Abstract
During interphase, recycling endosomes mediate the transport of internalized cargo back to the plasma membrane. However, in mitotic cells, recycling endosomes are essential for the completion of cytokinesis, the last phase of mitosis that promotes the physical separation the two daughter cells. Despite recent advances, our understanding of the molecular determinants that regulate recycling endosome dynamics during cytokinesis remains incomplete. We have previously demonstrated that Molecule Interacting with CasL Like-1 (MICAL-L1) and C-terminal Eps15 Homology Domain protein 1 (EHD1) coordinately regulate receptor transport from tubular recycling endosomes during interphase. However, their potential roles in controlling cytokinesis had not been addressed. In this study, we show that MICAL-L1 and EHD1 regulate mitosis. Depletion of either protein resulted in increased numbers of bi-nucleated cells. We provide evidence that bi-nucleation in MICAL-L1- and EHD1-depleted cells is a consequence of impaired recycling endosome transport during late cytokinesis. However, depletion of MICAL-L1, but not EHD1, resulted in aberrant chromosome alignment and lagging chromosomes, suggesting an EHD1-independent function for MICAL-L1 earlier in mitosis. Moreover, we provide evidence that MICAL-L1 and EHD1 differentially influence microtubule dynamics during early and late mitosis. Collectively, our new data suggest several unanticipated roles for MICAL-L1 and EHD1 during the cell cycle.
Keywords: MICAL-L1, EHD1, endocytic recycling, membrane trafficking, cytokinesis, mitosis, intercellular bridge, lagging chromosomes, midbody, centrosome
Introduction
The goal of mitosis is to uniformly segregate replicated DNA between two daughter cells. The equal distribution of DNA to each daughter cell is required for proper development and the maintenance of homeostasis in adult tissues. Indeed, the failure to accurately complete mitosis is a possible mechanism for promoting cellular transformation and oncogenesis(1-3). Recent studies have revealed essential roles for endocytic membrane trafficking pathways in the regulation of mitotic events(4). In particular, endocytic recycling is required for the successful completion of cytokinesis, the final stage of mitosis that generates two diploid daughter cells each containing a single nucleus(5, 6). Once chromosomes have been segregated to each centrosome, signals derived from the tubulin-rich central spindle orchestrate a series of events that promote the formation of an actino-myosin ring around the middle of the central spindle. This marks the site at which the cleavage furrow, and eventually, the midbody, is generated. Increased RhoA activity leads to myosin-based furrow ingression and the squeezing of the central spindle into a thin structure known as the intercellular bridge (ICB). Cytokinesis concludes with abscission, a poorly understood process that leads to the severing of the ICB and the separation of the two daughter cells(7).
Cumulative data from a number of laboratories suggest that recycling endosomes regulate cytokinesis mainly by two mechanisms: 1) Delivery of membranes to the ingressing furrow and then the ICB, thereby stabilizing these structures. 2) Controlling the transport of regulatory proteins that influence cytoskeletal and lipid dynamics to the furrow and ICB. Initial studies in Drosophila embryos demonstrated that endocytic regulatory proteins such as the GTP-binding protein Rab11 and its interacting partner Nuclear Fallout (the Drosophila homolog of the mammalian Rab11 Family Interacting Proteins 3, FIP3) are required for cellularization(8), a process akin to cytokinesis. In mammalian cells, depletion of Rab11 or FIP3 results in cytokinesis failure, leading to the accumulation of tetraploid cells with multiple nuclei(9).
A key component of endocytic trafficking is the tethering of endosomes at their target organelles. The exocyst, an octameric protein complex that mediates the tethering of secretory vesicles to the plasma membrane during cytokinesis(10), is also required for the tethering of Rab11/FIP3 endosomes at the ICB(11). FIP3 interacts with both Rab11 and the GTP-binding protein Arf6, forming a tertiary complex. Arf6 mediates the tethering of FIP3 endosomes to the ICB by interacting with the exocyst component Exo70. Expression of a dominant-negative GDP-locked Arf6 or siRNA-depletion of Exo70 impairs FIP3 recruitment to the ICB and thus disrupts cytokinesis(11). Additionally, Syntaxin 16, a soluble N-ethylmaleimide-sensitive factor attachment receptor (SNARE) protein, has been implicated in vesicle fusion during cytokinesis(12).
Recent studies have shed new light on additional factors that recruit FIP3 to the ICB and the role of FIP3 endosomes in regulating ICB actin dynamics. For example, FIP3 directly binds to Cyk-4/MgcRacGAP(13). Cyk-4 is a Rho GTPase-activating protein (GAP) and a component of the centralspindlin complex along with the microtubule bundling kinesin-like protein, MKLP1(14). Somewhat counterintuitively, Cyk-4 promotes activation of RhoA, presumably through its binding and activation of Ect2(15), a Rho guanine nucleotide exchange factor (GEF), which coordinates actino-myosin ring contraction during early cytokinesis. During late cytokinesis Cyk-4 acts as a tether for FIP3 containing endosomes and is required to prevent FIP3 endosome displacement from the ICB(13).
Changes in microtubule stability are also required for FIP3 localization to the ICB. Microtubule bending and depolymerization precede FIP3 endosome fusion at the ICB(16), which enhances ICB plasma membrane dynamics leading to secondary ingression, a rapid constriction of the ICB from a thickness of approximately 1-2 μm to 0.1 μm. While decreases in microtubule content are required for thinning of the ICB, depolymerization of cortical actin filaments is also required for both ICB thinning and abscission. FIP3 endosomes mediate the delivery of p50RhoGAP/ARHGAP1 and SCAMP2/3, which coordinate actin depolymerization at the ICB, allowing for secondary ingression (17) and eventually, abscission. Currently, there are two separate, but likely inter-connected models regarding the molecular machinery that mediates the abscission(18). Prekeris and colleagues suggest that FIP3 endosome-dependent secondary ingression mediates ICB thinning that creates a structure wherein the Endosomal Sorting Complex Required for Transport (ESCRT) is able to assemble and mediate abscission via a mechanism analogous to its function in membrane scission during multivesicular body formation or retroviral budding. (19-21). CEP55, a centrosomal protein that localizes to the midbody, binds to ESCRT component tumor susceptibility gene 101 (TSG 101) and the ESCRT accessory protein Alix and recruits them onto the midbody. Alix and TSG101 then recruit ESCRT III component CHMP4B, which mediates membrane constriction and abscission. Alternatively, FIP3 endosomes may not be required for the terminal event of abscission. The ESCRT complex alone, through CHMP4B polymerization into long spiral filaments(22), may mediate abscission. Thus, abscission may be ESCRT-dependent or recycling endosome-dependent. It is likely that the two models are not mutually exclusive(18).
In addition to Rab11/FIP3 endosomes, Rab35 recycling endosomes are also required for abscission(23, 24). Rab35 binds to the phosphoinositide phosphatase OCRL and regulates its delivery to the ICB(25). OCRL dephosphorylates phosphatidylinositol-4,5-bisphosphate, thereby causing actin depolymerization on the ICB likely through decreased Arp 2/3 activity. SiRNA-mediated depletion of Rab35 or OCRL impairs abscission, leading to daughter cells that remain connected by abnormally long and stable ICB with increased actin content.
Given the importance of recycling endosome delivery in controlling cytokinesis, it is crucial to understand the roles of regulatory proteins involved in recycling endosome dynamics during cytokinesis. Our lab has demonstrated that the protein Molecule Interacting with CasL-Like1 (MICAL-L1) acts as a membrane hub on tubular recycling endosomes (TRE) that emanate from the perinuclear region and is required for the recycling of several clathrin-dependent and clathrin-independent cargos(26, 27). MICAL-L1 interacts with and recruits membrane bending and vesiculating proteins such as Syndapin2 and the C-terminal Eps15 homology domain proteins 1 and 3 (EHD1/3) to TRE, thus regulating TRE dynamics(28, 29). Given the established role of MICAL-L1 and EHD1 in regulating recycling during interphase and the necessity of recycling endosome delivery to the ICB for the completion of cytokinesis, we hypothesized that MICAL-L1 and EHD1 are required for cytokinesis. We found that depletion of MICAL-L1 and EHD1 in HeLa cells resulted in cytokinesis failure and the generation of bi-nucleated and multi-nucleated cells. MICAL-L1 and EHD1 are required for the delivery of recycling endosomes to the ICB during late cytokinesis. Moreover, MICAL-L1 and EHD1 also regulate pre-cytokinetic events such as mitotic spindle orientation, chromosome alignment and microtubule dynamics, findings that supports recent studies by Doxsey and colleagues that implicate Rab11 and recycling endosomes in these pre-cytokinetic processes(30). Thus, we establish MICAL-L1 and EHD1 as novel regulators of mitosis and cytokinesis.
Results
MICAL-L1- or EHD1-depletion in HeLa cells impairs normal cell cycle
To test if MICAL-L1 and EHD1 are required for normal cell division, we used siRNA to deplete HeLa cells of MICAL-L1 and EHD1. By immunoblot analysis, we demonstrated >90% depletion of MICAL-L1 (Figure 1A) and EHD1 (Figure 1B) after 48 and 72 h of transfection. The MICAL-L1- and EHD1-depleted cells were then grown on glass coverslips, and stained with tubulin and DAPI to assess nuclear morphology (Figure 1C-H). In contrast to nearly all control-siRNA cells that contained a single, oval shaped nucleus (Figure 1C, and D), MICAL-L1-depleted cells displayed several abnormal nuclear phenotypes including bi-nucleation (Figure 1E and F; red arrow) and micro-nuclei/nuclei separated by ‘chromatin bridges’ (Figure 1E and F; purple arrows). EHD1-depletion led to a significant increase in bi-nucleated and multi-nucleated cells (Figure 1G and H; red arrows). We classified cells based on 5 nuclear/morphological phenotypes and quantified the number of cells corresponding to each phenotype in control, MICAL-L1-depleted, and EHD1-depleted cells (Figure 1I and J; for simplicity, quantitation of multi-nucleation and micronuclei are shown. Detailed phenotypic and statistical analysis is found in Supplemental Figure 1A). By quantifying nuclear area, we observed that MICAL-L1-depleted cells contained a significantly smaller nuclear area than control nuclei. On the other hand, EHD1-depleted cells often contained one or more large nuclei and thus had a significantly larger nuclear area/cell (Figure 1J; representative images exported from Image J are shown in Supplemental Figure 1 B-G).
Figure 1. Depletion of MICAL-L1 or EHD1 in HeLa cells causes cell cycle defects.
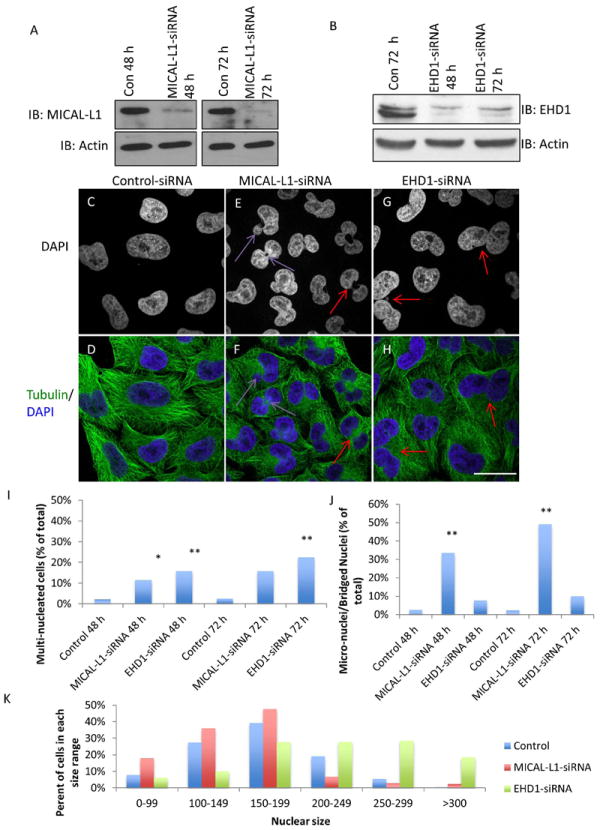
(A and B) Immunoblots demonstrating the efficacy of MICAL-L1 (A) and EHD1 (B) siRNA-mediated knockdown. (C-H) HeLa cells were transfected with control-siRNA (C and D), MICAL-L1-siRNA (E and F) or EHD1-siRNA (G and H) for 72 h and stained with α-tubulin (green) and DAPI (Blue). Purple arrows show micro-nucleation and bridged nuclei while red arrows mark bi-nucleated cells. (I and J) Quantification of nuclear phenotypes from three independent experiments (n=300/experiment) expressed as a percentage of cells/experiment in each phenotypic category. (K) Nuclear area per cell was quantified using the ImageJ ‘analyze particles’ function. n=100 cells/experiment, from three independent experiments. One-way ANOVA *p<0.05, **p<0.01. Scale bar=10 μm.
To rule out potential off-target effects for the pooled siRNA oligonucleotides, we depleted MICAL-L1 using two different individual oligonucleotides and found that both efficiently depleted MICAL-L1 (Supplemental Figure 2A). As shown, both MICAL-L1 siRNA oligonucleotides significantly increased the number of bi-nucleated and micro-nucleated cells compared to control-siRNA cells (Supplemental Figure 2B-E). The increase in bi/multi-nucleated cells was also specific to EHD1-depletion, as introduction of a siRNA-resistant EHD1 construct partially but significantly rescued the bi-nucleation phenotype induced upon EHD-depletion (Supplemental Figure 2 F-M; quantified in L). As an additional control, we analyzed the impact of EHD2-depletion. EHD2 is a paralog that displays ~67% amino acid identity to EHD1, but localizes to the plasma membrane(31) and regulates caveolar motility rather than membrane recycling(32-34). As demonstrated, EHD2-depletion did not induce bi-nucleation (Supplemental Figure 3A-D). Overall, these data indicate a role for MICAL-L1 and EHD1 in the regulation of cell cycle.
MICAL-L1 and EHD1 are required for cytokinesis and transport of recycling endosomes to the ICB
To determine at which stages MICAL-L1 and EHD1 are required in the cell cycle, we monitored control cells, MICAL-L1-depleted cells, and EHD1-depleted cells by live imaging analysis. Cells were imaged between 24-48 h post-transfection to observe the first rounds of cell division after efficient siRNA-mediated down-regulation (Figure 2). Control cells entered mitosis and completed cytokinesis in approximately 2-3 hours (Figure 2A; see Supplemental Video 1). MICAL-L1-depleted cells also entered mitosis, but these cells typically remained attached via their ICB for ~5 hours (Figure 2B; arrow; see Supplemental Video 2), and in some cases, remained connected until the next mitotic division and failed cytokinesis (Figure 2C; arrows at 60 min. mark the daughter cells from distinct mitoses that fuse to form a single bi-nucleated cell at 540 min.; see Supplemental Video 3). EHD1-depleted cells also displayed abnormal cytokinesis. Many cells displayed asymmetric cell spreading early in cytokinesis (Figure 2D; asterisks), a phenotype previously documented in Rab11-depleted HeLa cells(30). EHD1-depletion also led to cytokinesis failure resulting in bi-nucleated cells (Figure 2D; arrow).
Figure 2. MICAL-L1- or EHD1-depletion leads to cytokinesis failure.
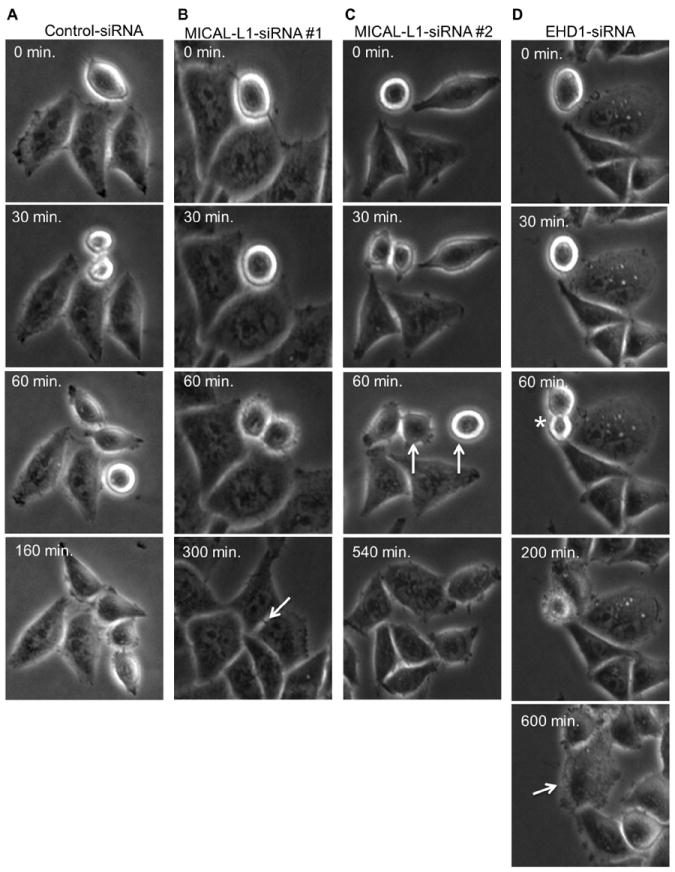
(A-D) HeLa cells growing on plastic dishes were transfected with control-siRNA (A), MICAL-L1-siRNA (B and C) or EHD1-siRNA (D) for 24 h. Live cells were then imaged by phase contrast every 10 minutes for ~36 h to follow progression through mitosis. (B) An arrow marks delayed abscission while in C, arrows (C) mark cytokinesis failure in MICAL-L1-depleted cells. In D, the asterisk denotes asymmetric division where there is unequal cell spreading during cytokinesis and the arrow points to cytokinesis failure in EHD1-depleted cells.
Given that MICAL-L1 and EHD1 are required for cytokinesis, we hypothesized that these two proteins localize to the ICB during cytokinesis. To test this notion, fixed HeLa cells were labeled with antibodies against endogenous EHD1 and MICAL-L1 to assess their localization during cytokinesis. As demonstrated, both proteins localized near the ingressing furrow during early cytokinesis, and to the ICB during late cytokinesis (Figure 3A-F; yellow arrows; see insets). Importantly, this localization is specific to the recycling regulator, EHD1, as EHD2 is excluded from the ICB (Supplemental Figure 4A-C). MICAL-L1 also partially co-localized with Rab11 on the ICB during late cytokinesis (Figure 3G-I). Interestingly, we noted that MICAL-L1 (and EHD1) and Rab11 frequently localized to tubules during late cytokinesis, with Rab11 tubules concentrated in the perinuclear region and MICAL-L1 tubules extending outward to the cell periphery. Lastly, MICAL-L1 is required for recruitment of EHD1 to the ICB, as MICAL-L1-depletion impaired EHD1 localization to the ICB during late cytokinesis (Supplemental Figure 4D-K).
Figure 3. MICAL-L1 and EHD1 are required for delivery of transferrin- containing recycling endosomes to the intercellular bridge (ICB).
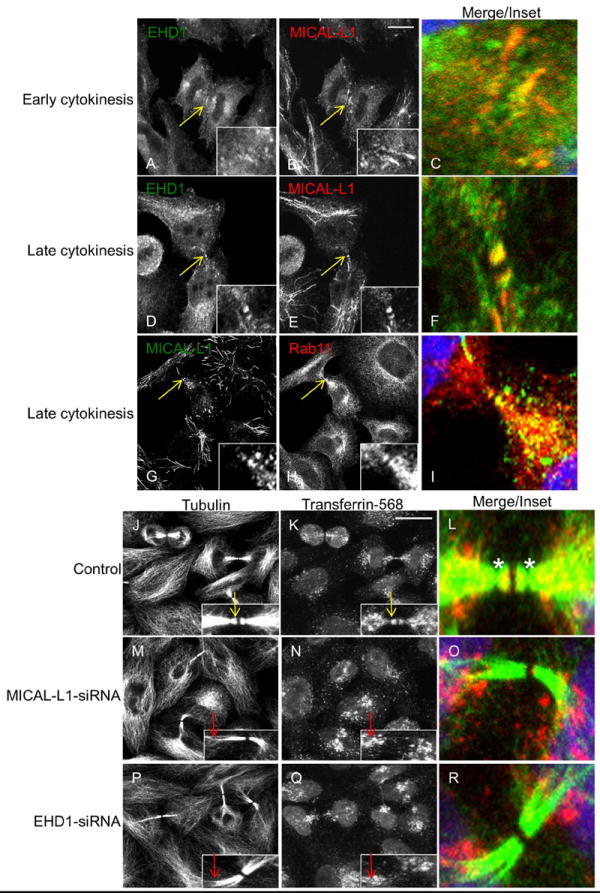
(A-F) HeLa cells were co-stained with antibodies to endogenous EHD1 (green) and MICAL-L1 (red). EHD1 and MICAL-L1 localized near the ingressing furrow during early cytokinesis (A-C, yellow arrows; see inset) and on the ICB during late cytokinesis (D-F, yellow arrows; see inset). (G-I) Hela cells co-stained with MICAL-L1 (green) and Rab11 (red) antibodies during late cytokinesis. (J-R) HeLa cells were pulse-labeled with transferrin-568 for 1 h to reach equilibrium, fixed and then stained with α-tubulin antibody to mark the ICB (green) and with DAPI to mark the nucleus (blue). In control-siRNA treated cells (J-L), transferrin-568 localized to the tubulin-rich ICB (yellow arrows in insets, asterisk in L marks secondary abscission). In contrast, depletion of MICAL-L1 (M-O) or EHD1 (P-R) caused transferrin-568 retention at the base of the ICB (red arrows in insets). Maximum projections of 1 μm optical sections are shown. Scale bar= 10 μm.
The delivery of recycling endosomes, containing marker proteins such as transferrin (Tf), to the ICB appears to be essential for cytokinesis(9). Since MICAL-L1 and EHD1 are both required for efficient recycling of transferrin and several other cargo proteins during interphase(27), we assessed the localization of fluorescently labeled Tf during cytokinesis in MICAL-L1- and EHD1-depleted cells. In agreement with previous data, we found that Tf-568 localized to the ICB during late cytokinesis in control cells (Figure 3J-L; yellow arrows; see inset). These Tf-labeled endosomes localized distal to a narrowing of the ICB, known as the site of secondary ingression (Figure 3L; asterisks). A number of studies support the notion that recycling endosome delivery to the distal ICB is required for secondary ingression, and ultimately abscission(16, 17). Depletion of either MICAL-L1 or EHD1 caused Tf-endosomes to be retained at the base of the ICB (Figure 3M-R; red arrows; see insets). Typically, during late telophase, MICAL-L1- and EHD1-depleted cells were characterized by a thick ICB lacking secondary ingression. This supports a role for each protein in mediating transport of recycling endosomes to the ICB, a process that is required for secondary ingression and the completion of cytokinesis.
We also assessed the effect of MICAL-L1 and EHD1-depletion on the localization of the Golgi apparatus (Supplemental Figure 5A-C) and early endosomes (Supplemental Figure 5D-F) during cytokinesis. During late cytokinesis, in control cells and MICAL-L1-depleted cells the Golgi was observed in two discrete areas: 1) at the base of the bridge, and 2) behind the nucleus, presumably near the centrosomes (Supplemental Figure 5A and B, asterisks). However, EHD1-depletion resulted in altered Golgi localization during late cytokinesis, predominantly to the base of the bridge with no peri-centrosomal staining observed in most of cells (Supplemental Figure 5C; yellow arrows). While the functional significance of this altered localization is unclear, we speculate that Golgi localized near the centrosome is required for delivery of exocytic vesicles to the plasma membrane opposite of the ICB, thus impairing cell spreading of EHD1-depleted cells during cytokinesis (Figure 2D, asterisk). Lastly, early endosome localization was not grossly affected by either MICAL-L1- or EHD1-depletion (Supplemental Figure 5D-F), although EHD1-depleted cells did display a more compact early endosome staining pattern at the base of the ICB and the peri-centrosomal areas. Our findings support a model in which MICAL-L1 recruits EHD1 to the ICB, which then facilitates the release of recycling endosomes from the base of the ICB.
Recruitment of MICAL-L1 to ICB is independent of EHD1, Rab11 and Rab35
Although MICAL-L1 and EHD1 are required for the completion of cytokinesis and the transport of Tf-containing endosomes to the ICB (this study), Rab11/FIP3 (9) and Rab35(24) have been previously implicated in the regulation of cytokinesis. How MICAL-L1/EHD1, Rab11 and Rab35 coordinate membrane recycling is not well understood, although EHD1 does directly bind to FIP2(35), a Rab11 effector that is not required for cytokinesis. Rab35 binds to MICAL-L1 through the MICAL-L1 C-terminal coiled-coiled domain. Studies in HeLa cells as well as neuronal cells suggest that MICAL-L1 acts as a membrane hub for Rab35 and other GTP-binding proteins to coordinate recycling during interphase(26, 36, 37). To address the relationship between MICAL-L1, EHD1, Rab11 and Rab35 during cytokinesis, we tested if EHD1-, Rab11- or Rab35-depletion impaired the recruitment of MICAL-L1 to the distal ICB during late cytokinesis. Immunoblot analysis demonstrated the specificity and efficacy of each siRNA treatment (Figure 4A). Surprisingly, Rab11-depletion decreased the protein levels of MICAL-L1, although it did not affect the localization of MICAL-L1 to the ICB (see below; Fig. 4E) or interphase tubular membranes. In control cells, MICAL-L1 localized to the distal ICB (Figure 4B). MICAL-L1-depletion resulted in a loss of MICAL-L1 signal at the ICB, demonstrating the specificity of the MICAL-L1 antibody (Figure 4C). EHD1-, Rab11, or Rab35-depletion did not appear to affect the recruitment of MICAL-L1 onto the ICB (Figure 4D-F) although all knockdowns affected MICAL-L1 distribution, likely because the depletion of each affects the morphology of the ICB.
Figure 4. Recycling regulatory protein requirements for their recruitment to the ICB.
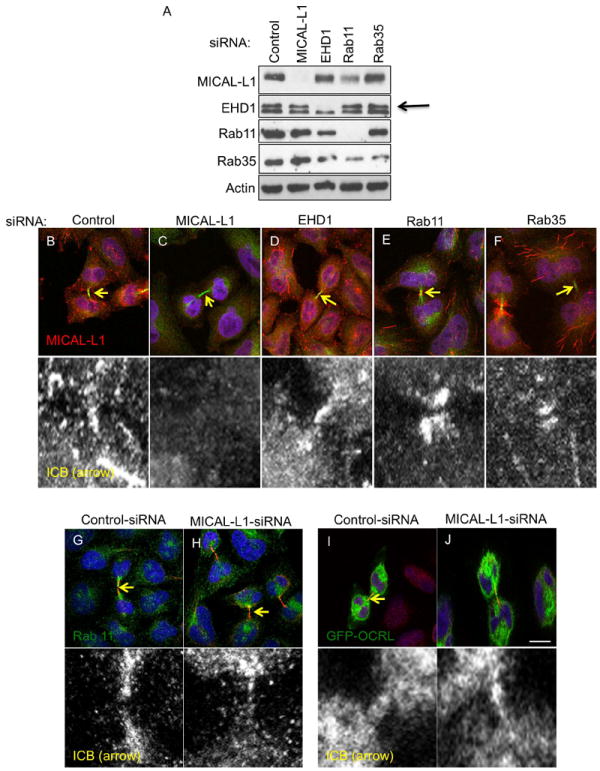
(A) Immunoblot analysis demonstrating efficacy and specificity of siRNA-knockdown of indicated recycling proteins. (B-F) HeLa cells were transfected with indicated siRNAs for 48 h, fixed and stained with MICAL-L1 (red), tubulin (green) antibodies, and DAPI (blue). (G and H) HeLa cells were transfected with control or MICAL-L1-siRNA for 48 h, fixed and stained with Rab11 (green), tubulin (red) antibodies and DAPI (blue). (I-J) HeLa cells were transfected with control or MICAL-L1-siRNA for 24 h and then transfected with the GFP-OCRL construct and incubated for another 24 h prior to fixation and immunostaining with tubulin antibody (red) and DAPI (blue). Yellow arrows mark the position of midbody on the ICB and the region of interest some in the insets. Scale bar=10 μm.
Given that MICAL-L1 localizes to the ICB independently of EHD1, Rab11 and Rab35, we next asked if MICAL-L1 is required for Rab11 or Rab35 recruitment onto the ICB. In control cells, endogenous Rab11 was found throughout the ICB but concentrated at the distal end of the ICB near the midbody (Figure 4G), in agreement with previous findings(9). However, in MICAL-L1-depleted cells, Rab11 recruitment onto the distal parts of the ICB was impaired (Figure 4H). MICAL-L1-depletion also impaired the recruitment of over-expressed FIP3-GFP to the distal ICB (data not shown). Due to technical difficulties, we could not assess the localization of endogenous Rab35, thus, we used GFP-OCRL as a marker for Rab35 endosomes, as its localization to the ICB is dependent on Rab35(25). Under these conditions, MICAL-L1-depletion did not drastically impair the recruitment of GFP-OCRL to the distal ICB(Figure 4I and J).
Overall, we stress that our evaluation of the relationship between each recycling protein must be interpreted with caution. Without a mechanistic link between MICAL-L1 and Rab11, we cannot conclude if MICAL-L1-depletion directly impairs Rab11 localization to the ICB (see Discussion). Moreover, our analysis was done on fixed cells. While we staged cells in cytokinesis based on previously reported morphological traits(13), the use of live-cell imaging will be crucial to assess the temporal recruitment of each recycling protein to the ICB.
EHD1- but not MICAL-L1-, Rab11- or Rab35- depletion affects central spindle formation
Our live cell imaging analysis demonstrated that EHD1-depletion frequently resulted in asymmetric cell divisions (Figure 2D; asterisk), suggestive of a defect during the furrowing process. Given that the centralspindlin complex, comprised of Cyk-4/MgcRacGAP and MKLP1, is key to regulating the site of RhoA-mediated contraction during the early parts of cytokinesis (14), we performed immunofluorescence analysis of MKLP1 localization during early cytokinesis in EHD1-depleted cells.
In control cells, punctate MKLP1 staining was observed in a narrow zone at the center of the central spindle (Figure 5A). While EHD1-depletion did not impair the localization of MKLP1 to central spindle microtubules, it did result in a drastic widening of MKLP1 staining (Figure 5B). Importantly, this phenotype is specific to EHD1-depletion, as MICAL-L1-, Rab11- or Rab35-depletion did not significantly impair MKLP1 localization (Figure 5C-E), which is in agreement with previous studies showing that Rab11/FIP3 and Rab35 are required for the later stages of cytokinesis rather than early cytokinesis(9, 13, 24, 25). We found similar localization patterns for other central spindle components such as Aurora B and Polo-Like-Kinase 1 (Supplemental Figure 6A-J). Using Structured Illumination Microscopy (SIM), we observed that MKLP1 puncta are found at the terminal ends of microtubules at the center of the central spindle (Figure 5F; single 0.110μm z-section shown; we refer reader to Supplemental Video 5 for entire z-stack). Also note the highly-ordered and bundled morphology of the central spindle microtubules. In EHD1-depleted cells, the central spindle microtubules are unorganized and do not appear to have ends that terminate at the center of the central spindle (Figure 5G; see also Supplemental Video 6). As a consequence, MKLP1 staining is elongated. This phenotype is reminiscent of KIF4-depletion (38). Further work is required to discern if EHD1 is directly involved in microtubule plus end dynamics or if EHD1 regulates the localization of KIF4 or other microtubule regulators to the central spindle. We speculate that the abnormal furrowing phenotype we observed upon EHD1-depletion is an indirect result of the elongated centralspindlin localization, which would likely broaden the RhoA activation zone. In support of this hypothesis, we found that bright cortical actin staining is concentrated at the ingressing furrow in control cells (Supplemental Figure K; white arrow), however, EHD1-depleted cells displayed uniform cortical actin along the plasma membrane (Supplemental Figure 6L).
Figure 5. Depletion of EHD1 but not MICAL-L1, Rab11 or Rab35 affects central spindle formation.
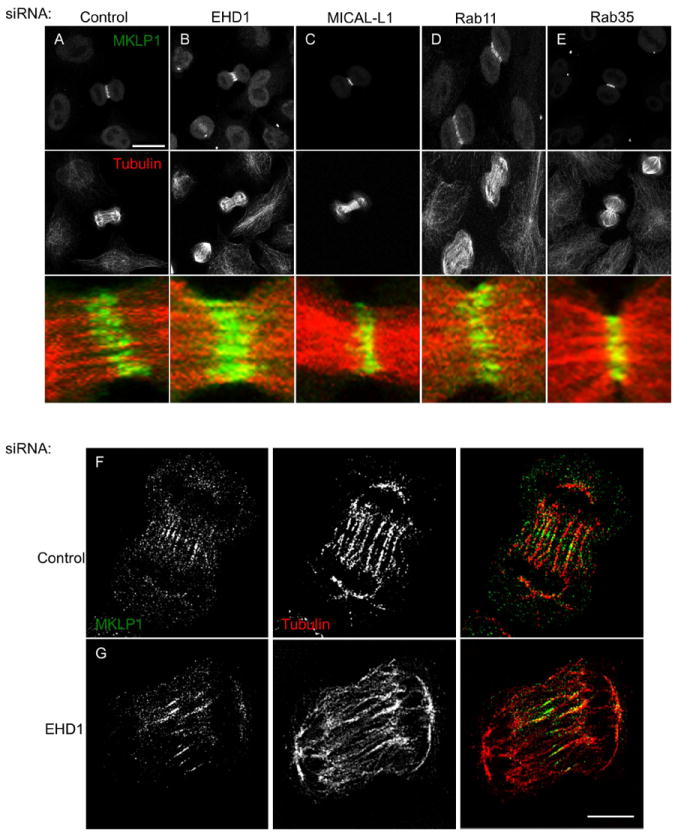
(A-E) HeLa cells were transfected with the indicated siRNA for 48 h and then fixed and stained with MKLP1 (green), tubulin (red) antibodies and DAPI (blue). Maximum projections of 0.5 μm optical sections are shown. (F and G) HeLa cells were transfected with control or EHD1-siRNA for 48 h and then fixed and stained with MKLP1 (green) and tubulin (red) antibodies and imaged by Structured Illumination Microscopy. Scale bar=10 μm.
EHD1 and Rab35 regulate mitotic spindle orientation while MICAL-L1 controls spindle length
While there is considerable evidence supporting a role for recycling endosomes in the completion of cytokinesis, new data also implicates Rab11 recycling endosomes in controlling pre-cytokinetic events such as microtubule dynamics, chromosome alignment and spindle orientation during metaphase(30). Accordingly, we used a well-characterized assay(39) to test if MICAL-L1, EHD1 and Rab35 are also required for spindle orientation (Figure 6). Control-, MICAL-L1-, EHD1- and Rab35-depleted cells were plated on fibronectin-coated coverslips for 48 h and then arrested in metaphase using MG132 for 2 h. Cells were then fixed and the centrosomes were labeled using anti-pericentrin antibodies. Confocal microscopy was used to acquire 0.5 μm optical sections through metaphase cells (Figure 6A; XZ sections), and LSM5 Pascal imaging software was used to measure the angle between centrosomes (Figure 6B and C; α-angle) and the distance between centrosomes (Figure 6D; pole-pole distance). Note that only bipolar cells with congressed chromosomes were analyzed and that the presence of other centrosomes in the XZ images are from surrounding interphase cells. While the majority of control cells and MICAL-L1-depleted cells had spindle angles of less than 5 degrees (Figure 6B and C), EHD1- and Rab35 depleted cells displayed a drastic increase in the number of cells with spindle angles greater than 10 degrees. The significant increase in spindle angle for EHD1-and Rab35-depleted cells suggests a general role for recycling proteins in regulating spindle orientation. Unexpectedly, whereas MICAL-L1-depletion did not affect spindle orientation, it did significantly increase the length of the mitotic spindle (Figure 6D). EHD1- and Rab35-depletion led to a small, but significant decrease in spindle length.
Figure 6. Role of recycling proteins in regulating mitotic spindle orientation and length.
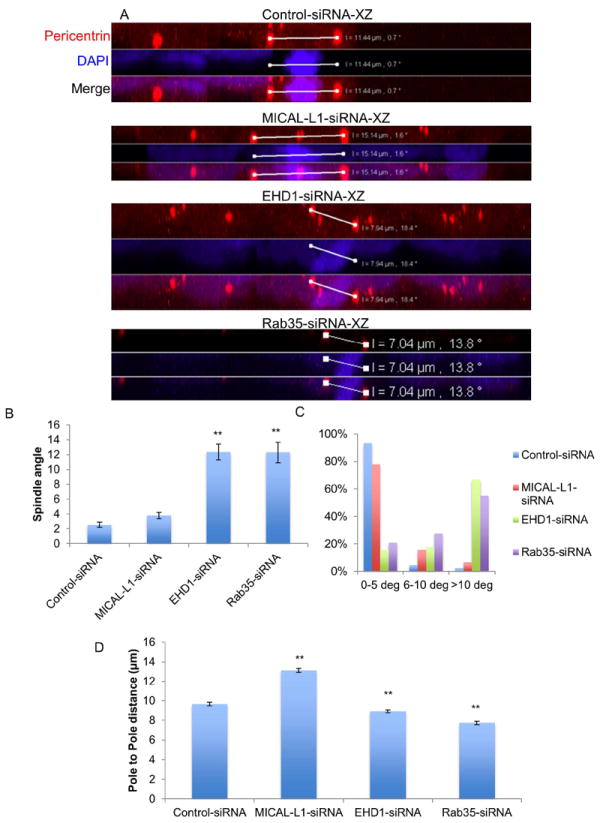
(A-D) HeLa cells were plated onto fibronectin-coated coverslips and transfected with control-, MICAL-L1-, EHD1-, or Rab35-siRNA for 48 h. Cells were treated with the proteasomal inhibitor MG132 (10 μM) for 2 h to prevent metaphase-anaphase transition and then fixed and stained with pericentrin (red) to mark centrosomes and DAPI (blue). 0.5 μm optical sections were acquired through the entire depth of metaphase cells. LSM5 Pascal software was used to measure the α-spindle angle (B and C) and the distance between the two pericentrin labeled centrosomes (D, pole-pole distance). Results are from three independent experiments, 45 cells/experiment. One-way ANOVA **p<0.05.
Effect of MICAL-L1-, EHD1-, and Rab35-depletion on kinetochores, inter-kinetochore tension and kinetochore fibers
Doxsey and colleagues recently demonstrated that Rab11 is required for chromosome alignment(30). In unsynchronized cells, control cells with congressed chromosomes (as assessed by DAPI staining-not shown) had an inter-kinetochore distance of 1.02 +/- 0.013 μm (Figure 7A; quantified in 7E). MICAL-L1-depleted metaphase cells with congressed chromosomes had significantly shorter inter-kinetochore distances compared to control cells 0.89 +/- 0.015 μm (Figure 7B). Interestingly, EHD1-depleted cells had significantly wider inter-kinetochore distances (1.25 +/- 0.017 μm) compared to control cells (Figure 7C) while Rab35-depletion had no significant effect (1.05 +/- 0.012 μm; Figure 7D). Lastly, endogenous MICAL-L1 localized to detergent-resistant microtubules (see Materials and Methods) during metaphase and anaphase in both HeLa cells (Figure7F and G; see insets) and BJ human foreskin fibroblasts (Figure 7H and I; see insets).
Figure 7. Role of recycling proteins in regulating inter-kinetochore tension.
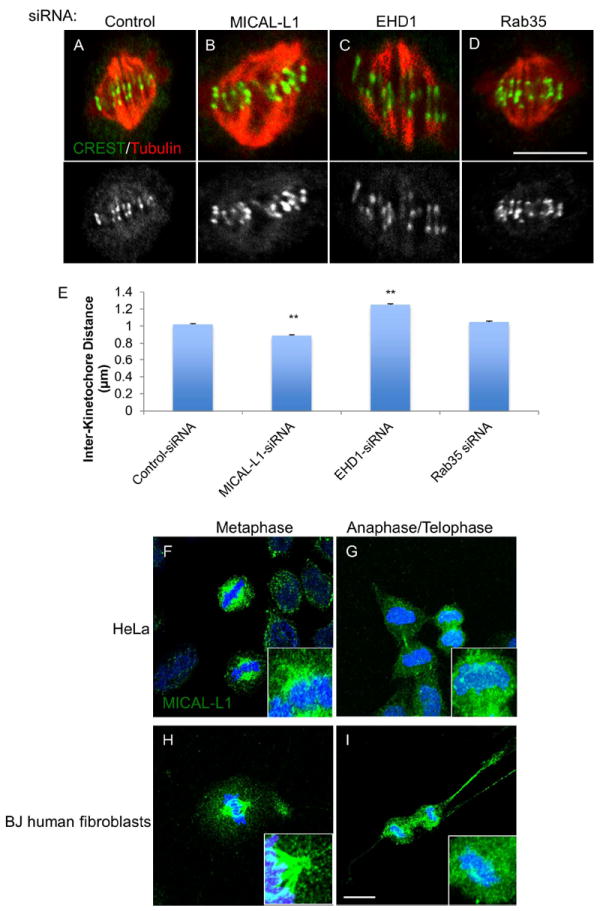
(A-D) Cells were transfected with indicated siRNAs for 48 h, fixed and stained with anti-centromere/kinetochore antisera (CREST, green) and tubulin (red) antibody. A single 0.2 μm optical section is shown for each condition. (E) Inter-kinetochore distance was measured in >100 kinetochores from 10 representative cells across three experiments. Asynchronous HeLa cells (F and G) or BJ human fibroblasts (H and I) were subjected to Triton X-100 extraction prior to fixation (see methods) and then stained with anti-MICAL-L1 antibody (green) and DAPI (blue). Maximum projections of 1 μm optical sections are shown. One-way ANOVA **p<0.01.
Inter-kinetochore distance is maintained by dynamic interactions between kinetochores and microtubules(40, 41). We hypothesized that the effects of MICAL-L1- and EHD1-depletion on inter-kinetochore distance might be a result of altered kinetochore fiber stability. We used a high calcium buffer to depolymerize all non-kinetochore microtubules(42). Figure 8A is a representative image of microtubules and kinetochores prior to calcium induced depolymerization in a cell treated with control-siRNA. Control, MICAL-L1-, EHD1-, or Rab35-depleted cells were then subjected to high calcium buffer for 10 min. prior to fixation and processing for immunofluorescence (Figure 8B-E). Single 0.2 μm optical sections were acquired by confocal microscopy and kinetochore fiber length was measured by manually drawing a line from the CREST labeled kinetochore to the end of the tubulin fluorescence at the spindle pole (Figure 8F). Compared to control cells, which had an average kinetochore fiber length of 3.05 +/- 0.06 μm, MICAL-L1-depleted cells had significantly longer kinetochore fibers (4.3 +/- 0.08 μm) while EHD1-depleted cells had significantly shorter kinetochore fibers (2.73 +/- 0.05 μm). Rab35-depleted cells did not display a significant difference in kinetochore fiber length (3.25 +/- 0.05 μm). We also found that MICAL-L1-depletion significantly increased cold-stable microtubules in the metaphase mitotic spindle (Supplemental Figure 7A-H). Using SIM, we found that MICAL-L1-depletion leads to abnormal chromosome attachments. In control cells treated with calcium buffer as described above, punctate kinetochore structures localized to the ends of kinetochore fibers in an ‘end-on’ orientation (Figure 8G; white arrow). However, in MICAL-L1-depleted cells, kinetochores were often connected to microtubules from both spindle poles (Figure 8H; merotelic attachments, yellow arrows). Lastly, we also observed lateral kinetochore attachments where microtubules failed to terminate at kinetochores (Figure 8H; white arrowheads).
Figure 8. MICAL-L1-depletion enhances kinetochore fiber length and leads to abnormal kinetochore-microtubule interactions while EHD1-depletion decreases kinetochore fiber length.
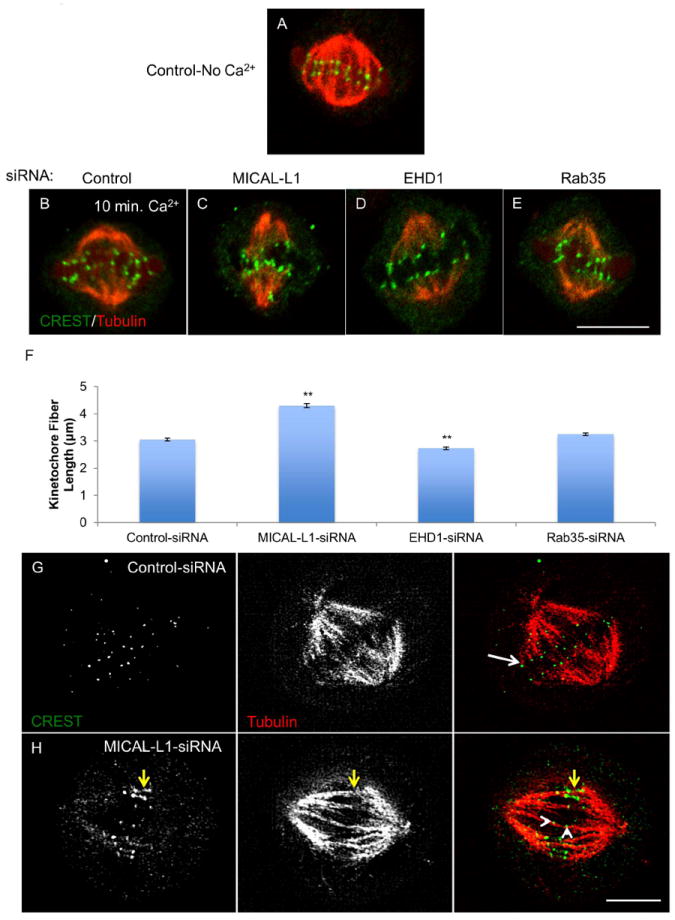
(A) HeLa cell fixed and stained with CREST anti-sera (green) and tubulin antibody (red) demonstrating presence of kinetochore and interpolar microtubules. (B-E) HeLa cells were transfected with indicated siRNAs for 48 h, incubated in calcium containing buffer (Ca2+) for 10 min, fixed and stained with CREST (green) and tubulin (red; a single 0.2 μm optical section is shown for each condition. (F) Quantitation of kinetochore fiber length. >50 kinetochore fibers were measured from 10 representative cells across three experiments. (G and H) Control and MICAL-L1-depleted cells were fixed and stained as above and analyzed by SIM. White arrow denotes ‘end-on’ attachment of microtubule to kinetochore. Yellow arrows in MICAL-L1-depleted cell shows abnormal kinetochore stretching while white arrowheads show ‘lateral’ microtubule-kinetochore attachment. One-way ANOVA**p<0.01.
Accordingly, our data provide support for the notion that MICAL-L1 is required for kinetochore microtubule dynamics, and that the depletion of MICAL-L1 can lead to a variety of mitotic aberrations, including longer and more stable kinetochore fibers, improper microtubule attachments to kinetochores and lagging chromosome strands (data not shown).
Discussion
The timing and precision of mitotic progression is paramount to insure genetic stability. Endocytic pathways are well suited to provide stringent spatiotemporal control over localization of mitotic regulators during each phase of mitosis. A variety of endocytic regulatory proteins control the steps of mitosis, and proteins such as Rab11/FIP3, Rab35, Arf6, ARH and dynamin are crucial for the successful completion of cytokinesis(9, 11, 23, 24, 43, 44). Rab11/FIP3, Rab35, and Arf6 are all regulators of membrane recycling, and by interacting with and localizing tethering proteins such as the exocyst, SNARES, or the Endosome Sorting Required for Transport (ESCRT) complex, influence membrane and cytoskeletal dynamics at the ICB.
We now provide evidence that the recycling proteins MICAL-L1 and EHD1 also regulate cytokinesis, as the depletion of either protein results in increased bi-nucleation and multi-nucleation. Given that MICAL-L1 and EHD1 are required for transport of recycling endosomes to the ICB, we speculate that the MICAL-L1/EHD1 pathway is responsible for the delivery of membranes and cytokinetic regulators to the ICB. We show that MICAL-L1 localizes to the ICB independent of EHD1, Rab11 or Rab35. MICAL-L1-depletion impairs recruitment of Rab11 onto the ICB, while having no effect on the Rab35-dependent pathway, which is in agreement with studies in interphase cells showing that MICAL-L1 is downstream of Rab35(26). Further studies will be required to elucidate the molecular mechanisms of how MICAL-L1 affects Rab11 recruitment. We stress that without direct evidence for an interaction between MICAL-L1 and Rab11, the impaired recruitment of Rab11 to the ICB may be secondary to defective microtubule depolymerization on the ICB or the presence of lagging chromosomes. Indeed, siRNA-depletion of the microtubule severing enzyme Spastin increases ICB microtubule content and impairs FIP3 recruitment onto the ICB(16). As shown in Figures 3 and 5, MICAL-L1 (and EHD1)-depleted cells often had enlarged, abnormally formed ICB. Furthermore, abscission (and possibly the recruitment of endosomes to the distal ICB) is inhibited in cells with lagging chromosomes in an Aurora B-dependent manner (45, 46). Given that we frequently observed lagging chromosomes in MICAL-L1-depleted cells, it is tempting to speculate that Rab11 transport is inhibited by an Aurora B-dependent mechanism in the presence of lagging chromosomes.
Our live cell imaging and morphological analysis of microtubules and the centralspindlin complex suggests that EHD1 is required for proper central spindle formation (Figure 5). Currently, it is thought that central spindle microtubules emanate from non-centrosomal locations distal to the separating chromosomes(47). Recent work by Doxsey and colleagues suggests that recycling endosomes may serve as sites for central spindle microtubule nucleation(30). If EHD1-depletion affects recycling endosome localization during early cytokinesis, this may explain the abnormal morphology of the central spindle microtubules. Alternatively, we predict that EHD1 may regulate the localization of the kinesin protein KIF4. KIF4-depletion results in a similar early cytokinetic phenotype characterized by unorganized and improperly bundled central spindle microtubules and elongation of the central spindle components MKLP1, Aurora B and PLK1(38).
In addition to the new roles for MICAL-L1 and EHD1 in cytokinesis, we found that EHD1 and MICAL-L1 are involved in pre-cytokinetic events. Why EHD1- and Rab35-depletion affects spindle orientation, whereas MICAL-L1-depletion elongates the mitotic spindle, presumably through stabilizing kinetochore microtubules (Figure 8), requires further study. At this point, we speculate that MICAL-L1 may perform ‘moonlighting’ or non-endocytic functions early in mitosis(4). Indeed, MICAL-L1 contains several modular domains such as a calponin homology (CH domain), a LIM domain as well as a coiled-coiled domain. It is noteworthy that these domains are often found on microtubule regulators (i.e., Nuf2, Hec1, NDC80, EB1) that affect the stability and/or dynamics of kinetochore microtubules(48). Given the frequency that we observed micronuclei and lagging chromosomes in MICAL-L1-depleted cells, we speculate that MICAL-L1 affects microtubule de-polymerase activity at improperly attached kinetochores. Indeed, microtubule de-polymerases such as MCAK are recruited to merotelic kinetochores, thereby releasing the improperly attached chromosomes and allowing for correct or amphitelic chromosome attachment(49, 50). MCAK is also part of the machinery required to fix lateral kinetochore attachments(51), which we frequently saw in MICAL-L1-depleted cells. Without MICAL-L1, kinetochore fibers become hyper-stable and the cells are unable to resolve abnormal, merotelic attachments, which occur frequently early in mitosis but are often corrected prior to the onset of anaphase(52).
Although the precise mechanisms by which MICAL-L1 and EHD1 regulate mitosis remains unclear, as do the exact functions of many endocytic proteins that affect mitosis, we provide evidence that both proteins affect cytokinesis and pre-cytokinetic events. We suggest a model whereby MICAL-L1 performs ‘moonlighting’ functions during early mitosis. At the transition from early-to-late cytokinesis, MICAL-L1 resumes its more typical function as an endocytic regulatory protein that controls the delivery of recycling vesicles to the ICB. MICAL-L1, which is likely directly attached to microtubules, would then recruit EHD1 to the base of the ICB. EHD1 would then promote the vesiculation and release of recycling endosomes from the base of the ICB. We propose that MICAL-L1 and EHD1 then transport these endosomes to the distal ICB, where they control the fusion of recycling endosomes to the plasma membrane. This is supported by recent work demonstrating that EHD1 interacts with Snapin(53), a SNARE protein with a well-described role in cytokinesis(10). Furthermore, EHD1 interacts with the exocyst (unpublished observations), which is also required for vesicle fusion at the ICB. Overall, our findings support roles for both EHD1 and MICAL-L1 at distinct steps of mitosis, widening their roles as cellular regulatory proteins.
Materials and Methods
Reagents and Antibodies
Fibronectin, monastrol and MG132 were purchased from Sigma. Transferrin conjugated to AlexaFluor 568, Phalloidin-568, DAPI, as well as all secondary antibodies used for immunofluorescence were purchased from Molecular Probes. The following primary antibodies were used: EHD1(54), MICAL-L1 (Novus Biologicals), affinity-purified rabbit polyclonal peptide antibody directed against the C-terminus of EHD2 (VERGPDEAMEDGEEGSDDEA) (AnaSpec), α-tubulin (Molecular Probes), anti-human anticentromere (ACA; Antibodies Inc.), pericentrin, giantin, MKLP1 and PLK1 (Abcam), Pan-actin (Millipore), Rab11 (US Biologicals), Rab35 (Protein Tech), Aurora B (Abnova; generous gift from Dr. Jixin Dong) and EEA1 (Cell Signaling Technologies).
Cell Culture, siRNA Transfection and Rescue
Cervical cancer cell line HeLa cells (ATCC-CCL2) and Normal human foreskin fibroblasts (BJ; ATCC-2522) were grown in DMEM (high glucose) containing 10% fetal bovine serum, penicillin/streptomycin (Invitrogen) and 2 mM glutamine. All siRNA and plasmid transfections were done in the absence of antibiotics. Pooled and individual oligonucleotides targeting human MICAL-L1, EHD2, Rab11a, Rab35, custom EHD1 siRNA(27) and non-targeting control siRNA were obtained from Dharmacon. HeLa cells were transfected with 50 nM of SmartPool oligonucleotides (MICAL-L1, EHD2, Rab11a, Rab35) or 100 nM of individual oligonucleotides (EHD1) using Lipofectamine RNAiMAX (Invitrogen). Efficiency of protein knockdown was measured 48-72 h post-transfection by immunoblot or immunofluorescence. For EHD1 rescue experiments, HeLa cells at ~60% confluency were transfected with siRNA-resistant-GFP-EHD1 construct(29) using Lipofectamine 2000. After 2.5 h, DNA complexes were removed and fresh antibiotic-free media was given to cells. Cells were then transfected with EHD1 siRNA as described above for 48 h and processed for immunofluorescence or immunoblot as described below. Control and MICAL-L1-depleted cells were transfected with FIP3-GFP or OCRL-GFP (generous gifts from Rytis Prekeris and Claudio Aguilar, respectively) 24 h after siRNA transfection using Lipofectamine 2000.
Immunoblotting
Cell lysates were prepared by washing cells two times in ice-cold PBS. Cells were then scraped off the plate with a rubber policeman into ice-cold RIPA buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA, 1% Triton, 0.5% sodium deoxycholate, 0.1% SDS, 1.8 mg/ml iodoacetamide, 1 mM orthovanadate, 2.5 mM sodium pyrophosphate, 1 mM glycerophosphate, 1 μg/ml leupeptin, 5 μg/ml aprotinin, 1 mM PMSF). Lysates were then clarified by centrifugation at 13,000 RPM at 4°C. Protein levels were quantified using the BCA assay (Biorad) and 20-30 μg protein lysate/sample was separated by 8% SDS-PAGE. Proteins were transferred onto nitrocellulose membranes. Membranes were blocked for 1 h at room temperature in TBST (TBS +0.1% Tween) plus 5% dry milk (TBST-B) and then incubated overnight in primary antibody in TBST-B at 4 degrees Celsius. Membranes were washed with TBST and then incubated with HRP-conjugated goat anti-mouse (Jackson Research Laboratories) or donkey anti-rabbit (GE healthcare) secondary antibody for 1 h at room temperature. After three washes in TBST, membranes were incubated in SuperSignal West Chemiluminescent Substrate (Pierce) and developed using standard film-based techniques.
Immunofluorescence
Cells were treated as indicated in the text and then fixed in 3.7% paraformaldehyde in PHEM (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 7.0) for 15 minutes at room temperature. For MICAL-L1 kinetochore fiber staining, cells were pre-extracted in PHEM buffer + 0.1% Triton-X (warmed to 37 degrees Celsius) for 1 minute. For calcium treatment, cells were incubated in calcium buffer containing 100 mM PIPES, 1 mM MgCl2, 1 mM CaCl2, pH 7.0 for 10 minutes. Cells were then fixed as described above. Cells were rinsed three times in PHEM and permeabilized in 0.5% Triton-X/PHEM for 15 minutes. The cells were incubated with primary antibody in PHEM/1% BSA/0.02% Triton-X for 1 h at room temperature. After three washes in PHEM, cells were incubated with appropriate fluorochrome-conjugated secondary antibodies (Molecular Probes) plus DAPI for 1 h at room temperature. Cells were washed 3X in PHEM and mounted in Fluoromount G.
Single plane or z-stack (slice size indicated in text) confocal images were collected using Zeiss LSM5 Pascal laser confocal microscope with a Plan-Apochromat 63X/1.4 oil objective or Plan-Apochromat 100X/1.4 oil objective (for kinetochore images). For quantification, images were imported into ImageJ or LSM Pascal Image Examiner and quantified as described in text. Images presented in figures were imported into Adobe Photoshop CS, where they were re-sized and formatted to 300 dpi resolution with minimal image manipulation (whole-image adjustment of brightness was done using ‘levels’ function).
Structured Illumination Microscopy (SIM)
Cells were fixed and immunostained as described for confocal microscopy, and mounted with Vectashield H-1000 mounting solution (Vector Laboratories). SIM images were collected with a Zeiss ELYRA PS.1 illumination system (Carl Zeiss MicroImaging) using a 63X oil objective lens with a numerical aperture of 1.46 at room temperature. Three orientation angles of the excitation grid were acquired for each Z plane, with Z spacing of 110 nm between planes. SIM processing was performed with the SIM module of the Zen BLACK software (Carl Zeiss MicroImaging).
Live cell imaging
Cells growing in 6-well plastic dishes were imaged in an environmental chamber (19% oxygen, 5% carbon dioxide, 37 degrees Celsius) using an Olympus 1×81 inverted microscope with a 20X objective (NA 0.45). Images were gathered using a Hamamatsu Orca-ER camera every 10 min. for 36 hours. Slidebook 5.5.2 was used for image acquisition and processing.
Statistics
Data from ImageJ or LSM 5 Pascal Image Examiner were imported into Microsoft Excel. Mean and standard error of the mean were calculated from data obtained from three independent experiments. Statistical significance was calculated by One-way ANOVA and Tukey test (when comparing more than two samples) or Student’s T-test using Vassar stats program (www.vassarstats.net).
Supplementary Material
Supplemental Figure 1: Raw data and source images for Figure 1 I-K. (A) Table showing percentages illustrated in Figure 1I. Data is a composite of three independent experiments. One-way ANOVA tests comparing the number of control (M1), MICAL-L1-depleted (M2) and EHD1-depleted cells (M3) containing each nuclear phenotype. (B-G) Examples of images exported from ImageJ that were used to quantify nuclear area/cell. For cells with multiple nuclei, as is the case for F and G (arrow), area from each nucleus was added to give total area/cell. Scale bar= 10 μm.
Supplemental Figure 2: Specificity of MICAL-L1- and EHD1-siRNA knockdown phenotypes. (A-E) HeLa cells were transfected with control-siRNA or MICAL-L1-siRNA individual oligonucleotides for 72 h. Immunoblotting demonstrates the efficacy of MICAL-L1-depletion by oligo #1 and oligo #2. Immunofluorescence analysis of control- (B), MICAL-L1-siRNA oligo #1- (C) or MICAL-L1-siRNA oligo#2- (D) transfected cells stained with α-tubulin (red) and DAPI (blue). (E) Nuclear phenotypes were quantified from three independent experiments (150 cells/experiment). Data is presented as percentage of abnormal nuclei (micro-nucleated and bi-nucleated). (F-K) HeLa cells were either mock transfected (Lipofectamine 2000 alone in F, I, and G, J) or transfected with siRNA-resistant-GFP-EHD1 (siResis-GFP-EHD1) for 2 h (H and K). Cells were then washed and transfected with control-siRNA (F and I) or EHD1-siRNA (G, J and H, K) for 48 h. Cells were fixed and stained with α-tubulin (red) and DAPI (blue). Yellow arrows mark multi-nucleated cells in G and H. (L) Quantification of multi-nucleation in control-siRNA, EHD1-siRNA and EHD1-siRNA + siRNA-Resis-GFP-EHD1 cells. Data is presented from three independent experiments, 200 cells/experiment. (M) Immunoblot analysis demonstrating depletion of endogenous EHD1 and exogenous expression of siResis-GFP-EHD1 in EHD1-depleted cells (black arrows). One-way ANOVA *p<0.05, **p<0.01. Scale bar= 10 μm.
Supplemental Figure 3: Depletion of the EHD family member, EHD2, has no gross effect on cell cycle. (A) HeLa cells were transfected with control- or EHD2-siRNA for 48 h and subjected to immunoblot analysis. (B-D) Cells treated as in A were processed for immunofluorescence (B and C) and multi-nucleated cells were quantified (D). Data is from three independent experiments (150 cells/experiment). Scale bar= 10 μm.
Supplemental Figure 4: MICAL-L1 is required for EHD1 localization to the ICB. (A-C) HeLa cells were fixed and stained with anti-EHD2 antibody (green), anti-α-tubulin (red) and DAPI (blue). (D-I) HeLa cells transfected with control-siRNA (D-F) or MICAL-L1-siRNA (G-I) were stained with anti-EHD1 antibody (green) and anti-α-tubulin (red). The area in the dashed yellow boxes in D and E is enlarged as an inset in panel F, while the dashed yellow boxes in G and H are enlarged as an inset in panel I. A yellow arrow shows EHD1 localization to the ICB in control cells (F). (J and K) Line-scan analysis of the ICB from control (J) and MICAL-L1-depleted cells (K). The black arrow denotes the midbody, which is devoid of EHD1 and tubulin staining. Scale bar= 10 μm.
Supplemental Figure 5: The effect of MICAL-L1- or EHD1-depletion on Golgi and early endosome positioning during cytokinesis. (A-F) HeLa cells transfected with control-siRNA (A and D), MICAL-L1-siRNA (B and E) or EHD1-siRNA (C and F) for 48 h were stained either with anti-giantin antibody to mark Golgi (green; A-C) or early endosome antigen 1 (EEA1) antibody to label early endosomes (green; D-F), α-tubulin (red) and DAPI (blue). The asterisks in A mark dual localization of Golgi in control cells to the base of the ICB and the pericentrosomal area. Yellow arrows in C demonstrate localization of the Golgi to the base of ICB in EHD1-depleted cells. Scale bar= 10 μm.
Supplemental Figure 6: Recruitment of Aurora B or PLK1 is not affected by depletion of MICAL-L1, EHD1, Rab11 or Rab35. Cells were transfected with the indicated siRNAs, fixed, and stained with Aurora B antibody (A-E) or PLK1 antibody (F-J). Control (K) and EHD1-depleted (L) cells stained with tubulin (green), phalloidin-568 (red) and DAPI (blue). Scale bar= 10 μm.
Supplemental Figure 7: MICAL-L1-depletion enhances kinetochore fiber cold-stability. (A-F) HeLa cells were transfected with either control-siRNA (A-C) or MICAL-L1-siRNA (D-F) for 48 h and arrested in metaphase using MG132 (10 μM, 2 h). Cells were then placed on ice in ice-cold DMEM for 10 min. (A and D), 15 min. (B and E) or 30 min. (C and F) and then fixed and stained with α-tubulin (green), pericentrin (red) and DAPI (blue). Maximum projections of 1 μm optical sections are shown. (G) Tubulin fluorescence of kinetochore fibers after 30 min. of cold-treatment was quantified using the ImageJ ‘measure’ function. Results are shown from three independent experiments (15 cells/experiment). (H) Pole-pole distance was measured as described in Figure 4 after cold treatment (three independent experiments, n=45 cells/experiment). Student’s t-test **p<0.01. Scale bar= 10 μm.
Cells were transfected with control-siRNA for 24 h and then imaged every 10 min. for 36 h as described in the Materials and Methods. Cluster of cells located in the center of the video show that control cells complete mitosis in less than 3h. Video is a wide-field of cells shown in Figure 2A.
Cells were transfected with MICAL-L1-siRNA for 24 h and then imaged every 10 min. for 36 h as described in the Materials and Methods. Cluster of cells located in bottom left of the video demonstrate the prolonged time spent in cytokinesis (presence of intercellular bridge) due to MICAL-L1-depletion. Video is a wide-field of cells shown in Figure 2B.
Cells were treated and imaged as described in Supplemental Video 2. Cluster of cells in the bottom right of the video show two daughter cells from distinct mitoses fusing to form one bi-nucleated cell. Video is a wide-field of cells shown in Figure 2C.
Cells were transfected with EHD1-siRNA for 24 h and then imaged every 10 min. for 36 h as described in Materials and Methods. Note the delay of cell spreading during early telophase compared to control cells shown in Supplemental Video 1. Specifically, cells located in bottom right of the video show impaired cell spreading which then results in eventual cytokinesis failure leading to bi-nucleated cells. Video is a wide-field of cells shown in Figure 2D.
Z-sections were acquired at 0.110 μm increments, and displayed at 10 frames/sec.
Z-sections were acquired at 0.110 μm increments, and displayed at 10 frames/sec.
Z-sections were acquired at 0.110 μm increments, and displayed at 10 frames/sec.
Z-sections were acquired at 0.110 μm increments, and displayed at 10 frames/sec.
Acknowledgments
The authors are indebted to Dr. Keith Johnson, Tom Dao and the Nebraska Center for Cellular Signaling for their expertise with live cell imaging (NCCS supported by COBRE grants P30GM106397 and P20GM103489), to Dr. Rytis Prekeris and Dr. Claudio Aguilar for the FIP3-GFP and OCRL-GFP constructs, respectively, to Dr. Jixin Dong for Aurora B antibody and to the UNMC Advanced Microscopy Facility for Structured Illumination Microscopy. The authors would also like to thank Sugandha Saxena for preparation of critical reagents and the rest of the Caplan laboratory for helpful discussions. JBR is supported by a UNMC graduate student fellowship.
Footnotes
Author Contributions
JBR carried out the majority of the experiments, prepared the figures and wrote the manuscript. DK also performed many of the experiments and prepared key reagents required for several figures in the manuscript. NN and SC jointly directed the study, assisted in figure preparation, manuscript preparation and editing.
Conflict of Interest
The authors have no conflicts of interest to declare.
References
- 1.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. The Journal of cell biology. 2008;180(4):665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Current biology : CB. 2010;20(6):R285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nature reviews Cancer. 2010;10(2):102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Royle SJ. Protein adaptation: mitotic functions for membrane trafficking proteins. Nature reviews Molecular cell biology. 2013;14(9):592–599. doi: 10.1038/nrm3641. [DOI] [PubMed] [Google Scholar]
- 5.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131(5):847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Montagnac G, Echard A, Chavrier P. Endocytic traffic in animal cell cytokinesis. Current opinion in cell biology. 2008;20(4):454–461. doi: 10.1016/j.ceb.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Schiel JA, Prekeris R. Making the final cut - mechanisms mediating the abscission step of cytokinesis. TheScientificWorldJournal. 2010;10:1424–1434. doi: 10.1100/tsw.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelissier A, Chauvin JP, Lecuit T. Trafficking through Rab11 endosomes is required for cellularization during Drosophila embryogenesis. Current biology : CB. 2003;13(21):1848–1857. doi: 10.1016/j.cub.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 9.Wilson GM, Fielding AB, Simon GC, Yu X, Andrews PD, Hames RS, Frey AM, Peden AA, Gould GW, Prekeris R. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Molecular biology of the cell. 2005;16(2):849–860. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123(1):75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 11.Fielding AB, Schonteich E, Matheson J, Wilson G, Yu X, Hickson GR, Srivastava S, Baldwin SA, Prekeris R, Gould GW. Rab11-FIP3 and FIP4 interact with Arf6 and the exocyst to control membrane traffic in cytokinesis. The EMBO journal. 2005;24(19):3389–3399. doi: 10.1038/sj.emboj.7600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neto H, Kaupisch A, Collins LL, Gould GW. Syntaxin 16 is a master recruitment factor for cytokinesis. Molecular biology of the cell. 2013;24(23):3663–3674. doi: 10.1091/mbc.E13-06-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon GC, Schonteich E, Wu CC, Piekny A, Ekiert D, Yu X, Gould GW, Glotzer M, Prekeris R. Sequential Cyk-4 binding to ECT2 and FIP3 regulates cleavage furrow ingression and abscission during cytokinesis. The EMBO journal. 2008;27(13):1791–1803. doi: 10.1038/emboj.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White EA, Glotzer M. Centralspindlin: at the heart of cytokinesis. Cytoskeleton (Hoboken) 2012;69(11):882–892. doi: 10.1002/cm.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao WM, Fang G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(37):13158–13163. doi: 10.1073/pnas.0504145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiel JA, Park K, Morphew MK, Reid E, Hoenger A, Prekeris R. Endocytic membrane fusion and buckling-induced microtubule severing mediate cell abscission. Journal of cell science. 2011;124(Pt 9):1411–1424. doi: 10.1242/jcs.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiel JA, Simon GC, Zaharris C, Weisz J, Castle D, Wu CC, Prekeris R. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nature cell biology. 2012;14(10):1068–1078. doi: 10.1038/ncb2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiel JA, Prekeris R. ESCRT or endosomes?: Tales of the separation of two daughter cells. Communicative & integrative biology. 2011;4(5):606–608. doi: 10.4161/cib.4.5.16789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458(7235):172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316(5833):1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 21.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(12):4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. The Journal of cell biology. 2008;180(2):389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chesneau L, Dambournet D, Machicoane M, Kouranti I, Fukuda M, Goud B, Echard A. An ARF6/Rab35 GTPase cascade for endocytic recycling and successful cytokinesis. Current biology : CB. 2012;22(2):147–153. doi: 10.1016/j.cub.2011.11.058. [DOI] [PubMed] [Google Scholar]
- 24.Kouranti I, Sachse M, Arouche N, Goud B, Echard A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Current biology : CB. 2006;16(17):1719–1725. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 25.Dambournet D, Machicoane M, Chesneau L, Sachse M, Rocancourt M, El Marjou A, Formstecher E, Salomon R, Goud B, Echard A. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nature cell biology. 2011;13(8):981–988. doi: 10.1038/ncb2279. [DOI] [PubMed] [Google Scholar]
- 26.Rahajeng J, Giridharan SS, Cai B, Naslavsky N, Caplan S. MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic. 2012;13(1):82–93. doi: 10.1111/j.1600-0854.2011.01294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma M, Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Molecular biology of the cell. 2009;20(24):5181–5194. doi: 10.1091/mbc.E09-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giridharan SS, Cai B, Vitale N, Naslavsky N, Caplan S. Cooperation of MICAL-L1, syndapin2, and phosphatidic acid in tubular recycling endosome biogenesis. Molecular biology of the cell. 2013;24(11):1776–1790. S1771–1715. doi: 10.1091/mbc.E13-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai B, Giridharan SS, Zhang J, Saxena S, Bahl K, Schmidt JA, Sorgen PL, Guo W, Naslavsky N, Caplan S. Differential roles of C-terminal Eps15 homology domain proteins as vesiculators and tubulators of recycling endosomes. The Journal of biological chemistry. 2013;288(42):30172–30180. doi: 10.1074/jbc.M113.488627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hehnly H, Doxsey S. Rab11 endosomes contribute to mitotic spindle organization and orientation. Developmental cell. 2014;28(5):497–507. doi: 10.1016/j.devcel.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simone LC, Caplan S, Naslavsky N. Role of phosphatidylinositol 4,5-bisphosphate in regulating EHD2 plasma membrane localization. PloS one. 2013;8(9):e74519. doi: 10.1371/journal.pone.0074519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoeber M, Stoeck IK, Hanni C, Bleck CK, Balistreri G, Helenius A. Oligomers of the ATPase EHD2 confine caveolae to the plasma membrane through association with actin. The EMBO journal. 2012;31(10):2350–2364. doi: 10.1038/emboj.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moren B, Shah C, Howes MT, Schieber NL, McMahon HT, Parton RG, Daumke O, Lundmark R. EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Molecular biology of the cell. 2012;23(7):1316–1329. doi: 10.1091/mbc.E11-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen CG, Howard G, Nichols BJ. Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. Journal of cell science. 2011;124(Pt 16):2777–2785. doi: 10.1242/jcs.084319. [DOI] [PubMed] [Google Scholar]
- 35.Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Molecular biology of the cell. 2006;17(1):163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi H, Fukuda M. Rab35 establishes the EHD1-association site by coordinating two distinct effectors during neurite outgrowth. Journal of cell science. 2013;126(Pt 11):2424–2435. doi: 10.1242/jcs.117846. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi H, Etoh K, Ohbayashi N, Fukuda M. Rab35 promotes the recruitment of Rab8, Rab13 and Rab36 to recycling endosomes through MICAL-L1 during neurite outgrowth. Biology open. 2014 doi: 10.1242/bio.20148771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu CK, Coughlin M, Field CM, Mitchison TJ. KIF4 regulates midzone length during cytokinesis. Current biology : CB. 2011;21(10):815–824. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. The EMBO journal. 2007;26(6):1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheeseman IM. The kinetochore. Cold Spring Harbor perspectives in biology. 2014;6(7):a015826. doi: 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarangapani KK, Asbury CL. Catch and release: how do kinetochores hook the right microtubules during mitosis? Trends in genetics : TIG. 2014;30(4):150–159. doi: 10.1016/j.tig.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weisenberg RC, Deery WJ. The mechanism of calcium-induced microtubule disassembly. Biochemical and biophysical research communications. 1981;102(3):924–931. doi: 10.1016/0006-291x(81)91626-0. [DOI] [PubMed] [Google Scholar]
- 43.Ishida N, Nakamura Y, Tanabe K, Li SA, Takei K. Dynamin 2 associates with microtubules at mitosis and regulates cell cycle progression. Cell structure and function. 2011;36(2):145–154. doi: 10.1247/csf.10016. [DOI] [PubMed] [Google Scholar]
- 44.Lehtonen S, Shah M, Nielsen R, Iino N, Ryan JJ, Zhou H, Farquhar MG. The endocytic adaptor protein ARH associates with motor and centrosomal proteins and is involved in centrosome assembly and cytokinesis. Molecular biology of the cell. 2008;19(7):2949–2961. doi: 10.1091/mbc.E07-05-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, Gerlich DW. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136(3):473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 46.Thoresen SB, Campsteijn C, Vietri M, Schink KO, Liestol K, Andersen JS, Raiborg C, Stenmark H. ANCHR mediates Aurora-B-dependent abscission checkpoint control through retention of VPS4. Nature cell biology. 2014;16(6):550–560. doi: 10.1038/ncb2959. [DOI] [PubMed] [Google Scholar]
- 47.Uehara R, Goshima G. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. The Journal of cell biology. 2010;191(2):259–267. doi: 10.1083/jcb.201004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varma D, Salmon ED. The KMN protein network--chief conductors of the kinetochore orchestra. Journal of cell science. 2012;125(Pt 24):5927–5936. doi: 10.1242/jcs.093724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Domnitz SB, Wagenbach M, Decarreau J, Wordeman L. MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. The Journal of cell biology. 2012;197(2):231–237. doi: 10.1083/jcb.201108147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Current biology : CB. 2006;16(17):1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 51.Shrestha RL, Draviam VM. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Current biology : CB. 2013;23(16):1514–1526. doi: 10.1016/j.cub.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. Journal of cell science. 2003;116(Pt 20):4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 53.Wei S, Xu Y, Shi H, Wong SH, Han W, Talbot K, Hong W, Ong WY. EHD1 is a synaptic protein that modulates exocytosis through binding to snapin. Molecular and cellular neurosciences. 2010;45(4):418–429. doi: 10.1016/j.mcn.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 54.Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. The EMBO journal. 2002;21(11):2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Raw data and source images for Figure 1 I-K. (A) Table showing percentages illustrated in Figure 1I. Data is a composite of three independent experiments. One-way ANOVA tests comparing the number of control (M1), MICAL-L1-depleted (M2) and EHD1-depleted cells (M3) containing each nuclear phenotype. (B-G) Examples of images exported from ImageJ that were used to quantify nuclear area/cell. For cells with multiple nuclei, as is the case for F and G (arrow), area from each nucleus was added to give total area/cell. Scale bar= 10 μm.
Supplemental Figure 2: Specificity of MICAL-L1- and EHD1-siRNA knockdown phenotypes. (A-E) HeLa cells were transfected with control-siRNA or MICAL-L1-siRNA individual oligonucleotides for 72 h. Immunoblotting demonstrates the efficacy of MICAL-L1-depletion by oligo #1 and oligo #2. Immunofluorescence analysis of control- (B), MICAL-L1-siRNA oligo #1- (C) or MICAL-L1-siRNA oligo#2- (D) transfected cells stained with α-tubulin (red) and DAPI (blue). (E) Nuclear phenotypes were quantified from three independent experiments (150 cells/experiment). Data is presented as percentage of abnormal nuclei (micro-nucleated and bi-nucleated). (F-K) HeLa cells were either mock transfected (Lipofectamine 2000 alone in F, I, and G, J) or transfected with siRNA-resistant-GFP-EHD1 (siResis-GFP-EHD1) for 2 h (H and K). Cells were then washed and transfected with control-siRNA (F and I) or EHD1-siRNA (G, J and H, K) for 48 h. Cells were fixed and stained with α-tubulin (red) and DAPI (blue). Yellow arrows mark multi-nucleated cells in G and H. (L) Quantification of multi-nucleation in control-siRNA, EHD1-siRNA and EHD1-siRNA + siRNA-Resis-GFP-EHD1 cells. Data is presented from three independent experiments, 200 cells/experiment. (M) Immunoblot analysis demonstrating depletion of endogenous EHD1 and exogenous expression of siResis-GFP-EHD1 in EHD1-depleted cells (black arrows). One-way ANOVA *p<0.05, **p<0.01. Scale bar= 10 μm.
Supplemental Figure 3: Depletion of the EHD family member, EHD2, has no gross effect on cell cycle. (A) HeLa cells were transfected with control- or EHD2-siRNA for 48 h and subjected to immunoblot analysis. (B-D) Cells treated as in A were processed for immunofluorescence (B and C) and multi-nucleated cells were quantified (D). Data is from three independent experiments (150 cells/experiment). Scale bar= 10 μm.
Supplemental Figure 4: MICAL-L1 is required for EHD1 localization to the ICB. (A-C) HeLa cells were fixed and stained with anti-EHD2 antibody (green), anti-α-tubulin (red) and DAPI (blue). (D-I) HeLa cells transfected with control-siRNA (D-F) or MICAL-L1-siRNA (G-I) were stained with anti-EHD1 antibody (green) and anti-α-tubulin (red). The area in the dashed yellow boxes in D and E is enlarged as an inset in panel F, while the dashed yellow boxes in G and H are enlarged as an inset in panel I. A yellow arrow shows EHD1 localization to the ICB in control cells (F). (J and K) Line-scan analysis of the ICB from control (J) and MICAL-L1-depleted cells (K). The black arrow denotes the midbody, which is devoid of EHD1 and tubulin staining. Scale bar= 10 μm.
Supplemental Figure 5: The effect of MICAL-L1- or EHD1-depletion on Golgi and early endosome positioning during cytokinesis. (A-F) HeLa cells transfected with control-siRNA (A and D), MICAL-L1-siRNA (B and E) or EHD1-siRNA (C and F) for 48 h were stained either with anti-giantin antibody to mark Golgi (green; A-C) or early endosome antigen 1 (EEA1) antibody to label early endosomes (green; D-F), α-tubulin (red) and DAPI (blue). The asterisks in A mark dual localization of Golgi in control cells to the base of the ICB and the pericentrosomal area. Yellow arrows in C demonstrate localization of the Golgi to the base of ICB in EHD1-depleted cells. Scale bar= 10 μm.
Supplemental Figure 6: Recruitment of Aurora B or PLK1 is not affected by depletion of MICAL-L1, EHD1, Rab11 or Rab35. Cells were transfected with the indicated siRNAs, fixed, and stained with Aurora B antibody (A-E) or PLK1 antibody (F-J). Control (K) and EHD1-depleted (L) cells stained with tubulin (green), phalloidin-568 (red) and DAPI (blue). Scale bar= 10 μm.
Supplemental Figure 7: MICAL-L1-depletion enhances kinetochore fiber cold-stability. (A-F) HeLa cells were transfected with either control-siRNA (A-C) or MICAL-L1-siRNA (D-F) for 48 h and arrested in metaphase using MG132 (10 μM, 2 h). Cells were then placed on ice in ice-cold DMEM for 10 min. (A and D), 15 min. (B and E) or 30 min. (C and F) and then fixed and stained with α-tubulin (green), pericentrin (red) and DAPI (blue). Maximum projections of 1 μm optical sections are shown. (G) Tubulin fluorescence of kinetochore fibers after 30 min. of cold-treatment was quantified using the ImageJ ‘measure’ function. Results are shown from three independent experiments (15 cells/experiment). (H) Pole-pole distance was measured as described in Figure 4 after cold treatment (three independent experiments, n=45 cells/experiment). Student’s t-test **p<0.01. Scale bar= 10 μm.
Cells were transfected with control-siRNA for 24 h and then imaged every 10 min. for 36 h as described in the Materials and Methods. Cluster of cells located in the center of the video show that control cells complete mitosis in less than 3h. Video is a wide-field of cells shown in Figure 2A.
Cells were transfected with MICAL-L1-siRNA for 24 h and then imaged every 10 min. for 36 h as described in the Materials and Methods. Cluster of cells located in bottom left of the video demonstrate the prolonged time spent in cytokinesis (presence of intercellular bridge) due to MICAL-L1-depletion. Video is a wide-field of cells shown in Figure 2B.
Cells were treated and imaged as described in Supplemental Video 2. Cluster of cells in the bottom right of the video show two daughter cells from distinct mitoses fusing to form one bi-nucleated cell. Video is a wide-field of cells shown in Figure 2C.
Cells were transfected with EHD1-siRNA for 24 h and then imaged every 10 min. for 36 h as described in Materials and Methods. Note the delay of cell spreading during early telophase compared to control cells shown in Supplemental Video 1. Specifically, cells located in bottom right of the video show impaired cell spreading which then results in eventual cytokinesis failure leading to bi-nucleated cells. Video is a wide-field of cells shown in Figure 2D.
Z-sections were acquired at 0.110 μm increments, and displayed at 10 frames/sec.
Z-sections were acquired at 0.110 μm increments, and displayed at 10 frames/sec.
Z-sections were acquired at 0.110 μm increments, and displayed at 10 frames/sec.
Z-sections were acquired at 0.110 μm increments, and displayed at 10 frames/sec.


