Abstract
This study investigates PCR analysis of immunoglobulin heavy chain (IgH) and T cell receptor (TCR) gene rearrangements on paraffin-embedded tissue sections and bone marrow aspirates of patients suspected to have lymphoproliferative disorders but with inconclusive diagnosis in histopathological examination. 130 samples of patients with inconclusive immunohistochemistry results were evaluated for clonal rearrangement of IgH and TCR genes. Based on histopathology examination, the patients were divided into three groups: the first group without any definite diagnosis of lymphoproliferative disorders (60 cases, 46.2 %), the second group suspected to have a lymphoproliferative disorder but in favor of benign disorders (19 cases, 14.6 %) and the third group suspect to lymphoproliferative disorders but relatively in favor of malignant disorders (51 cases, 39.2 %). After DNA extraction and quality control, semi-nested PCR was performed using consensus primers for amplification of TCR-γ and CDR-3 regions of IgH genes. PCR products were analyzed after heteroduplex analysis using polyacrylamide gel electrophoresis, and were subject to silver staining. Totally, in over half of the cases (55.4 %), a monoclonal pattern was found in IgH or TCR-γ genes rearrangements. Monoclonal IgH gene rearrangement was detected in 48.1 % of patients, whereas monoclonal TCR-γ gene rearrangement was found in 33.6 % of them, which was not statistically significant (P = 0.008). Only in 32 patients (24.6 %) were the results of TCR-γ and IgH gene rearrangements consistent with respect to the presence (2.3 %) or absence (22.3 %) of monoclonality. Finally, PCR analysis of TCR-γ and IgH gene rearrangements led to definite diagnosis in 105 patients (80.8 %), and only 25 cases (19.2 %) remained inconclusive. Our results emphasize the usefulness of gene rearrangement study in cases without a definite diagnosis in immunohistochemistry studies. Multiple PCR analysis results when combined with patient’s clinical course and immunohistochemistry can lead to early diagnosis and subsequent therapy.
Keywords: Immunoglobulin heavy chain, T-cell receptor-γ, Gene rearrangement
Introduction
Today, molecular analysis is regarded as a specific tool in diagnosis and classification of non-Hodgkin’s lymphoma (NHL) as well as traditional histopathology and immunohistochemistry assays. Detection of chromosomal translocations as well as T and B-cell clonality are the chief goals of such novel techniques [1, 2].
In most of the cases, NHL is diagnosed using usual diagnostic methods like histopathology and immunohistochemistry; however, in nearly 10–15 % of cases, definite diagnosis is impossible and requires molecular studies to distinguish benign reactive lymphoid proliferations from neoplastic lymphoproliferative disorders [3–5]. However, histopathological examination is already considered as the gold standard in primary diagnosis and classification of NHLs. Clonality assessments are particularly helpful in establishment of definite diagnosis of such malignancies [6, 7]. Moreover, even in cases where molecular assays are not a prerequisite for definite diagnosis, assessment of clonality may be useful in disease follow-up by identifying a tumor-specific marker.
A number of techniques including polymerase chain reaction (PCR) and southern blot analysis have been used to detect clonality in different patients [8]. PCR has become the molecular technique of choice due to higher sensitivity and lower cost compared to southern blot. Avoiding formalin-fixed and paraffin-embedded sections and the use of fresh specimens is another advantage of PCR [3, 4, 9].
Immunoglobulin (Ig) and T-cell receptor (TCR) gene rearrangement studies have been used to assess clonality in lymphoid malignancies [10]. Consensus primers directed to constant regions of immunoglobulin heavy chain (IgH) genes enable detection of monoclonality in polyclonal populations [11–16]. IgH PCR is able to distinguish monoclonal B-cells among 103–105 polyclonal cells [17]. The detection rate varies from 50 % to nearly 100 % in previous works [18–20]. Using IgH PCR, minute samples like biopsy from tissues, fine needle aspiration and specimens from cytological slides can be assessed for monoclonal rearrangement [21–24].
Few studies have dealt with IgH and TCR rearrangement in diagnosis of lymphoproliferative disorders, in which the sample size has been low. However, the advantage of molecular studies as complementary diagnostic techniques in various lymphoproliferative disorders has been discussed previously [25–30]. The use of such techniques for definite diagnosis in inconclusive cases of immunomorphological studies is arguable, and has been investigated just in some case-series studies. As far as we know, this is the largest study to assess the advantages of molecular studies in inconclusive cases of immunomorphological assays. This study was designed to investigate PCR analysis of IgH and TCR-γ gene rearrangements on paraffin-embedded tissue sections and bone marrow aspirates of patients suspected to have lymphoproliferative disorders with inconclusive diagnosis in immunomorphological studies.
Patients and Methods
Patients
In this prospective study, paraffin-embedded tissue sections or bone marrow aspirates of 130 patients without a definite diagnosis in histopathological examination were assessed for IgH and TCR gene clonal rearrangements. The specimens were extracted from the files in archive of Payvand Clinical and Specialty Laboratory between 2010 and 2012. The patients were divided to three groups in terms of morphology and immunology. Trephines were considered to be positive for B cell involvement if more than two large (at least six time the size of an adipocyte) aggregates mostly composed of B cells were present. Additional evidence supporting the diagnosis of lymphomatous involvement was infiltration of scattered solid B cell growth or large atypical cells. Samples with two small aggregates or one large or paratrabecular aggregate of B cells were classified in group 1, and did not meet the definitive criteria but were still suspicious of a diagnosis of bone marrow involvement by B cell NHL. Furthermore, biopsies showing an interstitial increase of B cells or paratrabecular, combined B and T cell aggregates were another finding in favor of NHL involvement. BM aspirates included <20 % phenotypically atypical lymphocytes. These samples showed homogeneous staining for CD20 as well. In group 2, a single tiny B cell aggregate or multiple heterogenous intertrabecular localized aggregates were observed, which morphologically indicated that the bone marrow was not involved; however, a mixed infiltrate of CD20 and CD45RO/CD3 positive cells suggested a reactive lymphoproliferative process. However, this approach is also of limited value due to increased number of reactive T cells associated with malignant B cell lymphoid nodules.
DNA Isolation and PCR Analysis of Samples
The samples in our assessment were paraffin-embedded tissue sections (46.2 %), EDTA anticoagulated peripheral blood (29.2 %) and EDTA anticoagulated bone marrow aspirates (24.6 %). After preparing required sections from paraffin-embedded tissue blocks (each 10–15 µm thick) using the standard technique [31], the sections were collected in sterile microtubes labeled with patient’s name. They were deparaffinized using xylol solution and washed using ethanol with different concentrations. DNA was extracted from bone marrow and from paraffinized samples using DNA Extraction Kit (Roche Molecular Biochemicals, Mannheim, Germany). Quality of extracted DNA was confirmed by determining DNA concentration and its purity (OD 260/280) using Eppendorf Biophotometer (Eppendorf, Hamburg, Germany).
After DNA extraction and quality control, semi-nested PCR was performed using consensus primers for amplification of the CDR-3 region. The reaction was performed with 1.5 mMol MgCl2, 200 µMol dNTP, 10–15 picoMol of primers, 1 unit Taq DNA polymerase and 100–200 ng patient’s DNA in the final volume of 25–50 µl according to the PCR program (Table 1).
Table 1.
PCR schedule for amplification of rearranged immunoglobulin heavy chain gene
| PCR stages | First reaction FR3A-LJH | Second reaction FR3A-VLJH |
|---|---|---|
| Primary denaturation | 94 °C/3 min | 94 °C/1 min |
| Denaturation | 94 °C/1 min | 94 °C/1 min |
| Primer annealing | 57 °C/45 s | 60 °C/45 s |
| Chain extension | 72 °C/2 min | 72 °C/2 min |
| Numbers of cycles | 30 | 20 |
| Final extension | 72 °C/10 min | 72 °C/10 min |
1 µl of PCR product was used as sample in the second semi-nested PCR without any dilution. The PCR was performed using model 96 PeqLab Primus thermal cycler (PeqLab, Erlangen, Germany). All PCR reactions were conducted in duplicate on patient’s samples. PCR products were analyzed after heteroduplex analysis (5 min in 95 °C and then 60 min in 4 °C) using polyacrylamide gel electrophoresis and silver staining (Fig. 1). Multiple positive and negative controls were used as mentioned in Table 2. The demonstration of TCR-γ gene rearrangements was performed by two rounds of multiplex PCR with a set of seven primers as previously described (Fig. 2) [32].
Fig. 1.
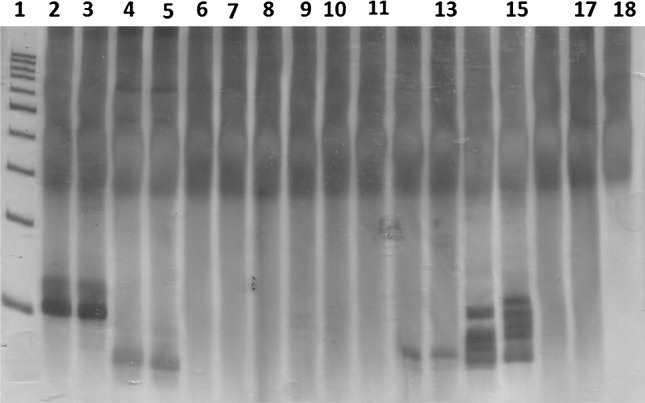
PAGE silver-stained gel which shows the immunoglobulin heavy chain (IgH) gene rearrangement. Lane 1 100 bp DNA ladder; Lane 2 and 3 IgH gene rearrangement monoclonal pattern (positive control I); Lane 4 and 5 IgH gene rearrangement monoclonal pattern (positive control II); Lane 6 and 7 IgH gene rearrangement polyclonal pattern (negative control I); Lane 8 and 9 IgH gene rearrangement polyclonal pattern (negative control II); Lane 10 and 11 IgH gene rearrangement polyclonal pattern (patient I); Lane 12 and 13 IgH gene rearrangement monoclonal pattern (patient II); Lane 14 and 15 IgH gene rearrangement polyclonal pattern (patient III); Lane 16 and 17 IgH gene rearrangement polyclonal pattern (patient IV); Lane 18 non-template control
Table 2.
Positive and negative controls used in PCR of IgH gene
| Controls | Number | Specimen | Diagnosis | PCR |
|---|---|---|---|---|
| Positive control | 1 | Paraffin-embedded tissue block | B-cell malignant lymphoma | Monoclonal |
| 2 | Paraffin-embedded tissue block | B-cell malignant lymphoma | Monoclonal | |
| Negative control | 1 | Paraffin-embedded tissue block | Follicular hyperplasia | Polyclonal |
| 2 | Peripheral blood mononuclear cell | Healthy | Polyclonal | |
| 3 | Sterile distilled water | Negative |
Fig. 2.
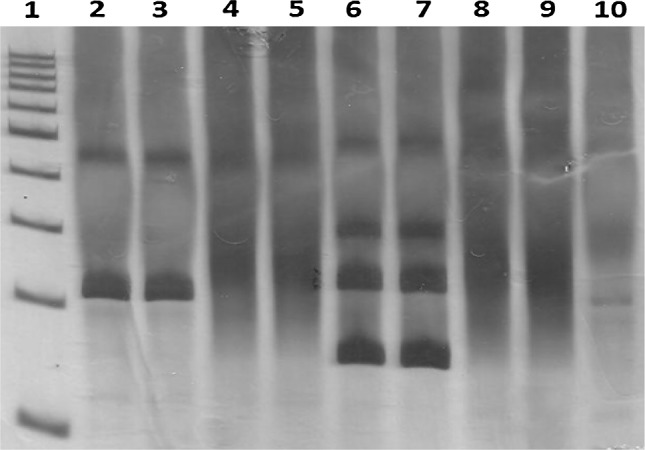
PAGE silver-stained gel which shows the TCR gene rearrangement. Lane 1 100 bp DNA ladder; Lane 2 and 3 TCR gene rearrangement monoclonal pattern (positive control); Lane 4 and 5 TCR gene rearrangement polyclonal pattern (negative control); Lane 6 and 7 TCR gene rearrangement monoclonal pattern (patient I); Lane 8 and 9 TCR gene rearrangement polyclonal pattern (patient II); Lane 10 non-template control
According to tissue of origin, sensitivity of IgH and TcR-γ PCR was different in detection of a clonally rearranged cell population [33–35]. Indeed, the number of polyclonal cells in the sample, which if present are coamplified with the clonal cell population, defines this sensitivity. Thus, the results should be interpreted considering the tissue of origin, since the number of polyclonal cells is different in various tissues and disease stages. In a background of lymph node–based polyclonal B-cells, a clonally rearranged B-cell population of approximately ≥1 % of total cells is detectable using seminested-PCR assay [31]. The sensitivity of this technique was sufficient to produce a detectable band from a single IgH gene-rearranged B-cell even in the absence of polyclonal cell populations [33, 34]. PCR amplifications for TCR-γ genes showed comparable results in the same conditions [31]. The sensitivity was intermediate between the above mentioned values when using both IgH and TCR-γ PCR gene amplifications for detection of clonality in bone marrow samples.
When the results of IgH and TCR-γ gene rearrangements are in agreement with both likely clinical and immunomorphologic findings, a definite diagnosis has been made. When PCR analysis of gene clonality is the same as one of the clinical or immunomorphologic findings, the most probable diagnosis (benign vs. malignant) has been obtained, but further follow-up is suggested. In cases without clinical features of malignancy with different results of gene clonality and immunomorphologic study or cases without a specific finding in immunomorphologic study with different results of gene clonality and clinical findings, the diagnosis remains inconclusive.
Statistical Analysis
The results were reported as mean ± standard deviation (SD) for quantitative variables and percentage for categorical variables. The groups were compared using Student’s t test for continuous variables and Chi square test (or Fisher’s exact test if required) for categorical variables. P values of 0.05 or less were considered statistically significant. All the statistical analyses were performed using SPSS version 13 (SPSS Inc, Chicago, IL, USA).
Results
In this study, 130 patients (mean age 47.5 ± 19.9 years, range 3–86 years) without definite diagnosis of lymphoproliferative disorders based on histopathological and immunohistochemistry studies were recruited for further evaluation by PCR analysis of IgH and TCR gene rearrangements.
The samples in our assessment were paraffin-embedded tissue sections (46.2 %), EDTA anticoagulated peripheral blood (29.2 %) and EDTA anticoagulated bone marrow aspirates (24.6 %). Tissue samples were taken from lymph nodes (55 %), skin biopsies (36.7 %) and gastrointestinal tract (8.3 %).
Totally, in over half of the cases (55.4 %), a monoclonal pattern was found in IgH or TCR-γ gene rearrangement. Monoclonal IgH gene rearrangement with an expected size of 80–120 bp was detected in 48.1 % of patients, whereas monoclonal TCR-γ gene rearrangement was found in 33.6 % of patients, which was not statistically significant (P = 0.008). Only in 32 patients (24.6 %) were the results of TCR-γ and IgH gene rearrangements consistent in view of the presence (2.3 %) or absence (22.3 %) of monoclonality. Based on the clinical features, 41.5 % of the patients were suspected to be malignant. IgH gene monoclonality pattern was in agreement with patients’ clinical course in favor of malignancy (P = 0.92), whereas TCR gene showed a different clonality pattern from patients’ clinical course (P = 0.032).
Based on histopathology examination, the patients were divided into three groups: the first group without definite diagnosis of lymphoproliferative disorders (60 cases, 46.2 %), the second group with suspicious lymphoproliferative disorders but in favor of benign disorders (19 cases, 14.6 %) and the third group with suspicious lymphoproliferative disorders but in favor of malignant disorders (51 cases, 39.2 %). Patients’ demographic and clinical data in each group is summarized in Table 3.
Table 3.
Patients demographic and clinical data in each group
| Variables | Group one (n = 60) | Group two (n = 19) | Group three (n = 51) | Total (n = 130) | |
|---|---|---|---|---|---|
| Age (Years) | 51.2 ± 2.0 | 42.3 ± 1.9 | 45.2 ± 1.6 | 47.5 ± 1.9 | |
| (3–86) | (15–80) | (8–78) | (3–86) | ||
| Male gender (%) | 56.7 | 63.2 | 68.8 | 59.5 | |
| TCR gene | Monoclonal pattern | 29 (48.3) | 1 (5.3) | 12 (33.5) | 42 (32.3) |
| Polyclonal pattern | 31 (51.7) | 18 (94.7) | 39 (76.5) | 88 (67.7) | |
| IgH gene | Monoclonal pattern | 28 (46.7) | 6 (31.6)* | 29 (56.9)† | 63 (48.5)‡ |
| Polyclonal pattern | 32 (53.3) | 13 (68.4) | 22 (43.1) | 67 (51.5) |
IgH Immunoglobulin heavy chain, TCR T-cell receptor
* P value = 0.042 for difference between TCR and IgH monoclonality in group two
† P value = 0.017 for difference between TCR and IgH monoclonality in group three
‡ P value = 0.008 for difference between TCR and IgH monoclonality in total
In group one (60 cases), monoclonality of TCR-γ and IgH genes was found in 48.3 and 46.7 % of cases, respectively, which was not statistically significant (P = 0.86). In 17 out of 60 patients (28.3 %), analysis of TCR-γ and IgH gene rearrangements for presence (5.0 %) or absence (23.3 %) of monoclonality showed similar results. 8 patients (13.3 %) with monoclonal pattern and clinical features in favor of malignancy were diagnosed with malignant proliferative disorders. 31 patients (51.7 %) with polyclonal gene rearrangement and clinical features of benign disorders were diagnosed as benign, but were recommended to be followed. In 21 patients (35.0 %) in whom clinical features and analysis of genes rearrangement were inconsistent, the final diagnosis remained ambiguous, and follow-up was recommended in them.
In group two (19 cases), although immunomorphologic assays were relatively in favor of benign disorders, 13 patients (68.2 %) had clinical features in favor of malignancy. Monoclonal pattern of IgH gene was more frequently seen in these patients compared to TCR gene (31.6 % vs. 5.3 %, P = 0.042). In this group, 7 patients (36.8 %) with clinical course in favor of malignancy and IgH or TCR-γ monoclonal gene rearrangements were considered as malignant. The other 12 patients (63.2 %) were considered benign, but 6 patients with clinical features in favor of malignancy were recommended for further investigations in the future.
In group three, among 51 patients with immunomorphologic features in favor of malignancy, TCR and IgH monoclonal patterns were seen in 33.5 and 58.9 % of patients (P = 0.017), respectively. 29 (56.9 %) patients suspected to have malignancy in clinical and immunomorphologic studies with a monoclonal pattern were considered as malignant. 4 patients (7.8 %) who were clinically suspect of malignancy but showed polyclonal pattern for IgH and TCR-γ gene rearrangements were recommended for further study, and their diagnosis remained indefinite. Other 18 patients (35.3 %) with polyclonal pattern in IgH gene rearrangement and without clinical features in favor of malignancy were considered as benign but were recommended for further follow-up.
Overall, among 130 patients without definite diagnosis after immunomorphologic assays, PCR analysis of TCR and IgH gene rearrangements led to a definite diagnosis in 105 patients (80.8 %), and only 25 patients (19.2 %) remained inconclusive. In cases without any impression in immunomorphologic study, IgH and TCR gene clonality study were not found to be significantly different, while in cases with no definite diagnosis but in favor of benign or malignant disorders, a significant difference was detected in clonality study of IgH and TCR genes.
Discussion
In some cases, immunomorphologic study of lymphoid populations does not lead to a definite detection of cell proliferation, and does not clarify the benign (reactive) or malignant state. In such cases, complementary diagnostic techniques like molecular clonality assays with analysis of IgH gene rearrangements are recommended [3, 6, 7].
In this study, EDTA or paraffin-embedded sections of 130 patients with inconclusive immunomorphologic diagnosis were assessed for clonal rearrangement of IgH and TCR-γ genes. In more than half of the cases (55.4 %), a monoclonal pattern was found in IgH or TCR gene rearrangements. Monoclonal IgH gene rearrangement with an expected size of 80–120 bp was detected in 48.1 % of patients, while clonal arrangement of TCR-γ gene was found to be less statistically significant (33.6 %, P = 0.008). Only in 32 patients (24.6 %) were the results of TCR-γ and IgH gene rearrangements consistent regarding the presence (2.3 %) or absence (22.3 %) of monoclonality. In our previous study on 12 patients with indefinite diagnosis or patients suspected to have to malignancy, 75 % showed monoclonal pattern of IgH gene [35]. Maes B et al. [7] studied paraffin-embedded bone marrow samples of patients with suspicious or indefinite diagnosis for IgH gene clonality using FR3 region primers, which revealed a monoclonal pattern in 43 % of cases. Maroto et al. [36] found 71.4 % of 14 patients with suspicious diagnosis of malignancy with clonal rearrangements of IgH gene.
In a well-conducted study, Theriault et al. [32] assessed PCR analysis of IgH and TCR chain gene rearrangements in 525 cases, among which 85 had no definite diagnosis like our ones. In 76 % of cases with no definite diagnosis, they were able to find the correlation of immunomorphologic features with presence or absence of clonal lymphoid cell population, which led to a definite diagnosis. In almost all these cases, the final diagnosis was found to be in agreement with clinical course [32]. In our study, based on clinical features, 41.5 % of patients were suspected to be malignant. IgH gene monoclonality pattern was in agreement with patients’ clinical course in favor of malignancy (P = 0.92), while TCR-γ gene showed a different clonality pattern from patients’ clinical course (P = 0.032). However, our patients had no definite diagnosis. Based on the histopathological examination, we divided our patients in three groups: group one without any definite diagnosis (60 cases, 46.2 %), group two suspicious but in favor of benign disorders (19 cases, 14.6 %) and group three suspicious but in favor of malignancy (51 cases, 39.2 %). In total, among 130 patients without definite diagnosis after immunomorphologic assays, PCR analysis of TCR-γ and IgH gene rearrangements led to a definite diagnosis in 105 patients (80.8 % benign or malignant), and only 25 cases patients (19.2 %) remained inconclusive. In cases without any impression in immunomorphologic study, clonality study of IgH and TCR-γ genes was found to have no significant difference, while in cases with no definite diagnosis but in favor of benign or malignant disorders, a significant difference was detected in clonality study of IgH and TCR-γ genes.
The samples in our assessment were paraffin-embedded tissue sections (46.2 %), EDTA anticoagulated peripheral blood (29.2 %) and EDTA anticoagulated bone marrow aspirates (24.6 %). Tissue samples were extracted from lymph nodes (55 %), skin biopsies (36.7 %) and gastrointestinal tract (8.3 %). A wide spectrum of skin disorders are associated with considerably increased amounts of lymphocytes in the skin. It is necessary to differentiate between reactive hyperplasia and neoplastic formations. Hughes et al. [9] in assessment of paraffin-embedded sections of 31 patients with cutaneous lymphoma for IgH gene rearrangement illustrated that the results of IgH gene clonality were consistent with histopathological examination and immunomorphologic diagnosis in all of the cases.
In assessment of IgH clonal rearrangement of cutaneous tissues suspicious to lymphoma, false negative cases are not common. In contrast, false positivity is common in investigation of clonality in suspicious cutaneous involvements as lymphocyte populations are located separately in suspicious cutaneous lesions, and there are limited numbers of lymphoid cells in these lesions [37–39]. Thus, the use of inflammatory dermatosis and natural cutaneous cells as negative controls in clonality assessment is recommended to prevent false positivity [38]. Moreover, in such cases, it is suggested to perform triplicate PCR and consider those with clonal rearrangements in all three groups as positive for clonality [38]. In this study, in patients with suspicious cutaneous lesions, triplicate PCR was done, which showed a considerable reproducibility.
Despite several morphologic and immunologic characteristics, which help distinguish lymphoid malignancies from reactive formation [40, 41], in some cases it is very hard or even impossible to differentiate between benign and malignant lymphocyte populations in bone marrow, especially when the clinical findings are ambiguous [42]. Ben-Ezra JM et al. investigated malignant lymphoid populations in bone marrow samples among 18 patients using IgH clonal rearrangement. They found monoclonality in 44 % of cases. All benign populations in bone marrow showed polyclonality. They suggested using PCR analysis of IgH gene rearrangement in cases without a definite diagnosis in order to reach a definite diagnosis using PCR results as well as clinical and morphologic findings [41]. We followed this suggestion in our study to achieve a definite diagnosis in cases with no definite diagnosis in cutaneous or bone marrow involvements.
However, investigation of IgH gene rearrangement using consensus primers of FR3 region showed acceptable specificity with limited number of false positivity. In order to prevent probable false cases, duplicate or triplicate PCR should be performed for each patient, and in case of reproducibility, it should be considered as positive. In order to improve sensitivity and prevent false negativity, PCR should be repeated at least two times for each patient. In cases of polyclonality in both reactions, the result should be considered as negative. Moreover, primers for distal regions (FR1 and FR2) should be used to avoid false negativity [11, 23, 27, 28].
Another important issue in assessment of tissue samples is their quality. When DNA is extracted from paraffin embedded or bone marrow samples without tumoral involvement or with limited tumoral cells, it is more probable to face false negativity. It is suggested to use microdissection in such cases [43] or increase the number of sections from paraffin blocks and frequency of DNA extraction.
It is important to take it into account that the existence of a monoclonal band does not always imply malignancy. Some benign disorders like CD8+ (sometimes CD4+) lymphocytosis, benign monoclonal gammopathies, early stages of EBV-positive lymphoproliferation, immunosuppressed patients and T-cell benign cutaneous proliferation like lymphomatoid papulosis may also have clonal sources. Thus, it is important to interpret the results of clonality studies with more caution and consider the clinical features of patients as well as morphologic and immunohistochemistry studies [32].
Conclusion
Among 130 patients without a definite diagnosis, only 25 patients remained inconclusive after applying PCR analysis of TCR-γ and IgH gene rearrangements.
Finally, the patients with a definite diagnosis of malignancy based on results of this study received the proper treatment, and our diagnostic method made any further biopsies redundant.
In total, our results emphasize the benefit of gene rearrangement study in cases without a definite diagnosis in immunomorphologic studies. Findings of multiple PCR-run approach with the same DNA samples, when added to patient’s clinical course and immunomorphologic findings, can lead to early diagnosis and subsequent appropriate therapy.
Acknowledgments
We wish to thank all our colleagues in Payvand Clinical and Specialty Laboratory.
Conflict of Interest
None of the authors has conflict of interest.
References
- 1.Wilkins BS. Molecular genetic analysis in the assessment of lymphomas. Curr Diagn Pathol. 2004;10:351–359. doi: 10.1016/j.cdip.2004.04.004. [DOI] [Google Scholar]
- 2.Macintyre E, Bahloul M, Asnafi V. Clinical impact of molecular diagnostics in low-grade lymphoma. Best Pract Res Clin Haematol. 2005;18(1):11–97. doi: 10.1016/j.beha.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Nikiforova M, His E, Braziel R, et al. Detection of clonal IGH gene rearrangements: summary of molecular oncology surveys of the college of American pathologists. Arch Pathol Lab Med. 2007;131:185–189. doi: 10.5858/2007-131-185-DOCIGR. [DOI] [PubMed] [Google Scholar]
- 4.Rezuke WN, Abernathy EC, Tsongalis GJ. Molecular diagnosis of B- and T-cell lymphomas: fundamental priiples and clinical applications. Clin Chem. 1997;43(10):1814–1823. [PubMed] [Google Scholar]
- 5.Weirich G, Funk A, Hoepner I, et al. PCR-based assays for the detection of monoclonality in non-Hodgkin’s lymphoma: application to formalin-fixed paraffin-embedded tissue and decalcified bone marrow samples. J Mol Med. 1995;73:235–324. doi: 10.1007/BF00189923. [DOI] [PubMed] [Google Scholar]
- 6.Krober SM, Horney HP, Greschniok A, Kaiserling E. Reactive and neoplastic lymphocytes in human bone marrow: morphological, immunohistological, and molecular biological investigations on biopsy specimens. J Clin Pathol. 1999;52:521–526. doi: 10.1136/jcp.52.7.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maes B, Achten R, Demunter A, et al. Evaluation of B cell lymphoid infiltrates in bone marrow biopsies by morphology, immunohistochemistry, and molecular analysis. J Clin Pathol. 2000;53:835–840. doi: 10.1136/jcp.53.11.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kocjan G. Cytological and molecular diagnosis of lymphoma. J Clin Pathol. 2005;1:561–567. doi: 10.1136/jcp.2004.019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes J, Weston S, Bennetts B, et al. The application of a PCR technique for the detection of immunoglobulin heavy chain gene rearrangements in fresh or paraffin-embedded skin tissue. Pathology. 2001;33:222–225. [PubMed] [Google Scholar]
- 10.Segal GH. Assessment of B-cell clonality by the polymerase chain reaction: a pragmatic overview. Adv Anat Pathol. 1996;3:195–203. doi: 10.1097/00125480-199603030-00006. [DOI] [Google Scholar]
- 11.Bagg A, Braziel RM, Arber D, et al. Immunoglobulin heavy chain gene analysis in lymphomas: a multi-center study demonstrating the heterogeneity of performance of polymerase chain reaction asssays. J Mol Diagn. 2002;4:81–89. doi: 10.1016/S1525-1578(10)60685-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deane M, Norton JD. Detection of immunoglobulin gene monoclonal rearrangement in B lymphoid malignancies by polymerase chain reaction gene amplification. Br J Haematol. 1990;74:251–256. doi: 10.1111/j.1365-2141.1990.tb02579.x. [DOI] [PubMed] [Google Scholar]
- 13.Trainor KJ, Brisco MJ, Wan JH, et al. Gene rearrangement in B- and T-lymphoproliferative disease detected by the polymerase chain reaction. Blood. 1991;78:192–196. [PubMed] [Google Scholar]
- 14.Ramasamy I, Brisco M, Morley AA. Improved PCR method for detecting monoclonal immunoglobulin heavy chain rearrangement in B cell neoplasms. J Clin Pathol. 1992;45:770–775. doi: 10.1136/jcp.45.9.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deane M, McCarthy KP, Wiedemann LM, Norton JD. An improved method for the detection of B-lymphoid clonality by polymerase chain reaction. Leukemia. 1991;5:726–730. [PubMed] [Google Scholar]
- 16.Aubin J, Davi F, Nguyen-Salomon F, et al. Description of a novel FR1 IgH PCR strategy and its comparison with three other strategies of clonality in B cell malignancies. Leukemia. 1995;9:471–479. [PubMed] [Google Scholar]
- 17.Medeiros L, Carr J. Overview of the role of molecular methods in the diagnosis of malignant lymphomas. Arch Pathol Lab Med. 1999;123:1189–1207. doi: 10.5858/1999-123-1189-OOTROM. [DOI] [PubMed] [Google Scholar]
- 18.Ilyas M, Jalal H, Linton C, Rooney N. The use of the polymerase chain reaction in the diagnosis of B-cell lymphomas from formalin-fixed paraffin-embedded tissue. Histopathology. 1995;26:333–338. doi: 10.1111/j.1365-2559.1995.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 19.Inghirami G, Szabolcs M, Yee HT, et al. Detection of immunoglobulin gene rearrangement of B cell non-Hodgkin’s lymphomas and leukemias in fresh, unfixed and formalin-fixed, paraffin-embedded tissue by polymerase chain reaction. Lab Invest. 1993;68:746–757. [PubMed] [Google Scholar]
- 20.Segal GH, Jorgensen T, Masih AS, Braylan RC. Optimal primer selection for clonality assessment by polymerase chain reaction analysis: I. Low grade B-cell lymphoproliferative disorders of nonfollicular center cell type. Hum Pathol. 1994;25:1269–1275. doi: 10.1016/0046-8177(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 21.Calvert RJ, Evans PA, Randerson JA, et al. The significance of B-cell clonality in gastric lymphoid infiltrates. J Pathol. 1996;180:26–32. doi: 10.1002/(SICI)1096-9896(199609)180:1<26::AID-PATH681>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 22.Sukpanichnant S, Vnencak-Jones CL, McCurley TL. Determination of B-cell clonality in paraffin-embedded endoscopic biopsy specimens of abnormal lymphocytic infiltrates and gastrointestinal lymphoma by polymerase chain reaction reaction. Am J Clin Pathol. 1994;102:299–305. doi: 10.1093/ajcp/102.3.299. [DOI] [PubMed] [Google Scholar]
- 23.Wan JH, Sykes PJ, Orell SR, Morley AA. Rapid method for detecting monoclonality in B cell lymphoma in lymph node aspirates using the polymerase chain reaction. J Clin Pathol. 1992;45:420–423. doi: 10.1136/jcp.45.5.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alkan S, Lehman C, Sarago C, et al. Polymerase chain reaction detection of immunoglobulin gene rearrangement and bcl-2 translocation in archival glass slides of cytologic material. Diagn Mol Pathol. 1995;4:25–31. doi: 10.1097/00019606-199503000-00006. [DOI] [PubMed] [Google Scholar]
- 25.Melotti CZ, Amary MF, Sotto MN, et al. Polymerase chain reaction-based clonality analysis of cutaneous B-cell lymphoproliferative processes. Clinics (Sao Paulo). 2010;65(1):53–60. doi: 10.1590/S1807-59322010000100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellan C, Lazzi S, Zazzi M, et al. Immunoglobulin gene rearrangement analysis in composite hodgkin disease and large B-cell lymphoma: evidence for receptor revision of immunoglobulin heavy chain variable region genes in Hodgkin-Reed-Sternberg cells? Diagn Mol Pathol. 2002;11(1):2–8. doi: 10.1097/00019606-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Child FJ, Woolford AJ, Calonje E, et al. Molecular analysis of the immunoglobulin heavy chain gene in the diagnosis of primary cutaneous B cell lymphoma. J Invest Dermatol. 2001;117(4):984–989. doi: 10.1046/j.0022-202x.2001.01482.x. [DOI] [PubMed] [Google Scholar]
- 28.Lukowsky A, Marchwat M, Sterry W, Gellrich S. Evaluation of B-cell clonality in archival skin biopsy samples of cutaneous B-cell lymphoma by immunoglobulin heavy chain gene polymerase chain reaction. Leuk Lymphoma. 2006;47(3):487–493. doi: 10.1080/10428190500305380. [DOI] [PubMed] [Google Scholar]
- 29.Bereczki L, Kis G, Bagdi E, Krenacs L. Optimization of PCR amplification for B- and T-cell clonality analysis on formalin-fixed and paraffin-embedded samples. Pathol Oncol Res. 2007;13(3):209–214. doi: 10.1007/BF02893501. [DOI] [PubMed] [Google Scholar]
- 30.Yosefian A, Poopak B, Abolghasemi H, et al. Molecular diagnosis of B cell Non-Hodgkin Lymphoma by evaluation of immunoglobulin heavy chain gene rearrangement. Sci J Blood Transfus Organ. 2008;5(3):157–166. [Google Scholar]
- 31.Al Saati T, Galoin S, Gravel S, et al. IgH and TcR-g gene rearrangements in Hodgkin’s disease by PCR demonstrate lack of correlation between genotype, phenotype and Epstein–Barr virus status. J Pathol. 1997;181:387–393. doi: 10.1002/(SICI)1096-9896(199704)181:4<387::AID-PATH781>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 32.Thériault C, Galoin S, Valmary S, et al. PCR analysis of immunoglobulin heavy chain (IgH) and TcR-gamma chain gene rearrangements in the diagnosis of lymphoproliferative disorders: results of a study of 525 cases. Mod Pathol. 2000;13(12):1269–1279. doi: 10.1038/modpathol.3880232. [DOI] [PubMed] [Google Scholar]
- 33.Segal GH, Hussey CE, Wittwer CT. PCR for T-cell rearrangements. Diagn Mol Pathol. 1996;5:297–298. doi: 10.1097/00019606-199612000-00012. [DOI] [PubMed] [Google Scholar]
- 34.Rockman SP. Determination of clonality in patients who present with diagnosis dilemmas: a laboratory experience and review of the literature. Leukemia. 1997;11:852–862. doi: 10.1038/sj.leu.2400678. [DOI] [PubMed] [Google Scholar]
- 35.Poopak B, Latifzadeh H, Abolghasemi H, Yousefian A, et al. Malignant B cell lymphomas vs benign lymphoproliferations and the role of immunoglobulin heavy chain gene rearrangement analysis. Sci J Blood Transfus Organ. 2008;4(4):285–295. [Google Scholar]
- 36.Maroto A, Rodri guez J, Martinez MA, et al. A single primer pair immunoglobulin polymerase chain reaction assay as a useful tool in fine-needle aspiration biopsy differential diagnosis of lymphoid malignancies. Cancer Cytopathol. 2003;99:196–205. doi: 10.1002/cncr.11060. [DOI] [PubMed] [Google Scholar]
- 37.Bouloc A, Delfau-Larue MH, Lenormand B, et al. Polymerase chain reaction analysis of immunoglobulin gene rearrangement in cutaneous lymphoid hyperplasias. Arch Dermatol. 1999;135:168–172. doi: 10.1001/archderm.135.2.168. [DOI] [PubMed] [Google Scholar]
- 38.Nihal M, Mikkola D, Wood GS. Detection of clonally restrited immunoglobulin heavy chain gene rearrangements in Normal and lesional skin. J Mol Diagn. 2000;2:5–10. doi: 10.1016/S1525-1578(10)60609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabath DE, Wood BL, Kussick SJ. PCR methods for determining B cell clonality. J Mol Diagn. 2000;2:92–96. doi: 10.1016/S1525-1578(10)60640-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenna RW, Hernandez JA. Bone marrow in malignant lymphoma. Hematol Oncol Clin North Am. 1988;2:617–635. [PubMed] [Google Scholar]
- 41.Ben-Ezra JM, King BE, Harris AC, et al. Staining for bcl-2 protein helps to distinguish benign from malignant lymphoid aggregates in bone marrow biopsies. Mod Pathol. 1994;7:560–564. [PubMed] [Google Scholar]
- 42.Ben-Ezra J, Hazelgrove K, Ferreira-Gonzalez A, Garrett CT. Can polymerase chain reaction help distinguish benign from malignant lymphoid aggregates in bone marrow aspirates? Arch Pathol Lab Med. 2000;124:511–515. doi: 10.5858/2000-124-0511-CPCRHD. [DOI] [PubMed] [Google Scholar]
- 43.Yamauchi A, Nakatsuka S, Miyanaga I, et al. Clonality analysis of follicular lymphoma using laser capture microdissection method. Int J Mol Med. 2002;10:649–653. [PubMed] [Google Scholar]


