMed] [Google Scholar]
- Oliver DC, Huang G, Fernandez RC. Identification of secretion determinants of the Bordetella pertussis BrkA autotransporter. J Bacteriol. 2003a;185:489–495. doi: 10.1128/JB.185.2.489-495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver DC, Huang G, Nodel E, Pleasance S, Fernandez RC. A conserved region within the Bordetella pertussis autotransporter BrkA is necessary for folding of its passenger domain. Mol Microbiol. 2003b;47:1367–1383. doi: 10.1046/j.1365-2958.2003.03377.x. [DOI] [PubMed] [Google Scholar]
- Oomen CJ, van Ulsen P, Van Gelder P, Feijen M, Tommassen J, Gros P. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 2004;23:1257–1266. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto BR, Sijbrandi R, Luirink J, Oudega B, Heddle JG, Mizutani K, et al. Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J Biol Chem. 2005;280:17339–17345. doi: 10.1074/jbc.M412885200. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pohlner J, Halter R, Beyreuther K, Meyer TF. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- Pohlner J, Langenberg U, Wölk U, Beck SC, Meyer TF. Uptake and nuclear transport of Neisseria IgA1 protease-associated α-proteins in human cells. Mol Microbiol. 1995;17:1073–1083. doi: 10.1111/j.1365-2958.1995.mmi_17061073.x. [DOI] [PubMed] [Google Scholar]
- Ramesh B, Sendra VG, Cirino PC, Varadarajan N. Single-cell characterization of autotransporter-mediated Escherichia coli surface display of disulfide bond-containing proteins. J Biol Chem. 2012;287:38580–38589. doi: 10.1074/jbc.M112.388199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn J, Junker M, Besingi R, Braselmann E, Clark P. ATP-independent control of autotransporter virulence protein transport via the folding properties of the secreted protein. Chem Biol. 2012;19:287–296. doi: 10.1016/j.chembiol.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn JP, Clark PL. A conserved stable core structure in the passenger domain β-helix of autotransporter virulence proteins. Biopolymers. 2008;89:420–427. doi: 10.1002/bip.20924. [DOI] [PubMed] [Google Scholar]
- Roman-Hernandez G, Peterson JH, Bernstein HD. Reconstitution of bacterial autotransporter assembly using purified components. eLife. 2014;3 doi: 10.7554/eLife.04234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel-Jazédé V, Grijpstra J, van Dam V, Tommassen J, van Ulsen P. Lipidation of the autotransporter NalP of Neisseria meningitidis is required for its function in the release of cell-surface-exposed proteins. Microbiology. 2013;159:286–295. doi: 10.1099/mic.0.063982-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of enterobacteriaceae. J Bacteriol. 2009;191:6571–6583. doi: 10.1128/JB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford N, Charbonneau MÈ, Berthiaume F, Betton JM, Mourez M. The periplasmic folding of a cysteineless autotransporter passenger domain interferes with its outer membrane translocation. J Bacteriol. 2006;188:4111–4116. doi: 10.1128/JB.01949-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurí A, Oreshkova N, Soprova Z, Jong WS, Sani M, Peters PJ, et al. Autotransporter β-domains have a specific function in protein secretion beyond outer-membrane targeting. J Mol Biol. 2011;412:553–567. doi: 10.1016/j.jmb.2011.07.035. [DOI] [PubMed] [Google Scholar]
- Saurí A, Soprova Z, Wickström D, de Gier JW, Van der Schors RC, Smit AB, et al. The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology. 2009;155:3982–3991. doi: 10.1099/mic.0.034991-0. [DOI] [PubMed] [Google Scholar]
- Saurí A, ten Hagen-Jongman CM, van Ulsen P, Luirink J. Estimating the size of the active translocation pore of an autotransporter. J Mol Biol. 2012;416:335–345. doi: 10.1016/j.jmb.2011.12.047. [DOI] [PubMed] [Google Scholar]
- Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuoiating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- Schuler B, Fürst F, Osterroth F, Steinbacher S, Huber R, Seckler R. Plasticity and steric strain in a parallel β-helix: Rational mutations in the P22 tailspike protein. Proteins. 2000;39:89–101. [PubMed] [Google Scholar]
- Sevastsyanovich YR, Leyton DL, Wells TJ, Wardius CA, Tveen-Jensen K, Morris FC, et al. A generalized module for the selective extracellular accumulation of recombinant proteins. Microb Cell Fact. 2012;11:69. doi: 10.1186/1475-2859-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shere KD, Sallustio S, Manessis A, D’Aversa TG, Goldberg MB. Disruption of IcsP, the major Shigella protease that cleaves IcsA, accelerates actin-based motility. Mol Microbiol. 1997;25:451–462. doi: 10.1046/j.1365-2958.1997.4681827.x. [DOI] [PubMed] [Google Scholar]
- Shimada K, Ohnishi Y, Horinouchi S, Beppu T. Extracellular transport of pseudoazurin of Alcaligenes faecalis in Escherichia coli using the COOH-terminal domain of Serratia marcescens serine protease. J Biochem. 1994;116:327–334. doi: 10.1093/oxfordjournals.jbchem.a124527. [DOI] [PubMed] [Google Scholar]
- Soprova Z, Sauri A, van Ulsen P, Tame JRH, den Blaauwen T, Jong WSP, Luirink J. A conserved aromatic residue in the autochaperone domain of the autotransporter Hbp is critical for initiation of outer membrane translocation. J Biol Chem. 2010;285:38224–38233. doi: 10.1074/jbc.M110.180505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr M, Benz I, Schmidt MA. Processing of the AIDA-I precursor: removal of AIDAC and evidence for the outer membrane anchoring as a β-barrel structure. Mol Microbiol. 1996;22:31–42. doi: 10.1111/j.1365-2958.1996.tb02653.x. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Lett MC, Sasakawa C. Extracellular transport of VirG protein in Shigella. J Biol Chem. 1995;270:30874–30880. doi: 10.1074/jbc.270.52.30874. [DOI] [PubMed] [Google Scholar]
- Szabady RL, Peterson JH, Skillman KM, Bernstein HD. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci USA. 2005;102:221–226. doi: 10.1073/pnas.0406055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima N, Kawai F, Park SY, Tame JR. A novel intein-like autoproteolytic mechanism in autotransporter proteins. J Mol Biol. 2010;402:645–656. doi: 10.1016/j.jmb.2010.06.068. [DOI] [PubMed] [Google Scholar]
- Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014;42:D553–D559. doi: 10.1093/nar/gkt1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh MY, Morona R. Identification of Shigella flexneri IcsA residues affecting interaction with N-WASP, and evidence for IcsA-IcsA co-operative interaction. PLoS ONE. 2013;8:e55152. doi: 10.1371/journal.pone.0055152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh MY, Tran ENH, Morona R. Absence of O antigen suppresses Shigella flexneri IcsA autochaperone region mutations. Microbiology. 2012;158:2835–2850. doi: 10.1099/mic.0.062471-0. [DOI] [PubMed] [Google Scholar]
- Thanassi DG, Stathopoulos C, Karkal A, Li H. Protein secretion in the absence of ATP: the autotransporter, two-partner secretion and chaperone/usher pathways of Gram-negative bacteria. Mol Membr Biol. 2005;22:63–72. doi: 10.1080/09687860500063290. [DOI] [PubMed] [Google Scholar]
- van den Berg B. Crystal structure of a full-length autotransporter. J Mol Biol. 2010;396:627–633. doi: 10.1016/j.jmb.2009.12.061. [DOI] [PubMed] [Google Scholar]
- van der Woude MW, Henderson IR. Regulation and function of Ag43 (Flu) Annu Rev Microbiol. 2008;62:153–169. doi: 10.1146/annurev.micro.62.081307.162938. [DOI] [PubMed] [Google Scholar]
- van Ulsen P, Rahman Su, Jong WS, Daleke-Schermerhorn MH, Luirink J. Type V secretion: From biogenesis to biotechnology. Biochim Biophys Acta. 2014;1843:1592–1611. doi: 10.1016/j.bbamcr.2013.11.006. [DOI] [PubMed] [Google Scholar]
- van Ulsen P, van Alphen L, Hopman CT, van der Ende A, Tommassen J. In vivo expression of Neisseria meningitidis proteins homologous to the Haemophilus influenzae Hap and Hia autotransporters. FEMS Immunol Med Microbiol. 2001;32:53–64. doi: 10.1111/j.1574-695X.2001.tb00534.x. [DOI] [PubMed] [Google Scholar]
- van Ulsen P, Van Alphen L, Ten Hove J, Fransen F, Van Der Ley P, Tommassen J. A neisserial autotransporter NalP modulating the processing of other autotransporters. Mol Microbiol. 2003;50:1017–1030. doi: 10.1046/j.1365-2958.2003.03773.x. [DOI] [PubMed] [Google Scholar]
- Velarde JJ, Nataro JP. Hydrophobic residues of the autotransporter EspP linker domain are important for outer membrane translocation of its passenger. J Biol Chem. 2004;279:31495–31504. doi: 10.1074/jbc.M404424200. [DOI] [PubMed] [Google Scholar]
- Walsh I, Martin AJM, Di Domenico T, Tosatto SCE. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012;28:503–509. doi: 10.1093/bioinformatics/btr682. [DOI] [PubMed] [Google Scholar]
- Wells TJ, Totsika M, Schembri MA. Autotransporters of Escherichia coli: a sequence-based comparison. Microbiology. 2010;156:2459–2469. doi: 10.1099/mic.0.039024-0. [DOI] [PubMed] [Google Scholar]
- Wilhelm S, Gdynia A, Tielen P, Rosenau F, Jaeger KE. The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J Bacteriol. 2007;189:6695–6703. doi: 10.1128/JB.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S, Tommassen J, Jaeger KE. A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J Bacteriol. 1999;181:6977–6986. doi: 10.1128/jb.181.22.6977-6986.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida N, Uozumi T, Beppu T. Specific excretion of Serratia marcescens protease through the outer membrane of Escherichia coli. J Bacteriol. 1986;166:937–944. doi: 10.1128/jb.166.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TH, Pan JG, Seo YS, Rhee JS. Use of Pseudomonas putida EstA as an anchoring motif for display of a periplasmic enzyme on the surface of Escherichia coli. Appl Environ Microbiol. 2004;70:6968–6976. doi: 10.1128/AEM.70.12.6968-6976.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen MR, Peabody CR, Partovi SM, Zhai Y, Tseng YH, Saier MH. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim Biophys Acta. 2002;1562:6–31. doi: 10.1016/s0005-2736(02)00359-0. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Zhang K, Huo Y, Zhu Y, Zhou Q, Lu J, et al. Autotransporter passenger domain secretion requires a hydrophobic cavity at the extracellular entrance of the β-domain pore. Biochem J. 2011;435:577–587. doi: 10.1042/BJ20101548. [DOI] [PubMed] [Google Scholar]