Abstract
Cancer cells form three dimensional (3D) multicellular aggregates (or spheroids) under non-adherent culture conditions. In ovarian cancer (OC), spheroids serve as a vehicle for cancer cell dissemination in the peritoneal cavity, protecting cells from environmental stress-induced anoikis. To identify new targetable molecules in OC spheroids, we investigated gene expression profiles and networks upregulated in three dimensional (3D) versus traditional monolayer culture conditions. We identified ALDH1A1, a cancer stem cell marker as being overexpressed in OC spheroids and directly connected to key elements of the β-catenin pathway. B-catenin function and ALDH1A1 expression were increased in OC spheroids vs. monolayers and in successive spheroid generations, suggesting that 3D aggregates are enriched in cells with stem cell characteristics. B-catenin knockdown decreased ALDH1A1 expression levels and β-catenin coimmunoprecipitated with the ALDH1A1 promoter, suggesting that ALDH1A1 is a direct β-catenin target. Both siRNA mediated β-catenin knockdown and A37, a novel ALDH1A1 small molecule enzymatic inhibitor described here for the first time, disrupted OC spheroid formation and cell viability (p<0.001). B-catenin knockdown blocked tumor growth and peritoneal metastasis in an OC xenograft model. These data strongly support the role of β-catenin regulated ALDH1A1 in the maintenance of OC spheroids and propose new ALDH1A1 inhibitors targeting this cell population.
Keywords: ovarian cancer, cancer stem cells, ALDH1, β-catenin, spheroids, small molecule inhibitors
Introduction
Epithelial ovarian cancer (OC) is the most lethal of all gynecologic malignancies; with the majority of cases being diagnosed at an advanced stage. OC metastasis is the primary cause of clinical complications and is characterized by several unique features (1). While in other epithelial tumors breakdown of the basement membrane is required for tumor invasion into lymphatics or vasculature and subsequent dissemination of cancer cells to distant sites (2), hematogenous metastasis is uncommon in OC. Tumor dissemination occurs directly in the peritoneal cavity; with most sites of secondary implants involving the mesentery, omentum, and bowel. This is facilitated by the fact that OC cells at the primary site are in direct anatomic contact with the overlying peritoneal surface and fluid. Their dislodgement from the primary tumor on the surface of the ovary or fallopian tube (3), allows cells to float in the peritoneal fluid. Importantly, after exfoliation from the primary tumor, OC cells form multicellular aggregates or spheroids (4). These 3D cellular aggregates serve as the vehicle for dissemination in the peritoneal cavity, protecting cells from anoikis induced by stress in the extracellular compartment (5, 6).
Within spheroid structures, cells adopt mesenchymal features (4, 7) that are regulated by cytokines and growth factors such as estrogen (8), TGF-β (9), or other proteins secreted in the peritoneal milieu (10, 11). The mesenchymal phenotype allows cells to invade when they come in contact with the mesothelium (7), leading to the establishment of peritoneal implants. We hypothesized that cells forming spheres are enriched in cancer stem cells (CSCs) (12, 13), allowing development of distant metastases and persistence after chemotherapy.
Recent reports suggest that cells grown as 3D structures behave differently compared to monolayer cultures and represent a better approximation of tumors developing in vivo. For instance, spheroids display distinct genetic expression profiles (14-16), specific intercellular signaling (17-20), and are subjected to different mechanical forces compared to monolayers (21-23). The cellular dimensionality and the resulting microenvironment exert a critical influence on cell survival, impacting drug sensitivity (24, 25) or resistance (26, 27). OC spheroids can be isolated directly from malignant ascites or cultured from OC cells by using non-adherent conditions or the hanging drop culture method (28). In this manuscript we set out to identify oncogenic pathways regulating formation of multicellular aggregates with the goal of identifying novel targets enriched in 3D culture models. We hypothesized that inhibition of such targets would disrupt spheroid formation and block cancer metastasis.
Microarray analysis comparing OC multicellular structures to monolayer cultures identified ALDH1A1, a known CSC marker, upregulated in spheroids. ALDH1A1 was part of a gene network with β-catenin and chromatin immunoprecipitation (ChIP) demonstrated that ALDH1A1 is a direct β-catenin target. Both β-catenin knockdown and a novel ALDH1A1 inhibitor (A37) prevented multicellular aggregation, supporting that inhibition of this pathway effectively disrupts spheroid formation.
Results
Gene expression profiles of OC spheroids
OC cell lines (IGROV1, SKOV3, A2780) and primary OC cells derived from malignant ascites were grown as monolayers, spheroids, or transitioned back from spheroid-to-monolayer cultures. When grown under non-adherent conditions, OC cells formed 3D aggregates with distinct features. For instance, SKOV3 adenocarcinoma cells, formed glandular structures with prominent extracellular matrix secretion; IGROV1 endometrioid cells formed branching spheroids, while A2780 and OVCA primary cells formed compact round multi-cellular aggregates. Spheroids derived from human primary cells displayed calcifications, similar to psammoma bodies formed in human tumors (Figure 1A). To identify genes and pathways upregulated in spheroids, we compared expression profiles of monolayers, spheroids, or spheroid-to-monolayer IGROV1 cultures using Affymetrix microarrays. Unsupervised hierarchical clustering demonstrated distinct profiles of 3D versus 2D cultures, with reversal of the spheroid genotype when cells were transitioned back to monolayers (Figure 1B) and ANOVA-based statistical analysis identified 473 transcripts differentially expressed in spheroids compared to monolayers (p<0.01 and FDR<0.01). Of those, 15 transcripts were upregulated > 4 fold and 25 transcripts were downregulated > 4-fold in spheroids versus monolayers (Tables 1 and 2). Validation of top differentially expressed genes using semi-quantitative RT-PCR confirmed up-regulation of aldehyde dehydrogenase 1A1 (ALDH1A1), angiopoietin-like 2 (ANGPTL2), thrombospondin type I (THSD), and neurotensin (NTS) and downregulation of family 25, member B (FAM25B), v-ets erythroblasosis virus homolog (ETS) in spheroids compared to monolayer cultures (Figure 1C).
Figure 1. Gene Expression Analysis of OC spheroids and monolayers.
A, Morphology of OC cells grown as spheroids and stained with methylene blue and fuchsin (400X magnification). Shown are spheroids derived from SKOV3, IGROV1, A2780 and primary human cells derived from OC ascites. Arrows point to extracellular matrix deposited by SKOV3 cells and calcifications (psammoma bodies) formed in the ascites derived spheroids. B, Hierarchical clustering displays differential expression profiles for IGROV1 cells grown as monolayer, spheroid, or spheroid to monolayer cultures (n=3 replicates). Rows represent individual samples and columns represent genes. Each cell corresponds to the level of expression of a particular gene in a given sample. A visual dual color code is utilized with red and blue indicating relatively high and low expression levels, respectively. The scale of color saturation, which reflects the gene expression levels, is included. C, Differentially expressed genes between spheroids and monolayer were validated by semi-quantitative RT-PCR. Densitometry shows relative gene expression normalized for GAPDH. D, Cell morphology of SKOV3, IGROV1, and OC primary cells grown as monolayers (m) and spheroids (s), 100X magnification (left panels). Semiquantitative RT-PCR assessed ALDH1A1, ALDH1A2, ALDH1A3, ALDH2, ALDH3A1 mRNA expression levels in SKOV3, IGROV1, and primary OC cells cultured as monolayers (m) compared with spheroids (s, right panels). E, Flow cytometry measures Aldefluor positive cells in SKOV3 and IGROV1 cells grown as spheroids compared to monolayers. DEAB-treated cells serve as negative controls. Measurements were performed in three replicates.
Table 1.
Genes preferentially expressed in OC cell monolayers compared to spheroids (>4.0)
| Probeset ID | Gene Symbol | Gene_assignment | Fold-Change (mono vs. sphere) | p-value (mono vs. sphere) |
|---|---|---|---|---|
| 7951271 | MMP1 | matrix metallopeptidase 1(interstitial collagenase) | 19.07 | 1.54E-06 |
| 7933423 | FAM25B | family with sequence similarity 25, member B | 14.55 | 1.31E-08 |
| 8037205 | CEACAM1 | carcinoembryonic antigen-related cell adhesion molecule 1 | 12.62 | 8.81E-09 |
| 7971461 | LCP1 | lymphocyte cytosolic protein 1 (L-plastin) | 8.93 | 5.16E-08 |
| 7930498 | ACSL5 | acyl-CoA synthetase long-chain family member 5 | 8.72 | 2.87E-08 |
| 7928429 | PLAU | plasminogen activator, urokinase | 7.62 | 1.46E-10 |
| 8015337 | KRT15 | keratin 15 | 7.08 | 3.08E-05 |
| 8134564 | MYH16 | myosin, heavy chain 16 pseudogene | 6.79 | 2.74E-06 |
| 8051322 | XDH | xanthine dehydrogenase | 6.59 | 9.41E-07 |
| 7983512 | SQRDL | sulfide quinone reductase-like (yeast) | 6.38 | 6.38E-11 |
| 8015060 | KRT24 | keratin 24 | 5.93 | 2.62E-08 |
| 8148184 | FAM83A | family with sequence similarity 83, member A | 5.86 | 1.61E-07 |
| 7952601 | ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 5.67 | 1.72E-10 |
| 7964834 | CPM | carboxypeptidase M | 5.64 | 1.19E-08 |
| 7961075 | CD69 | CD69 molecule | 5.21 | 2.45E-07 |
| 8135069 | SERPINE1 | serpin peptidase inhibitor, member 1 | 4.82 | 7.03E-09 |
| 8086517 | CDCP1 | CUB domain containing protein 1 | 4.6 | 3.46E-06 |
| 8095680 | IL8 | interleukin 8 | 4.45 | 2.14E-05 |
| 7970676 | SHISA2 | shisa homolog 2 (Xenopus laevis) | 4.45 | 8.25E-06 |
| 8026490 | UCA1 | urothelial cancer associated 1 (non-protein coding) | 4.39 | 8.68E-06 |
| 8054712 | IL1A | interleukin 1, alpha | 4.35 | 6.79E-06 |
| 7996264 | CDH5 | cadherin 5, type 2 | 4.29 | 2.32E-06 |
| 7946292 | CYB5R2 | cytochrome b5 reductase 2 | 4.17 | 1.31E-08 |
| 8072328 | SEC14L2 | SEC14-like 2 | 4.16 | 5.48E-09 |
| 8064904 | FERMT1 | fermitin family member 1 | 4.13 | 3.17E-05 |
Table 2.
Genes preferentially expressed in OC cell spheroids compared to monolayers (>4.0)
| Probeset ID | Gene Symbol | Gene_assignment | Fold-Change (mono vs. sphere) | p-value (mono vs. sphere) |
|---|---|---|---|---|
| 7957458 | NTS | neurotensin | 15.41 | 7.64E-09 |
| 8141094 | PDK4 | pyruvate dehydrogenase kinase, isozyme 4 | 11.61 | 1.84E-08 |
| 8138231 | THSD7A | thrombospondin, type I, domain containing 7A | 8.24 | 1.59E-07 |
| 8164200 | ANGPTL2 | angiopoietin-like 2 | 7.05 | 3.89E-09 |
| 7963054 | TUBA1A | tubulin, alpha 1a | 6.19 | 4.28E-07 |
| 7958262 | TCP11L2 | t-complex 11, testis-specific-like 2 | 5.48 | 3.38E-05 |
| 8095110 | KIT | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 5.42 | 2.12E-10 |
| 7968678 | FREM2 | FRAS1 related extracellular matrix protein 2 | 5.20 | 2.63E-05 |
| 8108716 | PCDHB16 | protocadherin beta 16 | 5.10 | 4.50E-10 |
| 8124492 | HIST1H2BK | histone cluster 1, H2bk | 4.89 | 5.24E-06 |
| 8161755 | ALDH1A1 | aldehyde dehydrogenase 1 family, member A1 | 4.45 | 9.69E-09 |
| 7962455 | NELL2 | NEL-like 2 | 4.37 | 5.08E-08 |
| 8131550 | SCIN | scinderin | 4.24 | 1.18E-07 |
| 8108744 | PCDHB14 | protocadherin beta 14 | 4.15 | 1.10E-06 |
| 8142997 | PLXNA4 | plexin A4 | 4.04 | 1.42E-07 |
ALDHA1 expression in spheroids
One of the upregulated transcripts in OC spheroids was ALDH1A1, a recently recognized stem cell marker (29). Semiquantitative RT-PCR validated increased mRNA expression levels of ALDH1A1 in SKOV3 and IGROV1 spheroids compared with monolayers (Figure 1D). Interestingly, the other ALDH1 isoforms (A2 and A3) or ALDH2 and ALDH3A1 were not upregulated in spheroids compared to monolayer cultures (Figure 1D), supporting that the main isoform upregulated in spheroids is ALDH1A1, as identified by gene expression analysis. Flow cytometry analysis showed an increased percentage of Aldefluor-positive cells in spheroids compared with monolayers: 25.05% vs. 6.68% for SKOV3 and 48.0% vs. 20.46% for IGROV1 cells (Figure 1E). The results indicate that culture under ultra-low adherent conditions selected for a population enriched in Aldefluor-positive cells which are known to possess self-renewal properties (29, 30).
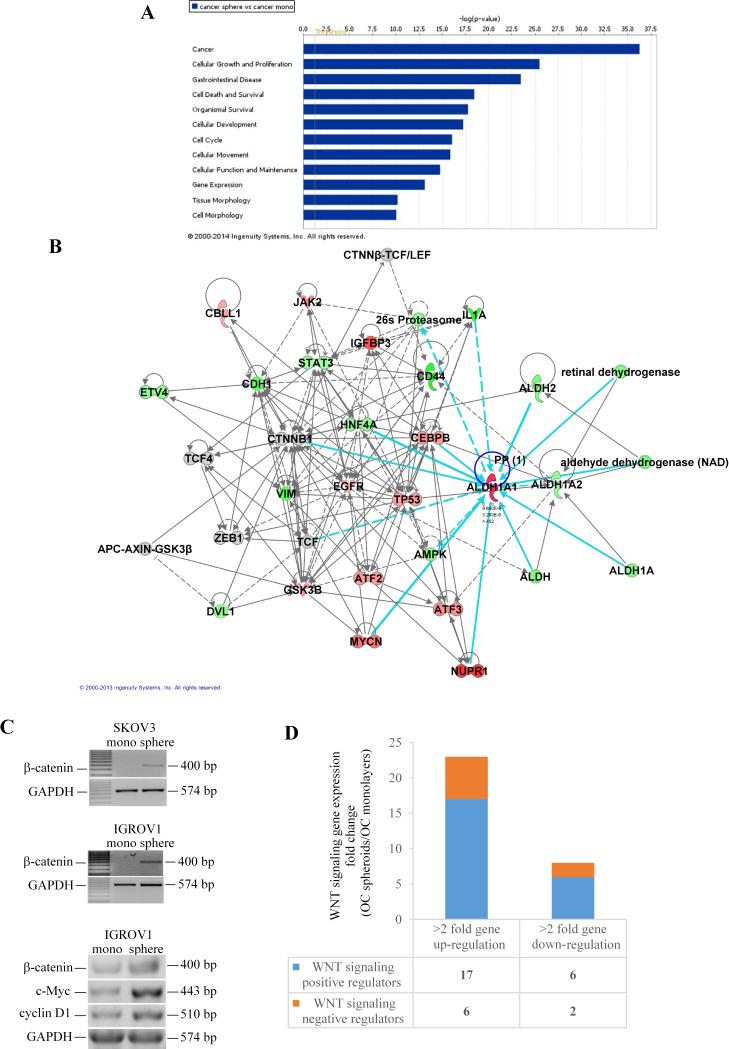
To investigate potential gene interactions in spheroid formation, the bioinformatics tool from IPA was used and identified 62 gene networks with statistically significant scores. The top gene networks represented in spheroids compared to monolayer cultures included: cancer, cellular growth and proliferation, gastrointestinal disease, cell death and survival, organismal survival, cellular development, cell cycle, and cellular movement (Figure 2A, ranking scores ranging from 16 to 36). Within the top networks (cancer and cellular growth and proliferation) ALDH1A1 was a central node directly connected to β-catenin, c-myc, nuclear protein transcriptional regulator 1 (NUPR1), T cell factor (TCF), hepatocyte nuclear factor 4α (HNF4A), and the enhancer binding protein CEBPB (Figure 2B), suggesting interactions with these molecules under 3D growth conditions. Because of the previously recognized role of the β-catenin pathway in the maintenance of CSCs (31-33), the role of ALDH1A1 as a CSC marker (29, 34), and the observed network connections between ALDH1A1 and several of the β-catenin pathway key elements (β-catenin, c-myc, TCF, CEBPB), we focused subsequent analyses on validating the β-catenin-ALDH1A1 interaction in the generation of OC spheroids.
Figure 2. Gene networks in OC monolayers versus spheroids.
A, Gene networks generated using the IPA bioinformatics tool were ranked by log p-values and compared spheroid versus monolayer cultures. Networks with larger log p-values are more significant. B, Analysis within the top ranked networks (log p value > 25) displays interconnected genes as nodes. Genes are colored according to expression level values; red symbols correspond to up-regulated genes, while green symbols indicate down-regulation. Dashed lines between nodes show indirect interactions, while continuous lines indicate direct interactions. C, Semiquantitative RT-PCR assessed mRNA expression levels for β-catenin and its targets (c-myc and cyclin D1) in SKOV3 and IGROV1 cells monolayers compared with spheroids. D, Stacked diagram representing number of genes upregulated >2 fold or downregulated >2-fold in IGROV1 spheroids vs. monolayers. Positive and negative Wnt pathway regulators are included (full list of genes is shown in Supplementary Tables 4 and 5). The RT2 Profiler PCR Array for human Wnt signaling pathway was used to compare expression profiles in spheroids vs. monolayer cultures.
B-catenin signaling regulates spheroid formation and self- renewal
Semi-quantitative RT-PCR validated upregulation of β-catenin and of its target genes c-myc and cyclin D1 in IGROV1 and SKOV3 spheroids compared to monolayers (Figure 2C). An additional exploratory analysis used quantitative RT-PCR for the human Wnt signaling pathway and demonstrated an overall upregulation of this pathway in spheroids compared to monolayers cultures. Specifically, 23 Wnt related transcripts were upregulated > 2-fold in spheroids vs. monolayers, with 17 genes being known positive regulators of the pathway (Figure 2D and Supplementary Tables 4 and 5). Notably, Frizzled receptors 1, 4, and 7 were upregulated > 4-fold in spheroids cultures, supporting enrichment in Wnt signals leading to increased β-catenin activity under 3D conditions.
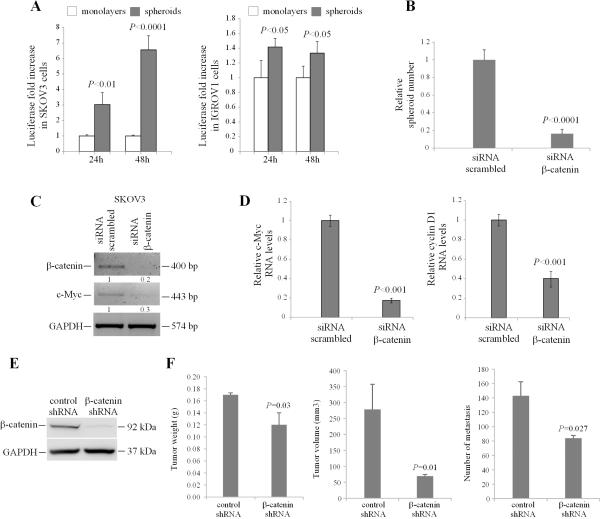
To measure β-catenin function in spheroids vs. monolayers, the TCF/LEF reporter assay was used. A greater than 3-fold increase in TCF/LEF activity was noted in SKOV3 spheroids compared to monolayers, and a lesser magnitude, but still significant increase was recorded in IGROV1 cells grown as multi-cellular aggregates (Figure 3A). The results indicate that the increase in spheroids proliferation was accompanied by an active β-catenin/TCF transcriptional activity.
Figure 3. β-catenin regulates OC spheroid and tumor formation.
A, SKOV3 and IGROV1 cells grown as monolayers were co-transfected with TCF/LEF1 luciferase reporter and Renilla control plasmid, prior to plating as monolayers or spheroids. Luciferase activity relative to renilla activity compared monolayers and spheroids at 24 and 48 hours and is expressed as fold increase. Data are shown as means of duplicate measurements +/− SD. Experiments were repeated at least three times. Significant differences are marked. B, SKOV3 cells were transfected with scrambled or β-catenin targeting siRNA prior to plating in ultra-low attachment plates. Sphere counts are shown as means +/− SD of quadruplicate measurements. C, Semiquantitative RT-PCR measures β-catenin and c-Myc expression levels in SKOV3 spheroid cells transfected with scrambled or β-catenin targeting siRNA. Densitometry shows relative gene expression normalized for GAPDH. D, Real-time PCR measures the expression levels of β-catenin target genes c-Myc and cyclin D1 in SKOV3 cells transfected with scrambled or β-catenin targeting siRNA. Data are shown as means +/− SD of 3 replicate measurements. E, Western blotting shows β-catenin expression levels in SKOV3 cells stably transduced with control- and β-catenin targeting shRNA and used for ip inoculation of nude mice. F, Tumor weights, volumes, and numbers of peritoneal metastases derived from SKOV3 cells stably transduced with control- and β-catenin targeting shRNA and injected ip in nude mice (n = 5 and 7, respectively). Data are shown as means +/− SEM. Significant differences are marked.
To further assess the role of β-catenin signaling in spheroid formation, SKOV3 cells were transiently transfected with siRNA targeting β-catenin or with scrambled siRNA prior to plating in ultra-low adherent plates to allow sphere formation. B-catenin knockdown prevented aggregation of cells as spheroids, as quantified by microscopic examination (Figure 3B and Supplementary Figure 1) and semiquantitative and real time RT-PCR confirmed β-catenin knockdown and corresponding downregulation of its targets c-myc and cyclin D1 (Figures 3C-D and Supplementary Figure 1). In all, these data support that β-catenin/TCF signaling regulates spheroids proliferation.
Because of the presumed role of spheroids as vehicles of ip dissemination and the observed effect of β-catenin knockdown disrupting multicellular aggregation, we next measured the effects of β-catenin knockdown on tumor formation and dissemination in an ip xenograft model. For this, SKOV3 cells were stably transduced with control or β-catenin targeting shRNA. Decreased β-catenin expression levels were confirmed in SKOV3 cells transduced with shRNA targeting β-catenin compared to control shRNA (Figure 3E). Tumor weights (0.17gms vs. 0.12 gms, p = 0.03) volumes (278.8 mm3 vs. 69.5 mm3, p = 0.01) and number of peritoneal implants (143 vs. 84, p = 0.002) were significantly decreased in xenografts derived from SKOV3 cells transduced with shRNA targeting β-catenin compared to those transduced with control shRNA (Figure 3F). Collectively, these data support the role of β-catenin in OC spheroid formation contributing to peritoneal dissemination.
ALDH1A1 is a β-catenin target gene
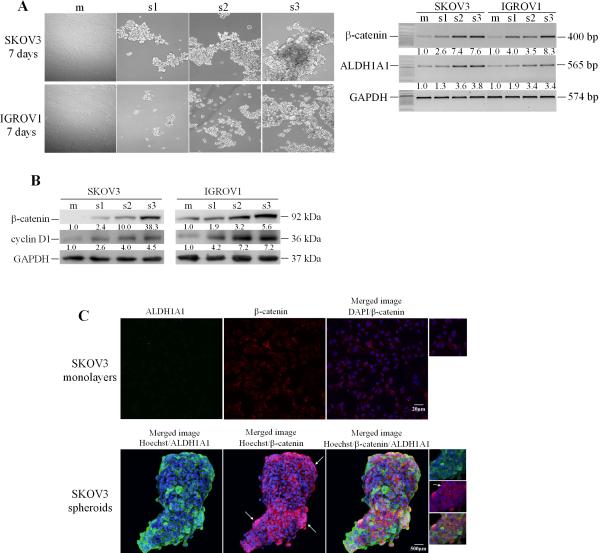
Further confirmation of β-catenin and ALDH1A1 expression used three sequential spheroids passages (s1-s3). Phase contrast microscopy demonstrated that OC cells formed more rapidly compact and large 3D structures after enzymatic dissociation and passage through several generations (Figure 4A). The spheroids’ self-renewing and growth corresponded to an increase in β-catenin gene expression. Compared with monolayer cultures, β-catenin expression levels were up-regulated 2.6- and 4.0-fold respectively in SKOV3 and IGROV1 first generation spheroids. Further increase in β-catenin mRNA and protein levels was observed during the second and third spheroid generations in both cell lines (Figures 4A-B), suggesting its role in the self-renewal and maintenance of spheroids. Furthermore, cyclin D1 expression levels, a β-catenin target (35, 36), were also increased in spheroids vs. monolayers and in subsequent spheroid passages. In parallel, ALDH1A1 mRNA expression levels increased during successive spheroid generations from ~1.3- and 1.9-fold for the first generation to 3.8- and 3.4-fold for third generation spheroids in SKOV3 and IGROV1 cells, respectively compared with monolayers (Figure 4A).
Figure 4. ALDH1A1 and β-catenin expression in OCspheroids.
A, Cell morphology of SKOV3 and IGROV1 cells grown as monolayers (m) and three spheroid generations (s1-s3, left panel). Semiquantitative RT-PCR for β-catenin and ALDH1A1 mRNA expression levels comparing monolayers and the three generation of spheroids (right panel). B, Western blotting measures expression levels of β-catenin and cyclin D1 in monolayer cultures and three spheroid generations. Densitometry quantifies β-catenin, ALDH1A1, and cyclin D1 expression levels normalized for GAPDH. C, IF staining for β-catenin (Cy5, red) and ALDH1A1 (AlexaFluor488, green) in SKOV3 cells grown as monolayers or spheroids (200X magnification). Nuclear β-catenin localization is identified by emergence of purple spectra on merged images.
Confirmation of ALDH1A1 and β-catenin expression and cellular localization in monolayers cultures and in spheroids was obtained by immunofluorescence (IF) based staining and confocal microscopy. ALDH1A1 was not clearly visualized in monolayer cultures, but was enriched in OC cells grown as spheroids (Figure 4C). Likewise, β-catenin expression was increased in cells grown as spheroids compared to monolayers. Furthermore, while in monolayer cultures, β-catenin was localized at the plasma membrane, translocation to the nucleus, was observed in cells aggregated as spheroids (Figure 4C), suggesting increased transcriptional activity. Increased ALDH1A1 expression was recorded in cells where β-catenin was expressed and localized in the nucleus.
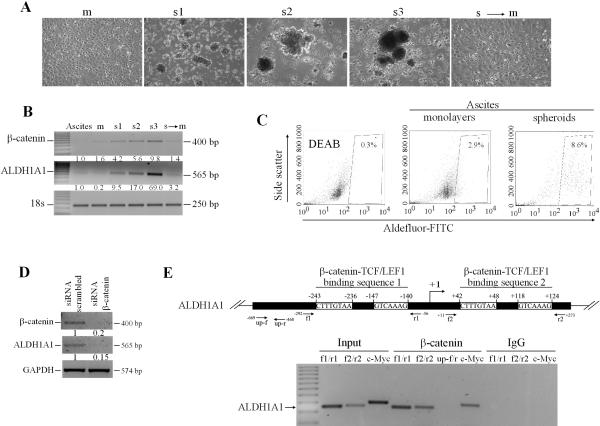
After establishing the importance of the β-catenin/ALDH1A1 gene interaction during the formation of spheroids from OC cell lines, we tested whether these genes also regulate the formation of native spheroids. For this purpose, we isolated spheroids from human OC ascites and maintained them in non-adherent conditions or as monolayers (n=5 specimens). To test β-catenin and ALDH1A1 expression during successive spheroid generations, we used enzymatic and mechanical dissociation prior to passage every 7 days. Figure 5A illustrates increasing number and more compact spheres formed with each successive passage. The basal ALDH1A1 and β-catenin mRNA expression levels increased through successive generations compared with monolayers and were repressed when spheroids were re-plated as monolayers (Figure 5B), supporting the significance of this pathway to multicellular aggregation in human primary cells. Flow cytometry analyzed ALDH1A1 enzymatic activity in monolayer cultures and spheroids derived from cells isolated from OC ascites, noting ~8.7% vs. 2.9% Aldefluor-positive cells in spheroids vs. monolayers, respectively, consistent with observations in cancer cell lines (Figure 5C).
Figure 5. ALDH1A1 is a β-catenin target in OC cells.
A, Morphology of primary cells derived from OC malignant ascites grown as monolayers (m), three spheroid generations (s1-s3), or spheroid to monolayer culture (s to m). B, Semiquantitative RT-PCR for β-catenin and ALDH1A1 mRNA expression levels comparing monolayers, three generation of spheroids, and spheroid to monolayer cultures. Densitometry quantified β-catenin and ALDH1A1 normalized with the house-keeping gene 18S. C, Flow cytometry quantifies Aldefluor positive cells derived from OC ascites and grown as spheroids compared with monolayers. DEAB-treated cells serve as negative controls. Measurements were performed in duplicates. D, Semiquantitative RT-PCR for ALDH1A1 expression levels in SKOV3 cells transfected with scrambled or β-catenin targeting siRNA. E, Scheme representing the TCF/LEF1 binding sequences within the ALDH1A1 promoter relative to the designed primers (top panel). ChIP assay used chromatin from IGROV1 cells immunoprecipitated with β-catenin or IgG (control). Results of PCR amplification are as follows: DNA ladder; chromatin from IGROV1 cells not subjected to IP (input) and amplified with 2 sets of primers corresponding to the two predicted TCF/LEF1 binding sequences on the ALDH1A1 promoter (lanes 1-2,f1/r1 and f2/r2) or with primers corresponding to the TCF/LEF binding site on the c-myc promoter (lane 3, positive control); chromatin immunoprecipitated with β-catenin antibody and amplified with ALDH1A1 promoter specific primers (lanes 4-5, f1/r1 and f2/r2); ALDH1A1 promoter nonspecific primers (lane 6, up f/r, negative control), or c-myc promoter specific primers (lane 7, positive control); or chromatin immunoprecipitated with IgG and amplified with ALDH1A1 and c-myc specific primers (lanes 8-10, negative controls).
The direct correlation between β-catenin and ALDH1A1 expression levels observed in spheroids along with the IPA network analysis suggested that ALDH1A1 may represent a β-catenin target. To test this hypothesis, we measured ALDH1A1 mRNA expression levels after β-catenin knockdown. SiRNA mediated β-catenin downregulation induced decreased ALDH1A1 expression, suggesting that ALDH1A1 is transcriptionally regulated by β-catenin (Figure 5D). To definitively demonstrate this concept we searched and identified potential TCF/LEF responsive elements at positions (-243 to -236), (-147 to-140), (+42 to+48) and (+118 to +124) within the ALDH1A1 promoter sequence (Figure 5E) by using a promoter motif searching software (PROMO). ChIP tested whether β-catenin interacts with the ALDH1A1 promoter. PCR amplified the ALDH1A1 promoter fragments corresponding to the TCF/LEF responsive regions in the chromatin pulled down by a β-catenin antibody (Figure 5E, lanes 4-5). Specificity of β-catenin antibody binding to ALDH1A1 promoter was demonstrated by observing no PCR product in chromatin immunoprecipitated with IgG (Figure 5E, lanes 8-10). These data demonstrate that ALDH1A1 is a direct β-catenin target in OC cells.
Targeting ALDH1A1 disrupts OC spheroid formation
Having shown that OC multicellular aggregates are enriched in ALDH1A1+ cells and that ALDH1A1 is a direct β-catenin target, we next explored whether ALDH1A1 enzymatic inhibitors disrupt spheroid formation and OC cell survival under non-adherent culture conditions. We used a novel ALDH1A1 inhibitor (A37) identified through high throughput screening of the ChemDiv library and the less selective inhibitor DEAB. A37 has a molecular weight of 431.6 Daltons and no structural similarity to any known aldehyde dehydrogenase inhibitors (Figure 6A). A37 has good potency (IC50 of 4.6 ± 0.8 μM; Ki of 300 ± 26 nM) and is selective for ALDH1A1 with no effect on the other members of the ALDH1A subfamily (ALDH1A2 and ALDH1A3), or toward ALDH2 and ALDH3A1 at concentrations up to 100 μM (Figure 6B). A37 has a competitive mode of inhibition with respect to varied substrate acetaldehyde (Figure 6C).
Figure 6. Structure, properties, and effects of A37 in OC cells.
A, Structure of A37 {(ethyl-2-((4-oxo-3-(3-(pryrrolidin-1-yl)propyl)-3,4-dihydrobenzo[4,5]thioeno[3,2-d]pyrimidin-2-yl)thio)acetate)}. B, Normalized residual activity of selected ALDH isoenzymes in the presence of 20 μM A37 in the presence of saturating concentrations of aldehyde substrate. C, Representative Lineweaver-Burk plot for the non-linear fit to the competitive inhibition equation for A37 inhibition of ALDH1A1 versus varied acetaldehydes. D, Aldefluor activity measured by flow cytometry in IGROV1 cells treated with control (DMSO and DEAB) and A37 (1-50μM) for 3 days. E-G, Morphology (E), number of spheres (F), and percentage of viable cells measured by the CCK-8 assay (G) after treatment with DMSO (control) and A37 (1, 5, 10, 25, and 50μM) for 3 days. Data are shown as means of triplicate measurements +/− SD. Significant differences are marked. H-I, Effects of cisplatin (25nM-5μM) and A37 (1μM) in OC cells grown as spheroids. Dose-response curves representing sphere numbers (H) and percentage of surviving cells (I) were plotted using GraphPad Prism against the logarithmic concentrations of cisplatin used during a 72h treatment period.
To test the activity of A37 in OC cells, IGROV1 cells grown under low attachment conditions were used. Under these conditions, the ALDH1A1+ population represents ~30-50% of the cell population. Treatment with A37 blocked ALDH1A1 activity as measured by flow cytometry in IGROV1 cells in a dose dependent manner and more significantly than DEAB (Figure 6D). A dose response experiment demonstrated dose-dependent inhibition of spheroid formation starting at 1μM concentration (Figure 6E-F) and dose-dependent decrease in cell viability at concentrations greater than 1μM (Figure 6G). A37 also induced moderate sensitization of IGROV1 cells to cisplatin, a common cytotoxic used for the treatment of OC (Figure 6H-I). Collectively these data support that targeting of ALDH1A1 with a novel small molecule blocks OC cell proliferation and survival under 3D culture conditions and synergizes with chemotherapy.
Discussion
Through genomic profiling we identified ALDH1A1 and β-catenin signaling, two known pathways regulating CSCs, as being upregulated and interconnected in OC spheroids compared to monolayer cultures. We demonstrated that ALDH1A1 is a direct β-catenin target and that its inhibition by a novel small molecule disrupts formation of multicellular aggregates. Our data point to novel pathways activated in anoikis-resistant spheroids and potential new strategies to target them. These results have several implications.
First, we identified a specific gene network activated under 3D conditions compared to monolayer cultures. Other studies have explored the molecular characteristics of 3D cultures, demonstrating differences in gene expression based on culture conditions (37-39). It is increasingly accepted that multicellular aggregates are a better representation of human tumors compared to standard cultures on plastic. Cell growth under 3D conditions replicates the mechanical forces and gradients of oxygen and nutrient existent in native tissues which regulate cellular polarity, differentiation, and activate various morphogenic signaling programs. Well established breast cancer 3D models have replicated the architecture of normal or transformed mammary tissue and helped understanding cellular differentiation and response to therapy (40-42). OC cells grown as spheroids display distinct response to chemotherapeutics compared to monolayers (43, 44) and adopt an invasive phenotype characterized by a TGF-βfibrotic response that may protect them from unfavorable external conditions (9, 45). These features suggest that cells grown as spheroids may be enriched in stem cells. The genomic signature identified in this study identifies several CSCs markers upregulated in spheroids, including c-KIT, β-catenin, and ALDH1A1.
Second, our data point to β-catenin as an important pathway activated in spheroids. B-catenin activation has been implicated in the self-renewal and survival of hematopoietic, cutaneous and gastrointestinal stem cells (33, 46-48), however its role in ovarian CSCs has not yet been defined. The wingless pathway, which is an upstream regulator of β-catenin, is required for the maintenance of somatic stem cells in the Drosophila ovary (49) and activation of β-catenin has found downstream of the stem cell factor receptor in c-kit positive ovarian tumor initiating cells (50). The data presented here identify β-catenin involvement in the formation of OC multicellular aggregates and demonstrate that its targeting disrupts sphere formation, cell proliferation under non-adherent conditions, tumor metastasis in vivo, and expression of the stem cell marker ALDH1A1. These results suggest that inhibition of this pathway, or of its key downstream elements, may be instrumental in eliminating OC spheroids containing the stem cell population.
Third, we identified ALDH1A1 upregulation in spheroids through a non-biased genome mining approach. ALDH1A1 is a member of the highly conserved ALDH family which includes 18 other enzymes involved in the metabolism of reactive aldehydes (51). Through their detoxification functions, ALDHs exert cytoprotective roles in various tissues. In addition, the enzymes catalyze retinol oxidation to retinal, a limiting step during the synthesis of retinoic acid, which regulates cellular differentiation. Recent reports have linked ALDHs, and particularly ALDH1A1, to stem cells, both in normal (52), and in malignant tissues (53). It remains unknown whether the enzyme represents only a CSC marker or whether it is implicated in regulating the functions of stem cells. While several other markers have been proposed to recognize OC stem cells (13), ALDH1A1 activity detectable through the aldefluor assay has been validated as a reproducible CSC phenotype (29, 30). ALDH1A1+ cells have tumor initiating capacity, are resistant to cisplatin, and express higher levels of stem cell transcription factors (Sox2, nanog)(30, 34). Here we show that ALDH1A1 expression is increased in spheroids vs. monolayers and in successive spheroid generations, consistent with the recognized capacity of stem cells to organize as spheres and to self-renew. Furthermore we demonstrate that ALDH1A1 is a direct target of β-catenin, a pathway required in CSCs’ self-renewal (46, 47). This is the first demonstration that ALDH1A1 expression is regulated by the TCF/LEF transcriptional complex and our observations strengthen the connection between the enzyme and an ovarian CSC phenotype.
Furthermore, we describe for the first time the activity of a new small molecule targeting ALDH1A1 in OC spheroids. A37 is a relatively potent first-generation selective inhibitor for ALDH1A1 with a Ki of 300 nM, without significant effect on related orthologs in the ALDH family of enzymes. The effects of this small molecule inhibitor were tested on cells growing as spheres, as the 3D culture system allows for enrichment in ALDH1A1+ population. This is the first proof of principle that selective inhibition of ALDH1A1 blocks survival of OC cells by targeting the ALDH1A1+ population and supports a functional role for ALDH1A1 in this population. The results support further study of ALDH1A1 inhibitors alone or in combination with cytotoxics in OC models aiming to eradicate chemotherapy-resistant and perpetually self-renewing cancer stem cells.
Materials and Methods
Chemicals and reagents
Unless stated otherwise, chemicals and reagents were from Sigma (St Louis, MO, USA). The antibody for cyclin D1 was from Cell Signaling Technology Inc. (Beverly, MA, USA), for ALDH1A1 from Abcam (Cambridge, MA, USA), for β-catenin from ECM Biosciences (Versailles, KY, USA), and for GAPDH from Biodesign International (Saco, ME, USA). Secondary HRP-conjugated antibodies were from Amersham Biosciences (San Francisco, CA, USA) and Santa Cruz Biotechnology Inc (Santa Cruz, CA, USA). The Aldefluor kit assay was from StemCell Technologies (Vancouver, BC Canada). The ALDH1A1 inhibitor (A37) was from ChemDiv (San Diego, CA, USA), having >95% purity.
Cell culture
The human OC cell lines SKOV3, IGROV1 and A2780 were from the American Type Culture Collection (Rockville, MD, USA). De-identified OC ascites samples were obtained through an IRB approved protocol of the Indiana University Simon Cancer Center Tissue Bank. Ascites tumor cells were collected by centrifugation at 200 × g for 3 min. Erythrocytes were lysed by re-suspending the cell pellet in a 1:4 mixture of cold Hank's balanced salt solution modified (StemCell Technologies) supplemented with 2% FBS and red blood cell lysis buffer (0.8% ammonium chloride, 0.1 mM EDTA, pH 7.4) for 5 min. After centrifugation at 350 × g for 5 min, 25,000 ascites derived tumor cells were cultured as monolayers or spheroids. SKOV3 and primary OC cells were cultured in media containing 1:1 MCDB 105 (Sigma) and M199 (Cellgro, Herndon, VA, USA) supplemented with 10% FBS and antibiotics, while IGROV1 and A2780 cells were grown in RPMI 1640 at 37°, under a humidified atmosphere containing 5% CO2.
Spheroid and successive spheroid generation cultures
OC cell lines or primary cells were seeded at a concentration of 25,000 cells/ml in Mammocult complete medium (StemCell Technologies) and ultra-low attachment plates (Corning, Corning, NY, USA). Spheroids were trypsinized every 7 days and re-plated to generate successive generations. To observe spheroid morphology, A2780, SKOV3, IGROV1, and primary cells were cultured in a rotating bioreactor (Synthecon, Houston, TX, USA) until 200-400 μM compact spheroids were visible (10-30 days). They were harvested, preserved with Histochoice MB Tissue Fixative (Amresco, Solon, OH, USA) for 1 hour, and embedded in Immuno-bed resin (Polysciences, Warrington, PA, USA). Two micron thick sections were cut on an ultramicrotome and stained with 0.1% methylene blue/0.15% basic fuchsin in 50% methanol.
Transfection
Stable gene knockdown was obtained by using lentiviral transduction particles containing shRNA targeting β-catenin or shRNA control (Mission Lentiviral Transduction Particles from Sigma-Aldrich, St Louis, MO, USA) into SKOV3 cells. Lentiviral transduced SKOV3 cells were selected with puromycin (1.5 μM/mL). Transient transfection used a pool of 4 short interfering RNAs (siRNA) targeting β-catenin (Dharmacon, Pittsburgh, PA, USA; siGenome SMART pool) or individual siRNA sequences (#1: GCGUUUGGCUGAACCAUCA and #2: UAAUGAGGACCUAUACUUA, Dharmacon) and DreamFECT transfection reagent (Oz Biosciences, Marseille, France). Scrambled siRNA pool (Dharmacon) was used as control.
Western blot analysis
Cells were lysed in ice-cold Radio-Immunoprecipitation Assay (RIPA) buffer containing protease and phosphatase inhibitor cocktail, EDTA-free (Thermo Scientific, Rockford, IL USA). After sonication and centrifugation, equal amounts of proteins were separated by SDS-PAGE. After electroblotting, the PVDF membranes were incubated with primary and HRP–conjugated secondary antibodies. Immunoreactive proteins were detected by enhanced chemiluminescence solution (Thermo Scientific). Images were captured by a luminescent image analyzer with a CCD camera (ImageQuant LAS 4000 mini, GE Helthcare, Pittsburgh, PA, USA) and quantified by densitometric analysis using Gel-Pro Analyzer 3.1 software (Media Cybernetics Inc., Rockville, MD, USA).
Reverse transcription-PCR (RT-PCR)
Total RNA was extracted using RNA STAT-60 (Tel-Test Inc., Friendswood, TX, USA) and reverse-transcribed using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Primers and probes used for ALDH1A1, ALDH1A2, ALDH1A3, ALDH2, ALDH3A1, ANGPTL2, ETS1, FAM25B, NTS, NOG, THSD7A, β-catenin, cyclin D1 and c-Myc expression are included in Supplemental Material (SM), Tables S1-S2. The reverse transcriptase product (1 μL) and primers were heated at 94°C for 3 min followed by 25 cycles of amplification for GAPDH and 28 cycles for the remaining genes. The RT-PCR products were separated on a 1.5% agarose gel and visualized by ethidium bromide staining under UV light. Real-time PCR was carried out on an ABI Prism 7900 platform (Applied Biosystems) using the FastStart Taqman Probe Master (Rox; Roche, Indianapolis, IN, USA). The relative expression of different transcripts (cyclin D1, c-myc) was calculated as ΔCt and normalized by subtracting the Ct of target genes from that of the housekeeping control (GAPDH). Results are presented as means +/− SD of replicates. Each measurement was performed in duplicate and experiments were run three times in independent conditions.
Wnt Pathway PCR-Array
The human Wnt Signaling Pathway RT2 Profiler PCR Array and RT2 Real-Timer SyBR Green/ROX PCR Mix were purchased from SA Bioscience Corporation (Frederick, MD, USA). PCR was performed on ABI Prism 7900 Sequence Detector (Applied Biosystems). For data analysis the ΔΔCt method was used; for each gene, fold-changes being calculated as difference in gene expression between spheroid and monolayer cultures.
Aldefluor Assay and Flow Cytometry
ALDH1 enzymatic activity was measured using the Aldefluor assay (Stemcell Technologies). Briefly, dissociated monolayer and spheroid single cells were resuspended in Aldefluor assay buffer containing the ALDH1 substrate, bodipyaminoacetaldehyde (BAAA) at 1.5 mM, and incubated for 45 min at 37 °C. The test ALDH1A1-positive population was gated using control cells incubated under identical condition in the presence of a 10-fold molar excess of the ALDH inhibitor, diethylamino benzaldehyde (DEAB). The relative increase in FITC signal of the ALDH-positive cells was determined by a FACS Aria II flow cytometer (BD Biosciences, San Jose, CA, USA) and analyzed three times in independent experiments.
Gene reporter assays
Dual-Luciferase Assay (Promega, Madison, WI, USA) was performed to quantify Wnt/β-catenin signaling through TCF/LEF1 promoter activity in SKOV3 and IGROV1cells grown as monolayers and spheroids. Cells were transiently co-transfected with TCF/LEF1 promoter luciferase and Renilla plasmids, at a ratio of 10:1 using DreamFect Gold transfection reagent (OZ Biosciences). Luminescence was measured by using TD-20/20 Luminometer (Turner Biosystems, Madison, WI, USA) 24 hours after transfection. Experiments were performed in triplicate and repeated twice in independent conditions. To control for transfection efficiency, luminescence was normalized to Renilla activity.
Immunofluorescence (IF)
SKOV3 cells grown as monolayers on fibronectin-coated chamber slides (BD Biosciences) were stained as previously described (54), using β-catenin and ALDH1A1 antibodies. OC spheroids were fixed, permeabilized, and stained following a previously described protocol (55). Secondary antibodies included Cy5-conjugated anti-mouse antibody (1:500; Zymed) and Alexa fluor-488 anti-rabbit antibody (1:1000; Molecular Probes, Oregon, USA). Isotype-specific IgG was a negative control. Nuclei were visualized by 4’,6-diamidino-2-phenylindole (DAPI) staining (Vectashield, Vector Laboratories) in monolayers and by Hoechst staining (Molecular Probes) in spheroids. Confocal images were acquired with a confocal/two-photon Olympus Fluoview FV-1000 MPE system (Olympus America, Central Valley, PA, USA) available at the Indiana Center for Biological Microscopy (ICBM) facility (Indianapolis, IN, USA), using an Olympus XLUMPLFL 20×, NA 0.95 water immersion objective. Images were collected in a sequential illumination mode using 405-, 488- and 633-nm laser lines while fluorescent emission was collected with three PMTs using 425-475nm, 500-600nm and 655-755nm emission filters. Series of optical sections through the depth of spheroids (Z-stacks) were collected using optimal step size settings (1.18 um/slice) with images comprised of 800×800 pixels (488.2 × 488.2μm2). 3D reconstitution used Voxx software at ICBM (Indianapolis, IN, USA).
Chromatin immunoprecipitation (ChIP) assay
To detect the interaction between the transcriptional complex β-catenin/TCF/LEF1 and the ALDH1A1 promoter, we used ChIP using a kit from EMD Millipore (Billerica, MA USA, see SM). The DNA was extracted from β-catenin or IgG immunoprecipitates by using the QIAquick PCR purification kit (Qiagen, Valencia, CA, USA) and was subjected to PCR amplification using primers designed for the TCF/LEF1 binding domain of the ALDH1A1 promoter (Table S3 SM). The PCR products were resolved by 2 % agarose-ethidium bromide gel electrophoresis, visualized by UV, and quantified by densitometric analysis using Gel-Pro Analyzer 3.1. As a positive control, DNA immunoprecipitated with β-catenin antibody was amplified using primers for c-Myc promoter, a known TCF/LEF1 target gene. As negative control, DNA immunoprecipitated with β-catenin antibody was amplified with primers designed for the ALDH1A1 promoter, upstream of the predicted TCF/LEF1 binding sites.
I.p. ovarian xenograft model
1×106 SKOV3 cells stably transduced with shRNA control or targeting β-catenin, were injected i.p. in 7–8 weeks old female nude mice (n= 5 and 7, respectively) from Harlan (Indianapolis, IN, USA). Four weeks after injection, mice were euthanized, tumors were harvested, measured bi-dimensionally if >5mm and peritoneal implants were counted. Tumor volume was calculated as L*W2/2; where L is length and W is width. Experiments were approved by the IU Animal Care and Use Committee, being in compliance with federal regulations.
Discovery and characterization of ALDH inhibitors
ALDH1A1, ALDH1A2, ALDH1A3, ALDH2, and ALDH3A1 were produced and purified as previously described (56). A high-throughput screen (HTS) of 64,000 compounds from the ChemDiv Corp. was performed to identify activators and inhibitors of ALDH1A1. The hydrolysis of para-nitrophenylacetate [27] was used as a measure ALDH1A1 activity (see SM). Selectivity for closely related orthologs was tested at 20 and 100 μM using purified recombinant human ALDH1A1, ALDH1A2, ALDH1A3, ALDH2, and ALDH3A1. Dehydrogenase activity of ALDH1A1, ALDH1A2, ALDH1A3, and ALDH2 were measured in a solution containing 100–200 nM enzyme, 200 μM NAD+, 1% DMSO, and 100 μM propionaldehyde in 50 mM sodium BES, pH 7.5. ALDH3A1 activity was measured using 25 nM enzyme, 200 μM NAD+, 1% DMSO, and 1 mM benzaldehyde in 100 mM sodium phosphate buffer, pH 7.5. All assays were performed at 25°C and were initiated by the addition of the aldehyde substrate following a 2 minute pre-incubation with compound and NAD+. IC50 values were calculated by fitting the data to the four parameter EC50 equation using SigmaPlot (StatSys v.12.3, San Jose, CA, USA). The values represent the average of three independent experiments (each n = 3) using at least two protein preparations. The mode of inhibition was determined via steady-state kinetics by co-varying inhibitor and substrate concentrations at fixed concentration of the second substrate. All data were fit to tight-binding competitive, noncompetitive, uncompetitive, and mixed inhibition models using SigmaPlot (StatSys v.12.3, San Jose, CA, USA).
Cell proliferation and viability
Cell viability was measured by the CCK-8 assay (Dojindo Molecular Technologies, Rockville, MD, USA). Cell proliferation was quantified by the MTT assay. Spheroids were counted after centrifugation at 300xg for 5 minutes. All assays were performed in four replicates. Data are presented as means ± SEM.
Gene expression profiling
RNA extracted from IGROV1 cells grown as monolayer, spheroids, or spheroids transferred to monolayer, was labeled using the standard Affymetrix protocol for the Whole Transcript Target Labeling and Control Reagents kit (Affimetrix, Santa Clara, CA, USA) according to the Affymetrix user manual: GeneChip® Whole Transcript Sense Target Labeling Assay GeneChip. Three biological replicates were used. Individual labeled samples were hybridized to the Human Gene 1.0 ST GeneChips® for 17 hours then washed, stained and scanned with the standard protocol using Affymetrix GCOS (GeneChip Operating System, Affimetrix). GCOS was used to generate CEL data files, which were imported into Partek Genomics Suite (PGS, Partek, Inc., St. Louis, Mo).
Data Processing and Statistical Analysis
Robust Multi-array Average (RMA) background correction with quantile normalization of data was performed to remove background noise (see SM). Genes that had a p<0.001, FDR<0.05, and fold difference exceeding +/−0.5 fold were considered dysregulated. Data are deposited in GEO under acquisition number GSE16931151. Hierarchical clustering was performed using PGS with Pearson's dissimilarity as row and column dissimilarity measures and average linkage as linkage method. Lists of differentially expressed genes between phenotypic groups were imported into Ingenuity Pathway Analysis (© Ingenuity Systems, Redwood City, CA, USA) software to identify differentially dysregulated gene pathways and networks.
Supplementary Material
Acknowledgments
We thank Dr. Jeanette McClintick from the Indiana University Center for Medical Genomics, Dr. Malgorzata Kamocha from IBCM, and Dr. Chirayu Goswami from the Bioinformatics Core for technical assistance. This work was made possible by funding from the US Department of Veterans Affairs to DM and from NIH to TDH (R01-AA018123), DM and JT (R01EB016582).
Footnotes
Conflicts of Interest: None
References
- 1.Naora H, Montell DJ. Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat Rev Cancer. 2005;5(5):355–66. doi: 10.1038/nrc1611. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12(8):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 3.Levanon K, Crum C, Drapkin R. New insights into the pathogenesis of serous ovarian cancer and its clinical impact. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(32):5284–93. doi: 10.1200/JCO.2008.18.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sodek KL, Ringuette MJ, Brown TJ. Compact spheroid formation by ovarian cancer cells is associated with contractile behavior and an invasive phenotype. Int J Cancer. 2008 doi: 10.1002/ijc.24188. [DOI] [PubMed] [Google Scholar]
- 5.Burleson KM, Boente MP, Pambuccian SE, Skubitz AP. Disaggregation and invasion of ovarian carcinoma ascites spheroids. J Transl Med. 2006;4:6. doi: 10.1186/1479-5876-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puiffe ML, Le Page C, Filali-Mouhim A, Zietarska M, Ouellet V, Tonin PN, et al. Characterization of ovarian cancer ascites on cell invasion, proliferation, spheroid formation, and gene expression in an in vitro model of epithelial ovarian cancer. Neoplasia. 2007;9(10):820–9. doi: 10.1593/neo.07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burleson KM, Hansen LK, Skubitz AP. Ovarian carcinoma spheroids disaggregate on type I collagen and invade live human mesothelial cell monolayers. Clin Exp Metastasis. 2004;21(8):685–97. doi: 10.1007/s10585-004-5768-5. [DOI] [PubMed] [Google Scholar]
- 8.Park SH, Cheung LW, Wong AS, Leung PC. Estrogen regulates Snail and Slug in the down-regulation of E-cadherin and induces metastatic potential of ovarian cancer cells through estrogen receptor alpha. Mol Endocrinol. 2008;22(9):2085–98. doi: 10.1210/me.2007-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao L, Shao M, Schilder J, Guise T, Mohammad KS, Matei D. Tissue transglutaminase links TGF-beta, epithelial to mesenchymal transition and a stem cell phenotype in ovarian cancer. Oncogene. 2012;31(20):2521–34. doi: 10.1038/onc.2011.429. [DOI] [PubMed] [Google Scholar]
- 10.Ray A, Meng E, Reed E, Shevde LA, Rocconi RP. Hedgehog signaling pathway regulates the growth of ovarian cancer spheroid forming cells. International journal of oncology. 2011;39(4):797–804. doi: 10.3892/ijo.2011.1093. [DOI] [PubMed] [Google Scholar]
- 11.Moss NM, Barbolina MV, Liu Y, Sun L, Munshi HG, Stack MS. Ovarian cancer cell detachment and multicellular aggregate formation are regulated by membrane type 1 matrix metalloproteinase: a potential role in I.p. metastatic dissemination. Cancer research. 2009;69(17):7121–9. doi: 10.1158/0008-5472.CAN-08-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer research. 2005;65(20):9328–37. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 13.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer research. 2008;68(11):4311–20. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cody NAL, Zietarska M, Filali-Mouhim A, Provencher DM, Mes-Masson AM, Tonin PN. Influence of monolayer, spheroid, and tumor growth conditions on chromosome 3 gene expression in tumorigenic epithelial ovarian cancer cell lines. Bmc Medical Genomics. 2008:1. doi: 10.1186/1755-8794-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang TT, Hughes-Fulford M. Monolayer and Spheroid Culture of Human Liver Hepatocellular Carcinoma Cell Line Cells Demonstrate Distinct Global Gene Expression Patterns and Functional Phenotypes. Tissue Engineering Part A. 2009;15(3):559–67. doi: 10.1089/ten.tea.2007.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaedtke L, Thoenes L, Culmsee C, Mayer B, Wagner E. Proteomic analysis reveals differences in protein expression in spheroid versus monolayer cultures of low-passage colon carcinoma cells. Journal of Proteome Research. 2007;6(11):4111–8. doi: 10.1021/pr0700596. [DOI] [PubMed] [Google Scholar]
- 17.Barbone D, Yang TM, Morgan JR, Gaudino G, Broaddus VC. Mammalian target of rapamycin contributes to the acquired apoptotic resistance of human mesothelioma multicellular spheroids. Journal Of Biological Chemistry. 2008;283(19):13021–30. doi: 10.1074/jbc.M709698200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howes AL, Chiang GG, Lang ES, Ho CB, Powis G, Vuori K, et al. The phosphatidylinositol 3-kinase inhibitor, PX-866, is a potent inhibitor of cancer cell motility and growth in three-dimensional cultures. Molecular Cancer Therapeutics. 2007;6(9):2505–14. doi: 10.1158/1535-7163.MCT-06-0698. [DOI] [PubMed] [Google Scholar]
- 19.Frankel A, Man S, Elliott P, Adams J, Kerbel RS. Lack of multicellular drug resistance observed in human ovarian and prostate carcinoma treated with the proteasome inhibitor PS-341. Clinical Cancer Research. 2000;6(9):3719–28. [PubMed] [Google Scholar]
- 20.Poland J, Sinha P, Siegert A, Schnolzer M, Korf U, Hauptmann S. Comparison of protein expression profiles between monolayer and spheroid cell culture of HT-29 cells revealed fragmentation of CK18 in three-dimensional cell culture. Electrophoresis. 2002;23(7-8):1174–84. doi: 10.1002/1522-2683(200204)23:7/8<1174::AID-ELPS1174>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 21.Wang N, Butler JP, Ingber DE. Mechanotransduction Across The Cell-Surface And Through The Cytoskeleton. Science. 1993;260(5111):1124–7. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 22.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276(5317):1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 23.Ingber DE. Cellular mechanotransduction: putting all the pieces together again. Faseb Journal. 2006;20(7):811–27. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 24.Serebriiskii I, Castello-Cros R, Lamb A, Golemis EA, Cukierman E. Fibroblast-derived 3D matrix differentially regulates the growth and drug-responsiveness of human cancer cells. Matrix Biology. 2008;27(6):573–85. doi: 10.1016/j.matbio.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.David L, Dulong V, Le Cerf D, Cazin L, Lamacz M, Vannier JP. Hyaluronan hydrogel: An appropriate three-dimensional model for evaluation of anticancer drug sensitivity. Acta Biomaterialia. 2008;4(2):256–63. doi: 10.1016/j.actbio.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Frankel A, Buckman R, Kerbel RS. Abrogation of taxol-induced G(2)-M arrest and apoptosis in human ovarian cancer cells grown as multicellular tumor spheroids. Cancer research. 1997;57(12):2388–93. [PubMed] [Google Scholar]
- 27.Hazlehurst LA, Landowski TH, Dalton WS. Role of the tumor microenvironment in mediating de novo resistance to drugs and physiological mediators of cell death. Oncogene. 2003;22(47):7396–402. doi: 10.1038/sj.onc.1206943. [DOI] [PubMed] [Google Scholar]
- 28.Lengyel E, Burdette JE, Kenny HA, Matei D, Pilrose J, Haluska P, et al. Epithelial ovarian cancer experimental models. Oncogene. 2013 doi: 10.1038/onc.2013.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer research. 2011;71(11):3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasuda K, Torigoe T, Morita R, Kuroda T, Takahashi A, Matsuzaki J, et al. Ovarian cancer stem cells are enriched in side population and aldehyde dehydrogenase bright overlapping population. PloS one. 2013;8(8):e68187. doi: 10.1371/journal.pone.0068187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai C, Zhu X. The Wnt/beta-catenin pathway regulates self-renewal of cancer stem-like cells in human gastric cancer. Mol Med Report. 2012;5(5):1191–6. doi: 10.3892/mmr.2012.802. [DOI] [PubMed] [Google Scholar]
- 32.Camper SA. Beta-catenin stimulates pituitary stem cells to form aggressive tumors. Proc Natl Acad Sci U S A. 2011;108(28):11303–4. doi: 10.1073/pnas.1108275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmeyer K, Raggioli A, Rudloff S, Anton R, Hierholzer A, Del Valle I, et al. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science. 2012;336(6088):1549–54. doi: 10.1126/science.1218370. [DOI] [PubMed] [Google Scholar]
- 34.Shank JJ, Yang K, Ghannam J, Cabrera L, Johnston CJ, Reynolds RK, et al. Metformin targets ovarian cancer stem cells in vitro and in vivo. Gynecologic oncology. 2012;127(2):390–7. doi: 10.1016/j.ygyno.2012.07.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin PJ. beta-catenin signaling and cancer. Bioessays. 1999;21(12):1021–30. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 36.Mackay HJ, Buckanovich RJ, Hirte H, Correa R, Hoskins P, Biagi J, et al. A phase II study single agent of aflibercept (VEGF Trap) in patients with recurrent or metastatic gynecologic carcinosarcomas and uterine leiomyosarcoma. A trial of the Princess Margaret Hospital, Chicago and California Cancer Phase II Consortia. Gynecologic oncology. 2011 doi: 10.1016/j.ygyno.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrenson K, Notaridou M, Lee N, Benjamin E, Jacobs IJ, Jones C, et al. In vitro three-dimensional modeling of fallopian tube secretory epithelial cells. BMC cell biology. 2013;14:43. doi: 10.1186/1471-2121-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JM, Mhawech-Fauceglia P, Lee N, Parsanian LC, Lin YG, Gayther SA, et al. A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Laboratory investigation; a journal of technical methods and pathology. 2013;93(5):528–42. doi: 10.1038/labinvest.2013.41. [DOI] [PubMed] [Google Scholar]
- 39.Cody NA, Zietarska M, Filali-Mouhim A, Provencher DM, Mes-Masson AM, Tonin PN. Influence of monolayer, spheroid, and tumor growth conditions on chromosome 3 gene expression in tumorigenic epithelial ovarian cancer cell lines. BMC Med Genomics. 2008;1:34. doi: 10.1186/1755-8794-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. The Journal of cell biology. 2004;164(4):603–12. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidi PA, Bissell MJ, Lelievre SA. Three-dimensional culture of human breast epithelial cells: the how and the why. Methods in molecular biology. 2013;945:193–219. doi: 10.1007/978-1-62703-125-7_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, Hansen RK, Radisky D, Yoneda T, Barcellos-Hoff MH, Petersen OW, et al. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. Journal of the National Cancer Institute. 2002;94(19):1494–503. doi: 10.1093/jnci/94.19.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvero AB, Chen R, Fu HH, Montagna M, Schwartz PE, Rutherford T, et al. Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle. 2009;8(1):158–66. doi: 10.4161/cc.8.1.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.L'Esperance S, Bachvarova M, Tetu B, Mes-Masson AM, Bachvarov D. Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids. BMC genomics. 2008;9:99. doi: 10.1186/1471-2164-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sodek KL, Ringuette MJ, Brown TJ. Compact spheroid formation by ovarian cancer cells is associated with contractile behavior and an invasive phenotype. Int J Cancer. 2009;124(9):2060–70. doi: 10.1002/ijc.24188. [DOI] [PubMed] [Google Scholar]
- 46.Sumi T, Tsuneyoshi N, Nakatsuji N, Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135(17):2969–79. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650–3. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jian H, Shen X, Liu I, Semenov M, He X, Wang XF. Smad3-dependent nuclear translocation of beta-catenin is required for TGF-beta1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev. 2006;20(6):666–74. doi: 10.1101/gad.1388806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song X, Xie T. Wingless signaling regulates the maintenance of ovarian somatic stem cells in Drosophila. Development. 2003;130(14):3259–68. doi: 10.1242/dev.00524. [DOI] [PubMed] [Google Scholar]
- 50.Chau WK, Ip CK, Mak AS, Lai HC, Wong AS. c-Kit mediates chemoresistance and tumor-initiating capacity of ovarian cancer cells through activation of Wnt/beta-catenin-ATP-binding cassette G2 signaling. Oncogene. 2013;32(22):2767–81. doi: 10.1038/onc.2012.290. [DOI] [PubMed] [Google Scholar]
- 51.Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, et al. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Human genomics. 2011;5(4):283–303. doi: 10.1186/1479-7364-5-4-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pierre-Louis O, Clay D, Brunet de la Grange P, Blazsek I, Desterke C, Guerton B, et al. Dual SP/ALDH functionalities refine the human hematopoietic Lin-CD34+CD38- stem/progenitor cell compartment. Stem cells. 2009;27(10):2552–62. doi: 10.1002/stem.186. [DOI] [PubMed] [Google Scholar]
- 53.Reuben JM, Lee BN, Gao H, Cohen EN, Mego M, Giordano A, et al. Primary breast cancer patients with high risk clinicopathologic features have high percentages of bone marrow epithelial cells with ALDH activity and CD44(+)CD24lo cancer stem cell phenotype. European journal of cancer. 2011;47(10):1527–36. doi: 10.1016/j.ejca.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Condello S, Cao L, Matei D. Tissue transglutaminase regulates beta-catenin signaling through a c-Src-dependent mechanism. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27(8):3100–12. doi: 10.1096/fj.12-222620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiswald LB, Guinebretiere JM, Richon S, Bellet D, Saubamea B, Dangles-Marie V. In situ protein expression in tumour spheres: development of an immunostaining protocol for confocal microscopy. BMC cancer. 2010;10:106. doi: 10.1186/1471-2407-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parajuli B, Kimble-Hill AC, Khanna M, Ivanova Y, Meroueh S, Hurley TD. Discovery of novel regulators of aldehyde dehydrogenase isoenzymes. Chemico-biological interactions. 2011;191(1-3):153–8. doi: 10.1016/j.cbi.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.